Abstract
Background and aim:
The pandemic caused by SARS-COV-2 has increased Semi-Intensive Care Unit (SICU) admission, causing an increase in healthcare-associated infection (HAI). Mostly HAI reveals the same risk factors, but fewer studies have analyzed the possibility of multiple coinfections in these patients. The study aimed was to identify patterns of co-presence of different species describing at the same time the association between such patterns and patient demographics and, finally, comparing the patterns between the two cohorts of COVID-19 patients admitted at Policlinico during the first wave and the second one).
Methods:
All the patients admitted to SICUs during two COVID-19 waves, from March to June 2020 months and from October to December 2020, were screened following the local infection control surveillance program; whoever manifested fever has undergone on microbiological culture to detect bacterial species. Statistical analysis was performed to observe the existence of microbiological patterns through DBSCAN method.
Results:
246 patients were investigated and 83 patients were considered in our study because they presented infection symptoms with a mean age of 67 years and 33.7% of female patients. During the first and second waves were found respectively 10 and 8 bacterial clusters with no difference regarding the most frequent species.
Conclusions:
The results show the importance of an analysis which considers the risk factors for the possibility of co- and superinfection (such as age and gender) to structure a good prognostic tool to predict which patients will encounter severe coinfections during hospitalization (www.actabiomedica.it)
Keywords: COVID-19, Hospital Acquired Infection, Subintensive Unit, DBSCAN
Introduction
The pandemic of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has and had caused an unprecedented medical crisis for all health services all over the world.
In Italy, at the end of December 2020, there were 2.107.166 confirmed cases and 74.159 deaths overall; as of the 26th of June 2022, the confirmed cases are 18.071.634 and deaths 167.967. Almost 10% of patients with COVID-19 experimented hospital admission and 9% of them needed to stay in intensive care units but with substantial differences over the Italian territory (1,2).
To face this huge request for intense use of hospitals the health management boards had to organize hospital areas devoted to the cure of COVID-19 patients (3). The COVID-19 patients admitted to intensive or sub-intensive care units (ICUs/SICUs) to the gravity of their conditions, like all the other patients in the hospital, faced also a considerable threat to their safety caused by healthcare-associated infections (HAIs) which might determine adverse clinical outcomes and aggressive antimicrobial therapies with further resistance selection (4-10).
Both kinds of patients could present HAIs caused by different bacteria, likely associated with ventilators, invasive ventilation, and the usage of empirical broad-spectrum antimicrobials (11). Many researchers had investigated the problem of bacterial and fungal infection in COVID-19 patients admitted to ICUs but there are very few studies devoted to studying COVID-19 patients admitted to SICUs (12).
In IRCCS Fondazione Ca’ Granda Policlinico di Milano, a research and teaching hospital located in the center of Milan with active 716 beds, 84 of them were turned up in sub-intensive care to admit patients who needed less intensive care in both the two waves of the epidemic. In total, during the first wave, from March 9th to June 6th, 2020, 246 patients were admitted to these 84 beds for sub-intensive care and 80 of whom perished (9). At the beginning of October started the second period of very high pressure to admit COVID-19 patients to the hospital, the second wave, and the same SICUs were used with the same number of beds (84), and from October 15th to December 15th, 2020 were admitted 172 patients and 78 died (administrative data from hospital records).
This study aims to evaluate, in two cohorts of patients resulted positive to microbiological culture examination (83 over 246 patients admitted during the first wave, and 73 over 172 during the second wave) in the IRCCS Fondazione Ca’ Granda Policlinico di Milano, the prevalence of infections by different species of bacteria, to identify patterns of co-presence of different species, to describe the association between such patterns and patient demographics (gender, age).
Data about the first and second waves were analyzed separately and compared to consider the different features of the outbreak waves (13). For these patients are also available the results of the local infection control surveillance program to detect colonization by multidrug-resistant bacteria, namely MRSA (Methicillin-resistant Staphylococcus aureus), multidrug-resistant Gram-negative bacteria, and VRE (Vancomycin-resistant enterococci).
Patients and methods
During 2020, 418 patients were admitted to our SICUs, 246 during the first wave from March to June and 172 during the second wave from October to December. All patients admitted to SICUs during the first and second waves were routinely followed with the procedure of the local infection control surveillance program to detect colonization by multidrug-resistant bacteria, namely MRSA (Methicillin-resistant Staphylococcus aureus), multidrug-resistant Gram-negative bacteria and VRE (Vancomycin-resistant enterococci) in addiction received all the microbiological investigation in case of infectious symptoms. An infection control surveillance program was performed through oral-nasal and rectal swabs on all hospitalized patients respectively to detect MRSA and multidrug-resistant Gram-negative bacteria (also considering MDR enterococci). Since fever could be considered one of the most important infection signs, in any patient manifesting fever were performed microbiological cultures to detect species of bacterium and antimicrobial resistance; depending on reported symptoms such as urgency or dysuria, shortness of breath or chest pain, urine, sputum, and blood culture were performed, through a non-selective agar media for culture. No specific data about microbiological load were available. Microbiological cultures were performed on 83 and 73 patients during the first wave and the second one respectively.
Statistical methods
According to the main goals of the analysis, i.e., to evaluate the prevalence and co-presences of the species of bacterium detected in the patients enrolled in the first wave (N=83) and the patients enrolled in the second wave (N=73), data were included in two distinct datasets and structured as follows. In the first wave, a total of 30 species of bacteria were detected (at least in one patient). Accordingly, 30 variables coded as presence/absence (0=absence, 1=presence) of the species above were included in the dataset. The same was done for the 23 species of bacteria detected in the second wave.
For each species of bacterium, the prevalence was defined as the percentage of infected over the total amount of patients. Since patients were submitted to several laboratory investigations, we also reported, for each species of bacterium, the percentage of positive diagnoses over the total number of investigations.
To evaluate the patterns of co-presence of different species, cluster analysis methods were used.
In presence of many species of bacteria with relatively very low prevalence (<2,5%), density-based clustering methods were considered. In particular the DBSCAN (14) was chosen, because of its appreciable performances in dealing with subjects with very uncommon coinfection patterns. Furthermore, for the reasons above, the clusters are not likely to show spherical shapes: therefore, DBSCAN should be preferred to other common clustering algorithms, such as K-means.
DBSCAN is based on the indexes of dissimilarity between subjects. For identifying subjects in the same cluster, the algorithm identifies, for each subject, the subjects who are more similar to him; this set is called “neighborhood”. To define the neighborhood two parameters are needed: the parameter ε (called the “radius” of the neighborhood) and MinPoints (the minimum number of subjects in the neighborhood).
In this work, similarities were calculated according to two indexes specific for presence/absence data: namely, the Jaccard index and the Dice-Sorensen index. These indexes are based on the number of species of bacterium shared by two subjects. If two subjects have no species in common both the indexes are equal to 0, and when they share the same species, both indexes are equal to 1. In the remaining cases, the values of the two indices are different, with the Dice-Sorensen index giving more weight to the number of common species (15). It is worth noting that both these indices were considered in this work because there is no general agreement on which one performs better.
To choose the values of the parameters ε and MinPoints, the procedure described by Schubert et al and the silhouette method were adopted (16,17). In particular, the chosen parameter values were those values that maximized the compactness of the clusters according to the silhouette.
The age of subjects within each cluster was described using a dotplot and gender by reporting the percentage of females within each cluster.
All the analyses were performed using the software R release 3.6.2 and package “dbscan” (18,19).
Results
Analysis of the first wave
Two hundred and forty-six patients were investigated during the first wave and 83 patients were considered in our study because they presented infection symptoms. The mean age of these patients was 67 years and ranged from 39 to 89 and female patients were 28 (33,7%).
The most frequent positive materials sent in microbiological laboratories were blood from peripheral blood vessels (107/284, 37,7%) and blood from venous catheters (80/284 - 28,2%). The most frequent investigation was blood culture (205/284 – 72,2%). According to the results of microbic cultures, 30 species of bacteria were found; the counts and proportions of patients affected by each species are reported in Table 1. The most frequent bacterium isolated was Staphylococcus epidermidis (39/284 – 13,7%) and Staphylococcus aureus (53/284 – 18,7%).
Table 1.
Infections of positive diagnoses detected during the first wave.
| Species of bacteria (abbreviation) | Infected patients: (N=83) | Positive diagnoses (N=284) |
|---|---|---|
| Staphylococcus epidermidis (S.ep) | 21 (25,3%) | 39 (13,7%) |
| Escherichia coli (E.co) | 21 (25,3%) | 28 (9,9%) |
| Enterococcus faecalis (E.fa) | 20 (24,1%) | 39 (13,7%) |
| Staphylococcus aureus (S.au) | 11 (13,2%) | 53 (18,7%) |
| Enterococcus faecium (E.fa1) | 11 (13,2%) | 18 (6,3%) |
| Staphylococcus hominis (S.ho) | 10 (12,0%) | 18 (6,3%) |
| Staphylococcus haemolyticus (S.ha) | 9 (10,8%) | 11 (3,9%) |
| Klebsiella pneumoniae (K.pn) | 8 (9,6%) | 18 (6,3%) |
| Proteus mirabilis (P.mi) | 5 (6,0%) | 9 (3,2%) |
| Pseudomonas aeruginosa (P.ae) | 4 (4,8%) | 10 (3,5%) |
| Bacillus clausii (B.cl) | 4 (4,8%) | 5 (1,8%) |
| Enterobacter cloacae (E.cl) | 2 (2,4%) | 2 (0,7%) |
| Morganella morganii (M.mo) | 2 (2,4%) | 2 (0,7%) |
| Providencia stuartii (P.st) | 2 (2,4%) | 2 (0,7%) |
| Stafilococco aureo Meticillino Resistente (S.au.Res) | 1 (1,2%) | 9 (3,2%) |
| Serratia marcescens (S.ma) | 1 (1,2%) | 3 (1,1%) |
| Corynebacterium urealyticum (C.ur) | 1 (1,2%) | 2 (0,7%) |
| Enterobacter aerogenes (E.ae) | 1 (1,2%) | 2 (0,7%) |
| Staphylococcus capitis (S.ca) | 1 (1,2%) | 2 (0,7%) |
| Staphylococcus cohnii (S.co) | 1 (1,2%) | 2 (0,7%) |
| Aerococcus viridans (A.vi) | 1 (1,2%) | 1 (0,3%) |
| cocco-bacilli Gram positivi (Cc.pos) | 1 (1,2%) | 1 (0,3%) |
| Corynebacterium amycolatum (C.am) | 1 (1,2%) | 1 (0,3%) |
| Corynebacterium striatum (C.st) | 1 (1,2%) | 1 (0,3%) |
| K. pneumoniae resistente ai carbapenemi (K.pn.res) | 1 (1,2%) | 1 (0,3%) |
| Stafilococco aureo Meticillino Sensibile (S.au.sen) | 1 (1,2%) | 1 (0,3%) |
| Staphylococcus pettenkoferi (S.pe) | 1 (1,2%) | 1 (0,3%) |
| Stenotrophomonas maltophilia (S.ma1) | 1 (1,2%) | 1 (0,3%) |
| Streptococcus parasanguis (S.pa) | 1 (1,2%) | 1 (0,3%) |
| Str. beta emol. Gr.F (Str.be) | 1 (1,2%) | 1 (0,3%) |
Legend: For each species of bacteria: the prevalence (percentage of infection) was defined as the proportion of infected patients over the total number of patients (n=83); the percentage of positive diagnoses was calculated by the proportion of positive diagnoses over the total number of laboratory investigations (n=284).
In cluster analysis, the results obtained adopting the Jaccard index and those obtained with the Dice-Sorensen index showed very few differences. Therefore, only the results obtained with the latter one are reported below. According to the silhouette method, the best values of ε and MinPoints parameters were 0,20, and 3, respectively.
Ten clusters were identified, which altogether include 50 (60,2%) of the 83 subjects considered (Table 2). In six clusters, subjects were infected only by one species (cluster numbers: 1, 2, 5, 6, 8, and 9). In two clusters (clusters 3 and 7) subjects presented at most three species of bacterium. In cluster 4, at most 4 species can “co-exist”, and in cluster 10 at most 5 species can be found in a single patient.
Table 2.
Description of clusters of patients in the first wave.
| Cluster | Size | bacteria: species label (n) |
|---|---|---|
| 1 | 6 | E.fa (6) |
| 2 | 14 | E.co (14) |
| 3 | 3 | E.fa (2), E.co (3), B.cl (2) |
| 4 | 3 | E.fa (3), S.ep (3), E.fa1(3), S.pa (1) |
| 5 | 6 | S.ho (6) |
| 6 | 3 | K.pn (3) |
| 7 | 3 | S.ep (3), S.au (3), S.au.res (1) |
| 8 | 3 | S.au (3) |
| 9 | 5 | S.ep (5) |
| 10 | 4 | E.fa (4), E.co (1), S.ep(4), S.ho (1), S.ha (4) |
Legend: size = cluster size, i.e. the number of subjects included in the cluster. In the third column, there is reported for each cluster, the description of species of bacterium: ‘species name’ is the abbreviation of the species, and n is the number of subjects within the cluster that present the species. E.fa= Enterococcus faecalis, E.co= Escherichia coli, B.cl= Bacillus clausii, S.ep= Staphylococcus epidermidis, E.fa1= Enterococcus faecium, S.pa= Streptococcus parasanguis, S.ho= Staphylococcus hominis, K.pn=Klebsiella pneumoniae, S.au= Staphylococcus aureus, S.au.res= Stafilococco aureo Meticillino Resistente, S.ha= Staphylococcus haemolyticus
Figure 1 presents the distribution of age and gender of patients within each cluster. Concerning age, clusters 3 and 6 present narrower ranges: from 63 to 76 years for cluster 3, and 78 to 86 years for cluster 6. Cluster 10 is composed of relatively younger patients, whose age ranges from 43 to 69 years. The highest percentages (about 67%) were found in clusters 1, 3, and 9, whereas in clusters 7 and 10 all subjects were male.
Figure 1.
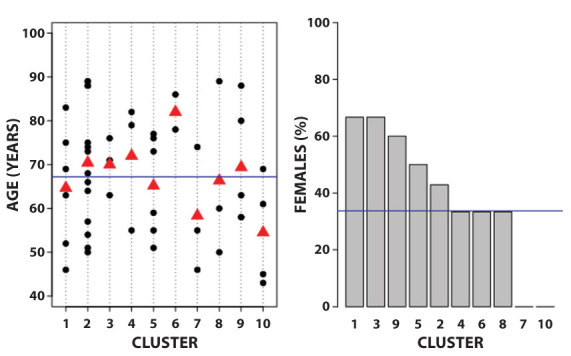
Mean of age and percentage of females in clusters about the first wave.
Left panel: age. The blue line is the mean age in the total sample; the red triangles are the mean age for each cluster. Black dots: age of each subject. Right panel: gender. The blue line is the percentage of females in the total sample; the grey bars are the percentage of females for each cluster.
Analysis of the second wave
All 172 patients were investigated during the second wave and 73 patients were considered in our study because they presented infection symptoms. Their age ranged between 42 and 96 years, with an average value of 73,4 years; female patients were 22 (30,1%).
The most frequent positive material sent in the microbiological laboratory was blood from peripheral veins (76/197 – 38,6%) and consequently, the most frequent investigation was blood culture (150/197 – 76,1%). The most frequent bacterium isolated was Staphylococcus epidermidis (42/197 – 21,3%).
Table 3 presents, for each species of bacteria, the number and percentage of infected subjects, and the number and percentage of positive diagnoses in the second wave. There are substantial differences between the distributions shown here and the distributions for the first wave (Table 1).
Table 3.
Amount of infections and of positive diagnoses detected during the second wave.
| Species of bacteria (abbreviation) | Infected patients: (n = 73) | Positive diagnoses (N=197) |
|---|---|---|
| Staphylococcus epidermidis (S.ep) | 19 (26,1%) | 42 (21,3%) |
| Staphylococcus hominis (S.ho) | 12 (16,4%) | 16 (8,1%) |
| Enterococcus faecalis (E.fa) | 11 (15,1%) | 13 (6,6%) |
| Staphylococcus aureus (S.au) | 10 (13,7%) | 23 (11,7%) |
| Enterococcus faecium (E.fa1) | 9 (12,3%) | 23 (11,7%) |
| Escherichia coli (E.co) | 9 (12,3%) | 13 (6,6%) |
| Pseudomonas aeruginosa (P.ae) | 7 (9,6%) | 16 (8,1%) |
| Staphylococcus haemolyticus (S.ha) | 6 (8,2%) | 11 (5,6%) |
| Bacillus clausii (B.cl) | 5 (6,8%) | 6 (3,0%) |
| Klebsiella pneumoniae (K.pn) | 4 (5,5%) | 4 (2,0%) |
| Staphylococcus capitis (S.ca) | 4 (5,5%) | 4 (2,0%) |
| Enterobacter hormaechei (E.ho) | 2 (2,7%) | 8 (4,1%) |
| Enterobacter aerogenes (E.ae) | 2 (2,7%) | 2 (1,0%) |
| Acinetobacter baumannii (A.ba) | 2 (2,7%) | 2 (1,0%) |
| Actinotignum schaalii (A.sc) | 1(1,4%) | 2 (1,0%) |
| Proteus mirabilis (P.mi) | 1 (1,4%) | 2 (1,0%) |
| Aerococcus urinae (A.ur) | 1 (1,4%) | 1 (0,5%) |
| Candida albicans (C.al) | 1 (1,4%) | 1 (0,5%) |
| Corynebacterium species (C.sx) | 1 (1,4%) | 1 (0,5%) |
| Corynebacterium spp (C.spp) | 1 (1,4%) | 1(0,5%) |
| Corynebacterium striatum (C.st) | 1 (1,4%) | 1(0,5%) |
| Hafnia alvei (H.al) | 1 (1,4%) | 1 (0,5%) |
| Stafilococco aureo Meticillino Sensibile (S.au.sen) | 1 (1,4%) | 1 (0,5%) |
| Staphylococcus schleiferi (S.sc) | 1 (1,4%) | 1 (0,5%) |
| Streptococcus mitis/oralis (S.m.o) | 1 (1,4%) | 1 (0,5%) |
| Streptococcus parasanguis (S.pa) | 1 (1,4%) | 1 (0,5%) |
Legend: For each species of bacteria: the percentage of infection was defined as the proportion of infected patients over the total number of patients (n=73); the percentage of positive diagnoses was calculated by the proportion of positive diagnoses over the total number of laboratory investigations n=197).
Concerning cluster analysis, according to the silhouette methods, the best values of ε and MinPoints parameters were 0,20, and 3, respectively. Eight clusters were identified, which altogether include 39 of the 73 subjects considered (53,4%). Table 4 describes the species detected in the subject for each cluster. Seven clusters of the eight above were formed by subjects infected by only one species (cluster numbers: 1, 2, 3, 4, 6, 7, and 8). In one cluster (cluster 5) subjects presented at most four species of bacterium.
Table 4.
Description of clusters of patients in the second wave.
| Cluster | Size | Bacteria: species label (n) |
|---|---|---|
| 1 | 3 | P.ae (3) |
| 2 | 7 | E.co (7) |
| 3 | 5 | S.au (5) |
| 4 | 5 | E.fa (5) |
| 5 | 6 | E.fa (2), S.ep (6), S.ca (2), S.ho (4) |
| 6 | 7 | S.ep (7) |
| 7 | 3 | S.ho (3) |
| 8 | 3 | K.pn (3) |
Legend: size = cluster size, i.e. the number of subjects included in the cluster. In the third column, there is reported for each cluster, the description of species of bacterium: ‘species name’ is the abbreviation of the species, n is the number of subjects within the cluster that present the species.
E.co= Escherichia coli, E.fa= Enterococcus faecalis, P.ae= Pseudomonas aeruginosa, S.au= Staphylococcus aureus, S.ep= Staphylococcus epidermidis, S.ca= Staphylococcus capitis, S.ho= Staphylococcus hominis, K.pn= Klebsiella pneumonia
Figure 2 presents the distribution of age and gender of patients within each cluster. Clusters 4 and 8 are formed by relatively older subjects: in fact, the age ranges from 75 to 88 in cluster 4, and from 73 to 84 years in cluster 8. The percentage of females in the sample was 69,9%. The highest percentage (100%) was found in cluster 3, whereas the lowest one was found in cluster 6.
Figure 2.
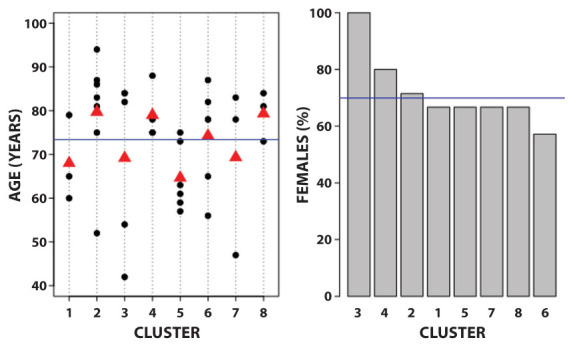
Mean of age and percentage of females in clusters about the second wave.
Left panel: age. The blue line is the mean age in the total sample; the red triangles are the mean age for each cluster. Black dots: age of each subject. Right panel: gender. The blue line is the percentage of females in the total sample; the grey bars are the percentage of females for each cluster.
Bacterial co-infections within the two waves
Table 5 presents the distribution of the patients according to the number of bacterial species found in the respective microbiologic cultures. Interestingly the majority of patients (respectively 57,8% and 61,6%) were affected by only one species of bacterium in both waves.
Table 5.
Number of species of bacteria found in patients.
| Number of species | First wave: frequency (%) | Second wave: frequency (%) |
|---|---|---|
| 1 | 48 (57,8%) | 45 (61,6%) |
| 2 | 18 (21,7%) | 19 (26,0%) |
| 3 | 9 (10,8%) | 7 (9,6%) |
| 4 | 6 (7,2%) | 1 (1,4%) |
| 5 | 1 (1,2%) | 0 (0,0%) |
| 6 | 1 (1,2%) | 1 (1,4%) |
| TOT | 83 | 73 |
Conclusions
Since its first appearance, Sars-CoV-2 is considered a public health issue, not only due to its infectivity and mortality rate but also to the increase in hospital admission. Indeed, severe respiratory infections, such as COVID-19, have increased the risk of admission to semi-intensive and intensive care units, especially for immunocompromised patients and patients with co-pathologies.
At the same time, several studies have investigated the role of respiratory infections in hospital-acquired infection (HAI) showing the link between the disruption of airways structure such as distortion of mucus secretion, cell death, lung edema, decreased mucosal clearance, reduced oxygen exchange through disrupted angiogenesis, and impaired surfactant secretion (20,21). These pathophysiological impairments and the increase in hospitalization, associated with combined therapy based on antibiotics and corticosteroids as suggested by several studies (22-24), affected the HAI rates with an increase not only in mortality and morbidity but also hospitalization, enhancing the length and the economic burden on the healthcare system (25).
In this setting, microbiological investigations assume a central role in the diagnostic workout of hospitalized patients. Microbiological investigations were performed on each patient who has been admitted to sub-intensive care wards for COVID-19 and who presented fever as an additional symptom. Among the 418 patients that were hospitalized in sub-intensive care wards during the year 2020, 156 were enrolled in the study due to at least one microbiological isolation.
To search for a specific pattern of the infection our team decided to describe coinfections clusters in semi-intensive care units during the first and second waves analyzing it also regarding patients’ demographic characteristics.
During the first wave of the pandemic, data from 83 patients were analyzed. The most represented microbiological investigation was blood culture, an inquiry routinely used in the diagnostic workup of persistent fever. S. aureus, a well-known hospital-acquired pathogen, was the commonest bacteria, along with coagulase-negative staphylococci (CoNS), species often considered contaminants. E. faecium and E. coli were among the most frequent, representing 13.7% and 9.8% respectively. Almost 60% of the patients studied had only one species of bacteria isolated from the samples collected.
Analyzing demographic characteristics of different clusters, it’s interesting to note as can’t be observed a specific link between the number of isolated species and cluster mean age or gender prevalence, setting out that the increase of species for a cluster could be linked to the specificity of a single patient rather than a demographic characteristic.
Considering the second wave, data collected, from 73 patients, confirmed that blood culture was the most frequent microbiological investigation performed. S.epidermidis was the commonest bacteria isolated, with S.aureus and E.faecium sharing the second place. Interestingly, P. aeruginosa, another known hospital-acquired pathogen, was the third most represented bacteria isolated accounting for 8% of all the isolated germs. Similarly, to the first wave, a little more than 60% of the patients showed only one detectable species of bacteria. As previously view for the first wave, the analysis of demographic characteristics for the second wave doesn’t show a specific link between the number of isolated species and demographic characteristics.
It’s interesting to note regarding isolate species as, considering the six most common isolates species they are the same in both waves, and S. epidermidis represents the commonest one, likely due to several invasive devices in sub-intensive care units (26)
These results emphasize the importance of an overall analysis of a single patient to avoid antimicrobial therapy mistakes based on non-specific characteristics that could invalidate antimicrobial-stewardship programs implemented in high-risk HAI areas, such as the emergency department or infectious disease ward.
As previously described, several studies have analyzed co- and superinfection in COVID-19 hospitalized patients and it’s interesting to note as none of the studies analyzed has considered the possibility of co-infection clusters simply describing the coinfection rates regardless of the number of different microbiological species isolate (27,28,29,30,31).
It’s evident from several studies that COVID-19 patients are an HAI high-susceptibility patient category during hospitalization and in HAI patients the number of microbiological species is directly linked to the severity of prognosis. The lack of studies that analyze co-infection clusters highlights the importance of our study as the first one exploring this topic. This study has several limitations; firstly, the retrospective design reduces control over especially data collection, and secondly, the study was limited to a single hospital despite it being carried out through two COVID-19 waves amounting to 150 days. In conclusion, the importance of HAI prevention, especially during the COVID-19 pandemic, highlights the importance of a tool to detect which colonized patients may develop infections to improve a specific prophylaxis procedure.
Fundings:
This research was supported by the Italian Ministry of Health.
Conflict of Interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article.
References
- https://opendatadpc.maps.arcgis.com/apps/dashboards/b0c68bce2cce478eaac82fe38d4138b1. (last view 26.06.2021) [Google Scholar]
- Rivieccio BA, Luconi E, Boracchi P, et al. Heterogeneity of COVID-19 outbreak in Italy. Acta Biomed. 2020;91(2):31–34. doi: 10.23750/abm.v91i2.9579. doi: 10.23750/abm.v91i2.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis G, Cavazzana L, Errico M, et al. After the COVID 19 outbreak in Italy: What have we learnt? Travel Medicine and Infectious Disease. 2020;38:101761. doi: 10.1016/j.tmaid.2020.101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auxilia F, Maraschini A, Bono P, et al. COVID-19: new scenario old problems. Acta Biomed. 2020;91(Suplement9):90–91. doi: 10.23750/abm.v91i9-S.10119. doi: 10.23750/abm.v91i2-S.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burriel MS, Keys M, Campillo-Artero C, et al. Impact of multi-drug resistant bacteria on economic and clinical outcomes of healthcare-associated infections in adults: Systematic review and meta-analysis. PLoS One. 2020;15(1):e0227139. doi: 10.1371/journal.pone.0227139. doi: 10.1371/journal.pone.0227139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maque M, Sartelli M, McKimm J, Bakar MA. Health care associated infections an overview. Infect Drug Resist. 2018;11:2321e–33. doi: 10.2147/IDR.S177247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LQ, Wang J, Huang A, Wang D, Wang J. COVID-19 and improved prevention of hospital-acquired infection. British Journal of Anaesthesia. 2020:e318–e319. doi: 10.1016/j.bja.2020.05.037. doi: 10.1016/j.bja.2020.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accardi R, Castaldi S, Marzullo A, Ronchi S, Laquintana D, Lusignani M. Prevention of healthcare associated infections: a descriptive study. Ann Ig. 2017 Mar-Apr;29(2):101–115. doi: 10.7416/ai.2017.2137. doi: 10.7416/ai.2017.2137. PMID: 28244579. [DOI] [PubMed] [Google Scholar]
- Ardoino I, Zangirolami F, Iemmi D, et al. Risk factors and epidemiology of Acinetobacter baumannii infections in a university hospital in Northern Italy: A case-control study. Am J Infect Control. 2016 Dec 1;44(12):1600–1605. doi: 10.1016/j.ajic.2016.05.005. doi: 10.1016/j.ajic.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Mellace L, Consonni D, Jacchetti G, et al. Epidemiology of Clostridium difficile-associated disease in internal medicine wards in northern Italy. Intern Emerg Med. 2013 Dec;8(8):717–23. doi: 10.1007/s11739-012-0752-6. doi: 10.1007/s11739-012-0752-6. [DOI] [PubMed] [Google Scholar]
- Vidal CG, Saniuan G, Moreno-Garcia E, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clinical Microbiology and Infection. 2021;27:83e88. doi: 10.1016/j.cmi.2020.07.041. https://doi.org/10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaldi S, Luconi E, Marano G, et al. Hospital acquired infections in COVID-19 patients in sub intensive care unit. Acta Biomed. 2020;91(3):e2020017. doi: 10.23750/abm.v91i3.10376. doi: 10.23750/abm.v91i3.10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi A, Golemi S, Carapella N, et al. Lombardy, Italy: COVID-19 second wave less severe than the first? A preliminary investigation. 03 November 2020, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-101345/v1. ]) [Google Scholar]
- Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Kdd. 1996, August;96(34):226–231. [Google Scholar]
- Seung-Seok C, Cha SH, Tappert CC. “A survey of binary similarity and distance measures.”. Journal of systemics, cybernetics and informatics 8. 2010;1:43–48. [Google Scholar]
- Schubert E, Sander J, Ester M, Kriegel HP, Xu X. DBSCAN revisited, revisited: why and how you should (still) use DBSCAN. ACM Transactions on Database Systems (TODS) 2017;42(3):1–21. [Google Scholar]
- Rousseeuw PJ. “Silhouettes: a graphical aid to the interpretation and validation of cluster analysis.”. Journal of computational and applied mathematics. 1987;20:53–65. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2021 URL https://www.R-project.org/) [Google Scholar]
- Hahsler M, Piekenbrock M, Doran D. “dbscan: Fast Density-Based Clustering with R.”. Journal of Statistical Software. 2019;91(1):1–30. doi: 10.18637/jss.v091.i01. [Google Scholar]
- Habashi NM, Camporota L, Gatto LA, Nieman G. Functional pathophysiology of SARS-CoV-2-induced acute lung injury and clinical implications. J Appl Physiol (1985) 2021 Mar 1;130(3):877–891. doi: 10.1152/japplphysiol.00742.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014 Apr;12(4):252–62. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow TAC, Longobardo A, Brealey D, et al. Beneficial ex vivo immunomodulatory and clinical effects of clarithromycin in COVID-19. J Infect Chemother. 2022 Apr 14 doi: 10.1016/j.jiac.2022.04.001. S1341-321X(22)00109-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020 Feb 15;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson LA, Rogers Van Katwyk S, Fafard P, Viens AM, Hoffman SJ. Lessons learned from COVID-19 for the post-antibiotic future. Global Health. 2020 Oct 8;16(1):94. doi: 10.1186/s12992-020-00623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscalzo J, Fauci A, Kasper D, Hauser S, Longo D, Jameson JL. McGraw Hill; 2022. Harrison’s Principles of Internal Medicine 21e. [Google Scholar]
- Arentz M, Yim E, Klaff L, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA [Internet] 2020 doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrasa H, Rello J, Tejada S, et al. SARS-Cov-2 in Spanish intensive care: early experience with 15-day survival in Vitoria. Anaesth Crit Care. Pain Med. 2020 doi: 10.1016/j.accpm.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Huang D, Ou P, et al. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020 doi: 10.1111/all.14309. ((Su, Fu) School of Medicine, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China) [DOI] [PubMed] [Google Scholar]
- Feng Y, Ling Y, Bai T, et al. COVID-19 with different severity: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020 doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, He W, Yu X, et al. Coronavirus disease 2019 in elderly patients: Characteristics and prognostic factors based on 4-week follow-up. J Infect. 2020 Jun;80(6):639–645. doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]


