Abstract
Multidrug-resistant (MDR) foodborne pathogens have created a great challenge to the supply and consumption of safe & healthy animal-source foods. The study was conducted to identify the common foodborne pathogens from animal-source foods & by-products with their antimicrobial drug susceptibility and resistance gene profile. The common foodborne pathogens Escherichia coli (E. coli), Salmonella, Streptococcus, Staphylococcus, and Campylobacter species were identified in livestock and poultry food products. The prevalence of foodborne pathogens was found higher in poultry food & by-product compared with livestock (p < 0.05). The antimicrobial drug susceptibility results revealed decreased susceptibility to penicillin, ampicillin, amoxicillin, levofloxacin, ciprofloxacin, tetracycline, neomycin, streptomycin, and sulfamethoxazole-trimethoprim whilst gentamicin was found comparatively more sensitive. Regardless of sources, the overall MDR pattern of E. coli, Salmonella, Staphylococcus, and Streptococcus were found to be 88.33%, 75%, 95%, and 100%, respectively. The genotypic resistance showed a prevalence of blaTEM, blaSHV, blaCMY, tetA, tetB, sul1, aadA1, aac(3)-IV, and ereA resistance genes. The phenotype and genotype resistance patterns of isolated pathogens from livestock and poultry had harmony and good concordance, and sul1 & tetA resistance genes had a higher prevalence. Good agricultural practices along with proper biosecurity may reduce the rampant use of antimicrobial drugs. In addition, proper handling, processing, storage, and transportation of foods may decline the spread of MDR foodborne pathogens in the food chain.
Keywords: AMR, MDR bacteria, foodborne bacteria, animal origin food, Bangladesh
1. Introduction
Foodborne illness is a key public health concern worldwide which occurs due to the ingestion of contaminated food products, mostly animal and poultry-derived food [1,2,3]. The prime cause of foodborne infections is the presence of bacteria in foods which will grow under favorable conditions and produce toxins in food [4]. Currently, foodborne illness caused by bacterial contamination is one of the foremost threats distressing public health [5]. Staphylococcus aureus, Salmonella paratyphoid, Campylobacter, Listeria monocytogenes (L. monocytogenes), and Escherichia coli (E. coli) are the most important zoonotic bacterial pathogens that cause foodborne illness worldwide, and deaths due to consumption of contaminated animal products [6]. In terms of biological threats, bacterial agents are the utmost severe concern regarding the issues of the supply of pathogen-free animal-derived foods to consumers [7,8]. Animal-derived food products, most commonly red meat, white meat including dairy products, and eggs are the important vehicles through which people may be exposed to foodborne pathogens, especially bacteria [9]. In addition, the poultry vendors at wet markets had poor knowledge of food safety, foodborne pathogens, and zoonoses in Bangladesh is also a key factor for transmitting foodborne pathogens [10].
Animal-source foods especially milk, meat, eggs, and their diversified products may become exposed with pathogenic bacteria during harvesting, slaughtering, processing, and marketing [11,12]. Environmental factors such as the nature of pathogens, their hosts (human or animal), host immunity, environmental temperature, etc. have evolved food-derived bacterial pathogens, making the population more susceptible to infection [13]. With rapidly changing human consumption habits, global food markets, and climate change, the fight against bacterial foodborne illness faces new challenges [14]. In this regard, foodborne bacterial infections may be prohibited and controlled by the proper cooking, preparation, and storage of food. In addition, multi-sectoral action is urgently needed to address AMR and to achieve the Sustainable Development Goals (SDGs) [15]. The World Health Organization (WHO) lists AMR as one of the top ten public health threats in the world. The unnecessary use of antibiotics in humans and agriculture has resulted in the widespread growth of antibiotic-resistant strains, and their outbreak in the environment has caused serious health hazards [16,17,18]. The Centers for Disease Control and Prevention (CDC) states that more than 2.8 million people in the United States develop serious infections with antibiotic-resistant bacteria each year, and at least 35,000 die each year from the direct effects of these resistant pathogens [19]. It is evidenced that AMR foodborne pathogens including E. coli, Campylobacter, Listeria, and Salmonella are closely linked with chicken meat, beef, and pork [20,21], as well as retail meat [22,23].
Extensive use of antibiotics in livestock and poultry production systems is known to contribute to the development of AMR [24]. To date, there is a growing concern about the potential for AMR to be transmitted through the food chain. AMR food pathogens in food-producing animals may infect humans through consumption of contaminated food or water as well as direct contact with animals [25]. In addition, bacteria in foods that are AMR may be more persistent in food processing environments. In this regard, monitoring antimicrobial resistance in the food chain is important for understanding the spread of resistance and making relevant risk assessment data. Previous studies showed the occurrences of foodborne pathogens in poultry meat collected from different sources and selected areas of Bangladesh [26,27,28]. However, no comprehensive research work was carried out on the determination of the antimicrobial drug susceptibility profile of common foodborne pathogens recovered from both livestock and poultry food products and by-products throughout the country. Therefore, considering the paramount importance of antimicrobial resistance globally, the present study was undertaken to isolate and identify foodborne pathogens along with the characterization of their antimicrobial drug susceptibility patterns with resistance genes in animal-derived food & by-product in Bangladesh. The study would help in addressing containment and intervention strategies of MDR foodborne pathogens in the food chain.
2. Results
2.1. Prevalence of AMR Pathogens in Animal-Derived Food Products
E. coli, Salmonella, and Staphylococcus species were isolated from both livestock and poultry food product & by-product samples throughout the country. In addition, Streptococcus species were isolated from only livestock-source food product & by-product samples. Furthermore, Campylobacter species was only isolated from poultry food products of the Chattogram Veterinary and Animal Sciences University (CVASU) component, i.e., from the Chittagong division. The overall prevalence of E. coli, Salmonella, Staphylococcus, and Streptococcus in livestock-source food products and by-products were found to be 38.47% (554/1440), 8.26% (119/1440), 14.67% (211/1440), and 4.79% (69/1440), respectively (details of prevalence pattern are presented in Table 1). Similarly, the overall prevalence of E. coli, Salmonella, Staphylococcus, and Campylobacter in poultry food products & by-products were 51.59% (454/880), 17.61% (155/880), 21.93% (193/880), and 4.43% (39/880), respectively (Table 2). Regardless of sources, the overall multidrug-resistant pattern of E. coli, Salmonella, Staphylococcus, and Streptococcus were found to be 88.33%, 75%, 95%, and 100%, respectively. The prevalence of E. coli was found higher in poultry food products & by-products compared with livestock (51.59% vs. 38.47%; p < 0.05). Similarly, a higher statistical association was found between the prevalence of Salmonella in livestock and poultry food products (17.61% vs. 8.26%; p < 0.05). In addition, an association was also found between the prevalence of Staphylococcus in livestock and poultry samples (21.93% vs. 14.67%; p < 0.05). On the other hand, Campylobacter was found in poultry food products & by-products collected from only the Chittagong division and the overall percentage was 4.43% (39/880).
Table 1.
Prevalence of AMR pathogens in livestock-source food products and by-products.
| Organisms | Positive Samples (Total Samples) |
Prevalence (%) | Confidence Interval (95% Cl) |
|---|---|---|---|
| E. coli | 554 (1440) | 38.47 | 35.99–41.01 |
| Salmonella | 119 (1440) | 8.26 | 6.95–9.80 |
| Staphylococcus | 211 (1440) | 14.65 | 12.92–16.57 |
| Streptococcus | 69 (1440) | 4.79 | 3.80–6.02 |
Table 2.
Prevalence of AMR pathogen in poultry source food products and by-products.
| Organisms | Positive Samples (Total Samples) |
Prevalence (%) | Confidence Interval (95% Cl) |
|---|---|---|---|
| E. coli | 454 (880) | 51.59 | 48.29–54.88 |
| Salmonella | 155 (880) | 17.61 | 15.24–20.27 |
| Staphylococcus | 193 (880) | 21.93 | 19.32–24.78 |
| Campylobacter | 39 (880) | 4.43 | 3.26–6 |
2.2. Phenotypic Resistance Pattern
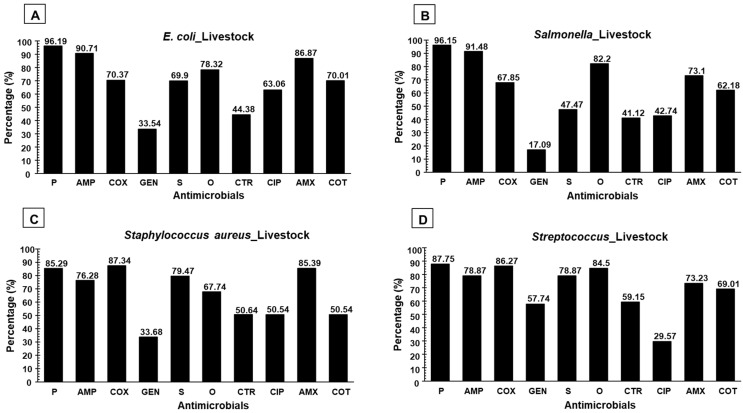
The AST (Antimicrobial Sensitivity Testing) pattern of E. coli showed the highest resistance to penicillin (P) (96.19%) followed by ampicillin (AMP) (90.71%), amoxicillin (AMX) (86.87%), oxytetracycline (O) (78.32%), cloxacillin (COX) (70.37%), and sulfamethoxazole-trimethoprim (COT) (70.01%). Among the antimicrobials, gentamicin (GEN) (66.46%) was found to be the most susceptible. The detailed AST pattern of E. coli is presented in Figure 1. The AST result of Salmonella showed the highest resistance to penicillin (96.15%), followed by ampicillin (AMP) (91.48%), oxytetracycline (O) (82.2%), amoxicillin (73.1%), and cloxacillin (67.85%) whilst the highest susceptibility was recorded to gentamicin (82.91%), followed by ceftriaxone (CTR) (58.88%). The detailed AST pattern of Salmonella is presented in Figure 1. On the other hand, the AST pattern of Staphylococcus showed the highest resistance to cloxacillin (87.345), followed by amoxicillin (85.39%), penicillin (85.29%), ampicillin (76.28%), streptomycin (S) (79.47%), and oxytetracycline (67.74%) whereas the highest susceptibility was found in gentamicin (66.32%), followed by ciprofloxacin (CIP) (49.46%) and ceftriaxone (49.36%). The detailed AST pattern of Staphylococcus is presented in Figure 1. Moreover, the AST pattern of Streptococcus indicated greater resistance to penicillin, followed by cloxacillin (86.27%), oxytetracycline (84.5%), streptomycin (78.87%), amoxicillin (73.23%), and sulfamethoxazole-trimethoprim (69.01%) whilst greater susceptibility was observed to ciprofloxacin (70.43%) and gentamicin (42.26%). The detailed AST pattern of Streptococcus is shown in Figure 1.
Figure 1.
Phenotypic resistance pattern to various antimicrobial agents, AST pattern of (A) E. coli, (B) Salmonella, (C) Staphylococcus, and (D) Streptococcus recovered from livestock food products & by-products.
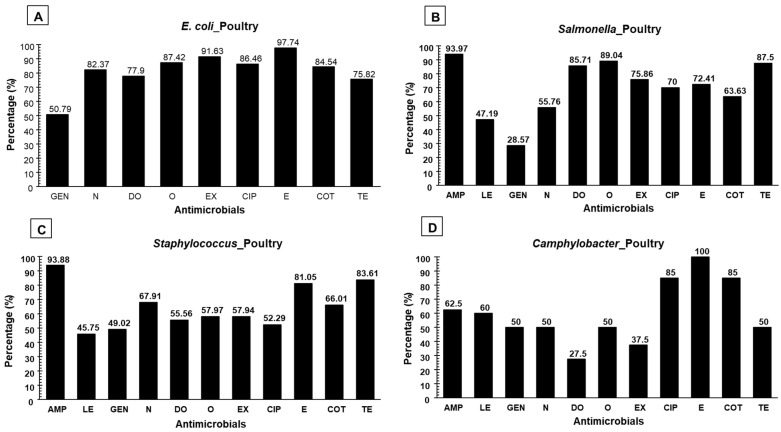
The AST result of E. coli showed higher resistance to erythromycin (97.74%), followed by enrofloxacin (EX) (91.63%), oxytetracycline (87.42%), ciprofloxacin (86.46%), sulfamethoxazole-trimethoprim (84.54%), and tetracycline (TE) (75.82%); however, the highest sensitivity was found to gentamicin (49.21%). The detailed AST pattern of E. coli is shown in Figure 2. The AST result of Salmonella showed higher resistance to ampicillin (93.97%), followed by oxytetracycline (89.04%), tetracycline (87.5%), doxycycline (DO) (85.71%), enrofloxacin (75.86%), erythromycin (E) (72.41%), ciprofloxacin (70%), and sulfamethoxazole-trimethoprim (63.63%); however, the highest sensitivity was found to gentamicin (71.43%) and levofloxacin (LE) (52.81%). The detailed AST pattern of Salmonella is outlined in Figure 2. Consecutively, the AST pattern of Staphylococcus predicted higher resistance to ampicillin (93.88%), followed by tetracycline (83.61%), erythromycin (81.05%), neomycin (N) (67.91%), sulfamethoxazole-trimethoprim (63.63%), oxytetracycline (57.97%), enrofloxacin (57.97%), and doxycycline (55.56%); however, the highest sensitivity was found to gentamicin (49.02%) and levofloxacin (45.75%). The detailed AST pattern of Staphylococcus is presented in Figure 2. Furthermore, the AST pattern of Campylobacter species anticipated higher resistance to erythromycin (100%), followed by ciprofloxacin (85%), sulfamethoxazole-trimethoprim (85%), ampicillin (62.5%), and levofloxacin (60%), whilst higher sensitivity was found to doxycycline (72.5%), oxytetracycline (50%), gentamicin (50%), and neomycin (50%). The AST pattern of Campylobacter species is presented in Figure 2.
Figure 2.
Phenotypic resistance pattern to various antimicrobial agents, AST pattern of (A) E. coli, (B) Salmonella, (C) Staphylococcus, and (D) Campylobacter recovered from poultry food products & by-products.
2.3. Genotypic Resistance Pattern of the Isolates Recovered from Both Livestock and Poultry Food Products & By-Products
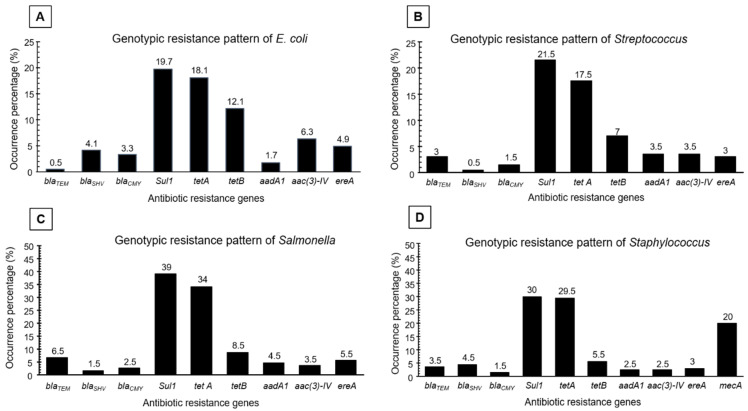
The genotypic resistance pattern of the isolates showed resistance to ESBL (blaTEM, blaSHV, and blaCMY), tetracycline (tetA and tetB), sulfonamide (sul1), streptomycin (aadA1), gentamicin (aac(3)-IV), and erythromycin (ereA). Similar genotypic trends, with some deviation, were found among the isolated foodborne pathogens of different species. Regardless of sources, all foodborne pathogens possessed antibiotic-resistant genes (blaTEM, blaSHV, blaCMY, tetA, tetB, aadA1, aac (3)-IV, and ereA) with different percentages.
The prevalence of antibiotic-resistant genes sul1, tetA, tetB, aac(3)-IV, ereA, blaSHV, blaCMY, aadA1, and blaTEM was found to be 19.7%, 18.1%, 12.1%, 6.3%, 4.9%, 4.1%, 3.3%, 1.7%, and 0.5%, respectively in the E. coil isolates. The prevalence of antibiotic-resistant genes sul1, tetA, tetB, aadA1, aac(3)-IV, ereA, blaTEM, blaCMY, and blaSHV was found to be 21.5%, 17.5%, 7%, 3.5%, 3.5%, 3%, 3%, 1.5%, and 0.5%, respectively in the Streptococcus isolates. The prevalence of antibiotic-resistant genes sul1, tetA, tetB, blaTEM, ereA, aadA1, aac(3)-IV, blaCMY, and blaSHV was found to be 39%, 34%, 8.5%, 6.5%, 5.5%, 4.5%, 3.5%, 2.5%, and 1.5%, respectively in the Salmonella isolates. The prevalence of antibiotic-resistant genes sul1, tetA, mecA, tetB, blaSHV, blaTEM, ereA, aadA1, aac(3)-IV, and blaCMY was found to be 30%, 29.5%, 20%, 5.5%, 4.5%, 3.5%, 3%, 2.5%, 2.5%, and 1.5%, respectively in the Staphylococcus isolates. The sul1 and tetA gene was found in higher percentages among the foodborne pathogens compared with other genotypic resistance genes regardless of the sources of collection of pathogens and the type of pathogens (Figure 3).
Figure 3.
Genotypic resistance pattern of (A) E. coli, (B) Streptococcus, (C) Salmonella, and (D) Staphylococcus recovered from both livestock and poultry food products & by-products.
There was a similar trend in the phenotypic resistance patterns of foodborne pathogens. The phenotypic resistance was found to be comparatively higher in the foodborne pathogens isolated from poultry than livestock-source foods and by-products (p < 0.05). The phenotypic and genotypic resistance profiles of various isolates of foodborne pathogens were shown to have a narrower range of variation and variability. The phenotypic and genotypic resistance results indicated that multidrug-resistant and ESBL-producing foodborne pathogens were prevailing in the livestock- and poultry-source food products & by-products in Bangladesh.
3. Discussion
Antimicrobials are frequently used to treat infectious diseases in both humans and animals [29]. Recently, the overuse and misuse of antibiotics in livestock have become a great concern for public health authorities. Contrarily, because withdrawal periods before harvesting or marketing livestock products have been ignored, antibiotic residues are now another rising concern to public health [30]. The main consequence of antibiotic residues in animal-derived foods is the enhancement of the development of antimicrobial resistance. The presence of antibiotic-resistant foodborne pathogens in food may lead to gastrointestinal disorders in human beings [31]. On the other hand, antibiotic-resistant pathogens may transfer the gene to other microorganisms through vertical and horizontal transmission [29,32] resulting in the spread of AMR pathogens. Several previous studies have shown the emergence of multi-resistant bacterial pathogens from a wide variety of sources in the food chain, increasing the need for proper use of antibiotics in both the veterinary and human health sectors [33,34,35]. MDR pathogens may cause difficult-to-treat illnesses, increased mortality, and financial burden. Furthermore, infections caused by MDR pathogens are considered a major global public health crisis by the World Health Organization, as the discovery of effective antibiotics has not kept pace with the increase in bacterial antibiotic resistance [36]. The demand for high-value animal products such as milk, meat, and eggs has increased due to economic solvency, rapid urbanization, and changing food habits of nations [37]. Foodborne pathogens can enter into the food cycle during production, processing, and marketing. Humans can get antibiotic-resistant bacterial infections in many ways, including ingestion of contaminated food or contact with colonized or diseased animals, body fluids, excretions, or secretions [38]. In addition, the pathogens can cause illness due to the consumption of undercooked food and produce illnesses either by their presence or by-production of toxins, or both.
The important foodborne pathogens of animal-source foods that have been globally identified are Salmonella, Campylobacter, E. coli, and Staphylococcus [20,22], and these trends are apparent in the current study in a similar fashion. Per the previous reports from home and abroad [39,40,41,42,43,44,45], the AST results of E. coli isolated from livestock and poultry showed a wider range of resistance to penicillin (100%), tetracycline (72–100%), oxytetracycline (78–93%), sulfamethoxazole-trimethoprim (51–88%), ampicillin (89.5–100%), amoxicillin (92–95%), streptomycin (19–70%), erythromycin (89%), ciprofloxacin (50%), chloramphenicol (43–50%), gentamicin (8–28%), enrofloxacin (55%), and norfloxacin (50%). In contrast, the phenotypic resistance pattern of E. coli to various antimicrobial agents recovered from both livestock and poultry in the present study also showed a similar trend where the AST pattern of E. coli showed the highest resistance to penicillin, followed by ampicillin, amoxicillin, oxytetracycline, cloxacillin, and sulfamethoxazole-trimethoprim. Among the antibiotics, gentamicin possessed the lowest resistance percentage, which is also comparable to the previous studies stated above. E. coli is one of the major pathogenic microorganisms that may reach animal-derived foods and is an indication of contamination by manure, soil, and contaminated water [46]. E. coli are commensal bacteria, and E. coli pathotypes can cause zoonotic disease that poses a public health risk [47]. In addition, Shiga toxin-producing E. coli is associated with the development of several life-threatening infections in humans [48]. In this regard, our recently published data showed that the most common class of antimicrobials used in large animal farms were Penicillin (61.79%), Oxytetracycline (55.66%), Sulfa drug (55.66%), Streptomycin (54.72%), followed by Ciprofloxacin (51.89%), Gentamicin (43.13%), and Ceftriaxone (34.91%) [30]. These data indicate that the bacteria became more resistant to such most commonly used antimicrobials in the study area.
Similarly, the AST result of Salmonella recovered from poultry revealed a wider range of resistance to ciprofloxacin (70–88%), ampicillin (66–75%), tetracycline (77–84%), gentamicin (33–68%), nalidixic acid (22–60%), and streptomycin (44–77%) [49]. While lower resistance (5–8%) was observed to chloramphenicol, azithromycin, imipenem, amikacin, and sulfamethoxazole-trimethoprim [49]. In contrast, in our present study, the AST result of Salmonella showed the highest resistance to penicillin, followed by ampicillin, oxytetracylcine, amoxicillin, and cloxacillin whilst the highest susceptibility was recorded to gentamicin, followed by ceftriaxone. Salmonella is widespread in nature [50] and is the most important pathogenic bacterium in both humans and animals [51]. Bacterial pathogens are more commonly found during outbreaks of foodborne disease [52] and are accountable for around 93.8 million foodborne illnesses and 155,000 fatalities annually worldwide [53].
According to the previous study reports, the AST result of Streptococcus in livestock and poultry showed a wider degree of resistance to streptomycin (70–100%), amoxicillin (30–100%), and ampicillin (60–100%) [39]. Sequentially, the AST result of Staphylococcus revealed a broader range of resistance to penicillin (82–100%), amoxicillin (42–100%), ampicillin (97%), streptomycin (70–100%), oxytetracycline (74–78%), ciprofloxacin (17–50%), sulfamethoxazole-trimethoprim (30%), gentamicin (18%) cefixime (73.9%), cloxacillin (82.6%), and oxacillin (56–98%) [44,54,55]. In contrast, the present study results from the AST pattern of Streptococcus indicated a greater resistance to penicillin, followed by cloxacillin, oxytetracycline, streptomycin, amoxicillin, and sulfamethoxazole-trimethoprim whilst greater susceptibility was observed to ciprofloxacin and gentamicin. The data from the present study suggested that gentamicin was found to be susceptible to isolated common foodborne pathogens.
Staphylococcus are commensal bacteria that are normal inhabitants of the skin, nose, and mucous membranes of healthy people and animals [56,57]. However, it is also known as an opportunistic foodborne pathogen [58] that may cause several infectious diseases with different degrees of severity [56]. It causes numerous infections in humans and animals [59]. The presence of S. aureus in food products is an alarming and serious threat to public health in terms of food safety when it releases toxins and causes illness [60]. Methicillin-resistant S. aureus (MRSA) has emerged due to the unnecessary use of antibiotics [61,62]. The presence of MRSA in farm animals and the potential for cross-contamination in humans have been a great concern [63].
Campylobacter are the leading cause of foodborne diarrhea in humans worldwide [64], which is mainly due to contamination of food of animal origin [65]. Campylobacter can colonize in warm-blooded animals and poultry [66]. The zoonotic nature of Campylobacter species makes it clinically and economically important worldwide [67]. It has accounted for 15% of food-related illnesses leading to hospital admissions and 6% resulting in death; about 400 million cases are reported each year due to foodborne infection [68,69]. The economic impact of Campylobacter infections is mainly related to the treatment cost, production loss, and pathogen control expenses [67].
The ESBL-producing foodborne pathogens were previously identified by researchers from a variety of sources of livestock and poultry [70,71,72]. More recent studies have shown that ESBLs-producing bacteria frequently colonized in poultry [73,74,75] and cattle [76,77]. On the other hand, tetracycline-resistant genes are commonly encoded by plasmids & transposons and are transmitted by conjugation. However, in some isolates, the corresponding gene is also found on the chromosome [78,79]. Mechanisms of resistance to tetracycline by the acquisition of the tet gene primarily include efflux pumping, ribosome protection, and enzymatic inactivation. The resistance of gram-negative bacteria to sulfonamides is associated with the presence of the sul gene, which encodes dihydropteroate synthase in a manner that the drug cannot inhibit. The sul gene has already been identified in Enterobacteriaceae, especially in the genera Escherichia and Salmonella [80]. In this regard, the present study finding showed that the sul1 and tetA genes were found in higher percentages among the foodborne pathogens compared with other resistance genes regardless of the sources of isolation of the pathogen and the type of pathogens.
To the best of our knowledge, for the first time, this study was conducted throughout the country and found that multidrug-resistant and ESBL-producing foodborne pathogens were prevailing in the livestock- and poultry-source food products & by-products in Bangladesh. However, the present study has some limitations. In this study, we did not collect the environmental samples which may be contaminated by the livestock- and poultry-source food products and by-products. In addition, the sampling area was limited in each division. Further detailed studies with larger samples size from each district of Bangladesh are needed. Details of further phenotypic & genotypic analysis in a wider range with 16S rRNA sequence profiling of these isolates would help the scientists in this field to combat AMR as well as to stop the spreading of MDR foodborne pathogens to humans.
4. Materials and Methods
4.1. Study Area
The study was conducted in thirty-three districts under the eight administrative divisions of Bangladesh. The seven components (educational institutes) of the project covered each division with at least four districts while the component (educational institute) Patuakhali Science and Technology University covered two divisions with nine districts. The seven components (educational institutes) of the project are the Bangladesh Agricultural University (BAU), Bangladesh Livestock Research Institute (BLRI), Rajshahi University (RU), Patuakhali Science and Technology University (PSTU), Chattogram Veterinary and Animal Sciences University (CVASU), Sylhet Agricultural University (SAU), and Haji Danesh Science and Technology University (HSTU). Figure 4 shows the study divisions followed by districts area covered by the seven components (educational institutes). The study protocol was authorized by the Animal Welfare and Experimentation Ethics Committee of the Bangladesh Agricultural University, Mymensingh, (approval number: AWEEC/BAU/2018(17)).
Figure 4.
AMR surveillance study areas covered by seven components (educational institutes).
4.2. Sampling Design
A total of 2320 samples were collected across the country, of which 880 were taken from poultry and 1440 from large & small ruminants in thirty-three districts of eight divisions of Bangladesh by the seven components of the project. Poultry source samples were broiler meat (n = 160), layer meat (n = 160), egg (n = 240), broiler feces (n = 160), and layer feces (n = 160). All poultry sources samples were directly collected from layer and broiler farms. On the other hand, large & small ruminant samples were cattle meat (n = 160), sheep or goat meat (n = 160), buffalo meat (n = 80), cattle raw milk (n = 200), sheep or goat raw milk (n = 200), buffalo raw milk (n = 120), cattle feces (n = 200), sheep or goat feces (n = 200), and buffalo feces (n = 80). Different types of meat samples were collected for local raw meat markets; however, raw milk and feces from different animals were collected from farms. All the samples were collected in aseptic condition using sterile instruments and carefully transferred into sterile Eppendorf tubes (for raw milk) or zipper bags (for solid samples) from animal and poultry farms. Immediately after collection, samples were kept in a transport box for maintaining a 4 °C temperature. The samples were then transported to the bacteriological laboratory of the Department of Microbiology and Hygiene, Faculty of Veterinary Science, BAU, Mymensingh for microbiological analysis. According to our previous study [30], the Raosoft sample volume calculation method was used to determine the sample size with a 5% margin of error and 95% confidence level.
4.3. Conventional Culture Method
E. coli, Salmonella, Streptococcus, and Staphylococcus were targeted for the isolation from livestock whilst E. coli, Salmonella, Staphylococcus, and Campylobacter were targeted for the isolation from poultry sources following the standard procedure as applied earlier [44,45,55,81,82,83,84,85]. Briefly, 0.5 g of each sample was inoculated in nutrient broth and incubated at 37 °C for 12 h for the initial growth of Escherichia coli, Salmonella, and Staphylococcus aureus. The cultures from nutrient broth were streaked on Eosin methylene blue (EMB) agar (Hi media, Maharashtra, India), Salmonella-Shigella (SS) agar (Hi media, Maharashtra, India), and Mannitol salt agar (MSA) (Hi media, India) plates using platinum loop for the isolation of E. coli, Salmonella, and S. aureus, respectively. Milk samples (200 µL) were also inoculated in Kenner Fecal (KF) Streptococcal broth (Hi media, India) for the initial growth of Streptococcus, then streaked on KF Streptococcal agar media for the isolation of Streptococcus. Then all the culture plates were incubated at 37 °C for 24 h. For the isolation of Campylobacter, all samples were directly inoculated on selective campylobacter base agar (Oxoid Ltd., Hampshire RG24 8PW, UK) containing antibiotics (Amphotericin B has been added to suppress the growth of yeast and fungal contaminants that may occur at 37 °C, and improved selectivity was achieved by adding cefoperazone) and 5–7% sheep blood [86]. The plates were incubated in an anaerobic jar (Oxoid™ AnaeroJar™ 2.5 L) under microaerophilic conditions with a CO2 sachet (Thermo Scientific TM Oxoid Anaero Gen 2.5 L sachet) (10% CO2, 95% humidity) at 42 °C for three days. After 72 h, single characteristic (small, round, creamy-gray, or whitish) colonies from each plate were selected and inoculated in tryptic soy broth (Oxoid Ltd., Hampshire RG24 8PW, UK) and incubated at 37 °C for three days under microaerophilic conditions. The single colonies of suspected bacteria were again inoculated in selective broth and streaked on selective agar media to obtain a pure culture.
4.4. Molecular Detection
Whole genomic DNA was extracted from each pure culture by a conventional boiling method [49,85]. Following the boiling method, the DNA was measured using spectrophotometers. PCR was performed for confirmatory detection of each isolate using specific primers (Table 3) and the PCR condition for each bacterial species was used following the standard operating procedure described by different authors (Table 3).
Table 3.
List of primers used for bacterial species identification.
| AMR Pathogens | Primers | Sequence (5′–3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| Salmonella | F | TCATCGCACCGTCAAAGGAACC | 284 | [87] |
| R | GTGAAATTATCGCCACGTTCGGGCAA | |||
| E. coli | F | CCCCCTGGACGAAGACTGAC | 401 | [88] |
| R | ACCGCTGGCAACAAAGGATA | |||
| Staphylococcus | F | CCTGAAACAAAGCATCCTAAAAA | 155 | [89] |
| R | TAAATATACGCTAAGCCACGTCCAT | |||
| Campylobacter | F | ATCTAATGGCTTAACCATTAAAC | 857 | [90] |
| R | GGACGGTAACTAGTTTAGTATT | |||
| Streptococcus | F | AGCGGGGGATAACTATTGGA | 569 | [84] |
| R | TACGCATTTCACCGCTACAC |
4.5. Determination of Phenotypic Resistance Pattern
Antimicrobial susceptibility testing was performed using the Kirby Bauer disk diffusion method in adherence with the guidelines of the Clinical and Laboratory Standards Institute [91]. Briefly, fresh colonies were suspended in saline and the turbidity of the suspension was measured in comparison with the 0.5 McFarland standards (approximately 1.5 × 106 CFU/mL). The bacterial suspension was smeared on the surface of Mueller-Hinton (MH) agar (Oxoid Ltd., Hampshire RG24 8PW, UK) and an antibacterial disc with a disc dispenser was placed on it within 15 min and the plate was incubated at 37 °C for 24 h. The zone of inhibition adjacent to the disks was measured and compared with the breakpoints of CLSI. A number of 16 different antimicrobial disks (Hi media, India) were used for AST of all four foodborne pathogens such as penicillin (P, 10 units), ampicillin (AMP, 10 μg), amoxicillin (AMX, 30 μg), cloxacillin (COX, 5 μg), ceftriaxone (CTR, 30 μg), tetracycline (TE, 30 μg), doxycycline (DO, 30 μg), oxytetracycline (O, 30 μg), sulfamethoxazole-trimethoprim (COT, 25 μg), gentamicin (GEN, 10 μg), erythromycin (E, 15 μg), ciprofloxacin (CIP, 5 μg), streptomycin (S, 10 μg), levofloxacin (LE, 5 μg), enrofloxacin (EX, 5 μg), and neomycin (N, 30 µg). Based on their common therapeutic usage at the field level in the study areas, different antimicrobial disks for livestock and poultry sources bacteria were chosen [30]. The criteria developed by Magiorakos et al. [92] were used to define multidrug-resistant (MDR) bacteria. Susceptible, intermediate, and resistant were defined according to the new DIN EN ISO 20776-1 standard [93], which is valid worldwide. The sensitivity of a bacterial strain to a given antibiotic is said to be intermediate when it is inhibited in vitro by a concentration of this drug that is associated with an uncertain therapeutic effect [94]. The Escherichia coli ATCC 25922 strain served as a validated positive control.
4.6. Determination of Genotypic Resistance Pattern
E. coli, Salmonella, Staphylococcus, Streptococcus, and Campylobacter isolates were screened by PCR for the detection of Extended Spectrum β-Lactamases (ESBL) genes (blaTEM, blaSHV, and blaCMY), tetracycline-resistant genes (tetA and tetB), sulfonamide-resistant gene (sul1), streptomycin-resistant gene (aadA1), gentamicin-resistant gene (aac(3)-II), neomycin-resistant gene (aph(3)-I), and erythromycin-resistant gene (ereA). The list of primers to detect resistance genes is given in Table 4.
Table 4.
List of primers used to detect resistant genes.
| Class | Target Gene | Primers | Sequence (5′–3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|
| Gentamicin | aac(3)-IV | F | CTTCAGGATGGCAAGTTGGT | 286 | [95] |
| R | TCATCTCGTTCTCCGCTCAT | ||||
| Tetracycline | tetA | F | GGTTCACTCGAACGACGTCA | 577 | [95] |
| R | CTGTCCGACAAGTTGCATGA | ||||
| tetB | F | CCTCAGCTTCTCAACGCGTG | 634 | ||
| R | GCACCTTGCTGATGACTCTT | ||||
| Beta lactams | blaTEM | F | ATAAAATTCTTGAAGAC | 1073 | [96] |
| R | TTACCAATGCTTAATCA | ||||
| Beta lactams | blaSHV | F | TCGCCTGTGTATTATCTCCC | 768 | [95] |
| R | CGCAGATAAATCACCACAATG | ||||
| Beta lactams | blaCMY | F | TGGCCAGAACTGACAGGCAAA | 462 | [95] |
| R | TTTCTCCTGAACGTGGCTGGC | ||||
| Erythromycin | ereA | F | GCCGGTGCTCATGAACTTGAG | 419 | [95] |
| R | CGACTCTATTCGATCAGAGGC | ||||
| Sulfonamide | sul1 | F | TTCGGCATTCTGAATCTCAC | 822 | [95] |
| R | ATGATCTAACCCTCGGTCTC | ||||
| Streptomycin | aadA1 | F | TATCCAGCTAAGCGCGAACT | 447 | [95] |
| R | ATTTGCCGACTACCTTGGTC | ||||
| Methicillin-resistant | mecA | F | AAAATCGATGGTAAAGGTTGGC | 533 | [97] |
| R | AGTTCTGGAGTACCGGATTTGC |
5. Conclusions
The phenotypic and genotypic resistance profiles uncovered by the present study indicated that MDR- and ESBL-producing foodborne pathogens were prevailing in the livestock- and poultry-source food products & by-products in Bangladesh. MDR foodborne pathogens are a current public health concern worldwide including in Bangladesh. Foodborne pathogens are usually spread due to improper handling, processing, preparation, and storage of food. The unnecessary use of antimicrobials in farm practices is the main driver for the emergence of antimicrobial-resistant pathogens in the livestock and poultry value chain. Good agricultural practices, good veterinary practices, good manufacturing practices, and proper farm biosecurity are important tools to curb the development of AMR pathogens in livestock and poultry food products. Moreover, policy intervention, stakeholder awareness, motivation, training, advocacy, and mass media dissemination are the imperative pathways to combat the spread of foodborne pathogens and AMR.
Acknowledgments
The authors appreciate the support of the Bangladesh Agricultural University (BAU), Bangladesh Livestock Research Institute, Chattogram Veterinary and Animal Sciences University, Rajshahi University, Sylhet Agricultural University, Patuakhali Science and Technology University, and Hajee Mohammad Danesh Science and Technology University to accomplish the research work. Finally, the authors would like to acknowledge the Bangladesh Agricultural University Research System (BAURES), BAU, Mymensingh for project management, monitoring, and evaluation for BAU component.
Author Contributions
K.R. and M.R.I. equally contributed to this work. Conceptualization and project administration by K.R., M.R.I., K.R., M.A.S., S.C., K.M.M.H., F.I.R. and M.K.H.; A.M.-E.-E. and M.T.H. performed all the experiments, prepared the draft of the manuscript; K.R. designed and supervised the research, revised and finalized the draft of the manuscript; N.A.S., K.R., M.Z.A., M.R. and M.T.H. performed the statistical analysis and prepared the graphs & tables; K.R., M.R.I., M.R.A., M.M., S.A. and N.A.R. revised the manuscript and prepare the graphs and tables. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study protocol was authorized by the Animal Welfare and Experimentation Ethics Committee of the Bangladesh Agricultural University, Mymensingh, (approval number: AWEEC/BAU/2018(17)).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have no competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Funding Statement
The research was funded by the Project Based Research Grant (PBRG) support from the National Agricultural Technology Project-2 (NATP-2), Bangladesh Agricultural Research Council (BARC), Dhaka, Bangladesh (Project ID-138).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Assefa A., Bihon A. A systematic review and meta-analysis of prevalence of Escherichia coli in foods of animal origin in Ethiopia. Heliyon. 2018;4:e00716. doi: 10.1016/j.heliyon.2018.e00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haile A.F., Kebede D., Wubshet A.K. Prevalence and antibiogram of Escherichia coli O157 isolated from bovine in Jimma, Ethiopia: Abattoirbased survey. Ethiop. Vet. J. 2017;21:109. doi: 10.4314/evj.v21i2.8. [DOI] [Google Scholar]
- 3.Tassew H., Abdissa A., Beyene G., Gebre-Selassie S. Microbial flora and food borne pathogens on minced meat and their susceptibility to antimicrobial agents. Ethiop. J. Health Sci. 2010;20:137–143. doi: 10.4314/ejhs.v20i3.69442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addis M., Sisay D. A review on major food borne bacterial illnesses. J. Trop. Dis. 2015;3:1–7. doi: 10.4176/2329891X.1000176. [DOI] [Google Scholar]
- 5.Wu S., Duan N., Gu H., Hao L., Ye H., Gong W., Wang Z. A review of the methods for detection of staphylococcus aureus enterotoxins. Toxins. 2016;8:176. doi: 10.3390/toxins8070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abebe E., Gugsa G., Ahmed M. Review on major food-borne zoonotic bacterial pathogens. J. Trop. Med. 2020;2020:4674235. doi: 10.1155/2020/4674235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelalem A., Sisay M., Vipham J.L., Abegaz K., Kebede A., Terefe Y. The prevalence and antimicrobial resistance profiles of bacterial isolates from meat and meat products in Ethiopia: A systematic review and meta-analysis. Int. J. Food Contam. 2019;6:1. doi: 10.1186/s40550-019-0071-z. [DOI] [Google Scholar]
- 8.Carrique-Mas J.J., Bryant J. A review of foodborne bacterial and parasitic zoonoses in Vietnam. EcoHealth. 2013;10:465–489. doi: 10.1007/s10393-013-0884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abunna F., Abriham T., Gizaw F., Beyene T., Feyisa A., Ayana D., Mamo B., Duguma R. Staphylococcus: Isolation, identification and antimicrobial resistance in dairy cattle farms, municipal abattoir and personnel in and around Asella, Ethiopia. J. Veter- Sci. Technol. 2016;7:1000383. doi: 10.4172/2157-7579.1000383. [DOI] [Google Scholar]
- 10.Alam Siddiky N., Khan S.R., Sarker S., Bhuiyan M.K.J., Mahmud A., Rahman T., Ahmed M.M., Samad M.A. Knowledge, attitude and practice of chicken vendors on food safety and foodborne pathogens at wet markets in Dhaka, Bangladesh. Food Control. 2022;131:108456. doi: 10.1016/j.foodcont.2021.108456. [DOI] [Google Scholar]
- 11.Mustafa A.S., Inanc A.L. Antibiotic resistance of Escherichia coli O157:H7 isolated from chicken meats. J. Agric. Nat. 2018;21:7–12. doi: 10.18016/ksudobil.289192.Makale. [DOI] [Google Scholar]
- 12.Tasbihullah T., Rahman S.U., Ali T., Saddique U., Ahmad S., Shafiq M., Ayaz S., Khan H., Ahmad I., Asadullah A., et al. High occurrence rate of multidrug-resistant ESBL-producing E. coli recovered from table eggs in district Peshawar, Pakistan. Pak. J. Zool. 2020;52:1231–1238. doi: 10.17582/journal.pjz/20190513220514. [DOI] [Google Scholar]
- 13.Hemalata V., Virupakshaiah D. Isolation and identification of food borne pathogens from spoiled food samples. Int. J. Curr. Microbiol. Appl. Sci. 2016;5:1017–1025. doi: 10.20546/ijcmas.2016.506.108. [DOI] [Google Scholar]
- 14.Faris G. Identification of campylobacter species and their antibiotic resistance patterns from raw bovine meat in Addis Ababa, Ethiopia. IJMIR. 2015;4:1–5. [Google Scholar]
- 15.Sustainable Development Goals (SDGs) AMR indicator Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. 2021. [(accessed on 1 October 2022)]. Available online: https://www.who.int/publications/i/item/9789240027336.
- 16.Alhashash F., Weston V., Diggle M., McNally A. Multidrug-resistant Escherichia coli bacteremia. Emerg. Infect. Dis. 2013;19:1699–1701. doi: 10.3201/eid1910.130309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornejo-Juárez P., Vilar-Compte D., Pérez-Jiménez C., Ñamendys-Silva S., Sandoval-Hernández S., Volkow-Fernández P. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int. J. Infect. Dis. 2015;31:31–34. doi: 10.1016/j.ijid.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Reinthaler F., Posch J., Feierl G., Wüst G., Haas D., Ruckenbauer G., Mascher F., Marth E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003;37:1685–1690. doi: 10.1016/S0043-1354(02)00569-9. [DOI] [PubMed] [Google Scholar]
- 19.Kwon J.H., Powderly W.G. The post-antibiotic era is here. Science. 2021;373:471. doi: 10.1126/science.abl5997. [DOI] [PubMed] [Google Scholar]
- 20.Mayrhofer S., Paulsen P., Smulders F.J., Hilbert F. Antimicrobial resistance profile of five major food-borne pathogens isolated from beef, pork and poultry. Int. J. Food Microbiol. 2004;97:23–29. doi: 10.1016/j.ijfoodmicro.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Shafiq M., Huang J., Rahman S.U., Shah J.M., Chen L., Gao Y., Wang M., Wang L. High incidence of multidrug-resistant Escherichia coli coharboring mcr-1 and blaCTX-M-15 recovered from pigs. Infect. Drug Resist. 2019;12:2135–2149. doi: 10.2147/IDR.S209473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uyttendaele M., De Troy P., Debevere J. Incidence of salmonella, campylobacter Jejuni, Campylobacter coli, and listeria monocytogenes in poultry carcasses and different types of poultry products for sale on the Belgian retail market. J. Food Prot. 1999;62:735–740. doi: 10.4315/0362-028X-62.7.735. [DOI] [PubMed] [Google Scholar]
- 23.Shafiq M., Huang J., Shah J.M., Ali I., Rahman S.U., Wang L. Characterization and resistant determinants linked to mobile elements of ESBL-producing and mcr-1-positive Escherichia coli recovered from the chicken origin. Microb. Pathog. 2021;150:104722. doi: 10.1016/j.micpath.2020.104722. [DOI] [PubMed] [Google Scholar]
- 24.Ferdous M.R.A., Ahmed R., Khan S.H., Mukta M.A., Anika T.T., Hossain T., Islam Z., Rafiq K. Effect of discriminate and indiscriminate use of oxytetracycline on residual status in broiler soft tissues. Vet. World. 2020;13:61–67. doi: 10.14202/vetworld.2020.61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oniciuc E.-A., Likotrafiti E., Alvarez-Molina A., Prieto M., López M., Alvarez-Ordóñez A. Food processing as a risk factor for antimicrobial resistance spread along the food chain. Curr. Opin. Food Sci. 2019;30:21–26. doi: 10.1016/j.cofs.2018.09.002. [DOI] [Google Scholar]
- 26.Uddin J., Hossain K., Hossain S., Saha K., Jubyda F.T., Haque R., Billah B., Talukder A.A., Parvez A.K., Dey S.K. Bacteriological assessments of foodborne pathogens in poultry meat at different super shops in Dhaka, Bangladesh. Ital. J. Food Saf. 2019;8:6720. doi: 10.4081/ijfs.2019.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam B., Uddin N., Mridha D., Akhter A.H.M.T., Islam S.K.S., Haque A.K.M.Z., Kabir S.M.L. Occurrence of Campylobacter spp. in selected small scale commercial broiler farms of Bangladesh related to good farm practices. Microorganisms. 2020;8:1778. doi: 10.3390/microorganisms8111778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoque M.N., Mohiuddin R.B., Khan MM H., Hannan A., Alam M.J. Outbreak of salmonella in poultry of Bangladesh and possible remedy. J. Adv. Biotechnol. Exp. Ther. 2019;2:87. doi: 10.5455/jabet.2019.d30. [DOI] [Google Scholar]
- 29.Granowitz E.V., Brown R.B. Antibiotic adverse reactions and drug interactions. Crit. Care Clin. 2008;24:421–442. doi: 10.1016/j.ccc.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Adzitey F. Incidence and antimicrobial susceptibility of Escherichia coli isolated from beef (meat muscle, liver and kidney) samples in Wa Abattoir, Ghana. Cogent Food Agric. 2020;6:1718269. doi: 10.1080/23311932.2020.1718269. [DOI] [Google Scholar]
- 31.Hossain T., Rafiq K., Islam Z., Chowdhury S., Islam P., Haque Z., Samad M.A., Sani A.A., Ferdous M.R.A., Islam R., et al. A survey on knowledge, attitude, and practices of large-animal farmers towards antimicrobial use, resistance, and residues in Mymensingh division of Bangladesh. Antibiotics. 2022;11:442. doi: 10.3390/antibiotics11040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Javadi A., Khatibi S.A. Effect of commercial probiotic (Protexin®) on growth, survival and microbial quality of shrimp (Litopenaeus vannamei) Nutr. Food Sci. 2017;47:204–216. doi: 10.1108/NFS-07-2016-0085. [DOI] [Google Scholar]
- 33.Abolghait S.K., Fathi A.G., Youssef F.M., Algammal A.M. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int. J. Food Microbiol. 2020;328:108669. doi: 10.1016/j.ijfoodmicro.2020.108669. [DOI] [PubMed] [Google Scholar]
- 34.Algammal A.M., Mabrok M., Sivaramasamy E., Youssef F.M., Atwa M.H., El-Kholy A.W., Hetta H.F., Hozzein W.N. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci. Rep. 2020;10:15961. doi: 10.1038/s41598-020-72264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enany M.E., Algammal A.M., Nasef S., Abo-Eillil S.A.M., Bin-Jumah M., Taha A.E., Allam A.A. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express. 2019;9:192. doi: 10.1186/s13568-019-0920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saraiva M.D.M.S., Lim K., Monte D.F.M.D., Givisiez P.E.N., Alves L.B.R., Neto O.C.D.F., Kariuki S., Júnior A.B., de Oliveira C.J.B., Gebreyes W.A. Antimicrobial resistance in the globalized food chain: A One Health perspective applied to the poultry industry. Braz. J. Microbiol. 2021;53:465–486. doi: 10.1007/s42770-021-00635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broglia A., Kapel C. Changing dietary habits in a changing world: Emerging drivers for the transmission of foodborne parasitic zoonoses. Vet. Parasitol. 2011;182:2–13. doi: 10.1016/j.vetpar.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Chang Q., Wang W., Regev-Yochay G., Lipsitch M., Hanage W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015;8:240–247. doi: 10.1111/eva.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Amin A., Hoque M.N., Siddiki A.Z., Saha S., Kamal M. Antimicrobial resistance situation in animal health of Bangladesh. Vet. World. 2020;13:2713–2727. doi: 10.14202/vetworld.2020.2713-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bag A.S., Khan S.R., Sami D.H., Begum F., Islam S., Rahman M., Rahman T., Hassan J. Virulence determinants and antimicrobial resistance of E. coli isolated from bovine clinical mastitis in some selected dairy farms of Bangladesh. Saudi J. Biol. Sci. 2021;28:6317–6323. doi: 10.1016/j.sjbs.2021.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bupasha Z.B., Begum R., Karmakar S., Akter R., Ahad A. Multidrug-resistant salmonella spp. isolated from apparently healthy pigeons in a live bird market in Chattogram, Bangladesh. World’s Vet. J. 2020;10:508–513. doi: 10.54203/scil.2020.wvj61. [DOI] [Google Scholar]
- 42.Das Gupta M., Islam M., Sen A., Sarker S., Das A. Prevalence and antibiotic susceptibility pattern of Escherichia coli in cattle on Bathan and intensive rearing system. Microbes Health. 2017;6:34062. doi: 10.3329/mh.v6i1.34062. [DOI] [Google Scholar]
- 43.Ievy S., Islam M.S., Sobur M.A., Talukder M., Rahman M.B., Khan M.F.R., Rahman M.T. Rahman molecular detection of avian pathogenic Escherichia coli (APEC) for the first time in layer farms in Bangladesh and their antibiotic resistance patterns. Microorganisms. 2020;8:1021. doi: 10.3390/microorganisms8071021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parvin M.S., Talukder S., Ali Y., Chowdhury E.H., Rahman T., Islam T. Antimicrobial resistance pattern of Escherichia coli isolated from frozen chicken meat in Bangladesh. Pathogens. 2020;9:420. doi: 10.3390/pathogens9060420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahman M., Husna A., Elshabrawy H.A., Alam J., Runa N.Y., Badruzzaman A.T.M., Banu N.A., Al Mamun M., Paul B., Das S., et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci. Rep. 2020;10:21999. doi: 10.1038/s41598-020-78367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Disassa N., Sibhat B., Mengistu S., Muktar Y., Belina D. Prevalence and antimicrobial susceptibility pattern of E. coli O157:H7 isolated from traditionally marketed raw cow milk in and around Asosa Town, Western Ethiopia. Vet. Med. Int. 2017;2017:7581531. doi: 10.1155/2017/7581531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messele Y.E., Abdi R.D., Tegegne D.T., Bora S.K., Babura M.D., Emeru B.A., Werid G.M. Analysis of milk-derived isolates of E. coli indicating drug resistance in central Ethiopia. Trop. Anim. Health Prod. 2018;51:661–667. doi: 10.1007/s11250-018-1737-x. [DOI] [PubMed] [Google Scholar]
- 48.Elmonir W., Abo-Remela E., Sobeih A. Public health risks of Escherichia coli and Staphylococcus aureus in raw bovine milk sold in informal markets in Egypt. J. Infect. Dev. Ctries. 2018;12:533–541. doi: 10.3855/jidc.9509. [DOI] [PubMed] [Google Scholar]
- 49.Siddiky N., Sarker S., Khan S.R., Begum R., Kabir E., Karim R., Rahman T., Mahmud A., Samad M. Virulence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from chicken at wet markets in Dhaka, Bangladesh. Microorganisms. 2021;9:952. doi: 10.3390/microorganisms9050952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemal J., Sibhat B., Menkir S., Terefe Y., Muktar Y. African journal of microbiology research antimicrobial resistance patterns of salmonella in Ethiopia: A review. Afr. J. Microbiol. Res. 2015;9:2249–2256. doi: 10.5897/AJMR2015. [DOI] [Google Scholar]
- 51.Addis Z., Kebede N., Sisay Z., Alemayehu H., Wubetie A., Kassa T. Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: A cross sectional study. BMC Infect. Dis. 2011;11:1–17. doi: 10.1186/1471-2334-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dhanalakshmi M., Balakrishnan S., Sangeetha A. Prevalence of Salmonella in chicken meat and its slaughtering place from local markets in Orathanadu, Thanjavur district, Tamil Nadu. J. Entomol. Zool. Stud. JEZS. 2018;6:2468–2471. [Google Scholar]
- 53.Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., International collaboration on enteric disease “burden of illness” studies The global burden of nontyphoidal salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 54.Hoque R., Ahmed S.M., Naher N., Islam M.A., Rousham E.K., Islam B.Z., Hassan S. Tackling antimicrobial resistance in Bangladesh: A scoping review of policy and practice in human, animal and environment sectors. PLoS ONE. 2020;15:e0227947. doi: 10.1371/journal.pone.0227947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jahan M., Rahman M., Parvej M.S., Chowdhury S.M.Z.H., Haque M.E., Talukder M.A.K., Ahmed S. Isolation and characterization of Staphylococcus aureus from raw cow milk in Bangladesh. J. Adv. Vet. Anim. Res. 2014;2:49–55. doi: 10.5455/javar.2015.b47. [DOI] [Google Scholar]
- 56.Lozano C., Gharsa H., Ben Slama K., Zarazaga M., Torres C. Staphylococcus aureus in animals and food: Methicillin resistance, prevalence and population structure. A review in the African continent. Microorganisms. 2016;4:12. doi: 10.3390/microorganisms4010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tessema D., Tsegaye S. Study on the prevalence and distribution of staphylococcus aureus in raw cow milk originated from Alage Atvet college dairy farm, Ethiopia. J. Nutr. Food Sci. 2017;7:2–5. doi: 10.4172/2155-9600.1000586. [DOI] [Google Scholar]
- 58.Wang B., Muir T.W. Regulation of virulence in Staphylococcus aureus: Molecular mechanisms and remaining puzzles. Cell Chem. Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haftay A., Geberemedhin H., Belay A., Goytom E., Kidane W. Antimicrobial resistance profile of Staphylococcus aureus isolated from raw cow milk and fresh fruit juice in Mekelle, Tigray, Ethiopia. J. Vet. Med. Anim. Health. 2018;10:106–113. doi: 10.5897/JVMAH2017.0664. [DOI] [Google Scholar]
- 60.Tsepo R., Ngoma L., Mwanza M., Ndou R. Prevalence and antibiotic resistance of staphylococcus aureus isolated from beef carcasses at abattoirs in north west province. J. Hum. Ecol. 2016;56:188–195. doi: 10.1080/09709274.2016.11907055. [DOI] [Google Scholar]
- 61.Adame-Gómez R., Toribio-Jimenez J., Vences-Velazquez A., Rodríguez-Bataz E., Dionisio M.C.S., Ramirez-Peralta A. Methicillin-resistant Staphylococcus aureus (MRSA) in artisanal cheeses in México. Int. J. Microbiol. 2018;2018:8760357. doi: 10.1155/2018/8760357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamzah A.M.C., Yeo C.C., Puah S.M., Chua K.H., Rahman N.I.A., Abdullah F.H., Othman N., Chew C.H. Tigecycline and inducible clindamycin resistance in clinical isolates of methicillin-resistant Staphylococcus aureus from Terengganu, Malaysia. J. Med. Microbiol. 2019;68:1299–1305. doi: 10.1099/jmm.0.000993. [DOI] [PubMed] [Google Scholar]
- 63.Karimi Dehkordi M., Ghasemi Shamsabadi M., Banimehdi P. The occurrence of Staphylococcus aureus, enterotoxigenic and methicillin-resistant strains in Iranian food resources: A systematic review and meta-analysis. Ann. Ig. 2019;31:263–278. doi: 10.7416/ai.2019.2289. [DOI] [PubMed] [Google Scholar]
- 64.Wieczorek K., Wołkowicz T., Osek J. Antimicrobial resistance and virulence-associated traits of campylobacter Jejuni isolated from poultry food chain and humans with diarrhea. Front. Microbiol. 2018;9:1508. doi: 10.3389/fmicb.2018.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kassa T., Gebre-Selassie S., Asrat D. Antimicrobial susceptibility patterns of thermotolerant Campylobacter strains isolated from food animals in Ethiopia. Vet. Microbiol. 2007;119:82–87. doi: 10.1016/j.vetmic.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 66.Mughal M.H. Campylobacteriosis: A global threat. Biomed. J. Sci. Technol. Res. 2018;11:8804–8808. doi: 10.26717/BJSTR.2018.11.002165. [DOI] [Google Scholar]
- 67.Skirrow M.B., Blaser M.J. Clinical and epidemiological considerations. In: Nachamkin I., Blaser M.J., Tompkins L.S., editors. Campylobacter jejuni: Current Status and Future Trends. American Society For Microbiology; Washington, DC, USA: 1992. pp. 3–8. [Google Scholar]
- 68.Hagos Y., Berhe M., Gugsa G. Campylobacteriosis: Emphasis on its status as foodborne zoonosis in Ethiopia. J. Trop. Dis. 2019;7:1000317. [Google Scholar]
- 69.Nigatu S., Mequanent A., Tesfaye R., Garedew L. Prevalence and drug sensitivity pattern of campylobacter jejuni isolated from cattle and poultry in and around Gondar town, Ethiopia. Glob. Vet. 2015;14:43–47. doi: 10.5829/idosi.gv.2015.14.01.9238. [DOI] [Google Scholar]
- 70.Dierikx C.M., Van Der Goot J.A., Smith H.E., Kant A., Mevius D.J. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: A descriptive study. PLoS ONE. 2013;8:e79005. doi: 10.1371/journal.pone.0079005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammerum A.M., Larsen J., Andersen V.D., Lester C.H., Skytte T.S.S., Hansen F., Olsen S.S., Mordhorst H., Skov R.L., Aarestrup F., et al. Characterization of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 2014;69:2650–2657. doi: 10.1093/jac/dku180. [DOI] [PubMed] [Google Scholar]
- 72.Moodley A., Guardabassi L. Transmission of IncN Plasmids Carrying bla CTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 2009;53:1709–1711. doi: 10.1128/AAC.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stuart J.C., Munckhof T.V.D., Voets G., Scharringa J., Fluit A., Hall M.L.-V. Comparison of ESBL contamination in organic and conventional retail chicken meat. Int. J. Food Microbiol. 2012;154:212–214. doi: 10.1016/j.ijfoodmicro.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 74.Kola A., Kohler C., Pfeifer Y., Schwab F., Kuhn K., Schulz K., Balau V., Breitbach K., Bast A., Witte W., et al. High prevalence of extended-spectrum-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 2012;67:2631–2634. doi: 10.1093/jac/dks295. [DOI] [PubMed] [Google Scholar]
- 75.Overdevest I., Willemsen I., Rijnsburger M., Eustace A., Xu L., Hawkey P., Heck M., Savelkoul P., Vandenbroucke-Grauls C., van der Zwaluw K., et al. Extended-spectrum B-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg. Infect. Dis. 2011;17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reist M., Geser N., Hächler H., Schärrer S., Stephan R. ESBL-producing Enterobacteriaceae: Occurrence, risk factors for fecal carriage and strain traits in the Swiss slaughter cattle population younger than 2 years sampled at abattoir level. PLoS ONE. 2013;8:e71725. doi: 10.1371/journal.pone.0071725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmid A., Hörmansdorfer S., Messelhäusser U., Käsbohrer A., Sauter-Louis C., Mansfeld R. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli on Bavarian dairy and beef cattle farms. Appl. Environ. Microbiol. 2013;79:3027–3032. doi: 10.1128/AEM.00204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guillaume G., Verbrugge D., Chasseur-Libotte M.-L., Moens W., Collard J.-M. PCR typing of tetracycline resistance determinants (Tet A–E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 2000;32:77–85. doi: 10.1111/j.1574-6941.2000.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 79.Oppegaard H., Steinum T.M., Wasteson Y. Horizontal transfer of a multi-drug resistance plasmid between coliform bacteria of human and bovine origin in a farm environment. Appl. Environ. Microbiol. 2001;67:3732–3734. doi: 10.1128/AEM.67.8.3732-3734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bai Y., Xu R., Wang Q.-P., Zhang Y.-R., Yang Z.-H. Sludge anaerobic digestion with high concentrations of tetracyclines and sulfonamides: Dynamics of microbial communities and change of antibiotic resistance genes. Bioresour. Technol. 2019;276:51–59. doi: 10.1016/j.biortech.2018.12.066. [DOI] [PubMed] [Google Scholar]
- 81.Hue O., Allain V., Laisney M.-J., Le Bouquin S., Lalande F., Petetin I., Rouxel S., Quesne S., Gloaguen P.-Y., Picherot M., et al. Campylobacter contamination of broiler caeca and carcasses at the slaughterhouse and correlation with Salmonella contamination. Food Microbiol. 2011;28:862–868. doi: 10.1016/j.fm.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 82.Sarker M.S., Mannan M.S., Ali M.Y., Bayzid M., Ahad A., Bupasha Z.B. Antibiotic resistance of Escherichia coli isolated from broilers sold at live bird markets in Chattogram, Bangladesh. J. Adv. Vet. Anim. Res. 2019;6:272–277. doi: 10.5455/javar.2019.f344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shome B.R., Bhuvana M., Das Mitra S., Krithiga N., Shome R., Velu D., Banerjee A., Barbuddhe S.B., Prabhudas K., Rahman H. Molecular characterization of Streptococcus agalactiae and Streptococcus uberis isolates from bovine milk. Trop. Anim. Health Prod. 2012;44:1981–1992. doi: 10.1007/s11250-012-0167-4. [DOI] [PubMed] [Google Scholar]
- 84.Shome B., Das Mitra S., Bhuvana M., Krithiga N., Velu D., Shome R., Isloor S., Barbuddhe S., Rahman H. Multiplex PCR assay for species identification of bovine mastitis pathogens. J. Appl. Microbiol. 2011;111:1349–1356. doi: 10.1111/j.1365-2672.2011.05169.x. [DOI] [PubMed] [Google Scholar]
- 85.Tawyabur M., Islam M.S., Sobur M.A., Hossain M.J., Mahmud M.M., Paul S., Hossain M.T., Ashour H.M., Rahman M.T. Isolation and characterization of multidrug-resistant Escherichia coli and Salmonella spp. from healthy and diseased turkeys. Antibiotics. 2020;9:770. doi: 10.3390/antibiotics9110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hutchinson D.N., Bolton F.J. Improved blood free selective medium for the isolation of campylobacter jejuni from faecal specimens. J. Clin. Pathol. 1984;37:956–957. doi: 10.1136/jcp.37.8.956-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rahn K., De Grandis S., Clarke R., McEwen S., Galán J., Ginocchio C., Curtiss R., Gyles C. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-F. [DOI] [PubMed] [Google Scholar]
- 88.Ferasyi T.R., Abrar M., Subianto M., Afrianandra C., Hambal M., Razali R., Ismail I., Nurliana N., Rastina R., Sari W.E., et al. Isolation, Identification, and Critical Points of Risk of Escherichia coli O157:H7 Contamination at Aceh Cattle Breeding Centre. E3S Web Conf. 2020;151:01021. doi: 10.1051/e3sconf/202015101021. [DOI] [Google Scholar]
- 89.Wang Z., Zuo J., Gong J., Hu J., Jiang W., Mi R., Huang Y., Chen Z., Phouthapane V., Qi K., et al. Development of a multiplex PCR assay for the simultaneous and rapid detection of six pathogenic bacteria in poultry. AMB Express. 2019;9:185. doi: 10.1186/s13568-019-0908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sabzmeydani A., Rahimi E., Shakerian A. Incidence and antimicrobial resistance of campylobacter species isolated from poultry eggshell samples. Egypt. J. Vet. Sci. 2020;51:329–335. doi: 10.21608/ejvs.2020.26303.1163. [DOI] [Google Scholar]
- 91.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100s; Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 92.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 93.Labormedizinische Untersuchungen und In-Vitro-Diagnostika-Systeme—Empfindlichkeitsprüfung von Infektionserregern und Evaluation von Geräten zur antimikrobiellen Empfindlichkeitsprüfung—Teil 1: Referenzmethode zur Testung der In-Vitro-Aktivität von antimikrobiellen Substanzen Gegen Schnell Wachsende Aerobe Bakterien, Die Infektionskrankheiten Verursachen (ISO 20776-1) Deutsche Fassung EN ISO 20776-1, DIN Deutsches Institut für Normung e.V.; Beuth Verlag; Berlin, Germany; Wien, Austria; Zürich, Switzerland: 2006. [Google Scholar]
- 94.Rodloff A., Bauer T., Ewig S., Kujath P., Müller E. Susceptible, intermediate, and resistant—The intensity of antibiotic action. Dtsch. Ärzteblatt Int. 2008;105:657–662. doi: 10.3238/arztebl.2008.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Momtaz H., Rahimi E., Moshkelani S. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet. Med. 2012;57:193–197. doi: 10.17221/5916-VETMED. [DOI] [Google Scholar]
- 96.Kim B., Kim J., Seo M.-R., Wie S.-H., Cho Y.K., Lim S.-K., Lee J.S., Kwon K.T., Lee H., Cheong H.J., et al. Clinical characteristics of community-acquired acute pyelonephritis caused by ESBL-producing pathogens in South Korea. Infection. 2013;41:603–612. doi: 10.1007/s15010-013-0441-z. [DOI] [PubMed] [Google Scholar]
- 97.Pournajaf A., Ardebili A., Goudarzi L., Khodabandeh M., Narimani T., Abbaszadeh H. PCR-based identification of methicillin–resistant Staphylococcus aureus strains and their antibiotic resistance profiles. Asian Pac. J. Trop. Biomed. 2014;4:S293–S297. doi: 10.12980/APJTB.4.2014C423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.