Abstract
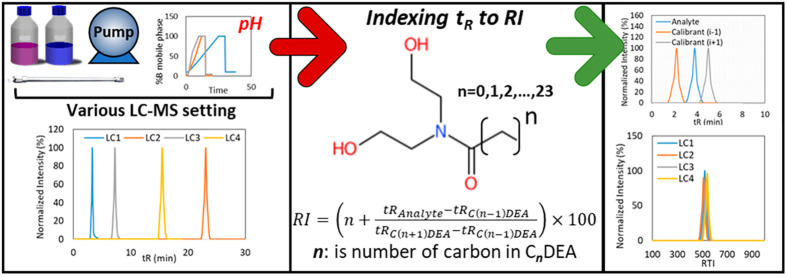
There is a growing need for indexing and harmonizing retention time (tR) data in liquid chromatography derived under different conditions to aid in the identification of compounds in high resolution mass spectrometry (HRMS) based suspect and nontarget screening of environmental samples. In this study, a rigorously tested, inexpensive, and simple system-independent retention index (RI) approach is presented for liquid chromatography (LC), based on the cocamide diethanolamine homologous series (C(n = 0–23)-DEA). The validation of the CDEA based RI system was checked rigorously on eight different instrumentation and LC conditions. The RI values were modeled using molecular descriptor free technique based on structural barcoding and convolutional neural network deep learning. The effect of pH on the elution pattern of more than 402 emerging contaminants were studied under diverse LC settings. The uncertainty associated with the CDEA RI model and the pH effect were addressed and the first RI bank based on CDEA calibrants was developed. The proposed RI system was used to enhance identification confidence in suspect and nontarget screening while facilitating successful comparability of retention index data between various LC settings. The CDEA RI app can be accessed at https://github.com/raalizadeh/RIdea.
Introduction
Coupling liquid chromatography (LC) to mass spectrometry (MS) and recently high-resolution mass spectrometry (LC-HRMS) were a breakthrough in the analysis of complex environmental samples.1,2 Not only the detection limit and sensitivity are improved for compounds at lower concentration, but also a new area in the screening strategies has been introduced.3 The conventional target screening is proceeded with two new screening approaches, so-called suspect and nontarget screening. In other words, a compound can be identified tentatively in the sample and, at a certain confidence, even if its reference standard is not available at the time of analysis.4 However, for a single mass to charge (m/z), many candidates can be assigned, and the bottleneck is to find the true positive compound among the pool of false positive candidates.1 In addition to the MS fragmentation pattern, it is common practice to use predicted and experimental retention time (tR) data and ionization behavior as well as other evidence to eliminate false positives.5 However, for many reasons, such as different mobile phase compositions, stationary phase chemistry (LC column), gradient or isocratic elution program, column temperature, and pump, the routine application of retention time data in LC-HRMS-based screening is challenging.6 Therefore, there is a demand to transform the retention time information into an indexing system to be less LC system-dependent.
Retention time data plays a key role in the gas chromatography (GC)-based identification workflows due to the existence of three main retention indexing (RI) systems, including Kovats-RI,7 Lee-RI,8 and Fiehn-RI,9 which are based on n-alkanes, polycyclic aromatic hydrocarbons (PAHs), and fatty acid methyl esters (FAMEs), respectively. The indexing system removes redundancies due to different temperature programs mainly. Unlike GC-RIs, there is not yet any suitable calibrants and RI systems for LC. Nevertheless, there have been few attempts to calibrate tR data and communicate the elution behavior of the analyte of interest across different LC conditions. Stanstrup et al.10 have introduced a method to directly project tR information between two different LC conditions (specially for reversed phase liquid chromatography (RPLC)) and thus enable communication of the tR data from one LC setting to another. However, this approach requires many chemicals to be measured and be common between different LC conditions to work appropriately. In our previous work,6 an index system was proposed that transfers tR data into a normalized tR value (RI values between 1 and 1000). The use of index values instead of tR values during LC-HRMS screening was easier, as it was less dependent on the LC conditions. This was only possible if the used calibrants had preserved linear and increasing elution order in the LC under investigation. Nevertheless, the main advantage of the University of Athens (UOA)-RI is the wide applicability domain and tR prediction models that can be used to exclude false positive candidates. UOA-RI is the first indexing method developed with a main application in environmental science and analysis of emerging contaminants. These two systems assume that the elution of analytes preserve in RPLC, and thus, a simple projection method can resolve the variabilities in the tR data.
Apart from projection methods, there have been efforts to create indexing system that are similar to n-alkanes used in GC. Hall and co-workers modeled an RI system based on n-nitroalkanes.11,12 Although this was a significant step toward indexing of tR values in LC and the metabolomic field, its validation with different LC settings as well as validation of RI formulation (logarithmic scale) was missing.11 Using logarithmic function in the RI formulation indicates that the changes in the polar area of the LC-chromatogram is not proportional to other medium and nonpolar segments. However, apart from difficulty in the synthesis (with alkyl halides and silver nitrite in water) of n-nitroalkanes and a low conversion yield (between 64 and 93%),12,13 the main drawback of this RI system is that n-nitroalkanes are not ionized properly at a reasonable concentration range in an electrospray ionization source. Another approach was proposed by Zheng et al.,14 which is based on a series of 2-dimethylaminoethylamine (DMED)-labeled fatty acids. Although Zheng-RI is a very promising strategy, it is proposed and tested in the case of DMED-labeled carboxylated compounds. In other words, the analytes should be chemically labeled first in order to fall into the correct elution order in contrast to analogous calibrants. The application of Zheng-RI would not be practical in the case of complex environmental samples because many different types of emerging pollutants exist and there is not any appropriate chemical labeling protocol. Recently, another interesting RI system was proposed based on series of N-alkylpyridinium sulfonates (NAPS),15 which is based on the patent filed by Michael Quilliam in 2016.16 Quilliam-RI can be ionized in both negative and positive ESI modes, and it is not yet tested for atmospheric pressure chemical ionization (APCI). From an elution pattern of NAPS (especially in the polar region of chromatogram, where NAPS (n = 1–3) elutes differently from the rest of the calibrants) it is evident that the possible logarithmic function is required in the formulation of Quilliam-RI.15 Therefore, the changes in the elution of NAPS in the polar region of RPLC is not linearly proportional to the rest of chromatogram. Moreover, the NAPS standards are complicated to synthesize and its application as RI is also patented until the next decade.16 Therefore, this restricts its use for routine application. Another fact about use of NAPS and other RIs such as nitroalkanes is the unresolved uncertainty associated with the effect of pH on the elution order.15 Since they are neutrally charged, their dissociation constant values (log D) may not change in proportion to mobile phase changes used in the gradient elution program. This might result in unchanged elution time for these RI systems in various pH values, while elution behavior of other analytes gets influenced by pH changes. In this regard, there is a need for a more accurate, cheap, stable, and open science strategy for indexing tR data from LC.
This work presents the development and validation of a new RI indexing system for RPLC-HRMS that is based on synthetic and naturally occurring amphiphilic homologous series of cocamide diethanolamine surfactant (C(n = 0–23)-DEA). Different LC settings, including various pH values, LC columns and temperatures, flow rates, mobile phase compositions, and gradient elution programs were tested. The applicability of this approach is evaluated in the LC-HRMS-based suspect and nontarget screening of a sewage sludge sample. The first RI database, including 3018 (2290 and 728 in +ESI and −ESI, respectively) emerging pollutants, is developed.
Experimental Section
Chemicals
Reference standards for pesticides were donated by Bruker Daltonics (Bremen, Germany) at a concentration of 1.0 mg L–1 in pure methanol (MeOH). The rest of the chemicals included in the study were purchased from Sigma–Aldrich. Individual stock solutions of these emerging pollutants were prepared in MeOH at a concentration of 1.0 mg L–1 and stored at −20 °C. The list of these chemicals can be found in the Supporting Information and in Tables S1 and S2 (the supplementary Excel file). Acetonitrile (ACN) and MeOH were purchased from Merck in LC-MS grade. The LC-MS grade isopropanol was purchased from Fisher Scientific. Ammonium acetate, ammonium formate, and formic acid, all LC-MS grade, were purchased from Fluka, Sigma–Aldrich. Distilled water used in the LC–MS analysis was provided by a Milli-Q purification apparatus (Millipore Direct-Q UV). The stock solution of atrazine-d5 (isotopically labeled standard (IS)) was prepared at 1.0 mg L–1 in MeOH (LC-MS grade). Ethylenediaminetetraacetic acid (EDTA, analytical grade, 99.0%) was purchased from Serva. Hexane and acetone (grade for pesticide analysis) were purchased from Carlo Erba Reagents (Spain). Sodium methoxide (reagent grade, 95.0%), methyl valerate (>99.0%), methyl propionate (>99.0%), methyl butyrate (>99.0%), heptanoic acid (>99.0%), henicosanoic acid, nonadecanoic acid, arachidic acid (>99.0%), pelargonic acid (96.0%), coconut oil (CAS 8001–31–8), and diethanolamine (reagent grade, >98.0%) were purchased from Sigma–Aldrich. Regenerated cellulose (RC) syringe filters (15 mm diameter, 0.22 μm pore size) were supplied from Phenomenex.
Instrumentation
An ultra-high-performance liquid chromatography (UHPLC) system, with a LPG-3400 pump (Dionex UltiMate 3000 RSLC, Thermo Fisher Scientific), coupled to a Quadrupole Time of Flight (Q-ToF) mass spectrometer (Maxis Impact, Bruker Daltonics) was used for the screening of analytes. Eight different LC settings were used to record the retention time of emerging contaminants and (C(n = 0–23)-DEA). The pH, mobile phase composition, LC column and its temperature, flow rate, and gradient elution program for each tested LC setting can be found in Table S3. LC conditions include reversed phase liquid chromatography (RPLC). The details of an analytical method and instrumentation for analysis of chemicals in atmospheric pressure chemical ionization (APCI) can be found elsewhere.17 The parameters of the electrospray ionization interface (ESI) were set as for the PI mode: capillary voltage, 2500 V; end plate offset, 500 V; nebulizer, 2 bar; drying gas, 8 L min–1; dry temperature, 200 °C; and for NI mode: capillary voltage, 3500 V; end plate offset, 500 V; nebulizer, 2 bar; drying gas, 8 L min–1; dry temperature, 200 °C. A QToF external calibration was performed daily with a sodium formate solution, and a segment (0.1–0.25 min) in every chromatogram was used for internal calibration, using a calibrant injection at the beginning of each run. The sodium formate calibration mixture consists of 10 mM sodium formate in a mixture of H2O/isopropanol (1:1). The theoretical exact masses of calibration ions in the range of 50–1000 Da were used for calibration. The instrument provided a typical resolving power of 36000–40000 during calibration (39373 at m/z 226.1593, 36953 at m/z 430.9137, and 36374 at m/z 702.8636).
Development of CDEA RI
The CDEA chemicals were synthesized by a condensation reaction at a 1:1 molar ratio of substrates (coconut oil, FAMEs and individual fatty acids) and diethanolamine in the presence of sodium methoxide (5% w/w of substrates) as catalyst. The synthesis was performed in a three neck round-bottom flask (250 mL), equipped with a condenser, a heating mantle, and a magnetic stirrer. The reaction took place in 9 h, and the reaction temperature was set gradually at 170 °C. The CDEA was produced as a light-yellow transparent liquid with a high viscosity. The product contained also a second phase that was glycerol, and it was removed from product by a separatory funnel and was further rotary-evaporated to yield pure CDEA products. More details about the synthesis of CDEA can be found elsewhere.18 A working solution of CDEA at 1.0 mg L–1 was prepared in ACN/H2O (50:50, v/v), MeOH/H2O (50:50, v/v), and acetone/hexane (50:50, v/v) and injected in the LC-(±ESI)-QToF and GC-APCI-QToF. The RI system was proposed according to eq 1 (eq 1).
| 1 |
where “n” is the number of carbons in the alkyl chain of CDEA and it is in the range of 0 to 23 carbons. tRC(n–1)DEA and tRC(n+1)DEA are the tR of CDEA calibrants eluting before and after analyte, respectively.
pH Effect
To assess the effect of pH on the elution pattern of emerging contaminants, the difference between retention time values of common compounds from two LC settings, LC1 (+ESI: pH = 3.6, from Table S1) and LC2 (−ESI, pH = 6.2 from Table S2), were derived. These two LC settings have the same gradient elution pattern, solvents (mobile phase composition), flow rate, and RPLC column, whereas the buffer system is changed (from 5 mM ammonium formate and 0.01% formic acid (pH = 3.6) to 5 mM ammonium acetate (pH = 6.2)) in order to monitor the tR shift due to pH values. The list of 402 common compounds between two LC settings can be found in Table S4. The compounds are classified into two groups, including “Level 1” and “Level 2”, which show the shifted tR values are less and more than 30 s, respectively. This threshold selected considering the acceptable tR shift of 20 s for target screening and an additional 10 s due to the possible effect of matrix.19 This shift was then modeled based on convolution neural network deep learning (CNN-DL) procedure19 for uncertainty calculation and nontarget screening.
Modeling of RI and pH Effect
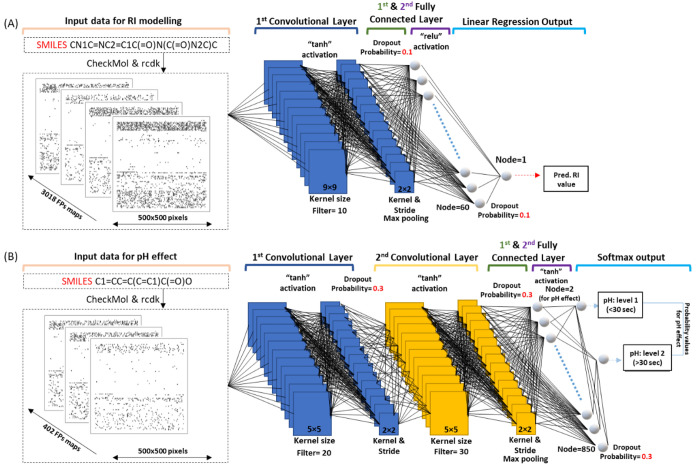
The canonical SMILES (simplified molecular input line entry system) were derived using Environmental Protection Agency’s (EPA) chemistry dashboard for 3018 chemicals.20 Overall, 10286 different types of fingerprints (FPs), such as the standard FP, graph FPs, circular FPs, chemical functional groups, and so on, were calculated for each SMILES entry using “rcdk v 3.6” and “checkmol” software. Then, the 500 × 500 pixels image of these FPs was created for each compound listed in Tables S1 and S4. If a FP had a value of 1.0, the pixel was given a black color (RGB color code: 0, 0, 0), otherwise, it left a white color (RGB color code: 255, 255, 255). A raster image of 40 × 40 pixels was then created from each FPs map and correlated to related RI or pH classes. Then, the CNN-DL method was used to model RI and pH effect data. The model architecture is shown in Figure 1. To create CNN-DL, the “mxnet” R package was used (https://github.com/apache/incubator-mxnet). The size of the convolution window (Kernel), output channels (filter), strides of convolution, activation functions, and hidden layers were optimized by leave-one-out cross validation analysis. More details about the description of CNN-DL parameters are provided in the Supporting Information.
Figure 1.
Convolutional neural network deep learner architecture for modeling of (A) RI and (B) pH effect.
Quality Control and Validation
A working solution of 83 compounds was prepared at 500 μg L–1 and injected in all LC-HRMS settings listed in Table S5. This list includes various types of chemicals such as illicit drugs, doping related substances, pesticides, herbicides, industrial chemicals, pharmaceuticals, and stimulants. This was used to evaluate the reproducibility of RI values under different LC conditions for various chemical classes. Atrazine-d5 was used as isotopically labeled internal standard along with this working solution and analyzed at 200 μg L–1 to control the MS sensitivity. The database for RI and pH effect were split into training (n = 1851 (RI modeling) and n = 321 (pH effect)) and test set (n = 439 (RI modeling) and n = 81 (pH effect)) by representative data selection method for model internal and external accuracy evaluation.6 In the case of pH effect, the receiver operating characteristics (ROC) curve was used to evaluate the classification capability of deep learner model built.21 Two classes were compared based on sensitivity, specificity, and accuracy. The accuracy of the RI model (regression case) was calculated by a root mean square error (RMSE).
Application in HRMS Screening of Environmental Sample
Activated sewage sludge samples collected from a wastewater treatment plant in Athens (WWTPs), Greece in 2019. The samples were freeze-dried upon arrival and stored in −70 °C. A total of 0.1 g of freeze-dried sample was weighed in a plastic tube (15 mL), spiked with surrogates (listed in Table S5), CDEA RI calibrants, and atrazine-d5 (isotopically labeled compound), and put back in the freezer at −20 °C overnight. Then, the sample was extracted with 2 mL of MeOH–Milli-Q water (pH 2.5, FA 0.5% and 0.1% EDTA; 50:50 (v/v)), vortexed for 1 min, and followed by ultrasonic extraction for 15 min at 40 °C. Then, the extract was centrifuged for 10 min (4000 rpm), and the supernatant was collected in a glass test tube. Overall, this procedure was repeated three times, and totally, 6 mL of supernatant was collected. Afterward, the extract was evaporated to dryness under a gentle steam of N2 at 40 °C. Reconstitution of the analytes was done with 0.2 mL of ACN/MeOH–water (50:50 (v/v)). Finally, the extract was filtered through a 0.2 μm RC syringe filter, and then the samples were transferred to a glass vial for LC-HRMS analysis. Similar procedure was repeated for a sample which was not spiked with CDEA RI and surrogates to understand background analyte. Additionally, an analytical procedural blank was created by repeating all the extraction procedure without additional of samples, surrogates, and CDEA to find any contamination due to analytical method. More details about the sample extraction procedure and its validation as well as HRMS based screening strategy can be found in our previous work.19,22
RI App
A shiny app is written in R, and it can be accessed at https://github.com/raalizadeh/RIdea. Through the app, RI values can be predicted or experimentally calculated for an analyte of interest. Moreover, uncertainty associated with the predictive RI models as well as reliability of RI under different pH values can be obtained for any compound.
Results and Discussion
Elution and MS Characteristics of CDEA
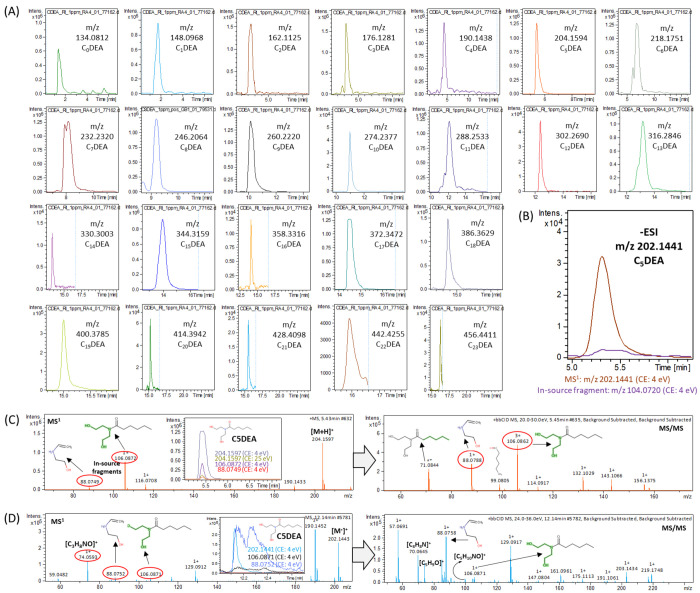
CDEA calibrants could be easily detected in positive ESI mode and when the pH of the mobile phase was acidic (pH < 4) at 1.0 mg L–1 (see Figure 2A), whereas they could give sharp and acceptable signal in negative ESI mode at higher concentration (8.0 mg L–1; depicted in Figure 2B). The most abundant adduct form of CDEAs are [M + H]+ and [M – H]− in ±ESI. They produce distinguished fragmentation pattern that is easy to be detected. CDEAs produce two fragments of m/z 88.0749 and m/z 106.0872, which are also in-source fragments (they can be ionized even at low collision energy (4 eV)) in +ESI (see Figure 2C) and one fragment (m/z 104.0720) in the −ESI mode (see Figure 2B). The MS and MS/MS spectrum are explained and exemplified for C5DEA in ESI in Figure 2B,C. CDEAs elute in a sequence relative to number of carbons in their structures. CDEA calibrants are also detectable in APCI ionization source. The MS and MS2 spectra recorded in GC-APCI-HRMS is exemplified for C5DEA. The MS spectra is recorded after increasing the concentration from 1.0 mg L–1 to 4.0 mg L–1 in APCI source. Therefore, it is also required to inject CDEAs relatively at high amount to be able to detect them easily in APCI source. The main adduct forms of CDEAs were [M•]+ and [M + H]+ in APCI source. Four CDEA calibrants (m/z 330.3003 (C14DEA), 358.3316 (C16DEA), 428.4098 (C21DEA), and 442.4255 (C22DEA)) show a relatively lower response than other calibrants. This could be due to the less abundance of their fatty acids in coconut oil. Therefore, it is crucial to either add pentadecylic acid, margaric acid, behenic acid, and tricosylic acid (or their ester form) to the reaction mix with 1:1 ratio to DEA or adjust dilution of final product to be able to detect them in MS. In addition, pelargonic acid (C8DEA) should be added to the mixture to enhance its response factor. Nevertheless, all CDEA RI can be detected at 1.0 mg L–1 with acceptable intensity. The conversion of FA/FAMEs to CDEAs by DEA and sodium methoxide amounted to 80.12% (v/v) after removing glycerol as major impurity in final product.
Figure 2.
(A) Elution of Cn=0–23DEA calibrants; MS and MS2 spectra exemplified for C5DEA in (B) −ESI, (C) +ESI, and (D) GC-APCI-MS (positive mode).
pH Effect on CDEA RI
When working with ionizable compounds, the mobile phase pH can influence retention behavior dramatically. pH is presumed to affect elution of compounds differently through their octanol–water dissociation constant (log D) and pKa values.23 To author’s knowledge, most of the existing methods for RI indexing did not address this effect in details.11,12,14,15 Out of 402 compounds measured in two different pH values (LC1, pH = 3.6 vs LC2, pH = 6.2), 253 compounds (63% of 402 compounds) showed the tR shift less than 30 s, whereas 149 compounds (37% of 402 compounds) showed a tR shift more than 30 s, as depicted in Figure 3A. This implies that the RI models and RI indexing values would be different for 37% of the data set when they are measured in different pH values. It is noteworthy that if RI indexing standards are affected by the pH value, then they would not be valid for the other 63% portion of the data set, as their RI values will be changed. This would be true also for any indexing system that the calibrants are neutrally charged homologous series.15 The best practice would be to limit application of such indexing system to compounds that the tR shift is negligible in terms of mobile phase pH. Therefore, RI system should be accompanied by an uncertainty measurement in relation to pH effect in order to avoid using RI values if pH affects the elution pattern significantly. On the other hand, the shifted values are not explainable by properties such as log D (Figure 3B), and this does not allow to easily quantify the uncertainty numerically and use of classification concept (or qualitative evaluation) is alternative approach. Here, the deep learning approach was used to model this shift so that the significance of pH effect on an unknown or suspect compound could be evaluated prior to use of RI indexing system. Nevertheless, two solutions can be proposed to avoid misinterpretation of RI values during the identification including; (1) the pH value should be reported while providing the RI values in the RI bank or database and use pH relevant RI models suitable for pH of working mobile phase; (2) use of a buffer system that provides similar pH ranges as the LC conditions 1 and 2 in order to benefit from the list of 3018 chemicals, available in Tables S1 and S2. The ROC curve for the measurement of selectivity, specificity, and accuracy of model for pH uncertainty measurement can be found in Figure 3C,D. It is noteworthy that uncertainty associated with pH effect is studied between pH ranges of 3.6–6.2, since it is the pH range used widely in environmental science6 and the CNN-DL model could be prone to error for any LC condition that the pH value of its mobile phase falls outside of this range. In addition, since tR is not estimated to change between pH range of 3.6 to 6.2 for compounds assigned to class 1, the direct comparison of experimental RIs from CDEA RI is acceptable for these pH ranges.
Figure 3.
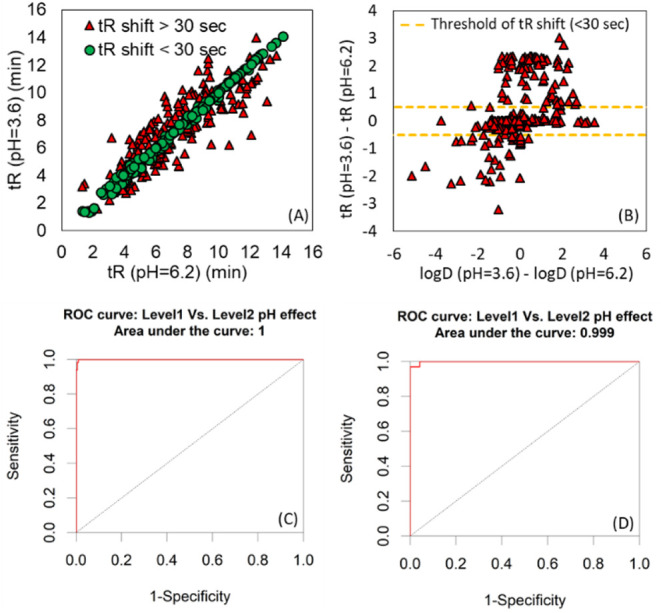
(A) Shift of tR data due to different pH values; (B) ΔtR vs Δlog D values; ROC curve for (C) train set and (D) test set.
Modeling of CDEA RI and Uncertainty
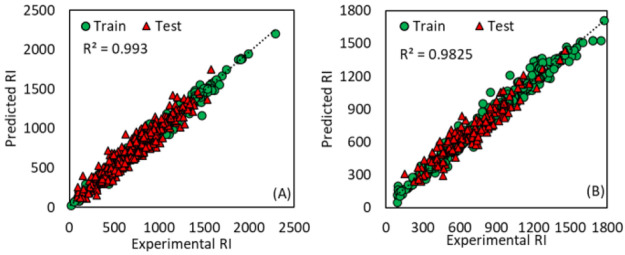
Since the pH effect was as high as 37% for overlapped data between LC 1 and LC 2, the RI data were modeled separately based on deep learning convolutional neural network. If the pH effect is calculated to be significant for an unknown compound, then a model would be selected which is closer to the pH of mobile phase of LC used to detect the analyte to decrease the error. For compounds in which the pH effect is not significant, any of the two models can be used. The predicted and experimental RIs values are provided for a list of overall 3018 compounds in Tables S1 and S2. The predicted RI values from models for pH < 4 (R2train = 0.993, RMSEtrain = 27.62, R2test = 0.913, RMSEtest = 85.90) and pH > 6 (R2train = 0.983, RMSEtrain = 47.50, R2test = 0.900, RMSEtest = 81.26) show high agreement with experimental RIs, as depicted in Figure 4. The histogram of the error from the RI models is depicted in Figure S1. Considering the 3σ, a threshold of 144.61 RI units could be used as an error threshold between acceptable predicted and experimental RIs. The acceptable error window to be applied when using the experimental RI values between different LC settings can be defined comparing the worst-case scenario such as comparing the result of LC 1 versus LC 8 from Table S5. These two LC settings are different and applying a restrict sigma value of 2σ (accounting for 95% data), a threshold of 232.11 RI unit could be assigned. Totally, this is 10% acceptable window of maximum experimental RI value recorded in Tables S1 and S2. The lower and upper limit for RI values can be derived from eq 1. In other words, the RI shift could be translated as degree of tR shift for an analyte of interest that is expected either due to pH or very diverse LC settings. For instance, using 1 min as expected shift in elution of analyte (for example eluting at 9.88 min (8.88–10.88 min), the analyte would elute n carbon number sooner (n – 1) or later (n + 1) in CDEA RIs elution pattern. Therefore, the RI value of 742 calculated for this analyte using Cn=7–8DEA, the lower (Cn=6–7DEA) and upper (Cn=8–9DEA) limit for the RI value, after applying a 1 min shift, would be 665.93 and 821.09, respectively. The application of the RI models is limited to the compounds that are covered by their chemical space domain provided by app.
Figure 4.
Predicted vs experimental RI for (A) pH 3.6 and (B) pH 6.2.
Evaluation of CDEA RI under Various LC Conditions
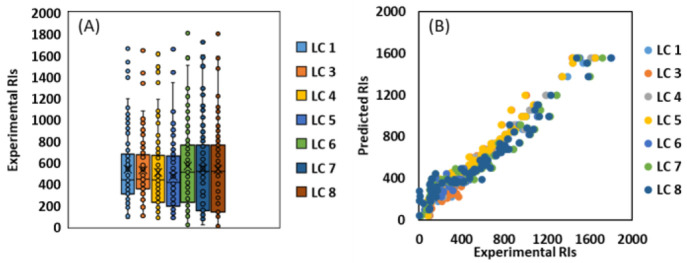
The tR values of CDEA calibrants are evaluated under eight different LC settings (provided in Table S3) and can be found in Table 1. The comparison of RI values for 83 compounds are provided in Figure 5 and Table S5. Figure 5A,B shows the distribution of RI values for 83 compounds analyzed by different LC settings and their correlation by predicted RI values, respectively. After a comparison of the LC 1 and LC 2 settings, which the only difference is buffer composition and pH of mobile phase, the CDEA RI calibrants do not show a significant tR shift (<30 s). This was expected as CDEA RI calibrants are neutral amphiphilic homologous series and their elution pattern and separation in LC should not be affected by pH. LC 2 was excluded from Figure 5, as most of the compounds were detectable only in +ESI mode. LC 3 has a similar gradient elution program, LC column and column temperature, gradient flow rate, and solvent types, as LC 1 and LC2 while instead of buffer solution, 0.1% of FA is used. This was done to compare the elution pattern of 83 and CDEA RI calibrants and its application domain from pH values of 6.2 to 2.6. Generally, not a big shift was observed in tR values recorded by LC 1 versus LC 3 due to pH changes (from 3.6 to 2.6). In other words, CDEA RIs can be safely used to index tR data of analytes from pH values of 3.6 to 2.6. As demonstrated in the previous section for higher pH values (LC 1 (pH = 3.6) versus LC 2 (pH = 6.2)), the chemical domain of pH effect needs to be investigated prior to RI indexing. LC 4 uses the same mobile phases used in LC 1 while using constant flow rate of 0.2 mL min–1 and different gradient elution program. The RI values created from LC 1 versus LC 4 are correlated highly (R2 = 0.991, Table S5), which proves that in addition to the distinguished characterization of the pH effect over analytes elution, CDEA RI is capable of indexing tR data under different gradient elutions and flow rate settings. This utility extends to other LC settings where the mobile phase is different (LC 1 vs LC 5 (R2 = 0.973), LC 1 vs LC 3 (R2 = 0.977) and LC 1 vs LC 6 (R2 = 0.969)). The most important results are the high correlation of RI values for LC 1 versus LC 6, LC 7, and LC 8, with R2 values of 0.967, 0.951, and 0.944 for 83 analytes, respectively. LC 6, LC 7, and LC 8 have different mobile phases (ACN/H2O), gradient elution programs, flow rate (0.35 mL min–1), and C18 column type (BEH C18 (2.1 × 100 mm, 1.7 μm) versus Acclaim RSLC C18 (2.1 × 100 mm, 2.2 μm)). Overall, using CDEA RIs, the tR information on chemicals, which is not affected significantly by pH, can be indexed. The compounds, where pH values may affect their elution pattern, can be distinguished before applying CDEA RI values, and pH-relevant RI models can be used during HRMS screening. Last but not least, after spiking the CDEA RI calibrants in the sludge samples, no significant shift in RI and tR values of CDEAs due to a matrix is observed (Table 1).
Table 1. Elution of CDEA under Different LC Setting and Related tR Valuesa.
| carbon (n) | name | [M + H]+ | [M – H]− | LC 1 | LC 2 | LC 3 | LC 4 | LC 5 | LC 6 | LC 7 | LC 8 | GC-APCI | LC 1 (spiked in sludge) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | C0DEA | 134.0812 | 132.0666 | 1.30 | 1.32 | 1.10 | 1.51 | 1.41 | 0.84 | 0.88 | 0.94 | 7.75 | 1.48 |
| 1 | C1DEA | 148.0968 | 146.0823 | 1.35 | 1.39 | 1.20 | 1.63 | 1.56 | 1.04 | 1.03 | 1.11 | 7.91 | 1.64 |
| 2 | C2DEA | 162.1125 | 160.0979 | 2.51 | 2.48 | 2.58 | 2.64 | 3.54 | 1.94 | 2.22 | 2.45 | 10.00 | 2.49 |
| 3 | C3DEA | 176.1281 | 174.1136 | 3.39 | 3.38 | 3.12 | 3.35 | 4.26 | 2.55 | 3.12 | 3.33 | 11.24 | 3.36 |
| 4 | C4DEA | 190.1438 | 188.1292 | 4.38 | 4.34 | 4.21 | 4.23 | 5.01 | 3.34 | 4.01 | 4.41 | 11.34 | 4.36 |
| 5 | C5DEA | 204.1594 | 202.1449 | 5.46 | 5.42 | 5.99 | 5.61 | 6.17 | 4.36 | 5.71 | 6.26 | 12.14 | 5.44 |
| 6 | C6DEA | 218.1751 | 216.1605 | 6.68 | 6.79 | 7.31 | 6.97 | 7.77 | 5.56 | 7.61 | 7.99 | 12.31 | 6.78 |
| 7 | C7DEA | 232.1907 | 230.1762 | 8.01 | 8.05 | 8.59 | 8.74 | 9.89 | 6.74 | 9.09 | 9.34 | 15.28 | 7.96 |
| 8 | C8DEA | 246.2064 | 244.1918 | 9.08 | 9.20 | 9.66 | 10.37 | 11.32 | 7.75 | 10.39 | 10.61 | 15.31 | 9.14 |
| 9 | C9DEA | 260.2220 | 258.2075 | 10.12 | 10.16 | 10.68 | 12.12 | 13.05 | 8.81 | 11.66 | 11.89 | 17.76 | 10.14 |
| 10 | C10DEA | 274.2377 | 272.2231 | 10.94 | 11.01 | 11.44 | 13.5 | 14.3 | 9.72 | 12.76 | 12.91 | 19.48 | 11.03 |
| 11 | C11DEA | 288.2533 | 286.2388 | 11.71 | 11.76 | 12.24 | 15.02 | 15.75 | 10.95 | 14.21 | 14.23 | 20.27 | 11.86 |
| 12 | C12DEA | 302.2690 | 300.2544 | 12.36 | 12.41 | 12.73 | 15.97 | 16.63 | 11.58 | 15.11 | 15.16 | 20.31 | 12.39 |
| 13 | C13DEA | 316.2846 | 314.2701 | 12.95 | 12.96 | 13.33 | 17.18 | 17.68 | 12.73 | 16.43 | 16.56 | 20.36 | 12.99 |
| 14 | C14DEA | 330.3003 | 328.2857 | 13.39 | 13.41 | 13.66 | 17.88 | 18.39 | 13.5 | 17.46 | 17.41 | 22.84 | 13.73 |
| 15 | C15DEA | 344.3159 | 342.3014 | 13.86 | 13.79 | 14.09 | 18.81 | 19.19 | 14.56 | 18.81 | 18.66 | 24.46 | 13.86 |
| 16 | C16DEA | 358.3316 | 356.3170 | 14.16 | 14.16 | 14.34 | 19.45 | 19.91 | 15.33 | 19.78 | 19.69 | 26.73 | 14.16 |
| 17 | C17DEA | 372.3472 | 370.3327 | 14.51 | 14.47 | 14.68 | 20.36 | 20.81 | 16.24 | 20.89 | 20.81 | ND | 14.46 |
| 18 | C18DEA | 386.3629 | 384.3483 | 14.74 | 14.74 | 14.96 | 21.26 | 21.87 | 16.96 | 21.93 | 21.66 | ND | 14.74 |
| 19 | C19DEA | 400.3785 | 398.3640 | 14.99 | 14.98 | 15.14 | 22.33 | 23.09 | 17.64 | 22.84 | 22.69 | ND | 15.01 |
| 20 | C20DEA | 414.3942 | 412.3796 | 15.34 | 15.31 | 15.51 | 23.69 | ND | 18.39 | 23.86 | ND | ND | 15.28 |
| 21 | C21DEA | 428.4098 | 426.3953 | 15.51 | 15.45 | 15.69 | 25.35 | ND | 18.97 | 24.79 | ND | ND | 15.56 |
| 22 | C22DEA | 442.4255 | 440.4109 | 15.79 | 15.74 | 16.01 | ND | ND | ND | 25.84 | ND | ND | 15.89 |
| 23 | C23DEA | 456.4411 | 454.4266 | 16.13 | 16.10 | 16.38 | ND | ND | ND | 27.23 | ND | ND | 16.26 |
ND: not detected.
Figure 5.
(A) Box plot elution pattern for 83 compounds under various LC settings after calibration with CDEA RI; (B) Correlation of experimental RIs vs predicted values.
Development of CDEA RI Bank
The experimental and predicted RI values as well as related pH effect were computed for a list of 3018 emerging contaminants (provided in Table S6). Researchers can use https://github.com/raalizadeh/RIdea and they provide observed m/z value at a given accuracy and retrieve the candidates with these data. Based on the uncertainty of absolute 232 RI units as well as pH effect, researchers can remove false positives during the identification. The utility of the RI bank in HRMS screening is discussed in the next section.
Application of CDEA RI in a Real Environmental Sample
Here, a proof-of-concept example about the utility of CDEA RI and RI bank is presented. The first example highlights the use of experimental RI values to verify a true positive match. As depicted in Figure S2, a peak was detected at a retention time of 9.88 min in sludge samples that were analyzed by a different LC setting (LC 8) than the one used to model and develop the CDEA RI system (identification case C01). For this peak with a m/z value of 237.1019, the unequivocal molecular formula of C15H12N2O was proposed based on the mass accuracy (Figure S2A). Using MetFrag for the given formula, 3538 candidates were retrieved from PubChem (accessed on 22/06/2022).24 Since m/z 237.1019 had a low intensity (9888) and it did not produce any MS2 spectra in the data-dependent acquisition mode, two fragments were deconvoluted by inspecting the MS/MS spectra recorded by data-independent acquisition mode. From 3538 candidates, carbamazepine is selected as the best candidate, as the experimental RI (742.52) matches the predicted RI one (763.92; Figure S2B). Since it was inside the application domain of the CDEA RI model and no significant pH effect was calculated for this candidate, the use of the CDEA RI model as well as a comparison of the experimental RI values were safe for this candidate. It is noteworthy that carbamazepine was ranked as candidate 17 in the results hit by MetFrag with a score of 0.783. After searching m/z 237.1019 in the CDEA RI bank (available in https://github.com/raalizadeh/RIdea), only one hit was found (Figure S2C), which was carbamazepine with an experimental RI value of 651.13. The error between the CDEA RI bank and the calculated RI from LC 8 for carbamazepine is 91.39, which is below the maximum error window of 232 RI units. The experimental RI match could support the identification confidence for LC for the first time, as it is routinely done in identification strategies, including the use of NIST RI in GC-MS.25 Finally, carbamazepine was confirmed and identified at level 2A (after matching with reference spectra in MassBank (AU112006), a spectrum similarity score of 0.892 (dot product), Figure S2D).
A second example highlights the application of the CDEA RI model in a chromatographic region where polar and ionizable compounds are eluting. A peak with a m/z value of 332.1405 is detected at a retention time of 5.81 min in sludge samples (identification case C02) that are analyzed in LC 8 (Figure S3A). Using a CDEA RI indexing system, a mass accuracy of 2 mDa, and a main adduct form of [M + H]+, a candidate is found in the CDEA RI bank (Figure S3B), which is ciprofloxacin. The error (72 RI unit) between the RI value in the CDEA RI bank (RI = 403.7) and the observed RI (RI = 475.7) is well below the uncertainty window (232 RI unit). In order to verify that this is a true positive match achieved by the CDEA RI system, the normal nontarget screening work is followed for this candidate. The unequivocal molecular formula of C17H18FN3O3 is proposed for this peak (Figure S3A). Using MetFrag with the given formula, 2687 candidates were retrieved from PubChem (accessed on 08/09/2022). A total of 1738 out of 2687 candidates show a predicted RI error above 144 RI units and therefore were deleted from the candidates list. In other words, the CDEA RI model could identify 65% of the total candidates list as potential false positives and decreased the identification effort to the remaining 949 compounds (Figure S3C,D). As it can be seen there is a 15% probability (Figure S3C) that this compound will be affected by pH (due to ionization property of this compound), and thus, there is a minimum uncertainty related to the pH effect. In addition, the compound is within the chemical space of the model, and therefore, CDEA RI can be used safely for this candidate. MetFrag excluded over 1000 candidates since they do not explain any of the provided three MS/MS fragments, and it ranked ciprofloxacin at 133 out of 1640 total processed candidates with a score of 0.7252 (Figure S3E). Finally, ciprofloxacin is confirmed and identified at level 2A (after matching with reference spectra in MassBank (AU102603), spectrum similarity score of 0.945 (dot product), Figure S3F). This proves that earlier RI values match between the CDEA RI bank and the candidate was correct to be a true positive match, and this can facilitate LC-HRMS screening, especially in the absence of good MS/MS data.
Another utility of CDEA RI is to assign several candidates to corresponding peaks extracted from isobaric compounds (same exact mass and often same molecular formula, but with different structural isomers). Two examples (m/z 163.1224 and 180.1013) are presented in Figure S4 (C03, C04 vs C05) to show how CDEA RI can be used to distinguish between isobaric substances under different LC settings (LC 8). In the case of C03, two peaks are observed in the extracted ion chromatogram with very low intensity. After searching the CDEA RI bank, nicotine and anabasine were the closest substances for these peaks (after comparing RI values), whereas nicotine has a lower RI value than anabasine. Although there was no clear MS/MS fragments for nicotine, anabasine could be verified by a diagnostic fragment of m/z 120.0808, proving that the CDEA RI system could correctly assign substances on the peaks. Both nicotine and anabasine were classified into class 1 (no significant pH effect), and therefore, the CDEA RI system was safe to use. Another interesting case is with m/z 180.1013, where two substances could be assigned to these peaks from the CDEA RI bank. According to the CDEA RI bank, 3,4-methylenedioxyamphetamine (MDA) elutes sooner than phenacetin. MDA is verified to be the first peak after a MS/MS match. Since, the MDA was not in class 1, the RI value with a relevant pH value, in contrast to a working mobile phase of LC 8, was used. Since a second peak in Figure S4 detected (case C05) at a very low intensity, no MS/MS fragments could be obtained. To verify that CDEA RI correctly assigned phenacetin, the reference standard was injected and compared to the observed peak in the sludge sample. This is experimental evidence (direct comparison of tR values between the reference standard and the detected peak), and it could increase the identification confidence to level 2b. Collectively, these examples verify that CDEA RI found a true positive match for isobaric substances and multiple peaks.
In addition to isobaric substances, the most important application of CDEA RI is when there is no clear MS/MS data available. This could be due to a low abundance of the substances or matrix effects in the sample. In this case, mass accuracy and molecular formulas could be derived from MS1 data, while the next available information is tR. Matching between RI values could be treated as extra experimental evidence, and it increases identification confidence to even a level of identification confidence of 2b. Through Figure S4, C06, C07, and C08, this scenario is presented. In these three cases, the observed peaks show an intensity lower than 3000, and no clear MS/MS data could be derived. Using the CDEA RI bank, fenbendazole, flumequine, and colchicine were matched (within acceptable maximum error window of 232 RI unit) to cases C06 (absolute RI error from RI bank is 72.54), C07 (absolute RI error from RI bank is 112.22), and C08 (absolute RI error from RI bank is 64.19), respectively. For case C07, the error is increased slightly, as the compound is not in class 1 (there is a pH effect expected, and RI values are compared at relevant pH values). All three compounds are verified after comparing them with corresponding reference standards, verifying that the initial match via CDEA RI is a true positive.
In addition to C01 and C02, the identification cases C09 and C10 in Figure S4 are two examples where true positive matches using CDEA RI are exemplified for various chromatographic regions. Lopinavir was obtained from the CDEA RI bank for m/z 629.3697 with a probability value of 0.312 for class 1 (pH effect is possible), and it is matched to the observed peak in case C09. The RI values were compared on the basis of closest pH value to working mobile phase of LC 8 (pH = 2.6) since the probability of class 1 was not >0.5. This is found to be a true positive match after evaluating the MS/MS fragments between MassBank and this m/z in sludge sample (dot product fit: 0.916). Similarly, the candidate (propranolol) that has been found through mass accuracy and RI match (absolute RI error from RI bank is 77.23) from the CDEA RI bank (Figure S4, C10) is proved to be a true positive match after comparing the MS/MS fragments between MassBank and sample (dot product fit: 0.803). C10 is also classified into class 1 (no significant pH effect), and therefore, the CDEA RI system was expected to provide the lowest uncertainty. All 10 cases show that CDEA RI statistically results correctly in a true positive match in LC-HRMS-based screening.
Safety and Stability of CDEA RI
It is known that N-nitrosodiethanolamine (NDELA), N-nitrosodiethylamine (NDEA), N-nitrosomorpholine (NMOR), N-nitrosodimethylamine (NDMA), and N-nitrosoethylmethylamine (NMEA) can be formed in the presence of nitrosating agents, such as sodium nitrite and DEA, which are carcinogenic to humans.26,27 These chemicals were included in the suspect list for the tentative identification. However, no nitrosamine derivatives were identified after synthesis (Figure S5). Therefore, the CDEA product synthesized does not pose a carcinogenic risk to humans due to these impurities. The pure CDEA is very stable and they can be kept in room temperature below 25 °C depending on the solvent used for their dilution. After their dilution with solvents, they should be kept in fridge under 8 °C. The three-month intralaboratory stability evaluation did not show any significant loss of the signal of CDEA calibrants (Figure S6).
Future Perspectives
In order to aid the ionization efficiency of CDEA calibrants in the negative mode, the primary alcohols in CDEAs could be oxidized to produce carboxylic acid using Jones oxidation or other environmentally friendly approaches.28 Such a transformation should be compatible also with an amide moiety. The drawback of turning primary alcohols to carboxylic acid in the CDEA structure would be the possible effect of pH on its elution pattern. If the elution of calibrants shifts significantly, as shown for other analytes in the pH Effect on CDEA RI section, the RI values would be varied and they would not be reproducible for the rest of the chemical space (as calculated, 63% of 402 compounds did not show significant tR shift due to different pH values). The best scenario would be to selectively oxidize one of the primary alcohols or to inject a high quantity of CDEAs in the case of negative ionization mode. The pH effect on the elution of analytes can be significantly increased if there is an ionic interaction between analytes and the stationary phase (LC column). In this case, no RI system can be used safely during HRMS screening. Such efforts are required to address the pH effect at the given LC system before their routine application, as discussed in this work. The elution of CDEAs in RPLC and GC are characterized in the current study, while future works may investigate the elution pattern of CDEAs or their carboxylic forms under various hydrophilic interaction liquid chromatography (HILIC).
Conclusions
A new, simple, and inexpensive retention time indexing system is developed and tested under various LC settings based on the Cocamide Diethanolamine Homologous Series (C(n)-DEA). Since CDEA calibrants are neutral, their elution pattern and separation in LC is not affected by pH. Although CDEA is detectable in both negative and positive polarity modes, it was observed that the acidic pH of the mobile phase is favorable for their ease of detection in MS. Nevertheless, by injecting a higher quantity of CDEA, they can be detected in the negative ESI mode. Moreover, CDEA provides an acceptable MS signal when using an atmospheric pressure chemical ionization source (APCI). This expands the application domain of CDEA RI to ESI and the APCI source. Uncertainty associated with the effect of pH on the elution of analytes was addressed by a deep learning method. This way, the question of how trustworthy is the calculated RI values under different pH values for an unknown compound was answered. It is concluded that, when using CDEA RI at pH > 6, the application of CDEA RI may be limited for compounds with an elution that could be affected by pH, and therefore, the experimental RI recorded under a certain pH should be used or compared with the CDEA RI bank. Nevertheless, this effect is not substituted to the whole chemical space of emerging contaminants, and it is shown that 63% of 402 chemicals did not show a significant change in their elution pattern due to different pH values. Although this is a first step to understanding the pH effect on elution of unknowns through deep learning models, they need to be improved by adding more compounds to the model under various pH values and extending the measurement of the pH effect to pH ranges beyond 3.6–6.2. The first CDEA-based RI bank was created for the overall 3018 emerging contaminants (2290 and 728 in +ESI (pH < 4) and −ESI (pH > 6), respectively) with the available information on the pH effect. Considering the rigorous evaluation of eight different LC conditions, it has been demonstrated that the CDEA RI experimental values as well as predictions are less LC-system-dependent, showing high accuracy and an acceptable uncertainty. No nitrosamine derivatives were detected or identified via mass spectrometry after the synthesis of CDEAs. These derivatives are known to be carcinogenic to humans, and they can be easily absorbed through the skin. Therefore, CDEA RI could be used safely.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c02893.
Author Contributions
# R.A. and V.N. contributed equally to this work. Conceptualization: R.A. Writing, original draft, and editing: R.A. and V.N. Experimental analysis: R.A. and V.N. Validation and environmental application: V.N. Software: R.A. and V.N. Review, editing, and formal analysis: N.S.T. Resources: N.S.T. Supervision: R.A. and N.S.T.
The authors declare no competing financial interest.
Supplementary Material
References
- Hollender J.; Schymanski E. L.; Singer H. P.; Ferguson P. L. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go?. Environ. Sci. Technol. 2017, 51, 11505–11512. 10.1021/acs.est.7b02184. [DOI] [PubMed] [Google Scholar]
- Andra S. S.; Austin C.; Patel D.; Dolios G.; Awawda M.; Arora M. Trends in the application of high-resolution mass spectrometry for human biomonitoring: An analytical primer to studying the environmental chemical space of the human exposome. Environ. Int. 2017, 100, 32–61. 10.1016/j.envint.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M.; Singer H.; Hollender J. LC–high resolution MS in environmental analysis: from target screening to the identification of unknowns. Anal. Bioanal. Chem. 2010, 397, 943–951. 10.1007/s00216-010-3608-9. [DOI] [PubMed] [Google Scholar]
- Schymanski E. L.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H. P.; Hollender J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Aalizadeh R.; Nika M.-C.; Thomaidis N. S. Development and application of retention time prediction models in the suspect and non-target screening of emerging contaminants. J. Hazard. Mater. 2019, 363, 277–285. 10.1016/j.jhazmat.2018.09.047. [DOI] [PubMed] [Google Scholar]
- Aalizadeh R.; Alygizakis N. A.; Schymanski E. L.; Krauss M.; Schulze T.; Ibáñez M.; McEachran A. D.; Chao A.; Williams A. J.; Gago-Ferrero P.; Covaci A.; Moschet C.; Young T. M.; Hollender J.; Slobodnik J.; Thomaidis N. S. Development and Application of Liquid Chromatographic Retention Time Indices in HRMS-Based Suspect and Nontarget Screening. Anal. Chem. 2021, 93, 11601–11611. 10.1021/acs.analchem.1c02348. [DOI] [PubMed] [Google Scholar]
- Kováts E. Gas-chromatographische Charakterisierung organischer Verbindungen. Teil 1: Retentionsindices aliphatischer Halogenide, Alkohole, Aldehyde und Ketone. Helv. Chim. Acta 1958, 41, 1915–1932. 10.1002/hlca.19580410703. [DOI] [Google Scholar]
- Lee M. L.; Vassilaros D. L.; White C. M. Retention indices for programmed-temperature capillary-column gas chromatography of polycyclic aromatic hydrocarbons. Anal. Chem. 1979, 51, 768–773. 10.1021/ac50042a043. [DOI] [Google Scholar]
- Kind T.; Wohlgemuth G.; Lee D. Y.; Lu Y.; Palazoglu M.; Shahbaz S.; Fiehn O. FiehnLib: Mass Spectral and Retention Index Libraries for Metabolomics Based on Quadrupole and Time-of-Flight Gas Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 10038–10048. 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanstrup J.; Neumann S.; Vrhovsek U. PredRet: prediction of retention time by direct mapping between multiple chromatographic systems. Anal. Chem. 2015, 87, 9421–9428. 10.1021/acs.analchem.5b02287. [DOI] [PubMed] [Google Scholar]
- Hall L. M.; Hall L. H.; Kertesz T. M.; Hill D. W.; Sharp T. R.; Oblak E. Z.; Dong Y. W.; Wishart D. S.; Chen M. H.; Grant D. F. Development of Ecom(5)(0) and retention index models for nontargeted metabolomics: identification of 1,3-dicyclohexylurea in human serum by HPLC/mass spectrometry. J. Chem. Inf. Model. 2012, 52, 1222–1237. 10.1021/ci300092s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogusz M.; Neidl-Fischer G.; Aderjan R. Use of Corrected Retention Indices Based on 1-Nitroalkane and Alkyl Arylketone Scales for HPLC Identification of Basic Drugs. J. Anal. Toxicol. 1988, 12, 325–329. 10.1093/jat/12.6.325. [DOI] [PubMed] [Google Scholar]
- Ballini R.; Palmieri A. Synthetic Procedures for the Preparation of Nitroalkanes. Adv. Synth. Catal. 2018, 360, 2240–2266. 10.1002/adsc.201800163. [DOI] [Google Scholar]
- Zheng S.-J.; Liu S.-J.; Zhu Q.-F.; Guo N.; Wang Y.-L.; Yuan B.-F.; Feng Y.-Q. Establishment of Liquid Chromatography Retention Index Based on Chemical Labeling for Metabolomic Analysis. Anal. Chem. 2018, 90, 8412–8420. 10.1021/acs.analchem.8b00901. [DOI] [PubMed] [Google Scholar]
- Stoffel R.; Quilliam M. A.; Hardt N.; Fridstrom A.; Witting M. N-Alkylpyridinium sulfonates for retention time indexing in reversed-phase-liquid chromatography-mass spectrometry–based metabolomics. Anal. Bioanal. Chem. 2022, 414, 7387–7398. 10.1007/s00216-021-03828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam M. A.Retention index standards for liquid chromatography. U.S. Patent US2017/0102367A, 2017.
- Aalizadeh R.; Nikolopoulou V.; Alygizakis N. A.; Thomaidis N. S. First Novel Workflow for Semiquantification of Emerging Contaminants in Environmental Samples Analyzed by Gas Chromatography–Atmospheric Pressure Chemical Ionization–Quadrupole Time of Flight–Mass Spectrometry. Anal. Chem. 2022, 94, 9766–9774. 10.1021/acs.analchem.2c01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amended Final Report on the Safety Assessment of Cocamide DEA. J. Am. Coll. Toxicol. 1996, 15, 527–542. 10.3109/10915819609008729. [DOI] [Google Scholar]
- Nikolopoulou V.; Aalizadeh R.; Nika M.-C.; Thomaidis N. S. TrendProbe: Time profile analysis of emerging contaminants by LC-HRMS non-target screening and deep learning convolutional neural network. J. Hazard. Mater. 2022, 428, 128194. 10.1016/j.jhazmat.2021.128194. [DOI] [PubMed] [Google Scholar]
- Williams A. J.; Grulke C. M.; Edwards J.; McEachran A. D.; Mansouri K.; Baker N. C.; Patlewicz G.; Shah I.; Wambaugh J. F.; Judson R. S.; Richard A. M. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J. Cheminform. 2017, 9, 61. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio D.; Consonni V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. 10.1039/c3ay40582f. [DOI] [Google Scholar]
- Gago-Ferrero P.; Borova V.; Dasenaki M. E.; Τhomaidis N. S. Simultaneous determination of 148 pharmaceuticals and illicit drugs in sewage sludge based on ultrasound-assisted extraction and liquid chromatography–tandem mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 4287–4297. 10.1007/s00216-015-8540-6. [DOI] [PubMed] [Google Scholar]
- Han S.-y.; Liang C.; Zou K.; Qiao J.-q.; Lian H.-z.; Ge X. Influence of variation in mobile phase pH and solute pKa with the change of organic modifier fraction on QSRRs of hydrophobicity and RP-HPLC retention of weakly acidic compounds. Talanta 2012, 101, 64–70. 10.1016/j.talanta.2012.08.051. [DOI] [PubMed] [Google Scholar]
- Ruttkies C.; Schymanski E. L.; Wolf S.; Hollender J.; Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok V. I.; Linstrom P. J.; Reed J. J.; Zenkevich I. G.; Brown R. L.; Mallard W. G.; Stein S. E. Development of a database of gas chromatographic retention properties of organic compounds. J. Chromatogr. A 2007, 1157, 414–421. 10.1016/j.chroma.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Miralles P.; Chisvert A.; Salvador A. Determination of N-nitrosamines in cosmetic products by vortex-assisted reversed-phase dispersive liquid–liquid microextraction and liquid chromatography with mass spectrometry. J. Sep. Sci. 2018, 41, 3143–3151. 10.1002/jssc.201800388. [DOI] [PubMed] [Google Scholar]
- Lim D. S.; Roh T. H.; Kim M. K.; Kwon Y. C.; Choi S. M.; Kwack S. J.; Kim K. B.; Yoon S.; Kim H. S.; Lee B.-M. Risk assessment of N-nitrosodiethylamine (NDEA) and N-nitrosodiethanolamine (NDELA) in cosmetics. J. Toxicol. Environ. Health Part A 2018, 81, 465–480. 10.1080/15287394.2018.1460782. [DOI] [PubMed] [Google Scholar]
- Balaraman E.; Khaskin E.; Leitus G.; Milstein D. Catalytic transformation of alcohols to carboxylic acid salts and H2 using water as the oxygen atom source. Nat. Chem. 2013, 5, 122–125. 10.1038/nchem.1536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.