Abstract
Objective
To discover a novel peptoid antagonist that targets the interleukin‐15 (IL‐15) receptor and to evaluate its therapeutic efficacy in the treatment of inflammation and arthritis.
Methods
A new compound (IFRA3, interleukin‐15 receptor antagonist 3) was discovered using a unique on‐bead two‐colour combinatorial cell screening of a peptoid library. The interaction of IFRA3 with IL‐15 receptor was assessed by in vitro pull‐down and thermal shift assays. The efficacy of IFRA3 in treating inflammation and arthritis was evaluated in mouse models.
Results
IFRA3Q1 (a tetrameric derivative of IFRA3) inhibited the activity of IL‐15 and suppressed CTLL‐2 cell proliferation (which depends on IL‐15 activity). IFRA3Q1 exhibited strong in vivo anti‐inflammatory activity in carrageenan‐induced inflammation in mice. Furthermore, IFRA3Q1 inhibited collagen‐induced arthritis in DBA/1J mice.
Conclusion
By binding to and inhibiting the function of IL‐15 receptor, IFRA3Q1 exhibited significant anti‐arthritis activity. Our findings suggest that IFRA3Q1 represents a new paradigm for arthritis therapy by targeting IL‐15 signalling.
Keywords: inflammation, interleukin‐15, peptoid, rheumatoid arthritis
IL‐15 plays an important role in rheumatoid arthritis and other autoimmune diseases. Using a combinatorial screening of a chemical library, we discovered a novel peptoid compound (IFRA3Q1) that binds to the IL‐15 receptor and blocks its function. Furthermore, IFRA3Q1 inhibits carrageenan‐induced inflammation and collagen‐induced arthritis in mice. These findings present an alternative strategy for blocking IL‐15, using peptoids instead of antibodies.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease that most commonly affects the joints of the hands, feet, wrists, elbows, knees and ankles, as well as the cardiovascular or respiratory systems. Rheumatoid arthritis affects about 1.3 million adults in the United States, with women having a 2–3 times greater predisposition for developing RA than men (Centers for Disease Control and Prevention). While there is no cure for RA, two major categories of drugs have been used to suppress inflammation, including conventional disease‐modifying antirheumatic drugs (DMARDs) and biologics. Among the conventional DMARDs, methotrexate (MTX) is the standard of care in the treatment of RA. However, only 28% of RA patients achieved a major clinical response [defined as the patient achieving an American College of Rheumatology (ACR 70) response for a continuous period of 6 months] when treated with MTX alone. 1 Adalimumab (Humira), a biologic that targets TNF‐α, alone was able to achieve a major clinical response in 25% of patients. Combining MTX with adalimumab (the gold standard for RA treatment) achieved a major clinical response in 49% of patients. 1 Thus, there are still a majority of RA patients for whom an effective treatment is not available.
IL‐15 is a cytokine that stimulates the generation of memory CD8 T cells and natural killer (NK) cells. 2 The IL‐15 receptor is a heterotrimeric complex consisting of a unique IL‐15 Receptor alpha subunit (IL‐15Rα) and the beta and the gamma subunits shared by interleukin‐2 (IL‐2) and IL‐15. 3 IL‐15 binds to the IL‐15Rα subunit with high affinity, subsequently activating the JAK–STAT signalling pathway and resulting in gene expression changes. 4 , 5 , 6 Both IL‐15 and IL‐15Rα deficient mice lack NK cells and have severely reduced numbers of natural killer T (NKT) cells and memory CD8+ T cells. 7 , 8 , 9 Indeed, IL‐15 has been placed at the apex of the proinflammatory cytokine cascade preceding the expression of TNF‐α and other inflammatory cytokines. 10 IL‐15 is constitutively up‐regulated in synoviocytes of RA patients. 11 , 12 Furthermore, abnormal T‐cell trafficking to the joints may be a key early step in RA. 13 As a potent T‐cell attractant, IL‐15 may play a major role in the pathogenesis of RA. 10 In addition, IL‐15 was shown to mediate T‐cell‐dependent regulation of TNF‐α production in RA. 14 IL‐15‐activated T cells from RA patients stimulated macrophage cell lines and primary monocytes/macrophages to produce TNF‐α in vitro. 14 In addition, IL‐15 promotes the activation of B cells, mast cells and dendritic cells, all of which play key roles in the RA pannus. Importantly, administration of soluble IL‐15Rα (which blocks the function of IL‐15) prevents collagen‐induced arthritis in mice. 15 In a phase I‐II clinical trial, HuMax‐IL‐15 (a human IgG1 anti‐IL‐15 monoclonal antibody) produced ACR70 response in 25% of RA patients. 16 Thus, strong evidence suggests the potential therapeutic value of targeting IL‐15 signalling in RA.
In this study, we discovered a novel peptoid compound, IFRA3Q1, that binds to IL‐15Rα and inhibits its activity. We showed that IFRA3Q1 is effective in suppressing inflammation and arthritis in mice.
Results
Discovery of new compounds that bind to IL‐15Rα
To identify new compounds that bind to IL‐15Rα, we performed an on‐bead two‐colour (OBTC) combinatorial cell screening of a peptoid library. We previously developed this OBTC screening technology, 17 , 18 , 19 , 20 a platform that has thus far led to the discovery of highly specific antagonist peptoids targeting VEGFR2, 17 , 21 , 22 , 23 , 24 , 25 , 26 , 27 T‐cell receptors, 28 lipid‐phosphatidylserine, 20 , 29 plectin, 19 ACE2 30 and vimentin. 31 Since OBTC has the ability to recognise differences between two cell surfaces with high accuracy, we applied this assay to specifically target the IL‐15Rα protein over all other cell surface proteins. As shown in Figure 1, we used IL‐15Rα positive HeLa cells (stained red) and HeLa cells with IL‐15Rα knocked down by siRNA (stained green) for this study. One‐bead one‐compound library of 50 000 peptoid‐carrying beads was incubated with equal ratio of red and green cells for 1 h. We identified three ‘hits’ in this screening experiment. We selected compound 3 for further study and named it IFRA3 (interleukin‐15 receptor antagonist 3). The structural characterisation of IFRA3 was shown in Supplementary figures 1 and 2.
Figure 1.

OBTC screening of the peptoid library. (a) OBTC cell screening was performed by incubating IL‐15Rα expressing HeLa cells stained with red Q‐dots and IL‐15Rα negative HeLa cells (created by siRNA knockdown) stained with green Q‐dots together with 50 000 peptoid library beads. One million cells of each colour were mixed at 1:1 ratio and incubated for 1 h with one‐bead one‐compound library composed of 50 000 compounds at 23°C. (b) Three hits were identified, one of which is named IFRA3. The structure of IFRA3 is shown in b.
Confirmation of IFRA3 binding to IL‐15Rα
We resynthesized the hit peptoids on TentaGel MB NH₂ beads and confirmed their interaction with HeLa cells expressing IL‐15Rα (Figure 2a). Using in vitro pull‐down assay, we confirmed the binding of the three compounds to IL‐15Rα (Figure 2b). To confirm a direct interaction between IFRA3 and IL‐15Rα, we performed a thermal shift assay using recombinant IL‐15Rα protein (R & D Systems). Thermal shift assays are routinely used in drug discovery to identify protein–ligand interactions. 32 , 33 The Applied Biosystems™ Protein Thermal Shift™ assay measures protein thermal stability using a fluorescent protein binding dye. The Protein Thermal Shift dye does not fluoresce in aqueous solutions but fluoresces in nonpolar environments. The protein is mixed with the dye and heated; as it unfolds or melts, hydrophobic parts of the protein are exposed and bind to the dye, resulting in fluorescence emission detected by qPCR. Binding of a ligand to the protein changes the stability of the protein, resulting in a change in fluorescence intensity. As shown in Figure 2c, when recombinant IL‐15Rα is mixed with IFRA3, a decrease in protein melting temperature was observed. This suggests that IFRA3 binds to IL‐15Rα directly and causes a conformation change (unfolding) of IL‐15Rα (that destabilises the protein and abolishes its function).
Figure 2.

Confirmation of IFRA3 binding to IL‐15Rα. (a) The identified peptoids were resynthesized on TentaGel MB NH₂ beads. IL‐15Rα‐positive HeLa cells were stained red and IL‐15Rα‐negative HeLa cells were stained in green. The red, green or red + green mixed (at 1:1 ratio) cells were incubated with the beads for 1 h and gently washed and visualised under the fluorescent microscope. Representative pictures are shown. (b) In vitro pull‐down assay. Cell lysates from IL‐15Rα‐positive or ‐negative cells (siRNA knockdown) were extracted using RIPA buffer, and subsequently incubated with IFRA1, IFRA2 or IFRA3 conjugated beads at 4°C overnight. The beads were then washed with RIPA buffer for three times, and the binding proteins were eluted with 1% SDS. The resulting lysates were then applied onto 12% SDS‐PAGE gel and subjected to Western blot analysis using an antibody against IL‐15Rα. (c) Recombinant IL‐15Rα was mixed with 100 μM IFRA3. A thermal shift assay was carried out using a QuantStudio 3 Real‐Time PCR System. Each bar represents the mean + standard deviation, *P < 0.05. All experiments were repeated at least three times.
IFRA3 tetramer inhibits the function of IL‐15
To determine whether IFRA3 can inhibit the function of IL‐15, we tested its effects on T‐cell proliferation. CTLL‐2 is a murine T‐cell line that expresses all three IL‐15 receptor subunits and depends on IL‐2/IL‐15 to proliferate. This cell line is routinely used to measure IL‐15 activity. 34 , 35 We synthesised the dimer (IFRA3D1), trimmer (IFRA3T1), tetramer (IFRA3Q1) derivatives of IFRA3 (Supplementary figures 3–5), as IL‐15 receptor is a multimer, and we may be able to enhance the binding of IFRA3‐multimers via the avidity effect. 36 As shown in Figure 3a, IFRA3 alone did not stimulate CTLL‐2 proliferation. While IL‐15 significantly stimulated CTLL‐2 proliferation, the IFRA3 tetramer (IFRA3Q1) completely inhibited the activity of IL‐15 at the concentration of 10 μM. The effects of the IFRA3 (monomer), IFRA3D1 (dimer), and IFRA3T1 (trimer) were less potent in inhibiting the activity of IL‐15 (data not shown). By contrast, IFRA3Q1 had no effect on IL‐2‐induced cell proliferation (Figure 3b). Furthermore, we performed a time course experiment to examine the ability of IFRA3Q1 to block IL‐15‐dependent signalling in CTLL‐2 cells. As shown in Figure 3c, IFRA3Q1 inhibited IL‐15‐induced activation of Stat3 and Jak3. However, IFRA3Q1 had no effect on IL‐2‐mediated activation of Stat3 and Jak3.
Figure 3.

IFRA3Q1 inhibits the activity of IL‐15. (a) CTLL‐2 cells were treated with various doses of IFRA3Q1 alone, IL‐15 (0.8 nM), or IL‐15 (0.8 nM) plus IFRA3Q1 for 7 days. Cell proliferation was measured using the WST‐1 assay. (b) CTLL‐2 cells were treated with IL‐15 (0.8 nM), or IL‐15 (0.8 nM) plus IFRA3Q1 (10 μM), IL‐2 (10 ng mL−1), or IL‐2 (10 ng mL−1) plus IFRA3Q1 (10 μM) for 7 days. Cell proliferation was measured using the WST‐1 assay. (c) CTLL‐2 cells were treated with IL‐15 (0.8 nM), or IL‐15 (0.8 nM) plus IFRA3Q1 (10 μM), IL‐2 (10 ng mL−1), or IL‐2 (10 ng mL−1) plus IFRA3Q1 (10 μM) for various lengths of time. Protein expression was determined by Western blotting with the antibodies. Each bar represents the mean + standard deviation, *P < 0.05. All experiments were repeated three times.
IFRA3 tetramer inhibits in vivo inflammation
Carrageenan is a potent inducer of acute local inflammation in vivo. It causes rapid recruitment of neutrophils to the site of administration and has been used extensively to study the mechanism of local inflammation. 37 Since IL‐15 plays a significant chemotactic role in cellular migration and infiltration, 38 , 39 we determined whether IFRA3Q1 can inhibit carrageenan‐induced inflammation. As shown in Figure 4a, while mice injected with λ‐carrageenan developed local footpad inflammation and swelling, co‐administration with IFRA3Q1 significantly reduced the swelling of the footpad. In addition, treatment with IFRA3Q1 significantly reduced the levels of inflammatory cytokines TNF‐α, INF‐γ and IL‐6 in the carrageenan‐treated paws of the mice (Figure 4b).
Figure 4.

IFRA3Q1 inhibits λ‐carrageenan‐induced inflammation in vivo. (a) BALB/c female mice were treated with 5 μg IFRA3Q1 or with 0.9% saline 30 min before they were injected on the left footpad with 300 μg λ‐carrageenan (10 mice per group). Footpad swelling was measured at various time points after injection using a dial calliper. (b) Levels of TNF‐α, INF‐γ and IL‐6 in the carrageenan‐treated paws of the mice were determined by ELISA. Each bar represents the mean + standard deviation, *P < 0.05, ***P < 0.001.
IFRA3 tetramer inhibits arthritis in mice
The collagen‐induced arthritis (CIA) mouse model 40 , 41 is one of the most used autoimmune models of rheumatoid arthritis. Since IL‐15 plays a key role in rheumatoid arthritis, 10 , 11 we evaluated the efficacy of IFRA3Q1 in this mouse model. DBA/1J mice were immunised with type II chicken collagen emulsion in complete Freund's adjuvant (CFA), and a boost of type II chicken collagen in incomplete Freund's adjuvant (IFA) at 21 days after the first injection. As shown in Figure 5a and b, while significant levels of arthritis developed in mice in the control group after collagen immunisation, treatment with IFRA3Q1 effectively reduced the symptoms of arthritis. Furthermore, in the spleens of the IFRA3Q1‐treated mice, we found that the numbers of NK cells, NKT cells, memory CD8+ T cells and naïve T cells were significantly reduced (Figure 5c and d). In addition, IFRA3Q1 significantly reduced the levels of inflammatory cytokines TNF‐α, INF‐γ and IL‐6 in the plasma (Figures 5e, 6).
Figure 5.

IFRA3Q1 inhibits collagen‐induced arthritis in mice. DBA1/J mice were immunised with a type II chicken collagen emulsion in complete Freund's adjuvant (CFA). Mice were treated with saline or 100 μg IFRA3Q1 twice per week, 10 mice per group. (a) Paw and wrist thickness was measured using a digital calliper. The measurements at 10th week after CFA immunisation is shown. Each bar represents the mean + standard deviation, *P < 0.05. (b) Histologic presentation of collagen‐induced arthritis in mice. Haematoxylin and eosin staining of paw sections from untreated arthritic mice showing mononuclear cell infiltration of the synovium with pannus formation (arrow) with subchondral bone erosions. Section of paws of IFRA3Q1‐treated mice showing a normal synovial lining of one joint (left) and cyst formation (arrow) with early pannus formation in another joint (right). Magnification is noted in the lower right‐hand corner. (c, d) The numbers of NK cells, NKT cells, memory CD8+ T cells and naïve T cells in spleen were analysed by flow cytometry. (e) The levels of TNF‐α, INF‐γ and IL‐6 in plasma were determined by ELISA. Each bar represents the mean + standard deviation, *P < 0.05, **P < 0.01, ***P < 0.001. These experiments were repeated three times.
Figure 6.

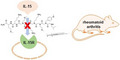
A peptoid binding to the Interleukin‐15 receptor may function as an antagonist that prevents IL‐15 binding to the cell surface receptors. When injected into a mouse model of arthritis, this peptoid subdues signs of arthritis.
Discussion
This discovery and validation study has led to the identification of a novel approach to target IL‐15 in inflammation, using peptoids, which possess several advantages over conventional antibodies or peptides. IFRA3 is not a protein, and its mechanism of action is distinct from that of antibodies targeting IL‐15R (as demonstrated by the thermal shift assay in Figure 2). Peptoids are nonimmunogenic in mice. 26 The minimum pharmacophore of peptoids can be found rapidly, and this knowledge can be used to further improve peptoid activity. 25 , 42 We have shown that peptoid multimerizations can be utilised to improve functional activity by several orders of magnitudes. 20 This is again confirmed in this study where the tetrameric IFRA3Q1 is more effective in inhibiting the IL‐15 receptor and inflammation, than the monomeric IFRA3.
Various immune and cytokine pathways are involved in the pathogenesis of RA. Our results show that by inhibiting IL‐15, IFRA3Q1 decreased the numbers of NK cells, NKT cells and memory CD8+ T cells in mice. In addition, inflammatory cytokines IFN‐γ, TNF‐α and IL‐6 were also suppressed by IFRA3Q1 treatment. Additional immune and cytokine pathways not examined here may also be affected by IFRA3Q1. For example, antigen‐presenting cells, particularly dendritic cells (DCs), are key players in the initiation and maturation of the autoimmune response in RA. 43 As IL‐15 regulates the survival of DCs, 44 IFRA3Q1 may also have impacted the function of DCs. In addition, joint inflammation in CIA is particularly mediated by Th17 cells. 45 Because Il‐15 maintains pathogenic memory Th17 cells (T helper 17) in autoimmunity, 46 it is possible that IFRA3Q1 may also have suppressed Th17 cells, and in turn decreased IL‐17 production. These predictions warrant systematic analysis in future studies.
This study has opened a novel approach to block the function of IL‐15 and potentially treat RA. The placement of IL‐15 at the apex of the proinflammatory cytokine cascade preceding the expression of TNF‐α and other inflammatory cytokines 10 raises hope that targeting this upstream perpetrator may be even more effective than targeting TNF‐α in RA. As IL‐15 is up‐regulated and potentially involved in many other autoimmune diseases, including multiple sclerosis, systemic lupus erythematosus, psoriasis and celiac disease, these findings also pave the path towards identifying novel therapeutics for these chronic and debilitating diseases.
The limitations of this study include the need for additional mouse models of arthritis to be evaluated for the therapeutic efficacy of IFRA3Q1. To ensure that the efficacy of IFRA3Q1 is not restricted to the CIA model, additional mouse models such as the collagen antibody‐induced arthritis (CAIA) can be used. In addition, peptoid agents can be administered orally instead of through injections. The efficacy of oral administration of IFRA3Q1 needs to be formally evaluated. Different doses and schedule of IFRA3Q1 can be tested to identify the optimal treatment regimen. Further study of this novel therapy compared with blocking antibodies will facilitate future clinical development.
Methods
General
TentaGel MB NH₂ resin (particle size: 140–170 μm, loading capacity: 0.2–0.3 mmol g−1, 520 000 beads g−1) was purchased from Rapp Polymere GmbH (Tuebingen, Germany). Rink amide resin (particle size: 100–200 mesh, loading capacity: 0.3–0.6 mmol g−1) was purchased from Chem‐Impex International, Inc. (Wood Dale, IL, USA). All Fmoc‐protected amino acids and 2‐(1H‐Benzotriazole‐1‐yl)‐1,1,3,3‐tetramethyluronium hexafluorophosphate (HBTU), Hydroxybenzotriazole (HOBt), all primary amines, bromoacetic acid, N,N‐diisopropylcarbodiimide (DIC), N,N‐diisopropylethylamine (DIPEA), piperidine, trifluoroacetic acid (TFA), cyanogen bromide (CNBr), Triisopropylsilane (TIS), α‐cyano‐4‐hydroxycinnamic acid, acetonitrile (ACN), hydrochloric acid (HCl), dichloromethane (DCM) and N,N‐dimethylformamide (DMF) were obtained from MilliporeSigma (Burlington, MA, USA). DMEM cell culture media and foetal bovine serum (FBS) were obtained from VWR (Radnor, PA, USA). GIBCO enzyme‐free cell dissociation buffer and Qtracker Cell Labelling Kits were obtained from Thermo Fisher Scientific (Waltham, MA USA). All chemical reagents and solvents from commercial sources were used without further purification. Five mL of disposable reaction columns (Intavis AG, Tuebingen, Germany) was used as reaction vessels for solid‐phase synthesis. Syntheses of peptoids under microwave conditions were performed in a 1000 W microwave oven with 10% power. All purifications were completed on a Waters HPLC system (Waters Corporation, MA, USA). Mass spectras were recorded on an Applied Biosystems Voyager DE Pro mass spectrometer using α‐cyano‐4‐hydroxycinnamic acid as the matrix.
Cells and siRNA transfection
The cell lines HeLa and CTLL‐2 were purchased from American Type Culture Collection (Manassas, VA, USA). HeLa cells were maintained in Dulbecco's modified eagle's medium (Corning, USA) supplemented with 100 U mL−1 penicillin, 100 μg mL−1 streptomycin and 10% foetal bovine serum at 37°C with 5% CO2. CTLL‐2 cells were maintained in RPMI‐1640 medium (Corning, USA) supplemented with 10% foetal bovine serum (FBS) and 10 ng mL−1 Recombinant Human IL‐2 (Peprotech, USA) at 37°C with 5% CO2. For siRNA transfection, HeLa cells were seeded (2.5 × 106/dish) in 10‐cm cell culture dishes 1 day before transfection and treated with human IL‐15Rα siRNA (Horizon, USA, Cat. #D‐007935‐21‐0005) for 24 h with X‐tremeGENE siRNA transfection reagent (MilliporeSigma, USA) according to the manufacturer's instructions.
On‐bead two‐colour binding assay for combinatorial library screen
50 000 peptoid library beads were washed twice in DMEM medium containing 10% FBS and then incubated in 1.0 mL media for 1 h in a polypropylene tube. IL‐15Rα‐positive HeLa cells and IL‐15Rα‐negative HeLa cells were removed from culture plates with GIBCO enzyme‐free cell dissociation buffer at 2.0 mL per plate for 20 min at 37°C. Cells were washed and suspended in DMEM + 10% FBS media. Cells were counted and distributed in 1.5‐mL microcentrifuge tubes with 1.0 × 106 cells in 1.0 mL of media. Cell labelling was then conducted as follows: 1.0 μL each of Qtracker reagent A and B was mixed in 1.5‐mL microcentrifuge tubes and incubated for 5 min at room temperature. 0.2 mL of media was added to each tube and vortexed for 30 s. 1.0 × 106 cells were added to each tube containing the labelling solution and incubated at 37°C for 60 min. IL‐15Rα‐positive Hela cells were labelled with Qtracker 655 (red colour) and IL‐15Rα‐negative HeLa cells labelled with Qtracker 565 (green colour). Cells were washed twice and suspended in DMEM +10% FBS media. Labelled cells were visualised with long‐pass DAPI filter of a BX‐51 fluorescence microscope (Olympus, PA) with a colour camera. Both cell types were mixed thoroughly and pipetted up and down several times to break the clumps. A volume of 2.0 mL of cell suspension mixture was added to the tube containing 50 000 beads and incubated at room temperature with gentle shaking for 1 h. During incubation, cell binding to the beads was checked at 15‐min intervals to ensure not to over equilibrate, which could increase non‐specific binding of cells to the beads. The beads were gently washed twice with media and visualised under the fluorescent microscope using a long‐pass DAPI filter.
Isolation and preparation of beads for sequencing
A single bead containing fluorescently tagged red cells was identified using a fluorescent microscope under 10× objective magnification and removed manually with a 20‐μL pipette with medium‐sized pipette tips. Selected beads were washed three times with 1.0% SDS and boiled in the same solution for 45 min to strip off bound cells and proteins. Finally, the beads were washed three times with water. To cleave the compound from the bead and prepare it for MS/MS sequencing, a cleaving solution was prepared, for which 30 μL of CNBr (5.0 m in ACN) was added to 1.0 mL of 0.1 N HCl. 50 μL of the cleaving solution was added to the 1.5‐mL tube containing the single isolated bead. The tube was incubated at 25°C for 4 h. The solution was evaporated using a freeze dryer (SP Scientific, USA), and the cleaved compound was suspended in 20 μL of water. MS/MS sequencing data were obtained using an AB Sciex TOF/TOF 5800 machine.
Validation of on‐bead two‐colour binding screening results
After identifying the compound (IFRA3) with MS/MS sequencing, it was resynthesized on TentaGel MB NH₂ beads. Three tubes of 25 000 beads (containing IFRA3 compounds) were prepared by washing and incubating for 1 h in DMEM +10% FBS. Two million cells each of IL‐15Rα‐positive HeLa cells were stained red in colour using Qtracker 655 and IL‐15Rα‐negative HeLa cells were stained in green colour using Qtracker 565. One million IL‐15Rα‐positive HeLa cells (red cells) were suspended in 1.0 mL of DMEM +10% FBS media and were added to 25 000 beads containing tube. One million of IL‐15Rα‐negative HeLa cells (green cells) were suspended in 1.0 mL of DMEM + 10% FBS media and were added to another 25 000 beads containing tube. 0.5 × 106 of red cells and 0.5 × 106 green cells were mixed and suspended in 1.0 mL of DMEM +10% FBS media and then added to the third tube containing 25 000 beads. The cells were incubated with the beads for 1 h at room temperature. The beads were gently washed two times with DMEM +10% FBS media and visualised under the fluorescent microscope using a long‐pass DAPI filter.
Synthesis of compound IFRA3
IFRA3 was synthesised on Rink amide resin. 100 mg of resin was loaded into a 5‐mL reaction column, the resin was swelled in DMF for 1 h prior to use and Fmoc group was deprotected by treating the resin with 2.0 mL of 20% piperidine solution in DMF twice for 10 min each time. The resin was first coupled to Fmoc‐Met‐OH (5.0 equivalent) using 5.0 equivalent (equiv.) HBTU and 5.0 equiv HOBt as coupling reagents in the presence of 10.0 equiv of DIPEA overnight. Fmoc was removed with the method described above. Subsequent amino acid Fmoc‐Lys(Boc)‐OH was introduced using the same peptide coupling protocol (HBTU/HOBt/DIPEA), washing 10 times with DMF in between the reactions. After removing the Fmoc group as described above, six peptoid residues were then coupled using a two‐step peptoid coupling procedure (acylation and amination) under a microwave‐assisted synthesis protocol. For the acylation step, beads were treated with 1.0 m bromoacetic acid (1.0 mL) and 1.5 m DIC (1.0 mL) and microwaved at 10% power (2 × 15 s) with gentle shaking in between for 30 s. After washing with DMF, beads were treated with 1.0 mL of 2‐methoxyethylamine (2.0 m), and coupling was performed by shaking at 25°C for 2 h. The procedure was repeated to attach the remaining five residues: N‐Boc‐1,4‐butanediamine, N‐Boc‐1,4‐butanediamine, 2‐methoxyethylamine, 4‐methoxybenzylamine and isobutylamine, respectively. At the end, beads were washed with DCM and dried under vacuum before cleavage. Beads were then treated with a cleavage cocktail of TFA/H2O/TIS (95%/2.5%/2.5%) for 2 h. The crude compound was then purified using HPLC and analysed by MALDI‐TOF.
Synthesis of compound IFRA1
The general synthesis procedure for IFRA1 was similar as described for IFRA3. Sequence of amino acids added in IFRA1 is Fmoc‐Met‐OH followed by Fmoc‐Lys(Boc)‐OH using peptide synthesis protocol. Next six peptoid residues were added using two‐step peptoid synthesis protocol, the sequence of amine residues in IFRA1 are β‐Alanine, N‐Boc‐1,4‐butanediamine, 3‐Isopropoxypropylamine, (R)‐(+)‐α‐Methylbenzylamine, N‐Boc‐1,4‐butanediamine and N‐Boc‐1,4‐butanediamine, respectively.
Synthesis of compound IFRA2
The general synthesis procedure for IFRA2 was similar as described for IFRA3. Sequence of amino acids added in IFRA2 is Fmoc‐Met‐OH followed by Fmoc‐Lys(Boc)‐OH using peptide synthesis protocol. Next six peptoid residues were added using two‐step peptoid synthesis protocol, the sequence of amine residues in IFRA2 are (R)‐(+)‐α‐Methylbenzylamine, Furfurylamine, 4‐methoxybenzylamine, Allylamine, N‐Boc‐1,4‐butanediamine and 3‐Isopropoxypropylamine, respectively.
Synthesis of compound IFRA3D1
The general synthesis procedure for IFRA3D1 was similar as described for IFRA3. Fmoc‐Lys(Fmoc)‐OH was coupled as the first amino acid onto the resin using the same peptide protocol described above. Both Fmoc were removed using 20% piperidine solution in DMF [2 × (2 mL × 10 min)]. The Fmoc deprotection of both amine groups produced two N‐terminals simultaneously to build two copies of IFRA3. The remaining IFRA3 was built as per procedure described for the synthesis of IFRA3.
Synthesis of compound IFRA3T1
To synthesise IFRA3T1, after removing Fmoc from the rink amide resin, orthogonally protected Fmoc‐Lys(ivDde)‐OH was coupled using peptide coupling protocol described previously. Selectively, the Fmoc group was removed by treating the resin with 2.0 mL of 20% piperidine solution in DMF twice for 10 min each. Fmoc‐Lys(Fmoc)‐OH was coupled using the method described above. Fmoc groups were removed as mentioned above, and ivDde group was removed by treating with 5.0% Hydrazine/DMF solution three times, each for 10 min. This afforded three copies of amines onto the remaining IFRA3 sequence were added using the methods mentioned above to obtain a trimer compound IFRA3T1.
Synthesis of compound IFRA3Q1
The general synthesis procedure of IFRA3Q1 is similar to the synthesis of IFRA3. After removing the Fmoc group from rink amide resin, Fmoc‐Lys(Fmoc)‐OH was coupled as a central linker, and both the Fmoc groups were removed simultaneously. Next, Fmoc‐Lys(Fmoc)‐OH was coupled to the two open amines, all four Fmoc groups were then removed simultaneously, allowing four copies of the sequence to be built on the amine groups. Next, the coupling sequence of IFRA3 was followed as described above to yield a tetrameric IFRA3Q1.
Western blot analysis
Cells were lysed in RIPA buffer (1% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS in PBS). Complete protease inhibitor cocktail (Roche, USA) was added to lysis buffer before use. Protein concentration was determined by Bio‐Rad DC protein assay (Bio‐Rad, USA). Protein samples were subjected to SDS‐PAGE and transferred to nitrocellulose membrane. The membrane was blocked in 5% nonfat milk in PBST overnight and incubated with primary antibody and subsequently with appropriate horse radish peroxidase‐conjugated secondary antibody. Signals were detected using the Immun‐Star HRP peroxide Luminol/Enhancer (Bio–Rad, Cat. #1705040) and recorded on a ChemiDoc Touch Imaging System (Bio‐Rad, USA). Anti‐IL‐15Rα antibody was purchased from Santa Cruz (Dallas, USA, Cat. #sc‐374023). Anti‐Stat3 (Cat. #A1192), anti‐Jak3 (Cat. #A0748), antiphospho‐Jak3‐Y980/981 (Cat. #AP0532) and antiphospho‐Stat3‐Y705 (Cat. #AP0705) were purchased from ABclonal (Woburn, MA, USA).
In vitro pull‐down assay
Cell lysates from HeLa cells with IL‐15Rα expression or IL‐15Rα knockdown were extracted using RIPA buffer (Alfa Aesar, USA) supplemented with protease inhibitors (Roche, USA), and subsequently incubated with IFRA1, IFRA2 or IFRA3 conjugated beads at 4°C overnight. The beads were then washed with RIPA buffer for three times, and the binding proteins were eluted with 1% SDS at 95°C for 5 min. The resulting lysates were then mixed with 2× Laemmli Sample Buffer. The samples were then applied onto 12% SDS‐PAGE gel. The gels were then subjected to Western blot analysis.
Thermal shift assay
Five μg of Recombinant Human IL‐15Rα protein (R&D Systems Inc, USA, Cat. #7194‐1R) and 100 μM of IFRA3 were used. The experiment was performed following the Thermal Shift Dye Kit protocol (Applied Biosystems Inc, USA, Cat. #4461146) to monitor thermal stability of proteins using a real‐time PCR machine (Quant Studio 3, Applied Biosystems, USA).
WST‐1 assay
The WST‐1 cell viability assay (Takara, USA) was performed according to the manufacturer's instructions. Briefly, CTLL‐2 cells were collected and washed with IL‐2‐free cell culture medium (RPMI‐1640 without FBS) five times. Cells were then seeded at 10000 cells per well into 96‐well plates and exposed to IFRA3 compounds (IFRA3D1, IFRA3T1 and IFRA3Q1), or recombinant Human IL‐15 (Peprotech, USA, Cat. #200–15‐2UG), or their combinations for 72 h. Then, 10 μL of WST‐1 Reagent was added into each well. After 4‐h incubation at 37°C, the absorbance at 450 nm was measured with EMax Plus Microplate Reader (Molecular Devices).
Collagen‐induced arthritis (CIA) in DBA/1J mice and administration of IFRA3Q1
DBA/1J mice were purchased from The Jackson Laboratory Inc (Bar Harbour, USA). The experiments were performed following the protocol approved by the Institutional Animal Care and Use Committee of University of Houston. The first immunisation of CFA containing a final concentration 0.5 mg mL−1 of M. tuberculosis (Chondrex Inc, Woodinville, USA, Cat. #7001) with 100 μg of chicken type II collagen (Chondrex, Cat. #20012) per mouse was injected subcutaneously at 2 cm from the base of the tail. On Day 21 from first immunisation, the second booster Incomplete Freund's Adjuvant (IFA) (Chondrex, Cat. #7002) with 100 μg of chicken type II collagen per mouse was injected at 3 cm from the tail base. On Day 28 after first immunisation of CFA, 100 μg of IFRA3Q1 per mouse was administered by intraperitoneal injection, twice a week until the end of study in the treatment group. In the control group, 100 μL of 0.9% saline was injected intraperitoneally. The body weight, front paw and hind paw were measured twice a week. The thickness of the paws was measured using an electronic digital calliper. Arthritis was first observed 31 days after the first immunisation in DBA/1J mice.
Cytokine measurements
The mouse joints were pulverised using a mortar and pestle after freezing in liquid nitrogen. Tissue was transferred to 15‐mL tubes, placed on dry ice and resuspended in 1 mL PBS per 200 mg of tissue (containing Complete™ Protease Inhibitor Cocktail) and homogenised using a homogeniser. Mouse joint homogenates were centrifuged for 10 min at 500 g at 4°C. Supernatants were transferred to Eppendorf tubes and centrifuged at 15000 g for 5 min and collected for cytokine analysis. Concentrations of IFN‐γ, IL‐6 and TNF‐α in joint lysates or plasma were measured using mouse ELISA kits from RayBiotech (Peachtree Corners, USA, Cat. #ELM‐IFNg‐1, ELM‐IL‐6‐1 and ELM‐TNFa‐1), following the manufacturer's instructions. Briefly, 100 μL standards or samples was pipetted into the wells and incubated for 2.5 h at room temperature to allow the cytokine in a sample to be bound to the wells by the immobilised antibody. The wells were washed three times, and 100 μL biotinylated anti‐Mouse cytokine antibody was added. After washing away unbound biotinylated antibody, 100 μL HRP‐conjugated streptavidin was added to the wells. The wells were again washed three times, and a 100 μL TMB substrate solution was added to the wells. A volume of 50 μL Stop Solution was added to the wells, and the OD was measured at 450 nm.
Flow cytometry
Splenocytes were isolated from fresh whole mouse spleen. Briefly, the spleen was smashed in PBS with the plunger from a 10‐mL syringe, and the homogenate was passed through a 70‐μm cell strainer. Erythrocytes were lysed with Red Blood Cell Lysis Buffer. Cell suspension was washed with 10–20 mL cold PBS, centrifuged at 400–600 g for 5 min at 4°C, and cell pellet was resuspended in PBS at 1 × 107 cells mL−1. For antibody staining, cells were incubated with antimouse CD16/32 (1 μL mL−1; Thermo Fisher, USA) for 5 min to block non‐specific Fc receptor–mediated antibody binding. 1 × 106 cells were then transferred into polystyrene tubes. Fluorochrome‐conjugated antibodies or isotype controls (0.5 μg/tube) were added, incubated at 4°C for 30 min and washed with PBS. After centrifugation at 300 g for 5 min, cell pellets were fixed with 1% paraformaldehyde, and analysed by flow cytometry on a BD Accuri C6 Plus System. Antibodies used for surface marker labelling included mouse CD3‐FITC, mouse NKp46‐PerCP/Cy5.5, CD8‐FITC from BioLegend (San Diego, USA) and CD44‐PE‐Cy5 from Thermo Fisher. Appropriate isotype‐specific Abs were used as controls. The gating strategy is described in Supplementary figure 6.
Inflammation induced by λ‐carrageenan in Balb/c mice
Balb/c mice were obtained from Charles River Laboratories Inc (Houston, USA). The experiments were performed following the protocol approved by the Institutional Animal Care and Use Committee of University of Houston. Before inducing inflammation, the right hind paw was measured using an electronic digital calliper. In the treatment group, 5 μg of IFRA3Q1 (total volume 10 μL) was injected into right hind paw of Balb/c mice, and in control group, 10 μL of 0.9% saline was injected. After 30 min of administration of 5 μg IFRA3Q1 or 0.9% saline, respectively, inflammation was induced by injection of 300 μg λ‐carrageenan (in a volume of 20 μL) per mouse. λ‐carrageenan (Carbosynth Ltd, batch #YC417821602) was dissolved in 0.9% saline solution at 50 °C and centrifuged briefly.
Histologic evaluation
Mice were sacrificed at the termination of the experiment and hind paws were removed, fixed in 10% phosphate‐buffered formaldehyde. Paws were decalcified and then embedded in paraffin blocks as described. 47 Tissues were sectioned, stained with haematoxylin and eosin and analysed for inflammation and bone erosions.
Statistical analysis
Data are expressed as mean ± SD. Data were analysed by the Student's t test and were considered statistically significant if P < 0.05. The survival rates of the two groups were analysed using a log‐rank test and were considered statistically significant if P < 0.05. P‐values are represented as precise P‐values or generally as *P < 0.05, **P < 0.01 and ***P < 0.001.
Conflict of interest
The authors declare no potential conflicts of interest.
Author contributions
Kwang Bog Cho: Formal analysis; investigation; methodology; writing – original draft. Satya Prakash Shukla: Formal analysis; investigation; methodology; validation; writing – original draft. Mahesh Kumar Kannan: Data curation; formal analysis; investigation; methodology; validation; visualization. Haowen Zhang: Investigation; methodology. Sundus Jabeen Amina: Investigation. Shuang Zhou: Formal analysis; investigation; methodology. Yanping Chen: Investigation; methodology. Jeremiah F Molligan: Investigation. Veena Taneja: Investigation; methodology. Chandra Mohan: Funding acquisition; investigation; methodology; supervision; writing – review and editing. Gomika Udugamasooriya: Conceptualization; formal analysis; investigation; project administration; supervision; writing – original draft; writing – review and editing. Bin Guo: Conceptualization; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; visualization; writing – original draft; writing – review and editing.
Supporting information
Supplementary figure 1
Supplementary figure 2
Supplementary figure 3
Supplementary figure 4
Supplementary figure 5
Supplementary figure 6
Acknowledgments
This work was supported by the Innovative Research Award from The Rheumatology Research Foundation (B Guo, DG Udugamasooriya, and C Mohan) and the University of Houston Drug Discovery Institute Research Seed Grant (B Guo, DG Udugamasooriya, and C Mohan).
Contributor Information
D Gomika Udugamasooriya, Email: gomika@uh.edu.
Bin Guo, Email: bguo3@uh.edu.
References
- 1. Breedveld FC, Weisman MH, Kavanaugh AF et al. The PREMIER study: a multicenter, randomized, double‐blind clinical trial of combination therapy with adalimumab plus methotrexate versus methotrexate alone or adalimumab alone in patients with early, aggressive rheumatoid arthritis who had not had previous methotrexate treatment. Arthritis Rheum 2006; 54: 26–37. [DOI] [PubMed] [Google Scholar]
- 2. Waldmann TA, Miljkovic MD, Conlon KC. Interleukin‐15 (dys)regulation of lymphoid homeostasis: implications for therapy of autoimmunity and cancer. J Exp Med 2020; 217: e20191062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tagaya Y, Bamford RN, DeFilippis AP, Waldmann TA. IL‐15: a pleiotropic cytokine with diverse receptor/signaling pathways whose expression is controlled at multiple levels. Immunity 1996; 4: 329–336. [DOI] [PubMed] [Google Scholar]
- 4. Waldmann TA. The biology of interleukin‐2 and interleukin‐15: implications for cancer therapy and vaccine design. Nat Rev Immunol 2006; 6: 595–601. [DOI] [PubMed] [Google Scholar]
- 5. Becknell B, Caligiuri MA. Interleukin‐2, interleukin‐15, and their roles in human natural killer cells. Adv Immunol 2005; 86: 209–239. [DOI] [PubMed] [Google Scholar]
- 6. Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood 2001; 97: 14–32. [DOI] [PubMed] [Google Scholar]
- 7. Kennedy MK, Glaccum M, Brown SN et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15‐deficient mice. J Exp Med 2000; 191: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lodolce JP, Boone DL, Chai S et al. IL‐15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 1998; 9: 669–676. [DOI] [PubMed] [Google Scholar]
- 9. Gordy LE, Bezbradica JS, Flyak AI et al. IL‐15 regulates homeostasis and terminal maturation of NKT cells. J Immunol 2011; 187: 6335–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McInnes IB, Al‐Mughales J, Field M et al. The role of interleukin‐15 in T‐cell migration and activation in rheumatoid arthritis. Nature Med 1996; 2: 175–182. [DOI] [PubMed] [Google Scholar]
- 11. Harada S, Yamamura M, Okamoto H et al. Production of interleukin‐7 and interleukin‐15 by fibroblast‐like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum 1999; 42: 1508–1516. [DOI] [PubMed] [Google Scholar]
- 12. Benito‐Miguel M, Garcia‐Carmona Y, Balsa A et al. A dual action of rheumatoid arthritis synovial fibroblast IL‐15 expression on the equilibrium between CD4+CD25+ regulatory T cells and CD4+CD25− responder T cells. J Immunol 2009; 183: 8268–8279. [DOI] [PubMed] [Google Scholar]
- 13. Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol 1996; 14: 397–440. [DOI] [PubMed] [Google Scholar]
- 14. McInnes IB, Leung BP, Sturrock RD, Field M, Liew FY. Interleukin‐15 mediates T cell‐dependent regulation of tumor necrosis factor‐alpha production in rheumatoid arthritis. Nat Med 1997; 3: 189–195. [DOI] [PubMed] [Google Scholar]
- 15. Ruchatz H, Leung BP, Wei XQ, McInnes IB, Liew FY. Soluble IL‐15 receptor alpha‐chain administration prevents murine collagen‐induced arthritis: a role for IL‐15 in development of antigen‐induced immunopathology. J Immunol 1998; 160: 5654–5660. [PubMed] [Google Scholar]
- 16. Baslund B, Tvede N, Danneskiold‐Samsoe B et al. Targeting interleukin‐15 in patients with rheumatoid arthritis: a proof‐of‐concept study. Arthritis Rheum 2005; 52: 2686–2692. [DOI] [PubMed] [Google Scholar]
- 17. Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. A Peptoid "antibody surrogate" that antagonizes VEGF receptor 2 activity. J Am Chem Soc 2008; 130: 5744–5752. [DOI] [PubMed] [Google Scholar]
- 18. Udugamasooriya DG, Kodadek T. On‐bead two‐color (OBTC) cell screen for direct identification of highly selective cell surface receptor ligands. Curr Protoc Chem Biol 2012; 4: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raymond AC, Gao B, Girard L, Minna JD, Gomika Udugamasooriya D. Unbiased peptoid combinatorial cell screen identifies plectin protein as a potential biomarker for lung cancer stem cells. Sci Rep 2019; 9: 14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matharage JM, Minna JD, Brekken RA, Udugamasooriya DG. Unbiased selection of peptide‐Peptoid hybrids specific for lung cancer compared to Normal lung epithelial cells. ACS Chem Biol 2015; 10: 2891–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lynn KD, Udugamasooriya DG, Roland CL, Castrillon DH, Kodadek TJ, Brekken RA. GU81, a VEGFR2 antagonist peptoid, enhances the anti‐tumor activity of doxorubicin in the murine MMTV‐PyMT transgenic model of breast cancer. BMC Cancer 2010; 10: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J, Udugamasooriya DG, Lim HS, Kodadek T. Potent and selective photo‐inactivation of proteins with peptoid‐ruthenium conjugates. Nature Chem Biol 2010; 6: 258–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roland CL, Lynn KD, Toombs JE, Dineen SP, Udugamasooriya DG, Brekken RA. Cytokine levels correlate with immune cell infiltration after anti‐VEGF therapy in preclinical mouse models of breast cancer. PLoS One 2009; 4: e7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Udugamasooriya DG, Ritchie C, Brekken RA, Kodadek T. A peptoid antagonist of VEGF receptor 2 recognizes a 'hotspot' in the extracellular domain distinct from the hormone‐binding site. Bioorg Med Chem 2008; 16: 6338–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Udugamasooriya DG, Dunham G, Ritchie C, Brekken RA, Kodadek T. The pharmacophore of a peptoid VEGF receptor 2 antagonist includes both side chain and main chain residues. Bioorg Med Chem Lett 2008; 18: 5892–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Astle JM, Udugamasooriya DG, Smallshaw JE, Kodadek T. A VEGFR2 antagonist and other peptoids evade immune recognition. Int J Pept Res Ther 2008; 14: 223–227. [Google Scholar]
- 27. De Leon‐Rodriguez LM, Lubag A, Udugamasooriya DG et al. MRI detection of VEGFR2 in vivo using a low molecular weight peptoid‐(Gd)8‐dendron for targeting. J Am Chem Soc 2010; 132: 12829–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gocke AR, Udugamasooriya DG, Archer CT, Lee J, Kodadek T. Isolation of antagonists of antigen‐specific autoimmune T cell proliferation. Chem Biol 2009; 16: 1133–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Desai TJ, Toombs JE, Minna JD, Brekken RA, Udugamasooriya DG. Identification of lipid‐phosphatidylserine (PS) as the target of unbiasedly selected cancer specific peptide‐peptoid hybrid PPS1. Oncotarget 2016; 7: 30678–30690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shukla SP, Cho KB, Rustagi V et al. "Molecular masks" for ACE2 to effectively and safely block SARS‐CoV‐2 virus entry. Int J Mol Sci 2021; 22: 8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shukla SP, Zhang H, Fang B, Minna JD, Gomika Udugamasooriya D. Unbiased peptoid cell screen identifies a peptoid targeting newly appeared cell surface vimentin on tumor transformed early lung cancer cells. Bioorg Med Chem 2022; 58: 116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cimmperman P, Baranauskiene L, Jachimoviciute S et al. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys J 2008; 95: 3222–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lo MC, Aulabaugh A, Jin G et al. Evaluation of fluorescence‐based thermal shift assays for hit identification in drug discovery. Anal Biochem 2004; 332: 153–159. [DOI] [PubMed] [Google Scholar]
- 34. Soman G, Yang X, Jiang H et al. MTS dye based colorimetric CTLL‐2 cell proliferation assay for product release and stability monitoring of interleukin‐15: assay qualification, standardization and statistical analysis. J Immunol Methods 2009; 348: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jonuleit H, Wiedemann K, Muller G et al. Induction of IL‐15 messenger RNA and protein in human blood‐derived dendritic cells: a role for IL‐15 in attraction of T cells. J Immunol 1997; 158: 2610–2615. [PubMed] [Google Scholar]
- 36. Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell‐surface interactions. Curr Opin Chem Biol 2000; 4: 696–703. [DOI] [PubMed] [Google Scholar]
- 37. Ianaro A, Xu D, O'Donnell CA, Di Rosa M, Liew FY. Expression of TGF‐beta in attenuated salmonella typhimurium: oral administration leads to the reduction of inflammation, IL‐2 and IFN‐gamma, but enhancement of IL‐10, in carrageenin‐induced oedema in mice. Immunology 1995; 84: 8–15. [PMC free article] [PubMed] [Google Scholar]
- 38. Wilkinson PC, Liew FY. Chemoattraction of human blood T lymphocytes by interleukin‐15. J Exp Med 1995; 181: 1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oppenheimer‐Marks N, Brezinschek RI, Mohamadzadeh M, Vita R, Lipsky PE. Interleukin 15 is produced by endothelial cells and increases the transendothelial migration of T cells in vitro and in the SCID mouse‐human rheumatoid arthritis model in vivo . J Clin Invest 1998; 101: 1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trentham DE, Townes AS, Kang AH. Autoimmunity to type II collagen an experimental model of arthritis. J Exp Med 1977; 146: 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Courtenay JS, Dallman MJ, Dayan AD, Martin A, Mosedale B. Immunisation against heterologous type II collagen induces arthritis in mice. Nature 1980; 283: 666–668. [DOI] [PubMed] [Google Scholar]
- 42. Singh J, Shukla SP, Desai TJ, Udugamasooriya DG. Identification of the minimum pharmacophore of lipid‐phosphatidylserine (PS) binding peptide‐peptoid hybrid PPS1D1. Bioorg Med Chem 2016; 24: 4470–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia‐Gonzalez P, Ubilla‐Olguin G, Catalan D, Schinnerling K, Aguillon JC. Tolerogenic dendritic cells for reprogramming of lymphocyte responses in autoimmune diseases. Autoimmun Rev 2016; 15: 1071–1080. [DOI] [PubMed] [Google Scholar]
- 44. Dubois SP, Waldmann TA, Muller JR. Survival adjustment of mature dendritic cells by IL‐15. Proc Natl Acad Sci USA 2005; 102: 8662–8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murphy CA, Langrish CL, Chen Y et al. Divergent pro‐ and antiinflammatory roles for IL‐23 and IL‐12 in joint autoimmune inflammation. J Exp Med 2003; 198: 1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y, Chauhan SK, Tan X, Dana R. Interleukin‐7 and ‐15 maintain pathogenic memory Th17 cells in autoimmunity. J Autoimmun 2017; 77: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taneja V, Behrens M, Mangalam A, Griffiths MM, Luthra HS, David CS. New humanized HLA‐DR4‐transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis Rheum 2007; 56: 69–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1
Supplementary figure 2
Supplementary figure 3
Supplementary figure 4
Supplementary figure 5
Supplementary figure 6


