Abstract
Carbapenem-resistant (CR) Klebsiella pneumoniae represents an urgent worldwide threat. We focused on CR K. pneumoniae in selected facilities of the University Hospital Bratislava (UHB) to investigate sequence types (STs), clonal relatedness, and antimicrobial resistance. Suspected carbapenem-nonsusceptible K. pneumoniae strains were obtained from hospitalized patients. Further examination included carbapenemase confirmation, cgMLST, and quantitative susceptibility testing. Of the total 41 CR K. pneumoniae strains, 26 (63.4%) were determined as ST11 in hospital No. 1; of these ST11, 22 (84.6%) were found in the first internal clinic. Six (14.6%) ST258 and three (7.3%) ST584 occurred in hospital No. 2; the most, i.e., four (66.7%), ST258 were detected in the geriatric clinic. In hospital No. 3, we found two (4.8%) ST584 and one (2.4%) ST258. Of the ST11 set, 24 (92.3%) produced NDM-1. Furthermore, seven (87.5) ST258 and five (100%) ST584 strains generated KPC-2. Antimicrobial resistance was as follows: ertapenem 97.6%, meropenem 63.4%, tigecycline 7.3%, eravacycline 7.3%, and colistin 2.5%. We revealed a presumably epidemiological association of ST11 with transmission, particularly in the first internal clinic of hospital No. 1, while ST258 and ST584 were related to interhospital dissemination between medical facilities No. 2 and No. 3. It is essential to prevent the circulation of these pathogens within and between healthcare facilities.
Keywords: carbapenem-resistant Klebsiella pneumoniae, cgMLST, antimicrobial resistance, healthcare facilities
1. Introduction
Carbapenem-resistant Enterobacteriaceae (CRE), which include mainly Klebsiella pneumoniae strains, represent a significant and constantly growing problem in healthcare facilities. This concern should be considered a state-wide priority due to the increase in difficult-to-treat diseases and the high mortality rate, which is estimated to be between 33% and 42% [1]. These multidrug-resistant (MDR, i.e., resistance to three or more classes of antimicrobials) pathogens evolve primarily in a hospital setting after the exposure of patients to antibiotics, resulting in selection pressure [2,3]. Consequently, the development of resistance is dependent on the epidemiological status of the corresponding medical environment, the consumption of carbapenems, and the cross-transmission of MDR strains between patients. On the other hand, the rapid dissemination of resistance is exceedingly facilitated by β-lactamase (bla) genes through transferrable genetic elements [4]. Notably, resistance to carbapenems may be often associated with the presence of genes related to other antimicrobial classes; therefore, CR strains frequently appear as extensively drug-resistant (XDR, i.e., remain susceptible to only one or two antimicrobial categories) microorganisms [5]. Resistance to carbapenems linked to carbapenemase production (CP) is of greatest concern from an epidemiological point of view because carbapenemase genes located on plasmids are readily spread to many other bacterial agents [6]. The acquisition of carbapenem resistance is also possible via non-CP enzymes, such as ESBLs (extended-spectrum β-lactamases) and AmpC cephalosporinases, combined with other resistance mechanisms (porin mutations or loss, production of efflux pumps, or alteration of penicillin-binding proteins). Accordingly, the CP of CR strains must be determined and differentiated from that caused by ESBL and/or AmpC hyperproduction supported by structural mutations [4,5]. The most common carbapenemases are KPC (Klebsiella pneumoniae carbapenemase) from molecular class A and NDM (New Delhi metallo-β-lactamase) from class B [7], geographically related to the Slovak region and the surrounding area [4].
Regarding the global development and spread of carbapenem resistance, particularly that of Klebsiella pneumoniae strains, diverse national genetic and/or sequence type variations have been recorded worldwide. Using multilocus sequence typing (MLST), CR K. pneumoniae strains can often be associated with emergence in the relevant regional area [1]. Molecular methods enabling the detection of bacterial pathogens and their transmission have become an important tool in the control of nosocomial infections when providing care in healthcare facilities, as well as for the identification of the infection source, including the dissemination routes of epidemiological cases in the medical environment [8].
Our objective in this study was to determine the emergence of CR Klebsiella pneumoniae strains, their genotypic characteristics, the participation of sequence types (STs), and their genetic relatedness in connection with probable epidemiological transfer in three selected medical facilities of the University Hospital Bratislava (UHB). An important goal of this work was to investigate genes encoding antimicrobial resistance and determine the resistance of the evaluated strains to the tested antimicrobial agents.
2. Results
2.1. Isolation of CR K. pneumoniae Strains
In general, 41 CR Klebsiella pneumoniae strains were investigated due to presumed epidemiological transmission between hospitalized patients within three healthcare facilities belonging to the UHB. In the study cohort, there were 24 (58.5%) female patients. Patients over the age of 60 represented 90.2% (37) of the group, with a mean age of 74.7 (range of 25 to 96) years, as indicated in Table 1. In the healthcare-related setting No. 1 with 28 (68.3%) patients, the majority (23; 56.1%) originated from the first internal clinic. In the medical facility No. 2, there were ten (24.4%) individuals; of them, five (12.2%) were from the geriatric clinic. Finally, three (7.3%) CR K. pneumoniae strains were obtained from the Institute of Pathological Anatomy and the postmortem of patients who were treated and died in the third healthcare-associated environment within UHB. Regarding the carriage in the gastrointestinal tract, the 23 specimens were obtained from a rectal swab or stool (56.1%). As for skin and soft tissue infections, five (12.2%) samples were obtained from a wound (twice), abscess (once), or organ swab (twice). Finally, eight (19.5%) urine samples (six from infection, two from colonization) and five (12.2%) specimens from the respiratory tract (four throat swabs and one sputum sample) were obtained, as shown in Table 1. The patient conditions were designated according to previously defined criteria, including microbiological findings, as 29 (70.7%) colonizations and 12 (29.3%) infections (Supplementary Materials: Table S1a–c).
Table 1.
Characteristics of the patients, healthcare settings, and specimens related to 41 CR Klebsiella pneumoniae strains.
| Gender | n | % | |
|---|---|---|---|
| Male | 17 | 41.5 | |
| Female | 24 | 58.5 | |
| Age (years) | |||
| <60 | 4 | 9.8 | |
| ≥60 | 37 | 90.2 | |
| Mean (range) | 74.7 | (25–96) | |
| Setting | |||
| No. 1 | First internal clinic | 23 | 56.1 |
| Surgical clinic | 2 | 4.9 | |
| Dermatovenerological clinic | 2 | 4.9 | |
| Neurological clinic | 1 | 2.4 | |
| No. 2 | Geriatric clinic | 5 | 12.2 |
| Long-term care department | 3 | 7.3 | |
| Aftercare department | 2 | 4.9 | |
| No. 3 | Institute of Pathological Anatomy | 3 | 7.3 |
| Specimen | |||
| Rectal swab or stool | 23 | 56.1 | |
| Wound, abscess, or organ swab | 5 | 12.2 | |
| Urine | 8 | 19.5 | |
| Throat swab or sputum | 5 | 12.2 | |
n—number of patients, cases, or specimens; %—percentage; No. 1, No. 2, and No. 3—designations of selected medical facilities belonging to the University Hospital Bratislava (UHB).
2.2. Genome Sequencing of CR K. pneumoniae Strains
Forty-one Klebsiella genomes were sequenced using the Illumina technology. We obtained high-quality whole genome contigs for all strains; the length of the genomes ranged from 5.23 to 5.66 Mbp with an average coverage of 20 to 590 times.
The clonality of the strains was determined using MLST, which revealed the presence of five sequence types (STs; Table 2). The majority of the strains belonged to the sublineage SL258, covering the ST11 (twenty-six), ST258 (eight), and ST340 (one) strains (Table 2). Five strains were assigned to ST584 of SL2004. The ST15 profile was observed once in the set.
Table 2.
Characterization of 41 CR K. pneumoniae strains.
| Sublineage | Clonal Group | ST | Serotype | No of Strains |
|---|---|---|---|---|
| SL258 | CG340 | 11 | K15:O4 | 25 |
| K105:O2 | 1 | |||
| 340 | K15:O4 | 1 | ||
| CG258 | 258 | K106:O2 | 7 | |
| K107:O2 | 1 | |||
| SL15 | CG15 | 15 | K112:O1 | 1 |
| SL2004 | CG584 | 584 | K38:O3 | 4 |
| K50:O3 | 1 |
ST—sequence type; K—capsular antigen; O—specific polysaccharide antigen.
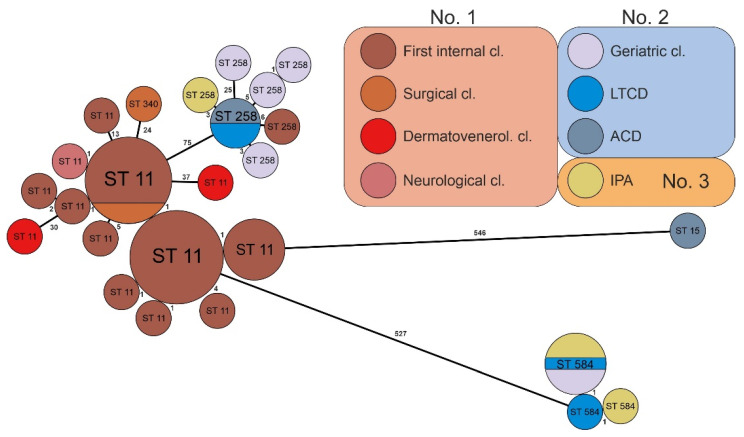
The mutual relatedness of strains assigned to the same ST was further assessed using core genome MLST (cgMLST). We observed that strains belonging to the same ST differed in 0–37 cgMLST alleles (Figure 1). Based on this analysis, the ST11 strains formed thirteen unique profiles, the majority of which differed from the main group by less than ten alleles. There were three strains (KMB-944, KMB-966, and KMB-953) in which the exceptions possessed 13–37 different alleles. Similar relations were observed in ST258, where only one strain (KMB-946) showed more than ten different alleles. Five ST584 strains were found to be very closely related (Figure 1).
Figure 1.
GrapeTree of 41 isolated CR Klebsiella pneumoniae strains based on cgMLST. Ring sizes correspond to the number of strains with the same genotype; the sequence types (STs) of the strains are marked inside the ring. The numbers on lines between rings correspond to the numbers of different alleles between groups. The color code depicts the origin of the strains—clinics, departments, or institutes, as well as the medical setting, labelled with the hospital number (No. 1, No. 2, or No. 3)—including clonal relatedness. Abbreviations: cl.—clinic; Dermatovenerol. cl.—dermatovenerological clinic; LTCD—long-term care department; ACD—aftercare department; IPA—Institute of Pathological Anatomy (strains isolated from patients who were treated and died in hospital No. 3).
2.3. Occurrence of ST in Hospital Facilities
We observed that some Klebsiella sequence types occurred predominantly in some hospital settings. Twenty-two of the twenty-six ST11 strains were isolated from the first internal clinic of medical facility No. 1, and the remaining four strains were detected in other clinics of the same hospital. The ST258 and ST584 strains were predominantly isolated from healthcare facility No. 2 and partially from medical facility No. 3 (Figure 1).
The tree was constructed in GrapeTree, and the numbers above/beside the lines show the counts of different alleles.
2.4. Antibiotic Resistance Genes of CR K. pneumoniae Strains
The presence of antibiotic resistance genes was analyzed using the PATRIC database. All strains contained at least three genes (Table 3). Strains belonging to ST11 showed a wide spectrum of β-lactamase genes. A notable finding regarding carbapenemase activity was that 24 (92.3%) of the 26 ST11 strains produced metallo-β-lactamase NDM-1, 26 (100%) produced SHV-11, and 23 (88.4%) produced CTX-M-15, while 21 (80.7%) also produced BSBL (broad-spectrum β-lactamase) OXA-1. On the other hand, the production of the KPC-2-encoded serine carbapenemase, which was recorded in seven of the eight ST258 strains (87.5%) and in the five ST584 CR K. pneumoniae strains (100%), should be highlighted. TEM-1 was present in five ST258 (63%) and five ST584 (100%) strains. Seven out of the eight ST258 strains (87.5%) were positive for SHV-12 genes, and all ST584 strains contained SHV-168, CTX-M-15, and OXA-1 β-lactamase genes (Table 3).
Table 3.
Presence of antimicrobial resistance genes (%) in 41 CR K. pneumoniae strains according to sequence type.
| ble MBL | 92 | 0 | 0 | 0 | 0 |
| qacEΔ1 | 92 | 0 | 75 | 100 | 0 |
| oqxA, oqxB20 | 0 | 100 | 0 | 0 | 0 |
| oqxA, oqxB14 | 0 | 0 | 0 | 0 | 100 |
| oqxA, oqxB | 100 | 0 | 100 | 100 | 0 |
| tet(D) | 0 | 0 | 0 | 100 | 0 |
| tet(A) | 11 | 0 | 0 | 0 | 100 |
| qnrB4 | 3 | 0 | 0 | 0 | 0 |
| qnrB1 | 0 | 0 | 0 | 0 | 100 |
| qnrA3 | 3 | 0 | 0 | 0 | 0 |
| mphA | 7 | 0 | 75 | 0 | 0 |
| catA1/catA4 | 7 | 0 | 75 | 0 | 0 |
| catB | 84 | 0 | 0 | 0 | 0 |
| fosA | 100 | 100 | 100 | 100 | 100 |
| Arr3 | 3 | 0 | 0 | 0 | 0 |
| Arr2 | 80 | 0 | 0 | 0 | 0 |
| aph(6)-Id | 3 | 100 | 0 | 100 | 100 |
| aph(3″)-Ib | 3 | 100 | 0 | 100 | 100 |
| aph(3′)-Ia | 0 | 0 | 75 | 0 | 0 |
| aac(6′)-Ib4 | 19 | 0 | 0 | 0 | 0 |
| aac(6′)-Ib | 57 | 0 | 0 | 0 | 0 |
| aac(3)-IId | 3 | 0 | 0 | 100 | 0 |
| aac(3)-IIa | 76 | 100 | 0 | 0 | 100 |
| rmtF1 | 76 | 0 | 0 | 0 | 0 |
| aadA16 | 7 | 0 | 0 | 0 | 0 |
| aadA2 | 88 | 0 | 75 | 100 | 0 |
| sul2 | 3 | 100 | 0 | 100 | 100 |
| sul1 | 92 | 0 | 75 | 100 | 0 |
| dfrA27 | 3 | 0 | 0 | 0 | 0 |
| dfrA12 | 88 | 0 | 75 | 0 | 0 |
| bla OXA-9 | 3 | 0 | 0 | 0 | 0 |
| bla OXA-1 | 80 | 100 | 0 | 100 | 100 |
| bla DHA-1 | 3 | 0 | 0 | 0 | 0 |
| bla NDM-1 | 92 | 0 | 0 | 0 | 0 |
| bla KPC-3 | 0 | 0 | 13 | 0 | 0 |
| bla KPC-2 | 0 | 0 | 87 | 0 | 100 |
| bla TEM-156 | 8 | 0 | 0 | 0 | 0 |
| bla TEM-116 | 4 | 0 | 0 | 0 | 0 |
| blaTEM-1 | 11 | 100 | 63 | 100 | 100 |
| bla CTX-M-15 | 88 | 100 | 0 | 100 | 100 |
| bla SHV-168 | 0 | 0 | 0 | 0 | 100 |
| bla SHV-28 | 0 | 100 | 0 | 0 | 0 |
| bla SHV-27 | 0 | 0 | 0 | 0 | 0 |
| bla SHV-12 | 0 | 0 | 87 | 0 | 0 |
| bla SHV-11 | 100 | 0 | 13 | 100 | 0 |
| ST11 | ST15 | ST258 | ST340 | ST584 |
The listed gene-encoding products confer resistance [mechanism] against the following agents: Ble-MBL—bleomycin [bleomycin-binding protein]; QacE delta 1—quaternary ammonium compounds; OqxA, OqxB20, OqxB14, OqxB—[efflux pumps] providing resistance to tetracyclines, nitrofurans, diaminopyrimidines, fluoroquinolones, and glycylcyclines; Tet(D), Tet(A)—tetracyclines; QnrB4, QnrB1, QnrA3—fluoroquinolone antibiotics (enoxacin, ciprofloxacin, levofloxacin, moxifloxacin); MphA—macrolides (erythromycin, roxithromycin, azithromycin); CatA1/CatA4, CatB—chloramphenicol [chloramphenicol O-acetyltransferase]; FosA—fosfomycin [fosfomycin thiol transferase]; Arr3, Arr2—rifamycin antibiotics such as rifampin; APH(6)-Id, APH(3″)-Ib—streptomycin; APH(3′)-Ia—kanamycin, neomycin, paromomycin; AAC(6′)-Ib4, AAC(6′)-Ib—tobramycin, amikacin; AAC(3)-IId; AAC(3)-IIa—tobramycin, gentamicin; Rmtf1—aminoglycosides (neomycin, amikacin, gentamicin); AadA16, AadA2—ANT(3″)-Ia [group of aminoglycoside nucleotidyl transferases], resistance to aminoglycosides (streptomycin, spectinomycin); Sul2, Sul1—sulfonamides; Dfra27, Dfra12—diaminopyrimidine antibiotics such as trimethoprim; OXA-9, OXA-1—betalactams (cephalosporins, penicillins); DHA-1—betalactams (cephalosporins, cephamycin, penicillins); NDM-1—betalactams (cephalosporins, carbapenems, penicillins, cephamycin); KPC-3, KPC-2—betalactams (cephalosporins, carbapenems, penicillins, cephamycin); TEM-156, TEM-116; TEM-1—betalactams (cephalosporins, penicillins, penems, monobactams); CTX-M-15, SHV-28, SHV-168, SHV-27, SHV-12, SHV-11—betalactams (cephalosporins, penicillins, monobactams).
The resistance of K. pneumoniae strains to antimicrobial agents was determined as follows: 100% were resistant to piperacillin/tazobactam and ceftazidime, 58.5% to ceftazidime/avibactam, 97.6% to ertapenem, 63.4% to meropenem, 78.1% to amikacin, 82.9% to gentamicin, 7.3% to tigecycline, 7.3% to eravacycline, and 2.5% to colistin, as listed in Table 4.
Table 4.
Antimicrobial resistance of 41 carbapenem-resistant Klebsiella pneumoniae strains (µg/mL).
| Antibiotics | MIC Range | MIC50 | MIC90 | %R | %S | ||
|---|---|---|---|---|---|---|---|
| Cefoperazone/sulbactam | 32 | - | 128 | 128 | 128 | 100 | 0 |
| Piperacillin/tazobactam | 64 | - | 128 | 128 | 128 | 100 | 0 |
| Cefuroxime | 32 | - | 64 | 64 | 64 | 100 | 0 |
| Cefotaxime | 16 | - | 64 | 64 | 64 | 100 | 0 |
| Ceftazidime | 32 | - | 64 | 64 | 64 | 100 | 0 |
| Ceftazidime/avibactam | 0.064 | - | 64 | 64 | 64 | 58.5 | 41.5 |
| Cefepime | 16 | - | 64 | 64 | 64 | 100 | 0 |
| Ertapenem | 0.25 | - | 8 | 8 | 8 | 97.6 | 2.4 |
| Meropenem | 0.125 | - | 32 | 32 | 32 | 63.4 | 21.9 |
| Amikacin | 0.5 | - | 128 | 128 | 128 | 78.1 | 21.9 |
| Gentamicin | 0.25 | - | 32 | 32 | 32 | 82.9 | 17.1 |
| Tobramycin | 0.5 | - | 32 | 32 | 32 | 97.6 | 2.4 |
| Ciprofloxacin | 2 | - | 8 | 8 | 8 | 100 | 0 |
| Tigecycline | 0.5 | - | 8 | 1 | 2 | 7.3 | 68.3 |
| Eravacycline | 0.25 | - | 2 | 0.5 | 0.5 | 7.3 | 90.2 |
| Colistin | 0.25 | - | 4 | 1 | 2 | 2.5 | 97.5 |
| Trimethoprim/sulfamethoxazole | 1 | - | 8 | 8 | 8 | 80.5 | 7.3 |
MIC—minimum inhibitory concentration; R—resistant; S—susceptible.
3. Discussion
CR K. pneumoniae strains account for a worldwide public health emergency and are designated as one of the major causes of hospital-acquired infection [9]. This is especially the case within the hospital; however, the interhospital spread of these worrisome pathogens is a very frequent and significant facilitator of the emergence of co-resistance to several antimicrobial classes [10].
We investigated 41 strains of CR K. pneumoniae from three UHB medical facilities and found a correlation between ST clonal similarity within these healthcare-associated settings. The mean age of the patients in our study (Table 1) was similar to that in the survey by Han et al. (2017) related to long-term acute care hospitals in the United States, where 25% of the K. pneumoniae clinical strains were CR K. pneumoniae; however, national surveillance and improved connections with acute care hospitals were decisive for reducing the spread of these pathogens [11]. The specimens used in this American observational study were from the respiratory system, urinary tract, and blood, slightly different from our specimens (Table 1); in addition, the authors used a notably larger sample size. An international multicenter cohort study [1] involving strains of CR K. pneumoniae isolated from patients (41% female) with a mean age of approximately 12 years or younger reported the opposite proportion to that in our research (Table 1). The authors of this intercontinental survey analyzed a large number of isolates with partially similar sample sources, including bloodstream infection, which we did not record. This research reported that most of the CR K. pneumoniae strains came from hospital transfers in China (64%), from long-term chronic care in the USA (28%), and even from the domestic environment in Australia (83%) and South America (74%) [1]. This diversity may be related to the variability associated with a particular region. Australian researchers [12] presented results on CR Klebsiella spp. causing bloodstream infections (BSIs) mainly detected in endemic areas, including Greece, Italy, and Israel. BSIs are commonly found in neutropenic and cancer patients, often requiring broad-spectrum antibiotics. This is a crucial factor for the colonization by ESBL and CR bacteria, which constitutes a reservoir for subsequent infection [12]. There were fewer patients with oncological disease in our investigated health facilities, which could be the reason why we did not obtain positive blood samples.
We found five sequence types; among these, ST11, ST15, ST258, and ST340 are widespread CR K. pneumoniae clones, while ST584 is rare [13]. Most strains (Table 2) were associated with ST11 in our investigation, which is in concordance with Chinese surveys of 27 provinces from 2016 to 2020 where ST11 was the main sequence type, accounting for 64.2% of the samples [14]. In our study, this clone was represented within CR K. pneumoniae strains at a slightly lower rate in hospital facility No. 1. High clonal relatedness was found in the first internal clinic in twelve (29.3%) and the next six (14.6%) strains (Figure 1), suggesting possible intrahospital cross-transmission within this clinic composed of three departments (A, B, and C). Other sporadic ST11 strains designated as KMB-965 from the surgical clinic and KMB-938 from the neurological clinic, were closely related to clones from the first internal clinic; this may indicate the potential dissemination of CR K. pneumoniae strains between the clinics in the same medical workplace—hospital No. 1 (Figure 1). The remaining ST11 strains, consisting of KMB-966, KMB-967, KMB-971, and KMB-960 from the first internal clinic, as well as KMB-944 and KMB-953 from the dermatovenerological clinic, were clonally different (Figure 1) and had less of a chance for nosocomial spreading. Raro et al. (2020) presented an investigation of carbapenemase-producing K. pneumoniae strains in transplanted patients and observed four epidemic clones: ST11, ST15, ST16, and ST437. ST11 was represented in 62.5% [9] of the patients, which is in line with our data. Additionally, in an Italian study of isolated CR K. pneumoniae strains in long-term care facilities, the sequence types ST307, ST512, and ST37 were prevalent; however, other lineages were also found (ST11, ST16, and ST253) [10]. In the long-term care department within medical facility No. 2, we found ST258 and ST584, while no ST11 was recorded. Consequently, the occurrence of sequence types can influence the transfer and variability of patients at a time and site, in a country or region, or even in a particular medical facility. CRE have achieved the remarkable endemicity and spreading of CR K. pneumoniae–producing KPC since 2010; the clonal group ST258 represented the majority of strains in Italy in one study [15], which is consistent with healthcare setting No. 2 in our study. Of the seven ST258 strains that were slightly mutually related, five were from hospital No. 2, including three from the geriatric clinic—KMB-942, KMB-963, and KMB-962—one from the aftercare department—KMB-940—and one from the long-term care department—KMB-948. This close clonal relatedness of the same sequence type (ST258) may indicate intrahospital cross-transmission of CR K. pneumoniae strains. Two ST258 clones designated as KMB-935, originating from hospital No. 3, and KMB-936, from the first internal clinic of hospital No. 1, were closely related to the ST258 clone lineage within hospital No. 2, which could suggest a conceivable interhospital transfer of the same sequence type. Furthermore, five highly related ST584 strains included KMB-968, KMB-954, and KMB-955 from healthcare facility No. 2 and KMB-969 and KMB-970 from hospital No. 3, which could illustrate the presumed dissemination of the assayed pathogens between the two medical facilities. ST584 is rare globally, while ST258 has frequently been described in the United States in previous years as the clonal expansion of dominant K. pneumoniae carbapenemase (KPC)-producing strains [1,11] and in Australia [16]. This K. pneumoniae ST258 clone is characterized by high plasticity, facilitating genetic variability [17].
The emergence of CR K. pneumoniae epidemics in distinct parts of the world varies along with the relation of carbapenemase and other resistance genes to sequence types [1]. Our results regarding the emergence of detected carbapenemases highlight the importance of the spread of these strains in our healthcare facilities, which may be related to the epidemiology of the respective STs as well as particular strains that generally produce carbapenemase in Slovakia and the central European region. In our examination, we determined a high association between ST11 and NDM-1 metallo-carbapenemase, including the ESBL enzymes SHV-11 and CTX-M-15. In a similar manner, a highly prevalent ST11 clone was detected in previous years in Italy, very frequently co-harboring NDM and other carbapenemases (OXA-48, VIM, and/or KPC), including ESBL, AmpC β-lactamase, aminoglycoside-modifying enzymes, fluoroquinolone resistance enzymes, and determinants of trimethoprim/sulfamethoxazole nonsusceptibility [18]. However, our results did not show the co-production of more than one carbapenemase per strain. Moreover, we noted that ST258 was more connected with KPC-2 and KPC-3 as well as SHV-12, while ST584 was completely bound to KPC-2, SHV-168, and CTX-M-15 (Table 3). In a Brazilian survey, ST11, ST16, and ST437 K. pneumoniae epidemic clones encompassed KPC-2 production, while ST15 created NDM-1, including carriership and a significant range of acquired resistance genes [9]. These authors reported that only 3.7% of their isolates were KPC-2 carbapenemase-producing K. pneumoniae ST258; however, in our results, these sequence types produced KPC-2 at a high rate (87.5%). Compared to our values, in the research from the USA, ST258 CR K. pneumoniae strains contained less blaKPC-2 (44%), and an even lower rate of this gene (39%) was reported in some countries of South America [1]. On the contrary, in China, ST11 was the main carrier of blaKPC-2 (94%) in the elderly [1,19] but of other carbapenemase genes (blaNDM-1) in neonates [20]. The latter corresponds to our data. Furthermore, in contrast to our results and the findings from the United States in correlation with ST258, authors from Panama published findings about NDM-producing ST258 CR K. pneumoniae strains (33.3%) isolated from the secretions and blood cultures of hospitalized patients [17]. The expansion of CR K. pneumoniae in various regions and countries has revealed more differences than similarities with regard to carbapenemase genes and additional genes encoding enzymes and/or mechanisms responsible for antimicrobial resistance against aminoglycosides, fluoroquinolones, tetracyclines, trimethoprim/sulfamethoxazole, fosfomycin, and others. The listed genetic determinants are transmitted by these clones and can be represented at a more diverse rate [9,10,21] than in our findings.
In a study by Ippolito et al. (2021), CRE, including CR K. pneumoniae strains, were isolated from patients with COVID-19 and ventilator-associated pneumonia (3%); however, in all Gram-negative bacteria, carbapenem resistance was achieved in up to 32% [22]. The dissemination of carbapenem resistance was facilitated by the SARS-CoV-2 pandemic; therefore, we were faced more often with the transmission of CR K. pneumoniae strains due to precautions focusing mainly on COVID-19 patients. Some studies reported that the prevalence of CR K. pneumoniae in COVID-19 patients ranged from 0.35 to 53%, and blaKPC, blaOXA-48, and blaNDM were determined to be among the predominant genes [23].
A study published by Yin et al. (2018) on antimicrobial resistance showed that resistance against ertapenem (100%) and meropenem (95.7%) was higher, especially for meropenem (greater than 32%), when compared to our values (Table 4). In this investigation, the authors mentioned that most of these CR K. pneumoniae strains were NDM-producing ST278; due to their homogeneity, the carbapenem resistance profile was high [20]. Regarding the observation from China, resistance to cephalosporins—cefoperazone/sulbactam, cefuroxime, cefotaxime, ceftazidime, and cefepime—was 100%, which is in agreement with our data. Nonetheless, it should be noted that nonsusceptibility to amikacin (8.5%), gentamicin (21.3%), and ciprofloxacin (21.3%) was several times lower [20] than that in our results.
With regard to another study on ST258 CR K. pneumoniae strains from selected long-term acute care hospitals in 16 countries in the USA, the rate of resistance to ciprofloxacin (98.1%) is in agreement with our profile and lower against amikacin (59.2%) and tigecycline (0.7%); however, it was considerably higher against gentamicin (97.8%) and colistin (16.1%) [11]. We can comment that the results reported by the American investigators referred to ST258, which we also found, and the resistance to colistin in our data was much lower. To our knowledge, resistance could result from the administration of colistin in general treatment; however, the horizontal transfer of genes related to colistin resistance (e.g., mediated by the colistin resistance gene mcr-1) via plasmids can also have an impact [24]. Another study mainly analyzed the resistance of CR K. pneumoniae NDM producers; compared to our results, nonsusceptibility to colistin (9.8%) and tigecycline (15.9%) was higher, while it was 100% for ceftazidime/avibactam [25], which is basically intended for strains producing KPC and OXA-48. In our outcomes, resistance to eravacycline was the same as resistance to tigecycline; however, susceptibility was improved (Table 4). This synthetic fluorocycline antibacterial agent was approved for treating serious intra-abdominal infections and has an antibacterial potential that is four times greater than that of tigecycline, with a preferable concentration in serum and tissue; however, it is not indicated for the treatment of complicated urinary tract infections. Eravacycline is efficient in vitro against CRE, including strains producing KPC, NDM, VIM, IMP, and OXA-48; however, the clinical data for treatment are insufficient [26,27].
While we could rely on the improved efficacy of other more current antibacterial agents [28,29] or maybe some natural substances [30] and their derivatives that are expected in our healthcare facilities, we cannot conclude that the resistance of CR K. pneumoniae strains will not increase over time.
Further research is necessary to analyze genotypic determinants and detect the development of antimicrobial resistance against these dangerous MDR pathogens spreading in the medical environment as well as to find and prevent the establishment of epidemiological relationships, including clonal relatedness.
4. Materials and Methods
4.1. Hospital Setting and Patients
CR Klebsiella pneumoniae strains were obtained from patients residing in the University Hospital Bratislava (UHB) in the period from April 2017 to May 2019. UHB is one of the largest Slovak hospitals and consists of five healthcare workplaces. The assayed strains originated from 3 selected medical facilities, which due to anonymity preservation, were referred to as hospital No. 1, No. 2 and No. 3. The colonization of the patients was determined by a positive culture without clinical proof of infection at the time of isolation. Infection was defined on the basis of the treating physician’s diagnosis, which was compatible with the signs of clinical infection associated with the isolation of CR K. pneumoniae in the relevant biological sample and significant findings for microorganisms.
4.2. Isolation and Selection of Bacterial Strains
The identification of Klebsiella pneumoniae strains was carried out using the biochemical ENTEROtest 24 kit (Erba Lachema s.r.o., Czech Republic) at the Institute of Microbiology, Faculty of Medicine, Comenius University and UHB. Only one non-repetitive isolate per patient was included in the survey. The samples were collected from the various body sites of the patients and cultured on COLOREX™ ESBL screening chromogenic medium (MkB Test a.s., Rosina, Slovak Republic) and standard culture medium. Possible carbapenem resistance was predetermined by MacConkey agar with ertapenem (ERT; 10 µg) discs and Mueller–Hinton agar with ERT and meropenem (MER; 10 µg) discs. In the case of suspicious K. pneumoniae growth on the abovementioned screening media and/or reduced susceptibility to carbapenem antibiotics according to EUCAST criteria [31], the phenotypic colorimetric Carba NP test was carried out to prove carbapenemase production [32]. The antimicrobial susceptibility was determined using a colorimetric micromethod [33].
4.3. Isolation of DNA and Preparation of Libraries for NGS
Bacterial strain culture was performed on Luria–Bertani (LB) medium at 37 °C under aerobic conditions for 12 h. The Higher PurityTM Bacterial Genomic DNA Isolation Kit (CanvaxBiotech; Cordoba, Spain) and the DNeasy® Blood & Tissue Kit (Qiagen; Hilden, Germany) were used for DNA isolation, and DNA was quantified using the QubitTM dsDNA HS Assay Kit (Thermofisher Scientific; Eugene, OR, USA). Sequencing libraries were preprepared using the Nextera XT DNA Library Prep Kit protocol (Illumina; San Diego, CA, USA) and purified on AMPure XP magnetic beads (Beckman Coulter Life Sciences; Indianapolis, IN, USA). The quality of the library was checked using a high-sensitivity DNA electrophoresis chip and a 2100 Bioanalyzer Instrument (Agilent Technologies; Santa Clara, CA, USA). Sequencing was carried out with 2 × 150 bp reads using the Illumina NextSeq 500TM platform (Illumina).
4.4. Bioinformatic Processing and Genome Analysis
The obtained sequencing data were assembled de novo with SPAdes using a standard setting of parameters (Center for Algorithmic Biotechnology; St. Petersburg State University, Russia) [34] and annotated using the online PATRIC program (Pathosystem Resource Integration Center) and RAST (Rapid Annotation using Subsystem Technology) software. The criterion of 100% identity and 100% coverage with the reference gene was used as the threshold for gene presence. The genomes were further analyzed using the CGE database (Center for Genomic Epidemiology; Kongens, Lyngby, Denmark) and the BIGSdb database (Institut Pasteur MLST). The standard MLST and cgMLST based on 694 genes available in the Klebsiella BIGSdb database (https://bigsdb.pasteur.fr/klebsiella/, (accessed on 6 May 2020) were used for the determination of strain relatedness [35].
GrapeTree was used for the visualization of strain clusters based on the Klebsiella cgMLST analysis [36]. The sequenced genomes were deposited in the Klebsiella MLST database (https://bigsdb.pasteur.fr/klebsiella/, (accessed on 6 May 2020) with accession numbers 12,559–12,599.
5. Conclusions
Although a small sample was employed in this examination, our thorough molecular analyses suggest a probable horizontal transfer with high clonal relatedness of CR K. pneumoniae ST11 strains in the same healthcare setting: in hospital No. 1, we observed the emergence and spread of two sequence types (ST258 and ST584) between two medical workplaces—hospitals No. 2 and No. 3—providing health care in the UHB complex. Therefore, it is enormously important to strengthen antibiotic control to prevent the development of antimicrobial resistance and to emphasize infection control and measures related to avoiding the transmission of these difficult-to-treat pathogens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11111538/s1, Table S1a, Sequencing parameters; Table S1b, Genome data; Table S1c, Antibiotic resistance.
Author Contributions
Conceptualization, J.K. and H.D.; data curation, J.K., M.A., H.D., Z.H. and A.L.; formal analysis, J.K., M.A., H.D. and A.L.; methodology, J.K., M.A. and H.D.; investigation, J.K., M.A. and Z.H.; software, M.A.; supervision, H.D., A.L. and T.M.; visualization, M.A.; writing—original draft, J.K. and H.D.; writing—review and editing, H.D. and A.L.; funding acquisition, J.K., A.L. and T.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This publication was created thanks to the support of the Operational Program Integrated Infrastructure for the project “Research and development in medical sciences—the way to personalize the treatment of serious neurological, cardiovascular, and cancer diseases” (code: ITMS: 313011T431), co-financed by the resources of the European Regional Development Fund, for the project “Long-term strategic research of prevention, intervention, and mechanisms of obesity and its comorbidities”, NFP313010V344, and finally for the project SRDA-20-0413.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang M., Earley M., Chen L., Hanson B.M., Yu Y., Liu Z., Salcedo S., Cober E., Li L., Kanj S.S., et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): A prospective, multicentre, cohort study. Lancet Infect. Dis. 2022;22:401–412. doi: 10.1016/S1473-3099(21)00399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray J., Oppenheim B., Mahida N. The Journal of Hospital Infection—A history of infection prevention and control in 100 volumes. J. Hosp. Infect. 2018;100:1–8. doi: 10.1016/j.jhin.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Ben-David D., Masarwa S., Fallach N., Temkin E., Solter E., Carmeli Y., Schwaber M.J. Success of a National Intervention in Controlling Carbapenem-resistant Enterobacteriaceae in Israel’s Long-term Care Facilities. Clin. Infect. Dis. 2019;68:964–971. doi: 10.1093/cid/ciy572. [DOI] [PubMed] [Google Scholar]
- 4.Logan L.K., Robert A., Weinstein R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durante-Mangoni E., Andini R., Zampino R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 2019;25:943–950. doi: 10.1016/j.cmi.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Humphries R.M., Hindler J.A., Epson E., Horwich-Scholefield S., Miller L.G., Mendez J., Martinez J.B., Sinkowitz J., Sinkowtiz D., Hershey C., et al. Carbapenem-Resistant Enterobacteriaceae Detection Practices in California: What Are We Missing? Clin. Infect. Dis. 2018;66:1061–1067. doi: 10.1093/cid/cix942. [DOI] [PubMed] [Google Scholar]
- 7.Federico Perez F., Bonomo R.A. Evidence to improve the treatment of infections caused by carbapenem-resistant Gram-negative bacteria. Lancet Infect. Dis. 2018;18:358–360. doi: 10.1016/S1473-3099(18)30112-9. [DOI] [PubMed] [Google Scholar]
- 8.Kluytmans-van den Bergh M.F.Q., John W.A., Rossen J.W.A., Bruijning-Verhagen P.C.J., Bonten M.J.M., Friedrich A.W., Vandenbroucke-Grauls C.M.J.E., Willems R.J.L., Kluytmans J.A.J.W. Whole-Genome Multilocus Sequence Typing of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2016;54:2919–2927. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raro O.H.F., da Silva R.M.C., Filho E.M.R., Sukiennik T.C.T., Stadnik C., Dias C.A.G., Iglesias J.O., Pérez-Vázquez M. Carbapenemase-Producing Klebsiella pneumoniae From Transplanted Patients in Brazil: Phylogeny, Resistome, Virulome and Mobile Genetic Elements Harboring blaKPC- 2 or blaNDM- 1. Front. Microbiol. 2020;11:1563. doi: 10.3389/fmicb.2020.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccirilli A., Cherubini S., Azzini A.M., Tacconelli E., Lo Cascio G., Maccacaro L., Bazaj A., Naso L., Amicosante G., LTCF-Veneto Working Group et al. Whole-Genome Sequencing (WGS) of Carbapenem-Resistant, K. pneumoniae Isolated in Long-Term Care Facilities in the Northern Italian Region. Microorganisms. 2021;9:1985. doi: 10.3390/microorganisms9091985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J.H., Goldstein E.J.C., Wise J., Bilker W.B., Tolomeo P., Lautenbach E. Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae in a Network of Long-Term Acute Care Hospitals. Clin. Infect. Dis. 2017;64:839–844. doi: 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Righi E., Peri A.M., Harris P.N.A., Wailan A.M., Liborio M., Lane S.W., Paterson D.L. Global prevalence of carbapenem resistance in neutropenic patients and association with mortality and carbapenem use: Systematic review and meta-analysis. J. Antimicrob. Chemother. 2017;72:668–677. doi: 10.1093/jac/dkw459. [DOI] [PubMed] [Google Scholar]
- 13.David S., Reuter S., Harris S.R., Glasner C., Feltwell T., Argimon S., Abudahab K., Goater R., Giani T., Errico G., et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019;4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C., Dong N., Chan E.W.C., Chen S., Zhang R. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in China, 2016–2020. Lancet Infect. Dis. 2022;22:167–168. doi: 10.1016/S1473-3099(22)00009-3. [DOI] [PubMed] [Google Scholar]
- 15.Arena F., Vannetti F., Di Pilato V., Fabbri L., Colavecchio O.L., Giani T., Marraccini C., Pupillo R., Macchi C., Converti F., et al. Diversity of the epidemiology of carbapenemase-producing Enterobacteriaceae in long-term acute care rehabilitation settings from an area of hyperendemicity, and evaluation of an intervention bundle. J. Hosp. Infect. 2018;100:29–34. doi: 10.1016/j.jhin.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 16.Gorrie C.L., Mirceta M., Wick R.R., Judd L.M., Wyres K.L., Thomson N.R., Strugnell R.A., Pratt N.F., Garlick J.S., Watson K.M., et al. Antimicrobial-Resistant Klebsiella pneumoniae Carriage and Infection in Specialized Geriatric Care Wards Linked to Acquisition in the Referring Hospital. Clin. Infect. Dis. 2018;67:161–170. doi: 10.1093/cid/ciy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vega S., Acosta F., Landires I., Morán M., Gonzalez J., Pimentel-Peralta G., Núñez-Samudio V., Goodridge A. Phenotypic and genotypic characteristics of carbapenemase—And extended spectrum β-lactamase-producing Klebsiella pneumoniae ozaenae clinical isolates within a hospital in Panama City. Ther. Adv. Infect. Dis. 2021;8:20499361211054918. doi: 10.1177/20499361211054918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perletti G., Magri V., Cai T., Stamatiou K., Trinchieri A., Montanari E. Resistance of uropathogens to antibacterial agents: Emerging threats, trends and treatments. Arch. Ital. Urol. Androl. 2018;90:85–96. doi: 10.4081/aiua.2018.2.85. [DOI] [PubMed] [Google Scholar]
- 19.Wyres K., Holt K. Regional differences in carbapenem-resistant Klebsiella pneumoniae. Lancet Infect. Dis. 2022;22:309–310. doi: 10.1016/S1473-3099(21)00425-4. [DOI] [PubMed] [Google Scholar]
- 20.Yin D., Zhang L., Wang A., He L., Cao Y., Hu F., Wang C. Clinical and molecular epidemiologic characteristics of carbapenem-resistant Klebsiella pneumoniae infection/colonization among neonates in China. J. Hosp. Infect. 2018;100:21–28. doi: 10.1016/j.jhin.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Da Silva Y., Ferrari R., Marin V.A., Junior C.A.C. A Global Overview of β-lactam Resistance Genes in Klebsiella pneumonia. Open Infect. Dis. J. 2019;11:22–34. doi: 10.2174/1874279301911010022. [DOI] [Google Scholar]
- 22.Ippolito M., Misseri G., Catalisano G., Marino C., Ingoglia G., Alessi M., Consiglio E., Gregoretti C., Giarratano A., Cortegiani A. Ventilator-Associated Pneumonia in Patients with COVID-19: A Systematic Review and Meta-Analysis. Antibiotics. 2021;10:545. doi: 10.3390/antibiotics10050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mędrzycka-Dąbrowska W., Lange S., Zorena K., Dąbrowski S., Ozga D., Tomaszek L. Carbapenem-Resistant Klebsiella pneumoniae Infections in ICU COVID-19 Patients-A Scoping Review. J. Clin. Med. 2021;10:2067. doi: 10.3390/jcm10102067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrosillo N., Taglietti F., Granata G. Treatment Options for Colistin Resistant Klebsiella pneumoniae: Present and Future. J. Clin. Med. 2019;8:934. doi: 10.3390/jcm8070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcone M., Daikos G.L., Tiseo G., Bassoulis D., Giordano C., Galfo V., Leonildi A., Tagliaferri E., Barnini S., Sani S., et al. Efficacy of Ceftazidime-avibactam Plus Aztreonam in Patients With Bloodstream Infections Caused by Metallo-β-lactamase-Producing Enterobacterales. Clin. Infect. Dis. 2021;72:1871–1878. doi: 10.1093/cid/ciaa586. [DOI] [PubMed] [Google Scholar]
- 26.Sheu C.C., Chang Y.T., Lin S.Y., Chen Y.H., Hsueh P.R. Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Front. Microbiol. 2019;10:80. doi: 10.3389/fmicb.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrissey I., Olesky M., Hawser S., Lob S.H., Karlowsky J.A., Corey G.R., Bassetti M., Fyfe C. In Vitro Activity of Eravacycline against Gram-Negative Bacilli Isolated in Clinical Laboratories Worldwide from 2013 to 2017. Antimicrob. Agents Chemother. 2020;64:e01699-19. doi: 10.1128/AAC.01699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mojica M.F., Rossi M.A., Vila A.J., Bonomo R.A. The urgent need for metallo-β-lactamase inhibitors: An unattended global threat. Lancet Infect. Dis. 2022;22:e28–e34. doi: 10.1016/S1473-3099(20)30868-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansone P., Giaccari L.G., Coppolino F., Aurilio C., Barbarisi A., Passavanti M.B., Pota V., Pace M.C. Cefiderocol for Carbapenem-Resistant Bacteria: Handle with Care! A Review of the Real-World Evidence. Antibiotics. 2022;11:904. doi: 10.3390/antibiotics11070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trajčíková E., Kurin E., Slobodníková L., Straka M., Lichváriková A., Dokupilová S., Čičová I., Nagy M., Mučaji P., Bittner Fialová S. Antimicrobial and Antioxidant Properties of Four Lycopus Taxa and an Interaction Study of Their Major Compounds. Molecules. 2020;25:1422. doi: 10.3390/molecules25061422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.EUCAST EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. [(accessed on 1 January 2022)]. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf.
- 32.Nordmann P., Poirel L., Dortet L. Rapid Detection of Carbapenemase-producing Enterobacteriaceae. Emerging Infect. Dis. 2012;18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gattringer R., Niks M., Ostertág R., Schwarz K., Medvedovic H., Graninger W., Georgopoulos A. Evaluation of MIDITECH automated colorimeter MIC reading for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2002;49:651–659. doi: 10.1093/jac/49.4.651. [DOI] [PubMed] [Google Scholar]
- 34.Prjibelski A.D., Antipov D., Meleshko D., Lapidus A.L., Korobeynikov A.I. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 35.Bialek-Davenet S., Criscuolo A., Ailloud F., Passet V., Jones L., Delannoy-Vieillard A.S., Garin B., Le Hello S., Arlet G., Nicolas-Chanoine M.H., et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z., Alikhan N.F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carriço J.A., Achtman M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.