Abstract
Tumor mutational burden (TMB), a surrogate for tumor neoepitope burden, is used as a pan-tumor biomarker to identify patients who may benefit from anti-program cell death 1 (PD1) immunotherapy, but it is an imperfect biomarker. Multiple additional genomic characteristics are associated with anti-PD1 responses, but the combined predictive value of these features and the added informativeness of each respective feature remains unknown. We evaluated whether machine learning (ML) approaches using proposed determinants of anti-PD1 response derived from whole exome sequencing (WES) could improve prediction of anti-PD1 responders over TMB alone. Random forest classifiers were trained on publicly available anti-PD1 data (n = 104), and subsequently tested on an independent anti-PD1 cohort (n = 69). Both the training and test datasets included a range of cancer types such as non-small cell lung cancer (NSCLC), head and neck squamous cell carcinoma (HNSCC), melanoma, and smaller numbers of patients from other tumor types. Features used include summaries such as TMB and number of frameshift mutations, as well as more gene-level features such as counts of mutations associated with immune checkpoint response and resistance. Both ML algorithms demonstrated area under the receiver-operator curves (AUC) that exceeded TMB alone (AUC 0.63 “human-guided,” 0.64 “cluster,” and 0.58 TMB alone). Mutations within oncogenes disproportionately modulate anti-PD1 responses relative to their overall contribution to tumor neoepitope burden. The use of a ML algorithm evaluating multiple proposed genomic determinants of anti-PD1 responses modestly improves performance over TMB alone, highlighting the need to integrate other biomarkers to further improve model performance.
Keywords: Checkpoint inhibitors, therapeutic response, algorithmic biomarker, patient stratification, anti-PD-1 immunotherapy
Introduction
Immune checkpoint inhibitor therapy targeting program cell death 1 (PD1) or its ligand is firmly established as a major pillar of cancer therapy, but it is estimated less than half of all patients are eligible for these therapies, and only a minority of those patients will respond to these agents. Identifying new, predictive biomarkers to identify patients most likely to respond to these immunotherapeutic agents could extend the number of patients who can receive anti-PD1 treatment, while minimizing treatment costs and avoiding immune-related toxicities in patients unlikely to derive clinical benefit. Biomarkers in clinical use today to select patients for anti-PD1 therapy fit broadly into 2 categories: (1) tumor genomic biomarkers related to neoepitope burden, including mismatch repair deficiency (MMRd) and microsatellite instability-high (MSI-H),1-5 and (2) biomarkers indicative of an inflamed tumor immune microenvironment, including expression of PD-L1, abundant immune infiltration, or other T cell factors such as mitochondrial biogenesis.6-11 These 2 classes of anti-PD1 biomarkers are only marginally correlated across the entire landscape of human cancers, and are uncorrelated within many tumor types. Therefore, each biomarker provides independent information.4
An elevated TMB has been associated with response to checkpoint therapies in multiple studies. Across the entire landscape of cancer types, it is estimated that approximately 50% of the variation in anti-PD1 response rates may be explained by the TMB alone. In 2021, a TMB of greater than 10 mutations/megabase (Mb) of DNA was approved in the United States as a pan-tumor biomarker to identify patients for the PD1 inhibitor pembrolizumab who have failed other approved systemic therapies.4,5 However, response prediction performance is somewhat limited because TMB is only about 40% to 60% sensitive for identifying pembrolizumab responders at a specificity of 90%.12 These diagnostic characteristics are consistent for TMB derived from both whole exome sequencing data as well as estimates of TMB derived from targeted gene panels.13 Significant overlap in TMB between responders and non-responders and low TMB associated with high response rates in cancers that express very immunogenic antigens (viral and intron derived antigens) precludes TMB alone from being a highly sensitive predictor of checkpoint blockade therapeutic efficacy.
Observations from patient groups with low TMB that exhibit anti-PD1 responses can provide some insight into additional factors that may predict therapeutic benefit. For example, renal-cell carcinoma (RCC) exhibits a relatively high objective response rate to anti-PD1 (~25%-40%), while usually exhibiting a low median TMB (~5 coding somatic mutations per Mb).14,15 Notably, top quintile TMB is not predictive of response in renal cell carcinoma.16 One possible explanation for the high anti-PD1 response rate in RCC in the context of relatively low mutation burden may be the quality of the neoantigens resulting from those mutations. In an analysis of the TCGA WES data, Turajlic et al17 found RCC to exhibit the highest proportion (0.12) of tumors with insertion or deletion (indel) mutations. This rate is more than double the median proportion of all other TCGA cancer types. Neoantigens derived from indels are expected to be more immunogenic than those derived from non-synonymous single nucleotide variants (SNVs) by virtue of increased “foreign-ness” from self-peptides. Where an SNV-derived neoantigen will have a single changed amino acid, an indel-derived neoantigen may have several if multiple codons are altered, or potentially many if a frameshift has occurred. An analysis of 3 distinct melanoma cohorts demonstrated a significant correlation of frameshift indel count with response to anti-PD1 therapy.18 Similarly, responders to anti-PD1 treatment of head and neck squamous cell carcinoma (HNSCC) demonstrated an enrichment for frameshift mutations over non-responders.18 Consistent with the concept of immunodominance, it is possible that a single highly immunogenic neoantigen derived from a frameshift mutation may drive the anti-tumor T cell response.19 In this case, a single potent frameshift mutation may be sufficient to drive an anti-PD1 response, regardless of the overall tumor mutational load. The same pattern is observed in Merkle-cell carcinoma (MCC) associated with MCPyV, which demonstrates a similar response rate to anti-PD1 therapy as MCPyV-negative MCC, despite exhibiting a markedly lower median neoepitope burden (<1 mut/Mb vs >10 mut/Mb).20 In the case of MCPyV-positive MCC, a single high quality clonal viral antigen may be sufficient to drive an anti-PD1 response. Therefore, other neoantigen features beyond TMB may be critical determinants of anti-PD1 responses in some tumor types.
While both subclonal passenger mutations and highly clonal oncogenes may contribute to mutational counts, emerging research indicates that responses against clonal oncogenes may be more important drivers of anti-PD1 responses because immunoediting and neoantigen loss is less likely to occur.21,22 For example, oncogene specific T cells have been identified in patients responding to anti-PD1 in low-TMB tumors.23 Other genomic determinants of anti-PD1 responses include HLA type and heterogeneity,24 mutations in key immune and antigen presentation pathways,25 loss of heterozygosity in key genes, and various assessments of neoantigen quality.25
We hypothesized that combining multiple genomic features in a random forest machine learning classifier for the prediction of anti-PD1 response could improve upon the performance of TMB alone in the pan-tumor setting. In this study, we evaluated over 90 proposed genomic and immune features for use in random forest classifiers to improve the prediction of anti-PD1 responders over TMB alone using publicly available Training Dataset of patients treated with anti-PD1 or anti-PDL1 therapy. We then confirmed the utility of this approach using an independent cohort of patients treated with anti-PD1 therapy for which genomic and clinical outcomes data were available. We show that multiple genomic tumor features may enrich for anti-PD(L)1 responses, and demonstrate that the use of multiple concurrent features derived from tumor sequencing data outperforms the use of TMB-alone.
Materials and Methods
Patient selection
Two pan-tumor cohorts of patients treated with anti-PD1 therapy with whole exome sequencing (WES) data were analyzed in this study (see Table 1). The first cohort (Training Dataset) was comprised of data and sequencing from publicly available patients consolidated by Miao et al (2018) (n = 104; anti-PD1/anti-PDL1 therapies only)26. The second cohort (Test Dataset) is a newly described clinically annotated cohort of anti-PD1 treated patients with paired sequencing from the Providence Health System. Both cohorts are obtained across cancer subtypes, with baseline disease and response characteristics are shown in Figure 1. RECIST radiological measurements are publicly available for the Training Dataset, while response categories for the Test Dataset were assigned by the treating oncologist who cared directly for the majority of the patients (author BC). Binary response assessment was performed using RECIST 1.1 criteria27 and locked prior to WES and classification with random forest. Whole exome sequencing was performed under study protocol 2016000083 approved by the Providence Institutional Review Board (IRB). All patients provided informed consent for genomics research.
Table 1.
Patient demographics. Cancer types and anti-PD1 therapeutic response are described for the training and test dataset. Training data “Other” cancers include 1 anal cancer and 1 sarcoma. Test data “Other” cancers include secondary malignancies (n = 4), female genital cancer (n = 2), ovarian cancer (n = 2), endometrial cancer (n = 1), small cell lung cancer (n = 1), Merkel cell carcinoma (n = 1), stomach cancer (n = 1), small intestine cancer (n = 1), metastatic squamous cell carcinoma (n = 1), thyroid cancer (n = 1), hepatocellular carcinoma (n = 1), and malignant thymoma (n = 1).
| Patient characteristics | ||
|---|---|---|
| Training dataset (column %) | Test dataset (column %) | |
| Total | 104 | 69 |
| Cancer type | ||
| NSCLC | 57 (55%) | 12 (17%) |
| Bladder | 27 (26%) | — |
| HNSCC | 12 (12%) | 10 (14%) |
| Melanoma | 6 (6%) | 12 (17%) |
| Other | 2 (2%) | 17 (25%) |
| Breast | — | 7 (10%) |
| Colorectal | — | 4 (6%) |
| Kidney | — | 4 (6%) |
| Prostate | — | 3 (4%) |
| Anti-PD1 therapeutic response | ||
| Responders (CR/PR) | 38 (37%) | 12 (17%) |
| Non-responders (SD/PD) | 66 (63%) | 57 (83%) |
Figure 1.
Project workflow. In this project, the training data was used for both feature selection out of the 92 features initially analyzed, as well as for model training. The test data was used to evaluate model performance.
Next generation sequencing
The wet lab methodology for the generation of the training dataset is described elsewhere.14 For the novel test cohort, briefly, formalin-fixed paraffin embedded (FFPE) tumor tissue slides were reviewed for tumor purity by a licensed molecular pathologist and tumor-rich regions were macrodissected. DNA extraction was performed using AllPrep kits on an automated Qiacube platform (Qiagen). Paired germline DNA was extracted from an EDTA blood collection tube on an automated Qiasymphony system. Library preparation was performed using KAPA HyperPlus library preparation kits (Roche) and whole exome hybrid capture was performed using IDT xGen Research Panel v1 kits. Samples were pooled and sequenced on an Illumina HiSeq 2500 or HiSeq 4000 instrument. Sequencing was performed under CLIA/CAP guidelines in the Providence clinical genomics laboratory.
Analysis of WES data and feature generation
Paired tumor-normal whole exome sequencing (WES) data was analyzed for over 90 features including tumor mutational burdens, clonality, HLA type, neoantigens, mutations known to interact with anti-PD1 response, frameshift mutations and consequent amino acid changes, and tumor sample purity (see Figure S1 for complete list of features). WES .bam files were prepared according to GATK best practices for somatic mutation and small indel calling with Mutect2 (v2.2, GRCh37 genome build).28 Ensembl’s VEP (v96) was used to annotate mutation type and consequence. TMB was calculated according to the Cancer Research TMB Harmonization Project.29 ClinVar (202012) annotation distinguished known pathogenic mutations. HLA typing was performed bioinformatically with Polysolver (v4) on germline data. Pvacseq (pvactools v1.5.9) was used for neoantigen analyses.30,31 DNACopy (v1.68.0) was used for copy number alterations and SciClone (v1.1.0) was used to determine both tumor sample purity and clonal versus subclonal mutations.32 A recent review on determinants of anti-PD1 response24 (see Table 1 in Keenan et al24) was used to set lists of mutations in the features related to known mechanisms of resistance or response (Figure S1B). Specific mutations known to result in gain-of-function and loss-of-function within the response and resistance genes were identified using cBioPortal. We then annotated our datasets with these lists of gain-of-function and loss-of-function mutations, along with nonsense and frameshift mutations identifying inactivating mutations. Only clonal mutations were assessed for these resistance- or response-associated mutations. In most cases, identical preprocessing pipelines were used for the test and training datasets. The following differences were used in the training data pipeline: somatic mutation calling was done with Mutect, small indel detection was done with Strelka, and ABSOLUTE was used to identify clonal versus subclonal mutations and tumor sample purity. Mutect2 covers small indel detection, which are not identified with Mutect. We found ABSOLUTE to be unmaintained to the extent that it was unusable.
Random forest classifier approach
Random forest classifiers were trained using the scikit-learn (v0.22.1) Python machine learning package to predict binary response to anti-PD1 therapy. Responders include both complete and partial responses, while all others (stable and progressive disease) are considered non-responders. As our dataset is imbalanced, the ADASYN (Adaptive Sythetic; imblearn v0.5.0) was used to generate more data for the minority class (responders). Model hyperparameter tuning was performed with GridSearchCV and optimized to ROC AUC scoring. Cross validation was performed with two 50/50 data divisions in the training data. The pipeline used to train and test the 3 models was identical except that the GridSearchCV for the TMB-only model only included a max_features parameter of 1. Model depths of both 1 and 2 were evaluated for all 3 models.
Intra-dataset features were not scaled per machine learning standards for random forest algorithms, which can handle features of differing scales. However, the distribution of values in some of the features differed significantly between the training and test datasets, due a combination of the very different cancer types in each group as well as slight difference in the bioinformatic pipelines. As a result, some continuous features (TMB and “Num_neo_<2000_DAI_0.1,” ie, the TMB surrogate in the “cluster” list) were rescaled within each dataset to the percentage of the total range of that feature within the dataset for better parity across the datasets. In order to evaluate TMB alone, a categorical variable (top quintile (“TMBhigh”) vs other (“TMBlow”)) was created consistent with the currently clinically used TMB >10 mut/Mb of DNA.
Random Forest classifier performance was evaluated using receiver-operator curve area under the curve (ROC-AUC). Clinical utility at a given specificity on the curves was also evaluated, as described below. A subgroup analysis by cancer type was performed to elucidate the effect of disease type presence in the Training Dataset to inform test data prediction.
Feature subset selection
Ninety-two features—genomic, neoantigen, immunologic, and literature review based—were evaluated in the Training Dataset for inclusion in the final classifier algorithms. Individual feature predictive value was evaluated by the AUC for a random forest model developed on each individual feature by itself, optimized from either a 1- or 2-tiered model. Ten different data splits within the Training Dataset were evaluated; the average test data AUC of the 10 models was used for feature selection to carry forward to the validation dataset (see Figure 2). Similarly, a 2-feature random forest model was generated in the Training Dataset for each feature in combination with totalTMB (TMB + feature) to further support feature selection (Figure S2).
Figure 2.
Predictive performance of individual features, and their relationship with TMB. Performance evaluated by receiver-operator curve area under the curve (ROC-AUC) is plotted against Pearson correlation with TMB. Feature marks are colored by biological association of feature clusters (see Figure S3). Feature names are included only for features utilized in the “cluster” and “human-guided” algorithms. Quadrant of interest indicates features that provide predictive information for anti-PD-1 response that are not highly correlated with TMB.
As many features evaluated are either highly correlated to TMB (Figure 2) or to other features, hierarchical clustering was used to group features (Figure S3). Clusters were defined by Pearson correlation; a dendrogram was then generated and the tree cut at a level that generated both biologically-driven clusters and a reasonable number of clusters for feature selection (around a dozen). Features were carried forward according to the following rules: (1) Features were only included if their individual AUC exceeded 0.575, (2) Within each of the 11 feature clusters, the feature with the highest AUC was selected, (3) If 2 features in the same cluster had the same AUC, then the 1 with the higher AUC from the TMB + feature model was selected, (4) If a feature other than the 1 already selected from the cluster based on individual AUC had the highest TMB + feature AUC, and its single feature AUC feature exceeded the threshold of 0.575 indicated above, that second feature was also included from the cluster. In this way, features that are less predictive on their own but additive to TMB are conserved; and (5) No feature was selected from a cluster if the highest individual feature AUC was not greater or equal to the threshold of 0.575.
The “human-guided” feature list (Figure 3) was selected by authors EMJ and MY, through consultation with field expert and evaluation of the AUC data upon which “cluster” features were selected. Cancer type was not considered as a potential feature in either dataset as (1) the goal of this study was to make a pan-tumor algorithm and (2) the cancer types in the training and test datasets were divergent. Subgroup analysis per cancer type was performed and analyzed to evaluate algorithm sensitivity to cancer type distribution in the Training Dataset (Figure 4).
Figure 3.
Performance of ML algorithms and TMB alone in predicting anti-PD1 responses. Receiver-operator curves of random forest models for 2 feature sets: (A) “Human-Guided,” and (B) “Cluster”—against TMB. The evaluation of TMB is performed by designating TMB values in the upper quintile in each dataset as “HighTMB.” The table lists features included in each of the 2 algorithms, as well as overlapping features included in both sets.
Figure 4.
Subgroup analysis of ML algorithm performance by cancer type for head and neck cancers (A), melanoma (B), NSCLC (C), and other cancers (D). The baseline “train” algorithm in each plot was trained on all 104 available training dataset patients. The table displays results for cancer types for which there are no responders in the test dataset.
Algorithm feature importance
Individual feature importance relative to the performance of the whole model was evaluated in both the “cluster” and “human-guided” models, in both the training and test datasets, using permutation importance. One at a time, a given feature’s values were randomly permutated among the patients. The resulting reduction (or in some cases improvement) in accuracy of the resulting model compared with a baseline Random Forest model is used to indicate importance of that permutated feature to the intact model’s performance. Variation across the 10 iterations is reported as standard deviation of importances. It should be noted that this method can falsely suggest important features as unimportant if that feature is highly correlated with other feature(s) in the model.
Statistical analysis
Additionally, significant differences in features between responders and non-responders were evaluated using Fisher’s exact test for categorical variables and Mann–Whitney U test for continuous variables. Features with a range of ⩽5 and integer values only (such as mutation counts in small gene sets) were evaluated as categorical variables as 0 versus ⩾1 mutations.
Data availability
All data, code, and materials used in the analysis is available upon request. Publicly available data can be found at dbGaP with the following accession numbers: phs000694.v3.p2, phs000980.v1.p1, phs001041.v1.p1, phs000452.v2.p1, and phs001075.v1.p1. Test dataset may be made available through material transfer agreement (MTA) with Providence Health.
Results
Patient selection
The Training Dataset was comprised of 104 patients with the following cancer types (Figure 1): non-small cell lung cancer (NSCLC; n = 57), bladder cancer (n = 27), head and neck squamous cell carcinoma (HNSCC; n = 12), melanoma (n = 6), anal cancer (n = 1), and sarcoma (n = 1). Of these patients, 37% (or n = 38) were responders to anti-PD1/anti-PDL1 therapy according to RECIST criteria (see Figure 1). The Test Dataset is comprised of 69 patients with the following cancer types: NSCLC (n = 14), melanoma (n = 13), HNSCC (n = 10), breast cancer (n = 6), colon or colorectal cancer (n = 4), kidney cancer (n = 3), prostate cancer (n = 3), ovarian cancer (n = 2). Patients included in the “other” disease category in Figure 1 had the following cancer types: include secondary malignancies (n = 4), female genital cancer (n = 2), ovarian cancer (n = 2), endometrial cancer (n = 1), small cell lung cancer (n = 1), Merkel cell carcinoma (n = 1), stomach cancer (n = 1), small intestine cancer (n = 1), metastatic squamous cell carcinoma (n = 1), thyroid cancer (n = 1), hepatocellular carcinoma (n = 1), and malignant thymoma (n = 1). The Test Dataset includes 12 responders to anti-PD1 (17% of patients).
“Cluster” feature subset selection
WES data was processed into a set of 92 total features. One group of 9 features was selected to reflect independent sources of variation in the data through clustering analysis (“cluster” list). The other was a set of 10 features selected based upon the anti-PD1 response literature and clinical practice (“human-guided” list). Individual feature performance and biological cluster are shown in Figure 2. Feature lists, and shared features, are shown in Figure 3. All features, classifier results, and clusters are available in data file S1. For the “cluster” list, 7 of the 11 clusters (Figure S3) contributed a feature, and 1 cluster (#2) contributed 2 features as the feature with the highest individual AUC (Num_neo_<2000_DAI_0.1) was not the same as the highest TMB + feature AUC (Total_oncogene_neoantigens_<2000). Seven features were not found to cluster with any others largely because each was a categorical variable and had insufficient distributions in the Training Dataset samples from which to derive any significant classification.
“Human-guided” feature subset selection
The “human-guided” list was selected based on consensus view of biological mechanism as expressed in the medical literature and expert experience. Both TMB high and clonal TMB (cTMB) were selected given the abundance of data that demonstrates a correlation with anti-PD1 response. Clonal TMB has been associated with improved correlation with hypotheses including (1) that clonal mutations represent epitopes targetable in all tumor cells or (2) that tumor clonality is in some way representative of patient treatment history.33,34
While the data-driven selection did not reveal any patient HLA related features as highly predictive, many studies suggest a link between HLA diversity or certain supertypes with anti-PD1 response given the consequent effect on tumor epitope diversity.34,35 Among these features, HLA-B homozygosity has been both well substantiated in other studies as well as has some predictive value in our Training Dataset. Sample purity was included as it has been implicated as interacting with measured TMB given variant allele frequency (VAF) cutoffs for somatic mutation filtering.36 Additionally, VAFs are implicated in clonality analysis and therefore could interact with features highly dependent on clonal counts (such as consideration of only clonal oncogene ClinVar mutants).
Both features counting known mutations that modify anti-PD1 response—one of mechanisms of response, the other of resistance—were included in the “human-guided” list, while only the mutations of response feature was included in the “cluster” list. Additionally, a count of pathogenic mutations (as annotated by ClinVar) found in genes that drive anti-PD1 response literature review (Lit_Rev_genes_ClinVar_mut) was included. This feature is distinct from the “response_litrev_mut” feature as the latter includes counts of any nonsense or frameshift mutations in cases in which gene loss of function was correlated with anti-PD1 response. As shown in the correlograms of features lists (Figure S4A), these 2 features are not highly correlated.
In choosing a feature to represent frameshift mutations and consequent potential neoantigens, we opted for the simple count of clonal frameshift mutations. In contrast, the data-driven approach chose the count of predicted amino acid changes due to frameshifts for the “cluster” list. We chose the simple count even though the range of consequent amino acid changes per individual frameshift mutation is wide since the amino acid changes in the tumor average out to a value highly correlated with the number of frameshift mutations (Figure S3). Additionally, many of the counted amino acid changes may not be transcribed or persist in the cytosol as nonsense mediated decay is likely to target some of these highly altered proteins.37 Therefore, we believe the count of clonal frameshift mutations is likely to be most robust across datasets.
Finally, pathological mutations in oncogenes (as annotated by ClinVar) was included for 2 reason: (1) pathological oncogene mutations may indicate the aggressiveness of a tumor and/or (2) such mutations may represent ideal neoantigen targets for the immune system as neoantigens derived from tumor-driving oncogenes will not easily undergo immunoediting. The distribution of this feature in the 2 datasets indicates that the responder groups are enriched for 1 or more ClinVar pathogenic mutations on the list of oncogenes (Data file S1 for gene list). The list of oncogenes included is quite broad as this algorithm is developed for pan-tumor use.
Random forest classifiers
Three distinct Random forest algorithms were trained; one on each of the 2 features lists—cluster and human-guided—and one on TMB alone. TMB as a single feature is evaluated in the context of a random forest classifier as multi-tiered models with multiple cut-off points may perform better than logistic regression, which will only allow for 1 cut-off value. All algorithms were trained on the publicly available dataset of 104 anti-PD1/anti-PDL1 recipients analyzed in Miao et al14 which drew patients from across several studies. All algorithms were tested on the new dataset of 69 anti-PD1 recipient patients in the TRISEQ Providence Health System cohort.
Shown in Figure 3, both the “cluster” and “human-guided” models outperform TMB alone in both the training and test datasets. Several versions of TMB were evaluated in both the TMB-only model, as well as the feature list models, to ensure that the difference in TMB value distributions in the 2 datasets did not put the TMB-only model at a significant disadvantage. Models were trained on only the categorical feature “High TMB” (positive for patients with TMB in the top quintile of values). A hard TMB cutoff of 10 mut/Mb in the test dataset resulted in high sensitivity (91.7%) at poor specificity (21.1%). Therefore, algorithmic selection of threshold or use of a categorical variable to describe TMB status was preferred. The poor performance of this established cutoff is likely due to both bioinformatic pipeline differences and cancer types in the test dataset. Similar outcomes for each model were achieved even with variations in the calculation of TMB, but all of these calculated features resulted in improved performance of the TMB-only model in the Test Dataset. Ultimately, for clinical use, such an approach should be able to handle continuous TMB values as the algorithm would be trained and utilized on data from the same sample preparation, sequencing, and bioinformatics pipeline.
A subgroup analysis by cancer type (Figure 4) reveals that performance in the Test Dataset is likely not driven by representation of cancer type in the Training Dataset. NSCLC, which makes up the majority of the training data, appears to underperform in the test data compared with all other cancer types. A highly varied set of “Other” cancers in the Test Dataset, none of which are represented in the training data, performs the best in test data. See “Patient Selection” results for specific disease types. Of the several more highly represented cancer types in the Test Dataset that aren’t in the training data—breast, prostate, colorectal, and kidney—none of the patients are responders. The table in Figure 4 displays positive identification of non-responders in these disease types.
Clinical utility
We next evaluated clinical utility of the “cluster” and “human-guided” algorithms by determining how many additional anti-PD1 responders are identified with algorithms over TMB alone. Probability thresholds for each Random Forest model were identified that achieved 80% specificity to match the specificity achieved by the “High TMB” model. Model performance was also evaluated on a per-patient basis to determine which algorithms correctly identified which patients as both responders and non-responders. A subgroup analysis by cancer type was performed to identify differences in patient identification between cancer types.
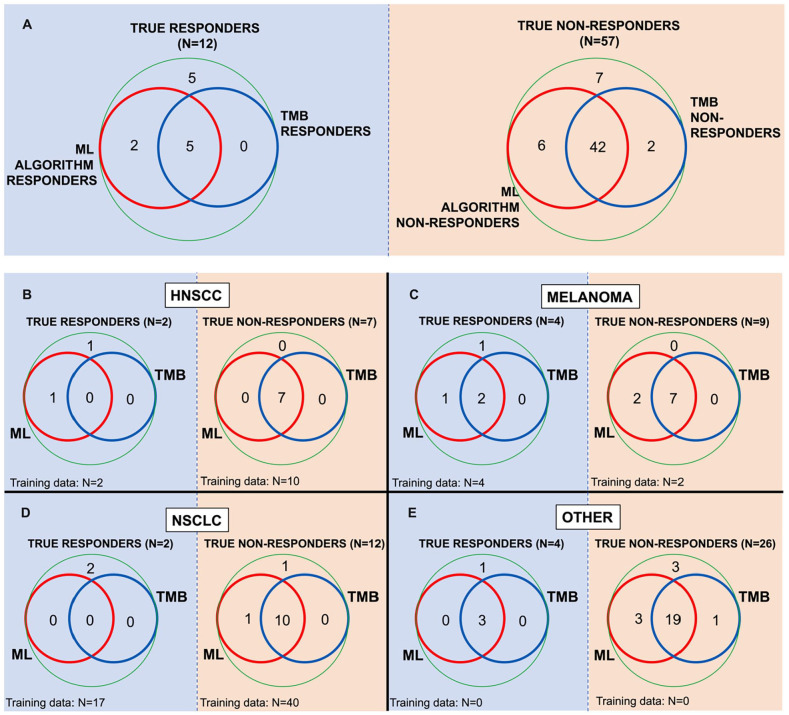
Both the “cluster” and “human-guided” algorithms were evaluated for their ability to detect low TMB patients who respond to anti-PD1 therapy (Figure 5A). The sensitivity for all 3 algorithms were evaluated at the random forest probability threshold that achieves 80% specificity. In the Test Dataset, the ML algorithms detected 2 additional responders over TMB-only algorithm. These 2 additional responders represent 29% of the responders in this dataset that would not be detected by TMB alone. Notably, no responders identified by the TMB-only algorithm were missed by the algorithm. There remains room for improvement in patient stratification as, at this specificity, all algorithms still missed 42% of all the responders (5 patients). There was no clear pattern among the other features between the low TMB responders who were correctly identified as compared with low TMB responders who were no identified. In the training data (data not shown), the algorithms detected 3 additional responders over TMB, although they missed 1 patient identified by TMB alone. These additional 3 responders account for 23% of the responders not detected by TMB alone.
Figure 5.
The clinical utility of the biomarker algorithms. (A) Algorithm ability to identify true responders and true non-responders compared with TBM alone. All comparisons are performed at the same specificity of 80%. (B-E) Subgroup analysis by cancer type for HNSCC (B), melanoma (C), NSCLC (D), and other cancers (E).
Abbreviations: HNSCC, head and neck squamous cell carcinoma; NSCLC, non-small cell lung cancer.
Subgroup analysis (Figure 5B–E) indicates that the 2 additional responders identified by ML algorithm are 1 HNSCC patient and 1 melanoma patient. The “other” disease group, both mixed in cancer type and not represented in the training data, represents the biggest improvement in identification of non-responders. Additional non-responders are identified in the melanoma and NSCLC patient groups.
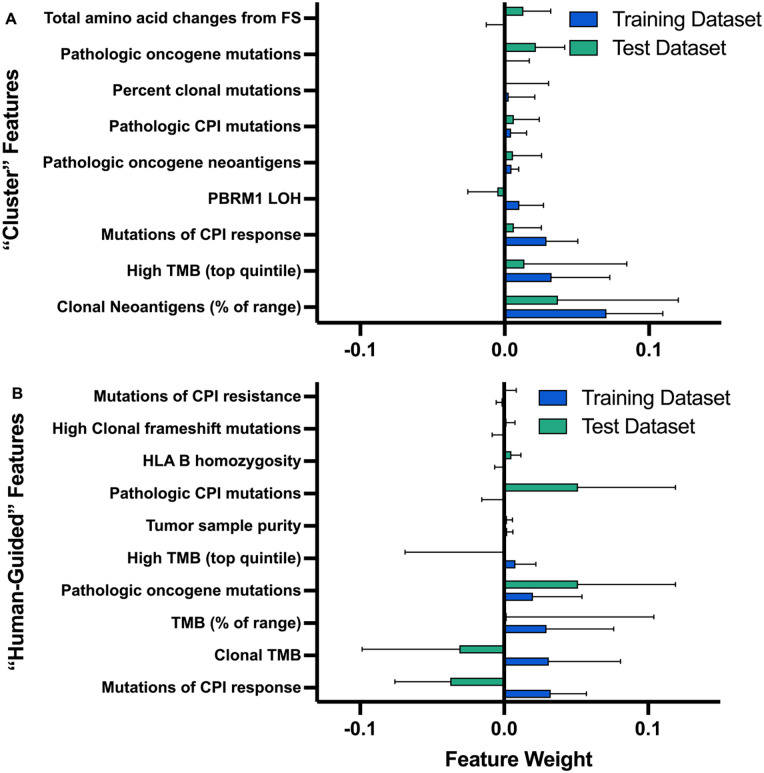
Feature importance
Feature importance was evaluated using permutation importance to assess the degree to which “loss” of a feature is detrimental to the performance of the classifier (Figure 6). Features selected by the “cluster” method are notably more predictive and have improved preserved performance across the train and test datasets when compared with the “human-guided” features. While the performance of “cluster” features is expected in the training data set, given the data-driven selection process, the maintained performance in the test data suggests that this approach identified translatable features.
Figure 6.
Algorithm feature importance. The contribution of individual features in the (A) “Cluster” and (B) “Human-Guided” algorithms is quantified by permutation importance, or the performance loss of a trained algorithm if the feature under evaluation is randomly permutated. Feature permutation importances indicate the mean reduction in model performance over 10 iterations; error bars represent standard deviation.
Widely changing performance of the features in the “human-guided” dataset may be attributable to higher correlation between some of the selected features when compared with the “cluster” features. If a feature is highly correlated with another feature, even if important to the model, permutating 1 feature only will erroneously not appear to significantly detract from the model’s performance.
The following 4 features in the “human-guided” list appeared to not significantly influence the classifier: (1) literature review mutations of CPI resistance, (2) categorical feature of clonal frameshift mutations, (3) HLA B homozygosity, or (4) tumor sample purity. All features were represented in both datasets (Figure S7), so lack of feature distribution doesn’t seem to drive insignificance. In an effort to evaluate how significant the role of overfitting due to inclusion of many features, we trained the “human-guided” and “cluster” algorithms each on only the top 4 features according to the training data permutation importance. While the training data performance was maintained, the test data performance dropped (data not shown), which suggests the eliminated features did provide some useful data and that the full feature lists did not include too many features to cause significant overfitting.
Features were additionally evaluated using standard statistics—Fisher’s exact test and Mann–Whitney U test—to detect differences in feature distributions between the responders and non-responders (Figure S6). Pathological oncogene mutations (aka the “oncogenes_ClinVar_mut” feature) demonstrated significant Test Dataset performance and statistical significance (test data P = .0278; Fisher’s exact). One or more pathologic oncogene mutations was associated with response to anti-PD1 (Figure 7). Notably, neither total TMB nor cTMB was significant for response in the Test Dataset (Figure S6).
Figure 7.

Oncogene pathogenic mutations. Pathogenic mutations (per ClinVar annotation) in an oncogene were statistically significant for differences between responders and non-responders in both the training and test datasets (Training: P = .0388, test: P = .0278, Fisher’s exact test). Total oncogene neoantigens (not restricted to ClinVar mutated genes) also achieved statistical significance in the test dataset (P = .0451, Mann–Whitney U test).
Discussion
Tumor mutational burden (TMB), a surrogate for tumor neoepitope burden, is clinically used as a pan-tumor biomarker to identify patients who may benefit from anti-PD1 immunotherapy. However, TMB is a highly imperfect biomarker since some patients with low-TMB tumors but that have other immunogenic tumor features may miss the opportunity to obtain clinical benefit from anti-PD1 therapy in clinical practice. Likewise, the majority of patients currently eligible to receive anti-PD1 therapy based on TMB in clinical practice do not all derive clinical benefit from these therapies and may be subjected to unnecessary toxicity. We have developed an additional and complementary approach for informing selection of patients who are most likely to benefit from anti-PD1 therapy. We chose to define response and therefore benefit as radiographic response using RECIST 1.1 criteria, which is a stringent definition of benefit after anti-PD1 administration. In the medical literature, other definitions of response and benefit have been used such as stability of disease, progression-free survival, overall survival, time to next treatment, or patient quality of life assessments. Analysis of these other benefit measures was not performed given the variety of tumor types in the training and test data sets and the small size of the data sets analyzed.
Both the “human-guided” and “cluster” ML algorithms demonstrated a modest improvement in ROC-AUC over a TMB-only algorithm. The “human-guided” feature ML algorithms identified 40% more responders than TMB-only at a clinically relevant specificity of 80%. The “human-guided” algorithm performs best for patient selection at 80% specificity, while the “cluster” algorithm has a better overall ROC-AUC. Together these data demonstrate that multiple components of neoepitope quality may influence antitumor immunity in the setting of anti-PD1 therapy. At the same time, the only modest improvement of these algorithms over TMB alone highlights both the high predictive value of TMB as a single feature, as well as the limitations of many other features derived from WES data and the unpredictable nature of responses to anti-PD1 therapy. All of the features evaluated in our model are derived exclusively from WES paired tumor-normal data, and do not incorporate host immune fitness, physiological factors, and the immune contexture of the tumor immune microenvironment. We and others have previously shown that biomarkers indicative of an inflamed tumor immune microenvironment are relatively uncorrelated with tumor genomic biomarkers related to neoepitope burden. Our results here suggest that the integration of such data, which can be derived from RNA or protein-based analysis of the tumor microenvironment, may be necessary to meaningfully improve the predictive value of TMB and other biomarkers derived from tumor WES. In the course of evaluating the potential for a combined approach of sequencing and tumor microenvironment data, we plan to study the importance of WES compared with targeted gene panel testing as well.
While machine learning algorithm performance is always expected to decrease when moving from training to test data, the drop in the present study was significant. We attribute this performance decrease to 2 major reasons: (1) significant differences in the cancer types in the 2 datasets, and (2) inter-institution sample preparation, sequencing, and bioinformatics pipelines. While the goal of this study was to create a pan-tumor biomarker of response, ideally cancer types would have been similarly represented in the 2 datasets. However, subgroup analyses indicate that a cancer type not being represented in the training data does not detract from the ability to make predictions within that disease. This finding suggests that achieving a pan-tumor algorithmic biomarker for anti-PD1 response is feasible. A hard cutoff of TMB >10 mut/Mb performed as expected in the training dataset (specificity 88%, sensitivity 47%), it did not maintain performance in the test dataset (specificity 21%, sensitivity 92%). To account for differences in bioinformatics that contributed to significant differences in the distribution of TMB values between the 2 datasets, several methods of TMB analysis were explored including a categorical high/low TMB feature, percentile of TMB range within a given dataset, and TMB decile. As the number of neoantigens effectively stood in for TMB in the “cluster” dataset, that feature was modified in accordance with the TMB variation. Harmonizing data in this way greatly improved the performance of the TMB-only algorithm and improved the feature list algorithms, although to a lesser extent. Normalizing and scaling TMB values, or all features, was considered undesirable given the difference in cancer types between the 2 datasets.
Limitations in this study include the limited size of both datasets, and particularly the low number of responders to anti-PD1 therapy in the Test Dataset. Differences in the bioinformatic pipelines for the Training and Test datasets necessitated by unmaintained publicly available code present an additional limitation. Future testing of our ML algorithm in other independent cohorts will provide additional information about the sensitivity and specificity of our algorithms.
Conclusion
This study describes the use of machine learning techniques to identify potential responders to anti-PD1 immunotherapy using whole exome genomic data. In the presented pan-tumor cohort, this approach of combining various genomics-based features demonstrated an incremental improvement over TMB alone. The development of ML algorithms incorporating novel predictive biomarkers of anti-PD1 response, including those derived from RNA or protein assessment of the tumor immune microenvironment, may be needed to further improve model performance.
Supplemental Material
Supplemental material, sj-docx-1-cix-10.1177_11769351221136081 for A Random Forest Genomic Classifier for Tumor Agnostic Prediction of Response to Anti-PD1 Immunotherapy by Emma Bigelow, Suchi Saria, Brian Piening, Brendan Curti, Alexa Dowdell, Roshanthi Weerasinghe, Carlo Bifulco, Walter Urba, Noam Finkelstein, Elana J Fertig, Alex Baras, Neeha Zaidi, Elizabeth Jaffee and Mark Yarchoan in Cancer Informatics
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-2-cix-10.1177_11769351221136081 for A Random Forest Genomic Classifier for Tumor Agnostic Prediction of Response to Anti-PD1 Immunotherapy by Emma Bigelow, Suchi Saria, Brian Piening, Brendan Curti, Alexa Dowdell, Roshanthi Weerasinghe, Carlo Bifulco, Walter Urba, Noam Finkelstein, Elana J Fertig, Alex Baras, Neeha Zaidi, Elizabeth Jaffee and Mark Yarchoan in Cancer Informatics
Acknowledgments
The authors thank the patients who participated and the clinical investigators and staff who assisted in the acquisition of tumor genomic data used in this study.
Footnotes
Author Contributions: Conceptualization: MY, EMJ, SS. Methodology: SS, BP, NF, EF, AB. Investigation: EB, BP, BC, AD. Visualization: EB. Funding acquisition: EMJ, MY, AB, WU. Project administration: EMJ, MY
Supervision: MY, EMJ. Writing – original draft: EB, MY. Writing – review & editing: EMJ, SS, BP, BC, CB, EF, NZ.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Stand Up to Cancer (EMJ, MY), Incyte Pharmaceuticals Young Investigator Award (MY), the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Gastrointestinal Cancers P50 CA062924 (Project 2 – EMJ), NIH Center Core Grant P30 CA006973 (EMJ), Providence Foundation of Oregon (WU).
Data and Materials Availability: All data, code, and materials used in the analysis is available upon request. WES raw .bam files for training dataset patients can be found at the database of Genotypes and Phenotypes (dbGaP) with the following accession numbers: phs000694.v3.p2, phs000980.v1.p1, phs001041.v1.p1, phs000452.v2.p1, and phs001075.v1.p1. Test dataset WES .bam files may be made available through material transfer agreement (MTA) with Providence Health.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le DT, Durham JN, Smith KN, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti–programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reck M, Rodríguez-Abreu D, Robinson AG, et al.; KEYNOTE-024 Investigators. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N Engl J Med. 2016;375:1823-1833. [DOI] [PubMed] [Google Scholar]
- 9. Chen PL, Roh W, Reuben A, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yarchoan M, Albacker LA, Hopkins AC, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019;4:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Akbari H, Taghizadeh-Hesary F, Bahadori M. Mitochondria determine response to anti-programmed cell death protein-1 (anti-PD-1) immunotherapy: an evidence-based hypothesis. Mitochondrion. 2022;62:151-158. [DOI] [PubMed] [Google Scholar]
- 12. Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353-1365. [DOI] [PubMed] [Google Scholar]
- 13. Kowanetz M, Zou W, Shames DS, et al. Tumor mutation load assessed by FoundationOne (FM1) is associated with improved efficacy of atezolizumab (atezo) in patients with advanced NSCLC. Ann Oncol. 2016;27:vi23. [Google Scholar]
- 14. Motzer RJ, Tannir NM, McDermott DF, et al.; CheckMate 214 Investigators. Nivolumab plus Ipilimumab versus Sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378:1277-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;806:801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18:1009-1021. [DOI] [PubMed] [Google Scholar]
- 18. Hanna GJ, Lizotte P, Cavanaugh M, et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69-74. [DOI] [PubMed] [Google Scholar]
- 20. Nghiem PT, Bhatia S, Lipson EJ, et al. PD-1 blockade with Pembrolizumab in advanced merkel-cell carcinoma. N Engl J Med. 2016;374:2542-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumor-specific antigens underlies cancer immunoediting. Physiol Behav. 2017;176:139-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith KN, Llosa NJ, Cottrell TR, et al. Persistent mutant oncogene specific T cells in two patients benefitting from anti-PD-1. J Immunother Cancer. 2019;7:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med. 2019;25:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luksza M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551:517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miao D, Margolis CA, Vokes NI, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-247. [DOI] [PubMed] [Google Scholar]
- 28. Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. Calling somatic SNVs and indels with Mutect2. Preprint. Posted online December 2, 2019. BioRxiv 861054. doi: 10.1101/861054 [DOI] [Google Scholar]
- 29. Merino DM, McShane LM, Fabrizio D, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the friends of cancer research TMB harmonization project. J Immunother Cancer. 2020;8:e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hundal J, Kiwala S, McMichael J, et al. PVACtools: a computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res. 2020;8:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hundal J, Carreno BM, Petti AA, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller CA, White BS, Dees ND, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10:e1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abed A, Calapre L, Lo J, et al. Prognostic value of HLA-I homozygosity in patients with non-small cell lung cancer treated with single agent immunotherapy. J Immunother Cancer. 2020;8:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anagnostou V, Niknafs N, Marrone K, et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat Cancer. 2020;1:99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindeboom RGH, Vermeulen M, Lehner B, Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet. 2019;51:1645-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-cix-10.1177_11769351221136081 for A Random Forest Genomic Classifier for Tumor Agnostic Prediction of Response to Anti-PD1 Immunotherapy by Emma Bigelow, Suchi Saria, Brian Piening, Brendan Curti, Alexa Dowdell, Roshanthi Weerasinghe, Carlo Bifulco, Walter Urba, Noam Finkelstein, Elana J Fertig, Alex Baras, Neeha Zaidi, Elizabeth Jaffee and Mark Yarchoan in Cancer Informatics
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-2-cix-10.1177_11769351221136081 for A Random Forest Genomic Classifier for Tumor Agnostic Prediction of Response to Anti-PD1 Immunotherapy by Emma Bigelow, Suchi Saria, Brian Piening, Brendan Curti, Alexa Dowdell, Roshanthi Weerasinghe, Carlo Bifulco, Walter Urba, Noam Finkelstein, Elana J Fertig, Alex Baras, Neeha Zaidi, Elizabeth Jaffee and Mark Yarchoan in Cancer Informatics
Data Availability Statement
All data, code, and materials used in the analysis is available upon request. Publicly available data can be found at dbGaP with the following accession numbers: phs000694.v3.p2, phs000980.v1.p1, phs001041.v1.p1, phs000452.v2.p1, and phs001075.v1.p1. Test dataset may be made available through material transfer agreement (MTA) with Providence Health.