Abstract
Preoperative prediction of recurrence outcome in hepatocellular carcinoma (HCC) facilitates physicians’ clinical decision-making. Preoperative imaging and related clinical baseline data of patients are valuable for evaluating prognosis. With the widespread application of machine learning techniques, the present study proposed the ensemble learning method based on efficient feature representations to predict recurrence outcomes within three years after surgery. Radiomics features during arterial phase (AP) and clinical data were selected for training the ensemble models. In order to improve the efficiency of the process, the lesion area was automatically segmented by 3D U-Net. It was found that the mIoU of the segmentation model was 0.8874, and the Light Gradient Boosting Machine (LightGBM) was the most superior, with an average accuracy of 0.7600, a recall of 0.7673, a F1 score of 0.7553, and an AUC of 0.8338 when inputting radiomics features during AP and clinical baseline indicators. Studies have shown that the proposed strategy can relatively accurately predict the recurrence outcome within three years, which is helpful for physicians to evaluate individual patients before surgery.
Keywords: recurrence prediction, efficient features, ensemble learning, hepatocellular carcinoma, surgery
1 Introduction
Hepatocellular carcinoma (HCC) accounts for 85%-90% of the main pathological types of primary liver cancer (1–3). It is easy to spread in the liver through the portal vein system to form intrahepatic metastasis, and it is also easy to form tumor thrombus in the portal vein and cause portal hypertension. HCC is mostly found in the middle and late stages, which leads to its generally poor prognosis (4–8). According to statistics, the recurrence rate of HCC after surgery is as high as about 70% (9), and the survival rate is only 15%-40% (10). Fortunately, treatment modalities represented by precision surgery have greatly improved patient prognosis. Liver resection with early diagnosis can improve the survival rate of patients within one year to 91%-98% (11, 12). Therefore, rational clinical decision-making is essential to reduce recurrence and improve survival.
Accurate preoperative prediction of recurrence can help doctors assess the necessity and risk of surgery, so that they can design rational clinical decisions. Early (1-2 years after surgery) (13) and long-term (5 years and beyond) (14) recurrence predictions have been performed in a small number of studies, with encouraging results. It is worth noting that the recurrence rate of HCC within 3 years after surgery is 50-55%, which accounts for about 71%-78% of the total recurrence (15). Three years after surgery is a critical period, and the absence of recurrence within 3 years indicates a relatively good prognosis. There is no doubt that preoperative prediction of the recurrence outcome in patients within 3 years after surgery is also of great significance for evaluating the illness and selecting treatment options.
The rise of artificial intelligence (AI) technology has brought new strategies for the prediction of HCC recurrence, especially novel data processing methods represented by machine learning and radiomics. Studies have shown that patients’ preoperative imaging, personal information and clinical manifestations are closely related to prognosis (16, 17). Because of this, some researchers have employed the preoperative performance of patients to predict postoperative recurrence through AI algorithms. Ji et al. (18) collected data on 480 patients undergoing HCC resection from 3 centers. Combined with radiomics characteristics and some biochemical indicators, a Cox-based recurrence risk prediction model was constructed, and the final C-index reached 0.633-0.699. Zeng et al. developed a random survival forest (RSF) model using the 15 characteristics of HCC patients. The model obtained a C-index of 0.725 on the validation set, which was encouraging. Huang et al. (19) developed a machine learning prognostic model to identify high-risk patients after surgical resection. The results show that the eXtreme Gradient Boosting tree (XGBoost) achieved the best discrimination in the internal validation queue. In reference (20), 143 features were extracted, including 26 preoperative clinical features, 5 postoperative pathological features, and 112 imaging features, for predicting early recurrence of HCC. As a result, the area under the receiver operating characteristic curve (AUC) of the preoperative model was 0.739, with relatively strong generalization ability.
Nevertheless, there is still room for improvement in the current related work. For example, the lesion area adopted to extract features in most studies needs to be manually segmented from the original image, which brings great challenges to improving work efficiency and reducing costs. In addition, the features of the input model are often not concise and efficient, which will lead to a decrease in accuracy. It is necessary to explore efficient feature representations and achieve automatic and accurate predictions.
This study aimed to develop an excellent predictive strategy for recurrence-free survival (RFS) outcomes in patients with HCC within 3 years after surgery. A 3-dimension deep learning framework was applied to automate lesion segmentation. Seven feature representation methods were compared to explore the most superior feature combinations, including clinical baseline indicators, radiomics features during arterial phase (AP), portal venous phase (PVP), and delayed phase (DP), and combination of clinical data with radiomics features during each phase. Four novel Boosting ensemble learning models were selected for prediction of recurrence outcome. This work has the following highlights:
● Deep learning was employed for automatic segmentation of regions of interest (ROI), which avoided the drawbacks of manual delineation.
● Seven feature representations were explored to find the best model input.
● The study compared novel Boosting ensemble learning methods to select the model with best performance, which may be applicable in the future.
2 Materials and methods
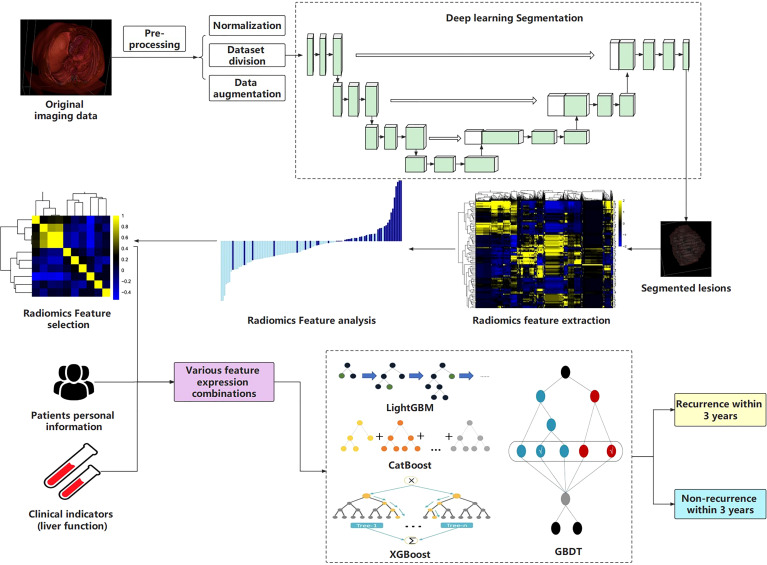
The workflow of this study is shown in Figure 1 .
Figure 1.
The workflow of this study.
2.1 Patients
HCC patients who underwent partial hepatectomy in Qingdao University Affiliated Hospital from January 2014 to December 2018 were followed after surgery regularly. The inclusion criteria were as follows: 1. The pathological diagnosis was HCC; 2. The first treatment was partial hepatectomy; 3. Enhanced CT examination was performed within 1 month before surgery, and all periods were completed; 4. The patient’s personal information and relevant clinical data were complete; 5. It has been confirmed that whether the recurrence occurred within 36 months after surgery. The following were the exclusion criteria: 1. Patients who have received chemotherapy, interventional therapy, targeted therapy, etc. before partial hepatectomy; 2. Patients with a history of other tumors; 3. Patients whose tumors have metastasized; 4. Imaging and clinical data were incomplete; 5. The follow-up data were incomplete or the recurrence outcome within 3 years couldn’t be judged. Additionally, all patients included in the study underwent radical hepatectomy. The criteria for radical hepatectomy were: (1) no residual tumor was found at the margin of resection, which was negative; (2) no tumor was found in the remaining liver; (3) tumor markers returned to normal within two months after surgery. Ultimately, 105 patients were selected for the study. RFS period is defined as the time from the date of liver resection to the date of recurrence and within 3 years after surgery is within 36 months from the date of liver resection.
It must be emphasized that the principles of the Declaration of Helsinki were followed and the study was approved by the hospital ethics committee (ethics number: 20001-01). All patients signed an informed consent certificate before surgery.
2.2 Imaging acquisition
The scanning equipment for the detection was the German CT (SOMATOM Definition Flash, Siemens) and the American Discovery CT (GE Healthcare). The scanning method was a three-level contrast-enhanced scan of the upper abdomen, and the scanning range was from the top of the liver to the lower edge of the two kidneys. During the scanning process, the voltage, current, scanning layer thickness, layer spacing, and pixel matrix size were set to 120 kV, 200-350 mA, 5 mm, 5 mm, and 512 × 512, respectively. Workers administered iohexol and 350 mg/m1 of iodine through a peripheral vein at a flow rate of 3.0 ml/s and a dose of 1.5 ml/kg under the action of a pressure syringe. Finally, AP, PVP, and DP images were obtained for the study.
2.3 Lesion segmentation
Generally, studies mostly segment lesions manually, which reduces work efficiency. Based on the previous manual annotation, we built a 3D U-Net deep learning model for automatic and accurate segmentation of lesions.
2.3.1 Manual annotation
This work adopted the supervised learning to automatically segment the ROIs, so manual annotation was required before model training. Two physicians with extensive experience in radiology were selected for this task, one of whom delineated the tumor area of each slice with the help of 3D Slicer (Boston, MA, USA) software without knowing any patient’s baseline data, and the other one was responsible for checking the annotation results. Once there was a dispute, return to discuss and re-mark if necessary. All CT images for the three periods were delineated and formed into volumes of interest (VOIs).
2.3.2 Data pre-processing
Considering that some slices in CT images do not contain ROIs, this will increase the computational complexity. Slices without lesions were cropped according to the annotated images and the remainders were studied. Moreover, we normalized the image format to 256×256×48 for better input to the model. In order to expand the amount of data, data augmentation operations were performed on the divided training set, including but not limited to image flipping, rotation, cropping, scaling, and blurring (21, 22).
2.3.3 Construction of segmentation model
CT images have 3D structures, and the traditional method convert them into 2D slices and then send into the 2D segmentation model, which results in the loss of spatial information. In this study, a 3D convolutional neural network (3D U-Net) was constructed to segment lesions directly, which comprehensively preserved the spatial information between slices (23, 24).
Similar to the classic U-Net, the 3D U-Net also consists of Encoder and Decoder, each of which contains four sub-modules. In the Encoder, each sub-module contains two 3 × 3 × 3 convolutional layers, and each convolutional layer is connected to an activation function. After completing the convolution operation, max-pooling with a stride of 2 is performed on each dimension. In Decoder, each sub-module contains an upsampling process (deconvolution operation) with a stride of 2, and then two 3 × 3 × 3 convolutional layers and activation functions are added in turn. It must be emphasized that the padding in the convolutional layer of this module is set to 1, which makes the convolution operation not change the size of the image. Changes in image size are completely controlled by pooling and upsampling. Additionally, the last sub-module of the Decoder consists of a 1 × 1 × 1 convolutional layer, which reduces the number of output feature maps. Batch normalization (BN) was introduced before each activation function.
This work aims to segment liver tumors from other tissues, where the input channel of the model was set to 256 × 256 × 48, and the activation function adopted ReLU. After the construction was completed, the total parameters and the trainable parameters of the neural network reached 4,122,466 and 4,117,570, respectively.
2.4 Radiomics feature extraction and selection
Feature extraction is an essential part of radiomics analysis. In this study, we performed radiomic feature extraction for segmented liver tumors. Using the Pyradiomics 3.0.1 library in Python, a total of 788 dimensional features including Shape, Firstorder, GLCM, GLRLM, GLSZM, and GLDM were extracted. Each type of features was performed 9 transformations including Original, Wavelet-LLH, Wavelet-LHL, Wavelet-LHH, Wavelet-HLL, Wavelet-HLH, Wavelet-HHL, Wavelet-HHH, and Wavelet-LLL. Among them, “Wavelet-XXX” represents the wavelet transform, followed by the corresponding basis function type.
Due to the high dimension of the extracted features, it is easy to cause “dimensionality disaster” and affect the model performance. Therefore, selecting features with large contributions can reduce the dimension as much as possible without affecting the comprehensiveness of the features. This work employed the Least Absolute Shrinkage Selector Operator (Lasso) algorithm to select the extracted features and ranked the contribution of each feature. By constructing a penalty function, Lasso can compress the coefficients of variables and make some regression coefficients 0, so as to achieve the purpose of variable selection. In addition, Lasso can also filter variables and reduce the complexity of the model. The variable screening here refers to not putting all the variables into the model for fitting, but selectively putting the variables into the model to get better performance parameters. Complexity adjustment refers to controlling the complexity of the model through a series of parameters to avoid overfitting. The optimal model was fit and the value of the penalty parameter α was determined based on the sklearn library in Python. For the dimensionality-reduced features, correlation coefficients and cluster heatmaps, as well as the coefficient distribution of each feature are visualized to better interpret the radiomics features.
2.5 Selection of clinical baseline features
This study collected clinical baseline data of HCC patients in addition to CT images, such as personal information and clinical indicators. The gender and age of patients were collected as personal information data. Clinical indicators here were mainly tumor markers and liver function indicators, including alpha-fetoprotein (AFP), hepatitis B surface antigen (HBsAg), albumin (ALB), the total bilirubin (T- BIL), alanine aminotransferase (ALT) and aspartate aminotransferase (AST), etc. It should be noted that positive and negative results were obtained for AFP and HBsAg, while other liver function indicators were represented as specific test results.
2.6 Construction of recurrence prediction models
A total of seven feature representations, including selected radiomics features during AP, PVP, and DP, clinical baseline features, and their combined features, were input into the recurrence prediction models. The Boosting ensemble learning algorithms were adopted to predict the RFS outcome within 3 years.
2.6.1 Light gradient boosting machine
Gradient Boosting Decision Tree (GBDT) is a classic ensemble algorithm in machine learning. Its main idea is to employ weak classifiers (decision trees) to iteratively train to obtain the optimal model, which has the advantages of good training effect and not easy overfitting. LightGBM (Light Gradient Boosting Machine) is a framework for implementing the GBDT algorithm. It supports efficient parallel training and has faster training speed, lower memory consumption, better accuracy, support for distributed and fast processing of massive data, etc. (25). Currently, this framework has been relatively widely used in the field of medical data processing (26–28), but it has not been attempted in the HCC recurrence prediction task.
A leaf-wise algorithm with a depth limit is adopted in LightGBM. This strategy finds the leaf with the largest split gain from all the current leaves each time, and then splits and loops, which reduce more errors and get better accuracy under the same number of splits. Moreover, the Gradient-based One-Side Sampling (GOSS) operation is proposed to reduce computation and improve accuracy. This method does not calculate the gradient through the sample points used, but calculates the gradient by partial sampling. The Exclusive Feature Bundling (EFB) is also proposed to bundle some features together to reduce the feature dimension, thereby reducing the time-consuming to find the best fork. This study implemented the LightGBM algorithm based on the sklearn library in Python to perform the binary classification task, that is, recurrence or not within 3 years.
2.6.2 Categorical boosting
Categorical Boosting (CatBoost), as a novel ensemble learning algorithm, has been applied to some medical data processing tasks, but has not been used to predict HCC recurrence (29, 30). Catboost adopts the oblivious tree as the base tree model, which is characterized by the same segmentation features in each layer. Leaf nodes can be converted to binary codes, and the value of the node is stored in a floating-point vector of length 2 to the power of d (d is the depth of the tree). One of the advantages of this tree is that the prediction performance is better, and this structure can also weaken the shortcomings of easy fitting in decision trees to a certain extent. When Catboost completes training, it stores the leaf node value of each tree into a vector. When predicting, it can quickly retrieve the corresponding leaf node value by judging which leaf node it is in, so it can improve the prediction efficiency and enhance the model performance. This work selected it for predicting HCC recurrence.
2.6.3 eXtreme gradient boosting
XGBoost has been widely used in the field of medical data analysis since it was proposed in 2014 (31, 32). In the HCC recurrence prediction task, this algorithm was also tried and achieved significant results (19). Its greedy algorithm-based split node calculation and missing value handling techniques are very suitable for data mining. The algorithm was trained to predict RFS outcomes and compared with other models such as LightGBM and CatBoost.
2.6.4 Gradient boosting decision tree
We also employed GBDT as the baseline model for comparison. It is an ensemble learning algorithm based on decision trees that iterates over new learners through gradient descent. In this paper, the classification task was performed, and the Classification And Regression Tree (CART) was selected.
2.7 Statistical analysis
For the analysis of clinical baseline data, the differences involved in this study were compared using student t-test or Mann-Whitney U-test, where the criterion of significant difference was set at P<0.05. Mean ± 95% confidence interval (CI) was calculated as results for continuous variables. To reflect the criticality of certain variables, the univariate Kaplan-Meier curve was introduced for survival analysis.
We calculated the mean Intersection overUnion (mIoU), accuracy (Acc), Kappa and Dice coefficients of 3D U-Net to reflect the segmentation effect. Additionally, Acc, recall, precision (Prec), F1 score, receiver operating characteristic curve (ROC) and corresponding AUC were introduced as performance evaluation criteria for the ensemble learning models. It should be emphasized that the classification threshold was set to 0.5.
2.8 Experimental setup
The image data during the three scanning periods were randomly divided into training set, validation set and test set according to the ratio of 8:1:1. The segmentation model was trained on the training set and validation set, and the test set was employed to demonstrate the performance. All lesions segmented by the model during three periods were acquired and their radiomic features were extracted. For the Lasso regression algorithm, the study obtained the best α value through 10-fold cross-validation to select key features. Considering the small sample size, this study selected the 5-fold cross-validation method to determine the features representation and predict the recurrence outcome, and calculated the mean value of five experiments and the corresponding 95% CI as the results. The relevant computing equipment for this experiment was configured with a CPU AMD Ryzen 7 5800H (16 GB memory) and a GPU NVIDIA® Tesla V100 (32 GB memory) with acceleration support of the compute unified device architecture (CUDA). All work was carried out in the Windows 10 operating system, and the programming language, deep learning framework and key libraries included Python 3.7, Pytorch, Pyradiomics, sklearn, VTK, etc.
3 Results
3.1 Analysis of patients’ basic data
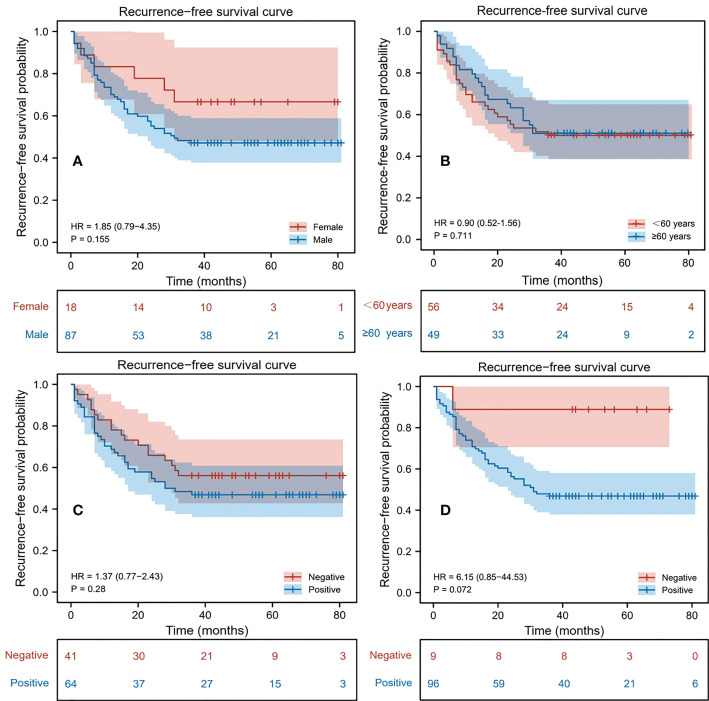
During follow-up, 52 patients (49.52%) were found to have recurrence within 3 years after surgery, of which 46 (88.46%) were male and 6 (11.54%) were female; 24 (46.15%) were aged 60 years or older and 28 (53.85%) were younger than 60 years old; 34 (65.38%) were AFP positive, and 18 (34.62%) were negative; 51 (98.08%) were HBsAg positive, and 1 (1.92%) were negative. 53 patients (50.48%) were found to have no recurrence within 3 years after surgery, of which 41 (77.36%) were male and 12 (22.64%) were female; 25 (47.17%) were aged 60 years or order and 28 (52.83%) were younger than 60 years old; 30 (56.60%) were AFP positive, and 23 (43.40%) were negative; 45 (84.91%) were HBsAg positive, and 8 (15.09%) were negative. Based on this, a univariate Cox proportional hazards model was established to judge the influence of different factors on RFS, and the related results were represented by the Kaplan-Meier curves ( Figure 2 ). Through the statistics of gender classification group (HR=1.85, P=0.155) and HBsAg result classification group (HR=6.15, P=0.072), it was found that gender and HBsAg affect RFS to some extent although the differences were not significant, followed by AFP (HR=1.37, P=0.280). Notably, age was not significantly associated with recurrence outcome from the age-categorized group in this study (HR=0.90, P=0.711). However, patient’s age is a key factor affecting prognosis from previous studies (20, 33), so we still regarded it as one of the features. Table 1 shows the statistical results of some continuous clinical indicators. It can be found that ALB, T-BIL, ALT, and AST (P=0.149, 0.377, 0.128, and 0.223, respectively) were relatively significantly different or not significantly different between the recurrence and non-recurrence groups.
Figure 2.
Kaplan-Meier survival analysis curve of patients, where the variables in (A–D) are gender, age, alpha-fetoprotein (AFP), hepatitis B surface antigen (HBsAg) respectively.
Table 1.
Statistical results of 4 clinical indicators.
| Clinicalindicator | Total dataset (N = 105) | P-value | |
|---|---|---|---|
| Recurrence (N =52) | Non-recurrence (N = 53) | ||
| ALB(g/L) | 40.36 ± 1.28 | 40.42 ± 1.26 | 0.1493 |
| T-BIL(mmol/L) | 20.32 ± 4.81 | 20.34 ± 4.69 | 0.3765 |
| ALT(u/L) | 51.12 ± 15.42 | 50.89 ± 15.02 | 0.1282 |
| AST(u/L) | 42.78 ± 12.23 | 42.20 ± 11.92 | 0.2233 |
all outcomes are based on recurrence within 3 years after surgery. ALB, T-BIL, ALT, and AST represent albumin, the total bilirubin, alanine aminotransferase and aspartate aminotransferase, respectively. Each indicator is represented by the mean of the sample and the corresponding 95% confidence interval (CI).
3.2 Results of lesion segmentation
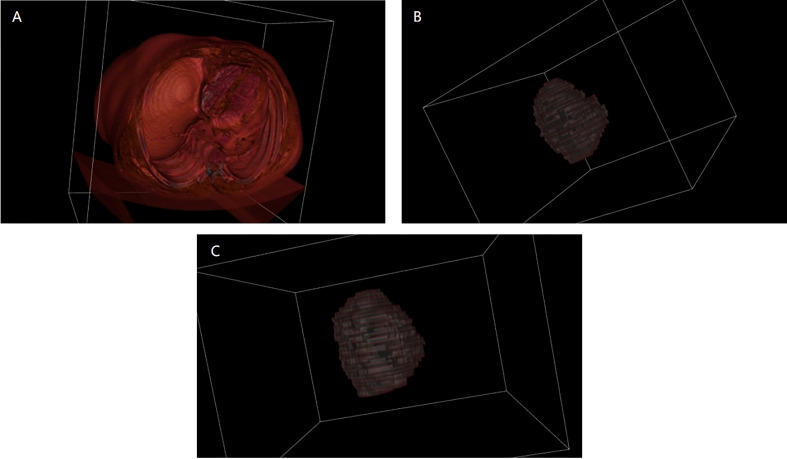
The training and validation sets during the three periods were input into 3D U-Net for training, and the model performance was optimized through parameter adjustment and continuous iteration. The key hyperparameters were set as follows: Momentum optimizer was selected and set to 0.9, initial learning rate, weight_decay and batch_size were set to 0.001, 4.0×10-3, 2, respectively. After the model iterated for 500 epochs, it fully converged (the loss value of the validation set was lower than 0.001). At this point, we stopped the training and saved the parameters. The performance on the test set was excellent, with mIoU of 0.8874, Acc of 0.9915, Kappa of 0.8738 and Dice coefficient of 0.9360, which indicates that the deep learning model has strong generalization ability for segmenting liver lesions. To visually compare the segmentation effects, this paper presents 3D reconstruction visualization images of the upper abdomen based on CT scans, manually annotated tumors, and deep learning-segmented tumors ( Figure 3 ). The VTK library in Python was adopted as the relevant drawing tool. It must be emphasized that the lesion areas involved in subsequent calculations were automatically segmented by the trained model.
Figure 3.
3D reconstruction visualization images before and after segmentation. (A) is the 3D visualization of the original CT image before segmentation; (B) is the 3D visualization after manually segmenting the tumor; (C) is the 3D visualization after segmenting the tumor using deep learning.
3.3 Results of radiomics feature extraction and selection
A total of 788 radiomic features were extracted in this study, including 100 features from original transform and 688 features from wavelet transform. In the original transform, the extracted contents were 14 shapes, 18 firstorder, 22 GLCM, 16 GLRLM, 16 GLSZM and 14 GLDM features. In the wavelet transform, the contents extracted by Wavelet-LLH, Wavelet-LHL, Wavelet-LHH, Wavelet-HLL, Wavelet-HLH, Wavelet-HHL, Wavelet-HHH, and Wavelet-LLL included 144 firstorder, 176 GLCM, 128 GLRLM, 128 GLSZM and 112 GLDM features. Since high-dimensional features may affect model performance, dimensionality reduction and selection of contributing features is significant.
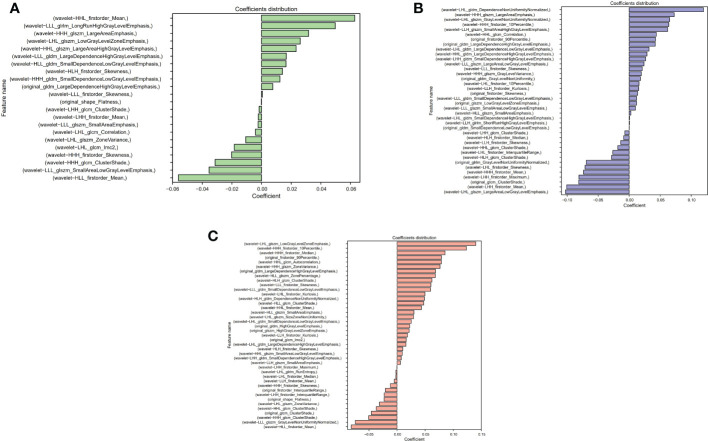
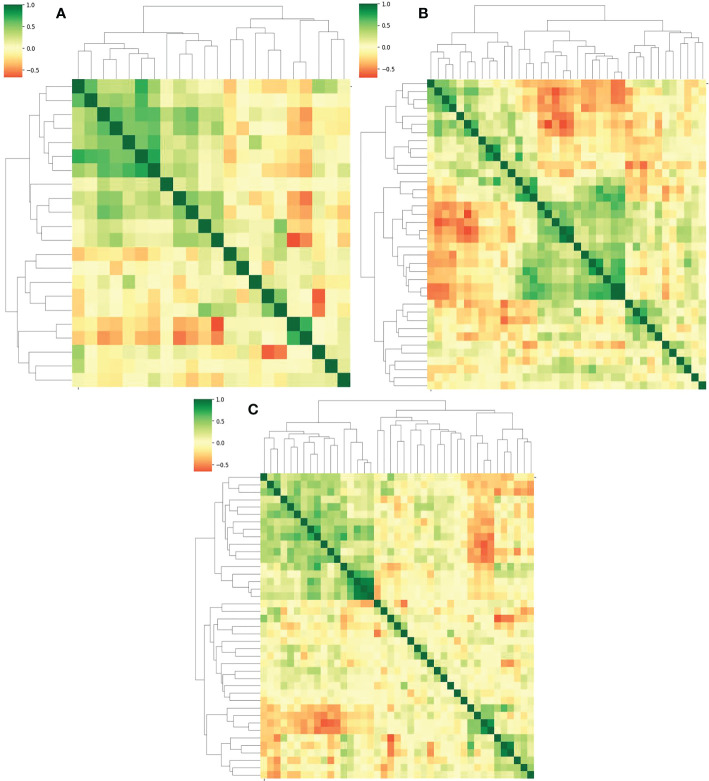
The Lasso algorithm was used for fitting to obtain the best α values during AP, PVP and DP, respectively. The model was fully converged after 10,000 iterations based on the 10-fold cross-validation. The optimized α values for AP, PVP and DP were calculated as 0.0518, 0.0244 and 0.0202, respectively. Meanwhile, 22, 38, and 41 features with contribution degrees were selected during the above three periods respectively. Figures 4A, B , and C show the selected feature names and the corresponding coefficients distribution in AP, PVP, and DP, respectively. Figure 5 shows the correlation coefficient between the features and the clustering results through heatmaps (the color depth represents the correlation strength).
Figure 4.
Distribution of selected radiomics feature coefficients. (A–C) show the features and their distributions during arterial phase (AP), portal venous phase (PVP) and delay period (DP), respectively.
Figure 5.
(A–C) represent the correlation and the clustering heatmaps between features during the AP, PVP and DP, respectively.
3.4 Results of recurrence prediction
3.4.1 Comparison of different feature representations
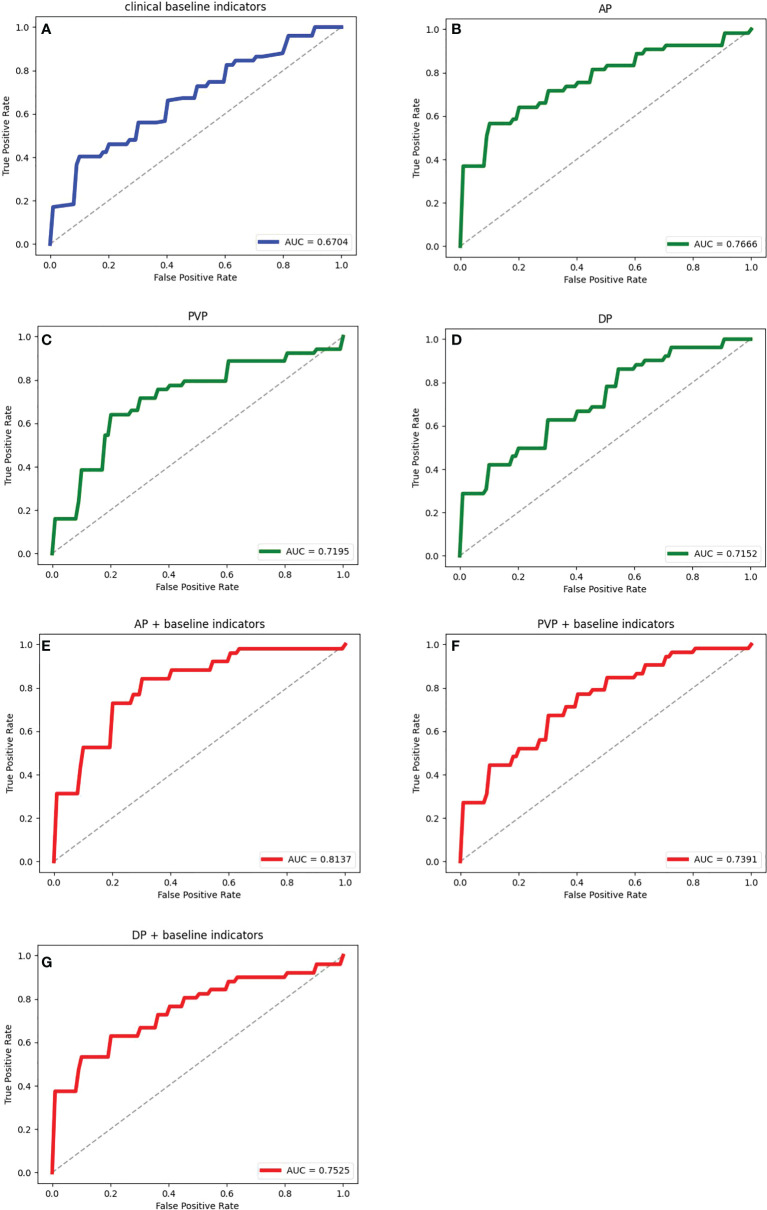
Seven feature representation methods for evaluating the prognosis of HCC were considered, including clinical baseline features, radiomics features of AP, radiomics features of PVP, radiomics features of DP, radiomics features of AP combined with clinical indicators, radiomics features of PVP combined with clinical indicators, and radiomics features of DP combined with clinical indicators. In order to explore the most excellent feature representation, we separately input the above features into the ensemble learning algorithms and optimized the training. Considering the randomness of the results based on the small sample size, the training process adopted 5-fold cross-validation, that is, the dataset was randomly divided into 5 equal parts, 4 of which were used for training and the remaining 1 was used for testing. This step was repeated 5 times. The average value of 5 experiments and the corresponding 95% CI were regarded as the evaluation standard. Meanwhile, the ROC curves and their AUC values reflected the generalization ability of the models. The ROC curves of the models with different features were drawn and their AUC values were calculated. Due to space limitations, we only show the results using the LightGBM algorithm in Table 2 and Figure 6 , and the rest of the results are in the Appendix. It can be seen that the effect of combining radiomics features with clinical baseline indicators was better than inputting radiomics features or clinical indicators alone, with AP combining obtaining the best effect, followed by DP combining and PVP combining. The effect of only inputting clinical indicators was the least satisfactory, which might be caused by too little information represented by the features.
Table 2.
Comparison of recurrence prediction results of ensemble learning models using different feature representations.
| Feature representation | Acc | Recall | Prec | F1 score |
|---|---|---|---|---|
| Personal and | 0.6062 | 0.6164 | 0.6019 | 0.6039 |
| clinical indicators | ± 0.0877 | ± 0.1429 | ± 0.0930 | ± 0.1027 |
| AP | 0.7224 | 0.6946 | 0.7528 | 0.7156 |
| ± 0.0834 | ± 0.0944 | ± 0.1231 | ± 0.0804 | |
| PVP | 0.6438 | 0.6782 | 0.6409 | 0.6570 |
| ± 0.1117 | ± 0.1122 | ± 0.1019 | ± 0.1014 | |
| DP | 0.6343 | 0.6909 | 0.6331 | 0.6560 |
| ± 0.0690 | ± 0.0413 | ± 0.0872 | ± 0.0520 | |
| AP+ | 0.7495 | 0.7673 | 0.7402 | 0.7502 |
| other indicators | ± 0.0629 | ± 0.1051 | ± 0.0525 | ± 0.0710 |
| PVP+ | 0.6824 | 0.6927 | 0.6844 | 0.6846 |
| other indicators | ± 0.0783 | ± 0.0941 | ± 0.0780 | ± 0.0771 |
| DP+ | 0.6819 | 0.6309 | 0.7168 | 0.6630 |
| other indicators | ± 0.0659 | ± 0.0912 | ± 0.1273 | ± 0.0756 |
Each result is represented by the mean of 5 experiments and 95% CI.
Figure 6.
The ROC curves and the corresponding AUCs of the ensemble learning model with different feature representations. (A) is the result of inputting personal information and clinical indicators; (B–D) are the results of inputting radiomic features during AP, PVP and DP respectively; (E–G) are the results of inputting radiomics features during AP, PVP and DP combined with clinical data respectively.
3.4.2 Comparison of prediction models
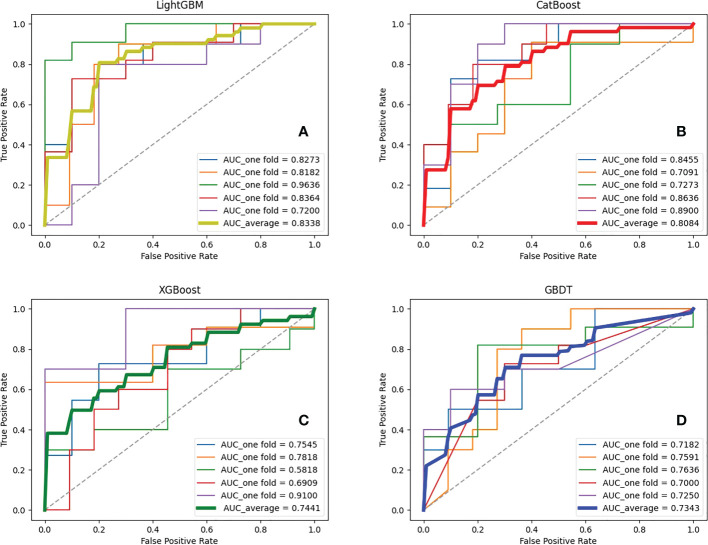
Seven feature representations were employed to compare the performance of ensemble learning models. Likewise, the study performed five-fold cross-validation on each model and calculated the associated evaluation metrics. During training, GridSearchCV method was adopted to adjust the model parameters and no overfitting occurred for each model. Due to space limitations, we only show the results inputting the most effective feature representations in this section, and the rest of the results are in the Appendix. It is found that for the four ensemble learning algorithms, different feature expressions input have similar laws, so the following only analyzes the models when radiomics features during AP and clinical indicators are input. Table 3 shows certain key parameters of each model. The test results of the Boosting ensemble models are shown in Table 4 . It can be found that the performance of LightGBM was the most excellent, with an average Acc of 0.7600, recall of 0.7673, Prec of 0.7733, and F1 score of 0.7553, which indicated that this algorithm can accurately predict recurrence outcome within 3 years after surgery. It is worth noting that XGBoost performed well in previous similar studies, but not as good as the former in this task. It had an Acc of 0.7224 and an F1 score of 0.6936, which was not as superior to LightGBM. Additionally, as the baseline model, GBDT only obtained an average Acc of 0.6543, recall of 0.6382, Prec of 0.6600 and F1 score of 0.6387. The per-fold and averaged ROC curves and corresponding AUC values are shown in Figure 7 . LightGBM had the strongest generalization, and its AUC reached 0.8338 (CI: ± 0.0680), followed by CatBoost (0.8084 ± 0.0650), XGBoost (0.7441 ± 0.0946), and GBDT (0.7343 ± 0.0214).
Table 3.
Key parameter settings for each ensemble learning model.
| Model | Parameter name | Parameter settings |
|---|---|---|
| LightGBM | n_jobs | -1 |
| n_estimators | 600 | |
| learning_rate | 0.01 | |
| max_depth | 5 | |
| num_leaves | 32 | |
| colsample_bytree | 0.51 | |
| subsample | 0.6 | |
| CatBoost | iterations | 5000 |
| learning_rate | 0.01 | |
| l2_leaf_reg | 3 | |
| bagging_temperature | 1 | |
| subsample | 0.6 | |
| random_strength | 1 | |
| depth | 6 | |
| border_count | 128 | |
| XGBoost | learning_rate | 0.001 |
| n_estimators | 1000 | |
| max_depth | 5 | |
| min_child_weight | 1 | |
| gamma | 0 | |
| subsample | 0.6 | |
| colsample_bytree | 0.8 | |
| seed | 27 | |
| GBDT | n_estimators | 1000 |
| learning_rate | 0.01 | |
| max_depth | 5 | |
| random_state | 4 |
Table 4.
5-fold cross-validation results for recurrence prediction using different ensemble learning models.
| Model | Acc | Recall | Prec | F1 score |
|---|---|---|---|---|
| LightGBM | 0.7600 | 0.7673 | 0.7733 | 0.7553 |
| ± 0.0579 | ± 0.1311 | ± 0.0771 | ± 0.0716 | |
| CatBoost | 0.6833 | 0.6164 | 0.7107 | 0.6511 |
| ± 0.0543 | ± 0.1313 | ± 0.0397 | ± 0.0879 | |
| XGBoost | 0.7224 | 0.6346 | 0.8032 | 0.6936 |
| ± 0.0834 | ± 0.1154 | ± 0.1521 | ± 0.0978 | |
| GBDT | 0.6543 | 0.6382 | 0.6600 | 0.6387 |
| ± 0.0463 | ± 0.1328 | ± 0.0286 | ± 0.0828 |
Each model was evaluated employing the mean of each fold result and corresponding 95% CI.
Figure 7.
ROC curves and corresponding AUCs of various ensemble learning models. (A–D) represent the results of ensemble learning models - Light Gradient Boosting Machine (LightGBM), Categorical Boosting (CatBoost), eXtreme Gradient Boosting (XGBoost) and Gradient Boosting Decision Tree (GBDT), respectively.
4 Discussion
In this study, the LightGBM model was constructed for the first time to accurately predict the recurrence outcome of HCC within three years after surgery. An efficient feature representation was explored, that is, the combination of radiomics features of tumor during AP, patient personal information, and clinical indicators. We trained the deep learning automatic segmentation model to make the process efficient. The results show that the proposed method was the most effective, achieving an accuracy of 0.7600 and an AUC of 0.8338.
Compared with manual segmentation, although the effect of deep learning segmentation is not as good as the former, it has higher efficiency and lower labor cost (34). In this paper, the mIoU of 3D U-Net reached 0.8874, which indicated that this algorithm can accurately segment the liver tumor region. It only took 1.22-1.85s to execute each sample on the local device, which was much faster than the manual way. It is undeniable that deep learning with excellent performance is the future trend of lesion segmentation methods (35).
This work selected 22 radiomics features during AP combined with 8 clinical baseline features from the seven feature representations and validated superiority. This feature combination eliminated dimensional redundancy, including tumor features with large contribution coefficients and clinical factors that affect prognosis. Notably, the present study found that the radiomics features during AP were superior to during PVP and DP, suggesting that AP might better capture features affecting recurrence. In addition, combined representations outperformed individual clinical or radiomics feature representations. Possibly the combination increased the amount of available information, making the model more likely to learn complex preoperative-prognostic associations (36).
Four novel Boosting ensemble models were adopted for comparison, among which LightGBM achieves the best performance (AUC=0.8338), outperforming CatBoost (AUC=0.8084), XGBoost (AUC=0.7441) and GBDT (AUC=0.7343) when inputting radiomics features during AP and clinical baseline indicators. Previous studies have confirmed the state-of-the-art of the XGBoost algorithm in the HCC prognosis prediction task (19). XGBoost belongs to the boosting family and is an engineering implementation of the GBDT algorithm. It focuses the residuals during training, uses a second-order Taylor expansion in the objective function and adds regularization. Meanwhile, the exact greedy idea is adopted in the generation process of the decision tree. When looking for the best split point, a pre-sort algorithm is adopted, that is, all features are pre-sorted according to the value of the feature, and then all the split points on all the features are traversed, and the total number of samples split according to these candidate split points is calculated. The objective function gain is to find the feature and candidate splitting point corresponding to the maximum gain, so as to split. XGBoost training is performed by addition, that is, each time a tree is trained by focusing residuals, and the final prediction result is the sum of all trees. However, XGBoost performs pre-sorting in the selection of optimal split points, and then calculates the objective function gain of all samples for all split points of all features. The space and time complexity of this process is very large, and to a certain extent affects the accuracy (31).
To address this issue, we adopted LightGBM for predicting recurrence. Based on XGBoost, LightGBM employs histogram algorithm to solve the problem of excessive number of split points. This method takes up less memory and reduces computation time. Secondly, it introduces the GOSS algorithm, which extracts according to the weight information of the samples to reduce a large number of samples with small gradients, and at the same time does not change the distribution of the dataset too much. Moreover, LightGBM also proposes the EFB mode, which reduces dimensionality by bundling features. Therefore, LightGBM can improve the model accuracy while reducing the computational effort (37), which leads to its better performance in the prognosis prediction task. In the future, it is necessary to further validate the applicability of the proposed method on larger datasets.
It should be emphasized that this study aimed to predict the postoperative recurrence risk of patients only through preoperative factors, including preoperative imaging examination and clinical indicators detection. Because only in this way can it help the doctor’s clinical decision-making. Although postoperative pathological examinations, such as microvascular invasion (MVI) are very meaningful for recurrence prediction (38), they were not considered in this study. The feasibility and effectiveness of this method have been demonstrated in reference (39, 40).
There are some other studies to predict the recurrence of HCC after surgery. Shen et al. (41) used the TCGA database and machine learning method to build a prediction model for recurrence of HCC patients, and optimized the recurrence prediction model. After the model was optimized, the prediction accuracy was 74.19%. Lee et al. (20) employed genetic algorithm to predict early recurrence of HCC, and extracted a total of 143 features, including 26 preoperative clinical features, 5 postoperative pathological features, and 112 imaging features. After training, the AUC of the preoperative and postoperative models were 0.781 and 0.767 on the training set, and 0.739 and 0.741 on the test set, respectively. Saito et al. (42) adopted support vector machine (SVM) to predict the recurrence outcome of HCC patients based on the postoperative pathological results. The patients were grouped according to the criteria of recurrence within 1 year, 1-2 years, and 4 years after resection. The final accuracy of ROI prediction in HCC and non-HCC regions was 80.6% and 68.1%, respectively. It must be emphasized that our work only collected 105 patients, but still obtained relatively remarkable performance, suggesting that the proposed method had more potential for predicting recurrence outcomes.
It is undeniable that the present study still has some shortcomings. For example, the small sample size from a single center challenges the applicability of the models. This work only focuses on the prediction of recurrence outcomes within 3 years, and further follow-up is required to predict at different times in the future. Moreover, the proposed method has not been tested in real clinical practice, which needs to be validated in the future. Zeng et al. (43) developed a machine learning method to predict the early recurrence of radical HCC hepatectomy using the data from two centers, and the effect was relatively significant. While we have mined the key features that influence the model, the interpretability issues of machine learning still need to be addressed.
5 Conclusion
This study aims to help physicians to evaluate the effectiveness of surgery and thus facilitate rational clinical decision-making. An ensemble learning strategy based on efficient feature representation was proposed for the recurrence outcome in HCC patients within three years after surgery. The 3D U-Net was used to automatically segment the lesions. Radiomics features during AP and clinical baseline features were selected as input and four ensemble models were trained. The results showed that LighGBM outperformed other ensemble algorithms, suggesting that it may be a novel model for predicting recurrence. In the future, the dataset will be expanded for early and late recurrence prediction and external clinical validation will be performed to validate the applicability of the method. When the generalization ability of the method is successfully verified, the relevant software (or web program) will be designed and applied to clinical practice.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: Since the dataset involved in this study involves patient privacy and has signed a non-disclosure agreement, it cannot be made public. Requests to access these datasets should be directed to Jiahong Dong, dongjiahong@mail.tsinghua.edu.cn.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
LW: Study concept, image preprocessing, experimental design, data analysis, writing of manuscript. MW: Experimental design, editing the manuscript, and data collection. CZ: Data analysis and data collection. RL: Data collection. SB: Experimental design. SY and JD: Study concept and funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (grant number: 82090052, 82090050, 81930119); CAMS Innovation Fund for Medical Sciences (grant number: 2019-I2M-5-056).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1019009/full#supplementary-material
References
- 1. Chen E, Xu X, Liu R, Liu T. Small but heavy role: MicroRNAs in hepatocellular carcinoma progression. BioMed Res Int (2018) 2018:6784607. doi: 10.1155/2018/6784607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am (2015) 24(1):1–17. doi: 10.1016/j.soc.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 3. Huang TE, Deng YN, Hsu JL, Leu W-J, Marchesi E, Capobianco ML, et al. Evaluation of the anticancer activity of a bile acid-dihydroartemisinin hybrid ursodeoxycholic-dihydroartemisinin in hepatocellular carcinoma cells. Front Pharmacol (2020) 11:599067. doi: 10.3389/fphar.2020.599067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng J, Dai W, Mao Y, Wu L, Li J, Chen K, et al. Simvastatin re-sensitizes hepatocellular carcinoma cells to sorafenib by inhibiting HIF-1α/PPAR-γ/PKM2-mediated glycolysis. J Exp Clin Cancer Res (2020) 39(1):24. doi: 10.1186/s13046-020-1528-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi J, Jo C, Lim YS. Tenofovir versus entecavir on recurrence of hepatitis b virus-related hepatocellular carcinoma after surgical resection. Hepatology (2021) 73(2):661–73. doi: 10.1002/hep.31289 [DOI] [PubMed] [Google Scholar]
- 6. Lee HA, Lee YS, Kim BK, Jung YK, Kim SU, Park JY, et al. Change in the recurrence pattern and predictors over time after complete cure of hepatocellular carcinoma. Gut Liver (2021) 15(3):420–9. doi: 10.5009/gnl20101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rattanasupar A, Chartleeraha S, Akarapatima K, Chang A. Factors that affect the surveillance and late-stage detection of a newly diagnosed hepatocellular carcinoma. Asian Pac J Cancer Prev (2021) 22(10):3293–8. doi: 10.31557/APJCP.2021.22.10.3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loi M, Comito T, Franzese C, Dominici L, Franceschini D, Mancosu P, et al. Stereotactic body radiotherapy in hepatocellular carcinoma: patient selection and predictors of outcome and toxicity. J Cancer Res Clin Oncol (2021) 147(3):927–36. doi: 10.1007/s00432-020-03389-2 [DOI] [PubMed] [Google Scholar]
- 9. Zhang W, Zhang B, Chen XP. Adjuvant treatment strategy after curative resection for hepatocellular carcinoma. Front Med (2021) 15(2):155–69. doi: 10.1007/s11684-021-0848-3 [DOI] [PubMed] [Google Scholar]
- 10. Gentile D, Donadon M, Lleo A, Aghemo A, Roncalli M, di Tommaso L, et al. Surgical treatment of hepatocholangiocarcinoma: A systematic review. Liver Cancer (2020) 9(1):15–27. doi: 10.1159/000503719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jia J, Zhang J, Shao Q, Wang Y, Qian B, Hu T, et al. Efficacy of surgical treatment on different sizes of hepatitis b virus-related hepatocellular carcinoma and prognostic analysis. J BUON (2020) 25(4):1866–74. [PubMed] [Google Scholar]
- 12. Yoon YI, Kim KH, Cho HD, Kwon JH, Jung DH, Park GC, et al. Long-term perioperative outcomes of pure laparoscopic liver resection versus open liver resection for hepatocellular carcinoma: a retrospective study. Surg Endosc (2020) 34(2):796–805. doi: 10.1007/s00464-019-06831-w [DOI] [PubMed] [Google Scholar]
- 13. Beumer BR, Takagi K, Vervoort B, Buettner S, Umeda Y, Yagi T, et al. Prediction of early recurrence after surgery for liver tumor (ERASL): An international validation of the ERASL risk models. Ann Surg Oncol (2021) 28(13):8211–20. doi: 10.1245/s10434-021-10235-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee IC, Lei HJ, Chau GY, Yeh YC, Wu CJ, Su CW, et al. Predictors of long-term recurrence and survival after resection of HBV-related hepatocellular carcinoma: the role of HBsAg. Am J Cancer Res (2021) 11(7):3711–25. [PMC free article] [PubMed] [Google Scholar]
- 15. He Q, Jiang JJ, Jiang YX, Wang WT, Yang L. Liver surgery group. health-related quality of life comparisons after radical therapy for early-stage hepatocellular carcinoma. Transplant Proc (2018) 50(5):1470–4. doi: 10.1016/j.transproceed.2018.04.041 [DOI] [PubMed] [Google Scholar]
- 16. Harding-Theobald E, Louissaint J, Maraj B, Cuaresma E, Townsend W, Mendiratta-Lala M, et al. Systematic review: radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment Pharmacol Ther (2021) 54(7):890–901. doi: 10.1111/apt.16563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El Jabbour T, Lagana SM, Lee H. Update on hepatocellular carcinoma: Pathologists' review. World J Gastroenterol (2019) 25(14):1653–65. doi: 10.3748/wjg.v25.i14.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ji GW, Zhu FP, Xu Q, Wang K, Wu MY, Tang WW, et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine (2019) 50:156–65. doi: 10.1016/j.ebiom.2019.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang Y, Chen H, Zeng Y, Liu Z, Ma H, Liu J. Development and validation of a machine learning prognostic model for hepatocellular carcinoma recurrence after surgical resection. Front Oncol (2021) 10:593741. doi: 10.3389/fonc.2020.593741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee IC, Huang JY, Chen TC, Yen CH, Chiu NC, Hwang HE, et al. Evolutionary learning-derived clinical-radiomic models for predicting early recurrence of hepatocellular carcinoma after resection. Liver Cancer (2021) 10(6):572–82. doi: 10.1159/000518728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chaitanya K, Karani N, Baumgartner CF, Erdil E, Becker A, Donati O, et al. Semi-supervised task-driven data augmentation for medical image segmentation. Med Image Anal (2021) 68:101934. doi: 10.1016/j.media.2020.101934 [DOI] [PubMed] [Google Scholar]
- 22. Kim J, Kim Y, Lee EK, Chae CK, Lee K, Kim WJ, et al. Rotational variance-based data augmentation in 3D graph convolutional network. Chem Asian J (2021) 16(18):2610–3. doi: 10.1002/asia.202100789 [DOI] [PubMed] [Google Scholar]
- 23. Hsu LM, Wang S, Walton L, Wang TW, Lee SH, Shih YI. 3D U-net improves automatic brain extraction for isotropic rat brain magnetic resonance imaging data. Front Neurosci (2021) 15:801008. doi: 10.3389/fnins.2021.801008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El Khoury K, Fockedey M, Brion E, Macq B. Improved 3D U-net robustness against JPEG 2000 compression for male pelvic organ segmentation in radiotherapy. J Med Imaging (Bellingham) (2021) 8(4):041207. doi: 10.1117/1.JMI.8.4.041207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yan J, Xu Y, Cheng Q, Jiang S, Wang Q, Xiao Y, et al. LightGBM: accelerated genomically designed crop breeding through ensemble learning. Genome Biol (2021) 22(1):271. doi: 10.1186/s13059-021-02492-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang C, Lei X, Liu L. Predicting metabolite-disease associations based on LightGBM model. Front Genet (2021) 12:660275. doi: 10.3389/fgene.2021.660275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rufo DD, Debelee TG, Ibenthal A, Negera WG. Diagnosis of diabetes mellitus using gradient boosting machine (LightGBM). Diagnostics (Basel) (2021) 11(9):1714. doi: 10.3390/diagnostics11091714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng C, Tian J, Wang K, Han L, Yang H, Ren J, et al. Time-to-event prediction analysis of patients with chronic heart failure comorbid with atrial fibrillation: a LightGBM model. BMC Cardiovasc Disord (2021) 21(1):379. doi: 10.1186/s12872-021-02188-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ambe K, Suzuki M, Ashikaga T, Tohkin M. Development of quantitative model of a local lymph node assay for evaluating skin sensitization potency applying machine learning CatBoost. Regul Toxicol Pharmacol (2021) 125:105019. doi: 10.1016/j.yrtph.2021.105019 [DOI] [PubMed] [Google Scholar]
- 30. Zhao QY, Wang H, Luo JC, Lou MH, Liu LP, Yu SJ, et al. Development and validation of a machine-learning model for prediction of extubation failure in intensive care units. Front Med (Lausanne) (2021) 8:676343. doi: 10.3389/fmed.2021.676343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hou N, Li M, He L, Xie B, Wang L, Zhang R, et al. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J Transl Med (2020) 18(1):462. doi: 10.1186/s12967-020-02620-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davagdorj K, Pham VH, Theera-Umpon N, Ryu KH. XGBoost-based framework for smoking-induced noncommunicable disease prediction. Int J Environ Res Public Health (2020) 17(18):6513. doi: 10.3390/ijerph17186513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y, Kuang S, Shan Q, Rong D, Zhang Z, Yang H, et al. Can IVIM help predict HCC recurrence after hepatectomy? Eur Radiol (2019) 29(11):5791–803. doi: 10.1007/s00330-019-06180-1 [DOI] [PubMed] [Google Scholar]
- 34. Tang P, Liang Q, Yan X, Xiang S, Sun W, Zhang D, et al. Efficient skin lesion segmentation using separable-unet with stochastic weight averaging. Comput Methods Programs Biomed (2019) 178:289–301. doi: 10.1016/j.cmpb.2019.07.005 [DOI] [PubMed] [Google Scholar]
- 35. Feng B, Ma XH, Wang S, Cai W, Liu XB, Zhao XM. Application of artificial intelligence in preoperative imaging of hepatocellular carcinoma: Current status and future perspectives. World J Gastroenterol (2021) 27(32):5341–50. doi: 10.3748/wjg.v27.i32.5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lewis S, Hectors S, Taouli B. Radiomics of hepatocellular carcinoma. Abdom Radiol (NY) (2021) 46(1):111–23. doi: 10.1007/s00261-019-02378-5 [DOI] [PubMed] [Google Scholar]
- 37. Zhu J, Su Y, Liu Z, Liu B, Sun Y, Gao W, et al. Real-time biomechanical modelling of the liver using LightGBM model. Int J Med Robot (2022) 18:e2433. doi: 10.1002/rcs.2433 [DOI] [PubMed] [Google Scholar]
- 38. Yanhan W, Lianfang L, Hao L, Yunfeng D, Nannan S, Fanfan L, et al. Effect of microvascular invasion on the prognosis in hepatocellular carcinoma and analysis of related risk factors: A two-center study. Front Surg (2021) 8:733343. doi: 10.3389/fsurg.2021.733343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei H, Jiang H, Qin Y, Wu Y, Lee JM, Yuan F, et al. Comparison of a preoperative MR-based recurrence risk score versus the postoperative score and four clinical staging systems in hepatocellular carcinoma: a retrospective cohort study. Eur Radiol (2022). doi: 10.1007/s00330-022-08811-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding DY, Liu L, Li HL, Gan XJ, Ding WB, Gu FM, et al. Development of preoperative prognostic models including radiological features for survival of singular nodular HCC patients. Hepatobiliary Pancreat Dis Int (2022) S1499-3872(22):00052–2. doi: 10.1016/j.hbpd.2022.04.002 [DOI] [PubMed] [Google Scholar]
- 41. Shen J, Qi L, Zou Z, Du J, Kong W, Zhao L, et al. Identification of a novel gene signature for the prediction of recurrence in HCC patients by machine learning of genome-wide databases. Sci Rep (2020) 10(1):4435. doi: 10.1038/s41598-020-61298-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saito A, Toyoda H, Kobayashi M, Yoshinori K, Fujii H, Fojita K, et al. Prediction of early recurrence of hepatocellular carcinoma after resection using digital pathology images assessed by machine learning. Mod Pathol (2021) 34(2):417–25. doi: 10.1038/s41379-020-00671-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeng J, Zeng J, Lin K, Lin H, Wu Q, Guo P, et al. Development of a machine learning model to predict early recurrence for hepatocellular carcinoma after curative resection. Hepatobiliary Surg Nutr (2022) 11(2):176–87. doi: 10.21037/hbsn-20-466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: Since the dataset involved in this study involves patient privacy and has signed a non-disclosure agreement, it cannot be made public. Requests to access these datasets should be directed to Jiahong Dong, dongjiahong@mail.tsinghua.edu.cn.