Abstract
The afa gene clusters, which encode proteins involved in adhesion to epithelial cells, from Escherichia coli strains associated with urinary and intestinal infections in humans have been characterized. Pathogenic isolates of bovine and porcine origin that possess afa-related sequences have recently been described. We report in this work the cloning and characterization of the afa-7 and afa-8 gene clusters from bovine isolates. Hybridization and sequencing experiments revealed that despite similarity in genetic organization, the afa-7 and afa-8 genes, and the well-characterized afa-3 operon expressed by human-pathogenic isolates, correspond to three different members of the afa family of gene clusters. However, like the afa-3 gene cluster, both the afa-7 and afa-8 gene clusters were found to encode an afimbrial adhesin (AfaE) and an invasin (AfaD). The AfaD peptides encoded by the three gene clusters were only 45% identical, but functional complementation experiments indicated that they belong to the same family of invasins. Hemagglutination and adhesion assays demonstrated that the AfaE-VII and AfaE-VIII adhesins bind to different receptors and that these receptors are not the human decay-accelerating factor recognized to be the receptor of all previously described AfaE adhesins. The AfaE-VIII adhesin is very similar to the M agglutinin of human-uropathogenic strains. We used PCR assays to screen 25 bovine strains for afaD and afaE genes of either the afa-7 or afa-8 gene cluster. The afa-8 gene cluster was highly prevalent in bovine isolates previously reported to carry afa-related sequences (23 of 24 strains), particularly in strains producing cytotoxic necrotizing factors (16 of 16 strains). The location of the afa-8 gene cluster on the plasmids or chromosome of these isolates suggests that it could be carried by a mobile element, facilitating its dissemination among bovine-pathogenic E. coli strains.
Escherichia coli is a major cause of bacterial diarrhea and urinary tract infection (UTI) in humans. It is also a major pathogen in agriculture, causing enteric and fatal septicemic colibacillosis in neonatal and young animals. Pathogenic E. coli strains have several virulence properties. One such property, adhesin production, plays an essential role in the colonization of the mucosal epithelium (14). The adhesins of pathogenic E. coli causing infections in humans are specific for a particular disease. The only known exception is that of afimbrial adhesins, which are encoded by the afa gene clusters and are produced by both uropathogenic and diarrhea-associated E. coli strains (14, 16, 17, 34). The afa family of gene clusters also includes the daa and dra operons, which encode the fimbrial F1845 and Dr adhesins, respectively (4, 47). Epidemiological studies have shown that afa-positive strains are enteric pathogens in children (17, 18, 35) and that the same afa-positive isolate may cause both diarrhea and cystitis in an individual child (16). Several studies have suggested that afa-positive strains play an important role in UTI pathogenesis. Such strains are common in E. coli isolates from pregnant woman (46) and from children (2, 3) with UTI and are associated with recurring UTI (10). An animal experimental chronic pyelonephritis model has also provided evidence that the expression of the dra gene cluster is important in persistent colonization of the urinary tract (19).
The afa gene clusters control the formation of an afimbrial sheath composed of the AfaE and AfaD proteins, an adhesin and an invasin, respectively (12, 26). The adhesin-encoding afaE gene is highly heterogeneous, producing antigenically different adhesins (31). The AfaE-I and AfaE-III adhesin subtypes mediate the mannose-resistant hemagglutination (MRHA) of human erythrocytes and specific attachment to epithelial cells via recognition of the decay-accelerating factor (DAF) molecule as a receptor (45).
afa-related sequences have been detected in isolates from diseased piglets and calves (21, 37). Hybridization and PCR analyses have suggested that the afa operons of animal-pathogenic E. coli are diverse and structurally different from the operons of human E. coli isolates (37, 38). In this study, we characterized the genes carried by the animal-pathogenic strains. We report here the cloning of two afa operons, designated afa-7 and afa-8, from bovine isolates and the analysis of similarity between these related gene clusters and the afa family of gene clusters from human-pathogenic E. coli. The binding properties of the AfaE-VII and AfaE-VIII adhesins were different from those of the AfaE adhesins produced by human-pathogenic strains. The afa-8 gene cluster appears to be widespread in bovine-pathogenic E. coli strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, tissue culture cells, and culture conditions.
Nineteen E. coli strains were isolated from intestinal or extraintestinal sites in calves with signs and/or lesions typical of septicemia or enteritis in European countries (Belgium, France, and Spain) and Canada. All 19 strains have been characterized (37). All carried sequences that hybridized with the afa family of gene clusters in colony hybridization experiments but test negative for afa gene clusters by PCR; the afa genes were located on the chromosome in 11 strains and on Vir plasmids in 8 strains (37). Four of the 19 strains produced the CNF1 (cytotoxic necrotizing factor 1) toxin, and 12 produced the CNF2 toxin. Sequences coding for P and F17 adhesins have been detected in six and four strains, respectively (36a, 37). Six F165-positive E. coli isolates carrying afa-related sequences were kindly provided by J. M. Fairbrother and J. Harel (Faculté de Medecine Vétérinaire, Université de Montréal, Saint-Hyacinthe, Québec, Canada). They were originally isolated from calves (five strains) or piglets (one strain) with diarrhea. Strain HB101(pSSS1) (4) was kindly provided by S. Moseley (University of Washington, Seattle). E. coli J96 (28) and A30 (31) were used as positive controls for the presence of pap (coding for P adhesins) and afa gene clusters, respectively. E. coli HB101 (6) and MC1061 (39) were used as recipients for the recombinant plasmids listed in Table 1.
TABLE 1.
Recombinant plasmids used in this study
| Plasmid | Antibiotic resistance markera | Vector | Insert
|
MRHA of human erythrocytes | Reference | ||
|---|---|---|---|---|---|---|---|
| Size (kb) | Origin | afa sequence | |||||
| pILL1101 | Sp | pILL570 | 11.6 | Sau3A partial digest of E. coli A30b DNA | afa-3 | + | 34 |
| pILL1189 | Cm | pACY184 | 10.5 | BamHI fragment of pILL1101 with a mutated afaD gene | afa-3 | + | 26 |
| pILL1191 | Sp | pILL570 | 7.7 | Sau3A partial digest of 262 KH 89 DNA | afa-7 | + | This study |
| pILL1194 | Cb | pUC18 | 5.5 | HindIII-EcoRI fragment of pILL1191 | afa-7 | − | This study |
| pILL1231 | Sp | pILL570 | 8.5 | pILL1191 with a mutated afaE gene by insertion of the kanamycin cassette | afa-7 | − | This study |
| pILL1211 | Cb | pHC79 | 45 | Sau3A partial digest of 239 KH 89 DNA | afa-8 | + | This study |
| pILL1222 | Cb | pUC18 | 5 | EcoRI fragment of pILL1211 | afa-8 | − | This study |
| pILL1226 | Cb | pBR322 | 9.5 | Sau3A partial digest of pILL1211 | afa-8 | + | This study |
| pILL1235 | Cb | pBR322 | 5.5 | pILL1226 deleted of EcoRI-SphI and SphI-SphI fragments | afa-8 | − | This study |
| pILL1244 | Cb | pBR322 | 6.5 | pILL1226 deleted of SphI fragments | afa-8 | + | This study |
Abbreviation: Cb, carbenicillin resistance; Sp, spectinomycin resistance; Cm, chloramphenicol resistance.
Isolated from a urine specimen from a patient with cystitis (34).
Vectors pBR322 (5), pUC18 (59), pACY184 (7), and pILL570 (30) and the cosmid vector pHC79 (8) were used in cloning experiments. The nonpolar kanamycin cassette was extracted from plasmid pUC18K. It contains the aphA-3 gene encoding resistance to kanamycin preceded by translation start codons in all three open reading frames (ORFs) and followed by a consensus ribosome-binding site (RBS). There is no promoter and no transcriptional terminator in this cassette, which is used to generate nonpolar mutations (40).
E. coli strains were grown in Luria broth without glucose (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter [pH 7.0]) or on Luria agar plates (1.5% agar) at 37°C. Antibiotic concentrations for the selection of transformants were as follows: carbenicillin, 100 mg/liter; tetracycline: 10 mg/liter; spectinomycin, 100 mg/liter; kanamycin, 20 mg/liter; and chloramphenicol, 20 mg/liter.
Human HeLa (ECACC 84121901) and Caco-2 cells were maintained as previously described (26, 51). Madin-Darby bovine kidney (MDBK) cells were maintained in Dulbecco modified Eagle medium–Ham’s F-12 medium (BIO MEDIA, Boussens, France) containing 1% glutamine and 10% fetal calf serum (GIBCO Laboratories, Eragny, France), and Madin-Darby canine kidney (MDCK) cells were maintained in minimal essential medium containing Earle’s salts and l-glutamine (GIBCO) and supplemented with 10% fetal calf serum. UROtsa cells, derived from human urothelium (49), were maintained in RPMI 1640 (GIBCO) containing 1% glutamine and 10% fetal calf serum. They were grown at 37°C in a 5% CO2–95% air atmosphere. Wild-type Chinese hamster ovary (CHO) cells and CHO cells stably transfected with the cDNA for human DAF (44) were cultured in Ham’s F-12 medium (GIBCO) containing 10% fetal calf serum. They were grown to confluence at 37°C in 10% CO2–90% air.
Hemagglutination and adhesion assays.
Hemagglutination of washed human or animal (cattle, cat, dog, horse, piglet, and sheep) erythrocytes, in phosphate-buffered saline (pH 7.4) containing 2% (wt/vol) α-methyl mannoside, was tested as previously described (2, 9). The DAF specificity of agglutination was tested with human erythrocytes previously pretreated with the DAF-specific monoclonal antibodies (MAbs) BRIC 230, BRIC 110, 1H4, and 8D11 (directed against the SCR-1, SCR-2, SCR-3, and SCR-4 domains of DAF, respectively) as previously described (50). The DAF-specific MAbs 1H4 and 8D11 were generously provided by D. M. Lublin (Washington University, St. Louis, Mo.). BRIC 230 and BRIC 110 were purchased from the International Blood Group Reference Laboratory (Bristol, Great Britain). Bacterial agglutination of P latex particles (P1 disaccharide latex; Chembiomed Ltd., Edmonton, Alberta, Canada) was used to determine P-adhesin-specific adhesion.
Adhesion assays were performed as previously described (26, 44). Monolayers were seeded with 4 × 105 cells in 35-mm-diameter tissue culture dishes (Corning Glass Works, Corning, N.Y.) and were incubated overnight (or for 2 days for Caco-2 cells). They were then infected by incubation for 3 to 5 h with bacteria. An inoculum ratio of 100 bacteria per cell was used for cell infection experiments.
DNA analysis and genetic techniques.
Plasmids were routinely isolated by alkaline lysis (39). Total plasmid DNA was extracted by the Kado method (27). Whole-cell DNA was prepared by cesium chloride gradient (31). Standard procedures were used for restriction endonuclease digestion and other common DNA manipulations (39). The afa and pap PCRs were as described previously (33). Four sets of primers were chosen, based on the sequences of the afaD and afaE genes from the afa-7 and afa-8 gene clusters. For afaD7 and afaE7 PCR assays, two sets of primers flanked segments of 377 and 618 bp, respectively. The upstream and downstream afaD primers were 5′-GCAGAGCTGAGTCTTGATGTCCGT-3′ and 5′-CGTAATTATTCCTGTGAACGGCTGTCCA-3′, respectively, and the upstream and downstream afaE primers were 5′-GCTAAATCAACTGTTGATGTT-3′ and 5′-GGACAATCCAAATGGCGAATTA-3′, respectively. For afaD8 and afaE8 PCR assays, two pairs of oligonucleotides flanking internal fragments of 354 and 302 bp of the afaD and afaE genes, respectively, were used; their sequences were 5′-GTTGAACTGAGTCTTAATACCAGTG-3′ and 5′-TGAGCATTCTCCGCTAACTGATAAT-3′ for afaD8 and 5′-CTAACTTGCCATGCTGTGACAGTA-3′ and 5′-TTATCCCCTGCGTAGTTGTGAATC-3′ for afaE8. PCR was carried out as previously described (33).
Cosmid and plasmid libraries.
Genomic DNA was isolated from E. coli 239 KH 89 and 262 KH 89 and was partially digested with the restriction endonuclease Sau3A. For the cosmid library, restriction fragments from 239 KH 89, 35 to 50 kb in size as determined by sucrose gradient (10 to 40%), were ligated into the BamHI-digested and alkaline phosphatase-treated cosmid vector pHC79 as previously described (39). Cosmids were packaged in vitro into phage lambda particles with the λ DNA in vitro packaging module (Amersham International). These particules were then used to infect E. coli HB101. For the plasmid library, restriction fragments from E. coli 262 KH 89, 6 to 12 kb in size, were fractionated by preparative agarose gel electrophoresis and ligated into pILL570 which had been BamHI digested and alkaline phosphatase treated. Recombinant plasmids were used to transform E. coli MC1061. Carbenicillin-resistant HB101 transductants and spectinomycin-resistant MC1061 transformants were screened by colony hybridization, using the 4.6-kb SphI fragment of pILL1101 containing the conserved region of the afa operons as a probe (Fig. 1A) (31), in low-stringency conditions.
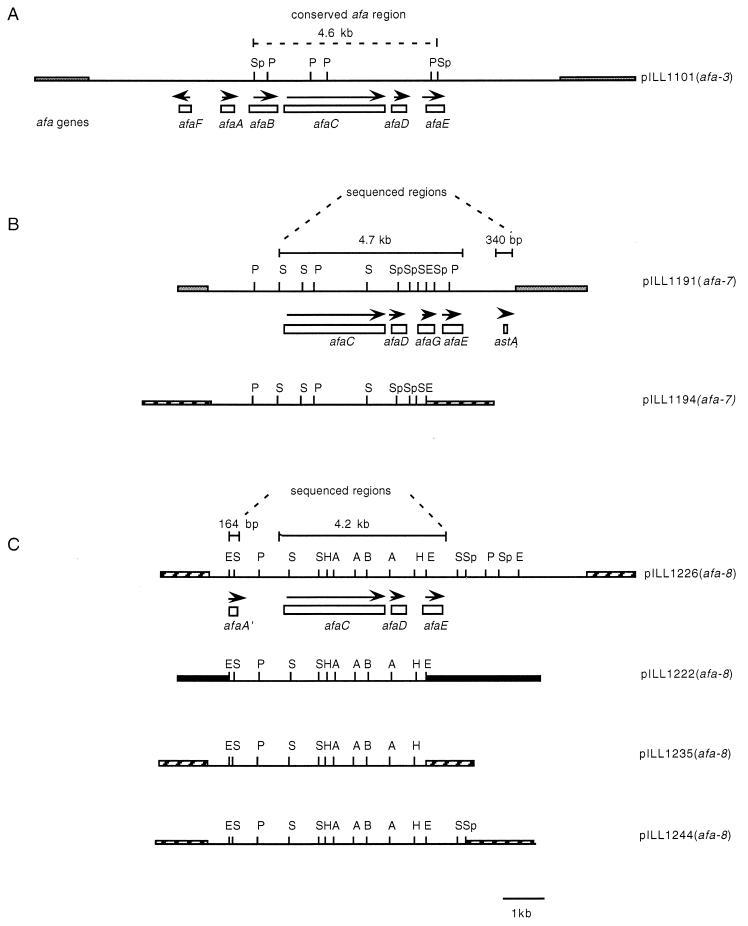
FIG. 1.
Genetic organization of the afa-7 and afa-8 gene clusters. (A) Physical and genetic maps of pILL1101, the plasmid carrying the afa-3 gene cluster (13). Locations of the six afa genes are shown by boxes and single-headed arrows indicating the direction of the transcription. The conserved region of the afa gene clusters from the human-pathogenic isolates, corresponding to the 4.6-kb SphI fragment, is represented by dashed lines (32). Only the restriction sites of interest are indicated. (B) Restriction maps of pILL1191 and pLLL1194 resulting from the cloning of the afa-7 gene cluster from total DNA of E. coli 262 KH 89. ORFs deduced from the partial nucleotide sequence are indicated by boxes. Sequenced regions are highlighted. (C) Restriction maps of pILL1226, pILL1222, pILL1235, and pILL1244 resulting from the cloning of the afa-8 gene cluster from total DNA of E. coli 239 KH 89. ORFs deduced from the partial nucleotide sequence are indicated by boxes. Sequenced regions are highlighted. Thin lines represent the inserts; thick lines represent pILL570, pBR322, and pUC18 vector DNAs. Abbreviations: A, AccI; B, BamHI; E, EcoRI; H, HincII; P, PstI; Pv, PvuI; S, SmaI; Sp, SphI.
Hybridization.
Bacteria grown for 3 h on nitrocellulose filters were used for colony hybridization as described by Grunstein and Hogness (20). For Southern blot hybridization, DNA restriction fragments were fractionated by agarose gel (0.7%) electrophoresis and transferred to nitrocellulose sheets (0.45-μm pore size; Schleicher & Schuell, Inc.) as previously described (56). Hybridization was performed under stringent (68°C) or nonstringent (50°C) conditions with probes labeled with 32P by using the Megaprime DNA labeling system (Amersham). The bound probes were detected by autoradiography with Amersham Hyperfilm-MP.
DNA sequencing.
Double-stranded DNA was sequenced by dideoxynucleotide chain termination with a Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham). If required, oligonucleotide primers were synthesized on the basis of the sequence acquired. Nucleotide and amino acid database searches were performed with BLAST, and sequences were aligned with the GAP and PILEUP programs of the Genetics Computer Group sequence analysis software package, version 7-UNIX.
Electron microscopy.
Negatively stained preparations of bacteria were examined for the presence of fimbriae on the bacterial surface as previously described (34). For transmission electron microscopy, infected HeLa cells were embedded as previously described (26), and ultrathin sections stained with uranyl acetate and lead citrate were examined with a JEOL 1010 transmission electron microscope operating at 80 kV. For scanning electron microscopy, glass coverslips coated with infected HeLa cells were processed as previously described (41) and examined with a JEOL JSM 35CF electron microscope.
Data imaging.
Photographic negatives and raw picture data were scanned into Adobe Photoshop with an AGFA Studio scanner, and the scanned images were printed with a Hewlett Packard Deskjet 870Cxi color printer.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported herein are AF072900, AF072901, and AF076152.
RESULTS
Characterization of the afa-positive strains 239 KH 89 and 262 KH 89.
Nineteen of the bovine clinical E. coli isolates in our collection were afa positive. Of these, strains 239 KH 89 and 262 KH 89 were selected for cloning experiments, based on the intestinal or extraintestinal site of isolation and the virulence factors carried. Strain 239 KH 89 was isolated from internal organs and contained sequences coding for the P adhesin and the CNF1 toxin (37). The gene coding for the CS31 adhesin was also detected in this strain by colony hybridization (data not shown). E. coli 239 KH 89 causes the MRHA of human and sheep erythrocytes but does not adhere to HeLa cells. In contrast, strain 262 KH 89 was isolated from feces and carried no sequences encoding P adhesin or CNF toxins (37). This strain causes the MRHA of human, bovine, and porcine erythrocytes and adheres to HeLa cells with a diffuse adhesion pattern.
Molecular cloning of afa-related sequences from strain 262 KH 89.
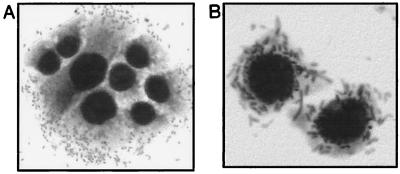
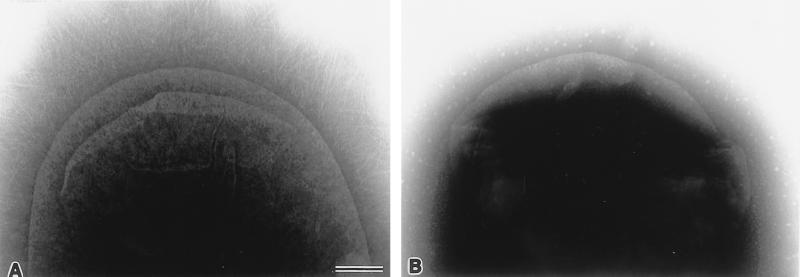
Recombinant plasmids containing portions (6 to 12 kb) of 262 KH 89 DNA were used to transform E. coli MC1061. Screening of the 322 clones of the library with the afa probe identified one clone which contained a hybrid plasmid, designated pILL1191 (Table 1). This plasmid resulted from the insertion of a 7.7-kb fragment (Fig. 1B). The insert of pILL1191 conferred a hemagglutination phenotype on the host bacteria and a HeLa cell adhesion pattern similar to that of strain 262 KH 89 (Fig. 2). Electron microscopy of negatively stained preparations of both 262 KH 89 and recombinant HB101(pILL1191) revealed no characteristic fimbriae on the bacterial surface (Fig. 3).
FIG. 2.
Photomicrographs of HeLa cells infected with E. coli 262 KH 89 (A) and HB101(pILL1191) (B). Bacteria adhered to the cell surface in a diffuse pattern.
FIG. 3.
Electron micrographs of E. coli strain preparations stained with ammonium molybdate. (A) HB101(pSSS1), used as a positive control for the detection of fimbriae; (B) HB101(pILL1191). Magnification, ×100,000; bar, 100 nm.
Molecular cloning of afa-related sequences from strain 239 KH 89.
Bacteriophage lambda particles carrying recombinant cosmid molecules with portions (35 to 50 kb) of 239 KH 89 DNA were prepared and used to infect E. coli HB101. Screening of 500 recombinant HB101 with the afa probe identified 10 positive clones. Eight of these clones agglutinated human erythrocytes. This MRHA phenotype was not due to the pap operon of strain 239 KH 89 being present on the recombinant cosmids because, unlike the parental strain, none of the clones gave positive amplification in the pap PCR assay or agglutinated P latex particles. The recombinant cosmids carried by these eight MRHA clones were analyzed by digestion with three restriction enzymes (BamHI, EcoRI, and HindIII). Comparison of the restriction profiles suggested that all of the cosmids carried the same region of 239 KH 89 genomic DNA. Hybridization with the afa probe showed that in three of the cosmids, including pILL1211 (Table 1), the sequences binding to the afa probe were located on a single 5-kb EcoRI fragment. pILL1222, which results from the insertion of this 5-kb EcoRI fragment from pILL1211 into pUC18 (Table 1; Fig. 1C), did not confer the property of MRHA on HB101. The MRHA-expressing DNA fragment was subcloned by partially cleaving pILL1211 with Sau3A such that 6- to 12-kb fragments were generated and ligated into BamHI-digested and alkaline phosphatase-treated pBR322. Recombinant plasmids were used to transform E. coli HB101. The carbenicillin-resistant transformants were tested for MRHA and were screened for afa-related sequences by using the 5-kb EcoRI fragment of pILL1222 as a probe. One positive clone carried the recombinant plasmid pILL1226, which has a 9.5-kb insert (Table 1; Fig. 1C). This clone was chosen for further characterization of the afa-related sequences. Unlike the parental strain, which agglutinates both human and sheep erythrocytes, HB101(pILL1126) causes the MRHA only of human erythrocytes. Electron microscopy of negatively stained preparations of HB101(pILL1226) showed that no fimbriae were present (data not shown).
Characterization of the afa-7 and afa-8 gene clusters.
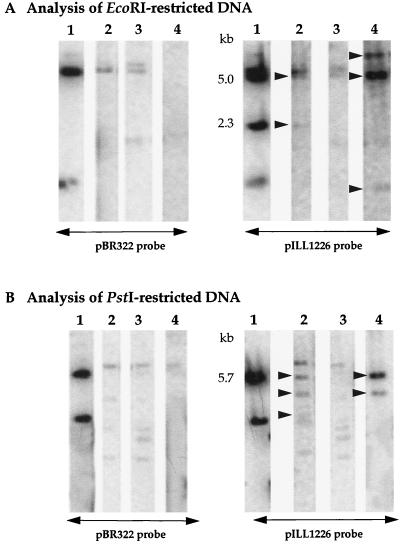
Restriction mapping and Southern analyses were performed with the cloned afa-related sequences from the two bovine strains. Comparison of the sequences of the insert from pILL1191 and total DNA from 262 KH 89 digested by PstI and SmaI (data not shown) and of the insert from pILL1226 and total DNA from 239 KH 89 digested by EcoRI and PstI (Fig. 4A and B, lanes 1 and 2) showed that the afa-related sequences were not rearranged during the molecular cloning processes. The restriction maps of pILL1191 and pILL1226 were compared to each other and to that of pILL1101, which carries the well-characterized and sequenced afa-3 gene cluster (Fig. 1). The restriction maps of pILL1191 and pILL1226 differed, and neither plasmid contained the three PstI fragments of 2.6, 1.1, and 0.4 kb characteristic of the conserved region of the afa gene clusters of human-pathogenic strains. Reciprocal hybridizations between restriction fragments from the inserts of the three plasmids indicated that (i) the inserts of both pILL1191 and pILL1226 contained, upstream from the conserved sequences of afa gene clusters, sequences related to the 1.9-kb PstI fragment of pILL1101, corresponding to the promoter region of the afa-3 operon, and (ii) the afa-related sequences in pILL1191 from strain 262 KH 89 were only slightly similar to those from strain 239 KH 89, which were inserted into pILL1226. These results suggest that the afa-related sequences inserted into pILL1191 and pILL1226 correspond to two different afa-related operons, designated afa-7 and afa-8, respectively.
FIG. 4.
Search for homology between the insert of pILL1226 and total and plasmid DNA from strain 239 KH 89 and between the insert of pILL1226 and plasmid DNA from strain 89.201.2/3. Plasmid or total DNA was digested with EcoRI (A) or PstI (B). Lanes: 1, pILL1226; 2 and 3, total and plasmid DNA from strain 239 KH 89, respectively; 4, plasmid DNA from strain 89.201.2/3. The resulting fragments were separated by electrophoresis in a 0.7% agarose gel, transferred to nitrocellulose filters, and probed with either pBR322 or pILL1226. The sizes indicated are those of the internal EcoRI and PstI fragments of the pILL1226 insert. The EcoRI and PstI fragments hybridizing specifically with the insert of pILL1226 are indicated by arrows.
Genetic organization of the afa-7 gene cluster.
Determination of the sequence of a 4.7-kb region of the pILL1191 insert confirmed that the genetic organization of the afa-7 gene cluster of strain 262 KH 89 was very similar to that of the afa operons of human-pathogenic strains (Fig. 1A and B). Computer analyses identified four ORFs transcribed in the same orientation (Fig. 1B). Two of the ORFs mapped to the same loci as the afaC and afaD genes in the afa-3 operon. The ORF designated afaC encoded a peptide of 838 amino acids (aa) similar (77% identity and 81% similarity) to AfaC, an outer membrane usher protein (data not shown). Following this ORF was a short intergenic region of 17 bp (rather than 15 bp as in the afa-3 gene cluster) and the afaD ORF, which encoded a peptide with 46% identity and 56% similarity to AfaD. The first ATG codon of this ORF was preceded by a Shine-Dalgarno sequence (data not shown) and followed by a probable signal peptide sequence with a classical cleavage site (Ser-Gln-Ala) that was recognized by signal peptidase I (Fig. 5). The predicted mature AfaD molecule contains 123 aa and has a deduced molecular mass of 13.6 kDa and a pI of 5.27.
FIG. 5.
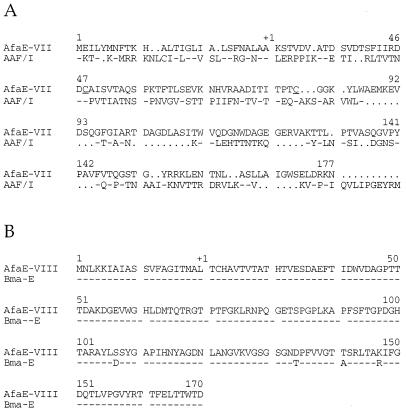
Sequence alignment of AfaD proteins from afa-3, afa-7, and afa-8 gene clusters. Dashes mark identical residues; gaps indicated by dots have been inserted to optimize the alignment. Numbers correspond to amino acid positions in the protein of the afa-3 gene cluster. The predicted translational start sites of AfaD-III, AfaD-VII, and AfaD-VIII proteins are indicated by +1.
The noncoding region (164 bp) downstream from the afaD gene was not similar to that of the corresponding region (482 bp) in the afa-3 gene cluster. The ORF, designated afaG, corresponds to a 717-bp coding sequence. It has three potential start codons, only the third of which is preceded by a probable RBS (23, 24), resulting in a putative 138-aa protein. No signal peptide was evident from the deduced sequence. A weak similarity (23 to 27% identity and 36 to 38% similarity) was found between this putative product and three proteins, from E. coli (accession no. Q47688), Marchantia polymorpha (48), and Saccharomyces cerevisiae (accession no. P03875), previously shown to be similar to group II intron maturases. This similarity principally involved the reverse transcriptase domain IV of group II intron maturases as defined by Mohr et al. (42). Downstream from afaG was a 123-bp intergenic region and the afaE ORF. Of the two potential start codons, only the second was preceded by a potential RBS, and a putative signal sequence was identified in the protein (Fig. 6A). The predicted mature peptide had a pI of 4.95 and consisted of 151 aa (116.21 kDa) including two cysteine residues in a position highly conserved in the AfaE-related adhesins (34). The deduced product was 31% identical (44% similar) to AggA, the fimbrial subunit of the AAF/I fimbriae produced by enteroaggregative E. coli (EAggEC) (43) (Fig. 6A). Nonpolar mutations in this ORF (created by insertion of the nonpolar kanamycin cassette in the same orientation as that of the ORF) abolished the agglutination and adhesion properties conferred on the host strain by the afa-7 sequences. All of these data strongly indicate that the afaE ORF is the adhesin-encoding gene.
FIG. 6.
Protein sequence alignments. Dashes mark identical residues. Numbers correspond to amino acid positions in the AfaE-VII and AfaE-VIII adhesins. The predicted translational start sites of AfaE-VII and AfaE-VIII adhesins are indicated by +1. (A) Protein sequence of AfaE-VII aligned with that of the AAF/I fimbrial adhesin. The two cysteine residues conserved among the afimbrial and fimbrial AfaE-related adhesins are underlined; gaps indicated by dots have been inserted to optimize the alignment. (B) Sequence alignment of AfaE-VIII and BmaE proteins.
Sequencing of 340 bp at the end of the pILL1191 insert revealed the presence of an ORF encoding a hypothetical 38-aa protein identical to the EAggEC heat-stable enterotoxin 1 (EAST1) encoded by the astA gene. The nucleotide sequence of the astA gene on pILL1191 was identical to that carried on pCS1 and the chromosome of enterotoxigenic E. coli isolates producing the CFA/I adhesin factor (57) and differed by only one base from that of EAggEC strain 17-2 (53), resulting in a single amino acid change (Thr to Ala).
Genetic organization of the afa-8 gene cluster.
Based on hybridization experiments, a 4.2-kb region of the insert of pILL1226 was presumed to contain the 3′ end of the afa-8 gene cluster. Computer analyses of the sequence of this region showed three ORFs transcribed in the same orientation. They were homologous to the afaC and afaD genes and to an adhesin-encoding gene (Fig. 1C), respectively, confirming that the genetic organizations of the afa-3, afa-7, and afa-8 gene clusters were similar. This similarity was confirmed by sequencing of the 5′ terminus of the insert of pILL1222 (Table 1; Fig. 1C), which detected a partial ORF (afaA′) with 75% identity (over 164 bp) to the afaA gene of the afa-3 gene cluster. The AfaA product has been shown to be similar to the PapB regulatory protein (13).
The afaC ORF of afa-8 was sequenced. It encodes a product of 861 aa similar to AfaC from the afa-3 gene cluster (71% identity and 76% similarity) and to AfaC from the afa-7 gene cluster (72% similarity and 77% identity) (data not shown). Following the afaC gene of afa-8 was an 18-bp intergenic region and an ORF, designated afaD. The ATG codon of this ORF was preceded by a Shine-Dalgarno sequence and followed by a probable signal peptide sequence (Fig. 5). The predicted mature protein consisted of 117 aa and had a deduced molecular mass of 12.74 kDa and a pI of 5.38. The AfaD peptide was similar to the AfaD proteins from the afa-3 (42% identity and 53% similarity) and afa-7 (46% identity and 56% similarity) gene clusters. Immediately downstream from the afaD gene of afa-8 was a noncoding region of 537 bp followed by an ORF, which had a putative cleavage site and coded for a predicted mature protein of 151 aa (15.99 kDa) and a pI of 5.19. There was a putative RBS upstream from the start codon. The MRHA phenotypes conferred on strain HB101 by two plasmids derived from pILL1226 by various deletions in the right part of the insert were compared. The product of this ORF, designated afaE, was found to be essential for MRHA expression: HB101(pILL1244) is MRHA positive and HB101(pILL1235) is MRHA negative (Table 1; Fig. 1C). The encoded protein, AfaE-VIII, was very similar (97% identity) to the M hemagglutinin encoded by the bmaE gene of uropathogenic E. coli strains (52) (Fig. 6B).
Comparison of the adhesion to various cell lines of E. coli strains expressing the afa-3, afa-7, and afa-8 gene clusters.
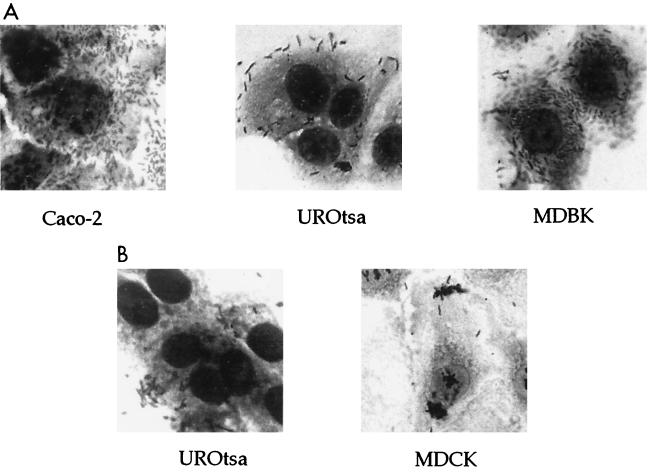
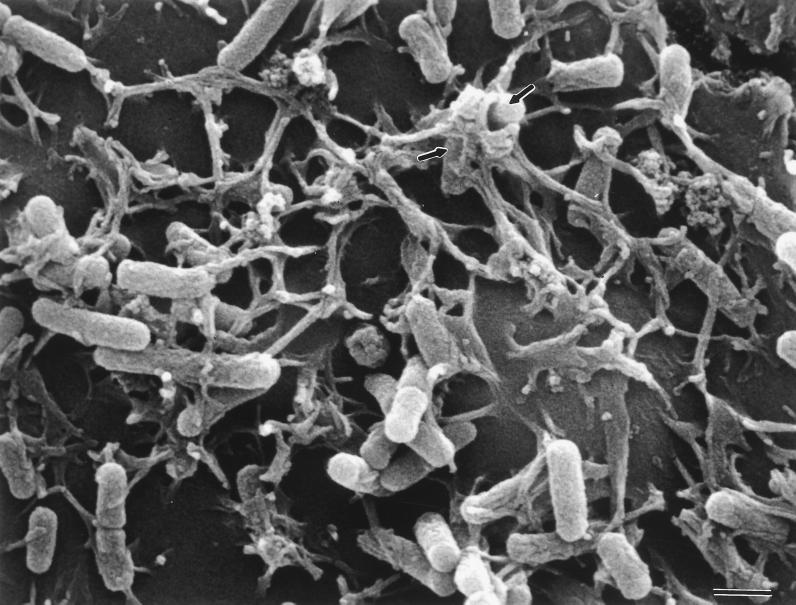
The production of AfaE adhesins by E. coli strains pathogenic for humans conferred on the bacteria the capacity to bind to HeLa cells, uroepithelial cells (32), and Caco-2 cells (29). The expression of the afa-7 gene cluster by HB101 strains conferred on the bacteria the capacity to bind to human HeLa, Caco-2, and UROtsa cell lines (Table 2; Fig. 2B and 7A). HB101 strains producing the AfaE-VII adhesin, but not those producing AfaE-III, adhered to bovine kidney-derived MDBK cells (Table 2; Fig. 7A). Scanning electron microscopy of cultured HeLa cells infected with HB101(pILL1191) showed bacteria adhering in a diffuse pattern and interacting with microvillar extensions of HeLa cells, with some bacteria enclosed by elongations (Fig. 8). These observations were similar to those previously reported (26) for interactions between HB101(pILL1101) bacteria expressing the afa-3 gene cluster and HeLa cells.
TABLE 2.
Binding to epithelial cell lines of HB101 strains expressing the afa-3, afa-7, and afa-8 gene clusters
| Strain (afa operon) | Binding to indicated epithelial cell line
|
||||
|---|---|---|---|---|---|
| HeLa | Caco-2 | UROtsa | MDBK | MDCK | |
| HB101 (afa-3) | + | + | + | − | − |
| HB101 (afa-7) | + | + | + | + | − |
| HB101 (afa-8) | − | − | + | − | + |
FIG. 7.
Photomicrographs of Caco-2, UROtsa, MDBK, and MDCK cells infected with HB101(pILL1191) (A) and HB101(pILL1244) (B). The infection time was 3 h except for UROtsa cells, which were infected for 5 h with HB101(pILL1244).
FIG. 8.
Scanning electron micrograph showing adhesion to HeLa cells of strain HB101(pILL1191) after 3 h of infection. Arrows indicated bacteria totally surrounded by microvillar extensions. Magnification, ×12,000; bar, 1 μm.
The results of the hemagglutination and adhesion assays suggest that the AfaE-VII adhesin recognizes as a receptor a molecule present in both human and bovine epithelial cells. It has been shown that AfaE-I and AfaE-III adhesins (45) and other subtypes of AfaE adhesins (AfaE-II, AfaE-V, and AfaE-X) produced by strains causing UTI and diarrhea (11) recognize as a receptor the SCR-3 domain of the human DAF. Additional binding and hemagglutination assays showed that faE-VII, unlike previously described AfaE adhesins, did not recognize human DAF as a receptor. Indeed, HB101(pILL1191), in contrast to HB101(pILL1101), which expresses the afa-3 gene cluster, did not bind to CHO cells transfected with the cDNA encoding human DAF (data not shown). Moreover, the MRHA capacity of HB101(pILL1191), unlike that of HB101(pILL1101), was not inhibited by prior incubation of the erythrocytes with MAbs directed against the SCR domains of human DAF.
The adhesion pattern of afa-8-expressing HB101 strains differed from that of afa-3 and afa-7-expressing bacteria (Table 2; Fig. 7B). HB101 strains producing the AfaE-VIII adhesin adhered to MDCK and UROtsa cells but not to HeLa, Caco-2, MDBK (Fig. 7B), or DAF-transfected CHO cells (data not shown).
Comparison of the invasion properties of E. coli strains expressing the afa-3, afa-7, and afa-8 gene clusters.
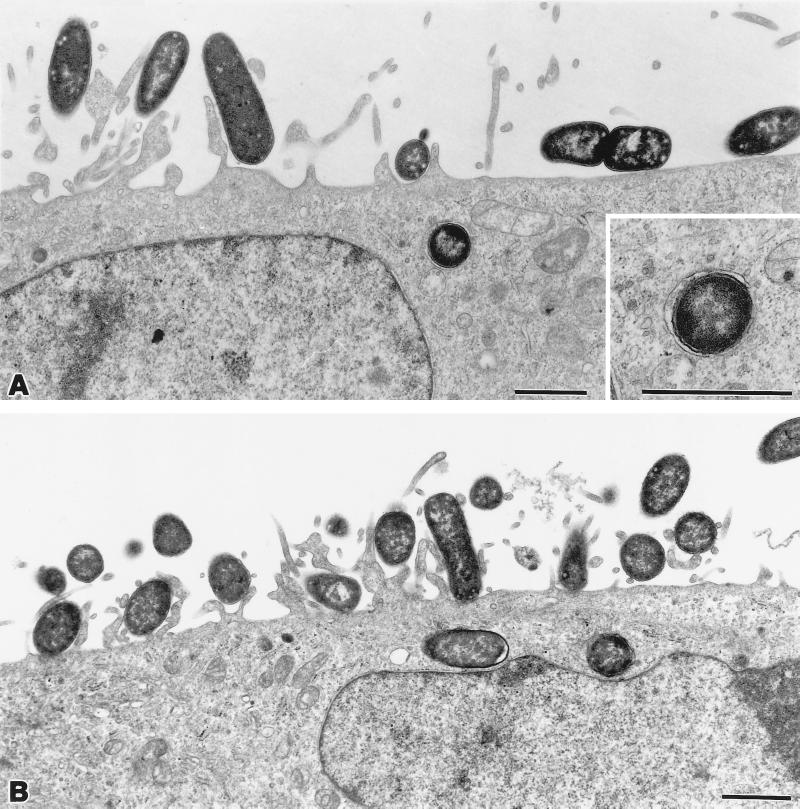
Examination of HeLa cells infected with HB101(pILL1191) showed intracellular bacteria within membrane-bound vacuoles (Fig. 9A). The AfaD-VII and AfaD-VIII products were also similar to the AfaD protein, which is encoded by the afa-3 gene cluster and has been reported to be an invasin (26). We investigated the role of AfaD from the afa-7 and afa-8 gene clusters in bacterial internalization by complementing the invasion deficiency of an afa-3-expressing HB101 with a mutated afaD gene by producing the AfaD-VII or AfaD-VIII protein. HeLa cell monolayers were infected with HB101 harboring two plasmids: one (pILL1189) containing the afa-3 gene cluster with a mutation in the afaD gene, and the other (pILL1194 or pILL1235) containing the afa-7 or afa-8 cluster deleted of the afaE gene. Electron microscopy showed that both HB101(pILL1189, pILL1194) (Fig. 9B) and HB101(pILL1189, pILL1235) (data not shown) adhered to HeLa cells because the AfaE-III adhesin was present, and it invaded HeLa cells due to the presence of AfaD-VII or AfaD-VIII proteins. As for internalized afa-3-expressing bacteria, internalized HB101(pILL1189, pILL1194) and HB101(pILL1189, pILL1235) were found within membrane-bound vesicles.
FIG. 9.
Transmission electron micrographs of recombinant strains interacting with HeLa cells 3 h postinfection. (A) HB101(pILL1191), expressing the afa-7 gene cluster. Intracellular bacteria were observed within membrane-bound vacuoles, as showed in the inset. (B) HB101(pILL1189, pILL1194), producing both AfaD-VII and AfaE-III. AfaE-III mediated adhesion of the bacteria to HeLa cells; AfaD-VII mediated internalization of the bacteria into the cells. Magnifications, ×15,000 (A), ×31,000 (inset), and ×14,000 (B); bar, 1 μm.
Distribution of afa-7 and afa-8 operons in bovine and porcine E. coli isolates reported to harbor afa-related sequences.
In addition to strains 262 KH 89 and 239 KH 89, 17 other bovine strains containing afa-related sequences (37) were screened for afa-7 and afa-8 sequences by colony hybridization in high-stringency conditions. The probes used were afaD and afaE PCR products obtained from strains 262 KH 89 and 239 KH 89 (see Materials and Methods for DNA amplification). None of the 17 strains hybridized with the afaD and afaE probes from the afa-7 gene cluster. In contrast, all 17 strains contained sequences that hybridized with both the afaD and afaE genes from the afa-8 gene cluster. We evaluated a PCR approach for the detection of afa-8 sequences, using the two sets of primers derived from the sequences of the afaD and afaE genes in amplification reactions to detect the presence of the corresponding sequences. All 17 strains positive by hybridization tested positive by PCR for both afaD8 and afaE8. The specificity of the PCR assays was validated by direct sequencing of the products obtained from nine isolates. The afaD amplification products differed by no more than 1 bp between any two sequences. Similar results were obtained for comparison of the sequences of the nine afaE amplification products.
Six F165-positive E. coli strains isolated in Canada from calves and piglets with diarrhea have also been reported to contain afa-related sequences (21). Structural analysis by DNA hybridization showed that the afa-related sequences of these isolates differ internally from those of human afa gene clusters (38). All of these clones, like the afa-8 gene cluster, have been reported to contain an EcoRI fragment of about 5 kb containing a sequence homologous to the afaC gene of human afa operons. These pathogenic strains (five of bovine and one of porcine origin) were tested for afa-8 sequences by PCR. All isolates tested positive for both afaD8 and afaE8 genes.
The afa-8 gene cluster is plasmid or chromosome borne.
Mainil et al. reported that the 19 isolates from cattle that he provided for this study carry afa-related sequences either on the chromosome or on Vir plasmids (37). The afa-related sequences carried by E. coli 239 KH 89 were described as chromosome borne (37). We confirmed that this was the case by DNA hybridizations of plasmid or total DNA from 239 KH 89 digested with EcoRI (Fig. 4A) and PstI (Fig. 4B), with pBR322 and pILL1226 used as probes. None of the plasmid-generated fragments had sequences similar to that of the pILL1226 insert (Fig. 4A and B, lanes 3). In contrast, two EcoRI (Fig. 4A, lane 2) and three PstI (Fig. 4B, lane 2) fragments of total DNA from 239 KH 89 specifically hybridized with the pILL1126 insert. Of these, the two EcoRI fragments and one PstI fragment had the same electrophoretic mobility as the EcoRI (5 and 2.3-kb) and PstI (5.7-kb) internal fragments carrying the afa-8 operon in the pILL1226 insert (Fig. 1C). Of the E. coli isolates testing positive for afa-8 by PCR that have been studied, E. coli 89.201.2/3 has been reported to carry afa-related sequences on the Vir plasmid (37). We investigated the location of the afa-8 operon in this strain by hybridization between each of the two probes described above and digested plasmid DNA from strain 89.201.2/3. Sequences similar to that of the pILL1226 insert were clearly detected in three EcoRI (Fig. 4A, lane 4) and two PstI (Fig. 4B, lane 2) plasmid fragments, indicating that the afa-8 determinants are plasmid borne in strain 89.201.2/3. However, although one hybridizing EcoRI fragment (5 kb) was similar to the internal EcoRI fragment of the pILL1226 insert that carried most of the afa-8 genes, we detected no hybridizing fragment that was the same size as the second internal EcoRI fragment (2.3 kb) of the pILL1226 insert that contained the afaE gene and flanking sequences. In the same way, one PstI fragment was similar in size to the internal PstI fragment (5.7 kb) of the pILL1226 insert that contained the afa-8 genes, but the second hybridizing fragment differed in size from the second hybridizing PstI fragment detected in total DNA from 239 KH 89. Thus, the plasmid and chromosomal sequences flanking the afa-8 gene cluster differ, suggesting that the presence of afa-8 sequences in the chromosome did not result from the insertion of a Vir plasmid.
DISCUSSION
Neonatal septicemia and diarrhea due to E. coli are common problems in farm animals, especially calves (15). Fimbrial adhesins, virulence factors produced by these pathogenic E. coli strains, have been extensively detected and studied. Recently, sequences related to the afa gene clusters that encode afimbrial adhesins in human-pathogenic E. coli associated with intestinal and extraintestinal infections have been detected in strains isolated from calves and piglets with diarrhea and septicemia (21, 37). We describe here two afa-related gene clusters from bovine E. coli isolates. We found that these two systems also code for afimbrial adhesins and demonstrated that the genetic organization of these two gene clusters, designated afa-7 and afa-8, is very similar to that of the afa gene clusters from human E. coli isolates. Hybridization and sequencing experiments indicated the presence in both gene clusters of regulatory sequences followed by an afaC gene encoding an outer membrane anchor protein, an afaD gene encoding an invasin, and an afaE gene encoding an adhesin. The afaC genes were very similar, the afaD genes were less similar, and the afaE genes were completely divergent. The afa-8 operon is very prevalent in a collection of bovine E. coli strains associated with intestinal and extraintestinal infections. These preliminary epidemiological results show a high prevalence of afa-8 genes in E. coli isolates from diseased animals. Like the afa-3 and afa-5 gene clusters from human strains (60), the afa-8 operon has been found on both large plasmids and the chromosome. We demonstrated that the chromosomal location of the afa-8 genes was not the result of chromosomal insertion of a virulence plasmid. The afa-3 gene cluster has been shown to translocate from a plasmid to the E. coli chromosome via IS1-specific recombination mediated by a recA-independent mechanism (13). Thus, all the available evidence strongly suggest that the gene clusters of the afa family may be carried by mobile genetic elements, facilitating their dissemination among E. coli strains.
Sequence analysis of the afa-7 gene cluster showed that directly upstream from the afaE gene is an ORF (afaG) with no homolog in the other afa gene clusters, which maps to the same locus as the newly described daaP gene in the daa operon (36). The DaaP polypeptide is involved in the regulation of expression of the daa operon via endoribonucleolytic processing of the polycistronic mRNA (36). Interestingly, afaG encodes a peptide with some similarity to the group II intron maturases involved in intron RNA processing, and the mRNA cleavage sequence described by Loomis et al. is also present in the afa-7 gene cluster. It is therefore possible that AfaG is involved in the regulation of expression of the afa-7 gene cluster.
The afa gene clusters characterized from human-pathogenic strains have the unique feature of encoding both an adhesin and an invasin, which seem to be involved in a two-stage process of interaction with epithelial cells (26). The results of this study demonstrate that the afa-related gene clusters expressed by animal-pathogenic strains are also involved in the adhesion to and invasion of epithelial cells. The AfaD proteins encoded by both the afa-7 and afa-8 gene clusters mediated internalization of bacteria into HeLa cells although they were only 46 and 42% identical, respectively to the AfaD invasin encoded by the afa-3 gene cluster. Thus, these three AfaD proteins may indeed be the first members of a new family of invasins.
Previous studies have demonstrated the great heterogeneity of the AfaE-related adhesins of E. coli strains isolated from humans. The AfaE-VII protein is genetically different from the AfaE adhesins produced by human-pathogenic strains and also has different binding specificities. Our data indicate that the receptor of the AfaE-VII adhesin is not human DAF, a species-specific molecule previously thought to be the receptor for ll of the AfaE adhesins produced by human-pathogenic strains (11, 45, 50). AfaE-VII mediates MRHA of human, bovine, and porcine erythrocytes and the adhesion of bacteria to human (HeLa, Caco-2, and UROtsa cells) and bovine (MDBK cells) epithelial cells. It therefore appears that its receptor is widely distributed in various tissues, at least in humans and cattle.
AfaE-VII is slightly similar to the fimbrial adhesin AAF/I produced by EAggEC isolates implicated in pediatric diarrhea in the developing world (43). Not only does AAF/I belong to a putative subfamily of AfaE adhesins, but the organization of the AAF/I biogenesis genes is typical of the afa family of operons (54), suggesting that the agg and afa operons may belong to the same phylogenetic family of adhesin-encoding gene clusters. The gene encoding EAST1 of E. coli is located approximately 1 kb downstream from the AAF/I (53)- and AfaE-VII-encoding structural genes, indicating that there may be a genetic association between such adhesin-encoding genes and this toxin-encoding gene. The EAST1-encoding gene has been also detected in enteropathogenic, enterohemorrhagic, and diffusely adherent E. coli strains associated with diseases in humans (53, 55, 57) and in enterotoxigenic E. coli isolates from humans, cattle, and pigs (57, 58). This widespread distribution among pathogenic E. coli led Yamamoto and Escheverria to suggest that the EAST1-encoding gene may be located on a transposon (57). Our results showing identity between the regions upstream (59 nucleotides) and downstream (147 nucleotides) from the EAST1-encoding gene on the chromosome of the afa-7-expressing strain, 262 KH 89, and on pCS1 in enterotoxigenic E. coli strains strongly support this notion.
The gene encoding AfaE-VIII, the second AfaE adhesin from animal-pathogenic strains characterized in this study, is also different from those of known AfaE adhesins produced by human-pathogenic strains, and AfaE-VIII binds to a different receptor, yet to be identified. AfaE-VIII is very similar (97% identity) to the M agglutinin, an afimbrial adhesin encoded by the bma gene cluster in human-uropathogenic strains (52). The bma gene cluster consists of five genes, the sequences of which, with the exception of that of the adhesin-encoding gene, are not available in databases. It is therefore not possible to compare the sequences of the bma and afa-8 gene clusters. As the prevalence of M-agglutinin-producing strains is estimated to be very low among human isolates (25), the biological function and significance of this adhesin in human diseases are unknown. In this study we have shown that the AfaE-VIII adhesin is frequent and highly conserved among E. coli strains producing either CNF1 or CNF2, factors that are pathogenic for calves. AfaE-VIII-producing strains have also been detected among non-CNF-producing strains isolated from calves and piglets. All of these afa-8-expressing bacteria are associated with diarrhea, septicemia, and internal organ infections in which the site of entry is the intestine. Bloodstream infections in which the bacteria are derived from the intestinal flora by bacterial translocation are common in patients with cancer (1). Preliminary results obtained with the afa-8-specific PCR assay in our laboratory (32a) show that afa-8 gene clusters are present in CNF1-producing E. coli strains recently associated with this type of bacteremia (22). It therefore seems likely that afa-8 is involved in the development of extraintestinal infections associated with a primary colonization of the intestine.
In conclusion, we have demonstrated that animal-pathogenic E. coli isolates express different afa gene clusters, all encoding products involved in adhesion to and internalization into epithelial cells. The role of the adhesins and invasins produced by afa-expressing strains in the pathophysiology of diarrheal and extraintestinal infections in young animals is unclear. We have described specific PCR assays that may be useful tools for the epidemiological studies of animals necessary to confirm the association of the afa-8 gene cluster with CNF-producing strains. The high degree of conservation of the AfaE-VIII adhesins between strains suggests that this antigen could be used as a component of vaccines in animal husbandry.
ACKNOWLEDGMENTS
We are grateful to A. Labigne, in whose unit this work was carried out, for her continuing interest and helpful discussions. We also thank Jacques Mainil (Bacteriology, Faculty of Veterinary Medecine, University of Liege, Liege, Belgium) and E. Oswald (ENVT-INRA, Toulouse, France) for the gift of E. coli strains isolated from calves. We thank John Fairbrother for carrying out the animal erythrocyte agglutination assays, A. Servin, in whose laboratory the assays of binding to CHO cell transfectants expressing human DAF were performed, R. W. John (Institute of Urology and Nephrology, London, Great Britain) for providing UROtsa cells, and C. Parsot (Pasteur Institute, Paris, France) for the gift of the nonpolar kanamycin cassette. We also thank P. Courcoux and H. Ohayon for technical assistance.
This work was supported by grant 1335 from the European Community program FAIR. L. Lalioui received a fellowship from the Marcel Mérieux Fondation. M. Jouve was a fellow of the Association pour les Journées de Biologie Clinique Institut Pasteur-CHU Necker Enfants Malades.
REFERENCES
- 1.Andremont A, Lancar R, Lê N A, Hattchouel J M, Baron S, Tavakoli T, Daniel M F, Tancrède C, Lê M G. Secular trends in mortality associated with bloodstream infections in 4268 patients hospitalized in a cancer referral center between 1975 and 1989. Clin Microbiol Infect. 1996;1:160–167. doi: 10.1111/j.1469-0691.1996.tb00547.x. [DOI] [PubMed] [Google Scholar]
- 2.Archambaud M, Courcoux P, Labigne-Roussel A. Detection by molecular hybridization of PAP, AFA, and SFA adherence systems in Escherichia coli strains associated with urinary and enteral infections. Ann Inst Pasteur Microbiol. 1988;139:575–588. doi: 10.1016/0769-2609(88)90156-1. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Johnson C E, Rubin R H, Arbeit R D, Campanelli C, Kim C, Steinbach S, Agarwal M, Wilkinson R, Goldstein R. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli. Infect Immun. 1989;57:303–313. doi: 10.1128/iai.57.2.303-313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilge S S, Clausen C R, Lau W, Moseley S L. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. 1989;171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins J. Escherichia coli plasmids packageable in vitro in lambda bacteriophage particles. Methods Enzymol. 1979;68:309–326. doi: 10.1016/0076-6879(79)68022-9. [DOI] [PubMed] [Google Scholar]
- 9.Fairbrother J M, Lariviere S, Lallier R. New fimbrial antigen F165 from Escherichia coli serogroup O115 strains isolated from piglets with diarrhea. Infect Immun. 1986;51:10–15. doi: 10.1128/iai.51.1.10-15.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foxman B, Zhang L, Palin K, Tallman P, Marrs C F. Bacterial virulence characteristics of Escherichia coli isolates from first-time urinary tract infection. J Infect Dis. 1995;171:1514–1521. doi: 10.1093/infdis/171.6.1514. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, M. I. Unpublished data.
- 12.Garcia M I, Gounon P, Courcoux P, Labigne A, Le Bouguénec C. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol Microbiol. 1996;19:683–693. doi: 10.1046/j.1365-2958.1996.394935.x. [DOI] [PubMed] [Google Scholar]
- 13.Garcia M I, Labigne A, Le Bouguénec C. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J Bacteriol. 1994;176:7601–7613. doi: 10.1128/jb.176.24.7601-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia M I, Le Bouguénec C. Role of adhesion in pathogenicity of human uropathogenic and diarrhoegenic Escherichia coli. Bull Inst Pasteur. 1996;94:201–236. [Google Scholar]
- 15.Gay C C, Besser T E. Escherichia coli septicaemia in calves. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 75–90. [Google Scholar]
- 16.Germani Y, Bégaud E, Duval P, Le Bouguénec C. An Escherichia coli clone carrying the adhesin-encoding afa operon is involved in both diarrhoea and cystitis in twins. Trans R Soc Trop Med Hyg. 1997;91:573. doi: 10.1016/s0035-9203(97)90031-6. [DOI] [PubMed] [Google Scholar]
- 17.Germani Y, Bégaud E, Duval P, Le Bouguénec C. Prevalence of enteropathogenic, enteroaggregative, and diffusely adherent Escherichia coli among isolates from children with diarrhea in New Caledonia. J Infect Dis. 1996;174:1124–1126. doi: 10.1093/infdis/174.5.1124. [DOI] [PubMed] [Google Scholar]
- 18.Giron J A, Jones T, Millan-Velasco F, Castro-Munoz E, Zarate L, Fry J, Frankel G, Moseley S L, Baudry B, Kaper J B, Schoolnik G K, Riley L W. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in mayan children in Mexico. J Infect Dis. 1991;163:507–513. doi: 10.1093/infdis/163.3.507. [DOI] [PubMed] [Google Scholar]
- 19.Goluszko P, Moseley S L, Truong L D, Kaul A, Williford J R, Selvarangan R, Nowicki S, Nowicki B. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae: mutation in the dra region prevented tubulointerstitial nephritis. J Clin Investig. 1997;99:1662–1672. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunstein M, Hogness D S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci USA. 1975;72:3961–3955. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harel J, Daigle F, Maiti S, Désautels C, Labigne A, Fairbrother J M. Occurrence of pap-, sfa-, and afa-related sequences among F165-positive Escherichia coli from diseased animals. FEMS Microbiol Lett. 1991;66:177–182. doi: 10.1016/0378-1097(91)90329-9. [DOI] [PubMed] [Google Scholar]
- 22.Hilali F, Saulnier P, Durand C, Barnabé C, Tibayrenc M, Le Bouguénec C, Andremont A. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Clustering of virulence genes and evolutionary relationship among strains of Escherichia coli causing bacteremia in cancer patients, abstr. R-27; p. 484. [Google Scholar]
- 23.Hui A, de Boer H A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob W F, Santer M, Dahlberg A E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci USA. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jokinen M, Ehnholm C, Väisänen-Rhen V, Korhonen T, Pipkorn R, Kalkkine N, Gahmberg C G. Identification of the major human sialoglycoprotein from red cells, glycophorin AM, as the receptor for Escherichia coli IH 11165 and characterization of the receptor site. Eur J Biochem. 1985;147:47–52. doi: 10.1111/j.1432-1033.1985.tb08716.x. [DOI] [PubMed] [Google Scholar]
- 26.Jouve M, Garcia M I, Courcoux P, Labigne A, Gounon P, Le Bouguénec C. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect Immun. 1997;65:4082–4089. doi: 10.1128/iai.65.10.4082-4089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellenius G, Molby R, Svenson S B, Windberg J, Lundblad A, Svenson S, Cedergren B. The Pk antigen as receptor for hemagglutination of pyelonephritic Escherichia coli. FEMS Microbiol Lett. 1980;7:297–302. [Google Scholar]
- 29.Kerneis S, Gabastou J M, Bernet-Camard M F, Coconnier M H, Nowicki B J, Servin A L. Human cultured intestinal cells express attachment sites for uropathogenic Escherichia coli bearing adhesins of the Dr adhesin family. FEMS Microbiol Lett. 1994;119:27–32. doi: 10.1111/j.1574-6968.1994.tb06862.x. [DOI] [PubMed] [Google Scholar]
- 30.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labigne-Roussel A, Falkow S. Distribution and degree of heterogeneity of the afimbrial-adhesin-encoding operon (afa) among uropathogenic Escherichia coli isolates. Infect Immun. 1988;56:640–648. doi: 10.1128/iai.56.3.640-648.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labigne-Roussel A F, Lark D, Schoolnik G, Falkow S. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect Immun. 1984;46:251–259. doi: 10.1128/iai.46.1.251-259.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Lalioui, L. Unpublished data.
- 33.Le Bouguénec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol. 1992;30:1189–1193. doi: 10.1128/jcm.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Bouguénec C, Garcia M I, Ouin V, Desperrier J M, Gounon P, Labigne A. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun. 1993;61:5106–5114. doi: 10.1128/iai.61.12.5106-5114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine M M, Ferreccio C, Prado V, Cayazzo M, Abrego P, Martinez J, Maggi L, Baldini M M, Martin W, Maneval D, Kay B, Guers L, Lior H, Wasserman S S, Nataro J P. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol. 1993;138:849–869. doi: 10.1093/oxfordjournals.aje.a116788. [DOI] [PubMed] [Google Scholar]
- 36.Loomis W P, Moseley S L. Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Mol Microbiol. 1998;30:843–853. doi: 10.1046/j.1365-2958.1998.01117.x. [DOI] [PubMed] [Google Scholar]
- 36a.Mainil, J. Personal communication.
- 37.Mainil J G, Jacquemin E, Hérault F, Oswald E. Presence of pap-, sfa- and afa-related sequences in necrotoxigenic Escherichia coli isolates from cattle: evidence for new variants of the AFA family. Can J Vet Res. 1997;61:193–199. [PMC free article] [PubMed] [Google Scholar]
- 38.Maiti S N, Harel J, Fairbrother J M. Structure and copy number analyses of pap-, sfa-, and afa-related gene clusters in F165-positive bovine and porcine Escherichia coli isolates. Infect Immun. 1993;61:2453–2461. doi: 10.1128/iai.61.6.2453-2461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maniatis T, Fritsh E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 40.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mengaud J, Ohayon H, Gounon P, Mege R M, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 42.Mohr G, Perlman P S, Lambowitz A M. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nataro J P, Yikang D, Giron J A, Savarino S J, Kothary M H, Hall R. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect Immun. 1993;61:1126–1131. doi: 10.1128/iai.61.3.1126-1131.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowicki B, Hart A, Coyne K E, Lublin D M, Nowicki S. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J Exp Med. 1993;178:2115–2121. doi: 10.1084/jem.178.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowicki B, Labigne A, Moseley S, Hull R, Hull S, Moulds J. The Dr hemagglutinin, afimbrial adhesins AFA-I and AFA-III, and F1845 fimbriae of uropathogenic and diarrhea-associated Escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect Immun. 1990;58:279–281. doi: 10.1128/iai.58.1.279-281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowicki B, Martens M, Hart A, Nowicki S. Gestational age-dependent distribution of Escherichia coli fimbriae in pregnant patients with pyelonephritis. Ann N Y Acad Sci. 1994;730:290–291. doi: 10.1111/j.1749-6632.1994.tb44268.x. [DOI] [PubMed] [Google Scholar]
- 47.Nowicki B, Svanborg-Eden C, Hull R, Hull S. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect Immun. 1989;57:446–451. doi: 10.1128/iai.57.2.446-451.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 49.Petzoldt J L, Leigh I M, Duffy P G, Sexton C, Masters J R. Immortalisation of human urothelial cells. Urol Res. 1995;23:377–380. doi: 10.1007/BF00698738. [DOI] [PubMed] [Google Scholar]
- 50.Pham T, Kaul A, Hart A, Goluszko P, Moulds J, Nowicki S, Lublin D M, Nowicki B J. dra-related X adhesins of gestational pyelonephritis associated Escherichia coli recognize SCR-3 and SCR-4 domains of recombinant decay-accelerating factor. Infect Immun. 1995;63:1663–1668. doi: 10.1128/iai.63.5.1663-1668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinto M, Robine-Léon S, Appay M-D, Kedinger M, Triadou N, Dussaulx E, Lacroix B, Simon-Assmann P, Haffen K, Fogh J, Zweibaum A. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line caco-2 in culture. Biol Cell. 1983;47:323–330. [Google Scholar]
- 52.Rhen M, Väisänen-Rhen V, Saraste M, Korhonen T K. Organization of genes expressing the blood-group-M-specific hemagglutinin of Escherichia coli: identification and nucleotide sequence of the M-agglutinin subunit gene. Gene. 1986;49:351–360. doi: 10.1016/0378-1119(86)90371-9. [DOI] [PubMed] [Google Scholar]
- 53.Savarino S J, Fasano A, Watson J, Martin B M, Levine M M, Guandalini S, Guerry P. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci USA. 1993;90:3093–3097. doi: 10.1073/pnas.90.7.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savarino S J, Fox P, Yikang D, Nataro J P. Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J Bacteriol. 1994;176:4949–4957. doi: 10.1128/jb.176.16.4949-4957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savarino S J, McVeigh A, Watson J, Cravioto A, Molina J, Echeverria P, Bhan M K, Levine M M, Fasano A. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J Infect Dis. 1996;173:1019–1022. doi: 10.1093/infdis/173.4.1019. [DOI] [PubMed] [Google Scholar]
- 56.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto T, Echeverria P. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect Immun. 1996;64:1441–1445. doi: 10.1128/iai.64.4.1441-1445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto T, Nakazawa M. Detection and sequences of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene in enterotoxigenic E. coli strains isolated from piglets and calves with diarrhea. J Clin Microbiol. 1997;35:223–227. doi: 10.1128/jcm.35.1.223-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, Foxman B, Tallman P, Cladera E, Le Bouguénec C, Marrs C F. Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coli isolates and identification of new subtypes. Infect Immun. 1997;65:2011–2018. doi: 10.1128/iai.65.6.2011-2018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]