Abstract
Nipah virus (NiV) and Hendra virus (HeV) are highly pathogenic paramyxovirus which belongs to Henipavirus family, causes severe respiratory disease, and may lead to fatal encephalitis infections in humans. NiV and HeV glycoproteins (G) bind to the highly conserved human ephrin-B2 and B3 (EFNB2 & EFNB3) cell surface proteins to mediate the viral entry. In this study, various molecular modelling approaches were employed to understand protein-protein interaction (PPI) of NiV and HeV glycoprotein (84% sequence similarity) with Human EFN (B2 and B3) to investigate the molecular mechanism of interaction at atomic level. Our computational study emphasized the PPI profile of both the viral glycoproteins with EFN (B2 and B3) in terms of non-bonded contacts, H-bonds, salt bridges, and identification of interface hotspot residues which play a critical role in the formation of complexes that mediate viral fusion and entry into the host cell. According to the reports, EFNB2 is considered to be more actively involved in the attachment with the NiV and HeV glycoprotein; interestingly the current computational study has displayed more conformational stability in HeV/NiV glycoprotein with EFNB2 complex with relatively high binding energy as compared to EFNB3. During the MD simulation, the number of H-bond formations was observed to be less in the case of EFNB3 complexes, which may be the possible reason for less conformational stability in the EFNB3 complexes. The current detailed interaction study on the PPI may put a path forward in designing peptide inhibitors to obstruct the interaction of viral glycoproteins with host proteins, thereby inhibiting viral entry.
Graphical abstract
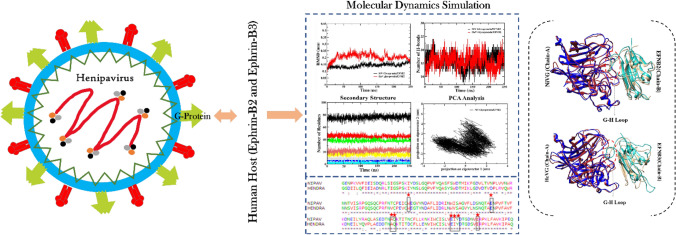
The viral attachment and fusion of Nipah and Hendra virus was explored through the interaction between viral glycoprotein and the host cell surface ephrin protein. The MD simulation results displayed more stability in Nipah and Hendra glycoprotein with EFNB2 as compared to EFNB3. The residue Glu533 in the central cavity of HeV/NiV glycoprotein protein identified as the potential hotspot in binding with the G-H loop of EFNB2.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12039-022-02110-9.
Keywords: Protein-protein interaction, NiV and HeV glycoprotein, EFNB2, EFNB3, MD simulations, hotspot residues, B-cell epitope
Introduction
The recent pandemic highlighted the need to study the vulnerability of humans due to viral outbreaks, and understandably, getting clarity on the primary step, which is viral entry into the host, is of outstanding importance. While there is an unprecedented activity in exploring the structural basis for the COVID-19 and other coronavirus infections, the indispensability of exploring various classes of viruses has become apparent in recent times. NiV and HeV viruses, known as the paramyxovirus, comprised of Paramyxoviridae and genus Henipavirus, are highly pathogenic and considered as zoonotic diseases.1 The viral entry is the significant target for developing antivirals, small peptide design, and different epitope-based antibody generation.2 The structural investigation of NiV and HeV virus glycoprotein in complex with the human receptor cell surface proteins Ephrin-B2 (EFNB2) and Ephrin-B3 (EFNB3) has been extensively studied to find the potential antivirals and vaccines to interrupt the viral interaction mediated fusion.3 The henipavirus virion contains two viral membrane proteins named glycoprotein and the membrane fusion (F) protein to mediate the viral entry and fusion into the host cell. In the human host, there are two cell surface receptor proteins EFNB2 and B3, from the large receptor tyrosine kinase family.4 The EFN proteins are normally bidirectional signaling proteins.2,5 The EFNB2 and B3 are reported as the potential receptors interacting with the henipavirus glycoprotein for mediating the viral interaction, followed by the activation of fusion protein for membrane fusion and replication.2,5 The henipavirus glycoprotein is a type II membrane protein that consists of a globular head, an N-terminus tail, a transmembrane domain, and a stalk domain; however, the EFN receptors are bound to the central cavity of the β-propeller head domain of both the NiV and HeV glycoproteins.4,5 Molecular characterization and various computer-aided drug discovery methods have been proposed to identify novel inhibitors to combat the NiV and HeV viral entry into the human cell.6,7 Furthermore, various neutralizing vaccines have been developed and reported to bind to the human EFN G-H loop to disrupt the attachment of Henipavirus glycoprotein followed by deactivating the fusion protein thereby stopping the membrane fusion and replication of the virus in the human cell.8,9
Although various structural and in vivo studies have been reported on the attachment of NiV and HeV glycoproteins with human EFNs, the detailed mechanism of viral glycoprotein attachment with the EFNB2 and EFNB3 proteins is not clear.10 The reported interface residues in the G-H loop (hydrophobic surface) of EFNs have shown potential binding towards the central cavity of the viral proteins by forming a few salt bridges and hydrogen bonds.2 Identifying the other probable interacting interface residues surrounded by the interface G-H loop and the central cavity of the virus is an important step; the available information in this regard is scarce. The current computational modelling study focuses on understanding the stability and conformational changes of protein-protein interaction (PPI) complexes (NiV and HeV glycoprotein with both EFNB2 and B3).
Various epitope-based peptides have been shown to have potential activity in developing vaccines against the NiV and HeV virus glycoprotein and the fusion proteins by applying both In vivo and In silico approaches.11 In vivo studies have shown that the human monoclonal antibody along with various developed models, is successful antibody therapy in targeting the henipavirus glycoproteins.12–14 In the computational study of human and NiV/HeV virus PPI pathways, various human cell surface proteins have been identified to have interactions with the six structural NiV and HeV proteins.15 Various molecular modelling studies have been applied to understand the molecular mechanism of PPI at an atomic level and to decipher the hotspot residues for drug designing and the effect of mutation on a protein’s structure and function.16–19 The study was evaluated by analysing the binding energy of the trajectory throughout the 250 ns MD simulations,20 the changes occur in the secondary structure of the PPI complex, identification of hotspot residues from the last 50 ns average structure extracted from the 250 ns MD trajectories.
Receptors Ephrin-B2 (EFNB2) and Ephrin-B3 (EFNB3)
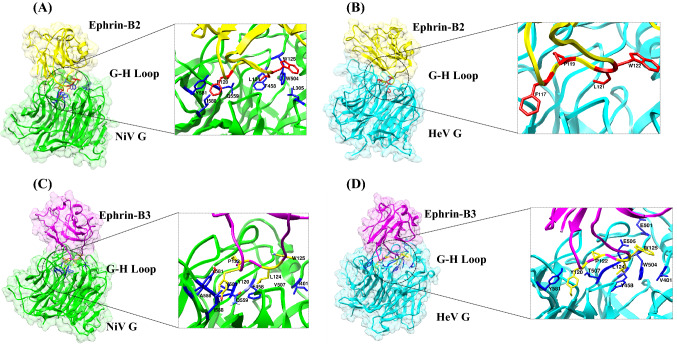
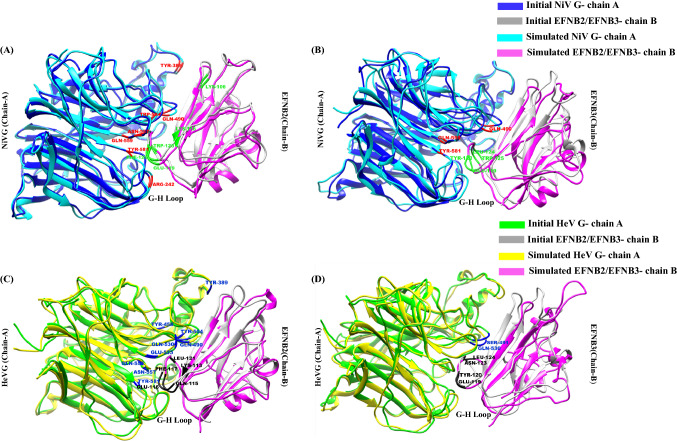
EFNB2 and EFNB3 belong to B-class ephrins, which are identified as potential henipaviral receptors in facilitating viral entry, followed by the activation of the fusion protein to mediate the virus-host fusion.21 Human EFNB2 and B3 are the cell surface membrane proteins that belong to the large receptor tyrosine kinase family and mediate bi-directional cell-cell signaling involved in tumorigenesis, different types of cancer, etc.22 The EFNB2 and B3 showed 38.7% identity and 53% similarity (Figure S2, SI) across amino acid sequences and can support the NiV and HeV infection in the host cell. The 3D structure of NiV/HeV glycoprotein in complex with EFN B2 and B3 is represented in Figure 1.
Figure 1.
Structures of (A) NiV glycoprotein/Ephrin-B2 (B) HeV glycoprotein/Ephrin-B2 and (C) NiV glycoprotein/Ephrin-B3 (D) HeV glycoprotein/Ephrin-B3 complexes with their respective interface regions. The interface residues are depicted in the square box. The NiV glycoprotein is colored in green in both the complexes and the HeV glycoprotein is colored in cyan in both the complexes. EFNB2 is colored in yellow and EFNB3 is colored in magenta.
The binding of NiV and HeV glycoprotein to the receptors EFNB2 and B3
The literature comprising structural studies on the viral glycoprotein attachment revealed that the monomeric glycoprotein head domain binds to the ephrin molecule in equal affinity and produces effective infection in the human body. Several reports have suggested that NiV and HeV glycoproteins procure the same binding affinity towards both EFNB2 and B3 proteins. The G-H loop of both the EFNs contains structural divergence and hence sits on the central cavity of the henipavirus glycoprotein.4 The G-H loop is the hydrophobic surface having 15 amino acid lengths and only a few selected residues such as Phe117, Pro119, Leu121, and Tyr122 of the EFNB2 and Tyr120, Tyr122, Leu124, and Tyr125 of EFNB3 shows interaction with respective NiV and HeV glycoproteins central cavity.4,22–24 The amino acid residue Tyr122 functions as a ‘latch’ to enable the glycoprotein receptor association.3 The formation of salt bridges in the hydrophobic surface of the G-H loop contributes more binding affinity towards the formation of the PPI complex. Several structural studies have reported that the residue Glu533 of NiV glycoprotein forms two salt bridges with Arg57 and Lys116 in EFN receptors that are found to have more affinity towards binding both proteins.24,25 Whereas, the residues Glu501 and Glu533 in the HeV glycoprotein involved in the salt bridge formation. With the guidance of the available structural information, the current study instigates the detailed investigation of PPIs to identify hotspots and may put a way forward in inhibiting the viral entry.
Computational
Protein complex setup
The crystal structure of NiV glycoprotein (PDB ID-3D11)3 and human Ephrin-B3 (PDB ID-4BKF)25 proteins were downloaded separately from Protein Data Bank.26 The protein complex was then prepared through docking in HADOCK webserver.27 The best-modelled protein complex was presented according to their respective HADDOCK scores, cluster size, and different energy parameters such as van der Waals energy, electrostatic energy, etc. along with the Z score. The PPI complex of NiV glycoprotein and human Ephrin-B2 complex structure (PDB ID-2VSM)4 was downloaded from Protein Data Bank (PDB).26 The complex was modelled using SWISS-Model Expasy28 due to missing residues, and the final model was validated using Ramachandran plot. The next PPI complex of Hendra virus (PDB ID – 6PD4)3 and human Ephrin-B3 (PDB ID-4BKF)25 were downloaded from PDB, and the complex was generated through docking in HADDOCK webserver. Then the HeV glycoprotein with Ephrin-B2 complex was downloaded from PDB (PDB ID- 6PDL)3 and modelled using SWISS Model Expasy due to presence of missing residues. Prior to MD simulation, all the complexes were cleaned by removing ions and solvents using UCSF chimera.29
Molecular dynamics MD simulations and MMPBSA calculations
GROMACS 5.0.4 package has been used to perform the MD simulations30 with CHARMM36 force field,31 and simple point charges (SPC) water model and systems were equilibrated.32 Periodic boundary condition (PBC) was imposed on the system to eliminate the boundary effect (Table S2, SI). A cutoff distance of 1Å was set. The system was energy minimized with the steepest descent algorithm with 25000 steps. All the systems were subjected to 500 ps pre-equilibration run under two different ensemble processes, namely NVT and NPT. All the bond lengths and angles were constrained during the simulation by employing the LINCS algorithm.33 MD run was conducted for 250 ns saving the data for every 500 picoseconds (ps). The simulation was conducted using periodic boundary conditions, under NVT ensemble, with a constant temperature of 300 K and 1 atm pressure. Trajectory by Trajectory analyses have been carried out by employing various modules of the GROMACS package, particularly RMSD and RMSF have been calculated to study the deviation, fluctuation, and radius of gyration of complexes with respect to their native un-equilibrated structures and plotted using Xmgrace tool.34 Binding free energy (BFE) estimation was obtained using the well-known Molecular Mechanics Poisson- Boltzmann Surface Area (MM-PBSA) method.35 The following equations illustrate the procedures employed to obtain the binding free energy values.
| 1 |
where the free energies for each species were evaluated by the following scheme:
| 2 |
| 3 |
| 4 |
| 5 |
In the above equations, ΔEMM corresponds to the molecular mechanical energy changes in the gas phase. ΔEMM includes ΔEbonded, also known as internal energy, and ΔEnonbonded, corresponding to the van der Waals and electrostatic contributions. Both polar and non-polar components of the solvation energy was calculated. ΔGsolvation is the sum of Gpolar and Gnon-polar. The formal surface area values have been obtained by employing the following equation and using the solvent accessible surface area (SASA), i.e., Gnon-polar = γSASA + b, where γ is the surface tension of 2.27 kJmol−1nm−2, and b is a constant with a value of 3.85 kJmol−1, which has been calculated by using the Poisson−Boltzmann (PB) model.36
Protein-Protein interaction profiling
Profiling of the PPI complexes was conducted using the PDBsum server.37 The analysis was performed for four sets of complexes (NiV glycoprotein/EFNB2, HeV glycoprotein/EFNB3, HeV glycoprotein/EFNB2, and HeV glycoprotein/EFNB3), the initial complex, and the average structure from the last 50 ns trajectory of the MD simulation. The PDBsum web server provides image-based structural information about proteins. The database provides a description of the bound molecules and a graphic showing interaction between the protein and secondary structure. The various interaction, including the number of H-bonds formed between the chains of the PPI complex, the salts bridges, and the non-bonded contacts, can be observed from the PDBsum server.
Hotspot residue identification
In the case of PPI, the hotspot residues play a crucial role by contributing more toward the binding energy. In a PPI complex, hotspot residues are considered as a small portion of amino acids in the interface, which is the main target for drug discovery and can be identified by alanine scanning mutagenesis.38 Hotspot identification of the four complexes in the study was carried out by using open-sourced servers Robetta,39,40 KFC2,41 and DrugScorePPI42 for cross-validation.
Principal Component Analysis (PCA) and Free Energy Landscape (FEL)
PCA is a common method used to study the dominant modes in the motion of the molecule. PCA extracts the dominant modes from the protein trajectories of the molecular dynamics simulation and observes the rotational motion throughout the MD trajectory. To get the average geometrical center of the molecule and the configurational changes in the protein structure can be constructed by using the covariance matrix. The covariant matrix forms a number of eigenvectors that give each component's motion along with the direction of motion. In the current study, GROMACS utilizes the g_covar and g_anaeig to construct PCA for analyzing the MD trajectories.43,44
Both PCA and FEL approaches are very much useful in studying the conformational changes and dynamics properties of the protein-protein complexes.44 The Gibbs free FEL was generated from the two principal components (PC1 and PC2) using g_sham module to capture the lowest energy conformer locations. The PCA-FEL analysis was performed for the 250 ns MD trajectory for the four sets of PPI complexes.
Protein sequence retrieval and B-cell epitope prediction for the NiV and HeV glycoproteins
Epitope prediction is essential for developing the antibody against the antigen (viral protein) that can detect and counteract the bioactive molecules to interrupt the viral interaction and entry into the host cell. In the case of henipavirus, the glycoprotein and the fusion envelope proteins are considered antigenic proteins as they show a more antigenic propensity towards epitope prediction.45 Immunoinformatics approaches are widely used to predict the epitopes by analyzing B-cell and cytotoxic T-cells by using various bioinformatics tools.46–48 The NiV glycoprotein (Accession ID: Q9IH62) and HeV glycoprotein (Accession ID: O89343)fasta sequences were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/protein).49 The sequence analysis of both NiV and HeV glycoproteins showed 89.2% sequence similarity and 78.5% sequence identity among both sequences (Figure S1, SI).
In the present study, B-cell epitope prediction was carried out using the ABCPred server50 (http://crdd.osdd.net/raghava/abcpred/) which uses an artificial neural network to predict the B-cell epitope(s) by taking the antigen sequence. The accuracy of the server in predicting the epitope is 65.93%. Another online server BepiPred51 (http://www.cbs.dtu.dk/services/BepiPred-2.0/) was also used for the B-cell epitope prediction. BepiPred-2.0 essentially works with the random forest algorithm with a reasonably high accuracy score.18 Then, by default, parameters were set for the analysis. The 20mer amino acid sequences that were commonly predicted from both servers were considered the predicted epitopes.52,53
Results and Discussion
Molecular dynamics simulations and MMPBSA analysis
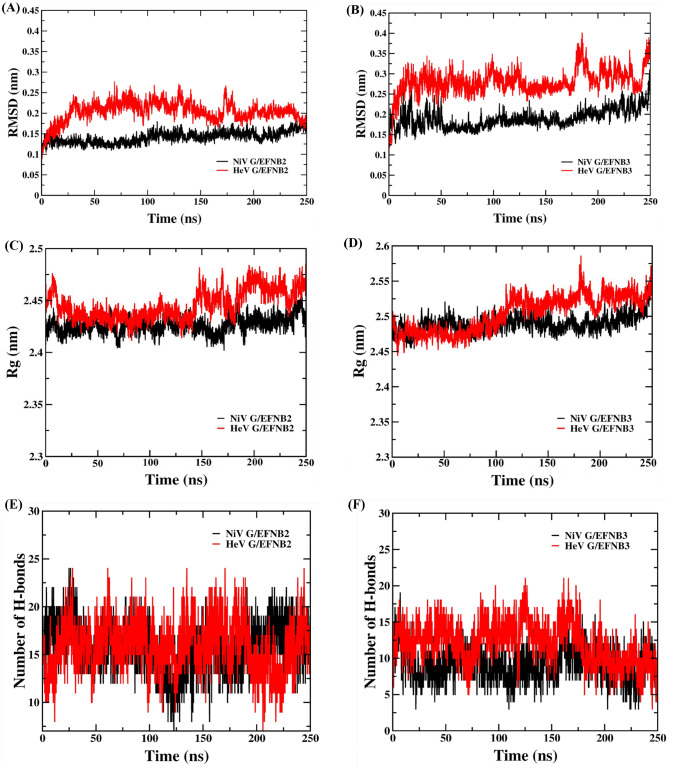
The MD simulations result for the four complexes of NiV and HeV G protein with EFNB2 and B3 were obtained from 250 ns MD trajectories. To study the stability and rigidity of the complexes, RMSD plot was examined for each complex. The RMSD graph was plotted in comparison between (A) NiV glycoprotein/EFNB2 and HeV glycoprotein/EFNB2 and (B) NiV glycoprotein/EFNB3 and NiV glycoprotein/EFNB3 (Figure 2). In the case of NiV glycoprotein/EFNB2 complex the RMSD showed a gradual increase between 0 to 100 ns (0.1-0.75 nm) and converged to a constant value around 0.14-0.15 nm (Figure 2A, B and Table S4, SI) in which the system was observed to be in a less fluctuating state. However, HeV G/EFNB2 complex seems to fluctuate more between 0 to 175 ns (0.25-0.27 nm) and converged to a constant value around 0.18-0.2 nm after 175 ns in comparison to the NiV glycoprotein/EFNB2 complex. In the other comparative analysis of NiV and HeV glycoprotein in complex with EFNB3, the NiV glycoprotein/EFNB3 complex showed a gradual increase in fluctuations from 0-50 ns (i.e., 0.15-0.25 nm) and converged to a stable value around 0.18nm-0.2nm after the 50 ns. Whereas, in case of HeV glycoprotein /EFNB3 complex the fluctuation was observed to be more starting from 0.25-0.4 nm, up to 175 ns trajectory. The system seems to be converged after 175 ns MD trajectory with 0.28 nm of RMSD.
Figure 2.
Protein backbone root mean square deviation (RMSD), radius of gyration and the number of hydrogen bonds graphs of NiV and HeV glycoproteins in complex with human EFN-B2 and B3 along 250 ns MD simulation. Black and red colour represent the Nipah and Hendra virus complexes, respectively.
To analyze the compactness of the PPI complexes and its conformational dynamics in course of 250 ns MD simulation, the radius of gyration (Rg) was analyzed. In the case of NiV glycoprotein/EFNB2 complex the Rg value showed fluctuation in the initial 50 ns MD simulation and the convergence observed after 50 ns with Rg value of 2.4 nm (see Figure 2C, D & Table S4, SI). However, in HeV glycoprotein/EFNB2 complex it has been observed that the continuous fluctuation started in the Rg after 135 ns of MD with a Rg value of 2.48 nm. In the next set of analysis, in NiV glycoprotein/EFNB3 PPI complex, the Rg showed fluctuation in the initial MD simulations, and the fluctuation declined after 50 ns of MD with an Rg value of 2.45 nm. In the case of HeV glycoprotein/EFNB3 the Rg value showed fluctuation till 175 ns of the MD run and obtained convergence after 175 ns with an Rg value of 2.5 nm. The lower value of Rg signify compactness in the complex structures, and high Rg value states low compactness, indicating that the PPI complex is highly fluctuating and less stable. The above comparative analysis showed a clear picture of the compactness of the NiV glycoprotein is more towards the binding with EFNB2 protein as compared to HeV glycoprotein. In the comparative analysis with the EFNB3, the NiV glycoprotein/EFNB3 complex showed more compactness than HeV glycoprotein/EFNB3. The Rg was analyzed for individual chains of the four sets of complexes of 250 ns MD trajectory. The individual Rg plots are depicted in Figure S3, SI.
Throughout the simulation time, the total intermolecular hydrogen bond formation was estimated to determine the conformational stability of the complex (Figures 2E & F). The analysis indicates that the NiV glycoprotein/EFNB2 complex had an average number of twelve H-bonds while the HeV glycoprotein/EFNB2 complex had ten H-bonds throughout the 250 ns simulations. While for NiV glycoprotein/EFNB3 complex, the average number of H-bonds decreased to nine, and in the case of HeV glycoprotein/EFNB3 complex was found to be ten. So, the comparative study indicates that the NiV and HeV glycoproteins are forming more H-bonds upon binding with EFNB2 as compared to EFNB3.
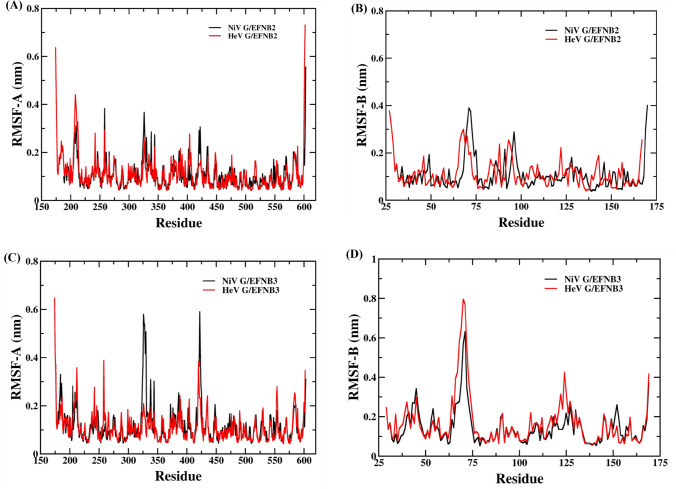
The RMSF value of chain A (NiV and HeV G protein) and chain B (EFNB2) of the first set of complexes was also analyzed to understand the residual flexibility throughout the MD run. The average RMSF value of NiV glycoprotein is 0.10 nm and EFNB2 is 0.10 nm (See Figure 3A, B & Table S4, SI). In case of HeV glycoprotein/EFB2 complex the average RMSF value of HeV glycoprotein is 0.11 nm and EFNB2 is also 0.11 nm. The comparative analysis indicates that the HeV glycoprotein/EFNB2 complex display more local fluctuation as compared to the NiV glycoprotein/EFNB2 complex (Figure 3C). In another set of complexes, the average RMSF value is 0.11 nm for the NiV glycoprotein (chain-A) while the value increased by 0.14 nm for the EFNB3 (chain-B) in the PPI complex. The average RMSF value is 0.16 nm for the HeV glycoprotein and the EFNB3 is 0.16 nm (Figure 3D). The current analysis emphasized the high residual fluctuation in the EFNB3 upon binding of both HeV and NiV as compared to the other set of complexes that binds with EFNB2.
Figure 3.
Root Mean Square Fluctuation (RMSF) graph of Nipah and Hendra virus complexes (A) chain-A of NiV glycoprotein/EFNB2 and HeV glycoprotein/EFNB2 (B) chain-B of NiV glycoprotein/EFNB2 and HeV glycoprotein/EFNB2 (C) chain-A of NiV glycoprotein/EFNB3 and HeV glycoprotein/EFNB3 and (D) chain-B of NiV glycoprotein/EFNB3 and HeV glycoprotein/EFNB3 complexes during 250 ns MD simulation.
Furthermore, the solvent accessible surface area (SASA) (Figure S4, SI) was analyzed in order to determine the bimolecular surface area of the PPI complexes from the 250 ns MD trajectories. The average SASA value (Table S4, SI) of the NiV glycoprotein/EFNB2 complex is observed to be 246.26 nm2 and HeV glycoprotein /EFNB2 is 249.93 nm2. In another set of complexes, the average values of the NiV G/EFNB3 is 251.5 nm2 while the HeV glycoprotein/EFNB3 the SASA value is 258.78 nm2. The comparative study indicates that the SASA value is more in case of the NiV and HeV glycoprotein while binds with the EFNB3 as compared to the binding with EFNB2 (Figure S3, SI). Likewise, the SASA was calculated for each individual chains of the four set of PPI complexes and the corresponding plots has been depicted in Figure S5, SI.
The total binding free energy (BFE) was calculated from the MMPBSA analysis for the four PPI complexes. The BFE was observed to be -389.6 KJ/mol for the NiV glycoprotein/EFNB2 complex and for HeV glycoprotein/EFNB2 it is -333.6KJ/mol. However, the BFE of NiV glycoprotein/EFNB3 is -301.2 KJ/mol which is slightly lower in comparison with HeV glycoprotein/EFNB3 i.e., -322.8 KJ/mol (Table 1). From the BFE of the four complexes it can be seen that the NiV glycoprotein and HeV glycoprotein displayed more binding energy in complex with the EFNB2 as compared to binding with EFNB3 throughout MD simulation. The van der Waals energy which gives the most significant contribution towards the interaction of atoms in both the proteins, thus upholding the structural integrity and stability of the complex throughout the MD run. The van der Waals energy was found to be higher in case of NiV glycoprotein/EFNB2 (-510.3 KJ/mol) and HeV glycoprotein/EFNB3 (-458 KJ/mol) as compared to the PPI complexes NiV glycoprotein/EFNB3 (-457.8KJ/mol) and HeV glycoprotein/EFNB3 (-356.4KJ/mol). Likewise, the electrostatic energy was also found to be higher in the NiV glycoprotein/EFNB2 (-1410.2KJ/mol) and HeV glycoprotein/EFNB2 (-1432.6KJ/mol) as compared to the PPI complexes NiV glycoprotein/EFNB3 (-872.2KJ/mol) and HeV glycoprotein /EFNB3(-901.3KJ/mol). Overall noncovalent interaction54 energies are observed to be higher in case of the NiV and HeV glycoprotein in complex with the EFNB2 as compared to the EFNB3.
Table 1.
MMPBSA analysis of Nipah and Hendra virus glycoprotein in complex with both human cell surface protein Ephrin-B3 and Ephrin-B2.
| Henipavirus | Complexes | Van der Waals energy | Electrostatic energy | Polar solvation energy | SASA energy | Binding energy |
|---|---|---|---|---|---|---|
| Nipah | Glycoprotein- Ephrin-B2 | -510.3 | -1410.2 | 1596.2 | -65.3 | -389.6 |
| Glycoprotein- Ephrin-B3 | -457.8 | -827.2 | 1039.5 | -55.7 | -301.2 | |
| Hendra | Glycoprotein- Ephrin-B2 | -458.0 | -1432.6 | 1620.5 | -63.5 | -333.6 |
| Glycoprotein- Ephrin-B3 | -356.4 | -901.3 | 982.8 | -47.9 | -322.8 |
The energy terms (in kJ/mol) were calculated from the data obtained from last 50 ns trajectory
Interaction profile analysis
The protein profiling of the four PPI complexes was analyzed for the initial structures (Table 2) and compared with the average structure from the last 50 ns of the 250 ns MD trajectory using PDBsum server which represents the interaction statistics showing the interacting area as well as residues, salt bridges, along with the non-bonded contacts. The protein profiling of all the four complexes was also analyzed for the average structure extracted from each 50 ns of 250 ns MD trajectory (Table S1, SI). A quick examination reveals that there is a significant reduction in the number of non-bonded contacts observed in all the four PPI complexes after the complete MD run.
Table 2.
Comparison of interface statistics of the initial NiV glycoprotein-EFNB2, HeV glycoprotein-EFNB2, NiV glycoprotein-EFNB3 and HeV glycoprotein-EFNB3 complexes (before MD) with the average complex structures extracted from last 50 ns of 250 ns Molecular dynamics trajectory.
| Nipah and Hendra Virus | Time | PPI complexes | No. of interface residues | Interface area (Ã2) | No. of salt bridges | No. of H-bonds | No. of non-bonded contacts |
|---|---|---|---|---|---|---|---|
| NiV Glycoprotein and Human EFNB2 | Initial structure | NiV Glycoprotein (A) | 32 | 1373 | 3 | 21 | 218 |
| EFNB2 (B) | 27 | 1473 | |||||
|
Average Str. from last 50 ns |
NiV Glycoprotein (A) | 24 | 1219 | 4 | 16 | 141 | |
| EFNB2 (B) | 23 | 1257 | |||||
| HeV Glycoprotein and Human EFNB2 | Initial structure | HeVGlycoprotein (A) | 30 | 1300 | 4 | 12 | 160 |
| EFNB2 (B) | 26 | 1389 | |||||
|
Average Str. From last 50 ns |
HeV Glycoprotein (A) | 20 | 1123 | 2 | 11 | 75 | |
| EFNB2 (B) | 16 | 1210 | |||||
| NiV Glycoprotein and Human EFNB3 | Initial structure | NiV Glycoprotein (A) | 24 | 997 | 4 | 11 | 127 |
| EFNB3 (B) | 19 | 1166 | |||||
|
Average Str. From last 50 ns |
NiV Glycoprotein (A) | 20 | 934 | 1 | 6 | 73 | |
| EFNB3 (B) | 12 | 1089 | |||||
| HeV Glycoprotein and Human EFNB3 | Initial structure | HeV Glycoprotein (A) | 26 | 1184 | 5 | 12 | 137 |
| EFNB3 (B) | 26 | 1272 | |||||
|
Average Str. From last 50 ns |
HeV Glycoprotein (A) | 17 | 820 | 1 | 8 | 61 | |
| EFNB3 (B) | 13 | 886 |
For each complex, interface statistics obtained from PDBsum server.
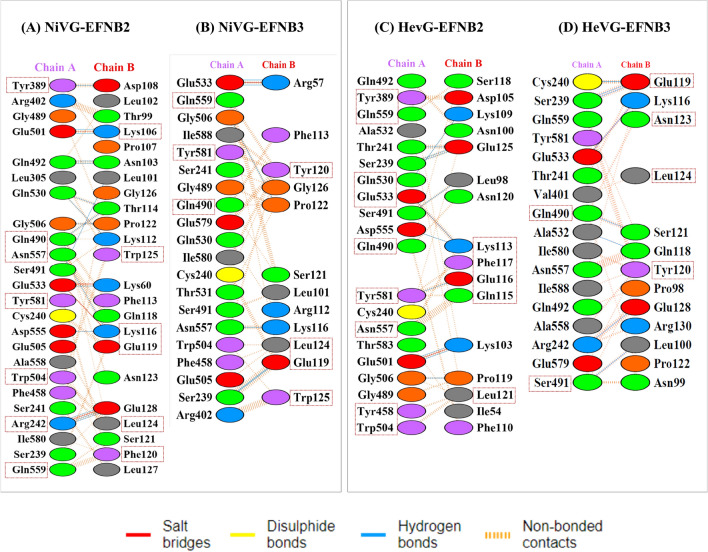
Hotspot residue prediction analysis and per residue energy contribution
The hotspot residue identification was performed using four server viz. Robetta, DrugScorePPI, KFC2 and pyDockEneRes server.55 pyDockEneRes server gives the per residue energy for the PPI complex to obtain the detailed understanding of the hotspot residues in the protein-protein interaction interface. Hotspots are a small portion of residues present across protein-protein interface which contribute the majority of the BFE towards the formation of PPI complex.56 Identification of such residues can provide essential information regarding the protein functions and will pave a way in identifying potential antiviral and peptides to obstruct the viral spread.16,57 The interface hotspot residues prediction was performed for four sets of complexes, the initial structure (Table S3, SI) and the average structure from the last 50 ns trajectory of MD simulations. The identified hotspot residues were scored higher value of per-residue energy contribution obtained by using pyDockEneRes online server.55 In case of the initial PPI complex of NiV G/EFNB2, a total of sixteen residues were predicted as hotspot by two and more servers. The predicted residues are Gln490-A, Gln530-A, Trp504-A, Arg242-A, Glu533-A, Asn557-A, Tyr581-A, Asn123-B, Phe113-B, Leu124-B, Trp125-B, Lys116-B, Gln118-B, Glu119-B, Glu128-B, Phe120-B (Table S3, SI). Whereas, NiV glycoprotein/EFNB2 average 50 ns trajectory complex there are a total of fourteen number of residues were predicted as hotspot by two or more servers. The predicted residues are Tyr389-A, Arg241-A, Gln420-A, Trp504-A, Glu533-A, Asn557-A, Gln559-A, Tyr581-A, Lys106-B, Lys116-B, Gln118-B, Glu119-B, Phe120-B, Leu124-B, Trp125-B (Table 3). In case of the initial PPI complex of HeV glycoprotein/EFNB2, a total of fourteen residues were predicted as hotspot. The predicted residues are Tyr581-A, Asn557-A, Thr531-A, Glu533-A, Trp504-A, Glu505-A, Tyr389-A, Gln490-A, Gln530-A, Lys113-B, Ser118-B, Pro119-B, Asn120-B, Leu124-B and the HeV glycoprotein/EFNB2 average 50ns structure presented a total of fourteen residues such as Tyr389-A, Gln559-A, Gln530-A, Glu533-A, Gln490-A, tyr581-A, Asn557-A, Tyr458-A, Trp504-A, Glu125-B, Lys113-B, Phe117-B, Glu116-B and Leu121-B. In the third set of PPI complex the initial NiV G/EFNB3 complex predicted eleven number of residues Thr531-A, Asn557-A, Gln490-A, Ser491-A, Tyr120-B, Glu119-B, Gln118-B, Leu124-B, Lys116-B, Trp125-B, Phe113-B as interface hotspot residues while a total of six residues Gln559-A, Trp504-A, Tyr120-B, Leu124-B, Glu119-B and Trp125-B are predicted as hotspots for the average 50 ns complex. In case of the fourth set of PPI complexes the initial complex of HeV glycoprotein/EFNB3 thirteen residues (Gln490-A, Glu533-A, Asn557-A, Tyr581-A, Thr531-A, Gln530-A, Glu119-B, Gln118-B, Lys116-B, Tyr120-B, Thr114-B, Leu124-B, Phe113-B) are predicted as hotspot residues and six residues Gln490-A, Ser491-A, Glu119-B, Tyr120-B, Asn123-B and Leu124-B are identified as hotspots in case of average 50 ns average structure (Figure 4).
Table 3.
List of interacting residues at protein-protein interacting interface of NiV and HeV glycoprotein (chain A) in complex with human EFNB3/B3 (chain B) each.
| Complex | Residues (Post MD) | KFC2 server | DrugScore PPI (kJ/mol) |
Robetta ΔΔG (kJ/mol) |
Per-residue energy contribution (kJ/mol) |
|---|---|---|---|---|---|
| NiV glycoprotein/EFNB2 | Arg242-A | - | 4.9 | 9.9 | -18.6 |
| Gln490-A | HS | 1.0 | 13.8 | -10.8 | |
| Trp504-A | - | 4.8 | 9.0 | -28.2 | |
| Glu533-A | HS | 2.2 | 19.7 | -11.0 | |
| Asn557-A | HS | 4.1 | 8.2 | -33.5 | |
| Gln559-A | HS | 4.9 | 7.4 | -22.0 | |
| Tyr581-A | HS | 6.16 | 7.4 | -47.6 | |
| Lys106-B | - | 4.6 | 4.1 | -17.7 | |
| Lys116-B | HS | 4.1 | 13.3 | 6.6 | |
| Gln118-B | HS | 2.9 | 2.6 | -17.3 | |
| Glu119-B | HS | 2.6 | 5.7 | -34.2 | |
| Phe120-B | HS | 8.1 | 19.0 | -78.4 | |
| Leu124-B | HS | 9.8 | 12.6 | -3.4 | |
| Trp125-B | HS | 3.2 | 9.3 | -63.2 | |
| NiV glycoprotein/EFNB3 | Gln559-A | HS | 3.7 | 9.4 | -8.7 |
| Trp504-A | - | 3.8 | 8.0 | -16.48 | |
| Tyr120-B | HS | 26.6 | 19.9 | -58.4 | |
| Leu124-B | HS | 6.9 | 10.7 | -31.96 | |
| Glu119-B | HS | 3.8 | 9.2 | 40.7 | |
| Trp125-B | HS | 6.3 | 13.0 | -56.1 | |
| HeV glycoprotein/EFNB2 | Tyr389-A | - | 51.4 | 46.8 | -119.6 |
| Gln559-A | - | 4.8 | 6.9 | -0.9 | |
| Gln530-A | HS | 0.3 | 6.3 | -0.19 | |
| Glu533-A | HS | 7.9 | 17.5 | -110.4 | |
| Gln490-A | HS | 6.61 | 24.6 | -7.5 | |
| Tyr581-A | HS | 11.6 | 17.7 | -84.8 | |
| Asn557-A | HS | 4.3 | 4.4 | 0.5 | |
| Tyr458-A | - | 5.5 | 3.5 | -1.4 | |
| Trp504-A | - | 2.6 | 10.0 | 48.4 | |
| Glu125-B | HS | 4.2 | 7.2 | -11.0 | |
| Lys113-B | HS | 4.1 | 8.32 | -2.5 | |
| Phe117-B | HS | 7.5 | 19.2 | -84.3 | |
| Glu116-B | HS | 0.8 | 6.3 | -3.58 | |
| Leu121-B | HS | 9.9 | 11.4 | -48.1 | |
| HeV glycoprotein/EFNB3 | Gln490-A | HS | 7.2 | 7.7 | -10.2 |
| Ser491-A | HS | 2.3 | 4.7 | -0.2 | |
| Glu119-B | HS | 1.2 | 11.0 | -38.6 | |
| Asn123-B | HS | 4.4 | 4.4 | -9.8 | |
| Leu124-B | HS | 1.0 | 2.3 | -7.2 | |
| Tyr120-B | HS | 24.2 | 17.9 | -63.4 |
Each pdb complex extracted from last 50 ns MD trajectory. The interface hotspot residues were predicted using three computational methods implemented in KFC2 server, and Robetta web server. The per-residue energy decomposition analysis was carried out using the pyDockEneRes Server.
Figure 4.
The interacting residues of the NiV and HeV glycoprotein in complex with both EFN-B2 and B3 using the average structure from the last 50 ns MD trajectory obtained from the PDBSum server. The squared box represents the predicted hotspots from four servers (KFC2, Robetta, DrugScorePPI and the pyDockEneRes Server
In the initial structure of NiV glycoprotein/EFNB2 complex the residue Glu533 of glycoprotein formed H-bonds and salt bridges with the residues Lys60 and Lys116 of EFNB2, while during the simulation H-bond with Lys116 was lost (Table S5, S9, S10, S14, SI). In case of HeV glycoprotein/EFNB2 initial complex the residue Glu533 of glycoprotein was found to be forming H-bond with Lys113 and salt bridges with Lys57 and Lys113 of EFNB2 (Table S7, S9, S12, S14, SI). Interestingly, in the hotspot analysis study for both the (NiV and HeV glycoprotein/EFNB2) complexes, Glu533 was identified as one of the potential hotspots in glycoprotein. However, the complex of both NiV and HeV with EFNB3 does not predict Glu533 as a hotspot (Table S6, S8, S11, S13, SI). In the current analysis Glu533 of both the glycoproteins has shown more significance towards the formation of complex with EFNB2. All the identified hotspots in the present study have been depicted in the superimposed initial and last 50 ns average MD simulation extracted structures in Figure 5.
Figure 5.
Superimposition of the initial structure of (A,B) NiV glycoprotein/EFN B2 & B3 (C,D) HeV glycoprotein/EFN B2&B3 with the average structure of the four complexes extracted from the last 50 ns of the 250 ns MD simulation trajectory. The highlighted residues in each complex display the identified hotspot residues in the present study. The RMSD value of superimposed initial and the final structure of NiV glycoprotein/EFNB2 and B3 is 1.219Å and 1.621Å, respectively. While for HeV glycoprotein/EFNB2 & B3 the values are 1.575 Å and 1.202 Å, respectively.
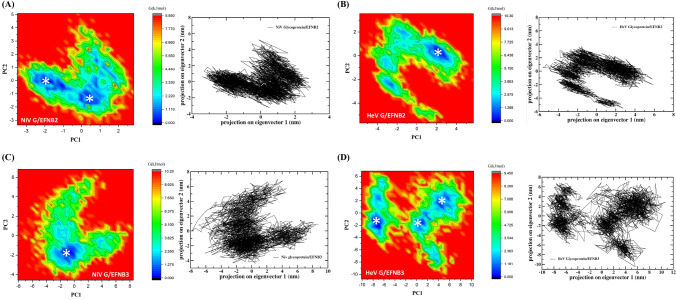
Principal component analysis (PCA) and free energy landscape (FEL)
Through PCA the motion of the trajectories can be computed through the eigen vector generated from the covariance matrix of the fluctuation of the atoms in the whole system. To find out the way how protein-protein interaction affects the motion has been depicted by PC1 and PC2 in the four set of PPI complexes.58 To demonstrate the effect of PPI on conformational redistributions, free energy landscape (FEL) for four systems were obtained as a function of the top PC1 and PC2. The conformational differences and the dominant modes in the protein-protein complexes were calculated and displayed in the Figure 6. The PCA-FEL was calculated for the backbone of the four complexes. In case of NiV glycoprotein/EFNB2 complex two distinct local basins containing lowest energy conformers were observed. However, HeV glycoprotein/EFNB2 and NiV glycoprotein/EFNB3 complexes have shown one distinct local basin. While, HeV glycoprotein/EFNB3 complex has shown multiple local minima basins (Figure 6).
Figure 6.
Free energy landscape (FEL) along projections onto the 1st and 2nd principal components of the 250 ns MD concatenated trajectory. Asterisks (*) indicate the localization on the FEL of the average structures of the different conformational ensembles. Two-dimensional principal component analysis (PCA) projection of trajectories obtained from 250 ns MD simulations of NiV and HeV glycoprotein in complex with both EFNB2/B3, respectively are shown in black color graphs. The free energy is given in kJ/mol and indicated by color.
A comparison of the FELs for the four set of complexes reveals that the FEL of HeV glycoprotein/EFNB2 and NiV glycoprotein/EFNB3 display a stronger free energy surface than that of the other two set of complexes having multiple number of local free energy minima in different cluster.
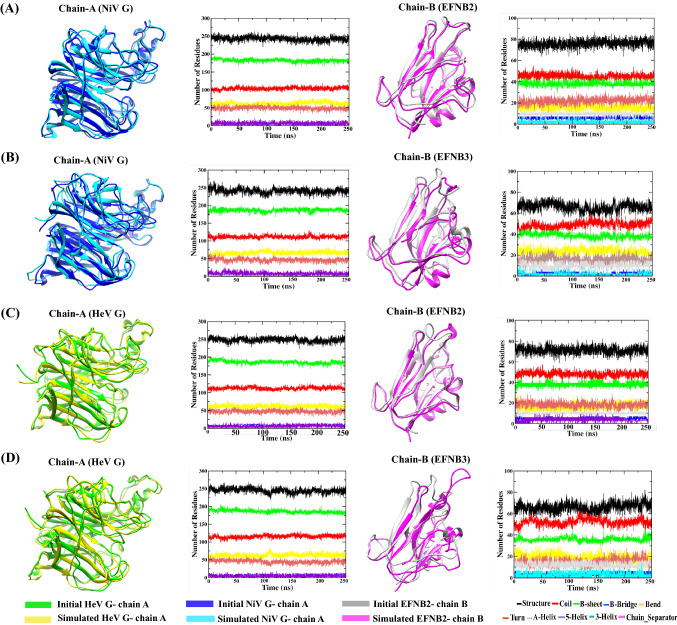
Exploring the variation in the secondary structure along with the MD trajectories
A database of secondary structure assignments (DSSP) analysis was carried out for the four PPI complexes to explore the variation in the content of the secondary structure elements along with the loss and gain of secondary structure, viz. alpha helix, beta sheets and coils, (Figure S6, SI) through the 250 ns trajectories. The DSSP, obtained using ‘do_dssp’ program in GROMACS (Figure 7), represent the secondary structure analysis of NiV glycoprotein/EFNB2 and EFNB3 and that of HeV glycoprotein/EFNB2 and EFNB3. From Figure 7 it can be observed that the secondary structure content was stably maintained in the complex structure with little fluctuations in the coils as well as beta sheets and alpha helix. During the course of MD simulations, the notable residual fluctuations have been observed in the secondary structure graph of EFNB2 and EFNB3 proteins. Consequently, this analysis provides clear evidence about the secondary and tertiary structure of the PPI complexes remained stable during 250 ns simulation.
Figure 7.
Time evolution of secondary structure elements during the course of 250 ns MD simulation in (A) NiV glycoprotein and human EFNB2 (B) NiV glycoprotein and human EFNB3 (C) HeV glycoprotein and human EFNB2 (D) HeV glycoprotein and human EFNB3.
B-cell epitope prediction
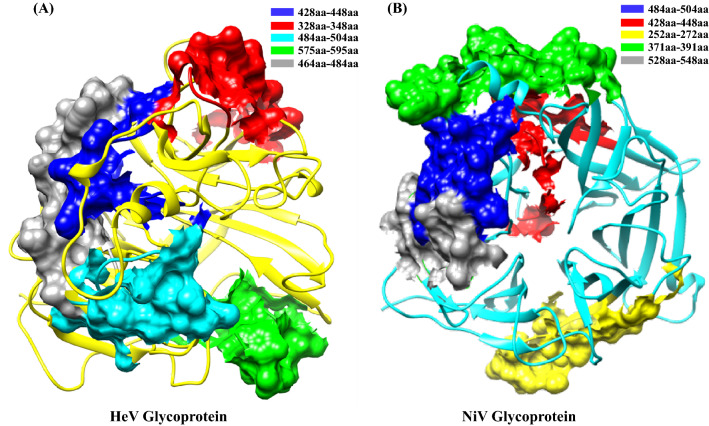
B-cell recognizes the portions within the connected antigen known as epitopes and identifying those helps in designing epitope-based vaccines, development of diagnostic assays, etc. By using in silico prediction methods, the 20 amino acid lengths epitopes were predicted for both NiV and HeV glycoproteins. The epitopes were predicted based on the specificity and sensitivity score towards the B-cell. Among the predicted linear epitopes, the top five epitopes were taken according to their high score for both the NiV and HeV glycoproteins. The predicted epitopes are also observed to be present in the conserved region of the aligned protein sequences (Table 4). The 20mer epitopes in different regions of NiV and HeV glycoproteins are structurally depicted in Figure 8 and Figure S7, SI. The residues present in the conserved regions are generally important towards understanding the pathophysiology of viral entry and replication.48,49 This study indicates that the identified epitope residues in the current study will help to develop inhibitors to decipher the viral interaction and entry to the host cell.
Table 4.
The linear B-Cell epitopes predicted for NiV and HeV glycoprotein.
| Protein | Position | Epitope | ABCPred Score | BepiPred (Average percentage) |
|
|---|---|---|---|---|---|
| NiV Virus | Glycoprotein | 484 | VISRPGQSQCPRFNTCPEIC | 0.93 | 0.52 |
| 428 | FIEISDQRLSIGSPSKIYDS | 0.93 | 0.44 | ||
| 252 | VGEVLDRGDEVPSLFMTNVW | 0.90 | 0.44 | ||
| 371 | VRTEFKYNDSNCPITKCQYS | 0.90 | 0.57 | ||
| 528 | SNQTAENPVFTVFKDNEILY | 0.89 | 0.46 | ||
| HeV Virus | Glycoprotein | 428 | FIEIADNRLTIGSPSKIYNS | 0.93 | 0.43 |
| 328 | GDYNQKYIAITKVERGKYDK | 0.93 | 0.52 | ||
| 484 | VISRPGQSQCPRFNVCPEVC | 0.90 | 0.52 | ||
| 575 | ISLVEIYDTGDSVIRPKLFA | 0.89 | 0.40 | ||
| 464 | IKLGDVDTVDPLRVQWRNNS | 0.89 | 0.48 |
The predicted epitopes are based on the BCPred score and the epitope percentage is given by the average generated from BepiPred server.
Figure 8.
Surface representation of linear predicted B cell epitopes highlighted in the protein structure of (A) HeV glycoprotein and (B) NiV glycoprotein. The stretch of epitopes (amino acid range) consisting of 20 amino acids are highlighted with different colors, respectively.
Conclusions
Computational studies have played a very important role in understanding the host-pathogen interactions. Recent years have highlighted the importance of gaining insights on the role of protein-protein interactions and their functional disparity due to the mutations. The current study focuses on performing computational analysis to bring insights into the viral entry, identification and validation of the hotspot residues, epitope analysis and MD simulations provides valuable information on the detailed structural and dynamic exploration of receptor binding domains (RBD) of the viral and host proteins and their interface, which may be helpful in devising strategies to restrict the viral entry. In this study, focus is mainly on the human EFN receptor mediated viral entry mechanism, as there is considerable focus in the recent literature on the viral attachment by alanine scanning mutagenesis along with other experimental approaches.
From the MD analysis (RMSD, Rg, RMSF, PCA and MM-PBSA) it has been observed that both the NiV and HeV virus glycoproteins/EFNB2 complexes attained conformational stability with less fluctuation throughout the simulation period with high binding affinity as compared to EFNB3 which is in agreement with the structural studies. Encouragingly, the current computational study observed that earlier observed hotspot residues display high stability in retaining at the interface throughout the 250 ns MD. Further, one residue (Glu533), in the central cavity of both the glycoprotein (NiV and HeV) is identified as potential hotspot in complex with EFNB2 and also found that it engages in the formation of the salt bridges and H-bonds throughout the MD simulation. The epitope prediction revealed the residues present are in the conserved regions of glycoproteins, and this information can be explored in the design of antivirals and vaccines. Further, targeting the hotspot residues that are identified in the receptor binding domain (RBD-RBD) of the glycoprotein interface (central cavity) and cell surface receptor proteins EFNB2 & EFNB3 (G-H loop) help in unraveling the viral entry mechanism. The current study reveal that a detailed structural analysis of the protein-protein interaction appears to through more profound questions, newer challenges to overcome, and more importantly on the atomistics exploration of the disease pathophysiology in a new dimension.
The massive knowledge generation in the area of exploring the structural details on the viral entry of different viruses, especially after the COVID-19 pandemic, will certainly be of great value in the vaccine and drug discovery efforts which are more directed towards host as well as host-viral interactions. Thus, the interplay between the modeling and experiment, in the exploration of PPI involving host and pathogen is interesting in its own right.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgement
The DBT is thanked for the center of excellence for Advanced Computation and Data Sciences (BT/PR40188/BTIS/137/27/2021). CSIR-NEIST and CSIR-Fourth Paradigm Institute is thanked for extending the computational facilities. GNS thanks JC Bose Fellowship of DST-SERB.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information (SI)
Figures S1-S5 and Tables S1-S14 are available at. http://www.ias.ac.in/chemsci.
References
- 1.Eaton B T, Broder C C, Middleton D, Wang L F. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 2006;4:23. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu K, Broder C C, Nikoloz D B. Ephrin-B2 and ephrin-B3 as functional henipavirus receptors. Semin. Cell. Dev. Biol. 2012;23:116. doi: 10.1016/j.semcdb.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu K, Chan Y P, Rajashankar K R, Khetawat D, Yan L, Kolev M V, Broder C C, Nikolov D B. New insights into the Hendra virus attachment and entry process from structures of the virus G glycoprotein and its complex with Ephrin-B2. PloS One. 2012;7:e48742. doi: 10.1371/journal.pone.0048742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden T A, Aricescu A R, Gilbert R J, Grimes J M, Jones E Y, Stuart D I. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 2008;15:567. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 5.Bradel-Tretheway B G, Zamora J L, Stone J A, Liu Q, Li J, Aguilar H C. Nipah and Hendra virus glycoproteins induce comparable homologous but distinct heterologous fusion phenotypes. J. Virol. 2019;93:e00577. doi: 10.1128/JVI.00577-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harcourt B H, Tamin A, Ksiazek T G, Rollin P E, Anderson L J, Bellini W J, Rota P A. Molecular characterization of Nipah virus a newly emergent paramyxovirus. Virol. 2000;271:334. doi: 10.1006/viro.2000.0340. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad F, Albutti A, Tariq M H, Din G, Tahir ul Qamar M, Ahmad S. Discovery of potential antiviral compounds against hendra virus by targeting its receptor-binding protein (G) using computational approaches. Molecules. 2022;27:554. doi: 10.3390/molecules27020554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar H C, Matreyek K A, Filone C M, Hashimi S T, Levroney E L, Negrete O A, Bertolotti-Ciarlet A, Choi D Y, McHardy I, Fulcher J A, Su S V, Wolf M C, Kohatsu L, Baum L G, Lee B. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 2006;80:4878. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowden T A, Crispin M, Harvey D J, Aricescu A R, Grimes J M, Jones E Y, Stuart D I. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: A template for antiviral and vaccine design. J. Virol. 2008;82:11628. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohammed A A, Shantier S W, Mustafa M I, Osman H K, Elmansi H E, Osman I A, Mohammed R A, Abdelrhman F A, Elnnewery M E, Yousif E M, Mustafa M M. Epitope-based peptide vaccine against glycoprotein G of Nipah henipavirus using immunoinformatics approaches. J. Immunol. Res. 2020;22:12. doi: 10.1155/2020/2567957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakib M S, Islam M, Hasan A K M, Nabi A H M. Prediction of epitope-based peptides for the utility of vaccine development from fusion and glycoprotein of nipah virus using in silico approach. Adv. Bioinform. 2014;2014:402492. doi: 10.1155/2014/402492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Bossart K N, Bishop K A, Crameri G, Dimitrov A S, McEachern J A, Feng Y, Middleton D, Wang L F, Broder C C, Dimitrov D S. Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J. Infect. Dis. 2008;197:846. doi: 10.1086/528801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Dimitrov A S, Bossart K N, Crameri G, Bishop K A, Choudhry V, Mungall B A, Feng Y R, Choudhary A, Zhang M Y, Feng Y, Wang L F, Xiao X, Eaton B T, Broder C C, Dimitrovn D S. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 2006;80:891. doi: 10.1128/JVI.80.2.891-899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossart K N, Zhu Z, Middleton D, Klippel J, Crameri G, Bingham J, McEachern J A, Green D, Hancock T J, Chan Y P, Hickey A C, Dimitrov D S, Wang L F, Broder C C. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009;5:e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Gil L, Vera-Velasco N M, Mingarro I. Exploring the human-Nipah virus protein-protein interactome. J. Virol. 2017;91:e01461. doi: 10.1128/JVI.01461-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarma H, Jamir E, Sastry G N. Protein-protein interaction of RdRp with its co-factor NSP8 and NSP7 to decipher the interface hotspot residues for drug targeting: A comparison between SARS-CoV-2 and SARS-CoV. J. Mol. Struct. 2022;1257:132602. doi: 10.1016/j.molstruc.2022.132602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamir E, Sarma H, Priyadarsinee L, Nagamani S, Kiewhuo K, Gaur A S, Rawal R K, Murugan N A, Subramanian V, Sastry G N. Applying polypharmacology approach for drug repurposing for SARS-CoV2. J. Chem. Sci. 2022;134:24. doi: 10.1007/s12039-022-02046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar N, Sarma H, Sastry G N. Repurposing of approved drug molecules for viral infectious diseases: a molecular modelling approach. J. Biomol. Struct. Dyn. 2021;22:7. doi: 10.1080/07391102.2021.1905558. [DOI] [PubMed] [Google Scholar]
- 19.Sarma S and Sastry G N 2022 A Computational Study on the Interaction of NSP10 and NSP14: Unraveling the RNA Synthesis Proofreading Mechanism in SARS-CoV-2 SARS-CoVand MERS-CoV ACS Omega 0000-00 (Accepted) [DOI] [PMC free article] [PubMed]
- 20.Dutta P, Siddiqui A, Botlani M, Varma S. Stimulation of nipah fusion: small intradomain changes trigger extensive interdomain rearrangements. Biophys. J. 2016;111:1621. doi: 10.1016/j.bpj.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffen D L, Xu K, Nikolov D B, Broder C C. Henipavirus mediated membrane fusion virus entry and targeted therapeutics. Viruses. 2012;4:2. doi: 10.3390/v4020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koolpe M, Burgess R, Dail M, Pasquale E B. EphB receptor-binding peptides identified by phage display enable design of an antagonist with ephrin-like affinity. J. Biol. Chem. 2005;280:17301. doi: 10.1074/jbc.M500363200. [DOI] [PubMed] [Google Scholar]
- 23.Xu K, Rajashankar K R, Chan Y P, Himanen J P, Broder C C, Nikolov D B. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Sci. U S A. 2008;105:9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillaume V, Aslan H, Ainouze M, Guerbois M, Wild T F, Buckland R, Langedijk J P. Evidence of a potential receptor-binding site on the Nipah virus G protein (NiV-G): Identification of globular head residues with a role in fusion promotion and their localization on an NiV-G structural model. J. Virol. 2006;80:7546. doi: 10.1128/JVI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seiradake E, Schaupp A, del Toro Ruiz D, Kaufmann R, Mitakidis N, Harlos K, Aricescu A R, Klein R, Jones E Y. Structurally encoded intraclass differences in EphA clusters drive distinct cell responses. Nat. Struct. Mol. Biol. 2013;20:958. doi: 10.1038/nsmb.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. The protein data bank. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Zundert G C P, Rodrigues J P G L M, Trellet M, Schmitz C, Kastritis P L, Karaca E, Bonvin A M J J. The HADDOCK2 2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen E F, Goddard T D, Huang C C, Couch G S, Greenblatt D M, Meng E C, Ferrin T E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Abraham M J, Murtola T, Schulz R, Páll S, Smith J C, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 31.Best R B, Zhu X, Shim J, Lopes P E, Mittal J, Feig M, MacKerell A D., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory. Comput. 2012;8:3257. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuhrmans M, Sanders B P, Marrink S J, de Vries A H. Effects of bundling on the properties of the SPC water model. Theor. Chem. Acc. 2010;125:335. doi: 10.1007/s00214-009-0590-4. [DOI] [Google Scholar]
- 33.Hess B, Bekker H, Berendsen H J, Fraaije J G. LINCS: a linear constraint solver for molecular simulations. J. Comp. Chem. 1997;18:1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 34.Turner P J 2005 XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton OR 2
- 35.Srivastava H K, Sastry G N. Molecular dynamics investigation on a series of HIV protease inhibitors: assessing the performance of MM-PBSA and MM-GBSA approaches. J. Chem. Info. Mod. 2012;52:3098. doi: 10.1021/ci300385h. [DOI] [PubMed] [Google Scholar]
- 36.Kumari R, Kumar R. Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zerbe BS, Hall DR, Vajda S, Whitty A, Kozakov D. Relationship between hot spot residues and ligand binding hot spots in protein–protein interfaces. J. Chem. Info. Model. 2012;52:2244. doi: 10.1021/ci300175u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D E, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kortemme T, Kim D E and Baker D 2004 Computational alanine scanning of protein-protein interfaces Sci. STKE.219 pl2 [DOI] [PubMed]
- 41.Darnell S J, LeGault L, Mitchell J C. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36:69. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krüger D M, Gohlke H. DrugScorePPI webserver: fast and accurate in silico alanine scanning for scoring protein–protein interactions. Nucleic Acids Res. 2010;38:W486. doi: 10.1093/nar/gkq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amadei A, Linssen A B, Berendsen H J. Essential dynamics of proteins. Proteins. 1993;17:425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 44.Altis A, Otten M, Nguyen P H, Hegger R, Stock G. Construction of the free energy landscape of biomolecules via dihedral angle principal component analysis. J. Chem. Phys. 2008;128:245102. doi: 10.1063/1.2945165. [DOI] [PubMed] [Google Scholar]
- 45.Majee P, Jain N, Kumar A. Designing of a multi-epitope vaccine candidate against Nipah virus by in silico approach: a putative prophylactic solution for the deadly virus. J. Biomol. Struct. Dyn. 2021;39:1480. doi: 10.1080/07391102.2020.1734088. [DOI] [PubMed] [Google Scholar]
- 46.Parvege M M, Rahman M, Nibir Y M, Hossain M S. Two highly similar LAEDDTNAQKT and LTDKIGTEI epitopes in G glycoprotein may be useful for effective epitope based vaccine design against pathogenic Henipavirus. Comput. Biol. Chem. 2016;61:280. doi: 10.1016/j.compbiolchem.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohammed A A, Shantier S W, Mustafa M I, Osman H K, Elmansi H E, Osman I A, Mohammed R A, Abdelrhman F A, Elnnewery M E, Yousif E M, Mustafa M M, Elfadol N M, Abdalla A I, Mahmoud E, Yagaub A A, Ahmed Y A and Hassan M A 2020 Epitope-Based Peptide Vaccine against Glycoprotein G of Nipah Henipavirus Using Immunoinformatics Approaches J. Immunol. Res. 2567957 [DOI] [PMC free article] [PubMed]
- 48.Kamthania M, Sharma D K. Screening and structure-based modeling of T-cell epitopes of Nipah virus proteome: an immunoinformatic approach for designing peptide-based vaccine. 3 Biotech. 2015;5:882. doi: 10.1007/s13205-015-0303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenuth JP. The NCBI. Publicly available tools and resources on the Web. Methods Mol. Biol. 2000;132:312. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- 50.Saha S, Raghava G P S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65:40. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 51.Jespersen M C, Peters B, Nielsen M, Marcatili P. BepiPred2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jamir E, Kiewhuo K, Priyadarsinee L, Sarma H, Nagamani S, Sastry G N. Structure-function relationships among selected human coronaviruses. Indian J. Biochem. Biophys. 2022;59:536. [Google Scholar]
- 53.El-Manzalawy Y, Honavar V. Recent advances in B-cell epitope prediction methods. Immunome Res. 2010;6:9. doi: 10.1186/1745-7580-6-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahadevi A S, Sastry G N. Cooperativity in noncovalent interactions. Chem. Rev. 2016;116:2825. doi: 10.1021/cr500344e. [DOI] [PubMed] [Google Scholar]
- 55.Romero-Durana M, Jiménez-García B, Fernández-Recio J. pyDockEneRes: per-residue decomposition of protein– protein docking energy. Bioinformatics. 2020;36:2285. doi: 10.1093/bioinformatics/btz884. [DOI] [PubMed] [Google Scholar]
- 56.Moreira I S, Fernandes P, Ramos M J. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins: Struct. Funct. Genet. 2007;68:803. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 57.Liu Q, Chen P, Wang B, Zhang J, Li J. Hot spot prediction in protein-protein interactions by an ensemble system. BMC Syst. Biol. 2018;12:89. doi: 10.1186/s12918-018-0665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandey P, Prasad K, Prakash A, Kumar V. Insights into the biased activity of dextromethorphan and haloperidol towards SARS-CoV-2 NSP6: in silico binding mechanistic analysis. J. Mol. Med. (Berl) 2020;98:1673 (PCA). doi: 10.1007/s00109-020-01980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.