Abstract
Glioblastoma is one of the most frequent primary brain tumors with a poor prognosis. Nevertheless, some patients show a prolonged survival. The aim of the present study was to compare the expression profiles of tumor derived microRNA (miR) of long-term survivors with those of short-term survivors in order to identify differentially expressed miRs as well as their target genes, which may elucidate mechanisms that play a role in varying tumor progression and, therefore, may influence survival. Formalin-fixed paraffin-embedded samples of 23 patients with glioblastoma were classified according to overall survival. Profiles of miR expression were determined using Nanostring technology. Expression levels of potential target genes of differentially expressed miRs were assessed using immunohistochemistry. MiR profiles of long-term survivors differed from those of short-term survivors. A total of three prominent differentially expressed miRs were highlighted: MiR-130b-3p, which is downregulated in long-term survivors, and miR-146b-5p and miR-148a-3p, which are upregulated in long-term survivors. Known tumor suppressor genes are among targets potentially affected by miR-130b-3p, whereas targets of miR-146b-5p and miR-148a-3p consist of several genes known to have a role in tumor invasion and aggressiveness. In conclusion, it was revealed that a type of miR-signature was associated with short- and long-term survival, potentially serving as biomarker for disease progression and providing a base for further functional studies.
Keywords: glioblastoma, overall survival, microRNAs, expression profile, molecular signature
Introduction
Glioblastoma (GBM) is the most frequent and fatal primary brain tumor. The common therapy regimen consisting of surgery, radiotherapy, and chemotherapy with Temozolomid (TMZ) (1,2) lack long lasting effects. The prognosis is dismal with a 5-year survival probability of 6.8% (varying with age at diagnosis) (3,4) which barely improved for decades (5), and a median overall survival (OS) of about 15–18 months after diagnosis which still remains at this low level despite several efforts (1–4).
Diverse therapeutic options are subject of actual or recent clinical studies, among them targeted approaches consisting of small-molecule kinase inhibitors (6–8) or antibodies (7), immunotherapy (9) or oncolytic viruses (10). But on the whole, the effects on OS remain rather low. Reasons for therapy failure include development of resistances caused by genetic heterogeneity or redundant signaling cascades, toxicity and other undesired side-effects, and low abundant drug availability on the tumor site due to inhibition by the blood-brain-barrier (6,7).
Nevertheless, some patients show a prolonged OS up to several years.
To date, several different factors, clinical and molecular, are discussed to cause or correlate with long-term survival (LTS). The possibility of gross tumor resection, a high Karnofsky performance status and a young age at diagnosis are considered as clinical factors for prolonged survival (11), also the location and whether the subventrical zone is affected or not might have an influence for progression and survival (12). Furthermore, for several patients, eligibility for a second surgery after tumor recurrence, is an option associated with prolonged OS (13).
On the molecular level, some expression signatures, mutations, chromosomal aberrations and MGMT methylation status are deemed to be factors influencing therapy response and survival (14–16), but no common profile indicating LTS is found for this highly heterogeneous tumor entity (17). In contrary, some biomarkers like MGMT methylation are controversially discussed for outcome prediction ability (18).
Although several studies describe potential molecular features in connection with survival or prognosis (14,15), studies analyzing microRNA (miR) profiles are quite rare and mostly not concordant, describing different sets of miRs to be differentially expressed and therefore considered as potential biomarkers for survival (19–23).
MiRs are short non-coding RNAs of ~22 nt length which derive from long primary transcripts which are matured in a multistep process. They influence the transcription and translation of targeted genes and are often deregulated in any kind of cancer (24), also in glioblastoma (25). Depending on the tissue, kind of dysregulation and affected targets, they have oncogenic and/or tumor suppressive character (26). They are considered as putative targets for directed therapies consisting of miR-mimics for upregulating repressed miRs, or anti-miRs or miR-sponges to suppress overexpressed miRs (27–29).
The aim of this study is to compare miR expression profiles of short-time and long-time survivors to determine, whether there are survival-associated signatures. miRs showing significant differential expression between the survival groups are deemed as candidates for further analysis regarding potential target genes and influence on tumor behavior. These findings could contribute to more knowledge and deeper insights on molecular features of this highly complex and heterogenous disease.
Identifying differential expressed miRs and their potential target genes can give clues to factors affecting OS, which, in turn, could be interesting candidates for developing specific therapeutic approaches.
Materials and methods
Patient samples
Formalin-fixed paraffin-embedded (FFPE) samples of GBM patients were retrieved from the archive of the Institute of Pathology of the University Medicine Rostock. Tissue was fixed in 4% neutral buffered formalin (Grimm) and incubated overnight at room temperature. Afterwards, it was embedded in molten paraffin (Merck).
Inclusion criteria were diagnosed GBM WHO Grade IV, sufficient availability of material and tumor content. For this study, the number of patient samples for array-based microRNA profile screening was limited, so from all potential available samples, initially 24 were arbitrarily selected to obtain a balanced distribution of sex and survival. One sample had to be removed afterwards due to issues of data quality and completeness, so a total of 23 patients were suitable for analysis.
Patient data (sex, age at diagnosis, OS, kind of surgery, therapeutic treatments) were obtained from the department of neurosurgery and patients were assigned to two groups: LTS and short-term survivor (STS) using the median OS of the cohort as discriminator. Molecular characteristics (EGFR amplification, mutation status of IDH1 and IDH2) were obtained by earlier projects and/or routine diagnostic procedures.
Specimen collection was conducted in accordance with the ethics guidelines for the use of human material, approved by the Ethics Committee of the University of Rostock (Reference no. A 2009/34) and with informed written consent from all patients prior to surgery.
MicroRNA extraction
For microRNA extraction from 10 µm FFPE sections, the miRNeasy FFPE-Kit (Qiagen) was used following the manufacturer's protocol. Concentration of extracted RNA was determined with a Nanodrop spectrometer (Peqlab).
miR screening arrays
Analysis of microRNA expression was performed using the Nanostring nCounter System with the Human v3 miRNA assay (Nanostring). The analysis procedure was performed with 250 ng per sample by a service provider lab (Transcriptome and Genome Analysis Laboratory (TAL), Microarray and Deep-Sequencing Facility, University Medicine Göttingen) and analyzed by the authors with the nSolver 4.0 software (Nanostring) using standard settings for background subtraction and housekeeping genes-based normalization. Group-wise comparison (LTS vs. STS) was performed, delivering fold change and p-values. Results were visualized as boxplots generated by BoxPlotR (http://shiny.chemgrid.org/boxplotr/).
Determination of miR-targets
For determination of potential target genes of miR-130b-3p, miR-146b-5p and miR-148a-3p, targeted literature research was performed looking up PubMed (https://pubmed.ncbi.nlm.nih.gov) for corresponding papers using search strings including the miR-name and ‘glioblastoma’. Furthermore, the online microRNA-target interaction database miRTarBase (http://mirtarbase.cuhk.edu.cn) was consulted for predicted potential targets of these three miRs.
Immunohistochemistry
Protein expression analysis for PTEN and TRAF6 was performed by immunohistochemistry using 2 µm thick FFPE sections on coated glass slides (Dako). For deparaffinization, rehydration and antigen demasking, slides were incubated for 20 min at 97°C and pH 9 in EnVision FLEX Target Retrieval Solution, high pH (pH 9) (Dako).
For slide processing, an automatic IHC system, AutostainerLink48 (Dako) was used, according to the following (routine) protocol. All steps were performed at room temperature. Slides were rinsed with EnVision FLEX Wash Buffer (Dako). To reduce unspecific background staining, slides were incubated for 5 min with 100 µl of EnVision FLEX Peroxidase-Blocking Reagent, readty-to-use (Dako), a phosphate buffer containing H2O2, 15 mMol NaN3 and detergent, and rinsed afterwards with wash buffer. For PTEN, a monoclonal (clone 6H2.1) mouse-anti human PTEN antibody (Dako, cat. no. M3627) was used, dilution 1:100 in EnVision FLEX Antibody Diluent (Dako). For TRAF6, a monoclonal (clone EP592Y) rabbit-anti human TRAF6 antibody (Abcam; cat. no. ab40675), dilution 1:50, was used. Slides were incubated with 100 µl of first antibodies for 20 min. and rinsed afterwards with wash buffer. For TRAF6, a signal enhancement step was inserted incubating slides for 15 min with 100 µl of EnVision FLEX+ Rabbit (LINKER), ready-to-use (Dako, cat. no. SM805). As secondary reagent, EnVision FLEX/HRP, ready-to-use (Dako, cat. no. SM802), was used, containing dextran coupled with peroxidase molecules and goat secondary antibodies against rabbit and mouse immunoglobulins. Slides were incubated with 100 µl thereof for 20 min and rinsed afterwards with wash buffer. As substrate working solution 1 drop EnVision FLEX DAB+ Chromogen (Dako, cat. no. DM827) was mixed with 1 ml EnVision FLEX Substrate Buffer (Dako, cat. no. SM803). 200 µl thereof were applied to each slide. Slides were incubated for 10 min and rinsed with wash buffer afterwards. Finally, for counter staining, 100 µl of EnVision FLEX Hematoxylin (Dako, cat. no. SM806) were applied and slides were incubated for 5 min. Slides were rinsed with deionized water, washed for 5 min with wash buffer and rinsed once more with water. Slides were covered after dehydration with ethanol and xylol with coverslips (epredia) and CV mount (Leica) as mounting medium using the automated Coverslipper (Dako).
Stained slides were scanned using a Pannoramic Desk DWII slide scanner (3DHistech) with Pannoramic Scanner 2.2.0 software (3DHistech). Visualization and analysis were performed using CaseViewer 2.4 software (3DHistech). For presentation, representative regions the magnification was set to 10×, and the chosen sections were exported as JPEG.
Statistical analyses
Significance of differential miR expression was determined using the nSolver Software 4.0 (Nanostring) applying a paired, two-tailed Student's t-test. Statistical analyses concerning OS and clinical data were performed using SPSS Statistics version 28 (IBM) using the Kaplan-Meier method. Significance was assumed for P≤0.05.
Results
Patients' characteristics
Samples of 23 GBM patients were finally included in this study (Table I). Median age at diagnosis was 59.96 years (47.36–80.82 years). Median OS was 380 days (30–2041 days). Patients with OS >median OS were classified as LTS, the others as STS. 11 patients were male, 12 were female. All but one were wild type for IDH1 R132 and 12 carried an EGFR amplification. Sex and EGFR amplification status were evenly distributed over both survival groups. 19 of 23 patients underwent gross-total resection and 4 had a subtotal resection. 22 patients received radiotherapy, for one patient it is unknown. 15 patients were treated with TMZ, 7 received no TMZ, for one patient it is unknown. Of the patients without TMZ therapy, five were grouped into the STS group and 2 into LTS. Regarding the clinical data, only for the type of surgery, a significant influence on OS could be observed (P=0.001). No significant differences in OS were seen for age at diagnosis (P=0.566), sex (P=0.927), EGFR status (P=0.437) and TMZ treatment (P=0.158) (Fig. S1, Table II).
Table I.
Characteristics of analyzed patients.
| Patient no. | Age at diagnosis (y) | OS (d) | Sex | IDH1 status | EGFR status | Surgery | Rtx | TMZ | Survival status |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 67.36 | 528 | M | Wt | Amp | Gross-total resection | Yes | Yes | LTS |
| 2 | 47.36 | 143 | F | Wt | Amp | Subtotal | Yes | Yes | STS |
| 3 | 47.72 | 427 | M | Wt | Amp | Gross-total resection | Yes | Yes | LTS |
| 4 | 73.00 | 297 | F | Wt | Amp | Gross-total resection | Yes | No | STS |
| 5 | 59.96 | 727 | M | Wt | Amp | Gross-total resection | Yes | Yes | LTS |
| 6 | 69.63 | 380 | F | Wt | Amp | Gross-total resection | Yes | Yes | STS |
| 7 | 50.05 | 2041 | M | Wt | Amp | Gross-total resection | Yes | Yes | LTS |
| 8 | 64.47 | 148 | M | Wt | Amp | Gross-total resection | Yes | Yes | STS |
| 9 | 76.46 | 883 | F | Wt | Amp | Gross-total resection | Yes | No | LTS |
| 10 | 69.35 | 148 | F | Wt | Amp | Gross-total resection | Yes | Yes | STS |
| 11 | 53.22 | 236 | M | Wt | Amp | Subtotal | N/A | N/A | STS |
| 12 | 57.18 | 687 | F | Wt | Norm | Gross-total resection | Yes | Yes | LTS |
| 13 | 59.75 | 357 | F | Wt | Norm | Gross-total resection | Yes | Yes | STS |
| 14 | 57.46 | 586 | F | Wt | Norm | Gross-total resection | Yes | Yes | LTS |
| 15 | 49.17 | 253 | M | Wt | Norm | Gross-total resection | Yes | Yes | STS |
| 16 | 77.23 | 777 | F | Wt | Norm | Gross-total resection | Yes | Yes | LTS |
| 17 | 53.63 | 129 | M | Wt | Norm | Gross-total resection | Yes | No | STS |
| 18 | 80.82 | 489 | F | Wt | Norm | Gross-total resection | Yes | Yes | LTS |
| 19 | 49.62 | 120 | F | Wt | Norm | Gross-total resection | Yes | No | STS |
| 20 | 66.29 | 55 | M | Wt | Norm | Subtotal | Yes | No | STS |
| 21 | 71.09 | 30 | M | R132H | Norm | Subtotal | Yes | No | STS |
| 22 | 70.50 | 397 | F | Wt | Amp | Gross-total resection | Yes | No | LTS |
| 23 | 56.74 | 871 | M | Wt | Norm | Gross-total resection | Yes | Yes | LTS |
y, years; OS, overall survival; d, days; wt, wild type; R132H, IDH1 R132H mutation; norm, normal copy number; amp, amplification; STS, short-term survivor; LTS, long-term survivor; Rtx, radiotherapy.
Table II.
Overview of clinical parameters and their influence on overall survival.
| Clinical parameter | Variables, n (%) | Influence on OS (P-value) |
|---|---|---|
| Age at diagnosis, years | 0.566 | |
| <60 | 11 (47.83%) | |
| >60 | 12 (52.17%) | |
| Median age, years | 59.96 | |
| Age range, years | 47.36-80.82 | |
| Sex | 0.927 | |
| Female | 12 (52.17%) | |
| Male | 11 (47.83%) | |
| EGFR status | 0.437 | |
| EGRF normal | 11 (52.17%) | |
| EGFR amplified | 12 (47.83%) | |
| IDH1/2 status | ND | |
| Wild-type | 22 (95.65%) | |
| Mutated | 1 (4.35%) | |
| Tmz | 0.158 | |
| Yes | 15 (65.2%) | |
| No | 7 (30.4 %) | |
| Unknown: | 1 (4.35%) | |
| Radiotherapy | ND | |
| Yes | 22 (95.65%) | |
| Unknown | 1 (4.35%) | |
| Type of surgery | 0.001 | |
| Gross-total | 19 (82.6%) | |
| Subtotal | 4 (17.4%) |
OS, overall survival; ND, not determined; TMZ, temozolomid; IDH1/2 status, mutation status of IDH1 and IDH2 genes.
miR expression profiling
The miR-expression profiling by the Nanostring nCounter software revealed on the whole just moderate changes in miR expression comparing profiles of LTS and STS (Table SI) for most of the miRs analyzed. But some miRs outperformed the moderate fold change values, rendering them as potential candidates as significant targets. Finally, three miRs were choosen for further analysis: miR-130b-3p, downregulated in LTS, miR-146b-5p and miR-148a-5p, both upregulated in LTS (Table III, Fig. 1). All three of them fulfilled the requirements for being further considered: a top fold change value with high significance, and robust absolute expression values (normalized counts) in both groups. MiRs showing high, significant fold change values, but only very low counts, weren't considered further.
Table III.
Top 10 significantly differentially expressed miRs showing fold-change values of the LTS group (‘true’) compared with the STS group (‘false’).
| Probe name | Accession no. | Fold-change LTS vs. STS | P-value |
|---|---|---|---|
| hsa-miR-146b-5p | MIMAT0002809 | 2.53 | 0.002 |
| hsa-miR-130b-3p | MIMAT0000691 | −2.72 | 0.007 |
| hsa-miR-148a-3p | MIMAT0000243 | 3.20 | 0.007 |
| hsa-miR-302b-3p | MIMAT0000715 | 1.78 | 0.011 |
| hsa-miR-3065-5p | MIMAT0015066 | 3.90 | 0.012 |
| hsa-miR-301b-3p | MIMAT0004958 | −3.17 | 0.012 |
| hsa-miR-887-3p | MIMAT0004951 | 2.24 | 0.013 |
| hsa-miR-23a-3p | MIMAT0000078 | 1.94 | 0.017 |
| hsa-miR-21-5p | MIMAT0000076 | 1.79 | 0.019 |
| hsa-miR-142-3p | MIMAT0000434 | 2.02 | 0.019 |
Fold-change data of all miRs analyzed are presented in Table SI. STS, short-term survivor; LTS, long-term survivor; miR, microRNA.
Figure 1.
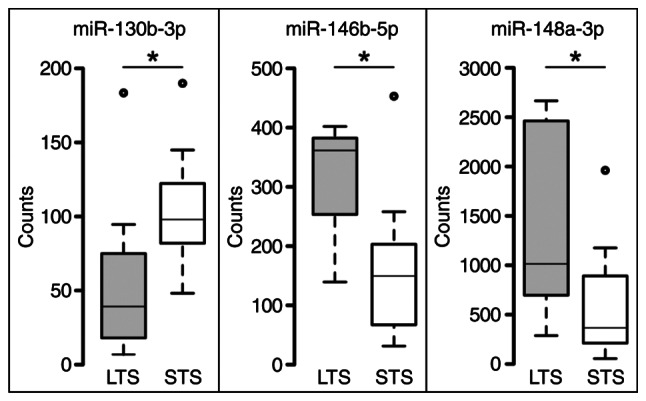
Boxplot representation of normalized expression values of miR-130b-3p, miR-146b-5p and miR-148a-3p, comparing LTS and STS groups. Center lines indicate medians, box limits indicate the 25 and 75th percentiles, whiskers extend 1.5× interquartile range from percentiles and dots represent outliers (generated using BoxPlotR; http://shiny.chemgrid.org/boxplotr/). *P<0.05. miR, microRNA; STS, short-term survivor; LTS, long-term survivor.
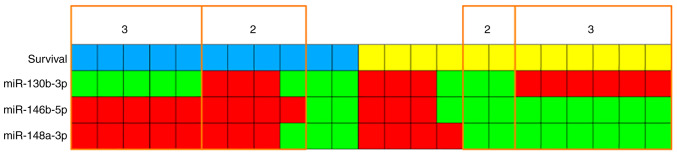
Although there are overlaps of expression values between the two survival groups (Fig. 1), a kind of signature can be seen: low miR-130b-3p expression and high expression of miR-146b-5p and miR-148a-3p for LTS; and vice versa for STS. High and low expression states are separated by the median expression values. Correlating survival and expression status for each case shows that 11/23 cases (5 LTS and 6 STS) completely correspond to this signature and another 6 cases (4 LTS and 2 STS) in 2 of 3 miRs. Another 6 cases (2 LTS and 4 STS) show just one miR but no case shows none (Fig. 2).
Figure 2.
Visualization of the signature model miR-130b-3p low, miR-146b-5p high, miR-148a-3p high or vice versa. Each column represents a patient, and the number indicates for how many of the three miRs the model statement is true. Blue=long-term survivor; yellow=short-term survivor; green=low expression; red=high expression. miR, microRNA.
Determination of potential target genes
Targeted PubMed search and specific investigation of miRTarBase revealed several genes proposed to be potential targets of miRs 130b-3p, 146b-5p and 148a-3p (Table IV). These two following were chosen to start further analyses with: PTEN as target of miR-130b-3p and important TSG in GBM, and TRAF6 as target of miR-146b-5b, an invasion promoting factor. For miR-148a-3p the situation is more complex, as it is described to function either as oncogene or as TSG in GBM, so due to its ambivalent role, target studies were delayed.
Table IV.
Literature and database predicted target genes for miR-130b-3p, miR-146b-5p and miR 148a-3p.
| miR | Literature | miRTarBase |
|---|---|---|
| miR-130b-3p | PTEN (31); PPARγ (30); MST, SAV1 (32) | RUNX3, ZEB1, IGF1, TP53INP1 |
| miR-146b-5p | TRAF6 (40,41); MMP16 (37); EGFR (34) | TRAF6, IRAK1, MMP16, KIT, EGFR |
| miR-148a-3p | ITGA9 (49); DLGAP1 (46); MIG, BIM (52) | DNMT3B, DNMT1, IKBKB, MMP7 |
miR, microRNA.
IHC analysis of PTEN and TRAF6
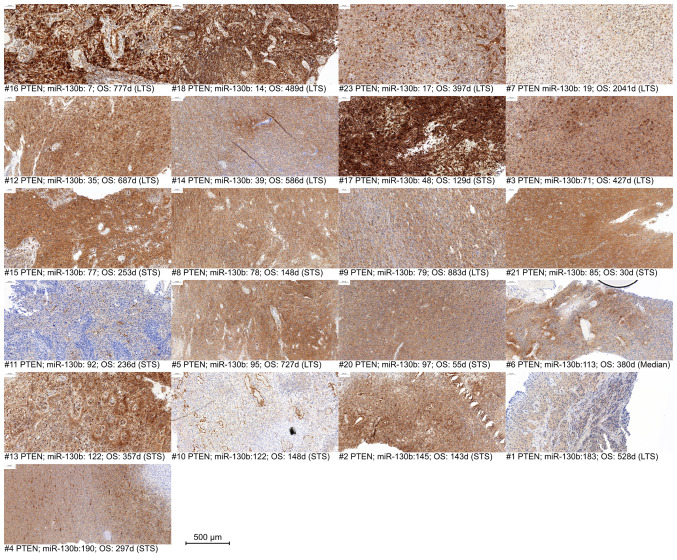
Immunohistochemistry of PTEN and TRAF6 was successful in 22 of 23 cases. Although IHC is not considered as a quantitative method, it is observable, that a partial loss or decrease of PTEN expression occurs in some cases, mostly in samples with a higher miR-130b expression (Fig. 3), especially samples #11, #10 and #1.
Figure 3.
PTEN immunohistochemistry, sorted, from left to right, in an ascending order of miR-130b-3p counts days in patients #1 to 23. miR, microRNA; OS, overall survival; STS, short-term survivor; LTS, long-term survivor; d, days.
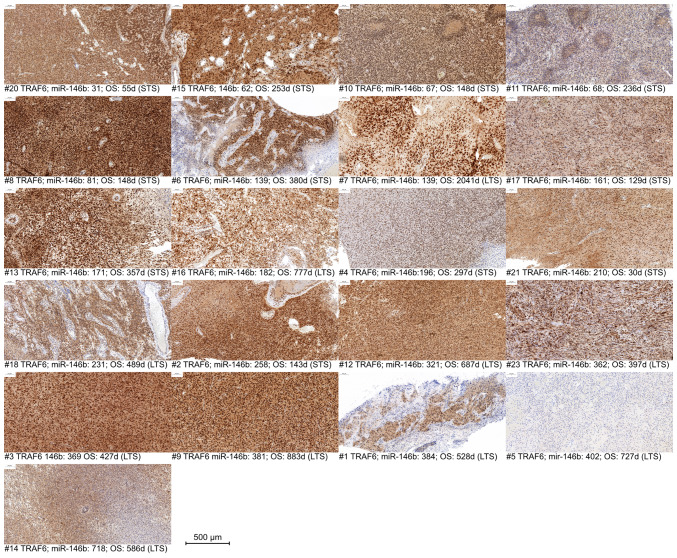
Similarly, for TRAF6, a reduction or partial loss in expression is observable predominantly in samples with higher miR-146b expression (Fig. 4), especially samples #4, #1 and #5.
Figure 4.
TRAF6 immunohistochemistry, sorted, from left to right, in an ascending order of miR-146b-5p counts in patients #1 to 23. miR, microRNA; OS, overall survival; STS, short-term survivor; LTS, long-term survivor; d, days; TRAF6, TNF receptor-associated factor 6.
In summary, PTEN and TRAF6 could be considered as putative targets of miR-130b and miR-146b, respectively, at least in cases with higher miR expression levels.
Discussion
This study shows a significant differential miR expression profile comparing LTS and STS, highlighting miR-130b-3p, miR-146b-5p and miR-148a-3p as most significantly differentially expressed miRs. As the definition of LTS is non-uniform throughout the literature, in our study, the median OS of the cohort (380 days) was used as delimiter to split the cohort in two more or less even groups. These data give the indication, that miR-130b-3p acts as oncomir, while miR-146b-5p and miR-148a-3p have tumor suppressor properties.
miR-130b is shown to be overexpressed in glioblastoma in contrast to non-neoplastic brain tissue, as well as in other tumor entities, and promotes cell proliferation by inhibiting PPARγ, leading to decreased E-cadherin levels (30). Furthermore, it was shown, that suppression of miR-130b inhibits GBM cell proliferation and invasion and induces apoptosis via the PTEN/AKT signaling pathway. In this context, PTEN was shown to be a direct target of miR-130b (31). Another study shows more oncogenic properties of miR-130b, demonstrating that its upregulation enhances a stem cell-like phenotype by inactivating the Hippo signaling pathway with MST1/2 and SAV1 as direct targets (32). Earlier, miR-130b expression was associated with progression from lower to high grade glioma (33). Together with these, our data, showing a higher expression in STS samples, underpin the oncogenic character of miR-130b.
miR-146b-5p is linked with tumor suppressive properties. It is shown to target EGFR and hence, at least in vitro, reduces invasion and migration of glioma cells (34). In the cohort analyzed in our study, immunohistochemistry showed, that the EGFR expression was only dependent on the EGFR amplification status, but not on the survival group (data not shown). But for in vitro models it is shown that EGFR amplification is often lost during cultivation (35), except for special cell culture conditions (36). Another target deemed to be regulated by miR-146b-5p is MMP16, whose inhibition leads to decreased migration and invasion (37,38). MMP16 is a member of matrix metalloproteases, important factors for migration and metastasis as they contribute to proteolysis of the extracellular matrix, enabling tumor cell migration (39). TRAF6 is another direct target of miR-146b-5p whose repression inhibits proliferation and progression, while promoting apoptosis. It is associated with a better prognosis (40). Furthermore, miR-146b-5p mediated TRAF6 repression is considered to suppress TMZ resistance (41). TRAF6 is correlated with a worse prognosis and oncogenic properties promoting invasion by upregulating another matrix-metalloprotease, MMP9 (42). In context with our data these studies underpin the correlation of miR-146-5p, TRAF6 expression and survival. Furthermore, miR-146b-5p inhibits GSCs and radioresistance by targeting SMARCA5 (43) and the β-catenin pathway (44). Although most studies attest miR-146b-5p tumor suppressive properties, some oncogenic potential cannot be totally excluded, as its upregulation is also associated with GBM recurrence (45). But the data acquired in our own study are rather concordant with the described tumor suppressive properties.
For miR-148a, its role in GBM seems to be ambivalent, according to the current literature. Several studies nominate different targets whose repression depict the oncogenic character of miR-148a, where others describe tumor suppressive properties. Repression of DLGAP1 leads to loss of cell polarity, therefore promoting growth, migration, invasion and EMT (46). Overexpression of miR-148a deemed to be induced by NFκB and targeting QKI and SKP1 activates TGFβ signaling and is associated with progression and augmented tumor aggressiveness (47). In a TCGA based study, it is shown that miR-148a targets FIH1, an HIF1 inhibitor and therefore promotes HIF1a and NOTCH1 signaling, enhancing vascularization, growth and survival (48). But also the opposite effect on vascularization is described, as miR-148a has shown to have tumor suppressive properties targeting ITGA9 (49), a cell adhesion factor involved in NOTCH1 controlled vascularization, leading to decreased angiogenesis (50). More oncogenic properties are demonstrated with targets like GADD45A, whose repression stimulates β-catenin and MMP9, promoting invasion, migration and stemness (51). Also positively influencing EGFR activity by targeting MIG2 and inhibiting apoptosis by targeting BIM is described (52). Even exosomal delivery of miR-148a seems to have a positive effect to proliferation and progression by targeting CADM1, leading to STAT3 activation (53). On the other side, more tumor suppressive properties are known. miR-148a targets ROCK-1, a factor promoting migration in several tumors (54). Even therapeutic effects are described, as in vivo experiments in mice show a prolonged survival when miR-148a is co-delivered with miR-296-5p by nanoparticles, targeting OCT4 and SOX2, reducing stemness (29). Although studies describing oncogenic properties of miR-148a in GBM predominate, our own data show, that it is higher expressed in LTS, rendering it more to the tumor suppressive side.
Considering the exemplarily selected targets PTEN and TRAF6, the impression is not that clear. The expression of these targets was assessed by IHC. This method is not considered as quantitative and not sensitive enough to identify minor changes in expression which is characteristic for miR-mediated regulation. Also, there are much more factors influencing genes' translational activity. But the data show, that at least at the upper ranges of miR expression, a decrease in target gene expression is observable, but there's no linearity.
To elucidate the downstream effects of differential miR expression, identify the real target genes in these tumors, and observe the effects on tumor cell behavior, functional approaches with GBM in vitro models using transfection of miR mimics or antagomiRs, followed by analyses on different levels are needed.
This study is limited by a quite low number of patients analyzed and a small selection of potential target genes. To improve the value of robustness of the data, a consecutive study based on the present data should include a higher number of patients. Furthermore, next to functional studies, the selection of potential target genes should be extended to elucidate more details of the molecular mechanisms of tumor behavior underlying the outcome. Also, the study is carried out on FFPE material, which offers lesser quality and opportunities for deeper analysis. Fresh material would serve as a source better quality nucleic acids and proteins for deeper molecular analysis, and would even offer the opportunity to be taken in culture to provide cell lines or xenografts for in vitro or in vivo studies.
In summary, this study shows a significant correlation of differential miR-expression and survival status. It emphasizes the role of regulatory components/epigenetic factors, in this case microRNAs, within the complex interactions determining tumor behavior and outcome. These results hopefully help to clarify the so far widely unconcordant approaches to determine outcome predicting biomarkers. Although the regulatory impact to the few selected, potential miR target genes is rather low, the connection between miR expression and survival is significant. These data provide a good basis for further, functional studies.
Supplementary Material
Acknowledgements
The authors would like to thank the technicians of the Institute of Pathology (University Medicine Rostock, Rostock, Germany), Mrs. Kerstin Westphal, Mrs. Heike Clasen and Mrs. Beate Krause for performing immunohistochemistry, and Mrs. Susanne Höffer for the sections.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BS designed the study, acquired, analyzed and interpreted data and wrote the manuscript. NL analyzed and interpreted immunohistochemistry data. AZ analyzed and interpreted immunohistochemistry data, contributed to the statistical analyses and critically edited the manuscript. CH acquired the patient data and substantially contributed to their analyses and interpretation, and critically edited the manuscript. AE provided patient material, substantially contributed to the concept of the study, and critically contributed to the manuscript. BS and AZ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the University of Rostock (approval no. A2009/34). Informed consent was obtained from all subjects involved in the study.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Authors' information
Dr Björn Schneider, ORCID-ID: 0000-0002-0282-7330.
References
- 1.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Wen PY, Weller M, Lee EQ, Alexander BM, Barnholtz-Sloan JS, Barthel FP, Batchelor TT, Bindra RS, Chang SM, Chiocca EA, et al. Glioblastoma in adults: A society for neuro-oncology (SNO) and european society of neuro-oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22:1073–1113. doi: 10.1093/neuonc/noaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21((Suppl 5)):v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller KD, Ostrom QT, Kruchko C, Patil N, Tihan T, Cioffi G, Fuchs HE, Waite KA, Jemal A, Siegel RL, Barnholtz-Sloan JS. Brain and other central nervous system tumor statistics, 2021. CA Cancer J Clin. 2021;71:381–406. doi: 10.3322/caac.21693. [DOI] [PubMed] [Google Scholar]
- 6.Heffron TP. Challenges of developing small-molecule kinase inhibitors for brain tumors and the need for emphasis on free drug levels. Neuro Oncol. 2018;20:307–312. doi: 10.1093/neuonc/nox179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Rhun E, Preusser M, Roth P, Reardon DA, van den Bent M, Wen P, Reifenberger G, Weller M. Molecular targeted therapy of glioblastoma. Cancer Treat Rev. 2019;80:101896. doi: 10.1016/j.ctrv.2019.101896. [DOI] [PubMed] [Google Scholar]
- 8.Perryman R, Renziehausen A, Shaye H, Kostagianni AD, Tsiailanis AD, Thorne T, Chatziathanasiadou MV, Sivolapenko GB, El Mubarak MA, Han GW, et al. Inhibition of the angiotensin II type 2 receptor AT2R is a novel therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2022;119:e2116289119. doi: 10.1073/pnas.2116289119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao TQ, Wainwright DA, Lee-Chang C, Miska J, Sonabend AM, Heimberger AB, Lukas RV. Next steps for immunotherapy in glioblastoma. Cancers (Basel) 2022;14:4023. doi: 10.3390/cancers14164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todo T, Ito H, Ino Y, Ohtsu H, Ota Y, Shibahara J, Tanaka M. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: A phase 2 trial. Nat Med. 2022;28:1630–1639. doi: 10.1038/s41591-022-01897-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anselmo P, Maranzano E, Selimi A, Lupattelli M, Palumbo I, Bini V, Casale M, Trippa F, Bufi A, Arcidiacono F, Aristei C. Clinical characterization of glioblastoma patients living longer than 2 years: A retrospective analysis of two Italian institutions. Asia Pac J Clin Oncol. 2020;17:273–279. doi: 10.1111/ajco.13457. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Yu K, Li M, Cui Y, Ren X, Yang C, Zhao X, Lin S. Classification of progression patterns in glioblastoma: Analysis of predictive factors and clinical implications. Front Oncol. 2020;10:590648. doi: 10.3389/fonc.2020.590648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualetti F, Montemurro N, Desideri I, Loi M, Giannini N, Gadducci G, Malfatti G, Cantarella M, Gonnelli A, Montrone S, et al. Impact of recurrence pattern in patients undergoing a second surgery for recurrent glioblastoma. Acta Neurol Belg. 2022;122:441–446. doi: 10.1007/s13760-021-01765-4. [DOI] [PubMed] [Google Scholar]
- 14.Jovčevska I. Genetic secrets of long-term glioblastoma survivors. Bosn J Basic Med Sci. 2019;19:116–124. doi: 10.17305/bjbms.2018.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gately L, McLachlan SA, Philip J, Rathi V, Dowling A. Molecular profile of long-term survivors of glioblastoma: A scoping review of the literature. J Clin Neurosci. 2019;68:1–8. doi: 10.1016/j.jocn.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Lei CG, Jia XY, Sun WJ. Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database. Yi Chuan. 2021;43:665–679. doi: 10.16288/j.yczz.20-428. [DOI] [PubMed] [Google Scholar]
- 17.Richardson TE, Kumar A, Xing C, Hatanpaa KJ, Walker JM. Overcoming the odds: Toward a molecular profile of long-term survival in glioblastoma. J Neuropathol Exp Neurol. 2020;79:1031–1037. doi: 10.1093/jnen/nlaa102. [DOI] [PubMed] [Google Scholar]
- 18.Butler M, Pongor L, Su YT, Xi L, Raffeld M, Quezado M, Trepel J, Aldape K, Pommier Y, Wu J. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer. 2020;6:380–391. doi: 10.1016/j.trecan.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriksen M, Johnsen KB, Andersen HH, Pilgaard L, Duroux M. MicroRNA expression signatures determine prognosis and survival in glioblastoma multiforme-a systematic overview. Mol Neurobiol. 2014;50:896–913. doi: 10.1007/s12035-014-8668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Zhang J, Yan W, You G, Bao Z, Li S, Kang C, Jiang C, You Y, Zhang Y, et al. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer. 2013;119:814–824. doi: 10.1002/cncr.27826. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasan S, Patric IRP, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niyazi M, Zehentmayr F, Niemöller OM, Eigenbrod S, Kretzschmar H, Schulze-Osthoff K, Tonn JC, Atkinson M, Mörtl S, Belka C. MiRNA expression patterns predict survival in glioblastoma. Radiat Oncol. 2011;6:153. doi: 10.1186/1748-717X-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Areeb Z, Stylli SS, Koldej R, Ritchie DS, Siegal T, Morokoff AP, Kaye AH, Luwor RB. MicroRNA as potential biomarkers in Glioblastoma. J Neurooncol. 2015;125:237–248. doi: 10.1007/s11060-015-1912-0. [DOI] [PubMed] [Google Scholar]
- 24.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolle K. miRNA Multiplayers in glioma. From bench to bedside. Acta Biochim Pol. 2015;62:353–365. doi: 10.18388/abp.2015_1072. [DOI] [PubMed] [Google Scholar]
- 26.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Kang C. miRNA interventions serve as ‘magic bullets’ in the reversal of glioblastoma hallmarks. Oncotarget. 2015;6:38628–38642. doi: 10.18632/oncotarget.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ananta JS, Paulmurugan R, Massoud TF. Tailored nanoparticle codelivery of antimiR-21 and antimiR-10b augments Glioblastoma Cell Kill by Temozolomide: Toward a ‘personalized’ anti-microRNA therapy. Mol Pharm. 2016;13:3164–3175. doi: 10.1021/acs.molpharmaceut.6b00388. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Bertoni H, Kozielski KL, Rui Y, Lal B, Vaughan H, Wilson DR, Mihelson N, Eberhart CG, Laterra J, Green JJ. Bioreducible polymeric nanoparticles containing multiplexed cancer stem cell regulating miRNAs inhibit Glioblastoma growth and prolong survival. Nano Lett. 2018;18:4086–4094. doi: 10.1021/acs.nanolett.8b00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu JJ, Zhang JH, Chen HJ, Wang SS. MicroRNA-130b promotes cell proliferation and invasion by inhibiting peroxisome proliferator-activated receptor-γ in human glioma cells. Int J Mol Med. 2016;37:1587–1593. doi: 10.3892/ijmm.2016.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu JJ, Fan KC, Zhang JH, Chen HJ, Wang SS. Suppression of microRNA-130b inhibits glioma cell proliferation and invasion, and induces apoptosis by PTEN/AKT signaling. Int J Mol Med. 2018;41:284–292. doi: 10.3892/ijmm.2017.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu G, Wang Y, Mijiti M, Wang Z, Wu PF, Jiafu D. Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem Biophys Res Commun. 2015;465:194–199. doi: 10.1016/j.bbrc.2015.07.149. [DOI] [PubMed] [Google Scholar]
- 33.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katakowski M, Zheng X, Jiang F, Rogers T, Szalad A, Chopp M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest. 2010;28:1024–1030. doi: 10.3109/07357907.2010.512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 36.William D, Mokri P, Lamp N, Linnebacher M, Classen CF, Erbersdobler A, Schneider B. Amplification of the EGFR gene can be maintained and modulated by variation of EGF concentrations in in vitro models of glioblastoma multiforme. PLoS One. 2017;12:e0185208. doi: 10.1371/journal.pone.0185208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S, Wang Q, Yan Y, Kang C, Jin S, et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 2013;339:260–269. doi: 10.1016/j.canlet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 38.Le Zhang, Wang J, Fu Z, Ai Y, Li Y, Wang Y, Wang Y. Sevoflurane suppresses migration and invasion of glioma cells by regulating miR-146b-5p and MMP16. Artif Cells Nanomed Biotechnol. 2019;47:3306–3314. doi: 10.1080/21691401.2019.1648282. [DOI] [PubMed] [Google Scholar]
- 39.Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu Rev Cell Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi C, Zhou X, Bian X, Ping Y, et al. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget. 2015;6:29129–29142. doi: 10.18632/oncotarget.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian Z, Zhou S, Zhou Z, Yang X, Que S, Lan J, Qiu Y, Lin Y. miR-146b-5p suppresses glioblastoma cell resistance to temozolomide through targeting TRAF6. Oncol Rep. 2017;38:2941–2950. doi: 10.3892/or.2017.5970. [DOI] [PubMed] [Google Scholar]
- 42.Sun J, Zhao B, Du K, Liu P. TRAF6 correlated to invasion and poor prognosis of glioblastoma via elevating MMP9 expression. Neuroreport. 2019;30:127–133. doi: 10.1097/WNR.0000000000001171. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Tan L, Dong X, Liu L, Jiang Q, Li H, Shi J, Yang X, Dai X, Qian Z, Dong J. MiR-146b-5p suppresses the malignancy of GSC/MSC fusion cells by targeting SMARCA5. Aging. 2020;12:13647–13667. doi: 10.18632/aging.103489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W, Yu H, Shen Y, Liu Y, Yang Z, Sun T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/β-catenin pathway. Oncotarget. 2016;7:41505–41526. doi: 10.18632/oncotarget.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khwaja SS, Cai C, Badiyan SN, Wang X, Huang J. The immune-related microRNA miR-146b is upregulated in glioblastoma recurrence. Oncotarget. 2018;9:29036–29046. doi: 10.18632/oncotarget.25528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Li W, Zeng X, Tang X, Zhang S, Zhong F, Peng X, Zhong Y, Rosol TJ, Deng X, et al. The role of microRNA-148a and downstream DLGAP1 on the molecular regulation and tumor progression on human glioblastoma. Oncogene. 2019;38:7234–7248. doi: 10.1038/s41388-019-0922-3. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Pan JQ, Luo L, Ning XJ, Ye ZP, Yu Z, Li WS. NF-κB induces miR-148a to sustain TGF-β/Smad signaling activation in glioblastoma. Mol Cancer. 2015;14:2. doi: 10.1186/1476-4598-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong HA, Fatimy RE, Onodera C, Wei Z, Yi M, Mohan A, Gowrisankaran S, Karmali P, Marcusson E, Wakimoto H, et al. The cancer genome atlas analysis predicts microRNA for targeting cancer growth and vascularization in Glioblastoma. Mol Ther. 2015;23:1234–1247. doi: 10.1038/mt.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu TJ, Qiu P, Zhang YB, Yu SY, Xu GM, Yang W. MiR-148a inhibits the proliferation and migration of glioblastoma by targeting ITGA9. Hum Cell. 2019;32:548–556. doi: 10.1007/s13577-019-00279-9. [DOI] [PubMed] [Google Scholar]
- 50.Guichet P-O, Guelfi S, Teigell M, Hoppe L, Bakalara N, Bauchet L, Duffau H, Lamszus K, Rothhut B, Hugnot JP. Notch1 stimulation induces a vascularization switch with pericyte-like cell differentiation of glioblastoma stem cells. Stem Cells. 2015;33:21–34. doi: 10.1002/stem.1767. [DOI] [PubMed] [Google Scholar]
- 51.Cui D, Sajan P, Shi J, Shen Y, Wang K, Deng X, Zhou L, Hu P, Gao L. MiR-148a increases glioma cell migration and invasion by downregulating GADD45A in human gliomas with IDH1 R132H mutations. Oncotarget. 2017;8:25345–25361. doi: 10.18632/oncotarget.15867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Zhang Y, Skalski M, Hayes J, Kefas B, Schiff D, Purow B, Parsons S, Lawler S, Abounader R. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 2014;74:1541–1553. doi: 10.1158/0008-5472.CAN-13-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai Q, Zhu A, Gong L. Exosomes of glioma cells deliver miR-148a to promote proliferation and metastasis of glioblastoma via targeting CADM1. Bull Cancer. 2018;105:643–651. doi: 10.1016/j.bulcan.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Fang Z, Weng Y, Xiao F, Yu J. LncRNA RP11-390F4.3 inhibits invasion and migration of glioblastoma cells by downregulating ROCK1. Neuroreport. 2021;32:888–893. doi: 10.1097/WNR.0000000000001676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.