Abstract
Background and aim:
Artificial intelligence was born to allow computers to learn and control their environment, trying to imitate the human brain structure by simulating its biological evolution. Artificial intelligence makes it possible to analyze large amounts of data (big data) in real-time, providing forecasts that can support the clinician’s decisions. This scenario can include diagnosis, prognosis, and treatment in anesthesiology, intensive care medicine, and pain medicine. Machine Learning is a subcategory of AI. It is based on algorithms trained for decisions making that automatically learn and recognize patterns from data. This article aims to offer an overview of the potential application of AI in anesthesiology and analyzes the operating principles of machine learning Every Machine Learning pathway starts from task definition and ends in model application.
Conclusions:
High-performance characteristics and strict quality controls are needed during its progress. During this process, different measures can be identified (pre-processing, exploratory data analysis, model selection, model processing and evaluation). For inexperienced operators, the process can be facilitated by ad hoc tools for data engineering, machine learning, and analytics. (www.actabiomedica.it)
Keywords: artificial intelligence, machine learning, data processing, software, anesthesia, intensive care
1 Introduction
1.1 Brief overview
Artificial intelligence (AI) was born to allow computers to learn and control their environment, trying to imitate the human brain structure by simulating its biological evolution (1). According to John McCarthy AI is “the science and engineering of making intelligent machines, especially intelligent computer programs. It is related to the similar task of using computers to understand human intelligence, but AI does not have to confine itself to methods that are biologically observable” (2). This article aims to analyze the operating principles of artificial intelligence and identify fundamental steps for the implementation of machine learning algorithms.
2 Background
2.1 Artificial intelligence history
The history of AI has now become a long one. Its birth coincides with the publication of the question “Can machines think?”. In fact, this phrase used by Alan Turing in the imitation game is considered the beginning of AI (3). On the other hand, the term owes its partnership to John McCarthy, a computer scientist who, in 1956, organized the Dartmouth conference in which the term was officially coined. The initial enthusiasm was followed by the so-called “AI winter”; a period identified from the 1970s to the 1990s, in which problems related to the capabilities of the available instrumentation have created an abrupt halt (4). Later, thanks to technological advancement, starting from the 2010s, AI is having a new renaissance. And in this new “AI spring”, AI in Medicine (AIM) had no exceptions. This was also possible thanks to the widespread health data digitalization, which made it possible to create big data systems capable of providing a solid basis for intelligent algorithms. Borges do Nascimento et al., analyzing the impact of big data analysis on health indicators and core priorities described in the World Health Organization (WHO) General Program of Work 2019/2023 and in the European Program of Work (EPW). The article highlighted how the accuracy and management of some chronic diseases can be improved by supporting real-time analysis for diagnostic and predictive purposes (5) (Figure 1).
Figure 1.
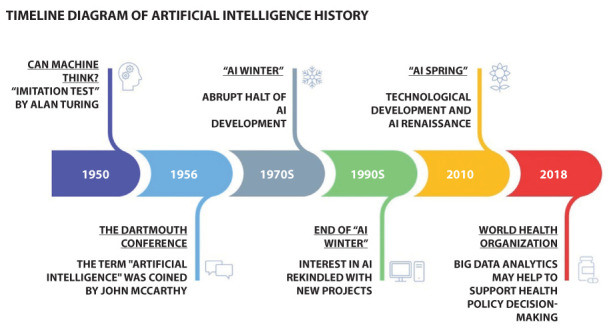
Timeline diagram showing the history of artificial intelligence.
2.2 Subtypes of artificial intelligence
AI includes various subtypes, among others: machine learning (ML), Computer Vision (CV), Fuzzy Logic (FL), and Natural Language Processing (NLP). ML is based on algorithms trained for decisions making that learn from the analyzed data. ML algorithms can be classified based on the type of feedback received. First is supervised learning which receives pre-catalogued data as input. Another category is unsupervised learning. The difference with the previous one is that the training data is not catalogued, and the system must recognize and label the same type of data. In semi-supervised learning, a combination of the two previous algorithms is performed. In this way, the system must consider both the tagged and the untagged elements. Another type of ML is represented by reinforcement learning which can learn from its successes and mistakes. Instead, deep learning (DL) is a subgroup of ML based on algorithms that use artificial neural networks (ANN) organized in several layers to imitate how the human brain interprets and draws conclusions from information. DL is characterized by multiple hidden node layers that learn representations of data by abstracting it in many ways. CV is the branch of AI that allows computers to recognize an image and distinguish the individual elements of a picture by assigning them meaning. Instead, FL uses non-binary values to solve problems that require dealing with more values that classical logic cannot solve. Finally, NLP is a subtype of AI that tries to understand natural language to communicate between machines and with humans (6, 7) (figure 2).
Figure 2.
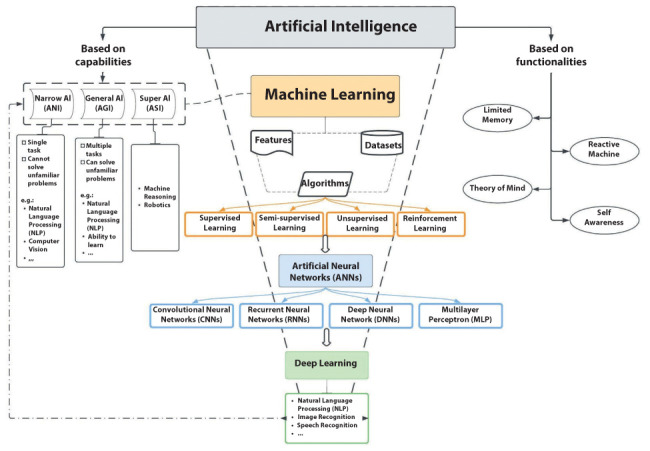
Diagram representing the relationships between Artificial Intelligence, Machine Learning, and Deep Learning.
2.2.1 The concept of data model
It has been considered a crucial point in supervised machine learning sincethe advent of DL in recent years. ML systems were not able to process data directly. It was almost always necessary to define parameters related to the human capabilities of representing the real world as numbers and codes for any specific problem in which the system had to learn from data to support decisions. This process was termed feature extraction and, in some way, the human could determine the success of the process by choosing the better feature set. Moreover, the process of feature extraction was slow and costly, as features in many cases were manually determined, and the amount of labelled data was always limited compared to what we can do nowadays in the big data era. DL has revolutionized this view: the necessity of managing the amounts of data of unimaginable size, the exponential increase of the hardware computational power, and the arrival on the scene of DL models such as BERT, Long-Short Term Memory (LSTM) models provided the AI designers with tools where features models importance is limited to the final stage of decision making: the fine-tuning of DL systems.
2.3 Artificial intelligence in medicine
AI has a major role in numerous sectors, from the financial industry to manufacturing. It also represents an essential part of our daily life. In particular, every time we are surfing the net, we scroll down the home page of our social networks, every time we use a smartphone or a smartwatch, we travel with a self-driving vehicle or simply every time we check our spam free e-mail, we are actually using AI (8).
2.3.1 Artificial intelligence in diseases diagnosis
AI is used in disease diagnosis. For example, Esteva and colleagues trained a deep convolutional neural network (CNN) using a dataset of 129,450 dermatological images and tested its performance against 21 dermatologists. Results showed that artificial intelligence is capable of classifying skin lesions with a level of competence comparable to dermatologists (9). AI can help in the early detection of lung nodules in thoracic imaging. It may also aid in characterizing liver lesions as benign or malignant in patients undergoing computed tomography (CT) and magnetic resonance imaging (MRI) (10).
2.3.2 Artificial intelligence in pre- intra- and post-operative settings
Ren et al. demonstrated that AI applied to a mobile device was able to predict postoperative complications with high sensibility and specificity, matching surgeons’ predictive accuracy (11). ML is also used successfully in airways evaluation for predicting intraoperative hypotension, ultrasound (US) anatomical structure detection, managing postoperative pain, and drug delivery (7).
2.3.3 Artificial intelligence in pharmaceutical industry
AI systems were also implemented in pharmaceutical industry, where DL helps to discover new drugs and to predict bioavailability at the benchIn this context, AI is able to predict 3D structure of target protein, predict drug-protein interactions, determine drug activity, predict toxicity, bioactivity and can identify target cells (12).
2.3.4 Artificial intelligence assistants
AI assistants are promising robots with the ability to support patients, elderly and disabled people, monitoring their physiological parameters while minimizing person-to-person contact and ensure proper cleaning and sterilization (13).
2.3.5 Artificial intelligence and SARS-CoV-2
The most recent application of AI in healthcare was during the pandemic of SARS-CoV-2, where it was used to improve diagnosis (14-16), to predict the epidemic trend (17,18), to predict patients at risk for more severe illness on initial presentation (19), and for faster drugs repurposing (20-24).
2.4 Artificial intelligence in intensive care medicine
With continuous patient monitoring and the enormous amount of data generated, intensive care units (ICU) represent the ideal field for the development and application of AI. Among the possible uses, some algorithms are able to predict the length of stay in ICU (25,26). Moreover, when a neural network algorithm was applied to the Medical Information Mart for Intensive Care III (MIMIC-III) database, ML was able to predict the risk of ICU readmission and mortality, outperforming standard scoring systems, such as sequential organ failure assessment (SOFA), APACHE-II, and Simplified Acute Physiology Score (SAPS) (27-29). An algorithm based on logistic regression and random forest models was able to predict patient instability in the ICU by measuring tachycardia on an electrocardiogram (30). Furthermore, an AI system commercialized as Better Care® can identify ineffective efforts during mechanical ventilation by acquiring, recording and analyzing data directly at patient’s bedside (31). Similar algorithms were even able to detect two types of asynchronies associated with flow asynchrony, dynamic hyperinflation, and double triggering (32,33). Moreover, Komorowski and colleagues developed a computational model using of reinforcement learning which is able to suggest the optimal fluid and vasopressor therapy in septic patients in ICUs, with a significant reduction in mortality (34). Finally, deep learning (DL) was used in ICU to assess delirium and agitation by continuously monitoring patient emotions, employing a camera and accelerometers to record facial expressions and movements (35).
2.5 Basic principles of functioning of Machine Learning
ML systems aim to generalize the results (predictions) in a particular scenario. This scenario can include diagnosis, prognosis, and treatment in intensive care medicine. Pattern recognition, extrapolation, and elaboration strategies can enhance the decision-making process and potentially lead to relevant improvements in clinical care (36).
Although the operating mechanisms are highly complex, a scheme for approaching the matter can be drawn. Every ML pathway starts from task definition and ends in the model application. By connecting the two extremes, different steps can be identified. Nevertheless, the system is not static. It can dynamically develop during the different stages of its progress, depending on potential issues to be addressed. High-performance characteristics and strict quality controls are needed.
The whole process can be summarized into four steps, as follows: (Figure 3)
Figure 3.
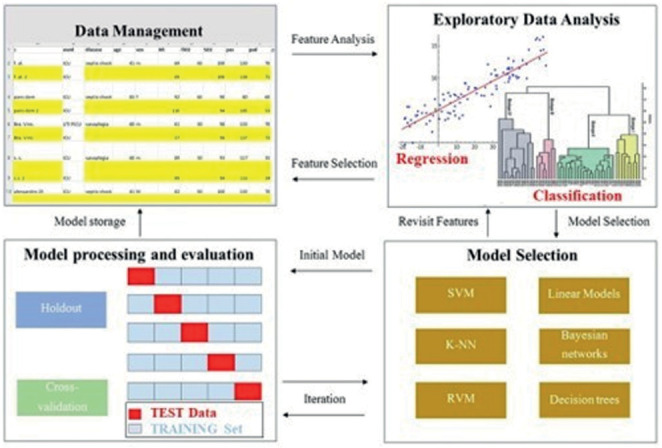
Schematic Machine Learning pathway. Abbreviation: SVM, Support Vector Machine; RVM, Relevance Vector Machine; K-NN, K-nearest neighbors.
Pre-processing
Exploratory Data Analysis (EDA)
Model selection
Model processing and evaluation
3 Pipeline
3.1 Pre-processing
After task definition and data collection, the pre-processing, or data preparation process, is implemented. This mandatory step is aimed at ensuring that the algorithm will easily interpret the dataset features. It includes data loading, normalization, and standardization of the dataset. Several procedures can be provided, but not all are always required (Table 1).
Table 1.
Data Preparation examples.
| Procedure | Process |
|---|---|
| Data Quality Assessment | It encompasses the analysis of missing values (e.g., raw elimination or estimation by interpolation methods), alongside inconsistent and duplicate values. |
| Feature Aggregation | This step is crucial for large amounts of data. Rows from multiple tables are combined through aggregators that perform calculations of sums, averages, medians, and the number of rows. It also combines data from different tables. |
| Feature Sampling | In some cases, it may be necessary to use only a portion of the dataset due to memory or time constraints. Nevertheless, the sample should represent the original (balanced dataset) dataset Sampling techniques include sampling with or without replacement and stratified sampling. |
| Dimensionality Reduction | Irrelevant features and noise can be eliminated to increase the dataset viewing. The most used methods are Principal Component Analysis and Singular Value Decomposition. |
| Feature Encoding | It is the transformation of the dataset to allow the input of machine learning algorithms while maintaining its original meaning. Different norms and rules for continuous or numeric variables are followed. |
Since outputs are strictly dependent on the data provided and AI-based predictions may influence clinical decisions, structuring and manipulating the pre-processing dataset must be remarkably accurate. At this critical stage, clinicians must work closely with bioinformaticians to avoid generating models that, although mathematically optimal, do not fit the clinical contexts in which they will operate (37).
3.2 Exploratory Data Analysis (EDA)
This phase aims to discover trends or patterns and is dynamically linked to the previous step. It is a crucial element of the whole process as most of the time is spent in feature engineering and pre-processing instead of model development and deployment. Data scientists implement EDA to analyze datasets and summarize their main characteristics, often adopting data visualization methods. It helps determine the optimal way to manipulate data, allowing researchers to detect patterns, potential anomalies, and test one or more hypotheses. In other words, this step is primarily used to evaluate what data can reveal beyond the formal modelling or hypothesis testing and provides a better understanding of dataset variables and their interrelationships. It can also help determine if the statistical techniques used for data analysis are appropriate.
Practically, EDA involves the analysis of the features through statistical summary and visual techniques such as histograms, scatter charts and other tools. According to the research to be performed, four types of EDA are described:
Univariate nongraphical. Valid for a single variable.
Univariate graphical. It includes stem-and-leaf plots, histograms, and box plots.
Multivariate nongraphical. Adopted for highlighting the relationship between two or more variables (cross-tabulation or statistics).
Multivariate graphical. Graphics (e.g., bar chart, scatter plot, bubble chart, and heat map) can illustrate links between two or more data sets.
Software for an automated EDA in a univariate or bivariate manner is available (e.g., AutoEDA from XanderHorn). Furthermore, the programming languages Python and R are commonly used for EDA purposes.
3.3 Model selection
Model selection and optimization (or training) are vital steps for selecting a hypothesis that will efficiently fit future examples. Available training models have evolved in various increasingly complex and sophisticated algorithms. Therefore, identifying the best model is a complicated and iterative procedure. In this stage, one or more algorithms are identified and implemented.
ML algorithms are elements of code that can be used to explore, analyze, and find meaning in complex datasets. By simplifying, each algorithm is a finite set of detailed instructions and analyzes data by following a concrete pathway. Algorithms are often grouped according to the ML techniques used (i.e., Supervised Learning, Unsupervised Learning, and Reinforcement Learning).
Although the ideal model does not exist, striving for the “most appropriate model” for both data and the context in which it will operate is necessary. Accordingly, clinicians should develop a dialogue with researchers in bioinformatics to ensure the appropriate choice of method(s).
The supervised Support Vector Machine (SVM) is one of medicine’s most used ML algorithms. For instance, it has been used for cancer genomics (38), biomarkers selection in Alzheimer’s Disease (39), and in classifying EEG signals (40). In anesthesia research, this algorithm was adopted to study the anesthesia awareness phenomenon (41), to classify ultrasound image features (42), and for a vast number of purposes (43).
Regarding other algorithms, Fakherpour et al. (44) implemented the Multinomial Logistic Regression to evaluate spinal anesthesia’s hemodynamic effects in elective cesarean section. In contrast, reinforcement learning algorithms were used to develop a closed-loop anesthesia method based on mean arterial pressure and bispectral index (BIS) data (45). Furthermore, the Decision Tree was chosen to predict the efficacy of patient-controlled analgesia (46), and the Fuzzy unsupervised logic is a critical element of EEG signal processing (47).
Several algorithms have been used to optimize decision processes for patient treatments. Reinforcement learning algorithms are used for addressing sequential decision problems. These algorithms could be of paramount importance in intensive care medicine, especially when combined with deep learning (e.g., Convolutional Neural Network) (48). For example, starting from two large ICU databases (MIMIC-III and Philips eRI), Komorowski et al. (34) adopted the Markov decision process reinforcement algorithm to identify patient-tailored strategies for managing hypotension in sepsis. Finally, Q learning is another Reinforcement learning algorithm mainly used in cognitive science (49), principally for memory investigations (50).
A selection of ML algorithms for medical research purposes is described in Table 2.
Table 2.
The most used algorithms of machine learning in medical research. Abbreviations: BIRCH, balanced iterative reducing and clustering using hierarchies.
| Algorithm | Brief Description |
|---|---|
| Supervised | |
| Support Vector Machine (SVM) | It divides the learning data into classes, enlarging the distance from all points. Used for both classification and regression problems. |
| Linear Regression | Allow estimating the value of a variable that depends on many others. |
| Multinomial Logistic Regression (MLR) | Classification approach that generalizes logistic regression (a binary regression model that uses the logistic function to model the binary dependence) with a multiclass task. It can be viewed as the modality of assignment to a definite class, adopting the one that ensures the best probability. |
| Bayesian networks (BN) | Graphic model indicates a probability distribution in a set of arbitrary variables. The algorithm encompasses a probability distribution of the variables and a graph illustrating the dependencies between variables. |
| K-nearest neighbors (kNN) | It classifies a point based on the known classification of other points (votes of the closest k neighbors). |
| Restricted Boltzmann machine (RBM) | Graphic model with proportional reciprocity between observable and hidden variables. Links between elements of the same layer are not permitted to facilitate the learning mechanism. |
| Relevance vector machine (RVM) and Gaussian process (GP) | Bayesian extensions of the SVM algorithm. The assessment is provided on the probability of being included in a class. |
| Decision Tree (DT) | Graphic model (rule-based model) shows the decision points as branching and the applicable prediction in terms of end-nodes or leaves. |
| Unsupervised | |
| Fuzzy C Means | Flat/Partitioning-based algorithm that assigns elements to each data point related to each cluster center. It is established on the distance between the the cluster’s center and the data point. |
| BIRCH | A hierarchical-based algorithm that works over large data sets, requiring a single database scan. |
| K-means | Clustering algorithm splits a set of points (with no external classification) into K sets (clusters). The points in a cluster are disposed near each other. It is one of the various possible methods for solving the k-NN problem. |
| Reinforcement algorithms | |
| Markov decision process | It dissects the environment (where the learner, or agent, interacts) as a grid by dividing it into states, actions, models/transition models, and rewards. The solution is a policy (rewards combinations) and the objective is to find the optimal approach. |
| Q learning | The value-based approach of supplying information to inform which action an agent should take. |
3.3.1 Model selection and Algorithms in Artificial Neural Networks
The schematic model of ML pathways is difficult to adopt when in-depth analyses must be carried out. It usually occurs when composite scenarios (and multicomponent data) are encountered in medical investigations. In these circumstances, ANN architectures can be implemented. Concerning their functioning, the “machine” autonomously manages to classify the data and structures them hierarchically, finding the most relevant and useful ones. The system improves its performance with continuous learning and data processing (knowledge). Because of these properties, the machine learns and improves even more complex functions.
In this context, the proper selection of variables is challenging, and the EDA stage cannot be performed. Thus, it is often necessary to manage a more simplified dataset by saving the original information as much as possible, allowing a more feasible pattern recognition.
Depending on the complexity of the investigated data, the pre-processing phase can take place differently. Factor Analysis can be adopted to reduce the sample size and, where necessary, to find macro variables such as comorbidities, risk factors, etc. Alternatively, a fully automatic Random Forest method can be used to select the covariates and explain the classification. Finally, based on one of the two paths, the ANN model (mainly the hidden layers) is chosen (Figure 4).
Figure 4.
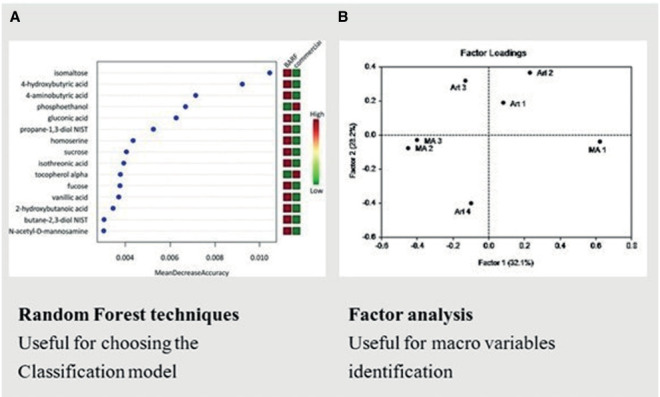
Two examples of Model Selection for artificial neural networks in medical investigations. Random Forest can help the screening of the covariates for the classification model (A). Factor analysis can be adopted to select macro variables (B).
Concerning algorithms, they have a fascinating functioning mechanism that mimics specific processes of brain functioning. Convolutional Neural Networks, for example, are biologically inspired by the retina. Furthermore, the backpropagation algorithm performs a “tuning” of the weights (of the previous layers) and develops a network process that allows the neural network to improve its performance progressively. It resembles most of the cognitive processes!
These algorithms are widely used in medical research, even in anesthesia and pain medicine. For instance, they recognize the EEG patterns and the analysis of linguistic and visual elements in pain studying (51). Moreover, they are of paramount importance for imaging investigation (52), brain monitoring (53), and have a broad application in the field of bioinformatics (54,55).
The most used algorithms are shown in Table 3.
Table 3.
Most used algorithms of deep learning in medical research.
| Algorithm | Brief Description |
|---|---|
| Multilayer perceptron (MLP) | Multiple layers system with a basic unit (perceptron). It allows a forward-directed flow of data from the input layer to the output one, passing through the hidden layer/layers, and with a learning algorithm of backward propagation. |
| Deep Learning | It provides a dissection of various levels that correlate to peculiar levels of abstraction. |
| Convolutional Neural Networks (CNNs) | An MLP development applied to two-dimensional matrices through a convolution operation. CNN neurons are linked to a limited number of inputs of a confined continuous region, used for computer vision tasks. |
| Recurrent neural networks (RNNs) | Uses sequential data or time-series data and develops loops between layers, storing memories for a short time. They are used for voice recognition. |
| Deep belief networks (DBNs) | Links between layers but not between elements within the same layer. |
| Long short-term memory (LSTM) | Advanced version of RNN that stores information for a more extended period (“long-term dependencies”). |
| Mixed Networks | Hybrid networks built by the correlation of two or more particular ANNs (e.g., CNNs and RNNs). |
3.3.2 Libraries
The various algorithms can be grouped into packages (libraries). Scikit-learn, for instance, is one of the most used libraries [56]. It is an open-source ML library for the Python programming language. It contains Supervised Learning (e.g., Linear Regression, Support Vector Machines, Naïve Bayes, K-Nearest Neighbor Classification) and Unsupervised Learning Estimators (e.g., PCA, and k-mean). The library is designed to work with NumPy and Pandas for data loading. Regardless of the model (algorithm), the Application Programming Interface (API) of Scikit-learn is always the same:
model.fit (X-train, Y_train)
predicted_test=model.predict(X_test)
the command fit trains the model and predicted is used to obtain outputs.
The command for the partition is:
X_train, X_test, y_train, y_test = train_test_split(X,y,random_state=0)
The random state hyperparameter in the train_test_split() function controls the shuffling process, influencing the model’s performance score. In particular, if the random_state=None (default value), different train and test sets across different executions are obtained. Consequently, the shuffling process is out of control. Integers are used and the function will produce the same results across distinct acquisitions. The most used integers are 0 and 42. When the random_state=0, the same train and test sets in different executions are achieved. Finally, with random_state=42, although the same train and test sets across other executions are produced, the train and test sets are not comparable with what was obtained with random_state=0. The Scikit-learn cross_val_score() function is commonly used to calculate values (and evaluate differences between values: root-mean-square errors) in the random state hyperparameter.
Libraries are obtained from other commonly used platforms. For instance, GitHub is a potent platform containing various open-source software implementations.
3.4 Model processing and evaluation
The system processing aims to generate a method (i.e., model) useful for future (unseen/out-of-sample) data and for prediction purposes. This phase can be achieved through holdout and Cross-validation (or k-fold cross-validation) methods.
Holdout. It is obtained through the dataset split into a ‘train’ and ‘test’ set. Usually, the division is managed by using 80% of the data for training and the remaining 20% for testing. It provides a single train-test split and is used to evaluate large datasets and for quick results. This method can also be the initial stage of a larger project.
Cross-validation. The dataset is randomly split up into ‘k’ groups. One of the groups is used as the test set and the rest works on the training set. The model is trained on the training set and scored on the test set. Subsequently, the operation is replicated until each group has been employed as the test set. For instance, for 4-fold cross-validation, the dataset is divided into four groups, and the model is trained and tested four partitioned times. Thus, each group has a chance to work as a test set.
If the model produces scarce performances, it can be re-built using improvement strategies. This process is called tuning.Subsequently, the method undergoes evaluation and tuning by using a combination of numerical (metrics:accuracy, precision, and recall) and visual tools such as ROC curves, residual plots, heat maps, and validation curves. When different models are tested, the model with the lowest validation error on the validation dataset can be selected. Models of increasing complexity are built using the model with the best empirical error rate.
For evaluation of the performances, the Confusion Matrix and the Accuracy Score can be used for Supervised Learning classification; Mean Absolute Error, Mean Squared Error, and R² Score for Supervised Learning regression. Furthermore, Homogeneity and V-measure are helpful for clustering in Unsupervised Learning approaches.
The model evaluation can also be performed with ANNs. For example, Hyperparameter tuning (or hyperparameter optimization) is a manual procedure that can facilitate the control of the training process by modifying the number of hidden layers and/or the representation of nodes in each layer.
3.4.1 Model Explainability
As models have increasing complexity, it is essential to understand the mechanisms that lead to the predicted outcomes. In this context, the so-called Model Explicability is a group of techniques designed to determine which model feature or combination led to a model-based decision. The purpose is not to explain how the model works but to answer the question “why an inference is given”. For this reason, we are moving from black boxes featuring mysterious or unknown internal functions or mechanisms to transparent model development.
Explainability can be viewed as an instrument in the toolbox that helps researchers understand their models’ decisions and the impact those decisions have on expected outcomes. Several methods, such as Shapley Additive exPlanations (SHAP) and Local Interpretable Model-Agnostic Explanations (LIME).
SHAP. It is a method used to break down the individual predictions of a complex model. The aim is to calculate each characteristic’s contribution to the forecast to identify each input’s impact. The basic principle is the study of cooperation between groups of players (“coalitions”) and the contribution to the final profit. Just as in game theory, some players contribute more to the outcome, so in ML some features contribute more to the prediction of the model and, therefore, have greater importance. Deep explainer, KernelSHAP, LinearSHAP are examples of SHAP-based versions.
LIME. It works on “black boxes” by developing an interpretable model for each forecast. LIME attempts to understand the relationship between the characteristics of a particular example and model prediction by training a more explainable model, such as a linear model, with examples derived from minor changes to the original input. The influences of perturbations in the model inputs on the final prediction of the model are evaluated. Through a training phase involving creating of a new set of data built on perturbing samples and from the correspondences in the outcome, the model selects the features (according to a qualitative and quantitative approach referred to normalized thresholds) that are the most important to explain model prediction. Each feature’s contribution in explaining the prediction developed by the model are considered. LIME can be used for tabular, image, or text datasets.
LIME is generally used to get a better explanation of a single prediction and SHAP to understand the whole model and feature dependencies.
3.4.2 Complexity, Model fitting, and Prediction errors
Each ML analysis aims to reduce differences between the model predictions and actual previsions. Consequently, a useful ML model can properly generalize any input of new data. Nevertheless, these differences are called errors and cannot be completely eliminated.
There are two types of errors, namely Bias and Variance. Notably, correcting or optimizing them will increase the model’s performance, avoiding model fitting errors, including overfitting and underfitting phenomena.
Prediction errors and model fitting performances are related to the complexity of the model. Latter refers to the complexity of the system is trying to learn, such as the degree of a polynomial. The optimal degree of complexity is usually based on the nature and quantity of the training data it contains. For instance, with a small amount of data, or if data are not distributed equally across several possible scenarios, a low-complexity model should be chosen. By using a small number of training data, a highly complex model will be ‘overfitted’.
Practically, overfitting is the learning of a function that adapts very well to the training data but is unable to generalize other information. In other words, our architecture is rigorously learning to produce the training data without discovering the actual trend or structure that drives this output. Prediction of the data that the system has never encountered will be poor!
Concerning underfitting, the model cannot capture the data’s underlying trend. It implies that our model or algorithm does not fit the data. It frequently occurs when there are scarce data or when a linear model with reduced non-linear data is trying to be built. In these cases, the rules of the ML model are too simple and flexible to be used for such data; accordingly, the model will likely produce many inaccurate predictions. On these premises, by summarizing:
Overfitting is high performance on the training set but not on the testing one. It resumes high Variance and low Bias.
Underfitting performs poorly on the training set. It expresses high Bias and low Variance.
Bias is the distance between expected and actual (mean) values. Thus, Bias and Variance represent opposite phenomena. On the contrary, Variance expresses the variability of the model prediction for a (single) given point (Figure 5).
Figure 5.
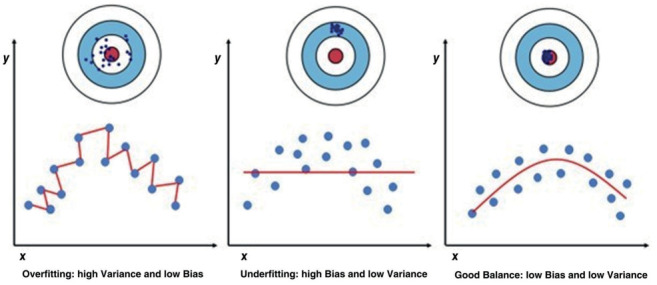
Model fitting errors. Variance is the variability (distance from the target center) of the model prediction for a single point. Bias is the distance between expected (target center) and means values. In overfitting, the model has high Variance and low Bias. It shows high performance on the training set but not on the test one. Underfitting is characterized by high Bias and low Variance and produces a poor performance on the training set.
To overcome high bias and underfitting and enhance the model’s performance, we can be followed two main strategies:
Increasing the number of layers or neurons in a layer (for ANN architectures). It increases the learning units that extract information from the previous neuronal elements, forwarding them to the next layer.
Using a new architecture (model/software).
To overcome high variance and overfitting, we can use several approaches:
Model complexity reduction.
Regularization (e.g., Ridge Regularization and Lasso Regularization). More robust regularization pushes the coefficients towards zero. It limits a model to avoid overfitting.
Dropout. The dropout rate is the parameter for the abandon function; it can be defined as the probability of training a given node in a layer: 1.0 means no dropout, and 0.0 means no output from the layer. In neural network models, it reduces interdependent learning between neurons.
Dataset size increment. However, adding more data would result in increased bias.
Ad hoc algorithms for early stopping in the training phase.
3.5 Practical suggestions
For inexperienced operators, the process can be facilitated by ad hoc tools for data engineering, ML, and analytics. They are component-based visual programming software packages that can allow simple data visualization, subset selection, and pre-processing until learning and predictive processes.
Among these instruments, KNIME is an open-source data tool usable for analytic processes, from data creation to production. Other easy-to-use data science software platforms are Orange and RapidMiner. More expert users can use Orange as a Python library to perform data manipulation and widget alteration.
Several data analytics platforms for data science and ML are available for more skilled operators. They are used to store and manage data with integrated tools and techniques capable of carrying out a variety of data analysis processes. The most implemented platforms include Microsoft Azure, Sumo Logic, Cloudera, and Google Cloud.
4 Conclusion
In conclusion, every Machine Learning pathway starts from task definition and ends in the model application. Different steps can be identified (pre-processing, exploratory data analysis, model selection, model processing and evaluation). High-performance characteristics and strict quality controls are needed during the different stages of its progress. For inexperienced operators, the whole process can be facilitated by ad hoc tools for data engineering, machine learning, and analytics.
Conflict of Interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article.
Author Contributions Statement:
VB, MC, FC, MR, RL have contributed significantly to: 1) the conception and design of the study, acquisition of the data, analysis and interpretation of the data; 2) drafting the article; and 3) the final approval of the version to be published. CC and EB have contributed significantly to: 1) substantial contributions to the conception and design of the study, data analysis and interpretation of the data; 2) revising the article critically for important intellectual content; and 3) the final approval of the version to be published.
References
- Melanie M. An Introduction to Genetic Algorithms (Fifth printing ed. Vol3). A Bradford Book The MIT Press, Cambridge, 221s. 1999 [Google Scholar]
- McCarthy J. What is artificial intelligence. 2004 accessible on:: http://www-formal.stanford.edu/jmc/whatisai.html . [Google Scholar]
- Amisha Malik P, Pathania M, Rathaur VK. Overview of artificial intelligence in medicine. J Family Med Prim Care. 2019;8(7):2328–2331. doi: 10.4103/jfmpc.jfmpc_440_19. doi:10.4103/jfmpc.jfmpc_440_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul V, Enslin S, Gross SA. History of artificial intelligence in medicine. Gastrointest Endosc. 2020 Oct;92(4):807–812. doi: 10.1016/j.gie.2020.06.040. doi: 10.1016/j.gie.2020.06.040. [DOI] [PubMed] [Google Scholar]
- Borges do Nascimento IJ, Marcolino MS, Abdulazeem HM, et al. Impact of Big Data Analytics on People’s Health: Overview of Systematic Reviews and Recommendations for Future Studies. J Med Internet Res. 2021 Apr 13;23(4):e27275. doi: 10.2196/27275. doi: 10.2196/27275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González García C, Núñez Valdéz ER, García Díaz V, Pelayo García-Bustelo BC, Cueva Lovelle JM. A review of artificial intelligence in the internet of things International Journal Of Interactive Multimedia And Artificial Intelligence. 2019;5:9–20. doi: 10.9781/ijimai.2018.03.004. [Google Scholar]
- Bellini V, Rafano Carnà E, Russo M, et al. Artificial intelligence and anesthesia: a narrative review. Ann Transl Med. 2022;10(9):528. doi: 10.21037/atm-21-7031. doi: 10.21037/atm-21-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu K, Sinha R, Ong A, Basu T. Artificial Intelligence: How is It Changing Medical Sciences and Its Future? Indian J Dermatol. 2020 Sep-Oct;65(5):365–370. doi: 10.4103/ijd.IJD_421_20. doi: 10.4103/ijd.IJD_421_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks? Nature. 2017;542:115–8. doi: 10.1038/nature21056. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018 Aug;18(8):500–510. doi: 10.1038/s41568-018-0016-5. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Loftus TJ, Datta S, et al. Performance of a Machine Learning Algorithm Using Electronic Health Record Data to Predict Postoperative Complications and Report on a Mobile Platform. JAMA Netw Open. 2022 May 2;5(5):e2211973. doi: 10.1001/jamanetworkopen.2022.11973. doi: 10.1001/jamanetworkopen.2022.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021 Jan;26(1):80–93. doi: 10.1016/j.drudis.2020.10.010. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Hossain ME, Qureshi MJU, et al. Design and Implementation of an IoT Based Medical Assistant Robot (Aido-Bot). 2020 IEEE International Women in Engineering (WIE) Conference on Electrical and Computer Engineering (WIECON-ECE) 2020:17–20. doi: 10.1109/WIECON-ECE52138.2020.9397958. [Google Scholar]
- Hurt B, Kligerman S, Hsiao A. Deep Learning Localization of Pneumonia: 2019 Coronavirus (COVID-19) Outbreak. J Thorac Imaging. 2020 May;35(3):W87–W89. doi: 10.1097/RTI.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang D, Dong J, Wang N, et al. False-Negative Results of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2: Role of Deep-Learning-Based CT Diagnosis and Insights from Two Cases. Korean J Radiol. 2020 Apr;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Qin L, Xu Z, et al. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology. 2020 Aug 19;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zeng Z, Wang K, et al. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J Thorac Dis. 2020 Mar;12(3):165–174. doi: 10.21037/jtd.2020.02.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Najjar HN, Al-Rousan N. A classifier prediction model to predict the status of Coronavirus COVID-19 patients in South Korea. Eur Rev Med Pharmacol Sci. 2020 Mar;24(6):3400–3403. doi: 10.26355/eurrev_202003_20709. [DOI] [PubMed] [Google Scholar]
- Jiang X, Coffee M, Bari A. Towards an Artificial Intelligence Framework for Data-Driven Prediction of Coronavirus Clinical Severity. 2020 Mar 30;63(1):537–551. [Google Scholar]
- Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadioglu O, Saeed M, Greten HJ, Efferth T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Bull World Health Organ. 2020 Mar 21:1–29. doi: 10.1016/j.compbiomed.2021.104359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. The Lancet. 2020 Feb;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A, Gentile F, Hsing M, Ban F, Cherkasov A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol Inform. 2020 Aug 11;39(8):e2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wu K, Zhang X, Deng S, Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. 2020 Mar;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthooft R, Ruyssinck J, van der Herten J, et al. Predictive modelling of survival and length of stay in critically ill patients using sequential organ failure scores. Artif Intell Med. 2015 Mar;63(3):191–207. doi: 10.1016/j.artmed.2014.12.009. doi: 10.1016/j.artmed.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Sotoodeh M, Ho JC. Improving length of stay prediction using a hidden Markov model. AMIA Jt Summits Transl Sci Proc 2019 May 6. 2019:425–434. [PMC free article] [PubMed] [Google Scholar]
- Lin YW, Zhou Y, Faghri F, Shaw MJ, Campbell RH. Analysis and prediction of unplanned intensive care unit readmission using recurrent neural networks with lon short term memory. PLoS One. 2019;14:e0218942. doi: 10.1371/journal.pone.0218942. doi: 10.1371/journal.pone.0218942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad A, Bader-El-Den M, McNicholas J, Briggs J. Early hospital mortality prediction of intensive care unit patients using an ensemble learning approach. Int J Med Inform. 2017;108:185–195. doi: 10.1016/j.ijmedinf.2017.10.002. doi: 10.1016/j.ijmedinf.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Holmgren G, Andersson P, Jakobsson A, Frigyesi A. Artificial neural networks improve and simplify intensive care mortality prognostication: a national cohort study of 217,289 first-time intensive care unit admissions. J Intensive Care. 2019;7:44. doi: 10.1186/s40560-019-0393-1. doi: 10.1186/s40560-019-0393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Mu L, Chen L, et al. Predicting tachycardia as a surrogate for instability in the intensive care unit. J Clin Monit Comput. 2019 Dec;33(6):973–985. doi: 10.1007/s10877-019-00277-0. doi: 10.1007/s10877-019-00277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanch L, Sales B, Montanya J, et al. Validation of the Better Care¯ system to detect ineffective efforts during expiration in mechanically ventilated patients: a pilot study. Intensive Care Med. 2012 May;38(5):772–80. doi: 10.1007/s00134-012-2493-4. doi: 10.1007/s00134-012-2493-4. [DOI] [PubMed] [Google Scholar]
- Rehm GB, Han J, Kuhn BT, et al. Creation of a Robust and Generalizable Machine Learning Classifier for Patient Ventilator Asynchrony. Methods Inf Med. 2018 Sep;57(4):208–219. doi: 10.3414/ME17-02-0012. doi: 10.3414/ME17-02-0012. [DOI] [PubMed] [Google Scholar]
- Adams JY, Lieng MK, Kuhn BT, et al. Development and Validation of a Multi-Algorithm Analytic Platform to Detect Off-Target Mechanical Ventilation. Sci Rep. 2017 Nov 3;7(1):14980. doi: 10.1038/s41598-017-15052-x. doi: 10.1038/s41598-017-15052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018;24(11):1716–1720. doi: 10.1038/s41591-018-0213-5. doi: 10.1038/s41591-018-0213-5. [DOI] [PubMed] [Google Scholar]
- Davoudi A, Malhotra KR, Shickel B, et al. Intelligent ICU for Autonomous Patient Monitoring Using Pervasive Sensing and Deep Learning. Sci Rep. 2019 May 29;9(1):8020. doi: 10.1038/s41598-019-44004-w. doi: 10.1038/s41598-019-44004-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med. 2019;380(14):1347–1358. doi: 10.1056/NEJMra1814259. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- Char DS, Shah NH, Magnus D. Implementing Machine Learning in Health Care - Addressing Ethical Challenges. N Engl J Med. 2018;378(11):981–983. doi: 10.1056/NEJMp1714229. doi: 10.1056/NEJMp1714229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics. 15(1):41–51. doi: 10.21873/cgp.20063. doi: 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Petersen M, Johnson L, Hall J, O’Bryant SE. Recursive Support Vector Machine Biomarker Selection for Alzheimer’s Disease. J Alzheimers Dis. 2021;79(4):1691–1700. doi: 10.3233/JAD-201254. doi: 10.3233/JAD-201254. [DOI] [PubMed] [Google Scholar]
- Kumar B, Gupta D. Universum based Lagrangian twin bounded support vector machine to classify EEG signals. Comput Methods Programs Biomed. 2021;208:106244. doi: 10.1016/j.cmpb.2021.106244. doi: 10.1016/j.cmpb.2021.106244. [DOI] [PubMed] [Google Scholar]
- Tacke M, Kochs EF, Mueller M, Kramer S, Jordan D, Schneider G. Machine learning for a combined electroencephalographic anesthesia index to detect awareness under anesthesia. PLoS One. 2020;15(8):e0238249. doi: 10.1371/journal.pone.0238249. doi: 10.1371/journal.pone.0238249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Tan KK, Sng BL, Li S, Sia AT. Lumbar Ultrasound Image Feature Extraction and Classification with Support Vector Machine. Ultrasound Med Biol. 2015;41(10):2677–89. doi: 10.1016/j.ultrasmedbio.2015.05.015. doi: 10.1016/j.ultrasmedbio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Hashimoto DA, Witkowski E, Gao L, Meireles O, Rosman G. Artificial Intelligence in Anesthesiology: Current Techniques, Clinical Applications, and Limitations. Anesthesiology. 2020;132(2):379–394. doi: 10.1097/ALN.0000000000002960. doi:10.1097/ALN.0000000000002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakherpour A, Ghaem H, Fattahi Z, Zaree S. Maternal and anaesthesia-related risk factors and incidence of spinal anaesthesia-induced hypotension in elective caesarean section: A multinomial logistic regression. Indian J Anaesth. 2018;62(1):36–46. doi: 10.4103/ija.IJA_416_17. doi: 10.4103/ija.IJA_416_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R, Meskin N, Haddad WM. Closed-loop control of anesthesia and mean arterial pressure using reinforcement learning. Biomed Signal Process Control. 2015;22:54–64. [Google Scholar]
- Hu YJ, Ku TH, Jan RH, Wang K, Tseng YC, Yang SF. Decision tree-based learning to predict patient controlled analgesia consumption and readjustment. BMC Med Inform Decis Mak. 2012;12:131. doi: 10.1186/1472-6947-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M. Mechanisms underlying brain monitoring during anesthesia: limitations, possible improvements, and perspectives. Korean J Anesthesiol. 2016;69(2):113–20. doi: 10.4097/kjae.2016.69.2.113. doi: 10.4097/kjae.2016.69.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson A. Deep Reinforcement Learning in Medicine. Kidney Dis (Basel) 2019;5(1):18–22. doi: 10.1159/000492670. doi: 10.1159/000492670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasoveanu A, Dotlačil J. Reinforcement Learning for Production-Based Cognitive Models. Top Cogn Sci. 2021;13(3):467–487. doi: 10.1111/tops.12546. doi: 10.1111/tops.12546. [DOI] [PubMed] [Google Scholar]
- Roscow EL, Chua R, Costa RP, Jones MW, Lepora N. Learning offline: memory replay in biological and artificial reinforcement learning. Trends Neurosci. 2021;44(10):808–821. doi: 10.1016/j.tins.2021.07.007. doi: 10.1016/j.tins.2021.07.007. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhang H, Kavitha PT, et al. Scalp EEG-Based Pain Detection Using Convolutional Neural Network. IEEE Trans Neural Syst Rehabil Eng. 2022;30:274–285. doi: 10.1109/TNSRE.2022.3147673. doi: 10.1109/TNSRE.2022.3147673. [DOI] [PubMed] [Google Scholar]
- Hasan N, Bao Y, Shawon A, Huang Y. DenseNet Convolutional Neural Networks Application for Predicting COVID-19 Using CT Image. SN Comput Sci. 2021;2(5):389. doi: 10.1007/s42979-021-00782-7. doi: 10.1007/s42979-021-00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madanu R, Rahman F, Abbod MF, Fan SZ, Shieh JS. Depth of anesthesia prediction via EEG signals using convolutional neural network and ensemble empirical mode decomposition. Math Biosci Eng. 2021;18(5):5047–5068. doi: 10.3934/mbe.2021257. doi: 10.3934/mbe.2021257. [DOI] [PubMed] [Google Scholar]
- Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017 Sep 1;18(5):851–869. doi: 10.1093/bib/bbw068. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- Liu X, Liu C, Huang R, et al. Long short-term memory recurrent neural network for pharmacokinetic-pharmacodynamic modeling. Int J. Clin Pharmacol Ther. 2021;59(2):138–146. doi: 10.5414/CP203800. doi: 10.5414/CP203800. [DOI] [PubMed] [Google Scholar]
- Python Scikit-Learn Cheat Sheet. accessable on:: https://intellipaat.com/blog/tutorial/python-tutorial/scikit-learn-cheat-sheet/ [Google Scholar]


