Abstract
Gibberellin and cytokinin synergistically regulate the stalk development in flowering Chinese cabbage. KNOX proteins were reported to function as important regulators of the shoot apex to promote meristem activity by synchronously inducing CTK and suppressing GA biosynthesis, while their regulatory mechanism in the bolting and flowering is unknown. In this study, 9 BcKNOX genes were identified and mapped unevenly on 6 out of 10 flowering Chinese cabbage chromosomes. The BcKNOXs were divided into three subfamilies on the basis of sequences and gene structure. The proteins contain four conserved domains except for BcKNATM. Three BcKNOX TFs (BcKNOX1, BcKNOX3, and BcKNOX5) displayed high transcription levels on tested tissues at various stages. The major part of BcKNOX genes showed preferential expression patterns in response to low-temperature, zeatin (ZT), and GA3 treatment, indicating that they were involved in bud differentiation and bolting. BcKNOX1 and BcKNOX5 showed high correlation level with gibberellins synthetase, and CTK metabolic genes. BcKONX1 also showed high correlation coefficients within BcRGA1 and BcRGL1 which are negative regulators of GA signaling. In addition, BcKNOX1 interacted with BcRGA1 and BcRGL1, as confirmed by yeast two-hybrid (Y2H) and biomolecular fluorescence complementation assay (BiFC). This analysis has provided useful foundation for the future functional roles’ analysis of flowering Chinese cabbage KNOX genes
Keywords: flowering Chinese cabbage, knotted1-like Homebox, expression analysis, hormone response, cold response
Introduction
The KNOX genes of Arabidopsis thaliana are clustered into three classes, class I, class II, and class M (Magnani and Hake, 2008; Mukherjee et al., 2009). Class I consists of four genes: SHOO-TMERISTMELESS (STM), KNAT1 (BP), KNAT2, and KNAT6 and specifically expresses in the shoot apical meristem (SAM) (Hake et al., 2004). Four class II KNOX (KNAT3, KNAT4, KNAT5, KNAT7) genes express more broadly compared with that of class I (Hake et al., 2004). Class I and class II KNOX proteins contain four conserved domains (Reiser et al., 2000; Hamant and Pautot, 2010). The KNOX1 and KNOX2 domains also known as the MEINOX domain are located at the N-terminal portion (Bellaoui et al., 2001; Müller et al., 2001; Smith et al., 2002). The ELK domain functioned as nuclear localization signal is located on the upstream of the homologous domain (Hofer et al., 2001). Moreover, the C-terminal region possess a DNA binding HD domain (Scofield and Murray, 2006). However, class M represents by KNATM which is characterized with missing the ELK and HD domains (Magnani and Hake, 2008).Numerous studies have indicated that class I KNOXs play vital roles in meristem maintenance, hormone homeostasis, internode elongation, and inflorescence architecture (Semiarti et al., 2001; Tsuda et al., 2011; Tsuda and Hake, 2015). In maize, loss-of-function mutations of homeobox gene, knotted1, shows shorter meristems (Kerstetter et al., 1997; Vollbrecht et al., 2000). Similar phenotype is observed in rice loss-of-function mutants of OSH1 (Tsuda et al., 2011). Moreover, another rice homeobox gene OSH15 plays a dispensable role in the architecture of internodes (Sato et al., 1999). Recent studies have showed that KNOX played significant role in abscission. In litchi, KNAT1 represses fruitlet abscission (Zhao et al., 2020). Meanwhile, the silencing of KNOTTED1-LIKE HOMEOBOX PROTEIN1 (KD1) in tomato also shows a repression of pedicel and petiole abscission (Ma et al., 2015). In model plant Arabidopsis, KNOX2 and KNOX6 play a role in carpel development and lateral root formation, respectively (Pautot et al., 2001; Dean et al., 2004). Whereas the functions of the Arabidopsis class II KNOXs are largely unknown which is in contrary to the well-defined class I KNOX genes (Hay and Tsiantis, 2010). In legume, KNOX4 would directly bind to the promoter of a cuticle biosynthetic gene and regulate seed physical dormancy (Chai et al., 2016). Besides that, class II KNOX genes involve in legume root nodule development (Di Giacomo et al., 2017). In addition, KNOX7 is a vital regulator of secondary wall biosynthesis in Arabidopsis, rice, and populus (Zhong et al., 2008; Li et al., 2012; Yu, 2019).
Additionally, numerous studies have shown KNOX genes affect the metabolism and signaling transduction pathway of hormones, such as ethylene, gibberellin, cytokinin and abscisic acid (Kim et al., 2013). KNAT1 would suppress ethylene biosynthesis by directly binding to the promoters of ethylene biosynthetic genes (Zhao et al., 2020). KD1 of tomato is involved in regulating the genes related to auxin transport and signaling (Ma et al., 2015). The isopentenyl transferase (IPT) and cytokinin dehydrogenase (CKX) are key genes in biosynthesis and metabolic of cytokinins (Frébort et al., 2011). Moreover, KNOX proteins activate both CTK biosynthesis and signaling transduction (Kusaba et al., 1998; Ori et al., 1999; Frugis et al., 2001; Jasinski et al., 2005). Class I KNOX proteins were known to promote expression of IPT7 (Jasinski et al., 2005), while no studies have shown that KONX proteins would regulate the expression of CKX. KNOX proteins are also reported to suppress the accumulation of GA (Sakamoto et al., 2001; Chen et al., 2004). Previous studies have shown that overexpression of POTH1 (potato homeobox 1) results in lower levels of GA20ox1 (a key rate-limiting enzyme in the synthesis of GA) transcription (Rosin et al., 2003) Moreover, in Arabidopsis, GA3ox1 (GA-biosynthesis gene) transcript shows no difference in KNOX1-overexpression plants (Hay et al., 2002), indicating that GA20ox1 is a specific target in the GA-biosynthesis pathway for KNOX proteins. Several recent studies have revealed that gibberellin and cytokinin play vital roles throughout bolting and flowering (Guan et al., 2021; Ou et al., 2022). Meanwhile, our preliminary results indicated that GA and cytokinin exert antagonistic effect on bolting in flowering Chinese cabbage (Ou et al., 2022). We may speculate that BcKNOX family have significant functions in bolting and flowering of flowering Chinese cabbage yet to be discovered.
Despite extensive studies within some model plants, little is known about the features of KNOX genes in flowering Chinese cabbage. In this study, nine BcKNOXs were identified and characterized in Brassica campestris genome. The subcellular locations of the majority of BcKNOXs were further analyzed. The expression patterns in various tissues at different stages and in response to cold stress and flowering-related hormones were also explored. The results provide insight into KNOX gene family of flowering Chinese cabbage and a foundation for further research on their function.
Materials and methods
Identification of KNOX genes and phylogenetic analysis
To identify genes encoding KNOX protein in Brassica campestris genome, the amino acid sequences of Arabidopsis thaliana KNOX genes were used as queries by using BLASP program. Meanwhile, we used the NCBI Conserved Domains database (Marchler-Bauer et al., 2016) and Pfam database (Mistry et al., 2020) to examine genes that have the conserved domains. Protein basic physicochemical characteristics and subcellular location were predicted with ExPASy program (Gasteiger et al., 2003) and WoLF PSORT (Horton et al., 2007) respectively. Phylogenetic tree including KNOXs from Arabidopsis thaliana, Brassica rapa and Brassica campestris was constructed utilizing MEGA11 (Tamura et al., 2021)
Chromosomal location and multiple sequence alignment of KNOX genes
TBtools software was used to map the Chromosomal position of BcKNOX genes. Multiple alignment was conducted by ClustalW based on the full-length amino acid sequences of KNOX proteins from Arabidopsis and Brassica campestris.
Synteny analysis
Synteny analysis was carried out using the Toolkits of TBtools software (Chen et al., 2020). MCScanX was applied to identify syntenic blocks and distinct duplication events within Brassica campestris and between Arabidopsis and Brassica campestris.
Gene structure and conserved motif analysis
Conserved motifs in BcKNOX protein sequence were identified with the MEME tools with the parameters as described elsewhere (Li et al., 2022). The TBtools software was used to visualize structural information (Chen et al., 2020).
Promoter sequence analysis
To analyze the cis-elements in the putative promoter regions, the 2,000-bp upstream sequences of the translation start site of BcKNOX genes were selected using the PlantCARE database (Lescot et al., 2002).
Plant materials and treatments
Flowering Chinese cabbage cultivar ‘Youlv501’ was used to collected samples, which is planted in the greenhouse at South China Agriculture University. To analysis the expression pattern, roots, stem tip, leaves and flowers were collected. Seedlings with three true leaves were transplanted and sprayed with ZT (40mg/L), GA3 (200mg/L) or water (control), followed by sampling at 12h and flowering stage after treatment. All samples were immediately frozen in liquid nitrogen and stored at -80°C.
RNA extraction, gene cloning and quantitative real-time PCR
The HiPure RNA Mini Kit (Magen, Guangzhou, China) was used to isolate total RNA following the manufacturer’s instructions. Primer pairs for quantitative real-time PCR (qRT-PCR) were designed using Primer3 (Untergasser et al., 2012) ( Supplementary Table S1 ). First-strand cDNA was synthesized using the Hiscript QRT SuperMix (with gDNA Wiper; Vazyme, Nanjing, China). qRT-PCR was conducted on a LightCycler 480 real-time PCR system (Roche, Basel, Switzerland) using the SYBR ® Green Premix Pro Taq HS qRT-PCR Kit (Accurate Biotechnology, AG11718, Hunan, China). The PCR protocol was described by (Ou et al., 2022). The comparative 2-ΔΔCT method was used to calculate relative expression values for each gene (Livak and Schmittgen, 2001).
Subcellular localization analysis
The coding region lacking the stop codon of BcKNOX1, BcKNOX3, BcKNOX4, BcKNOX5, BcKNOX7, and BcSTM were fused to the N-terminus of the hGFP gene. The primers used for cloning are listed in Supplementary Table S2 . The recombinant plasmids and localization signal (DsRed) were co-infiltrated into Agrobacterium tumefaciens strain GV3101 and then infiltrated Nicotiana benthamiana leaves (Sparkes et al., 2006). After two days, a laser scanning confocal microscopy (Axioimager.D2) was used to detect green fluorescent protein and DsRed protein.
Correlation analysis of gene expression
The correlation analysis among transcription factors, GA metabolic genes, CTK metabolic genes, and some genes encoding cell wall structural proteins was conducted via Pearson’s correlation coefficients. To visualize the relationship, the R soft was used.
Yeast two-hybrid assay
The coding sequences (CDS) of BcRGA1 and BcGRL1 were cloned in pGADT7 vector. Similarly, the BcKNOX1 coding region was cloned into pGBKT7 vector. The primers used for cloning are listed in Supplementary Table S3 . The vectors were co-transformed into yeast strain Y2HGold to generate fusion proteins.
Bimolecular fluorescence complementation assay
The coding region of BcRGA1, BcRGL1, and BcKNOX1 lacking stop codon were amplified by PCR. The primers used for cloning are listed in Supplementary Table S4 . The Agrobacterium stain harboring the recombinant plasmid and DsRed were co-infiltrated into young Nicotiana benthamiana leaves (Ding et al., 2015). Three days after infiltration, a laser scanning confocal microscopy (Axioimager.D2) was used to detect green fluorescent protein and DsRed protein.
Statistical analysis
Data was subjected to analysis of variance and the means were compared using Duncan’s test considering P<0.05 as the significance threshold using SPSS software (SPSS, Chicago, IL, USA). All results represented as the mean ± standard error of the mean of at least three replications.
Results
Genome-wide identification of flowering Chinese cabbage KNOX genes
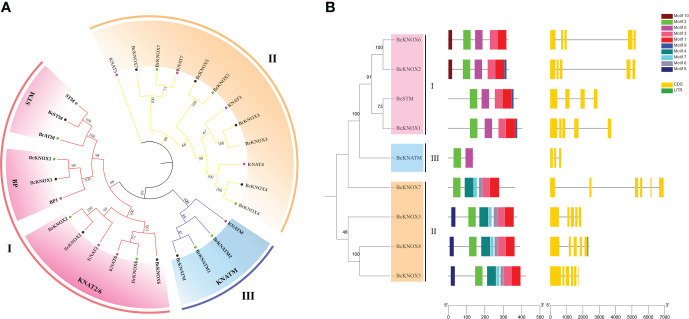
In Arabidopsis thaliana genome, nine KNOX gene were identified. With the nine AtKNOX proteins as queries, putative flowering Chinese cabbage KNOX genes were identified using BLASTP program based on conserved domain and named according their orthology with the reported isoform in Arabidopsis thaliana. The results of sequence analysis showed that the polypeptide lengths of the putative BcKNOXs ranged from 139 to 423 amino acids, isoeletric point (pI) values ranged from 4.68 to 6.28 and molecular weights ranged from 15.84 kDa to 46.62 kDa ( Table 1 ). WoLF PSORT was used to analyze the subcellular location, which exhibited that all BcKNOXs were nuclear proteins ( Table 1 ). Additionally, we selected BcKNOX1, BcKNOX3, BcKNOX4, BcKNOX5, BcKNOX7, and BcSTM to confirm the subcellar localization. As showed in Figure S1 , the six proteins were nuclear proteins. The full-length sequences of the proteins were used to investigate the phylogenetic relationship of KNOX among A. thaliana, B. rapa, and B. campestris by constructing a rootless neighbor-joining phylogenetic tree. The tree contained 9 Arabidopsis, 9 B. rapa, and 9 B. campestris KNOX proteins ( Figure 1A ). The family was grouped into three subfamilies ( Figure 1A ). BcKNOX1, BcKNOX2, BcKNOX6, and BcSTM were ascribed to subfamily I. BcKNOX3, BcKNOX4, BcKNOX5, and BcKNOX7 were clustered into subfamily II. In addition, BcKNATM belonged to subfamily III ( Figure 1A ).
Table 1.
Physicochemical properties of KNOX genes in flowering Chinese cabbage.
| Gene | CDS (bp) | AA | pI | MW (KD) | Subcellular localization |
|---|---|---|---|---|---|
| BcKNOX1 | 1209 | 403 | 6.18 | 46.41 | nucl: 14 |
| BcKNOX2 | 984 | 328 | 4.91 | 36.93 | nucl: 13, pero: 1 |
| BcKNOX3 | 1269 | 423 | 5.81 | 46.62 | nucl: 14 |
|
BcKNOX4
BcKNOX5 BcKNOX6 BcKNOX7 BcSTM BcKNATM |
1170 1125 981 1086 1152 417 |
390 375 327 362 384 139 |
6.28 5.77 5.15 5.58 6.12 4.68 |
43.61 41.91 36.58 40.09 43.10 15.84 |
nucl: 14 nucl: 14 nucl: 14 nucl: 12, plas: 1, cysk: 1 nucl: 14 nucl: 6, cyto: 4, chlo: 2, extr: 2 |
Figure 1.
Phylogenetic analysis, Schematic representation of protein and gene structure. (A) Phylogenetic analysis of the KNOX families in A thaliana, B rapa, and B campestris. The tree was clustered into three subfamilies. (B) Schematic representation of protein and gene structure of Brassica campestris KNOX gene.
Conserved motifs gene structure of BcKNOX genes
The BcKNOX gene family differed in the number of introns and exons ( Figure 1B ). We used the NCBI-CDD database and Pfam database to analyze the conserved domain of BcKNOXs respectively. Multiple sequence alignment exhibited that the four conserved domains: KNOXI, KNOXII, ELK, and HD domain were present in all the BcKNOX proteins except for BcKNATM ( Figure S2 ). BcKNOXs contained the KNOXI and KNOXII domains in their N-terminal region, while the C terminus possed ELK and HD domains. In addition, BcKNATM protein lacked ELK and HD domains. MEME program was used to investigated the conserved motifs. A total of 10 conserved motifs were predicted in BcKNOXs ( Figure 1B ). All BcKNOXs contained motifs 2; motif 1 and 3 were found in majority of BcKNOXs except for BcKNATM which belonged to subfamily III; motif 5 existed in members of subfamily I and III; motif 4, 6, and 7 were present in subfamily II. Furthermore, motif 8 only existed in BcKNOX3, BcKNOX4, and BcKNOX5; motif 9 was only present in BcKNOX1, BcKNOX2, and BcSTM; motif 10 only were found in BcKNOX2 and BcKNOX6.
Chromosomal distribution and collinearity analysis of BcKNOX genes
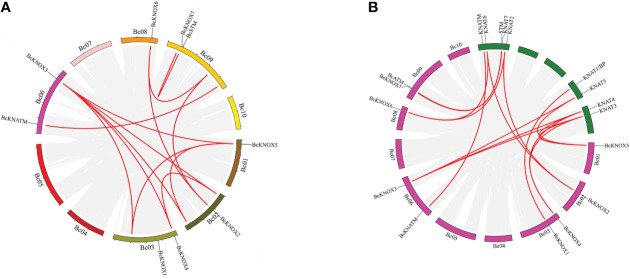
The chromosomal locations of the BcKNOX genes in Brassica campestris was mapped, and the physical mapping of BcKNOXs on 10 chromosomes revealed that an uneven distribution ( Figure S3 ). The BcKNOXs were distributed on just 6 chromosomes. Chromosomes 3, 6, and 9 contained 2 genes. Only one gene was mapped onto chromosomes 1, 2, and 8, while no KNOX genes were located in the other chromosomes. To detect the evolution of the BcKNOX gene family in B. campestris, segmental and tandem duplication were analyzed. As shown in Figure 2A , more than ten pairs of BcKNOX genes were segmentally duplicated, for example, BcKNOX3/BcKNOX4, BcKNOX3/BcKNOX5, BcKNOX2/BcKNOX6. Chromosomes 4, 5, 7, and 10 did not exist any duplicated genes. Moreover, the results of synteny map showed that fifteen pairs of syntenic orthologous genes were matched between A. thaliana and B. campestris ( Figure 2B ).
Figure 2.
Collinearity analysis of among KNOX gene family members. (A) Distribution of segmentally duplicated BcKNOX genes on B campestris chromosomes. Gray lines indicate collinear blocks in the whole B. campestris genome and red lines indicate duplicated BcKNOX gene pairs. (B) Collinearity of Arabidopsis thaliana and Brassica campestris. Red lines indicate the syntenic gene pairs.
Analysis of cis-acting elements in the promoter regions of BcKNOX gene
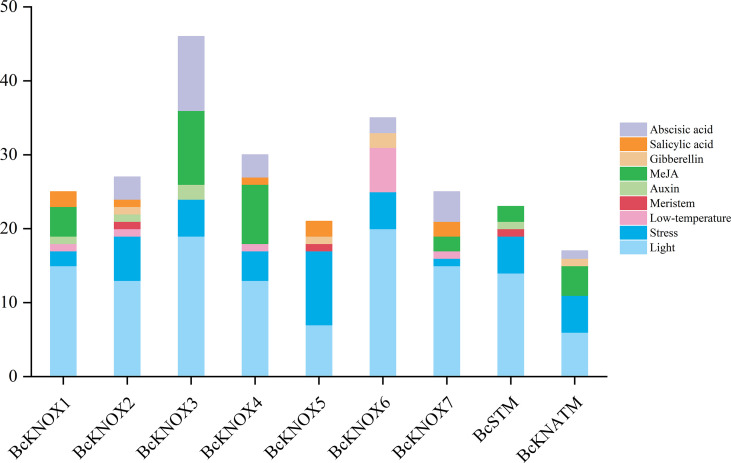
The cis-regulatory elements in promoter regions are closely associated with responses to environmental factors and phytohormones. To explore the regulatory mechanisms and potential roles of BcKNOXs, the 2.0 Kb upstream sequences from the transcription start site were downloaded and detected using the Plant CARE database (Lescot et al., 2002). As show in the Figure 3 , stress-related and light elements were found in the promoters of all BcKNOX genes. Low-temperature-related elements located in the promoters of BcKNOX1, BcKNOX2, BcKNOX4, BcKNOX6, and BcKNOX7. BcKNOX2, BcKNOX5, and BcSTM contained meristem-related cis-elements. Furthermore, several cis-elements were active in response to hormone, including ABA, SA, GA, methyl jasmonate, and auxin. Among these hormone-related cis-elements, the GA-responsive elements were existed in BcKNOX2, BcKNOX5, BcKNOX6, and BcKNATM; the auxin-responsive elements were present in BcKNOX1, BcKNOX2, BcKNOX3, and BcATM.
Figure 3.
Analysis of cis-elements in the BcKNOX promoters.
Expression patterns of the BcKNOX genes during flowering Chinese cabbage development
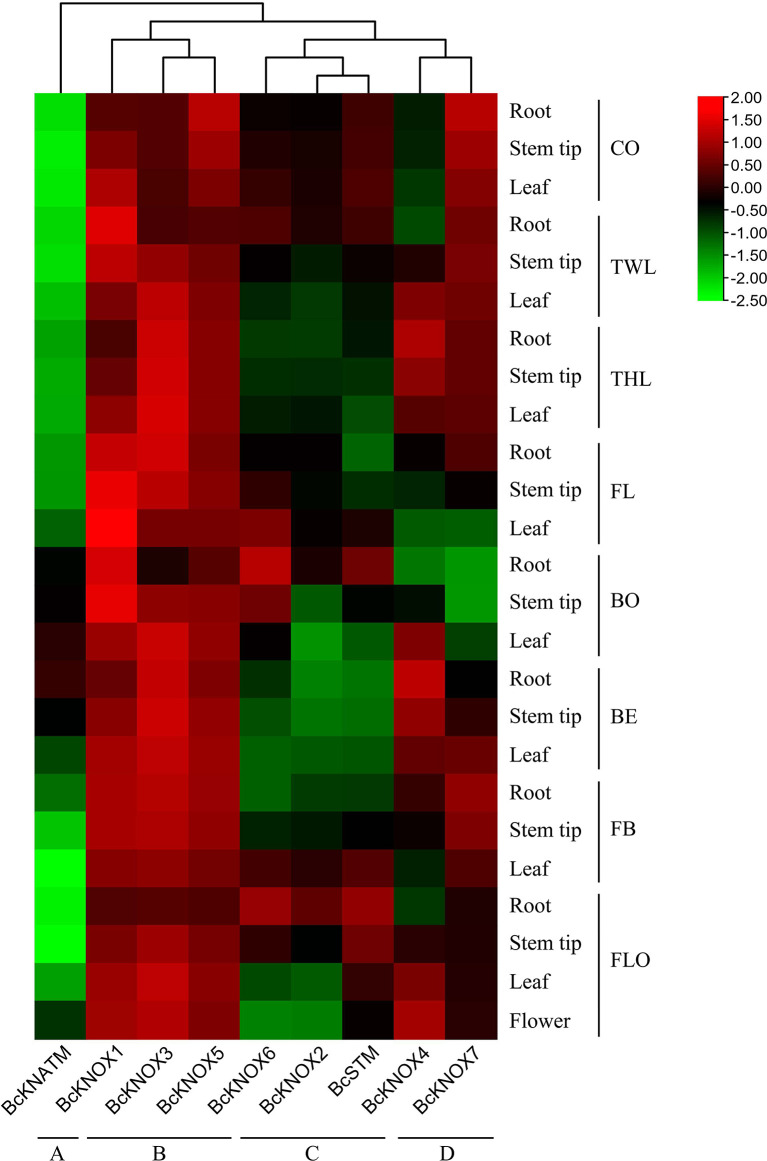
To preliminarily analyze the functional roles of BcKNOXs in stalk development of flowering Chinese cabbage, the expression profiles of BcKNOX genes were investigated in different tissue at several stages. A heatmap was generated to visualize the expression patterns of individual BcKNOX genes according to the qRT-PCR data. The heatmap displayed that these BcKNOX genes could be divided into four groups according to their expression profiles ( Figure 4 ). Group A contained BcKNATM, which showed low transcription level among the tested tissues at seven stages. BcKNOX1, BcKNOX3, and BcKNOX5 belonged to Group B and they exhibited high expression levels in four tested tissues during the whole development stage. Group C had BcKNOX2, BcKNOX6, and BcSTM, which belonged to subfamily I. Their expression profiles showed that they had high transcript levels in roots at bolting and flowering stage, especially BcKNOX6 and BcSTM, suggesting that these genes had strong preferential expression. Group D included BcKNOX4 and BcKNOX7. Among the two genes, BcKNOX4 showed high expression level in root, leaf, and stem tip during three-leaf, bud emergence, and flowering stage. In addition, during cotyledon, two-leaf, three-leaf, and fast bolting stages, BcKNOX7 exhibited high expression level, while showed low transcript level at bolting stage.
Figure 4.
Heatmap of BcKNOX genes during flowering Chinese cabbage development. The BcKNOXs were divided into four groups. CO, cotyledon stage; TWL, two-leaf stage; THL, three-leaf stage; FL, four-leaf stage; BO, bolting stage; BE, bud emergence stage; FB, fast bolting stage; FLO, flowering stage.
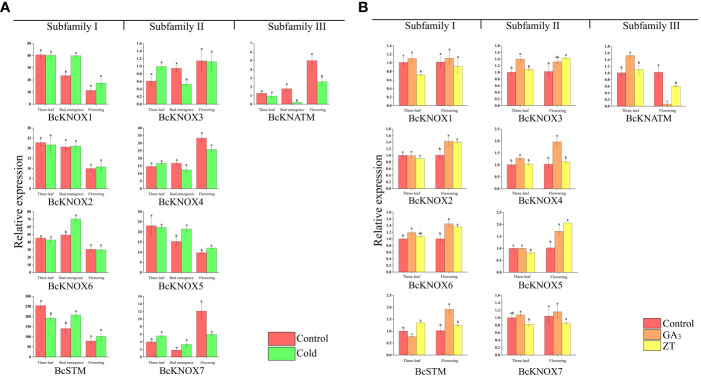
Expression of BcKNOX genes in response to cold stress
Low temperature is an important environmental factor for stalk elongation and flowering in many winter annuals and biennials (Hébrard et al., 2013; Mathieu et al., 2014; Liang et al., 2018). Meanwhile, our previous studies have shown that cold treatment promoted bolting and flowering of flowering Chinese cabbage by inducing the GA levels in the shoot apical meristems (Song et al., 2019; Wang et al., 2022). To determine whether cold stress have effect on BcKNOX gene transcription, we investigated and compared the expression profiles of BcKNOX genes in response to cold tress among three-leaf, bud emergence, and flowering stages (Huang et al., 2017). As shown in Figure 5A , among the three stages, these BcKNOX genes exhibited different patterns upon cold treatment. It was worth noting that cold treatment displayed no significant effect on the transcript levels of BcKNOX2 and BcKNOX4 compared with control at three stages. The transcription level of BcSTM was considerably repressed at three-leaf stage. Moreover, BcKNOX1 BcKNOX5, BcKNOX6, and BcSTM were significantly upregulated at bud emergence stage. The transcription of BcKNOX5 was also induced by cold treatment at flowering stage. Meanwhile, BcKNOX3 and BcKNATM were significantly repressed at bud emergence stage. At three-leaf stage, cold treatment induced the expression of BcKNOX7, while BcKNATM was suppressed at flowering stage.
Figure 5.
The expression patterns of BcKNOX genes were analyzed under cold, ZT and GA3 treatment. (A) Transcript levels of BcKNOXs in response to cold stress at three-leaf stage, bud emergence and flowering stage. (B) BcKNOX transcript levels of stem tip in response to ZT and GA3. Samples were collected at three-leaf and flowering stage after exogenous hormone treatment, with water used as a control. The lowercase letters indicate significance at p < 0.05.
Effect of phytohormone treatment on BcKNOX’s expression During the bud differentiation and flowering period
To investigate the potential effect of plant hormones on the transcript abundance of BcKNOXs at the bud differentiation and flowering stage, we estimated the transcript level after treatment with ZT and GA3 using RT-qPCR. As shown in Figure 5B , BcKNOXs showed various expression patterns over time in response to ZT and GA3. At three-leaf stage, ZT treatment significantly decreased the transcript level of BcKNOX1. Furthermore, the transcript abundance of BcKNOX2 and BcKNOX5 were downregulated in response to ZT at three-leaf stage, whereas treatment with GA3 and ZT promoted the expression level of BcKNOX2 and BcKNOX5 at flowering stage. No matter at three-leaf or flowering stage, the transcription of BcKNOX4 was strongly induced by GA3 treatment. GA3 considerably induced the expression of BcKNOX6 at three-leaf and flowering stage. Furthermore, BcKNOX6 was induced in response to ZT treatment at flowering stage. BcKNOX3 was remarkably upregulated by GA3 at three-leaf stage and was also promoted in response to ZT at flowering stage. The transcript abundance of BcSTM remained at lower levels compared with control in response to GA3 treatment at three-leaf stage, while ZT significantly increased the expression level of BcSTM. At flowering stage, GA3 treatment also induced the expression of BcSTM. BcKNOX7 expression showed no significant difference under ZT and GA3 treatment. With regard to BcKNATM, GA3 considerably increased its expression level at three-leaf stage, but BcKNATM was suppressed by GA3 and ZT treatment at flowering stage ( Figure 5B ). In leaf, all of BcKNOX genes exhibited lower transcript levels in response to ZT treatment compared with that of control at three-leaf stage ( Figure S4 ). The transcription of BcKNOX2, BcKNOX5, BcKNOX6, BcKNOX7, and BcSTM were also repressed by GA3 treatment at the same stage. In addition, at flowering stage, ZT decreased the expression levels of BcKNOX1, BcKNOX2, BcKNOX3, BcKNOX4, BcKNOX5, and BcKNOX7, whereas BcSTM and BcKNATM transcription were promoted ( Figure S4 ).
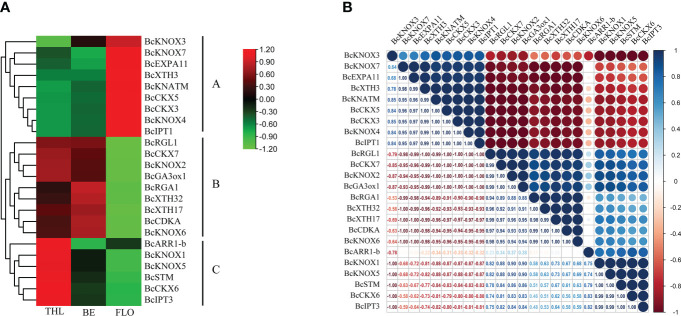
Correlation analysis of BcKNOX genes
The stalk of flowering Chinese cabbage is the main product organ, whose development consists of bolting and flowering. Both of them are closely associated with the product quality and yield. Therefore, for understanding the possible correlation of the BcKNOXs and bolting-related genes, we investigated the transcription level of the nine BcKNOXs and the bolting-related genes. In addition, a comprehensive correlation analysis of these genes was performed. As shown in Figure 6A , nine BcKNOXs showed preferential expression at three-leaf, bud emergence, and flowering stages, which was consistent with the expression pattern of the bolting-related genes, exhibiting that these BcKNOXs had relevance to bolting in flowering Chinese cabbage. According to the correlative expression patterns, there were three different clusters (clusters A-C). The genes of gibberellins synthetase (BcGA3ox1), negative regulator of GA signaling (BcRGA1 and BcRGL1), cytokinin oxidase (BcCKX7), cyclin-related gene (BcCDKA), and xyloglucan endotransglycosidase/hydrolase (XTH17 and XTH32) clustered in cluster B and were highly expressed at three-leaf and bud emergence stages compatible with the expression pattern of BcKNOX2 and BcKNOX6 ( Figure 6A ). Moreover, BcKNOX2 and BcKNOX6 were strongly associated with these genes. Remarkably, BcKNOX1, BcKNOX5, and BcSTM, which clustered with cytokinin biosynthesis and metabolic genes (BcIPT1 and BcCKX6) and cytokinin response regulator gene (BcARR1-b) (cluster C) were highly expressed at three-leaf stage. As shown in Figure 6B , BcKNOX1 and BcKNOX5 also showed high correlation level with gibberellins synthetase (BcGA3ox1). Besides, BcKNOX1, BcKNOX2, and BcKNOX5 showed high correlation coefficients within the CTK metabolic genes (BcCKX6 and BcIPT3). Furthermore, BcKNOX3, BcKNOX4, BcKNOX7, and BcKNATM were divided in cluster A, and highly expressed at flowering stage ( Figure 6A ). In addition, these BcKNOXs had high correlation with structural genes (EXPA11 and XTH3) and CTK metabolic genes (BcIPT1, BcCKX3, and BcCKX5).
Figure 6.
Expression patterns analysis and correlation of KNOX genes in flowering Chinese cabbage. (A) Heatmap of RNA-seq transcript abundance patterns of BcKNOXs and bolting related genes at three developmental stages. THL, three leaf stage; BE, bud emergence; FLO, flowering stage. (B) Correlative analysis of BcKNOX genes and bolting-related genes in three representative stages. R > 0.5 indicates a positive correlation; R < -0.5 indicates a negative correlation.
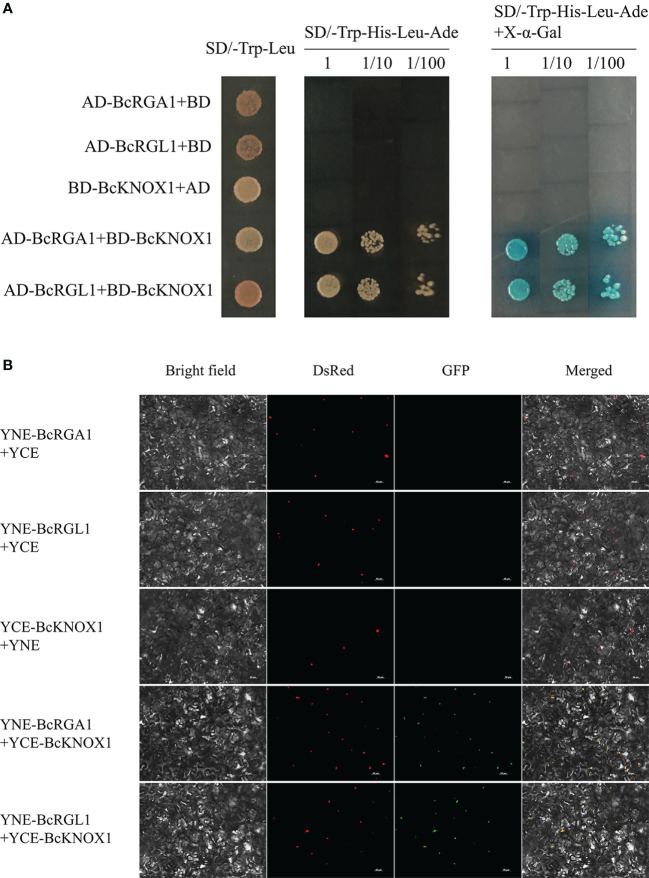
BcKNOX1 interacts with BcRGA1 and BcRGL1
Gibberellins are important orchestrator of plant growth and development. DELLAs are negative regulators of GA signaling, which function as coactivators or repressors by interacting with other transcription factors (Levesque et al., 2006; Cui et al., 2007; Fukazawa et al., 2014). Previous study have showed that BP/KNAT1 physically interacts with DELLA (Felipo-Benavent et al., 2018). To determine the protein interactions among BcKNOX1 and DELLAs in flowering Chinese cabbage, we conducted Y2H experiments. In yeast, BcKNOX1 interacted with BcRGA1 and BcRGL1 as shown in Figure 7A . To further verify the Y2H result in vivo, a biomolecular fluorescence complementation assay was performed. As shown in Figure 7B , the green fluorescent protein (GFP) fluorescence located in the nuclei of plant cells which confirmed that the interactions among BcKNOX1, BcRGA1 and BcRGL1.
Figure 7.
Yeast two-hybrid and biomolecular fluorescence complementation assays to determine protein interaction. (A) Interactions between BcKNOX1 and BcRGA1 and BcRGL1 in yeast cells. BcKNOX1 coding region was fused in frame to the coding sequence of the GAL4 DNA binding domain (BD), and coding sequences of BcRGA1 and BcRGL1 were fused to the coding sequence of GAL4 activation domain (AD). The numbers above the images represent cell densities obtain by 0, 10-, and 100-times dilutions. Positive bacteria were stained using X-α-Gal. (B) Bimolecular fluorescence complementary assay was used to detect the interactions of BcKNOX1 (fused with C-terminal fragment of GFP) with BcRGA1 and BcRGL1 (fused with N-terminal fragment of GFP). The nuclei were stained with DsRed. Bar = 50μm.
Discussion
The KNOX gene was first identified in maize and has since been characterized in many species (Vollbrecht et al., 1991; Hay and Tsiantis, 2010; Frangedakis et al., 2017). In this study, 9 BcKNOXs were obtained in flowering Chinese cabbage genome which was consistent with the number of KNOX genes in Arabidopsis thaliana. While, 13 Rice KNOX genes, 15 Poplar KNOX genes, and 22 Apple KNOX genes had been characterized up to date (Jain et al., 2008; Jia et al., 2020). The number of species harboring KNOX family in Rice, Poplar, and Apple was higher than the number in Arabidopsis and Brassica campestris, which may result from gene duplication. This suggests that segmental and tandem duplications are also known to contribute mainly in duplication modes during gene family expansion (Xu et al., 2020). In flowering Chinese cabbage genome, more than ten pairs of BcKNOX genes were segmentally duplicated. Chromosomes 4, 5, 7, and 10 did not exist any duplicated genes. For the evolution of plants, gene duplication is not only the motivation, but it has a prominent role in the process of genomic rearrangements (Vision et al., 2000). The KNOX genes in Arabidopsis are divided into three classes, class I, class II, and class M (Mukherjee et al., 2009). A rootless neighbor-joining phylogenetic tree was constructed using the full-length sequences of the KNOX proteins form Arabidopsis, B. rapa, and B. campestris. BcKNOXs homologous can also be clustered into three subfamilies based on sequences and gene structure. In addition, the majority of BcKNOXs except for BcKNATM contained four conserved domains ( Figure S2 ). That implied KNOX is an ancient gene family and conserved highly.
The abundances of BcKNOXs differed at various developmental stages and BcKNOXs displayed different expression profiles suggesting that they have distinct roles in different tissues during different stages. In flowering Chinese cabbage, bolting is the most obvious characteristic. Previous studies had showed that stalk development can be classified into five stages based on bolting (Huang et al., 2017; Song et al., 2019). In this study, we primarily focused on three representative periods, three-leaf, bud emergence, and flowering stage. Among them, three-leaf and bud emergence stages were the key stage for bud differentiation and stalk development respectively (Huang et al., 2017; Guan et al., 2021). The results showed that BcKNOX1, BcKNOX3, and BcKNOX5 were highly expressed in all tested tissues at each stage. Furthermore, BcKNOX2, BcKNOX6, and BcSTM had comparatively high expression levels in root of bolting and flowering stage, whereas BcKNOX4 not only showed high abundance level at three-leaf and bud emergence stage but also in leaf and flower tissues of flowering stage. In addition, correlation analysis showed that BcKNOX1, BcKNOX2, BcKNOX5, and BcKNOX6 had comparatively high correlation with gibberellins synthetase gene (GA3ox1), and some cytokinin metabolic genes. Expansin protein (ExPA) and xyloglucan endotransferases (XTH) are essential factors in cell elongation (Wang et al., 2022). In previous study, BcExPA11 and BcXTH3 showed a significant upregulation by low temperature and GA3 (Huang et al., 2017; Guan et al., 2021; Wang et al., 2022). BcKNOX3, BcKNOX4, and BcKNOX7 showed high correlation coefficients within BcExPA11 and BcXTH3. Meanwhile, the targets of KNOX showed a statistically significant enrichment of categories involved in cell wall metabolism and response to hormones (Felipo-Benavent et al., 2018). KNOX proteins can bind targets through a cis-regulatory elements containing a TGAC core (TGACTGAC or TGACAGG/CT) (Bolduc et al., 2012; Zhao et al., 2020). We conducted cis-regulatory element analysis which showed that more than 5 cis-regulatory elements containing TAGC motif were predicted within the promoters of BcEXPA11, BcXTH3, BcXTH32, BcGA3ox1, BcIPT1, BcIPT3, and BcCKX6 ( Table 2 ). We speculate that BcKNOXs may regulate the expression of these gene to involve in stalk development of flowering Chinese cabbage.
Table 2.
The number of cis-regulatory elements containing ATGC of important genes in flowering Chinese cabbage.
| Gene | Number of cis-regulatory elements containing ATGC |
|---|---|
| BcEXPA11 | 9 |
| BcXTH3 | 9 |
| BcXTH17 | 3 |
| BcXTH32 | 9 |
| BcGA3ox1 | 6 |
| BcIPT1 | 6 |
| BcIPT3 | 5 |
| BcCKX3 | 3 |
| BcCKX5 | 3 |
| BcCKX6 | 16 |
| BcCKX7 | 4 |
| BcARR1-b | 3 |
| BcRGA1 | 4 |
| BcRGL1 | 6 |
KNOX genes affected the metabolic and signaling transduction pathway of CTK and GA, while their expressions were also regulated by CTK (Truernit and Haseloff, 2007). At three-leaf stage, BcKNOX1 and BcKNOX5 were significantly suppressed upon ZT treatment, whereas BcSTM was remarkably induced by ZT. The transcript abundances of BcKNOX2, BcKNOX3, BcKNOX5, and BcKNOX6 were upregulated by ZT treatment at flowering stage. At three-leaf stage, the transcription level of BcSTM was increased in response to ZT. Several cis-elements associated with responses to low-temperature and gibberellin were predicated within the BcKNOXs promoters ( Figure 3 ). Among them, BcKNOX1, BcKNOX4, BcKNOX6, and BcKNOX7 shared low-temperature-responsive elements. Gibberellin-responsive elements located in BcKNOX2, BcKNOX5, BcKNOX6, and BcKNATM. Additionally, we investigated the expression patterns under cold and GA3 treatment. Low temperature is a vital environmental factor for stalk elongation and flowering in many winter annuals and biennials (Hébrard et al., 2013; Mathieu et al., 2014; Liang et al., 2018). The cold treatment promotes bolting and flowering by accelerating flower and bud emergence in flowering Chinese cabbage (Huang et al., 2017; Song et al., 2019). The expression level of BcSTM were downregulated in stem tip at three-leaf stage in response to cold treatment. At bud emergence stage, BcKNOX subfamily I members except for BcKNOX2 and subfamily II member BcKNOX5 were induced upon cold treatment. Besides, BcKNOX4, BcKNOX7, and BcKNATM were repressed by cold treatment at flowering stage. It has also been revealed that GA shows a significant impact on plant bolting and flowering. Exogenous GA3 treatment promotes stem elongation and bolting in brassica napus (Dahanayake and Galwey, 1999). Furthermore, GA3 treatment accelerates bolting and flowering of flowering Chinese cabbage (Song et al., 2019; Wang et al., 2022). BcKNOX2, BcKNOX5, and BcKNOX6 were significantly upregulated in response to GA3 at flowering stage. No matter three-leaf or flowering stage, the transcription of BcKNOX4 was strongly promoted by GA3. At three-leaf stage, the expression level of BcSTM was decreased under GA3 treatment. Furthermore, treatment with GA3 also induced BcKNATM expression at preceding stage, however, this gene was inhibited at flowering stage. Whereas, a previous study have shown that gibberellin did not affect KNAT expression (Truernit et al., 2006). The transcription regulation of BcKNOXs need to be further analyzed.
In Arabidopsis, DELLA proteins function as negative transcriptional regulators of GA signaling transduction by directly interacting with specific transcription factors (Locascio et al., 2013; La Rosa et al., 2014). In this study, BcKNOX1 showed high correlation coefficients within BcRGA1 and BcRGL1 which play a vital role in the bud differentiation and bolting (Guan et al., 2021). Moreover, BcKNOX1 could interact with BcRGA1 and BcRGL1 ( Figure 7 ) similar to a previous study (Felipo-Benavent et al., 2018). DELLA interaction with transcription factors has been shown to either impair their ability or to promote targets transaction (de Lucas et al., 2008; Gallego-Bartolomé et al., 2012; Fukazawa et al., 2014; Marin-de la Rosa et al., 2015). Meanwhile, DELLA proteins had been reported to be recruited by type-B ARRs that act as positive regulators of cytokinin signal transduction (Marin-de la Rosa et al., 2015). Given KNOX and CTK play a vital role in regulating meristematic activity. We hypothesized that BcKNOX1 would compete with type-B ARRs for binding to DELLA and control different targets depending on whether DELLA proteins were present. Taken together, through systematic analyses, we speculated that BcKNOX1, BcKNOX3, BcKNOX5, and BcSTM played important roles in stalk development.
Conclusion
In summary, we obtained 9 KNOX genes in flowering Chinese cabbage genome. A phylogenetic analysis exhibited that the BcKNOXs are divided into three subfamilies. Four conserved domains were widely present in the proteins. The subcellular location assay further confirmed that the majority of BcKNOXs were nuclear proteins. Three key genes (BcKNOX1, BcKNOX3, and BcKNOX5) showed high transcript abundance on tested tissues at various stages. The major part of BcKNOX genes displayed preferential expression patterns in response to low-temperature, ZT, and GA3 treatment, implying that they were involved in bud differentiation and bolting. BcKONX1 had a high correlation with BcRGA1 and BcRGL1. BcKNOX1 and BcKNOX5 also showed high correlation level with gibberellin synthetase (BcGA3ox1). Besides, BcKNOX1, BcKNOX2, and BcKNOX5 showed high correlation coefficients with CTK metabolic genes (BcCKX6 and BcIPT3). BcKNOX4 also showed high correlation coefficients within BcExPA11 and BcXTH3. In addition, BcKNOX1 interacted with BcRGA1 and BcRGL1, as confirmed by Y2H and biomolecular fluorescence complementation assay. This study has provided valuable information for the future functional roles analysis of flowering Chinese cabbage KNOX genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author contributions
XO performed the experiments and analyzed the data. JZ, ZX, JL, and BH collected the materials for the experiment and provided useful suggestions. YH, YudW and RC participated in the design of the study. XO and YH wrote the manuscript. YH, RC, SS, WS and YW assisted in revising the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (32072656) and (31972481), Key-Area Research and Development Program of Guangdong Province, China (2020B0202010006), and the China Agriculture Research System of MOF and MARA.
Acknowledgments
We show many thanks to the group members of protected horticulture in South China Agriculture University for providing the laboratory and every week group meeting.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1019884/full#supplementary-material
References
- Bellaoui M., Pidkowich M. S., Samach A., Kushalappa K., Kohalmi S. E., Modrusan Z., et al. (2001). The arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13 (11), 2455–2470. doi: 10.1105/tpc.010161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra MK., Morohashi K., O'Connor D., Grotewold E., et al. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26 (15), 1685–1690. doi: 10.1101/gad.193433.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai M., Zhou C., Molina I., Fu C., Nakashima J., Li G., et al. (2016). A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. U.S.A 113 (25), 6997–7002. doi: 10.1073/pnas.1601256113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Banerjee A. K., Hannapel D. J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38 (2), 276–284. doi: 10.1111/j.1365-313X.2004.02048.x [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang HR.Thomas Y., Frank MH., He Y., Xia R., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Cui H., Levesque MP., Vernoux T., Jung JW., Paquette AJ., Gallagher KL., et al. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Sci. (New York N.Y.) 316 (5823), 421–425. doi: 10.1126/science.1139531 [DOI] [PubMed] [Google Scholar]
- Dahanayake S. R., Galwey N. W. (1999). Effects of interactions between low-temperature treatments, gibberellin (GA3) and photoperiod on flowering and stem height of spring rape (Brassica napus. Var. annua). Ann. Bot. 84 (3), 321–327. doi: 10.1006/anbo.1999.0920 [DOI] [Google Scholar]
- Dean G., Casson S., Lindsey K. (2004). KNAT6 gene of arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol. Biol. 54 (1), 71–84. doi: 10.1023/B:PLAN.0000028772.22892.2d [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J-M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J-M., Lorrain S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 (7177), 480–484. doi: 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Di Giacomo E., Laffont C., Sciarra F., Iannelli MA., Frugier F., Frugis G., et al. (2017). KNAT3/4/5-like class 2 KNOX transcription factors are involved in medicago truncatula symbiotic nodule organ development. New Phytol. 213 (2), 822–837. doi: 10.1111/nph.14146 [DOI] [PubMed] [Google Scholar]
- Ding L., Yan S., Zhao L., Jiang W., Ning K., Zha J., et al. (2015). HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in arabidopsis. PloS Genet. 11 (9), e1005479. doi: 10.1371/journal.pgen.1005479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipo-Benavent A., Úrbez C., Blanco-Touriñán N., Serrano-Mislata A., Baumberger N., Achard P., et al. (2018). Regulation of xylem fiber differentiation by gibberellins through DELLA-KNAT1 interaction. Development 145 (23), dev164962. doi: 10.1242/dev.164962 [DOI] [PubMed] [Google Scholar]
- Frangedakis E., Saint-Marcoux D., Moody L. A., Rabbinowitsch E., Langdale J. A. (2017). Nonreciprocal complementation of KNOX gene function in land plants. New Phytol. 216 (2), 591–604. doi: 10.1111/nph.14318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frébort I., Kowalska M., Hluska T., Frébortová J., Galuszka P. (2011). Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62 (8), 2431–2452. doi: 10.1093/jxb/err004 [DOI] [PubMed] [Google Scholar]
- Frugis G., Giannino D., Mele G., Nicolodi C., Chiappetta A., Bitonti MB., et al. (2001). Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 126 (4), 1370–1380. doi: 10.1104/pp.126.4.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J., Teramura H., Murakoshi S., Nasuno K., Nishida N., Ito T., et al. (2014). DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in arabidopsis. Plant Cell 26 (7), 2920–2938. doi: 10.1105/tpc.114.125690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet EG., Grau-Enguix F., Abbas M., Locascio A., Thomas SG., et al. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in arabidopsis. Proc. Natl. Acad. Sci. U.S.A 109 (33), 13446–13451. doi: 10.1073/pnas.1119992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel RD., Bairoch A., et al. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31 (13), 3784–3788. doi: 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H., Huang X., Zhu Y., Xie B., Liu H., Song S., et al. (2021). Identification of DELLA genes and key stage for GA sensitivity in bolting and flowering of flowering Chinese cabbage. Int. J. Mol. Sci. 22 (22), 12092. doi: 10.3390/ijms222212092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S., Smith HMS., Holtan H., Magnani E., Mele G., Ramirez J., et al. (2004). The roke of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20 (1), 125–151. doi: 10.1146/annurev.cellbio.20.031803.093824 [DOI] [PubMed] [Google Scholar]
- Hamant O., Pautot V. (2010). Plant development: A TALE story. Comptes Rendus Biologies 333 (4), 371–381. doi: 10.1016/j.crvi.2010.01.015 [DOI] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M., et al. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12 (18), 1557–1565. doi: 10.1016/S0960-9822(02)01125-9 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137 (19), 3153–3165. doi: 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Hébrard C., Trap-Gentil M-V., Lafon-Placette C., Delaunay A., Joseph C., Lefèbvre M., et al. (2013). Identification of differentially methylated regions during vernalization revealed a role for RNA methyltransferases in bolting. J. Exp. Bot. 64 (2), 651–663. doi: 10.1093/jxb/ers363 [DOI] [PubMed] [Google Scholar]
- Hofer J., Gourlay C., Michael A., Ellis TH.. (2001). Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol. Biol. 45 (4), 387–398. doi: 10.1023/A:1010739812836 [DOI] [PubMed] [Google Scholar]
- Horton P., Park K-J., Obayashi T., Fujita N., Harada H., Adams-Collier CJ., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 (suppl_2), W585–W587. doi: 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Lei Y., Guan H., Hao Y., Liu H., Sun G., et al. (2017). Transcriptomic analysis of the regulation of stalk development in flowering Chinese cabbage (Brassica campestris) by RNA sequencing. Sci. Rep. 7 (1), 15517. doi: 10.1038/s41598-017-15699-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Tyagi A. K., Khurana J. P. (2008). Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 275 (11), 2845–2861. doi: 10.1111/j.1742-4658.2008.06424.x [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. biol.: CB 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Jia P., Zhang C., Xing L., Li Y., Shah K., Zuo X., et al. (2020). Genome-wide identification of the MdKNOX gene family and characterization of its transcriptional regulation in malus domestica. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R. A., Laudencia-Chingcuanco D., Smith LG., Hake S.. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124 (16), 3045–3054. doi: 10.1242/dev.124.16.3045 [DOI] [PubMed] [Google Scholar]
- Kim D., Cho Y-H., Ryu H., Kim Y., Kim T-H., Hwang I., et al. (2013). BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in arabidopsis. Plant J. 75 (5), 755–766. doi: 10.1111/tpj.12236 [DOI] [PubMed] [Google Scholar]
- Kusaba S., Kano-Murakami Y., Matsuoka M., Tamaoki M., Sakamoto T., Yamaguchi I., et al. (1998). Alteration of hormone levels in transgenic tobacco plants overexpressing the rice homeobox gene OSH1. Plant Physiol. 116 (2), 471–476. doi: 10.1104/pp.116.2.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa N. M. D., Sotillo B., Miskolczi P., Gibbs DJ., Vicente J., Carbonero P., et al. (2014). Large-Scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166 (2), 1022–1032. doi: 10.1104/pp.114.244723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M. P., Vernoux T., Busch W., Cui H., Wang JY., Blilou I., et al. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in arabidopsis. PLoS Biol. 4 (5), e143. doi: 10.1371/journal.pbio.0040143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang N., Cheng D., Liu Q., Cui J., Luo C. (2018). Difference of proteomics vernalization-induced in bolting and flowering transitions of beta vulgaris. Plant Physiol. Biochem. 123, 222–232. doi: 10.1016/j.plaphy.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Li E., Bhargava A., Qiang W., Friedmann MC., Forneris N., Savidge RA., et al. (2012). The class II KNOX gene KNAT7 negatively regulates secondary wall formation in arabidopsis and is functionally conserved in populus. New Phytol. 194 (1), 102–115. doi: 10.1111/j.1469-8137.2011.04016.x [DOI] [PubMed] [Google Scholar]
- Li J., Liu S., Chen P., Cai J., Tang S., Yang W., et al. (2022). Systematic analysis of the R2R3-MYB family in camellia sinensis: Evidence for galloylated catechins biosynthesis regulation. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.782220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Locascio A., Blázquez M. A., Alabadí D. (2013). Genomic analysis of DELLA protein activity. Plant Cell Physiol. 54 (8), 1229–1237. doi: 10.1093/pcp/pct082 [DOI] [PubMed] [Google Scholar]
- Ma C., Meir S., Xiao L., Tong J., Liu Q., Reid M.S., et al. (2015). A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167 (3), 844–853. doi: 10.1104/pp.114.253815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E., Hake S. (2008). KNOX lost the OX: The arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20 (4), 875–887. doi: 10.1105/tpc.108.058495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2016). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 (D1), D200–D203. doi: 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-de la Rosa N., Marín-de la Rosa N., Pfeiffer A., Hill K., Locascio A., Bhalerao R. P., Miskolczi P., et al. (2015). Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet. 11 (7), e1005337. doi: 10.1371/journal.pgen.1005337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu A.-S., Lutts S., Vandoorne B., Descamps C., Périlleux C., Dielen V., et al. (2014). High temperatures limit plant growth but hasten flowering in root chicory (Cichorium intybus) independently of vernalisation. J. Plant Physiol. 171 (2), 109–118. doi: 10.1016/j.jplph.2013.09.011 [DOI] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., et al. (2020). Pfam: The protein families database in 2021. Nucleic Acids Res. 49 (D1), D412–D419. doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Brocchieri L., Bürglin T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26 (12), 2775–2794. doi: 10.1093/molbev/msp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Wang Y., Franzen R., Santi L., Salamini F., Rohde W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein–protein associations in the regulation of Knox gene function. Plant J. 27 (1), 13–23. doi: 10.1046/j.1365-313x.2001.01064.x [DOI] [PubMed] [Google Scholar]
- Ori N., Juarez M. T., Jackson D., Yamaguchi J., Banowetz G. M., Hake S., et al. (1999). Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 11, 1073–1080. doi: 10.1105/tpc.11.6.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Wang Y., Zhang J., Xie Z., He B., Jiang Z., et al. (2022). Identification of BcARR genes and CTK effects on stalk development of flowering Chinese cabbage. Int. J. Mol. Sci. 23 (13), 7412. doi: 10.3390/ijms23137412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot V., Dockx J., Hamant O., Kronenberger J., Grandjean O., Jublot D., et al. (2001). KNAT2: evidence for a link between knotted-like genes and carpel development. Plant Cell 13 (8), 1719–1734. doi: 10.1105/TPC.010184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L., Sánchez-Baracaldo P., Hake S. (2000). Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42 (1), 151–166. doi: 10.1023/A:1006384122567 [DOI] [PubMed] [Google Scholar]
- Rosin F. M., Hart J. K., Horner H. T., Davies P. J., Hannapel D. J. (2003). Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol. 132 (1), 106–117. doi: 10.1104/pp.102.015560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 (5), 581–590. doi: 10.1101/gad.867901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Sentoku N., Miura Y., Hirochika H., Kitano H., Matsuoka M. (1999). Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18 (4), 992–1002. doi: 10.1093/emboj/18.4.992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Murray J. A. H. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60 (6), 929–946. doi: 10.1007/s11103-005-4478-y [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y., et al. (2001). The ASYMMETRIC LEAVES2 gene of arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 (10), 1771–1783. doi: 10.1242/dev.128.10.1771 [DOI] [PubMed] [Google Scholar]
- Smith H. M. S., Boschke I., Hake S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. U.S.A 99 (14), 9579–9584. doi: 10.1073/pnas.092271599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.-W., Lei Y., Huang X., Su W., Chen R., Hao Y., et al. (2019). Crosstalk of cold and gibberellin effects on bolting and flowering in flowering Chinese cabbage. J. Integr. Agric. 18 (5), 992–1000. doi: 10.1016/S2095-3119(18)62063-5 [DOI] [Google Scholar]
- Sparkes I. A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1 (4), 2019–2025. doi: 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Siemering K.R., Hodge S., Grbic V., Haseloff J. (2006). A map of KNAT gene expression in the arabidopsis root. Plant Mol. Biol. 60 (1), 1–20. doi: 10.1007/s11103-005-1673-9 [DOI] [PubMed] [Google Scholar]
- Truernit E., Haseloff J. (2007). A role for KNAT class II genes in root development. Plant Signaling Behav. 2 (1), 10–12. doi: 10.4161/psb.2.1.3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Ito Y., Sato Y., Kurata N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23 (12), 4368–4381. doi: 10.1105/tpc.111.090050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Hake S. (2015). Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 27, 91–96. doi: 10.1016/j.pbi.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40 (15), e115–e115. doi: 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision T. J., Brown D. G., Tanksley S. D. (2000). The origins of genomic duplications in arabidopsis. Sci. (New York N.Y.) 290 (5499), 2114–2117. doi: 10.1126/science.290.5499.2114 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S. (1991). The developmental gene knotted-1 is a member of a maize homeobox gene family. Nature 350 (6315), 241–243. doi: 10.1038/350241a0 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 (14), 3161–3172. doi: 10.1242/dev.127.14.3161 [DOI] [PubMed] [Google Scholar]
- Wang Y., Huang X., Huang X., Su W., Hao Y., Liu H., et al. (2022). BcSOC1 promotes bolting and stem elongation in flowering Chinese cabbage. Int. J.Mol. Sci. 23 (7), 3459. doi: 10.3390/ijms23073459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Pu X., Gao R., Demurtas O.C., Fleck S.J., Richter M., et al. (2020). Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 18 (1), 63. doi: 10.1186/s12915-020-00795-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. (2019). OsKNAT7 bridges secondary cell wall formation and cell growth regulation. Plant Physiol. 181 (2), 385–386. doi: 10.1104/pp.19.01018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Li C., Ma X., Xia R., Chen J., Liu X., et al. (2020). KNOX protein KNAT1 regulates fruitlet abscission in litchi by repressing ethylene biosynthetic genes. J. Exp. Bot. 71 (14), 4069–4082. doi: 10.1093/jxb/eraa162 [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.-H., et al. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in arabidopsis. Plant Cell 20 (10), 2763–2782. doi: 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .