Abstract
The znuA gene of Haemophilus ducreyi encodes a 32-kDa (mature) protein that has homology to both the ZnuA protein of Escherichia coli and the Pzp1 protein of H. influenzae; both of these latter proteins are members of a growing family of prokaryotic zinc transporters. Inactivation of the H. ducreyi 35000 znuA gene by insertional mutagenesis resulted in a mutant that grew more slowly than the wild-type parent strain in vitro unless ZnCl2 was provided at a final concentration of 100 μM. Other cations tested did not restore growth of this H. ducreyi mutant to wild-type levels. The H. ducreyi ZnuA protein was localized to the periplasm, where it is believed to function as the binding component of a zinc transport system. Complementation of the znuA mutation with the wild-type H. ducreyi znuA gene provided in trans restored the ability of this H. ducreyi mutant to grow normally in the absence of exogenously added ZnCl2. The wild-type H. ducreyi znuA gene was also able to complement a H. influenzae pzp1 mutation. The H. ducreyi znuA isogenic mutant exhibited significantly decreased virulence (P = 0.0001) when tested in the temperature-dependent rabbit model for experimental chancroid. This decreased virulence was not observed when the znuA mutant was complemented with the wild-type H. ducreyi znuA gene provided in trans.
Chancroid is one of the most prevalent sexually transmitted diseases and a major cause of morbidity in the resource-poor countries of Asia, Africa, and Latin America (53). This disease remains relatively uncommon in the United States and Western Europe (58). Prospective longitudinal and cross-sectional case control studies in Africa have provided substantial evidence that genital ulcer disease, either as a clinical syndrome or as an etiological diagnosis, is a significant risk factor for the heterosexual spread of the human immunodeficiency virus type 1 (HIV-1) even when sexual behavior is controlled for in the statistical analyses (11, 24, 49). In view of the epidemiological synergy that exists between chancroid and HIV-1 infection (63), there has been renewed research effort to elucidate the pathogenic mechanisms and virulence factors of Haemophilus ducreyi.
Several potential H. ducreyi virulence factors have been reported in the literature, and these include lipo-oligosaccharide (6, 13, 25, 59), a novel pilus (10), a soluble cytolethal distending toxin (16, 17), a hemoglobin-binding outer membrane protein (20, 57), a hemolysin capable of cytotoxicity involving direct contact between the bacterium and the eukaryotic cell (2, 45), and a copper-zinc superoxide dismutase which protects H. ducreyi from exogenous superoxide (55). In addition, gene products which may be involved in regulating the expression of H. ducreyi virulence factors have been described (14, 46). In contrast, much less is known about H. ducreyi gene products involved in the acquisition of essential nutrients and trace elements from the host environment.
Zinc is essential for all organisms due to its various physiological roles, which include maintaining both the structural stability and catalytic activity of several proteins (60), as well as stabilizing motifs in some transcriptional regulatory proteins (15). Zinc, at high concentration, is inherently toxic through inhibition of the aerobic respiratory chain (8, 34), and bacterial cells must regulate their zinc content over a small concentration range. Three factors have made study of zinc transport in bacteria difficult, including the extremely low bacterial requirement for zinc, the nonspecific binding of zinc to bacterial surfaces, and the rapid exchange of zinc between intracellular and extracellular compartments. An export system for cations, including zinc, was recently identified in Escherichia coli (7, 51), and a putative zinc uptake system (adcABC) was found to be essential for genetic competence in Streptococcus pneumoniae (18). The Pzp1 protein of H. influenzae was the first specific bacterial zinc transporter to be identified at the biochemical level (39). More recently, Patzer and Hantke (48) described the znuABC high-affinity zinc uptake system in E. coli.
In the present study, an H. ducreyi gene (znuA) encoding a zinc transport protein was identified and shown to complement a mutation in the equivalent H. influenzae pzp1 gene. Inactivation of the H. ducreyi 35000 znuA gene resulted in a mutant exhibiting decreased in vitro growth rates in broth compared to the wild-type parent strain. This growth defect was overcome by the addition of 100 μM ZnCl2. In addition, the isogenic znuA mutant exhibited significantly reduced virulence in the temperature-dependent rabbit model for experimental chancroid.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. H. ducreyi strains were routinely cultivated on chocolate agar (CA) plates containing GC agar base (36 g/liter) (Difco, Detroit, Mich.), bovine hemoglobin (20 g/liter) (Sigma, St. Louis, Mo.), 1% (vol/vol) IsoVitalex (BBL Microbiological Systems, Cockeysville, Md.) supplemented with 100 μM ZnCl2 (Zn). GC-heme agar (56) supplemented with kanamycin (30 μg/ml) was used to select H. ducreyi transformants after electroporation. Nontypeable H. influenzae (NTHI) strains were grown on CA-Zn plates or on BHIs plates (brain heart infusion [BHI; 37 g/liter] [Difco], Bacto Agar [15 g/liter] [Difco], 5% [vol/vol] Levinthal’s base [1]) with or without 100 μM ZnCl2 (Zn). E. coli strains were grown on Luria-Bertani medium (LB) (54). For antimicrobial supplementation, kanamycin (Sigma) was used at 30 μg/ml, and chloramphenicol (Sigma) was used at 2 μg/ml (for Haemophilus species) or 30 μg/ml (for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Host strain for cloning experiments | 54 |
| HB101 | Host strain essential for propagating plasmids carrying mutated H. ducreyi DNA inserts used in electroporation | 6, 54 |
| XL1-Blue | Host strain used for screening of the ZAP Express bacteriophage library | Stratagene |
| XLOLR | Host strain used for excision of recombinant pBK-CMV plasmids from the ZAP Express bacteriophage | Stratagene |
| H. influenzae | ||
| NTHI 6564 | Wild-type parent strain | 39 |
| NTHI 6564 pzp1 mutant | pzp1 mutant of NTHI 6564 | 39 |
| NTHI 6564.pzp1(pCW170) | NTHI 6564 pzp1 mutant containing the vector pCW170 | This study |
| NTHI 6564.pzp1(pDL6-2) | NTHI 6564 pzp1 mutant containing pDL6-2 with the H. ducreyi znuA gene | This study |
| H. ducreyi | ||
| 35000 | Wild-type strain isolated in Winnipeg, Manitoba, Canada | 29 |
| 35000.901 | Isogenic mutant with a kan2 cartridge inserted into the MscI site in the znuA gene | This study |
| 35000.901(pCW170) | 35000.901 containing the vector pCW170 | This study |
| 35000.901(pDL6-2) | 35000.901 containing pDL6-2 with the H. ducreyi znuA gene | This study |
| RO18 | Wild-type strain from Kenya | 3 |
| Cha-1 | Wild-type strain from Dallas, Tex. | 31 |
| A77 | Type strain from Institut Pasteur | 44 |
| 1151 | Wild-type strain from the Gambia | 57 |
| 1153 | Wild-type strain from the Gambia | Allan Ronald |
| 511 | Wild-type strain from Thailand | Allan Ronald |
| 512 | Wild-type strain from Thailand | 57 |
| Hd12 | Wild-type strain from Korea | 43 |
| 1352 | Wild-type strain from Kenya | 57 |
| 78226 | Wild-type strain from Winnipeg, Manitoba, Canada | 3 |
| 6V | Wild-type strain from Atlanta, Ga. | 3 |
| BG411 | Wild-type strain from Kenya | 3 |
| O41 | Wild-type strain from Sweden | 57 |
| 1145 | Wild-type strain from Amsterdam | 57 |
| Plasmids | ||
| pUC18 | Cloning vector, Ampr | 54 |
| pUC18K2 | pUC18 with the kan2 cartridge containing the promoterless aphA-3 gene | James Kaper |
| pBK-CMV | Phagemid excised from the ZAP Express cloning vector | Stratagene |
| pJKT913 | pBK-CMV with a 3.7-kb H. ducreyi DNA insert containing the znuA gene | This study |
| pBluescript II KS(−) | Cloning vector, Amp | Stratagene |
| pJKT933 | pBluescript II with a 3.7-kb HindIII-SacI fragment from pJKT913 containing the znuA gene | This study |
| pDL5-3 | pJKT933 with a kan2 cartridge inserted into the MscI site within the H. ducreyi znuA ORF | This study |
| pLS88 | Cloning vector capable of replication in H. ducreyi; Kanr Smr Sulr | 19 |
| pCW170 | pLS88 with a cat cartridge inserted into the EcoRI site of this vector | This study |
| pDL6-2 | pCW170 with a 402 nt PvuI-XhoI fragment removed from the kan gene and a 1.8-kb PCR-derived, XhoI-PvuI DNA fragment with the H. ducreyi znuA gene inserted in its place | This study |
For measurement of bacterial growth in vitro, H. ducreyi strains were inoculated into a filter-sterilized medium containing Columbia broth (35 g/liter) (Difco), Trizma base (1 g/liter) (Sigma), equine hemin (25 μg/ml), 2.5% (vol/vol) heat-inactivated fetal calf serum (HyClone, Logan, Utah), and 1% (vol/vol) IsoVitalex (62). NTHI strains were grown in filter-sterilized BHI broth containing 10% (vol/vol) Levinthal’s base. In some experiments, cations including ZnCl2 were added to these broths at various concentrations. Growth was monitored by serial absorbance measurements (λ = 600 nm) by using a Spectronic 20D+ spectrophotometer (Spectronic Instruments, Inc., Rochester, N.Y.). Cultures were incubated at 33°C (H. ducreyi) or 37°C (NTHI) at gyrotory speeds of 90 rpm (H. ducreyi) or 160 rpm (NTHI). All growth experiments were performed at least twice.
Monoclonal antibody (MAb) production.
Concentrated culture supernatant fluid was prepared from cultures of H. ducreyi 35000 grown in the presence of human foreskin fibroblasts and collagen in serum-free medium (26, 36a). Splenocytes from mice immunized with this preparation were used in a standard hybridoma fusion protocol (52) to obtain a hybridoma cell line that secreted the H. ducreyi ZnuA-directed immunoglobulin G MAb 3F1.
H. ducreyi genomic library construction and screening.
An H. ducreyi 35000 genomic library (37) constructed in the ZAP Express vector (Stratagene, La Jolla, Calif.) was screened for the presence of recombinant bacteriophages forming MAb 3F1-reactive plaques. The relevant pBK-CMV plasmids containing the H. ducreyi DNA inserts were excised by using the manufacturer’s protocol. One such plasmid, pJKT913, containing a 3.7-kb H. ducreyi DNA insert with the znuA gene, was selected for further study. This 3.7-kb H. ducreyi insert was removed from pJKT913 by digestion with HindIII and SacI (which are present in the pBK-CMV multicloning site on either side of the BamHI site); this fragment was ligated into HindIII- and SacI-digested pBluescript II KS(−) to obtain pJKT933.
Recombinant DNA techniques.
Standard techniques, including restriction enzyme digests, ligation, transformation, and plasmid purification, have been described elsewhere (4, 54). A cartridge (kan2) containing a promoterless aphA-3 gene encoding kanamycin resistance was constructed by Karen Jarvis and James B. Kaper, Center for Vaccine Development, University of Maryland, and was utilized for construction of nonpolar mutations as described by Menard et al. (42). This 0.85-kb cartridge was excised from plasmid pUC18K2 by digestion with SmaI. The pUCΔECAT-derived chloramphenicol acetyltransferase (cat) cartridge used to construct pCW170 was obtained from Bruce A. Green, Wyeth-Lederle Vaccines and Pediatrics, West Henrietta, N.Y. PCR was performed according to manufacturer instructions with either Taq DNA polymerase (Promega, Madison, Wis.) or Pfu DNA polymerase (Stratagene). Boiled bacterial cell preparations (33) or purified H. ducreyi chromosomal DNA (6) were used as templates for PCR.
Nucleotide sequence analysis.
Nucleotide sequence analysis was accomplished as described earlier (62). For the purpose of interstrain comparisons, a 1.9-kb PCR product containing the entire znuA gene was amplified from the chromosomal DNA of five other H. ducreyi strains (RO18, Cha-1, A77, 1151, and 512) with Pfu polymerase and the oligonucleotide primers P3 and P4 (see Fig. 2).
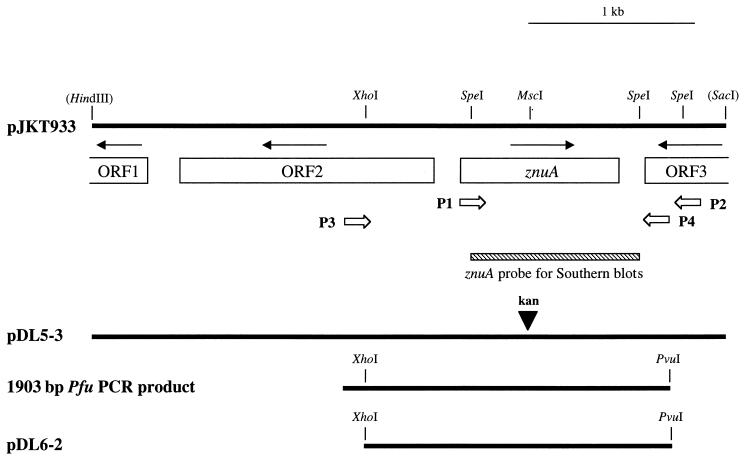
FIG. 2.
Partial restriction map of the H. ducreyi 35000 chromosomal DNA insert in pJKT933 and related plasmids. The two complete ORFs and two incomplete ORFs in this insert are indicated together with arrows designating the proposed direction of transcription. Restriction sites in parentheses indicate vector cloning sites. The open arrows indicate various oligonucleotide primers used in PCR. The cross-hatched bar beneath the znuA gene indicates the 1-kb probe used for Southern blot analysis in Fig. 4. The kan2 cartridge was ligated into the MscI site of pJKT933 to construct pDL5-3. Plasmid pDL6-2 is a modified pCW170 vector (see Materials and Methods) containing the 1.8-kb XhoI- and PvuI-digested PCR product with the znuA gene and flanking DNA.
N-terminal amino acid sequence analysis.
The 32-kDa protein reactive with MAb 3F1 was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride membrane by the method of Matsudaira (41), and subjected to N-terminal amino acid sequence analysis as described elsewhere (32).
Construction of an isogenic H. ducreyi znuA mutant.
Plasmid pJKT933 was linearized with MscI, which cut once within the znuA gene. The kan2 cartridge was then blunt-end ligated into pJKT933 to obtain pDL5-3. DNA sequence analysis confirmed the proper construction of the nonpolar mutation in the znuA gene. E. coli HB101 was transformed with pDL5-3, and plasmid purified from this strain was linearized by digestion with PstI and used to electroporate H. ducreyi 35000 (30); transformants were selected on GC-heme agar plates supplemented with kanamycin (30 μg/ml). Transformant colonies were screened by direct PCR-based amplification of chromosomal DNA from single colonies by using oligonucleotide primers P1 (5′-GCGATGTTAGGCGTAACGACAG-3′) and P2 (5′-GCGTGAAAAAATCGTCAGTTCG-3′) and by loss of reactivity with MAb 3F1 in a colony blot radioimmunoassay (28).
Southern blot analysis.
Purified chromosomal DNA preparations from wild-type H. ducreyi 35000 and its isogenic znuA mutant 35000.901 were digested to completion with SpeI, subjected to electrophoresis in a 0.7% (wt/vol) agarose gel, transferred to nitrocellulose paper, and probed via Southern blot analysis as previously described (54). A 1-kb DNA fragment obtained by SpeI digestion of pJKT933 (see Fig. 2) was used as a probe for the znuA gene, and the 0.85-kb kan2 cartridge was used as a probe for this antibiotic resistance cartridge. This same znuA DNA probe was used to screen SpeI digests of chromosomal DNA from 10 other H. ducreyi isolates (strains 1352, 1153, 511, Hd12, 78226, 6V, BG411, Cha-1, 041, and 1145).
Complementation of the znuA mutation in H. ducreyi and the pzp1 mutation in NTHI 6564.
A 1.9-kb DNA fragment containing the H. ducreyi znuA gene (see Fig. 2) was amplified from strain 35000 chromosomal DNA by using Pfu DNA polymerase and the oligonucleotide primers P3 (5′-TGGACCTTGTGTAATCGTGAG-3′) and P4 (5′-TACGATCGGGTGGATCACCCGAATATCG-3′); the underlined sequence indicates a PvuI site. This 1.9-kb PCR product was digested with both PvuI and XhoI to produce a 1.8-kb fragment containing the znuA gene and flanking H. ducreyi DNA sequences (see Fig. 2). Next, the 6.2-kb vector pCW170 was digested with both PvuI and XhoI; this digestion removed a 0.4-kb portion of the kanamycin resistance gene. The znuA-containing fragment was ligated into the 5.8-kb PvuI-XhoI fragment of pCW170. The ligation mixture was used to transform E. coli DH5α; transformants were selected by the ability to grow on LB agar containing chloramphenicol and screened for lack of growth on LB agar containing kanamycin. Transformants were screened for possession of the H. ducreyi znuA gene by PCR by using primers P1 and P4 and for reactivity with MAb 3F1 in a colony blot radioimmunoassay. Plasmid pDL6-2, obtained from one of these transformants, was purified utilizing the Wizard Plus Miniprep system. This plasmid was used to electroporate both the H. ducreyi znuA mutant 35000.901 and the NTHI 6564 pzp1 mutant.
Preparation of cell fractions for ZnuA localization studies.
Whole-cell lysates of H. ducreyi were prepared from CA-Zn plate-grown cells as described earlier (47). H. ducreyi cells grown in the same manner were used for the preparation of cell envelopes and their Sarkosyl-insoluble extracts as described previously (22). To prepare periplasmic fractions from H. ducreyi strains, the osmotic shock method described by Hultgren and coworkers (38) was modified slightly. Bacterial growth from 20 CA-Zn plates was scraped into 10 ml of cold phosphate-buffered saline (PBS) and subjected to centrifugation at 8,000 × g for 10 min. The wet weight of the pellet was determined, and the cells were resuspended to a final concentration of 25% (wt/vol) in cold, filter-sterilized 20 mM Tris-HCl (pH 8.0) containing 20% (wt/vol) sucrose (Sigma). To this suspension, held at 4°C, was added 0.1 M pH 8.0 EDTA (200 μl per g of cells) and lysozyme (600 μg per g of cells). After a 40-min incubation on ice, 0.5 M MgCl2 (160 μl per g cells) was added, and the suspension was shaken gently. Spheroplasts were pelleted by centrifugation at 23,000 × g for 20 min (4°C), and the resultant supernatant fluid was centrifuged at 200,000 × g for 90 min (4°C). The final supernatant fluid represented the periplasmic fraction and was shown to contain periplasmic contents by Western blot analysis with polyclonal antiserum raised against the H. ducreyi periplasmic Cu-Zn superoxide dismutase (55) as the primary antibody.
Concentration of bacterial culture supernatant fluids.
H. ducreyi strains were grown in the Columbia broth-based medium containing 100 μM ZnCl2 for 24 h. The culture was centrifuged at 8,000 × g for 10 min (4°C), and the supernatant fluid was filter sterilized by using Acrodisc syringe filters (0.2-μm pore size) (Gelman Sciences, Ann Arbor, Mich.). The supernatant fluid was centrifuged at 215,000 × g for 90 min (4°C) prior to being concentrated 40-fold by using a Centriprep-10 centrifugal concentrator (Amicon, Inc., Beverly, Mass.).
Colony blot radioimmunoassay, SDS-PAGE, and Western blot (immunoblot) methods.
These procedures were accomplished as described previously (28, 35).
Virulence testing.
The relative virulence of the H. ducreyi strains used in this study was determined by using the temperature-dependent rabbit model for experimental chancroid (50). The mean scores of the lesions resulting from injection of 105 and 104 CFU were analyzed statistically as described earlier (6). After lesion scoring on day 7, the rabbits were euthanized and the lesions resulting from injection of an inoculum containing 105 CFU were excised; the pustular material from these lesions was cultured on CA-Zn plates.
Nucleotide accession number.
The nucleotide sequence of the H. ducreyi 35000 znuA gene was assigned GenBank accession number AF141971.
RESULTS
Identification and cloning of the H. ducreyi znuA gene.
Polyclonal rabbit antiserum to H. ducreyi 35000 was used in Western blot analysis to probe supernatant fluid from cocultures of H. ducreyi 35000 and human foreskin fibroblasts embedded in a matrix of collagen (26, 36a). Among the immunoreactive antigens detected in these preliminary experiments was a 32-kDa molecule (data not shown). An MAb (3F1) reactive with this 32-kDa molecule also bound this protein in concentrated culture supernatant fluids of H. ducreyi alone (Fig. 1A, lane 1). The use of MAb 3F1 to screen a H. ducreyi genomic library yielded the recombinant plasmid pJKT913 which contained a 3.7-kb H. ducreyi DNA insert that expressed this MAb 3F1-reactive antigen. This H. ducreyi DNA insert together with a small amount of flanking vector DNA was subcloned into pBluescript II to form pJKT933 (Fig. 2). Nucleotide sequence analysis revealed the presence of two complete and two incomplete ORFs (Fig. 2). The first complete ORF (ORF 2) encoded a predicted protein that had homology to a putative oxidoreductase in E. coli (9), whereas the other complete ORF encoded a predicted protein that was 43% identical to the E. coli ZnuA (YebL) zinc-binding protein (48) and 54% identical to the H. influenzae (NTHI 6564) periplasmic zinc-binding protein Pzp1 (39). One of the incomplete open reading frames (ORFs) (ORF 1) encoded a partial protein with homology to the hypothetical H. influenzae protein HI0318 (23), and the other (ORF 3) encoded a partial protein with similarity to the H. influenzae ribose-5-phosphate isomerase A (23).
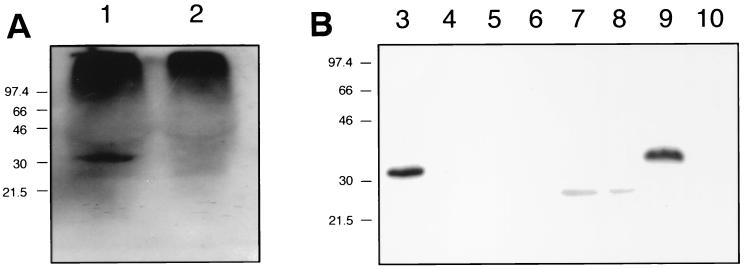
FIG. 1.
Western blot-based detection of ZnuA in culture supernatant fluid and subcellular fractions of H. ducreyi. The H. ducreyi ZnuA-reactive MAb 3F1 was used as the primary antibody. (A) Concentrated culture supernatant fluid from wild-type 35000 (lane 1) and the znuA mutant 35000.901 (lane 2). (B) Subcellular fractions from wild-type 35000 (lanes 3, 5, 7, and 9) and the znuA mutant 35000.901 (lanes 4, 6, 8, and 10). Lanes 3 and 4, whole-cell lysate; lanes 5 and 6, total cell envelopes; lanes 7 and 8, Sarkosyl-insoluble proteins from cell envelopes; lanes 9 and 10, periplasmic fraction. The aberrant migration of the ZnuA protein in the periplasmic fraction in lane 9 was caused by the presence of a high concentration of sucrose derived from the preparation of the periplasmic fraction; when this sample was diluted 1:4 in PBS, the ZnuA protein migrated at the same rate as the ZnuA protein seen in lane 3.
Features of the H. ducreyi 35000 znuA gene and its protein product.
A putative promoter region immediately upstream of the znuA ORF contained predicted −35 and −10 regions, as well as a potential ribosomal binding site. The H. ducreyi znuA ORF contained 930 nucleotides encoding a predicted protein with a calculated molecular weight of 34,289. The existence of a signal peptide was confirmed by N-terminal amino acid sequence analysis of the MAb 3F1-reactive protein band obtained from H. ducreyi 35000 cells. The N-terminal amino acid sequence (DVLTSIKPLGFIANAITDGV) derived from this approach matched the predicted N-terminal amino acid sequence of the mature protein exactly. The calculated molecular weight of the mature protein was 32,198. This mature protein also contained the central histidine-rich region as well as the two cysteine residues in the C-terminal region characteristic of the NTHI 6564 Pzp1 protein (39, 40).
Detection and characterization of the znuA gene in H. ducreyi clinical isolates.
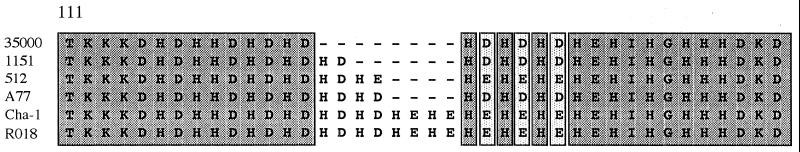
Southern blot analysis of SpeI-digested chromosomal DNA from 11 geographically diverse H. ducreyi clinical isolates (including strain 35000) was performed with a 1-kb znuA-containing DNA fragment derived from pJKT933 by SpeI digestion (Fig. 2). All of these isolates possessed a 1-kb SpeI fragment that bound this znuA probe (data not shown). Nucleotide sequence analysis of the znuA gene from five H. ducreyi strains (RO18, Cha-1, A77, 1151, and 512) revealed that the encoded ZnuA proteins had almost complete identity with the ZnuA protein of strain 35000, the only variation being seen within the histidine-rich central area (Fig. 3).
FIG. 3.
Comparison of the histidine-rich regions from the ZnuA proteins of six H. ducreyi strains using the CLUSTAL-W Alignment program in MacVector version 6. Dark shading indicates residues that are identical. Light shading indicates residues that are similar. The amino acid sequences begin with residue 111 in each strain. The upstream and downstream amino acid sequences of all six proteins (not shown) were identical.
Construction of an isogenic H. ducreyi znuA mutant.
The inactivated H. ducreyi 35000 znuA gene containing the kan2 cartridge in pDL5-3 was used to electroporate H. ducreyi 35000. Five kanamycin-resistant H. ducreyi transformants were tested initially by PCR (with primers P1 and P2) to detect allelic exchange. All five yielded a single 2.3-kb PCR product which was 0.85 kb larger than the PCR product from the wild-type strain; this finding was consistent with the presence of the kan2 cartridge in the znuA gene (data not shown). In addition, all five transformants failed to react with the H. ducreyi ZnuA-reactive MAb 3F1 in a colony blot assay (data not shown). One of these transformants was selected for further study and designated strain 35000.901. Western blot analysis indicated that strain 35000.901 did not express detectable ZnuA in concentrated culture supernatant fluid (Fig. 1A, lane 2).
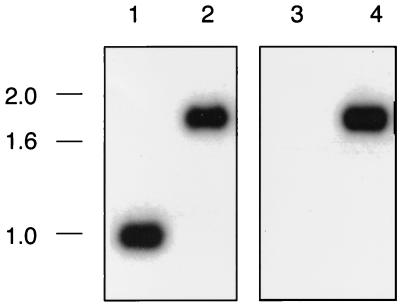
Southern blot analysis confirmed the occurrence of the desired allelic exchange in strain 35000.901 (Fig. 4). When chromosomal DNA from wild-type parent strain 35000 was probed with the znuA probe described above (Fig. 2), a 1-kb SpeI fragment hybridized with this probe (Fig. 4, lane 1). This same probe hybridized with a 1.85-kb SpeI fragment of chromosomal DNA from strain 35000.901 (Fig. 4, lane 2). A 1.85-kb fragment from strain 35000.901 also hybridized with a kan2 cartridge-based probe (Fig. 4, lane 4). Chromosomal DNA from the wild-type parent strain 35000 failed to hybridize with the kan2 probe (Fig. 4, lane 3).
FIG. 4.
Southern blot analysis of chromosomal DNA preparations from H. ducreyi wild-type and mutant strains. Chromosomal DNAs were digested with SpeI, resolved by agarose gel electrophoresis, and probed with either a 1-kb SpeI fragment derived from the H. ducreyi znuA gene (lanes 1 and 2) or with the kan2 cartridge (lanes 3 and 4). Lanes 1 and 3, strain 35000; lanes 2 and 4, znuA mutant 35000.901. Size markers (in kilobases) are on the left side of the figure.
In vitro growth characteristics of the isogenic H. ducreyi znuA mutant.
The znuA mutant strain 35000.901 grew more slowly than the wild-type strain 35000 in broth culture (Fig. 5A). The addition of ZnCl2 to a final concentration of 100 μM almost completely corrected the growth deficiency of strain 35000.901 in vitro. This growth deficiency was not reversed by the addition of ZnCl2 to a final concentration of 5 μM or by 100 μM concentrations of FeCl3, MnCl2, CuSO4, NiCl2, or MgCl2 (data not shown), a result which suggested that the growth defect in strain 35000.901 was zinc specific.
FIG. 5.
Growth of wild-type, mutant, and recombinant strains of H. ducreyi and H. influenzae in vitro. These data are from representative experiments. (A) H. ducreyi strains grown overnight on CA-Zn plates were inoculated into broth with (solid symbols) and without (open symbols) 100 μM ZnCl2, and growth was monitored for 36 h. Wild-type strain 35000 is indicated by squares, and znuA mutant 35000.901 is indicated by circles. (B) H. ducreyi strains grown overnight on CA-Zn plates were inoculated into broth without supplemental zinc, and growth was monitored for 32 h. Symbols: squares, wild-type H. ducreyi 35000; circles, H. ducreyi znuA mutant 35000.901; diamonds, H. ducreyi znuA mutant 35000.901(pCW170); triangles, H. ducreyi znuA mutant 35000.901(pDL6-2). (C) H. influenzae strains grown overnight on BHIs plates containing 100 μM ZnCl2 were inoculated into BHIs broth without supplemental zinc, and growth was monitored for 14 h. Symbols: squares, wild-type NTHI 6564; circles, NTHI 6564 pzp1 mutant; diamonds, NTHI 6564 pzp1 mutant containing pCW170; triangles, NTHI 6564 pzp1 mutant containing pDL6-2.
Complementation of the znuA mutation in H. ducreyi.
To eliminate the possibility that an undetected secondary mutation was responsible for the altered growth phenotype of the H. ducreyi znuA mutant, complementation analysis was performed. The wild-type znuA gene was amplified by PCR and cloned into pCW170 to yield the recombinant plasmid pDL6-2 (Fig. 2). Both the pCW170 vector and pDL6-2 were introduced into the H. ducreyi znuA mutant by electroporation. Southern blot analysis indicated that there had been no rearrangement between the mutated znuA gene in the chromosome and the vector-borne wild-type znuA gene in the recombinant strain 35000.901(pDL6-2) (data not shown). The presence of the wild-type H. ducreyi znuA gene in trans in strain 35000.901(pDL6-2) resulted in restoration of wild-type growth characteristics (Fig. 5B). In contrast, the presence of the vector alone in strain 35000.901(pCW170) did not correct the growth phenotype of the znuA mutant (Fig. 5B).
Complementation of the H. influenzae pzp1 mutant with pDL6-2.
The pzp1 (znuA) mutant of NTHI 6564 (39) was electroporated with pDL6-2 and with the pCW170 vector. Numerous transformants grew within 24 h on BHIs-Zn plates containing chloramphenicol, and six transformants from each electroporation were further characterized. All six pzp1 mutants containing pDL6-2 expressed H. ducreyi ZnuA, as evidenced by their reactivity with the H. ducreyi ZnuA-reactive MAb 3F1 (data not shown). The presence of the H. ducreyi znuA gene in these six transformants was confirmed by PCR with the oligonucleotide primers P1 and P4 (data not shown). The six pzp1 mutants containing pCW170 were not reactive with MAb 3F1 and did not yield a PCR product with primers P1 and P4 (data not shown). One recombinant strain from each group of six was selected for in vitro growth studies. The presence of the wild-type H. ducreyi znuA gene in trans in the NTHI 6564 pzp1 mutant resulted in restoration of the ability to grow in broth without additional zinc supplementation (Fig. 5C). In contrast, the presence of the pCW170 vector alone in the NTHI 6564 pzp1 mutant did not correct the growth deficiency of this strain (Fig. 5C).
Localization of ZnuA in H. ducreyi.
To localize ZnuA in H. ducreyi, whole-cell lysates, total cell envelopes, Sarkosyl-insoluble cell envelope material, and periplasmic extracts were prepared from cells of the wild-type strain 35000 and of its isogenic znuA mutant 35000.901 grown on CA plates supplemented with ZnCl2 at 100 μM. In addition, concentrated (40-fold) supernatant fluids were prepared from 24-h broth cultures of both strains grown in the presence of 100 μM ZnCl2. Western blot analysis with the H. ducreyi ZnuA-reactive MAb 3F1 detected ZnuA in both the whole-cell lysate and periplasmic extract of the wild-type strain (Fig. 1B, lanes 3 and 9, respectively) but not in the same preparations from the znuA mutant (Fig. 1B, lanes 4 and 10). ZnuA was not detectable in either the total cell envelope preparation or Sarkosyl-insoluble material of both the wild-type strain (Fig. 1B, lanes 5 and 7) and the mutant strain (Fig. 1B, lanes 6 and 8). A small amount of ZnuA was detected in the wild-type culture supernatant fluid (Fig. 1A, lane 1) but not in culture supernatant fluid from the mutant (Fig. 1A, lane 2).
Virulence testing of the H. ducreyi znuA mutant.
Four H. ducreyi strains were tested in the temperature-dependent rabbit model (Table 2): wild-type strain 35000, the znuA mutant 35000.901, strain 35000.901(pCW170), and strain 35000.901(pDL6-2). In the first of two experiments (Table 2, experiment A), statistical analysis showed that the znuA mutant was less virulent than both the wild-type parent strain (P = 0.0001) and the complemented mutant (P = 0.0001). There was no difference between the wild-type strain 35000 and the complemented mutant strain 35000.901(pDL6-2) (P = 0.1334). Viable H. ducreyi organisms were recovered at the same frequency (i.e., eight of eight) from lesions produced by both the wild-type strain 35000 and the complemented mutant 35000.901(pDL6-2). In contrast, viable H. ducreyi organisms were recovered from only one of eight lesions produced by the znuA mutant 35000.901.
TABLE 2.
Lesion formation by wild-type, znuA mutant, and complemented znuA mutant strains of H. ducreyi in the temperature-dependent rabbit modela
| Strain | Inoculum size | Mean lesion score (SD) at:
|
P valueb | ||
|---|---|---|---|---|---|
| Day 2 | Day 4 | Day 7 | |||
| Expt A | |||||
| 35000 (wild-type) | 105 | 4.00 (0) | 4.00 (0) | 4.00 (0) | |
| 35000.901 (znuA mutant) | 105 | 3.25 (0.46) | 2.75 (0.46) | 2.75 (0.46) | 0.0001c |
| 35000.901(pDL6-2) | 105 | 4.00 (0) | 4.00 (0) | 4.00 (0) | 0.1334 |
| 35000 | 104 | 3.13 (0.35) | 3.13 (0.35) | 3.63 (0.52) | |
| 35000.901 | 104 | 3.00 (0) | 2.13 (0.64) | 2.00 (0.93) | |
| 35000.901(pDL6-2) | 104 | 3.38 (0.52) | 3.50 (0.54) | 3.88 (0.35) | |
| Expt B | |||||
| 35000 | 105 | 3.88 (0.35) | 4.00 (0) | 4.00 (0) | |
| 35000.901(pCW170) | 105 | 3.13 (0.35) | 2.75 (0.71) | 3.13 (0.35) | 0.0002 |
| 35000.901(pDL6-2) | 105 | 3.38 (0.52) | 3.88 (0.35) | 4.00 (0) | 0.0366 |
| 35000 | 104 | 3.13 (0.35) | 3.38 (0.52) | 3.50 (0.54) | |
| 35000.901(pCW170) | 104 | 3.00 (0) | 2.63 (0.52) | 2.25 (0.46) | |
| 35000.901(pDL6-2) | 104 | 3.00 (0.54) | 3.00 (0.54) | 3.13 (0.64) | |
Eight rabbits were used in each experiment.
P value calculated for the difference between wild-type and test strain lesion scores. P values were calculated by using the lesion scores from both inoculum sizes and from all 3 days.
The complemented znuA mutant 35000.901(pDL6-2) was significantly more virulent than both the znuA mutant containing the vector pCW170 (P = 0.0061) and the znuA mutant alone (P = 0.0001).
In the second experiment (Table 2, experiment B), the znuA mutant containing the pCW170 vector was less virulent than both the wild-type strain (P = 0.0002) and the complemented mutant (P = 0.0061). The difference in lesion scores between the wild-type strain 35000 and the complemented mutant strain 35000.901(pDL6-2) just achieved significance (P = 0.0366). Viable H. ducreyi organisms were recovered from seven of eight lesions produced by the wild-type strain 35000 and from five of eight lesions resulting from inoculation with the complemented mutant 35000.901(pDL6-2). In contrast, no viable H. ducreyi organisms were recovered from the eight lesions produced by the znuA mutant 35000.901 containing the plasmid vector.
DISCUSSION
In the present study, a putative zinc transport protein (ZnuA) was identified in H. ducreyi which had homology to both the Pzp1 protein of H. influenzae (54% identity) and the ZnuA (YebL) protein of E. coli (43% identity). The gene (znuA) encoding this H. ducreyi protein appears to be well conserved among strains of this pathogen. Lu et al. (39) demonstrated that the H. influenzae Pzp1 protein functions as a periplasmic zinc binding protein based on results of protein localization studies, direct binding of 65Zn to recombinant Pzp1, neutron activation analysis, and atomic absorption spectroscopy. The observation that the H. ducreyi ZnuA protein could complement the in vitro growth defect of the NTHI 6564 pzp1 mutant suggests that the two proteins have identical functions in zinc transport. The localization of ZnuA to the periplasmic compartment in H. ducreyi is also compatible with its postulated role as the binding component of a zinc transport system. Whereas H. influenzae Pzp1 was undetectable in culture supernatant fluid (39), H. ducreyi ZnuA was detected in a Western blot analysis of a concentrated 24-h culture supernatant fluid (Fig. 1A). This finding probably reflects leakage of ZnuA into the medium from dying bacteria, although we have not formally excluded the less likely possibility of active secretion of this protein into the culture medium.
Inactivation of the znuA gene in H. ducreyi 35000 resulted in the isogenic mutant 35000.901, which exhibited decreased growth in broth compared to the wild-type parent strain (Fig. 5A). This growth defect of the mutant was overcome by complementation with the wild-type znuA gene in trans (Fig. 5B) or by the addition of ZnCl2 to the broth at a final concentration of 100 μM (Fig. 5A). In addition, the znuA mutant exhibited significantly reduced virulence in the temperature-dependent rabbit model for experimental chancroid (Table 2). It must be noted that provision of the wild-type H. ducreyi znuA gene in trans in the H. ducreyi znuA mutant restored virulence in the animal model.
Although the znuA mutant clearly produced lower lesion scores than did the wild-type strain in the rabbit model, interpretation of these virulence data is not straightforward. We have no information on the physiological concentration ranges of zinc in the skin of male New Zealand White rabbits but assume that it is very low since the normal plasma zinc concentration in this species is approximately 23 μM (36). The decreased virulence observed with the znuA mutant may therefore simply reflect limited in vivo growth of the mutant compared to that of the wild-type parent strain or complemented mutant. This hypothesis is substantiated by the observation that the in vitro growth defect of the znuA mutant is almost completely abolished by adding zinc chloride to a concentration of 100 μM or by complementing the znuA chromosomal mutation with a plasmid expressing H. ducreyi ZnuA. However, we cannot eliminate the possibility that the znuA mutation may affect virulence indirectly, either by affecting regulation of other virulence factors or because zinc is an essential cofactor for one or more virulence-related proteins.
The H. ducreyi znuA gene belongs to the recently described family of proteins involved in metal cation binding and subsequent transport into the bacterial cell (18). The MntABC Mn2+ transport system of the cyanobacterium Synechocystis sp. strain 6803 was the first member identified in which mntC encodes for a Mn2+ binding protein (5). A putative ABC-type zinc permease complex (Adc) and a putative ABC-type manganese permease complex (Psa) were subsequently identified in S. pneumoniae (18). The most recent addition to this family is the ZnuABC transport system of E. coli in which ZnuA performs the role of the zinc-binding protein (48). The znuA, znuB, and znuC genes correspond to the yebL, yebI, and yebM genes identified in the E. coli K-12 genome sequencing project (9). The Pzp1 zinc-binding protein of H. influenzae also belongs to this family but, interestingly, both in this species and in H. ducreyi adjacent genes encoding for hydrophobic membrane proteins or ATP-binding proteins that would be typical in an ABC transporter operon appear to be absent. It is possible that genes with these functions could be located at another site(s) on their respective chromosomes. In support of this hypothesis, the predicted protein products of ORFs HI0407 and HI0408 identified in the H. influenzae Rd genome (23) have 60 and 48% identity to the ZnuB (YebI) and ZnuC (YebM) proteins of E. coli, respectively.
Both ZnuA of H. ducreyi and ZnuA of E. coli have two conserved cysteine residues in their C-terminal region, making it likely that they have a C-terminal disulfide-bonded domain similar to that in Pzp1 of H. influenzae (39, 40). In support of this, the H. ducreyi ZnuA protein migrates more slowly in SDS-PAGE when in the presence of 2-mercaptoethanol than it does under nonreducing conditions (data not shown). These cysteines are not present in the pneumococcal AdcA protein (18). The amino acids histidine (H), aspartic acid (D), and glutamic acid (E) appear to be characteristic for zinc-binding proteins (18); all four proteins also possess a central histidine- and acidic amino acid-rich region, which is most pronounced in Pzp1 of H. influenzae (39). In contrast, the putative manganese-binding proteins MntC of Synechocystis sp. strain 6803 (5) and PsaA of S. pneumoniae (18) both lack this histidine- and acidic amino acid-rich central region. The ZnuA proteins from five other H. ducreyi isolates showed variation in amino acid sequence only in this histidine-rich area (Fig. 3). In eukaryotes, a metal binding motif (HX)3 has been proposed as the metal binding site of the high-affinity zinc transporter (Zrt1) of Saccharomyces cerevisiae (64) and of two of the four putative zinc transporter proteins described in Arabidopsis thaliana (21, 27). These proteins form part of the 15-member Zrt- and Irt-related protein (ZIP) family of eukaryotic proteins reviewed by Eng et al. (21) which includes functionally uncharacterized homologues in Caenorhabditis elegans, Mus musculus, and Homo sapiens.
Preliminary experiments using the H. ducreyi ZnuA-reactive MAb 3F1 to probe lysates of H. ducreyi 35000 cells grown in broths containing increasing zinc concentrations have failed to provide evidence for regulation of ZnuA protein expression, at least in vitro (data not shown). This is in contrast to the situation in E. coli, in which the first regulatory protein (i.e., Zur) to affect zinc uptake was described (48). In vivo-expressed genes with similarity to the E. coli zur gene have been identified in Vibrio cholerae (iviXI) (12) and Pseudomonas aeruginosa (np20) (61) and support the hypothesis that zinc could regulate expression of one or more gene products important for virulence in the mammalian host. Whether H. ducreyi possesses a homologous zur gene that is expressed solely in the in vivo environment remains to be determined.
ACKNOWLEDGMENTS
This study was supported by U.S. Public Health Service grant AI32011 to E.J.H. and by the Wellcome Research Training Fellowship in Clinical Tropical Medicine (reference number 049246/Z/96) to D.A.L. under the joint sponsorship of J. N. Weber (Department of Genitourinary Medicine and Communicable Disease) and D. B. Young (Department of Microbiology) at Imperial College School of Medicine, St. Mary’s Campus, London, United Kingdom.
C. A. Lingwood kindly provided H. influenzae NTHI 6564 and its pzp1 isogenic mutant. The kan2 cartridge used in this study was constructed by Karen Jarvis and James B. Kaper. The cat cartridge was constructed by Bruce A. Green. Beth A. Bauer assisted with the temperature-dependent rabbit model experiments. We thank Marla K. Stevens, Lani R. San Mateo, and Thomas H. Kawula for helpful discussions concerning the preparation of periplasmic fractions and the latter two individuals for providing polyclonal antiserum to the H. ducreyi periplasmic Cu-Zn superoxide dismutase.
REFERENCES
- 1.Alexander H E. The Haemophilus group. In: Dubos R J, Hirsch J G, editors. Bacterial and mycotic infections of man. J. B. Philadelphia, Pa: Lippincott Co.; 1965. pp. 724–741. [Google Scholar]
- 2.Alfa M J, Degagne P, Totten P A. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect Immun. 1996;64:2349–2352. doi: 10.1128/iai.64.6.2349-2352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa M J, Stevens M K, Degagne P, Klesney-Tait J, Radolf J D, Hansen E J. Use of tissue culture and animal models to identify virulence-associated traits of Haemophilus ducreyi. Infect Immun. 1995;63:1754–1761. doi: 10.1128/iai.63.5.1754-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1990. [Google Scholar]
- 5.Bartsevich V V, Pakrasi H B. Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 1995;14:1845–1853. doi: 10.1002/j.1460-2075.1995.tb07176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer B A, Stevens M K, Hansen E J. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect Immun. 1998;66:4290–4298. doi: 10.1128/iai.66.9.4290-4298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beard S J, Hashim R, Membrillo-Hernandez J, Hughes M N, Poole R K. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (0732) encodes a cation transport ATPase. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 8.Beard S J, Hughes M N, Poole R K. Inhibition of the cytochrome bd-terminated NADH oxidase system in Escherichia coli K-12 by divalent metal cations. FEMS Microbiol Lett. 1995;131:205–210. doi: 10.1111/j.1574-6968.1995.tb07778.x. [DOI] [PubMed] [Google Scholar]
- 9.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 10.Brentjens R J, Ketterer M, Apicella M A, Spinola S M. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J Bacteriol. 1996;178:808–816. doi: 10.1128/jb.178.3.808-816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron D W, Simonsen J N, D’Costa L J, Ronald A R, Maitha G M, Gakinya M N, Cheang M, Ndinya-Achola J O, Piot P, Brunham R C, Plummer F A. Female to male transmission of human immunodeficiency virus type 1: risk factors for seroconversion in men. Lancet. 1989;ii:403–407. doi: 10.1016/s0140-6736(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 12.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campagnari A A, Wild L M, Griffiths G E, Karalus R J, Wirth M A, Spinola S M. Role of lipopolysaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect Immun. 1991;59:2601–2608. doi: 10.1128/iai.59.8.2601-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carson S D B, Thomas C E, Elkins C. Cloning and sequencing of a Haemophilus ducreyi fur homolog. Gene. 1996;176:125–129. doi: 10.1016/0378-1119(96)00236-3. [DOI] [PubMed] [Google Scholar]
- 15.Coleman J E. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- 16.Cope L D, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortes-Bratti X, Chaves-Olarte E, Lagergard T, Thelestam M. The cytolethal distending toxin from chancroid bacterium Haemophilus ducreyi induces cell-cycle arrest in the G2 phase. J Clin Invest. 1999;103:107–115. doi: 10.1172/JCI3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dintilhac A, Alloing G, Granadel C, Claverys J-P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 19.Dixon L G, Albritton W L, Willson P J. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid. 1994;32:228–232. doi: 10.1006/plas.1994.1060. [DOI] [PubMed] [Google Scholar]
- 20.Elkins C, Chen C-J, Thomas C E. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eng B H, Guerinot M L, Eide D, Saier M H., Jr Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol. 1998;166:1–7. doi: 10.1007/s002329900442. [DOI] [PubMed] [Google Scholar]
- 22.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback R C, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 24.Fleming D T, Wasserheit J N. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex Transm Infect. 1999;75:3–17. doi: 10.1136/sti.75.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson B W, Campagnari A A, Melaugh W, Phillips N J, Apicella M A, Grass S, Wang J, Palmer K L, Munson R S., Jr Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J Bacteriol. 1997;179:5062–5071. doi: 10.1128/jb.179.16.5062-5071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grinnell F, Lamke C R. Reorganization of hydrated collagen lattices by human skin fibroblasts. J Cell Sci. 1984;66:51–63. doi: 10.1242/jcs.66.1.51. [DOI] [PubMed] [Google Scholar]
- 27.Grotz N, Fox T, Connolly E, Park W, Guerinot M L, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulig P A, Patrick C C, Hermanstorfer L, McCracken G H, Jr, Hansen E J. Conservation of epitopes in the oligosaccharide portion of the lipooligosaccharide of Haemophilus influenzae type b. Infect Immun. 1987;55:513–520. doi: 10.1128/iai.55.3.513-520.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond G W, Lian C J, Wilt J C, Ronald A R. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob Agents Chemother. 1978;13:608–612. doi: 10.1128/aac.13.4.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen E J, Latimer J L, Thomas S E, Helminen M E, Albritton W L, Radolf J D. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J Bacteriol. 1992;174:5442–5449. doi: 10.1128/jb.174.16.5442-5449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen E J, Lumbley S R, Richardson J A, Purcell B K, Stevens M K, Cope L D, Datte J, Radolf J D. Induction of protective immunity to Haemophilus ducreyi in the temperature-dependent rabbit model of experimental chancroid. J Immunol. 1994;152:184–192. [PubMed] [Google Scholar]
- 32.Hansen E J, Pelzel S E, Orth K, Moomaw C R, Radolf J D, Slaughter C A. Structural and antigenic conservation of the P2 porin protein among strains of Haemophilus influenzae type b. Infect Immun. 1989;57:3270–3275. doi: 10.1128/iai.57.11.3270-3275.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennessy K J, Iandolo J J, Fenwick B W. Serotype identification of Actinobacillus pleuropneumoniae by arbitrarily primed polymerase chain reaction. J Clin Microbiol. 1993;31:1155–1159. doi: 10.1128/jcm.31.5.1155-1159.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasahara M, Anraku Y. Succinate- and NADH oxidase systems of Escherichia coli membrane vesicles: mechanism of selective inhibition of the systems by zinc ions. J Biochem. 1974;76:967–976. [PubMed] [Google Scholar]
- 35.Kimura A, Gulig P A, McCracken G H, Jr, Loftus T A, Hansen E J. A minor high-molecular-weight outer membrane protein of Haemophilus influenzae type b is a protective antigen. Infect Immun. 1985;47:253–259. doi: 10.1128/iai.47.1.253-259.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby K A, Rothenburg B A, Victery W, Vander A J, Kluger M J. Urinary excretion of zinc and iron following injection of bacteria in the unanesthetized rabbit. Miner Electrolyte Metab. 1982;7:250–256. [PubMed] [Google Scholar]
- 36a.Klesney-Tait J. Ph.D. Dissertation. Graduate School of Biomedical Sciences. Dallas: University of Texas Southwestern Medical Center; 1996. [Google Scholar]
- 37.Klesney-Tait J, Hiltke T J, Maciver I, Spinola S M, Radolf J D, Hansen E J. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179:1764–1773. doi: 10.1128/jb.179.5.1764-1773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuehn M J, Jacob-Dubuisson F, Dodson K, Slonim L, Striker R, Hultgren S J. Genetic, biochemical, and structural studies of biogenesis of adhesive pili in bacteria. Methods Enzymol. 1994;236:282–306. doi: 10.1016/0076-6879(94)36022-7. [DOI] [PubMed] [Google Scholar]
- 39.Lu D, Boyd B, Lingwood C A. Identification of the key protein for zinc uptake in Haemophilus influenzae. J Biol Chem. 1997;272:29033–29038. doi: 10.1074/jbc.272.46.29033. [DOI] [PubMed] [Google Scholar]
- 40.Lu D, Boyd B, Lingwood C A. The expression and characterization of a putative adhesin B from H. influenzae. FEMS Microbiol Lett. 1998;165:129–137. doi: 10.1111/j.1574-6968.1998.tb13137.x. [DOI] [PubMed] [Google Scholar]
- 41.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 42.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oberhofer T R, Back A E. Isolation and cultivation of Haemophilus ducreyi. J Clin Microbiol. 1982;15:625–629. doi: 10.1128/jcm.15.4.625-629.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odumeru J A, Wiseman G M, Ronald A R. Role of lipopolysaccharide and complement in susceptibility of Haemophilus ducreyi to human serum. Infect Immun. 1985;50:495–499. doi: 10.1128/iai.50.2.495-499.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer K L, Goldman W E, Munson R S., Jr An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol Microbiol. 1996;21:13–19. doi: 10.1046/j.1365-2958.1996.00615.x. [DOI] [PubMed] [Google Scholar]
- 46.Parsons L M, Limberger R J, Shayegani M. Alterations in levels of DnaK and GroEL result in diminished survival and adherence of stressed Haemophilus ducreyi. Infect Immun. 1997;65:2413–2419. doi: 10.1128/iai.65.6.2413-2419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrick C C, Kimura A, Jackson M A, Hermanstorfer L, Hood A, McCracken G H, Jr, Hansen E J. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypable Haemophilus influenzae. Infect Immun. 1987;55:2902–2911. doi: 10.1128/iai.55.12.2902-2911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patzer S I, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 49.Plummer F A, Simonsen J N, Cameron D W, Ndinya-Achola J O, Kreiss J K, Gakinya M N, Waiyaki P, Cheang M, Piot P, Ronald A R, Ngugi E N. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- 50.Purcell B K, Richardson J A, Radolf J D, Hansen E J. A temperature-dependent rabbit model for production of dermal lesions by Haemophilus ducreyi. J Infect Dis. 1991;164:359–367. doi: 10.1093/infdis/164.2.359. [DOI] [PubMed] [Google Scholar]
- 51.Rensing C, Mitra B, Rosen B P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson S M, Frisch C F, Gulig P A, Kettman J R, Johnston K H, Hansen E J. Monoclonal antibodies directed against a cell surface-exposed outer membrane protein of Haemophilus influenzae type b. Infect Immun. 1982;36:80–88. doi: 10.1128/iai.36.1.80-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronald A R, Albritton W. Chancroid and Haemophilus ducreyi. In: Holmes K K, Mardh P-A, Sparling P F, Wiesner P J, editors. Sexually transmitted diseases. New York, N.Y: McGraw-Hill; 1990. pp. 263–271. [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.San Mateo L R, Hobbs M M, Kawula T H. Periplasmic copper-zinc superoxide dismutase protects Haemophilus ducreyi from exogenous superoxide. Mol Microbiol. 1998;27:391–404. doi: 10.1046/j.1365-2958.1998.00687.x. [DOI] [PubMed] [Google Scholar]
- 56.Stevens M K, Cope L D, Radolf J D, Hansen E J. A system for generalized mutagenesis of Haemophilus ducreyi. Infect Immun. 1995;63:2976–2982. doi: 10.1128/iai.63.8.2976-2982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevens M K, Porcella S, Klesney-Tait J, Lumbley S, Thomas S E, Norgard M V, Radolf J D, Hansen E J. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect Immun. 1996;64:1724–1735. doi: 10.1128/iai.64.5.1724-1735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tullius M V, Munson R S, Jr, Wang J, Gibson B W. Purification, cloning, and expression of a cytidine 5′-monophosphate N-acetylneuraminic acid synthetase from Haemophilus ducreyi. J Biol Chem. 1996;271:15373–15380. doi: 10.1074/jbc.271.26.15373. [DOI] [PubMed] [Google Scholar]
- 60.Vallee B L, Falchuk K H. The biochemical basis of zinc physiology. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward C K, Lumbley S R, Latimer J L, Cope L D, Hansen E J. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J Bacteriol. 1998;180:6013–6022. doi: 10.1128/jb.180.22.6013-6022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasserheit J N. Epidemiological synergy: interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex Transm Dis. 1992;19:61–77. [PubMed] [Google Scholar]
- 64.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]