Abstract
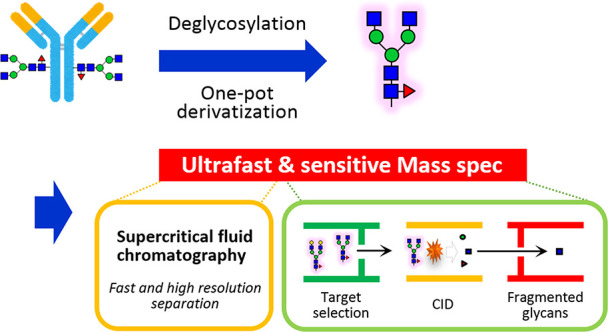
The glycoform of a therapeutic monoclonal antibody (mAb) has a significant impact on its effector function as well as its safety and pharmacokinetics. Glycoform heterogeneity is influenced by various factors, including the producing cells and cell culture processes. Therefore, accurate glycoform characterization is essential for drug design, process optimization, manufacturing, and quality control of therapeutic mAbs. In this study, we developed a fast, quantitative, and highly sensitive analytical platform for glycan profiling by supercritical fluid chromatography–tandem mass spectrometry (SFC-MS/MS) and applied the technique to the glycan structural analysis of mAbs. To achieve both the highest sensitivity and the most comprehensive glycan profiling, we integrated our energy-resolved oxonium ion monitoring (Erexim) method with SFC-MS to construct a new SFC-Erexim technology. An 8 min analysis of bevacizumab, nivolumab, ramucirumab, rituximab, and trastuzumab by SFC-Erexim detected a total of 102 glycoforms, with a detection limit of 5 attomoles. The dynamic range of glycan abundance was over 6 orders of magnitude for bevacizumab analysis by SFC-Erexim compared to 3 orders of magnitude for conventional fluorescence HPLC analysis. This method revealed the glycan profile characteristics and lot-to-lot heterogeneity of various therapeutic mAbs. We were also able to detect a series of structural variations in pharmacologically important glycan structures. The SFC-MS-based glycoform profiling method will provide an ideal platform for the in-depth analysis of precise glycan structure and abundance.
Introduction
Therapeutic monoclonal antibodies (mAbs) have become a powerful therapeutic modality for various diseases, including cancer, inflammatory diseases, and autoimmune diseases. As of 2020, the US Food and Drug Administration (FDA) has approved 84 therapeutic mAbs, and more than 500 therapeutic mAbs are being studied in clinical trials worldwide.1 Most of these mAbs are IgG isotypes, with two N-glycans attached to the heavy chain at Asn297 in the Fc region. Importantly, the use of mammalian expression systems results in significant heterogeneity of these glycans. Such variation in N-linked glycans can affect the biological activity, pharmacokinetics, stability, and immunogenicity of mAbs. Therefore, it is particularly important to fully characterize the N-glycan profile and routinely monitor it to ensure the consistent quality of the therapeutic antibodies.
Due to its complex structure, the complete analysis of the N-glycan structure on glycoproteins is a very challenging task. In conventional glycan structural analysis, high-performance liquid chromatography (HPLC), capillary electrophoresis (CE), and lectin microarray are used to detect fluorescence-labeled glycans released from glycoconjugates. However, the depth of analysis, throughput, and reproducibility have been considered inadequate for the evaluation of drug quality, i.e., chemistry, manufacturing, and control (CMC).2,3 In the last few decades, various mass spectrometry techniques have emerged as attractive alternatives to these technologies. In fact, existing mass spectrometry platforms, such as liquid chromatography (LC)–mass spectrometry (MS) and CE-MS, were able to analyze detailed glycan structures rapidly and with high sensitivity. Typically, these methods can detect glycans on the order of femtomoles (fmol) to nanomoles (nmol) within 1 to 2 h of mass spectrometric analysis.4−6 Here, we developed a fast, quantitative, and highly sensitive analytical platform for mAb glycan profiling for routine CMC procedures and basic research.
Experimental Section
Reagents
Bevacizumab (Avastin) and trastuzumab (Herceptin) were purchased from Chugai Pharmaceutical (Tokyo, Japan). Nivolumab (Opdivo) was purchased from Ono Pharmaceutical (Osaka, Japan), and ramucirumab (Cyramza) was purchased from Eli Lilly Japan (Hyogo, Japan). Rituximab (Rituxan) was obtained from Zenyaku Kogyo (Tokyo, Japan). Rapid PNGase F was purchased from New England Biolabs (Tokyo, Japan). The BlotGlyco kit was from Sumitomo Bakelite Co. Ltd. (Tokyo, Japan), and PA-glucose oligomer was obtained from TaKaRa (Kyoto, Japan). Sepharose CL-4B, ribonuclease B (RNase B), bovine fetuin, and LC/MS-grade ammonium formate were obtained from Sigma-Aldrich (St. Louis, MO). All other unspecified reagents were products of FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan).
N-Glycan Release and Derivatization
N-glycans were released from proteins using Rapid PNGase F reagent according to the manufacturer’s protocol. To extract the glycans, the digest mixture was adjusted to 85% acetonitrile (ACN) (v/v). Sepharose CL-4B beads (50 μL of 50% slurry) were aliquoted onto a well of a MultiScreen Solvinert filter plate (Millipore, Billerica, MA), conditioned with water (200 μL), equilibrated with 85% (v/v) ACN (200 μL), and then loaded with the sample. During adsorption, the plate was gently shaken for 15 min at room temperature. The adsorbed samples were subsequently washed twice in 200 μL volumes of a solution containing 0.1% trifluoroacetic acid (TFA) (v/v) in 85% (v/v) ACN and twice with 85% (v/v) ACN. Last, the enriched glycans were eluted in 100 μL of water. The extracted N-glycans were dried in vacuo. The released N-glycans were acetylated with 5 μL of dehydrated pyridine and 5 μL of acetic anhydride at 50 °C for 4 h and dried by SpeedVac. The progress of the reaction was monitored by MALDI-TOF-MS analysis (Figures S1 and S2). For HPLC analysis, the released N-glycans were pyridylaminated (PA-labeling) by a BlotGlyco kit according to the manufacturer’s protocol.
MALDI-TOF-MS Analysis
Approximately 4 pmol of peracetylated N-glycan was dissolved in 10 μL of methanol, of which 0.5 μL was applied onto an Opti-TOF 384-well plate (AB Sciex, Foster City, CA) together with 0.5 μL of CHCA matrix solution (4 mg/mL concentration in 70% acetonitrile, 0.1% TFA, and 0.08 mg/mL ammonium citrate). The samples were left to dry in air. MALDI-TOF-MS analysis was performed using a 4800 Plus MALDI TOF/TOF Analyzer (AB Sciex) operated using 4000 Series Explorer software version 3.5. The instrument was calibrated with the peptide calibration standard (AB Sciex) prior to analysis of the samples. MS measurements were performed in reflectron positive ion mode. For each spot, data were accumulated from 1000 laser shots in a randomized raster of 400 μm diameter over a mass range of m/z 1200–5000. The laser repeat rate was 200 Hz, and the laser power was fixed at 3500 units throughout the experiment.
SFC-MS Analysis
The acetylated glycans were dissolved in methanol and analyzed using a Shimadzu Nexera UC supercritical fluid chromatography system coupled with a Shimadzu LCMS-8050 triple-quadrupole mass spectrometer (Shimadzu, Kyoto, Japan). CO2 (99.99% grade, Iwatani Corporation, Osaka, Japan) was used as the mobile phase of SFC. The SFC-MS analytical conditions were as follows: modifier, methanol; injection volume, 2.5 μL; flow rate, 1.0 mL/min; make-up solvent, methanol containing 0.1% (v/v) ammonium formate; make-up flow rate, 0.1 mL/min; column oven temperature, 40 °C; and column, Shim-pack UC-Phenyl (2.1 × 150 mm, 3 μm; Shimadzu, Kyoto, Japan). Ammonium formate was added into the make-up solvents to promote ionization.7 The gradient conditions were as follows: 10% B (0 min), 40% B (4 min), 40% B (7 min), 10% B (7.1 min), and then stop (8.5 min). The parameters of LCMS-8050 were set as follows: interface voltage = 4000 V, interface temperature = 225 °C, heat block temperature = 400 °C, desolvation temperature = 225 °C, CID gas pressure = 270 kPa (argon gas), Q1 resolution = unit, and Q3 resolution = unit. For the relative quantification of IgG-Fc glycan microheterogeneity, Q1 transitions were set so that they covered all possible glycan compositions. The Q3 transition was set at m/z 210, indicating the dehydrated fragment ion of acetylated N-acetylglucosamine (GlcNAc) at the reducing terminus (i.e., [N-acetylhexosamine (HexNAc) + Ac – 2H2O]+), which was utilized as a quantitative reporter ion. The exact masses and compositions of other acetylated glycan-derived oxonium ions (Figure S3) are listed in Table S1. The collision energy for the detection of m/z = 210 was scanned and adjusted so that the intensity of this acetylated glycan-derived fragment ion reached the maximum (Figure S4). The optimized MRM parameters are listed in Table S2.
Fluorescent HPLC Analysis
Reverse-phase HPLC was performed using an Inertsil ODS-3 column (2.1 mm × 150 mm, 3 μm; GL Sciences, Tokyo, Japan). The elution conditions were as follows: solvent A, 0.1 M ammonium acetate buffer (pH 4.0); solvent B, 0.1 M ammonium acetate buffer (pH 4.0, containing 0.5% 1-butanol). The column was equilibrated with 5% solvent B at a flow rate of 200 μL/min. The injected samples were separated with a linear gradient from 5 to 55% solvent B over 100 min. The fluorescence of the labeled glycans was detected at the excitation wavelength (320 nm) and the emission wavelength (400 nm). The retention times were normalized and expressed as glucose units using PA-glucose oligomer, and the glycan structures were verified according to the HPLC mapping data as described previously.8
SFC-MS Data Processing
The acquired raw data were processed with LabSolutions LCMS software (Shimadzu), and the area of the product ion chromatogram for m/z = 210 was utilized for glycoform quantification. For relative quantification of the Fc-glycan profile, the percentage composition of each glycoform was calculated with respect to the sum of all glycoforms combined. To exclude cross-talk between transitions where the precursor ion masses are very close, a peak area curve for correction was created, and the correction value was subtracted for the corresponding glycan (Figure S5). The glycan composition is shown as a glycan ID, and the five digits correspond to HexNAc, Hexose, Fucose, Neu5Ac (N-acetylneuraminic acid), and Neu5Gc (N-glycolylneuraminic acid).
Statistical Analysis
Principal component analysis (PCA) was performed using Genedata Analyst software (Basel, Switzerland). Other statistical calculations for the clinical data were performed using GraphPad Prism 8 software (version 8.4.3). Intergroup differences were statistically compared by Student’s t test. When the means of more than two groups were compared, Tukey’s multiple comparisons test was used. In all these analyses, P < 0.05 was considered statistically significant.
Results and Discussion
Development of an Ultrafast Glycoform Profiling Method for Antibodies
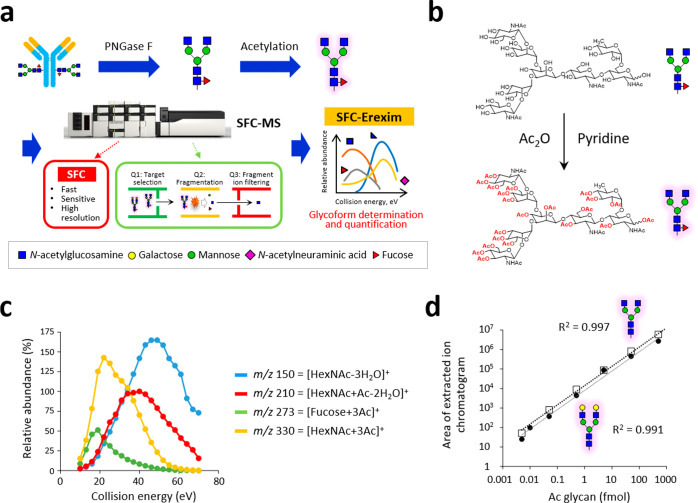
To establish a more time-efficient and sensitive glycoform analysis method, we employed supercritical fluid chromatography (SFC)-MS for the first time and developed an ultrafast and ultrasensitive glycoform profiling technique (Figure 1a). As the mobile phase of SFC, supercritical fluid carbon dioxide (CO2) was used in this study. CO2 has a low viscosity and a high diffusion coefficient comparable to that of a gas, allowing for fast separation of analytes at a much higher resolution and lower pressure than HPLC.9 Despite these great advantages, SFC has not been used for glycan analysis, which is due to the low solubility of high-polarity compounds in supercritical fluid CO2.7 In other words, the native form of glycans is almost insoluble in supercritical fluid CO2 due to its high polarity. To overcome this issue, we established a one-pot, cleanup-free protocol for peracetylation of glycans (Figure 1b). This method can replace all polar hydroxyl groups in the released N-glycans with nonpolar acetyl groups. Regardless of the glycan structure, the labeling efficiency was higher than 95% (Figures S1 and S2). A reaction time of 1 h was sufficient for glycans with simple structures, such as high mannose-type glycans, while the derivatization of highly branched glycans with multiple sialic acids took longer. On the other hand, when the reaction time was longer than 4 h (6–24 h), the yield decreased. The optimal reaction time for complete acetylation of all structures was 4 h. Eventually, the peracetylation pretreatment successfully enabled us to solubilize glycans in supercritical fluid CO2 and analyze them with SFC-MS.
Figure 1.
Novel glycoform profiling using SFC-MS. (a) Scheme of mass spectrometric glycan structure analysis by SFC-Erexim (energy-resolved oxonium ion monitoring) technology. N-glycans are released from glycoprotein samples (immunoglobulin G), acetylated, and directly subjected to SFC-MS analysis. In the quadrupole mass spectrometer, the first quadrupole (Q1) isolated glycan ions with unique intact mass, which were subsequently guided into the second quadrupole (Q2), where they underwent collision-induced dissociation, with continuously changed collision energies. The third quadrupole (Q3) filters the product ions to selectively detect oligosaccharide-derived ions. (b) Reaction scheme for the peracetylation of N-glycans. (c) Energy-resolved product ion profiles of a representative per-acetylated N-glycan (glycan ID: 43100, the most abundant glycoform in IgG). (d) Quantitative linearity evaluation for two N-glycan standards (glycan IDs: 43000 and 45000). Correlation coefficients between experimentally measured concentrations and expected concentrations are shown. Error bars represent the standard deviation (n = 6, independent technical experiments).
To achieve both the highest sensitivity and the most comprehensive glycan profiling, we integrated our energy-resolved oxonium ion monitoring (Erexim) method with SFC-MS to construct a new SFC-Erexim technology. The Erexim method utilizes the multiple reaction monitoring (MRM) method with collision energy scanning, which enables qualitative and quantitative glycan profiling.10,11 Collision-induced dissociation (CID) fragmentation of glycans/glycopeptides generates glycan-derived fragment ions called oxonium ions.12 We have previously found that the original glycan structure can be determined by continuously changing the collision energy and monitoring the variety and quantity of oxonium ions. Based on this Erexim technology, we first observed oxonium ions generated from acetylated glycans (Figure S3 and Table S1). As in the usual Erexim method, we scanned oxonium ions at various collision energies and plotted the ion yield against the collision energy (Figure 1c and Figure S6). Figure 1c shows the SFC-Erexim curves obtained from the glycan composed of 4[HexNAc]-3[Hexose]-1[Fucose]-0[Neu5Ac]-0[Neu5Gc] (glycan ID: 43100). The oxonium ion m/z = 210 derived from acetylated GlcNAc at the reducing end was utilized as a reporter ion for quantification. The collision energy for the detection of m/z = 210 was scanned and adjusted so that the intensity of this oxonium ion reached the maximum (Figure S4) since we have previously reported that the amount of oxonium ions produced from GlcNAc at the reducing end in this collision energy quantitatively correlates with the concentration of the original glycan regardless of the glycan structure.10 The optimized parameters for the SFC-Erexim analysis are listed in Table S2. Acetylated glycans were detected as H+- and/or NH4+-adduct forms in the SFC-MS analysis.
The absolute sensitivity of the SFC-MS method was assessed by analyzing the peracetylated glycan standards (glycan IDs: 43000 and 45000). The lower limit of detection (LOD) and lower limit of quantification (LOQ) were 5 attomoles (amol) and 10 amol, respectively. The quantitative dynamic range was over 5 orders of magnitude (R2 > 0.99), highlighting the broad dynamic range of this platform (Figure 1d).
SFC-MS measurements of permethylated13 or peracetylated N-glycans with the reducing end labeled14 were already performed three decades ago, but their sensitivity was quite low. Indeed, although the LOD was not determined in these studies, the authors concluded that glycans derived from approximately 1 pg of sheep urine protein samples were detectable.14 Similarly, in the other study, only eight high mannose-type glycan structures were identified based on the masses of precursor ions and their chromatographic properties from 200–400 ng protein samples derived from mannosidosis patients’ urine.13 After these trials, the idea of using SFC-MS for glycan structural analysis has been unfortunately suspended. Compared to them, the analytical depth of our SFC-MS-based glycan structure profiling has been significantly improved by employing Erexim-assisted sensitive and high-throughput scanning as well as a high-end triple quadrupole mass spectrometer, allowing attomolar scale detection.
Sensitivity and Quantitative Accuracy of a Novel Glycoform Profiling Method
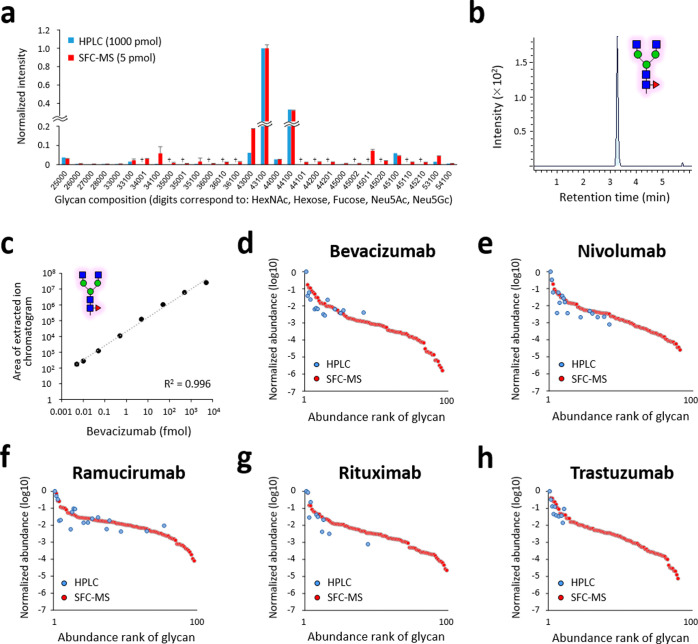
Next, we applied the SFC-Erexim method to the evaluation of glycan microheterogeneity for five therapeutic antibodies (i.e., bevacizumab, nivolumab, ramucirumab, rituximab, and trastuzumab) (Table S3). To confirm the quantitative reliability of the SFC-Erexim method, the relative abundance of each glycan quantified by this method was statistically compared with that from the traditional fluorescent HPLC method. The quantitative profiles of the major glycan structures were similar between the two methods, indicating that the outcome of SFC-Erexim analysis reflects the actual glycan profile of the therapeutic antibodies (Figure 2a and Figure S7). The accuracy of the quantification was maintained even though the glycans released from the antibodies equivalent to 1000 pmol by HPLC and 5 pmol by SFC-MS were measured. On the other hand, the quantitative profiles did not always match with respect to glycan structures with low abundance (Figure 2a). As shown in Figure S8, the HPLC results showed poor peak separation and may not be quantitatively accurate, especially for structures with low abundance. In contrast, our SFC-Erexim method employs the MRM method, which allows accurate quantification even at low concentrations without contamination of components with close retention time (Figure 2b). Furthermore, the SFC-Erexim method was highly reproducible between experiments even for low-abundance glycan structures (Figure S9). These results clearly show that the SFC-Erexim method is much more sensitive and robust than the conventional fluorescence HPLC method.
Figure 2.
Glycoform profiling of therapeutic antibodies by SFC-MS. (a) Relative abundances of the 30 most abundant glycoforms of nivolumab are displayed. The released N-glycans from nivolumab were analyzed after PA labeling or peracetylation by fluorescence HPLC (1000 pmol, blue bar) or SFC-MS (5 pmol, red bar), respectively. Error bars represent the standard deviation (n = 3, independent technical experiments). †, not detected. (b) Extracted ion chromatogram of glycan 43100 after injection of 10 amol of bevacizumab-derived peracetylated N-glycans. (c) Quantitative linearity evaluation of a peracetylated N-glycan (glycan ID: 43100, the most abundant glycoform in bevacizumab). Correlation coefficients between experimentally measured concentrations and expected concentrations are shown. Error bars represent the standard deviation (n = 3, independent technical experiments). (d–h) Rank plots of detected glycoform relative abundance for (d) bevacizumab, (e) nivolumab, (f) ramucirumab, (g) rituximab, and (h) trastuzumab. The released N-glycans from the five therapeutic antibodies were analyzed after PA labeling or peracetylation by fluorescence HPLC (1000 pmol, blue) or SFC-MS (5 pmol, red), respectively.
A total of 102 glycan structures (i.e., non-redundant number of structures detected from five antibodies), even from the 0.1% content structures, were quantitatively detected in 8 min SFC-Erexim runs (Table S4), indicating that therapeutic mAbs have huge glycan microheterogeneities. In addition, our method shows an advantage in the depth of analysis compared to previous reports, which detected 21 glycoforms from bevacizumab by a 60 min LC-MS run15 or 19 glycoforms from rituximab and 14 glycoforms from trastuzumab by a 45 min LC-MS/MS run.16 The lower LOD and lower LOQ of the most abundant N-glycan, glycan ID: 43100, released from bevacizumab were 5 and 10 amol, respectively (Figure 2b,c). It is known that various bioprocess conditions, such as incubation temperature and pH, affect glycan formation.17 Therefore, there is a great possibility that the balance of glycan content may be disrupted during the manufacturing process. Since monitoring only major glycan structures may miss abnormal glycan balance, it is worthwhile to obtain more information rather than monitoring only a limited number of major glycoforms.
To assess the dynamic range of the SFC-Erexim method, the detected glycan structures from the five therapeutic mAbs were ranked in order of their relative abundances and displayed with the measured results by the fluorescence HPLC method (Figure 2d–h). These abundance plots showed that the dynamic range of the SFC-Erexim method covered 4 to 6 orders of magnitude, whereas that of the fluorescent HPLC method was 2 to 3 orders of magnitude. The number of glycan structures detected was 96 for bevacizumab by the SFC-Erexim method, compared to 15 structures by the HPLC method. Thus, the depth of our SFC-Erexim method is much greater than that of the conventional HPLC method.
Glycoform Characteristics of the Five Therapeutic Antibodies
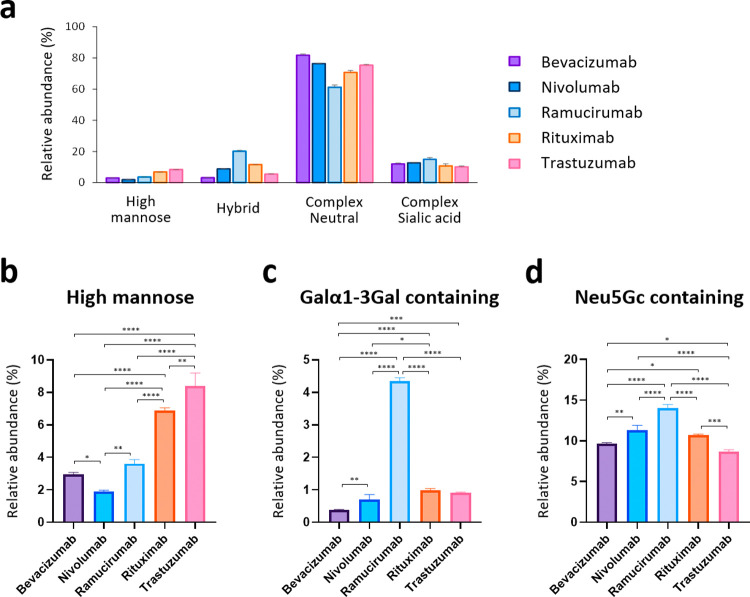
The detected N-glycans were then classified according to structural characteristics and compared among the five therapeutic mAbs (Figure 3a and Figure S10). The most abundant N-glycan type was neutral (i.e., not sialylated) complex-type structures for all antibodies. The relative abundance of high mannose-type glycans was found to vary widely, from 1.9% (nivolumab) to 8.4% (trastuzumab) (Figure 3b). Since high mannose-type glycans in the Fc region have been reported to significantly affect the circulating half-life of therapeutic antibodies,18,19 monitoring and manipulating this parameter may be valuable for controlling the pharmacokinetics of mAbs.
Figure 3.
Comparison of N-glycans from the five therapeutic antibodies. (a) Category comparison of N-glycomes obtained from five therapeutic antibodies. Error bars represent the standard deviation determined from three technical replicates (for statistical analysis, see Figure S10). (b–d) Relative abundances of (b) high mannose, (c) Galα1-3Gal-containing, and (d) Neu5Gc-containing glycans of the five therapeutic antibodies. Error bars represent the standard deviation (n = 3, independent technical experiments). The P values were calculated by Tukey’s multiple comparisons test. Only statistically significant differences are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Therapeutic monoclonal antibodies produced in murine myeloma cells (e.g., SP2/0 or NS0) contain terminal α1,3-linked galactose (Galα1-3Gal) structures on their Fc glycans.20 The Galα1-3Gal structure in the Fab region of the cetuximab heavy chain produced by SP2/0 cells has been associated with IgE-mediated anaphylactic reactions;21,22 however, anti-α1-3Gal IgE in allergic patients does not bind to α1,3-galactosylated glycans on the Fc domain,23 and therefore, antibodies with N-glycans only in the Fc region are considered to have a low risk of adverse events due to this structure.24 Therapeutic mAbs usually lack Fab glycosylation and carry glycans only on the Fc domain25 but need to be monitored and controlled for Galα1-3Gal levels during the manufacturing process. Of the five mAbs used in this study, only ramucirumab was produced in mouse NS0 cells, while the others were produced in CHO cells (Table S3). Glycoform profiling clearly showed that Galα1-3Gal structures were highly expressed in ramucirumab but negligible for other CHO-derived antibodies (Figure 3c). Our results also corroborated previous findings that terminal nonhuman sialic acid Neu5Gc is often found in the N-glycans of recombinant IgG produced in murine myeloma and, to a much lesser extent, CHO cells24 (Figure 3d). It is essential to determine and control the exact proportion of these nonhuman immunogenic components.
Lot-to-Lot Heterogeneity of Therapeutic Antibodies
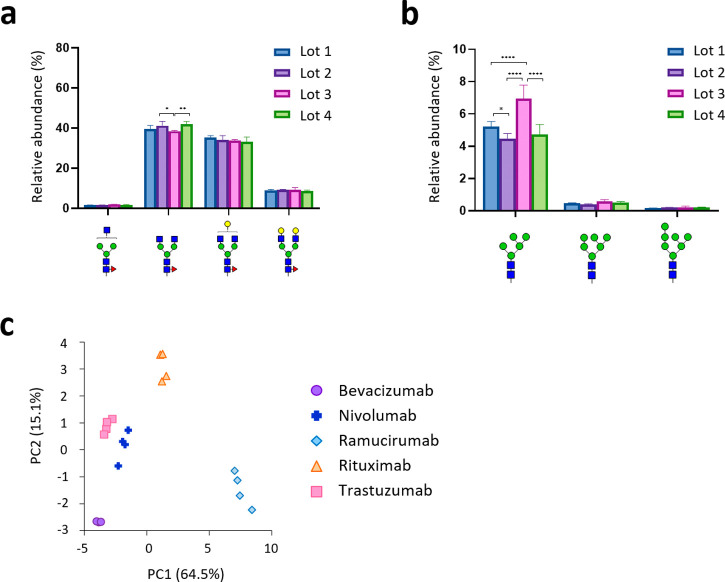
To demonstrate the robustness and sensitivity of this method, we next analyzed four lots of each therapeutic antibody and evaluated the lot-to-lot heterogeneity (Figure 4a,b and Figures S11 and S12). Interestingly, among the 96 glycan structures quantified in this experiment, the abundance of the M5 isoform (high mannose-type N-glycan containing five mannose residues) showed significant lot-to-lot variation for all mAbs (Figure 4b and Figure S12). Therapeutic IgGs containing high mannose-type N-glycans are known to be selectively cleared by a mannose receptor-mediated mechanism.18,19,24,26 Since high mannose-type glycans in the Fc region can have a great impact on therapeutic mAb pharmacokinetics,17 monitoring and controlling high mannose-type N-glycan levels in manufacturing processes is important. Finally, principal component analysis (PCA) of the SFC-Erexim dataset clearly illustrated glycomic landscape-dependent classification of therapeutic mAbs (Figure 4c). In this PCA plot, the glycan profile of ramucirumab was significantly different from the others, which may be due to differences in the producing cells (Table S3). On the other hand, the other four mAbs were clustered separately, even though they were all produced in the same cell line, CHO. This observation indicates that the glycan profile of each therapeutic mAb is unique and is influenced not only by the type of producing cells but also by the culture conditions.
Figure 4.
Evaluation of lot-to-lot heterogeneity. (a) Evaluation of lot-to-lot heterogeneity of ramucirumab for major fucosylated N-glycans. Relative abundances of glycoforms 33100, 43100, 44100, and 45100 of four lots of ramucirumab are displayed. (b) Evaluation of lot-to-lot heterogeneity of ramucirumab for high mannose-type N-glycans. Relative abundances of glycoforms 25000, 26000, and 27000 of four lots of ramucirumab are displayed. Error bars represent the standard deviation (n = 3). The P values were calculated by Tukey’s multiple comparisons test. Only statistically significant differences are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (c) Principal component analysis (PCA) of the five therapeutic antibodies based on N-glycomic data from four lots of each antibody.
Conclusions
In this study, we established a highly sensitive, quantitative, and fast profiling technology for glycan structure analysis using SFC-MS. The SFC-Erexim method is considered to be a breakthrough achievement in glycan analysis because it can provide a detection sensitivity of >5 amol within an 8 min measurement. Furthermore, our sample preparation method for SFC-MS is a simple and low-cost procedure that does not require expensive fluorescent labeling reagents or laborious purification steps. With these advantages, our SFC-Erexim are expected to enable not only the routine quality evaluation of biopharmaceuticals but also the development of new drugs and evaluation of generic drugs at a reasonable throughput. In addition to the antibodies targeted in this study, regulating glycosylation is very important in the development of any biologics as glycans play an essential role in both safety and effector functions such as antibody-dependent cellular cytotoxicity (ADCC). In therapeutic mAbs alone, there are currently three commercially available glycosylated antibodies (mogamulizumab, obinutuzumab, and benralizumab), and more than 20 antibodies have been evaluated in clinical trials.27 Our SFC-Erexim method can also contribute to the acceleration of various innovative basic studies, such as the analysis of single-cell glycan profiles and the identification of glycan biomarkers from small amounts of clinical specimens. Based on the knowledge obtained in this report, further improvement of analytical performance and expansion of applications (e.g., O-glycosylation) are expected in the future by altering the type of supercritical fluid as the mobile phase, make-up solvent, and SFC column.
Acknowledgments
This work was supported by JSPS KAKENHI (JP19K06553) and the Project for Utilizing Glycans in the Development of Innovative Drug Discovery Technologies from the Japan Agency for Medical Research and Development (AMED).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c01721.
(Figure S1) MALDI-TOF-MS spectrum of time course experiments under acetylation of N-glycans; (Figure S2) MALDI-TOF-MS spectrum of acetylated N-glycans; (Figure S3) MS/MS fragmentation spectra of peracetylated N-glycans; (Figure S4) energy-resolved product ion profiles of various peracetylated N-glycans; (Figure S5) identification of resolution of LCMS-8050 to evaluate the cross-talk; (Figure S6) energy-resolved product ion profiles of peracetylated N-glycans; (Figure S7) comparison of therapeutic IgG N-glycan analysis of fluorescence HPLC and SFC-MS; (Figure S8) elution profile of N-glycans from therapeutic antibodies analyzed by fluorescent HPLC; (Figure S9) inter- and intraday repeatability of low-abundance glycan analysis by SFC-Erexim; (Figure S10) comparison of N-glycans from five therapeutic antibodies; (Figure S11) evaluation of lot-to-lot heterogeneity for major fucosylated N-glycans; (Figure S12) evaluation of lot-to-lot heterogeneity for high mannose-type N-glycans (PDF)
(Table S1) Masses and identities of acetylated glycan-derived fragment ions; (Table S2) MRM parameters for quantification of antibody drug glycoforms; (Table S3) list of therapeutic antibodies analyzed in this study; (Table S4) relative abundance (%) of detected glycoforms of each therapeutic antibody (XLSX)
Author Contributions
Y. Haga, M.Y., Y. Hayakawa, and K.U. designed the research. Y. Haga, M.Y., and R.F. performed experiments and analyzed the data. Y. Haga and N.S. performed the statistical analysis. T.Y. and T.H. provided critical reagents. Y. Haga and K.U. wrote the manuscript.
The authors declare the following competing financial interest(s): M.Y. and Y. Hayakawa are employees of Shimadzu Corporation. The other authors have no financial relationships to disclose.
Supplementary Material
References
- Kaplon H.; Muralidharan M.; Schneider Z.; Reichert J. M. Antibodies to watch in 2020. mAbs 2020, 12, 1703531. 10.1080/19420862.2019.1703531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk B.; Tharmalingam T.; Rudd P. M. Glycans as cancer biomarkers. Biochim. Biophys. Acta 2012, 1820, 1347–1353. 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Reusch D.; Haberger M.; Maier B.; Maier M.; Kloseck R.; Zimmermann B.; Hook M.; Szabo Z.; Tep S.; Wegstein J.; Alt N.; Bulau P.; Wuhrer M. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles--part 1: separation-based methods. mAbs 2015, 7, 167–179. 10.4161/19420862.2014.986000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch D.; Haberger M.; Falck D.; Peter B.; Maier B.; Gassner J.; Hook M.; Wagner K.; Bonnington L.; Bulau P.; Wuhrer M. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 2: Mass spectrometric methods. mAbs 2015, 7, 732–742. 10.1080/19420862.2015.1045173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.; Zhao J.; Peng W.; Banazadeh A.; Williamson S. D.; Goli M.; Huang Y.; Mechref Y. Advances in mass spectrometry-based glycoproteomics. Electrophoresis 2018, 39, 3104–3122. 10.1002/elps.201800272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageveen-Kammeijer G. S. M.; de Haan N.; Mohaupt P.; Wagt S.; Filius M.; Nouta J.; Falck D.; Wuhrer M. Highly sensitive CE-ESI-MS analysis of N-glycans from complex biological samples. Nat. Commun. 2019, 10, 2137. 10.1038/s41467-019-09910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Velde B.; Guillarme D.; Kohler I. Supercritical fluid chromatography - Mass spectrometry in metabolomics: Past, present, and future perspectives. J Chromatogr B Analyt Technol Biomed Life Sci 2020, 1161, 122444 10.1016/j.jchromb.2020.122444. [DOI] [PubMed] [Google Scholar]
- Tomiya N.; Awaya J.; Kurono M.; Endo S.; Arata Y.; Takahashi N. Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal. Biochem. 1988, 171, 73–90. 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- Sethi N.; Anand A.; Jain G.; Srinivas K.; Chandrul K. Supercritical fluid chromatography-A Hybrid of GC and LC. Chron. Young Sci. 2010, 1, 12–22. [Google Scholar]
- Toyama A.; Nakagawa H.; Matsuda K.; Sato T. A.; Nakamura Y.; Ueda K. Quantitative structural characterization of local N-glycan microheterogeneity in therapeutic antibodies by energy-resolved oxonium ion monitoring. Anal. Chem. 2012, 84, 9655–9662. 10.1021/ac3023372. [DOI] [PubMed] [Google Scholar]
- Haga Y.; Uemura M.; Baba S.; Inamura K.; Takeuchi K.; Nonomura N.; Ueda K. Identification of Multisialylated LacdiNAc Structures as Highly Prostate Cancer Specific Glycan Signatures on PSA. Anal. Chem. 2019, 91, 2247–2254. 10.1021/acs.analchem.8b04829. [DOI] [PubMed] [Google Scholar]
- Halim A.; Westerlind U.; Pett C.; Schorlemer M.; Rüetschi U.; Brinkmalm G.; Sihlbom C.; Lengqvist J.; Larson G.; Nilsson J. Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 2014, 13, 6024–6032. 10.1021/pr500898r. [DOI] [PubMed] [Google Scholar]
- Leroy Y.; Lemoine J.; Ricart G.; Michalski J. C.; Montreuil J.; Fournet B. Separation of oligosaccharides by capillary supercritical fluid chromatography and analysis by direct coupling to high-resolution mass spectrometer: application to analysis of oligomannosidic N-glycans. Anal. Biochem. 1990, 184, 235–243. 10.1016/0003-2697(90)90674-X. [DOI] [PubMed] [Google Scholar]
- Sheeley D. M.; Reinhold V. N. Characterization of N-linked glycans by supercritical fluid chromatography-mass spectrometry. Anal. Biochem. 1991, 193, 240–247. 10.1016/0003-2697(91)90016-M. [DOI] [PubMed] [Google Scholar]
- Yang N.; Goonatilleke E.; Park D.; Song T.; Fan G.; Lebrilla C. B. Quantitation of Site-Specific Glycosylation in Manufactured Recombinant Monoclonal Antibody Drugs. Anal. Chem. 2016, 88, 7091–7100. 10.1021/acs.analchem.6b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Wang G.; Song T.; Lebrilla C. B.; Heck A. J. R. Resolving the micro-heterogeneity and structural integrity of monoclonal antibodies by hybrid mass spectrometric approaches. mAbs 2017, 9, 638–645. 10.1080/19420862.2017.1290033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangeli R.; Audino M. C.; Palinsky W.; Broly H.; Bierau H. The Formidable Challenge of Controlling High Mannose-Type N-Glycans in Therapeutic mAbs. Trends Biotechnol. 2020, 38, 1154–1168. 10.1016/j.tibtech.2020.05.009. [DOI] [PubMed] [Google Scholar]
- Goetze A. M.; Liu Y. D.; Zhang Z.; Shah B.; Lee E.; Bondarenko P. V.; Flynn G. C. High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology 2011, 21, 949–959. 10.1093/glycob/cwr027. [DOI] [PubMed] [Google Scholar]
- Yu M.; Brown D.; Reed C.; Chung S.; Lutman J.; Stefanich E.; Wong A.; Stephan J. P.; Bayer R. Production, characterization, and pharmacokinetic properties of antibodies with N-linked mannose-5 glycans. mAbs 2012, 4, 475–487. 10.4161/mabs.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeley D. M.; Merrill B. M.; Taylor L. C. Characterization of monoclonal antibody glycosylation: comparison of expression systems and identification of terminal alpha-linked galactose. Anal. Biochem. 1997, 247, 102–110. 10.1006/abio.1997.2036. [DOI] [PubMed] [Google Scholar]
- Chung C. H.; Mirakhur B.; Chan E.; Le Q. T.; Berlin J.; Morse M.; Murphy B. A.; Satinover S. M.; Hosen J.; Mauro D.; Slebos R. J.; Zhou Q.; Gold D.; Hatley T.; Hicklin D. J.; Platts-Mills T. A. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N. Engl. J. Med. 2008, 358, 1109–1117. 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts-Mills T. A.; Schuyler A. J.; Tripathi A.; Commins S. P. Anaphylaxis to the carbohydrate side chain alpha-gal. Immunol. Allergy Clin. 2015, 35, 247–260. 10.1016/j.iac.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bueren J. J.; Rispens T.; Verploegen S.; van der Palen-Merkus T.; Stapel S.; Workman L. J.; James H.; van Berkel P. H.; van de Winkel J. G.; Platts-Mills T. A.; Parren P. W. Anti-galactose-α-1,3-galactose IgE from allergic patients does not bind α-galactosylated glycans on intact therapeutic antibody Fc domains. Nat. Biotechnol. 2011, 29, 574–576. 10.1038/nbt.1912. [DOI] [PubMed] [Google Scholar]
- Reusch D.; Tejada M. L. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology 2015, 25, 1325–1334. 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R. Glycosylation of recombinant antibody therapeutics. Biotechnol. Prog. 2005, 21, 11–16. 10.1021/bp040016j. [DOI] [PubMed] [Google Scholar]
- Falck D.; Thomann M.; Lechmann M.; Koeleman C. A. M.; Malik S.; Jany C.; Wuhrer M.; Reusch D. Glycoform-resolved pharmacokinetic studies in a rat model employing glycoengineered variants of a therapeutic monoclonal antibody. mAbs 2021, 13, 1865596. 10.1080/19420862.2020.1865596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira N. A.; Chan K. F.; Lin P. C.; Song Z. The ″less-is-more″ in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. mAbs 2018, 10, 693–711. 10.1080/19420862.2018.1466767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.