ABSTRACT
In animals with germ plasm, specification of the germline involves ‘germ granules’, cytoplasmic condensates that enrich maternal transcripts in the germline founder cells. In Caenorhabditis elegans embryos, P granules enrich maternal transcripts, but surprisingly P granules are not essential for germ cell fate specification. Here, we describe a second condensate in the C. elegans germ plasm. Like canonical P-bodies found in somatic cells, ‘germline P-bodies’ contain regulators of mRNA decapping and deadenylation and, in addition, the intrinsically-disordered proteins MEG-1 and MEG-2 and the TIS11-family RNA-binding protein POS-1. Embryos lacking meg-1 and meg-2 do not stabilize P-body components, misregulate POS-1 targets, mis-specify the germline founder cell and do not develop a germline. Our findings suggest that specification of the germ line involves at least two distinct condensates that independently enrich and regulate maternal mRNAs in the germline founder cells.
This article has an associated ‘The people behind the papers’ interview.
Keywords: Germ plasm, P-bodies, Germline, Primordial germ cells, RNP granules, C. elegans
Highlighted Article: This paper describes a new condensate in the C. elegans germ plasm, ‘germline P-bodies’, which contain regulators of RNA translation and decay and are essential to specify germ cell fate.
INTRODUCTION
The germ plasm is a specialized cytoplasm, found in the eggs of certain insects, nematodes and vertebrates, which serves as a vehicle to segregate maternal proteins and RNAs to the nascent embryonic germline (Kulkarni and Extavour, 2017). Germ plasm assembly is a derived trait that arose independently several times in evolution as an alternative to the ancestral mode of germ cell fate specification by cell-to-cell signaling (Kemph and Lynch, 2022). A convergent characteristic of germ plasm in both vertebrate and invertebrate species is the presence of ‘germ granules’, micron-size ribonucleoprotein assemblies that contain RNAs coding for factors that promote germ cell development (Kulkarni and Extavour, 2017). Germ granules segregate with the germ plasm to the germline founder cells and are thought to contribute to their specification as primordial germ cells (PGCs). Germ granules were initially described using electron microscopy as mostly amorphous, electron-dense, micron-sized structures not surrounded by membranes (Arkov and Ramos, 2010). Fluorescence microscopy studies and proteomics in Drosophila, zebrafish, Xenopus, Caenorhabditis elegans and mice have revealed the presence of different types of condensates in germ cells, some with a complex sub-structure (Gallo et al., 2008; Wang et al., 2014; Vo et al., 2019; Eichler et al., 2020; Wan et al., 2018; Uebel et al., 2020, 2021; Roovers et al., 2018; Neil et al., 2021; Yang et al., 2022; Aravin et al., 2009). These studies have hinted that germ cells contain multiple condensates that compartmentalize different RNA-centered activities that collectively specify germ cell fate. For example, polar granules and founder granules are distinct granules in the germ plasm of Drosophila melanogaster that harbor mRNAs that need to be translated (polar granules) or degraded (founder granules) for proper germline development (Eichler et al., 2020). Here, we demonstrate that the C. elegans germ plasm also contains two condensate types that make distinct contributions towards germ cell fate.
The first condensates to be described in the C. elegans germ plasm were named P granules for their segregation with P (posterior) blastomeres through a series of four asymmetric divisions that eventually give rise to the germline founder cell P4 (Strome and Wood, 1982; Fig. 1E). P granules are scaffolded by the nematode-specific RGG-domain proteins PGL-1 and PGL-3, which form dense liquid-like condensates in vitro and in vivo (Brangwynne et al., 2009; Hanazawa et al., 2011; Updike et al., 2011; Saha et al., 2016; Putnam et al., 2019). In zygotes, the PGL condensates become covered on their surface by nanoscale solid clusters assembled by a pair of paralogous and redundant intrinsically-disordered proteins MEG-3 and MEG-4. MEG-3/4 form an essential protective layer that controls the dynamics and asymmetric segregation of PGL condensates into the P blastomeres in part by reducing the surface tension of PGL condensates (Folkmann et al., 2021). MEG-3/4 also recruit maternal mRNAs to P granules. MEG-3 binds RNA in vitro and co-precipitates with ∼500 maternal mRNAs in embryonic lysates, including the Nanos homolog nos-2 and the predicted E3 ubiquitin ligase Y51F10.2 that are required redundantly for fertility (Lee et al., 2020). Incorporation into P granules enriches RNAs in the P4 blastomere as much as 5-fold compared with what would have been achieved by equal segregation to all embryonic cells (Schmidt et al., 2021). nos-2 and Y51F10.2 are translationally repressed in the P0 to P3 blastomeres and become translationally activated in P4, the germline founder cell (Subramaniam and Seydoux, 1999; Lee et al., 2020). Despite their role in enriching mRNAs required for germ cell development, P granules are not essential for germ cell fate. In meg-3 meg-4 mutants, the germline founder cell P4 inherits no PGL condensates and reduced levels of nos-2 and Y51F10.2 transcripts (Lee et al., 2020; Schmidt et al., 2021). These transcripts, however, are still translationally activated in P4, and meg-3 meg-4 animals are mostly (∼70%) fertile (Lee et al., 2020). These observations indicate that the C. elegans germ plasm maintains proper regulation of maternal mRNAs in the absence of P granules.
Fig. 1.
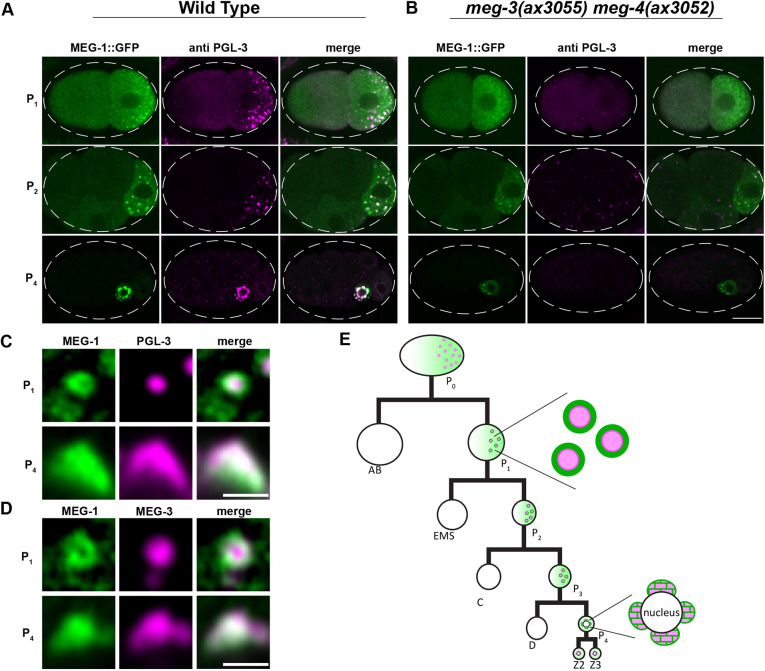
MEG-1 puncta are distinct from P granules. (A,B) Representative Airyscan photomicrographs of wild-type (A) and meg-3 meg-4 mutant (B) embryos expressing endogenous MEG-1::GFP and co-stained for GFP and PGL-3. MEG-1, but not PGL-3, enriches in P blastomeres in meg-3 meg-4 embryos. Dashed white line indicates embryo boundary. (C,D) Higher resolution images of MEG-1::GFP and PGL-3 (C) and MEG-1::GFP and MEG-3::OLLAS (D) in P1 and P4. In P1, MEG-1 enriches at the periphery of PGL-3 and MEG-3. In P4, P granules become perinuclear and MEG-1 and PGL-3/MEG-3 overlap. See Fig. S1A for quantification. (E) Abbreviated cartoon lineage summarizing the distribution of MEG-1 (green) and P granules (pink) in the germline (P) blastomeres. In the zygote P0, MEG-1 is present in a cytoplasmic gradient as well as small granules that are difficult to visualize at this stage. MEG-1 enriches at the periphery of P granules in the P1-3 blastomeres, and merges with P granules in P4. In the primordial germ cells Z2 and Z3, MEG-1 becomes cytoplasmic and is degraded, while P granules remain. Scale bars: 10 µm (A,B); 1 µm (C,D).
The C. elegans germ plasm contains a second condensate type that contains proteins characteristic of P-bodies, ubiquitous RNP granules implicated in mRNA storage and decay (Gallo et al., 2008; Ivanov et al., 2019). P-body-like condensates associate with P granules in germ plasm in tight assemblies containing a central P granule surrounded by several P-body-like condensates (Gallo et al., 2008). Dozens of proteins have been reported to enrich in granules in the C. elegans germ plasm (Updike and Strome, 2010; Phillips and Updike, 2022) and, in most cases, it is not known whether these localize to P granules proper (as defined by PGL-3 and MEG-3) or to the closely apposed P-body-like condensates described in Gallo et al. (2008), or to both. In particular, MEG-1 and MEG-2 are two intrinsically-disordered proteins, distantly related to MEG-3 and MEG-4, and originally described as P granule proteins (Leacock and Reinke, 2008). In this study, we demonstrate that MEG-1 and MEG-2 associate with canonical P-body proteins and stabilize P-body-like condensates in P4. Our findings indicate that, unlike P granules, ‘germline P-bodies’ are essential for maternal mRNA regulation and specification of P4 as the germline founder cell.
RESULTS
MEG-1 enriches in puncta distinct from P granules
To characterize the localization of MEG-1, we used a MEG-1::GFP fusion where GFP is inserted at the C-terminus of the MEG-1 open reading frame (ORF) in the meg-1 locus. Consistent with a previous report that used a polyclonal antibody raised against MEG-1 (Leacock and Reinke, 2008), MEG-1::GFP segregated with germ plasm in early embryos, distributing between a cytoplasmic pool and bright puncta in P blastomeres that overlapped with P granules (Fig. 1A). High resolution images revealed that the MEG-1 puncta localize to the periphery of P granules (visualized with PGL-3 or MEG-3) in P1 blastomeres (Fig. 1C,D; Fig. S1A). By the P4 stage, when P granules are fully perinuclear, the MEG-1::GFP signal was distributed throughout P granules (Fig. 1C,D; Fig. S1A). In Z2 and Z3, MEG-1::GFP dispersed back into the cytoplasm (Fig. S1B) and turned over by mid-embryogenesis (Leacock and Reinke, 2008).
Leacock and Reinke (2008) reported that MEG-1 enrichment in P blastomeres is independent of P granule components and vice versa. Consistent with these results, we found that MEG-1 still enriched preferentially in P blastomeres in meg-3(ax3055) meg-4(ax3052) mutants (Fig. 1B). MEG-1 puncta, however, remained cytoplasmic and did not associate with the nuclear envelope in P4 of meg-3(ax3055) meg-4(ax3052) mutants (Fig. 1A,B). Leacock and Reinke (2008) used a partial deletion of the meg-1 locus and RNAi of the meg-1 paralog meg-2 to generate embryos depleted of both meg-1 and meg-2. To complement these analyses, we created a deletion that removed the entire meg-1 meg-2 operon. meg-1 meg-2(ax4532) hermaphrodites were 100% maternal effect sterile as reported for meg-1(vr10) meg-2(RNAi) (Table S1). We found that MEG-3 and PGL-3 still assembled into puncta that segregated with P blastomeres in meg-1 meg-2(ax4532) embryos, confirming that P granule assembly does not require meg-1 and meg-2 (Fig. S1C). We noticed, however, that P granule enrichment in P blastomeres was not as robust in meg-1 meg-2 embryos (Fig. S1D) as previously reported (Leacock and Reinke, 2008; Wang et al., 2014), suggesting a minor contribution of MEG-1/2 to P granule segregation.
We conclude that MEG-1 localizes to assemblies that are distinct from P granules. MEG-1 puncta and P granules interact but assemble independently in the cytoplasm of P blastomeres.
MEG-1 immunoprecipitates with P-body components and several RNA-binding proteins, including POS-1
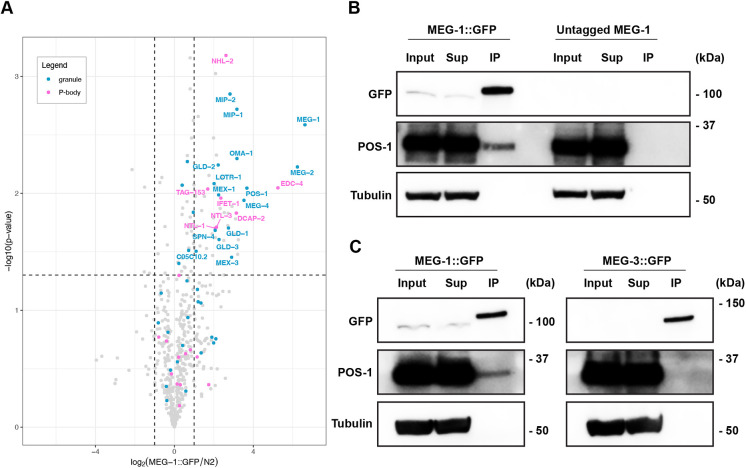
As we show here for MEG-1, we have previously reported that P-body markers enrich at the periphery of P granules in early P blastomeres (Gallo et al., 2008). Furthermore, Wu et al. (2017) identified MEG-1 and MEG-2 among immunoprecipitates of the P-body scaffold NTL-1 (also known as LET-711; CNOT1 in human) and identified seven CCR4-NOT subunits in MEG-2 immunoprecipitates. To complement these studies, we performed mass spectrometry on MEG-1::GFP immunoprecipitated from early embryo lysates using anti-GFP antibodies. As controls, we used lysates from wild-type worms expressing untagged MEG-1. We identified 54 proteins that were enriched at least 2-fold over untagged controls in two biological replicates (Fig. 2A; Table S2).
Fig. 2.
MEG-1 immunoprecipitates with P-body and RNA-binding proteins, including POS-1. (A) Volcano plot showing on the x-axis the log2 fold enrichment of proteins (dots) in MEG-1::GFP immunoprecipitates over ‘N2’ (wild-type lysates containing untagged MEG-1) as a function of the log10 P-value calculated from two independent immunoprecipitation experiments (y-axis). Of the 54 proteins enriched in MEG-1::GFP immunoprecipitates (top right quadrant), 13% correspond to P-body proteins (labeled in pink) and 28% correspond to proteins previously reported to localize to granules in P blastomeres (blue). (B) Representative western blots from two independent experiments confirm that GFP immunoprecipitates pull down MEG-1::GFP and POS-1, but not tubulin. (C) Western blots from MEG-1::GFP and MEG-3::GFP immunoprecipitates. Unlike MEG-1::GFP, MEG-3::GFP does not pull down POS-1. Full western blot images are shown in Fig. S2.
Among the proteins in MEG-1::GFP immunoprecipitates, we observed an enrichment for canonical P-body proteins (7 out of 36 canonical P-body proteins in the C. elegans genome/WormBase, P<0.0001, Fisher's exact test), including the decapping factors DCAP-2 (DCP2) and EDC-4 (EDC4), the TRIM-NHL family member and miRISC cofactor NHL-2 (TRIM45), the CCR4-NOT complex subunits NTL-1, TAG-153 (CNOT2), NTL-3 (CNOT3) and the translational repressor and DDX6-binding partner IFET-1 (EIF4ENIF1) (Table S2). In addition to P-body proteins, we also observed eight RNA-binding proteins including the translational repressor GLD-1, the poly-A polymerase GLD-2/GLD-3, the zinc finger proteins MEX-1, OMA-1 and POS-1, the KH domain protein MEX-3, and the RRM domain protein SPN-4. All of these have been reported to regulate maternal mRNAs and to enrich in germ plasm and ‘P granules’ (because P-bodies and P granules are closely linked in wild-type embryos, most studies have not distinguished between the two). Among these, POS-1 scored as one of the most highly enriched proteins in MEG-1::GFP precipitates after MEG-1 and MEG-2 (Fig. 2A; Table S2).
POS-1 regulates the poly-adenylation of thousands of maternal mRNAs containing AU-rich elements (AREs) in their 3′ untranslated region (UTR) (Farley et al., 2008; Elewa et al., 2015). ARE-binding proteins have been reported to recruit P-body components, including decapping enzymes and the deadenylation machinery (Ciais et al., 2013). To confirm the interaction between POS-1 and MEG-1, we probed the MEG-1::GFP immunoprecipitates with a polyclonal serum against POS-1 (Barbee and Evans, 2006) (Fig. 2B). This experiment confirmed that MEG-1::GFP precipitates contain POS-1, but not the control protein tubulin (Fig. 2B; Fig. S2A,B). POS-1 was not immunoprecipitated by a MEG-3::GFP fusion, further confirming the specificity of the MEG-1-POS-1 interaction (Fig. 2C; Fig. S2C,D). We conclude that MEG-1 exists in a complex that contains P-body components and RNA-binding proteins, including POS-1, a protein predicted to recruit P-body proteins to maternal mRNAs.
MEG-1 and POS-1 colocalize in P-body-like puncta in P4
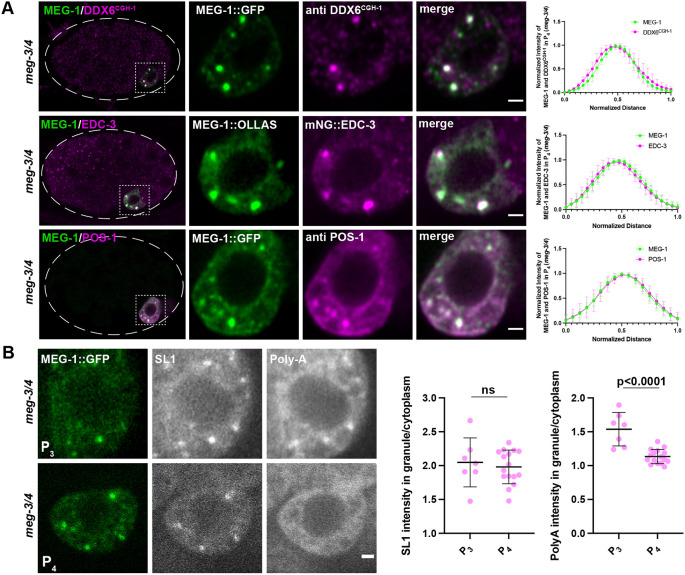
To examine the distribution of POS-1 and P-body components relative to MEG-1 and P granules, we used antibodies against POS-1 (Barbee and Evans, 2006) and P-body marker CGH-1 (DDX6) (Alessi et al., 2015) and a mNeonGreen::3×FLAG fusion to P-body marker EDC-3 (abbreviated mNG::EDC-3; DeMott et al., 2021). In P1 blastomeres, POS-1, CGH-1 and EDC-3 enriched in condensates at the periphery of PGL-3 puncta (Fig. S3A). The POS-1, CGH-1 and EDC-3 condensates overlapped but were not perfectly coincident with MEG-1 (Fig. S3B). In P4, MEG-1, POS-1, CGH-1 and EDC-3 appeared to mix more extensively with each other and PGL-3 (Fig. S3A,B). We reasoned that if P-body components associate with MEG-1, they might still form condensates in the absence of P granules. As expected, we found that in meg-3 meg-4 embryos, which lack P granules, POS-1, CGH-1 and EDC-3 enriched in cytoplasmic puncta most prominently in P4, and these colocalized with MEG-1 (Fig. 3A).
Fig. 3.
MEG-1 puncta in P4 correspond to germline P-bodies. (A) Airyscan photomicrographs of meg-3 meg-4 embryos expressing MEG-1::GFP and co-stained for GFP and CGH-1 (DDX6CGH-1), expressing MEG-1::OLLAS and mNG::3×FLAG::EDC-3 and co-stained for OLLAS and FLAG, and expressing MEG-1::GFP and co-stained for GFP and POS-1. Dashed white line indicates embryo boundary. Dashed square indicates P4. Inset shows P4 blastomere. Graphs plotting the mean intensities through the center of a granule indicate colocalization. For MEG-1 and CGH-1 n=7 granules from two embryos; for MEG-1 and EDC-3 n=9 granules from two embryos; for MEG-1 and POS-1 n=10 granules from two embryos. (B) Photomicrographs of meg-3 meg-4 embryos expressing MEG-1::GFP and probed for SL1 and poly-A. MEG-1 foci enrich SL1 to similar levels in P3 and P4, but show higher enrichment of poly-A in P3 compared with P4. The ratio of SL1 or poly-A intensity in MEG-1 granules over cytoplasm in P3 (n=7) was compared with P4 (n=16). Significance calculated by unpaired two-tailed t-test. ns, not significant. Quantification for each genotype is from one experiment in which several mutant and control animals were processed in parallel. Data are mean±s.d. Scale bars: 1 µm.
C. elegans mRNAs can be detected using an oligo-dT probe that detects poly-adenylated mRNAs and a probe against SL1, the splice leader found on the 5′ end of ∼60% of C. elegans mRNAs (Seydoux and Fire, 1994). Consistent with enriching maternal mRNAs, P granules are positive for both SL1 and poly-A (Seydoux and Fire, 1994). We reasoned that, as P-bodies are thought to enrich deadenylated mRNAs (Ivanov et al., 2019), P-bodies might be positive for SL1 but not poly-A. P-bodies also assemble in somatic blastomeres, becoming most prominent at the four-cell stage, when degradation of maternal mRNAs begins in somatic lineages (Gallo et al., 2008). Consistent with harboring deadenylated mRNAs, somatic P-bodies marked by EDC-3 showed a high SL1 signal but no poly-A enrichment (compared with the surrounding cytoplasm, Fig. S3C). Similarly, we found that MEG-1::GFP puncta in P4 of meg-3 meg-4 embryos were positive for SL1 but not poly-A (Fig. 3B). Interestingly, MEG-1::GFP puncta in P3 were positive for both SL1 and poly-A (Fig. 3B), suggesting that at this stage MEG-1 puncta do not yet correspond to mature P-body-like structures.
Taken together, these observations suggest that, in early P blastomeres, MEG-1 and P-body proteins form overlapping, but not perfectly coincident, assemblies at the periphery of P granules. In P4, MEG-1 and P-body components come together into condensates that contain deadenylated mRNAs. We refer to these P4-specific condensates as ‘germline P-bodies’ to distinguish these from somatic P-bodies which form in somatic blastomeres and do not contain MEG-1 or POS-1.
meg-1 and meg-2 are required to maintain CGH-1 and EDC-3 and assemble robust germline P-bodies in P4
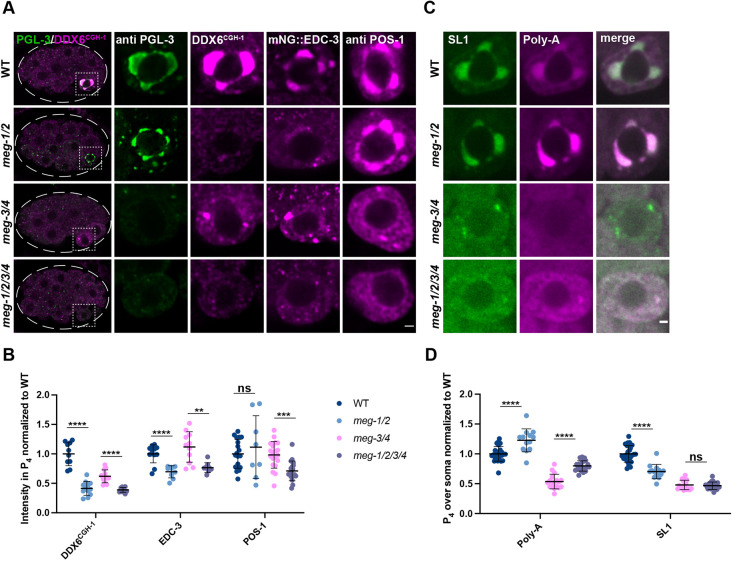
Unlike P granule proteins, such as PGL-3, which are asymmetrically segregated from the zygote stage (Fig. S1D), CGH-1 and EDC-3 are inherited by all blastomeres during early cleavages. After the eight-cell stage, CGH-1 is turned over in somatic blastomeres (Boag et al., 2005) and remains at high levels only in P4 (Fig. S4A-C). EDC-3 is maintained in somatic blastomeres throughout embryogenesis but enriches in P4 (Fig. S4D-F). In meg-1 meg-2 mutants, CGH-1 and EDC-3 distributions were unchanged through the eight-cell stage, but CGH-1 was not maintained and EDC-3 was not enriched in P4 (Fig. S4). In contrast, POS-1, which enriches with germ plasm from the zygote stage (Han et al., 2018), was not affected in meg-1 meg-2 (Fig. 4A). To quantify these observations, we compared the levels in P4 of CGH-1, EDC-3 and POS-1 in meg-1 meg-2, meg-3 meg-4 and embryos depleted of all four MEG proteins [meg-1(vr10) meg-2(RNAi) meg-3(tm4259) meg-4(RNAi) embryos] (Fig. 4A). CGH-1 and EDC-3 levels were significantly reduced in P4 of meg-1 meg-2 embryos compared with wild-type and in meg-1 meg-2 meg-3 meg-4 embryos compared with meg-3 meg-4 embryos (Fig. 4A,B). In contrast, POS-1 levels were not significantly affected in either meg-1 meg-2 or meg-3 meg-4 mutants and were reduced only in the quadruple mutant. We conclude that MEG-1/2 are essential to maintain high levels of CGH-1 and EDC-3 in P4 and are required redundantly with MEG-3/4 to maintain high levels of POS-1 in P4.
Fig. 4.
MEG-1/2 are required for maintenance of germline P-bodies in P4. (A) Airyscan photomicrographs of embryos of the indicated meg genotypes co-stained for PGL-3 and CGH-1 (DDX6CGH-1) (whole embryo and P4 inset), or expressing mNG::3×FLAG::EDC-3 and stained for FLAG, or stained for POS-1. meg-1 meg-2 are not essential for localization of PGL-3 or POS-1 to P4 but are required for maintenance of CGH-1 and EDC-3. Dashed white line indicates embryo boundary. Dashed square indicates P4. (B) Intensity of CGH-1, EDC-3 and POS-1 in P4 relative to wild type. Quantification of CGH-1 for each genotype is from one experiment in which mutant and control animals were processed in parallel. Wild type n=10; meg-1/2 n=12; meg-3/4 n=12; meg-1/2/3/4 n=10. Quantification of EDC-3 for each genotype is from one experiment in which mutant and control animals were processed in parallel. Wild type n=12; meg-1/2 n=9; meg-3/4 n=11; meg-1/2/3/4 n=9. Quantification of POS-1 for meg-1 meg-2 embryos is from one experiment and for meg-3 meg-4 and meg-1 meg-2 meg-3 meg-4 from two experiments in which mutant and control animals were processed in parallel. Wild type n=19; meg-1/2 n=8; meg-3/4 n=20; meg-1/2/3/4 n=19. (C) Photomicrographs of P4 in the indicated genotypes probed for SL1 and poly-A. Poly-A levels are increased in meg-1 meg-2 mutants, despite SL1 levels decreasing or not changing. (D) Quantification of poly-A and SL1 in P4 over soma normalized to wild type. Quantification for meg-1 meg-2 embryos is from two experiments and for meg-3 meg-4 and meg-1 meg-2 meg-3 meg-4 from three experiments in which mutant and control animals were processed in parallel. Wild type n=26; meg-1/2 n=13; meg-3/4 n=17; meg-1/2/3/4 n=20. Data are mean±s.d. ****P≤0.0001; ***P≤0.001; **P≤0.01; ns, not significant (unpaired two-tailed t-test). Scale bars: 1 µm.
The reduction in CGH-1 and EDC-3 levels in P4 suggests that germline P-body activity might be compromised in meg-1 meg-2 mutants. Consistent with this hypothesis, in situ hybridization against poly-A and SL1 revealed that poly-A levels were higher in P4 of meg-1 meg-2 embryos compared with wild-type and in meg-1 meg-2 meg-3 meg-4 embryos compared with meg-3 meg-4 embryos, despite either a reduction or no significant change in SL1 levels (Fig. 4C,D). We observed SL1+ puncta in P4 in 14/17 meg-3 meg-4 embryos and in 4/20 meg-1 meg-2 meg-3 meg-4 embryos (Fig. S5A). The SL1+ puncta did not enrich poly-A over the cytoplasm in meg-3 meg-4 embryos but did in meg-1 meg-2 meg-3 meg-4 embryos (Fig. S5A). Together, these observations indicate that MEG-1 and MEG-2 are required to maintain robust levels of P-body proteins and robust activation of mRNA deadenylation in P4.
meg-1 meg-2 embryos fail to turnover transcripts targeted for deadenylation by POS-1
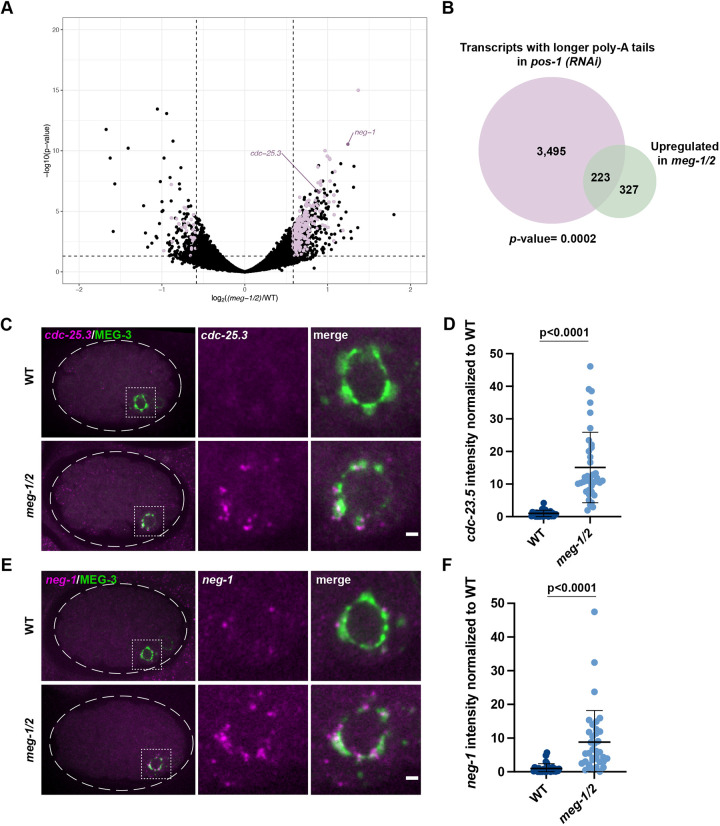
To examine directly whether meg-1 meg-2 mutants exhibit defects in maternal mRNA regulation, we performed RNA-seq to compare the transcriptomes of meg-1 meg-2 mutant embryos with that of wild-type. Two independent RNA-seq libraries were analyzed for each genotype [wild type and meg-1(vr10) meg-2(RNAi)]. This analysis identified 550 upregulated mRNAs and 230 downregulated mRNAs in meg-1 meg-2 embryos compared with wild-type (±1.5 fold change, P<0.05; Fig. 5A; Table S3).
Fig. 5.
meg-1/2 are required for the turnover of a subset of POS-1 targets. (A) RNA-seq from two independent experiments comparing meg-1 meg-2 (RNAi) and wild-type embryos identified 230 downregulated and 550 upregulated genes (±1.5 fold change). Purple dots correspond to genes significantly down/upregulated in meg-1 meg-2 embryos that also exhibited longer poly-A tails in pos-1(RNAi) embryos (Elewa et al., 2015). (B) A total of 223 genes upregulated in meg-1 meg-2 embryos overlap with genes with poly-A tails extended in pos-1(RNAi) embryos. P=0.0002 (Fisher's exact test; Materials and Methods). (C,E) Photomicrographs of cdc-25.3 and neg-1 smFISH in embryos expressing the P granule marker MEG-3::GFP. Inset shows P4. cdc-25.3 and neg-1 are turned over less efficiently in meg-1 meg-2 P4 blastomeres. Dashed white line indicates embryo boundary. Dashed square indicates P4. (D,F) Intensity of cdc-25.3 and neg-1 in P4 normalized to wild type. In situs for cdc-25.3 and neg-1 were done in the same embryos in two independent experiments in which mutant and control animals were processed in parallel. Wild type n=29; meg-1/2 n=38. Data are mean±s.d. Unpaired two-tailed t-test was used to make comparisons between genotypes. Scale bars: 1 µm.
Elewa et al. (2015) identified 3726 transcripts that display longer poly-A tails in pos-1(RNAi) embryos compared with wild-type (‘deadenylated POS-1 targets’), of which 3718 were detected in our RNA-seq. Of those genes upregulated in meg-1 meg-2 embryos, 40% (223/550) were among these deadenylated POS-1 targets (Fig. 5B; Table S3). Assuming a total pool of 11,121 transcripts that can be detected by these analyses in early embryos (see Materials and Methods), we found this overlap to be significant (Fisher's exact test, P=0.0002). In comparison, the overlap between transcripts downregulated in meg-1 meg-2 embryos and deadenylated POS-1 targets (30/3718 transcripts; P=1) or adenylated POS-1 targets [transcripts with shorter poly-A tails in pos-1(RNAi); 17/1307; P=0.99] was not significant (see next section). We conclude that MEG-1 and MEG-2 contribute to the turnover of a subset of maternal mRNAs also targeted by POS-1 for deadenylation.
neg-1 and cdc-25.3 are two transcripts among the 223 potential targets shared between POS-1 and MEG-1. neg-1 and cdc-25.3 are maternally deposited and turned over in all lineages by the 28-cell stage (Fig. S6A,B; Tintori et al., 2016; Elewa et al., 2015). In meg-1 meg-2 embryos, but not in meg-3 meg-4 embryos, neg-1 and cdc-25.3 transcripts were still detected in P4 in the 28-cell stage (Fig. 5C-F; Fig. S6C,D). These observations confirm that meg-1/2 activity is required for the efficient turnover of a subset of POS-1-regulated transcripts.
meg-1 meg-2 embryos fail to express efficiently transcripts activated by POS-1 for translation in P4
In addition to promoting deadenylation of a subset of maternal transcripts, POS-1 is also required to extend the poly-A tail of a different group of maternal transcripts that are translationally activated in embryos, including nos-2, Y51F10.2 and xnd-1 (Elewa et al., 2015). These transcripts code for factors required for germ cell fate and are translationally repressed in the P0, P1, P2 and P3 blastomeres and translationally activated in P4 (Subramaniam and Seydoux, 1999; Lee et al., 2020; Mainpal et al., 2015). It has been confirmed that translational activation of nos-2 and Y51F10.2 requires POS-1 (D'Agostino et al., 2006; Jadhav et al., 2008; Lee et al., 2020).
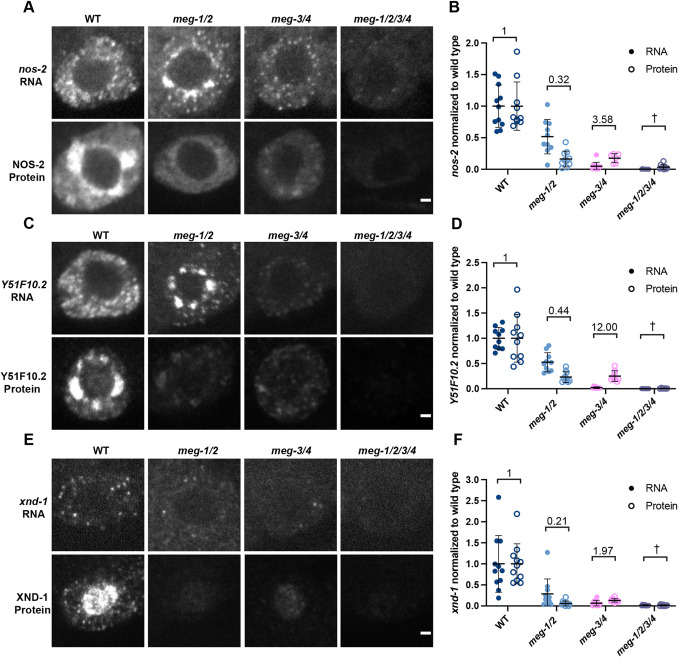
We used in situ hybridization and immunofluorescence to examine transcript and protein levels in P4 of wild-type, meg-1 meg-2, meg-3 meg-4 and meg-1 meg-2 meg-3 meg-4 embryos (Fig. 6). We found that for all three transcripts, RNA levels were lowest in the meg-3 meg-4 mutants, consistent with a dependence on P granules for enrichment in P4. RNA levels were also reduced in meg-1 meg-2 mutants compared with wild-type, suggesting that MEG-1/2 also contribute to RNA enrichment either directly or indirectly through an effect on P granule segregation, as P granules are also inefficiently segregated in these mutants (Fig. S1D). Adjusting for RNA levels, we found that protein output was reduced in meg-1 meg-2 and elevated in meg-3 meg-4 compared with wild-type (Fig. 6B,D,F). These differences did not correlate with POS-1 protein levels in P4, which were similar in these mutants (Fig. 4A,B). Consistent with meg-1 meg-2 and meg-3 meg-4 acting in parallel, protein levels were lowest in embryos depleted of all four meg genes compared with either double combination. Together, these observations suggest that meg-1 meg-2 and meg-3 meg-4 contribute independently to expression of maternal transcripts in P4, with MEG-3/4 acting primarily by boosting RNA levels and MEG-1/2 primarily by boosting protein output.
Fig. 6.
meg-1/2 are required for efficient translation of maternal mRNAs coding for germ cell fate determinants. (A,C,E) Photomicrographs of P4 in embryos of the indicated genotypes comparing nos-2, Y51F10.2 and xnd-1 RNA and protein levels. In all cases, the RNA is partially reduced in meg-1 meg-2 mutants, and significantly reduced in meg-3 meg-4 and meg-1 meg-2 meg-3 meg-4. In contrast, the protein levels of meg-1 meg-2 and meg-3 meg-4 are similar. In A and C, nos-2 and Y51F10.2 RNAs enrich in bright perinuclear puncta in meg-1 meg-2 mutants; however, the total RNA levels in P4 were lower. (B,D,F) Intensity of RNA and protein, normalized to wild type. The ratio of protein to RNA levels in each genotype is indicated. In meg-1 meg-2, the ratio is decreased, while in meg-3 meg-4 it is increased. Due to the very low levels of RNA present in meg-1 meg-2 meg-3 meg-4 embryos we were unable to calculate the protein/RNA ratio (†). Quantification for each genotype is from one experiment in which mutant and control animals were processed in parallel. For nos-2 RNA: wild type n=11, meg-1/2 n=10, meg-3/4 n=12, meg-1/2/3/4 n=12. For NOS-2 protein: wild type n=10, meg-1/2 n=10, meg-3/4 n=6, meg-1/2/3/4 n=9. For Y51F10.2 RNA: wild type n=10, meg-1/2 n=10, meg-3/4 n=10, meg-1/2/3/4 n=9. For Y51F10.2 protein: wild type n=10, meg-1/2 n=10, meg-3/4 n=9, meg-1/2/3/4 n=6. For xnd-1 RNA: wild type n=11, meg-1/2 n=11, meg-3/4 n=10, meg-1/2/3/4 n=10. For XND-1 protein: wild type n=11, meg-1/2 n=11, meg-3/4 n=10, meg-1/2/3/4 n=11. Data are mean±s.d. Scale bars: 1 µm.
In wild type, nos-2 and Y51F10.2 RNAs enrich in P granules through P3 and become cytoplasmic in P4 coincident with translational activation (Lee et al., 2020). xnd-1 is a much less abundant transcript which precluded us from evaluating its partitioning between P granules and the cytoplasm (Fig. 6E). Consistent with reduced translational activation in P4, we observed that nos-2 and Y51F10.2 remained enriched in a perinuclear pattern in meg-1 meg-2 embryos, as also observed in pos-1 embryos (Lee et al., 2020; Parker et al., 2020) (Fig. 6A,C; Fig. S7). As mentioned above, nos-2 and Y51F10.2 exhibited a higher protein output in P4 in meg-3 meg-4 embryos compared with wild-type and meg-1 meg-2 embryos (Fig. 6B,D), suggesting that assembly into P granules dampens translational activation. We could not determine translational output in meg-1 meg-2 meg-3 meg-4 owing to the extremely low levels of RNA in P4 in these mutants. We conclude that meg-1 meg-2 are required for maximal translation activation of POS-1 targets in P4, which is antagonized by meg-3 meg-4.
P4 adopts a mixed fate that resembles a muscle progenitor in meg-1 meg-2 mutants
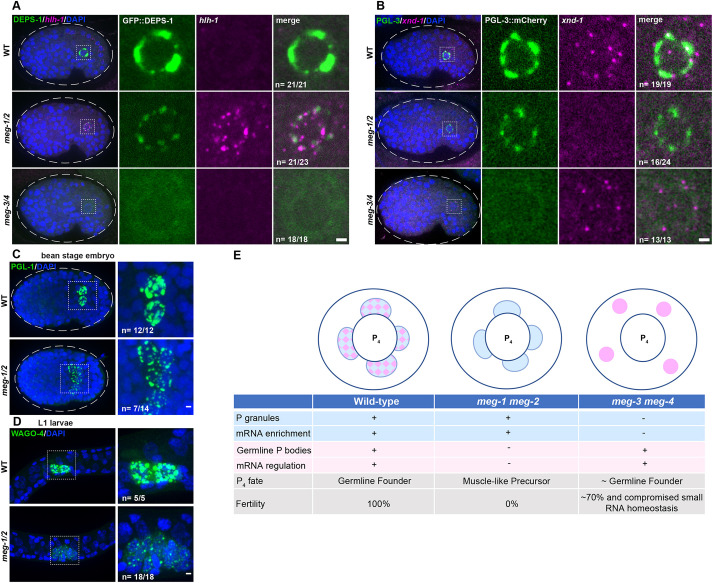
In pos-1 mutants, P4 descendants develop as muscle precursor cells that express the myoD homolog hlh-1 (Tabara et al., 1999). To determine whether a similar cell fate transformation occurs in meg-1 meg-2 mutants, we examined the expression of hlh-1 and the PGC zygotic transcript xnd-1 (Mainpal et al., 2015) by in situ hybridization using a P granule marker to identify P4 descendants. We observed hlh-1 transcripts in P4 descendants in 21/23 bean-to-comma-stage meg-1 meg-2 embryos examined, compared with 0/21 wild-type embryos examined (Fig. 7A). In addition, we failed to observe robust activation of xnd-1 in 16/24 meg-1 meg-2 embryos (Fig. 7B). P4 descendants, however, did not express detectable muscle myosin, suggesting that they are not fully transformed to muscle (Fig. S8A).
Fig. 7.
Primordial germ cells exhibit somatic-like characteristics in meg-1 meg-2 mutants. (A) Photomicrographs of bean-stage embryos of the indicated genotypes expressing DEPS-1::GFP and probed for hlh-1 RNA. Inset depicts a primordial germ cell. Embryos were scored from one independent experiment in which mutant and control animals were processed in parallel. All wild-type (21/21) and meg-3 meg-4 (18/18) bean-to-comma-stage embryos did not express hlh-1, while 21/23 meg-1 meg-2 did express hlh-1. (B) Photomicrographs of bean-stage embryos of the indicated genotypes expressing PGL-3::mCherry and probed for xnd-1 RNA (which is transcribed in PGCs at this stage). Inset depicts a primordial germ cell. Embryos were scored from two independent experiments for meg-1 meg-2 and one experiment for meg-3 meg-4 in which mutant and control animals were processed in parallel. All wild-type (19/19) and meg-3 meg-4 (13/13) bean-stage embryos expressed xnd-1, while 16/24 meg-1 meg-2 embryos did not express xnd-1. (C) Maximum projections of bean-stage embryos of the indicated genotypes stained for PGL-1. Inset shows the primordial germ cells. Embryos were scored from one experiment in which mutant and control animals were processed in parallel. All wild-type embryos (12/12) had two PGL-1-positive cells and 7/14 meg-1 meg-2 embryos had more than two PGL-1-positive cells. Dashed white line indicates embryo boundary. Dashed square indicates P4 descendants. (D) Maximum projections of germ cells from unfed L1 larvae expressing the germ granule marker 3×FLAG::GFP::WAGO-4. Embryos were scored from one experiment in which mutant and control animals were processed in parallel. All wild-type embryos (5/5) had two WAGO-4-positive cells and all meg-1 meg-2 embryos (18/18) had more than two WAGO-4-positive cells. (E) Working model: schematic and table summarizing P4 phenotypes based on this study and on Wang et al. (2014) and Ouyang et al. (2019). P granules are depicted in blue, germline P-body in pink and their merge in a checkered pattern. Note that P granule and germline P-body proteins also exist in a more dilute state in the cytoplasm. See text for additional details. Scale bars: 1 µm.
In wild type, P4 divides symmetrically to generate the primordial germ cells Z2 and Z3 by the 100-cell stage. These cells remain non-proliferative during embryogenesis and only divide in L1 larvae after the onset of feeding. In meg-1 meg-2 mutants, we observed more than two P granule-positive cells in 50% of bean-to-comma-stage embryos (Fig. 7C) and in 100% of non-fed L1 larvae stage (Fig. 7D). The extra P granule-positive cells were not due to mis-segregation of P granules to the D blastomere (Fig. S8B) and were first detected at around the 35-45 cell stage, consistent with premature division of P4 (Fig. S8C). We conclude that, in meg-1 meg-2 mutants, P4 adopts a mixed fate that resembles that of a muscle progenitor.
The meg-1 meg-2 phenotype contrasts with that of meg-3 meg-4 embryos, in which P4 does not proliferate prematurely and Z2 and Z3 express xnd-1 and do not express hlh-1 despite the absence of maternal P granules (Fig. 7A,B; Wang et al., 2014). Approximately 70% of meg-3 meg-4 mutants are fertile, in contrast to meg-1 meg-2 mutants, which are 100% sterile (Leacock and Reinke, 2008; Wang et al., 2014).
DISCUSSION
In this study, we demonstrate that the germ plasm of C. elegans contains two condensate types, P granules and germline P-bodies. Each rely on a different pair of intrinsically-disordered proteins for efficient accumulation in the germline founder cell P4: P granules depend on MEG-3 and MEG-4 and germline P-bodies depend on MEG-1 and MEG-2. We used these distinct genetic requirements to distinguish the contribution of each condensate to germ cell fate (Fig. 7E). P granules enrich regulators of small RNA homeostasis (Ouyang et al., 2019; Dodson and Kennedy, 2019) and maternal mRNAs but are not required for maternal mRNA regulation (Lee et al., 2020 and this study). mRNA regulation depends on ‘germline P-bodies’, which promote the translation of mRNAs coding for germline determinants and the turnover of mRNAs coding for somatic determinants. We propose that the germ cell fate-specifying ‘germ granules’ of C. elegans are assemblies of at least two distinct condensates, P granules and germline P-bodies, which enrich and regulate, respectively, maternal mRNAs in the germline founder cells.
Germline P-bodies and P granules are two types of condensates that require MEG proteins for stabilization in the embryonic germ lineage
P granules were the first characterized condensates in the C. elegans germ plasm (Strome and Wood, 1982). P granules consist of a dense liquid core, assembled by PGL proteins, surrounded by interfacial nanoscale RNA-rich solid clusters assembled by intrinsically-disordered proteins MEG-3 and MEG-4 (Folkmann et al., 2021). In this study, we describe a second condensate type, germline P-bodies, that contains regulators of mRNA adenylation and decapping, the RNA-binding protein POS-1, and MEG-1 and MEG-2, two intrinsically-disordered proteins related to MEG-3 and MEG-4. Germline P-body components assemble in complex patterns around P granules in early P blastomeres and merge with each other and P granules in P4. In embryos lacking P granules (meg-3 meg-4 mutants), germline P-bodies can be visualized in P4 as discrete SL1+ poly-A− cytoplasmic puncta that are also positive for MEG-1, POS-1 and the canonical P-body markers CGH-1 (DDX6) and EDC-3. In the absence of meg-1 meg-2, CGH-1 and EDC-3 levels are reduced and maternal mRNA regulation fails, despite normal P granule assembly and POS-1 levels (Fig. 7E).
How MEG-1/2 stabilize germline P-body components remains unclear. Unlike MEG-3/4 which are required for the asymmetric segregation of P granules from the zygote stage onward, MEG-1/2 do not appear to affect the distribution of germline P-body components until after the eight-cell stage. P-body components (CGH-1 and EDC-3) are initially segregated to all cells and coalesce into puncta in somatic cells coincident with the onset of maternal mRNA degradation (Gallo et al., 2008). MEG-1/2 do not affect P-body assembly in somatic cells but are required for stabilization of CGH-1 and EDC-3 specifically in P4 at the embryonic stage when CGH-1 is rapidly cleared from somatic lineages. In Drosophila embryos, the DDX6/4-ET-like complex (ME31B/Cup) is targeted for degradation by the E3 ubiquitin ligase complex CTLH and Marie Kondo (UBC-E2H), an E2 conjugating enzyme (Cao et al., 2020; Zavortink et al., 2020). It will be interesting to determine whether homologs of these factors promote CGH-1 turnover in C. elegans and how MEG-1/2 might oppose these activities in P4.
In contrast to somatic blastomeres, which activate zygotic transcription by the four-cell stage, P blastomeres remain transcriptionally silent until the birth of the daughters of P4, the primordial germ cells Z2 and Z3 (100-cell stage). We suggest that MEG-enhanced condensation of P granules and germline P-bodies serves as a mechanism to concentrate maternally-provided mRNAs and their regulators in germ plasm to ensure that P4 inherits sufficient machinery to initiate the maternal-to-zygotic transition. The MEG-1/2 and MEG-3/4 paralog pairs appear to have diverged such that MEG-1/2 interact preferentially with P-body components and MEG-3/4 interact preferentially with P granule components. MEG-3/4, but not MEG-1/2, contain an HMG-like domain essential for MEG-3/4 clusters to associate with the surface of PGL condensates (Schmidt et al., 2021). MEG-3/4 stabilize PGL condensates by lowering their surface tension (Folkmann et al., 2021); it remains to be determined whether MEG-1/2 function similarly or by another mechanism.
Germline P-body proteins control maternal mRNA regulation in the germline founder cell P4
The birth of the P4 blastomere appears to coincide with a major transition in maternal mRNA regulation in the P lineage as evidenced by: (1) coalescence of germline P-bodies containing deadenylated mRNAs, (2) degradation of transcripts coding for somatic factors, and (3) translation of transcripts coding for germ cell fate determinants. We suggest that regulators of mRNA adenylation and decapping that enrich in P-bodies drive this transition in P4 by targeting maternal mRNAs for de-adenylation/degradation or adenylation/translation, depending on the combination of RNA-binding proteins, including POS-1, bound to 3′ UTRs. The poly-A polymerase subunits GLD-2 and GLD-3 are enriched in MEG-1 immunoprecipitates and have been reported to enrich in granules in germ plasm (Wang et al., 2002; Eckmann et al., 2002). It will be interesting to determine whether GLD-2/3 also localize to germline P-bodies and are responsible for the translational activation of transcripts such as nos-2, Y51F10.2 and xnd-1.
The birth of P4 also coincides with the apparent mixing of germline P-bodies and P granules and the release of nos-2 and Y51F10.2 mRNAs from P granules coincident with their translational activation. This is also the stage where Z granules and SIMR-1 foci appear to de-mix from P granules to form the multi-condensate nuage characteristic of pre-gametic germ cells (Wan et al., 2018; Uebel et al., 2021). These observations suggest a dramatic switch in the material properties of condensates in the transition from P3 to P4. We do not know whether these changes arise as a cause, or consequence, of the changes in mRNA regulation that also occur at this stage. In principle, segregation of maternal mRNAs and their regulators into distinct condensates that eventually merge in P4 could be used as a physical mechanism to control RNA-protein interactions. Alternatively, changes in condensation patterns could derive from changes in the composition and solubility of complexes dispersed throughout the cytoplasm. We favor the latter as: (1) RNAs and proteins enriched in P granules and P-bodies are also found dispersed throughout the cytoplasm and (2) failure to assemble P granules does not prevent timely translational regulation of mRNAs enriched in P granules. We suggest that the complex condensation patterns of germline P-body components in early P blastomeres, and apparent ‘mixing’ with P granules in P4, are mesoscale manifestations of molecular-scale rearrangements that occur throughout the cytoplasm and eventually culminate in the targeting of the P-body machinery onto maternal mRNAs in P4. What regulates these changes during developmental time remains a mystery. The significance of the close association of germline P-bodies with P granules is also unclear and may reflect the fact that the two condensate types likely share some components such as POS-1, which depends on both MEG-1/2 and MEG-3/4 for maximal segregation to P4 (Fig. 4B).
A conserved role for P-body proteins in specifying germ cell fate
In meg-1 meg-2 mutants, P4 descendants divide precociously, fail to activate the transcription of the germ cell transcript xnd-1 and activate instead the transcription of the muscle transcription factor MYOD homolog hlh-1. These observations suggest a transformation to a muscle progenitor fate, such as that normally adopted by the sister of P4, the somatic blastomere D. This fate transformation occurs despite maintenance of P granules in Z2 and Z3 and their descendants, confirming that P granules are neither sufficient nor required to specify germ cell fate in primordial germ cells (Gallo et al., 2010; Strome et al., 1995). A similar P4→D fate transformation was reported for pos-1 mutants (Tabara et al., 1999). The apparent P4→D fate transformation is likely incomplete as Z2 and Z3 descendants do not express muscle myosin, remain in their normal central position in embryos and first-stage larvae, and stall proliferation during the first larval stage. meg-1 meg-2 fail to efficiently translate NOS-2 and Y51F10.2, two proteins implicated, respectively, in mRNA and protein turnover (Subramaniam and Seydoux, 1999; Kipreos, 2005). We have shown previously that the sterility of embryos lacking Nanos could be rescued by reducing the activity of maternal LIN-15B, a soma-promoting transcription factor expressed in oocytes (Lee et al., 2017). Similarly, the germ cell proliferation defect of meg-1 meg-2 larvae could be rescued partially by reducing GLD-1 activity (Kapelle and Reinke, 2011), an RNA-binding protein required for oocyte development and expressed in early P blastomeres (Francis et al., 1995; Jones et al., 1996). Together, these observations suggest that a key step to specify P4 as the germline founder cell is to program germline P-bodies to eliminate maternal factors that function during oogenesis.
The germline P-bodies we describe here share several features with the recently described ‘founder granules’ in Drosophila germ plasm. Founder granules contain mRNA for ME31B, the decapping factor DCP1 and Oskar, which, although required for germ plasm assembly in oocytes, must be degraded in embryos for proper germline development (Eichler et al., 2020). ME31B has been proposed to enrich in germ plasm independently of the canonical Oskar polar granule assembly pathway (McCambridge et al., 2020), as we demonstrate here for germline P-bodies, which assemble independently of P granules. Founder granules, however, have not yet been implicated in the translational activation of Nanos and other mRNAs enriched in polar granules, as we suggest here for germline P-bodies.
A role for P-bodies in early germ cell development has also been suggested by studies in mice. The mammalian Nanos homolog NANOS2 localizes to P-bodies, interacts with the CCR4-NOT1 deadenylation complex, and promotes mRNA degradation and the male germ cell fate program in mice (Suzuki et al., 2010; Shimada et al., 2019; Wright et al., 2021). DDX6/Me31B RNA helicases have also been implicated in the differentiation of various stem cell populations in human, mouse and Drosophila (Di Stefano et al., 2019; Nicklas et al., 2015; Jensen et al., 2021). Together, these studies suggest a conserved role for P-bodies as essential regulators of cell fate transitions in progenitors of the germline and beyond.
Limitations of the study
We inferred a requirement for P-body activity in embryonic germ cells through our analyses of meg-1 meg-2 mutants, which fail to stabilize germline P-bodies and regulate maternal mRNAs in P4. We did not test directly, however, for a requirement for P-body enzymatic activity, as mutants in key P-body proteins arrest development before the birth of P4. For example, RNAi reduction of the scaffold NTL-1 leads to early embryonic division defects, presumably because P-bodies also regulate the fate of mRNAs in somatic blastomeres (Gallo et al., 2008). The helicase CGH-1 stabilizes translationally repressed mRNAs during oogenesis and is essential for the production of mature oocytes that support normal embryogenesis (Boag et al., 2008; Noble et al., 2008). A CGH-1 temperature-sensitive mutant is available (Scheckel et al., 2012), which could potentially allow us to bypass an earlier requirement for CGH-1, but initial experiments proved inconclusive. Although we demonstrate that MEG-1 can be immunoprecipitated from lysates in a complex with POS-1 and a subset of P-body proteins, we have not investigated whether MEG-1 binds directly to these proteins or interacts indirectly by binding RNA for example. We also do not address whether MEG-1/2 or germline P-bodies are merely required (permissive) or are sufficient (instructive) to specify germ cell fate. MEG-1/2 enrich preferentially into P blastomeres from the zygote-stage onward; mutations that prevent this localization may help determine whether MEG-1/2 play a permissive or instructive role in germ cell fate specification.
MATERIALS AND METHODS
Worm handling, RNAi and sterility counts
C. elegans were cultured according to standard methods (Brenner, 1974). Strains used in this study are listed in Table S4. RNAi knockdown experiments were performed by feeding on HT115 bacteria (Timmons and Fire, 1998). The empty pL4440 vector was used as a negative control. Bacteria were grown at 37°C in LB+ampicillin (100 µg/ml) media for 5 h, induced with 5 mM IPTG for 30 min, plated on NNGM (nematode nutritional growth media)+ampicillin (100 µg/ml)+IPTG (1 mM) plates, and grown overnight at room temperature. L4 hermaphrodites were put onto RNAi plates and fed overnight at 25°C, and then shifted back to 20°C for at least 1 h before proceeding with further experiments. Effectiveness of knocking down meg genes was verified by scoring the sterility of adult progeny of the worms exposed to RNAi.
To culture larger numbers of worms, worm cultures were started from synchronized L1s (hatched from embryos incubated in M9 overnight) onto NA22 or RNAi bacteria containing plates and grown to gravid adults at 20°C. Early embryos were harvested from gravid adults.
To measure maternal-effect sterility of the meg-1 meg-2(ax4532) strain, 20 gravid adults from a mixed heterozygous population were singled out onto individual OP50 plates. Worms were allowed to lay eggs for 5 h, then removed and genotyped by PCR. Adult progeny were scored for empty uteri (white sterile phenotype).
CRISPR genome editing
Genome editing was performed using CRISPR/Cas9 as described in Paix et al. (2017). The meg-1 meg-2 ORF was deleted with two guide RNAs targeting the following sequences: 1, TGAGCGGCGATGGATAATCG; 2, AGTCAAAATTAGTTGCTGGG. Deletion of meg-1 meg-2 was confirmed by Sanger sequencing. This strain (JH3875) is maintained as a heterozygote because the homozygous meg-1 meg-2 deletion is 100% maternal effect sterile.
RNA extraction and preparation of mRNA-seq library
For each replicate, 26,000 synchronized L1 worms were plated on HT115 bacteria transformed with either L4440 (control) or meg-2 RNAi and grown at 20°C until the young adult stage. Adult worms were collected by filtering and the embryos were harvested by bleaching. Embryo pellets were flash frozen in liquid nitrogen. RNA was extracted with TRIzol reagent and chloroform. RNA was then concentrated and purified using a Zymo RNA Clean & Concentrator kit.
For mRNA-seq library preparation, 1 µg of total RNA was treated with Ribo-Zero Gold rRNA Removal Kit. A 1:100 dilution of ERCC RNA Spike-in Mix was added. Libraries were prepared using the TruSeq stranded total RNA library Prep Kit with 12 cycles of PCR amplification. All sequencing was performed using the Illumina HiSeq2500 at the Johns Hopkins University School of Medicine Genetic Resources Core Facility.
mRNA-sequencing analysis
Sequencing reads were aligned to the UCSC ce10 C. elegans reference genome using HISAT2 (Kim et al., 2015). Reads aligning to genetic features were then counted using HTSeq-count (Anders et al., 2015) and analyzed for differential expression analysis using DESeq2 (Love et al., 2014). Genes differentially expressed in wild-type versus meg-1 meg-2 embryos are listed in Table S3.
Immunoprecipitation
For each replicate for mass spectrometry analysis, 1×106 synchronized L1 worms were grown on NA22 bacteria at 20°C until the young adult stage. For immunoprecipitation (IP) to compare MEG-1::GFP and MEG-3::GFP by western blotting, four times as many MEG-3::GFP embryos were collected as MEG-1::GFP embryos, because MEG-1 is approximately four times more abundant than MEG-3 (Saha et al., 2016). Adult worms were collected by filtering and the embryos were harvested by bleaching. Embryos were washed and flash frozen in IP buffer [300 mM KCl, 50 mM HEPES (pH 7.4), 1 mM EGTA, 1 mM MgCl2, 1% glycerol, 0.1% NP-40] with 2× freshly prepared protease inhibitor mix #1 and mix #2 (100× protease inhibitor mix #1 contained 3 mg/ml antipain, 5 mg/ml leupeptin, 10 mg/ml benzamidine, 25 mg/ml AEBSF and 1 mg/ml phosphoramidon diluted in PBS; 100× protease inhibitor mix #2 contained 5 mg/ml aprotinin, 4 mM bestatin, 1 mg/ml E64 and 1 mg/ml trypsin inhibitor diluted in water). Thawed embryos were sonicated on ice using a Branson Digital Sonifier SFX 250 with a microtip (15 s on, 45 s off, 15% power, 6 min total on time or until embryos were completely lysed) and cleared by centrifugation at 4°C for 30 min at 21,000 g.
For the IP, 150 µl of anti-GFP nanobody conjugated to magnetic beads (Chromotek; gtma-10) were incubated with the lysates at 4°C for 90 min. The unbound fraction was removed and the beads were washed five times with ice-cold IP buffer. The bound fraction was eluted by boiling the beads in 1% SDS with 50 mM Tris-HCL (pH 7.4) for 5 min.
Western blotting
For western blotting, 1 M DTT and NuPAGE LDS sample buffer (4×) were added to lysates to a final concentration of 200 mM DTT and 1× NuPAGE LDS sample buffer. Samples were boiled for 5 min and run on 4-12% Bis-Tris gels in MES buffer. Samples were transferred to a PVDF membrane. Membranes were blocked in PBS with 0.1% Tween 20 and 5% non-fat dry milk (PBST+5% milk). Membranes were incubated in primary antibodies diluted in PBST+5% milk overnight at 4°C. Membranes were washed three times for 10 min in PBST and then incubated with secondary antibodies diluted in PBST+5% milk at room temperature for 1 h. Membranes were washed again three times for 10 min in PBST and visualized with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific; 32106) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific; 34095) and the KwikQuantTM Imager (Kindle Biosciences).
Primary antibodies and concentrations used were: mouse anti-GFP Living Colors (JL-8) (Takara Biosciences; 632381; 1:500), mouse anti-α-Tubulin (Sigma-Aldrich; T6199; 1:1000) and rabbit anti-POS-1 (a gift from Tom Evans; Barbee and Evans, 2006; 1:500).
Mass spectrometry
Mass spectrometry was performed by the Johns Hopkins Medical Institute (JHMI) Mass Spectrometry and Proteomics Facility. Samples were reduced with DTT, alkylated with iodoacetamide, TCA/acetone precipitated, and in solution digested with trypsin. Samples were analyzed by liquid chromatography (LC) tandem mass spectrometry (MS) (LC-MS-MS) on Q-Exactive Plus (Thermo Fisher Scientific) in FTFT at resolution 140K/35K with total 120 min gradient.
Mass spectrometry data analysis
Raw data were processed and analyzed using MaxQuant (2.0.3.0) software (Tyanova et al., 2016a). Default settings were used except that ‘Match between runs’ was turned on. Search parameters were as follows: cysteine carbamidomethyl was included as a fixed modification, and variable modifications included oxidation of methionine, protein N-terminal acetylation, deamidation of glutamine and asparagine, and phosphorylation of serine, threonine and tyrosine, and the maximum number of modifications per peptide was set to four. Trypsin was used as the digestion enzyme, a maximum of two missed cleavages were allowed, and the minimal peptide length was set to seven amino acids. Database search was performed against Uniprot C. elegans database (UP000001940_6239.fasta). False discovery rate (FDR) was set to 1% at peptide spectrum match (PSM) and protein level. Minimum peptide count required for protein quantification was set to two. Protein groups were further analyzed using Perseus (Tyanova et al., 2016b). Common contaminants, reverse proteins and proteins only identified by site were filtered out. Label free quantitation (LFQ) values were log2 transformed. Unpaired two-tailed t-tests were performed.
Immunostaining
Embryos were extruded from adult animals and subjected to freeze-crack on 0.01% poly-lysine coated slides followed by fixation in −20°C methanol ≥15 min. Slides were blocked in PBS with 0.1% Tween 20 (PBST) and 0.1% bovine serum albumin (BSA) (PBST+BSA) for 1 h. Slides were incubated in primary antibodies diluted in PBST+BSA at 4°C in a humidity chamber overnight. Slides were washed three times in PBST for 5 min and then incubated in secondary antibodies diluted in PBST+BSA for 1 h at room temperature. Slides were washed again three times in PBST for 5 min, then two quick washes in PBS. Samples were mounted in ProLong Glass Antifade mountant and cured overnight. When co-staining with OLLAS antibody, the OLLAS primary and secondary were applied first to avoid cross reactions.
Primary antibodies and concentrations used were: mouse anti-FLAG M2 (Sigma-Aldrich; F1804; 1:500), rat anti-OLLAS L2 (Novus; 06713; 1:50), rabbit anti-CGH-1 (a gift from John Kim; Alessi et al., 2015; 1:1000), rabbit anti-POS-1 (a gift from Tom Evans; Barbee and Evans, 2006; 1:100), guinea pig anti-XND-1 (a gift from Judith Yanowitz; Wagner et al., 2010; 1:2000), mouse anti-PGL-3 KT3 [Developmental Studies Hybridoma Bank (DSHB); 1:100], mouse anti-PGL-1 OIC1D4 (DSHB; 1:10), mouse anti-UNC-54 mAB 5-8 (DSHB; 1:10), anti-GFP nanobody conjugated to Alexa Fluor 488 (Chromotek; gb2AF488-10; 1:500). Antibody staining in this paper was consistent with that of previously published works.
Single molecule fluorescence in situ hybridization
Single molecule fluorescence in situ hybridization (smFISH) probes were designed using Biosearch Technologies Stellaris Probe Designer. Fluorophores used in this study were Quasar570 and Quasar670. For sample preparation, embryos were extruded from adult animals and subjected to freeze-crack on 0.01% poly-lysine coated slides followed by fixation in −20°C methanol for ≥15 min. Slides were washed five times in PBST and fixed in 4% paraformaldehyde (PFA) in PBS for 1 h at room temperature. Slides were again washed four times in PBST, twice in 2× SSC, and once in wash buffer (10% formamide, 2× SSC). Slides were then blocked in hybridization buffer (10% formamide, 2× SSC, 200 µg/ml BSA, 2 mM Ribonucleoside Vanadyl Complex, 0.2 mg/ml yeast total RNA, 10% dextran sulfate) for 30 min at 37°C in a humid chamber. For hybridization, slides were incubated in 50-100 nM probe in hybridization buffer at 37°C overnight. Slides were then washed twice in wash buffer at 37°C for 30 min, twice in 2× SSC, once in PBST and twice in PBS. Samples were mounted in ProLong Glass Antifade mountant and cured overnight.
Combined in situ hybridization/immunofluorescence
Combined in situ hybridization with immunofluorescence was carried out by first doing the in situ protocol as described above. After the last wash in PBS, the slides were then re-fixed in 4% PFA for 1 h at room temperature. The immunofluorescence protocol was then carried out as described above except 1 mg/ml UltraPure BSA (Thermo Fisher Scientific, AM2616) was used in the blocking and antibody incubation steps. The primary antibody used was KT3 (DSHB; 1:100). The secondary antibody used was goat anti-mouse IgA conjugated to FITC (Abcam; ab97234; 1:500).
Laser scanning confocal microscopy
Super-resolution microscopy was performed using a Zeiss LSM 880 microscope with a 63×-1.4 numerical aperture objective (Figs 1, 3A, 4A; Figs S1A,C, S3A,B). The raw data were processed using default Airyscan settings with ZEN software. For Fig. 4A, representative high-resolution images were shown. The images used for quantification in Fig. 4B were collected by spinning disk confocal microscopy. All images shown are single z-slices.
Spinning disk confocal microscopy
All other microscopy was performed using a Zeiss Axio Observer equipped with a CSU-W1 SoRA spinning disk scan head (Yokogawa). Images were taken using Slidebook software (Intelligent Imaging Innovations) with a 63× objective with a 2.8× relay lens (Yokogawa). All images shown are single z-slices, except in Fig. 7C,D.
Image quantification
All images were quantified in Fiji. For profile plots to show colocalization of granule components, a line was drawn through the center of a granule and the intensity along that line was measured using the plot profile tool in Fiji. As the size of each granule varied slightly, the length of each plot was normalized to the smallest granule size. The intensities were then binned using the averageifs function in Excel. The background signal was subtracted and the intensities were normalized to the highest intensity.
For quantification of conditions that included sparse or asymmetrically localized RNAs/proteins (i.e. POS-1, cdc-25.3, neg-1, nos-2/NOS-2, Y51F10.2/Y51F10.2, xnd-1/XND-1) in P4, the total intensity in the entire P4 cell above threshold was measured and normalized to wild-type controls. The threshold was defined as being 1.5× the mean intensity of the entire embryo. To minimize background, the smooth function in Fiji was used, which replaces each pixel with the average of its 3×3 neighbors.
For quantification of symmetrically localized RNA/proteins in P4 (i.e. CGH-1, EDC-3, poly-A, SL1), the ratio of the mean intensity in the P4 blastomere over the mean intensity of a same sized region in the soma was measured. A background measurement was taken from outside the embryo and subtracted from the germline and soma intensities. The ratios were then normalized to wild type.
To assess the segregation of PGL-3 (Fig. S1D), CGH-1 and EDC-3 (Fig. S4) into P blastomeres, the mean intensity was measured in each P blastomere and then normalized to the average P0 intensity. To measure the ratio of RNA inside/outside of granules, the granule (labeled by MEG-1::GFP in Fig. 3B, SL1 in Fig. S5 or PGL-3 in Fig. S7) was defined as being 1.5× above the mean intensity of the signal within the P blastomere. The mean intensity inside and outside the granule in the cytoplasm was measured. A background signal was taken from a region outside the embryo and subtracted.
Statistical analysis and plotting
Perseus (Tyanova et al., 2016b) was used for unpaired two-tailed t-tests on mass spec data. To determine the significance of the enrichment of P-body proteins in MEG-1 immunoprecipitates, we assumed a total pool of 6000 proteins, which is roughly the size of the embryonic proteome (Saha et al., 2016).
Statistics for differential expression analysis were carried out using DESeq2 (Love et al., 2014). To determine the significance of the overlap between predicted POS-1 targets (Elewa et al., 2015) and meg-1 meg-2 differentially expressed genes, we assumed a total pool of 11,121 transcripts. We arrived at this number by setting an FPKM threshold in our RNA-seq analysis of 0.002178852 FPKM, which was the lowest FPKM in meg-1 meg-2 animals for which we were able to detect a significant increase in gene expression. Any non-protein coding genes were also identified and removed from the list by using the SimpleMine tool on WormBase.
All other statistical analysis was conducted using R or Graphpad Prism 9 software. Data were plotted with either Graphpad Prism 9 or ggplot2 (Wickham, 2016).
Supplementary Material
Acknowledgements
We thank John Kim and Amelia Alessi, Tom Evans, and Judith Yanowitz for the CGH-1, POS-1 and XND-1 antibodies; Dominique Rasoloson and Helen Schmidt for the MEG-1::GFP and MEG-1::OLLAS alleles; Tu Lu for the strains JH3472, JH3410, JH3404 and JH3352; the Johns Hopkins Microscope Facility (S10OD023548) for microscopy support; the Johns Hopkins University School of Medicine Genetic Resources Core Facility for sequencing support, and the JHMI Mass Spectrometry and Proteomics Facility for mass spectrometry support. We also thank the Seydoux lab and the Baltimore Worm Club for their insights during this project. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Author contributions
Conceptualization: M.C., G.S.; Methodology: M.C., G.S.; Validation: M.C.; Formal analysis: M.C.; Investigation: M.C.; Resources: M.C., G.S.; Data curation: M.C.; Writing - original draft: M.C., G.S.; Writing - review & editing: G.S.; Visualization: M.C., G.S.; Supervision: G.S.; Project administration: M.C., G.S.; Funding acquisition: M.C., G.S.
Funding
Funding was provided by the National Institutes of Health (G.S.: R37HD037047; M.C.: T32 GM007445) and the National Science Foundation (M.C.: DGE-1746891). G.S. is an investigator of the Howard Hughes Medical Institute. Open Access funding provided by Johns Hopkins University School of Medicine. Deposited in PMC for immediate release.
Data availability
Sequencing data have been deposited onto the Gene Expression Omnibus (GEO) and can be found using the accession number GSE211555. Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE repository and can be found with the dataset identifier PXD036610.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.200920.reviewer-comments.pdf.
References
- Alessi, A. F., Khivansara, V., Han, T., Freeberg, M. A., Moresco, J. J., Tu, P. G., Montoye, E., Yates, J. R., III, Karp, X. and Kim, J. K. (2015). Casein kinase II promotes target silencing by miRISC through direct phosphorylation of the DEAD-box RNA helicase CGH-1. Proc. Natl. Acad. Sci. USA 112, E7213-E7222. 10.1073/pnas.1509499112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S., Pyl, P. T. and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A. A., Van Der Heijden, G. W., Castañeda, J., Vagin, V. V., Hannon, G. J. and Bortvin, A. (2009). Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 5, e1000764. 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov, A. L. and Ramos, A. (2010). Building RNA-protein granules: insight from the germline. Trends Cell Biol. 20, 482-490. 10.1016/j.tcb.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee, S. A. and Evans, T. C. (2006). The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Dev. Biol. 291, 132-143. 10.1016/j.ydbio.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Boag, P. R., Nakamura, A. and Blackwell, T. K. (2005). A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132, 4975-4986. 10.1242/dev.02060 [DOI] [PubMed] [Google Scholar]
- Boag, P. R., Atalay, A., Robida, S., Reinke, V. and Blackwell, T. K. (2008). Protection of specific maternal messenger RNAs by the P-body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182, 543-557. 10.1083/jcb.200801183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., Jülicher, F. and Hyman, A. A. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729-1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, W. X., Kabelitz, S., Gupta, M., Yeung, E., Lin, S., Rammelt, C., Ihling, G., Pekovic, F., Low, T. C. H., Siddiqui, N. U.et al. (2020). Precise temporal regulation of post-transcriptional repressors is required for an orderly Drosophila Maternal-to-Zygotic transition. Cell Rep. 31, 107783. 10.1016/j.celrep.2020.107783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais, D., Cherradi, N. and Feige, J.-J. (2013). Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cell. Mol. Life Sci. 70, 2031-2044. 10.1007/s00018-012-1150-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, I., Merritt, C., Chen, P.-L., Seydoux, G. and Subramaniam, K. (2006). Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 292, 244-252. 10.1016/j.ydbio.2005.11.046 [DOI] [PubMed] [Google Scholar]
- DeMott, E., Dickinson, D. J. and Doonan, R. (2021). Highly improved cloning efficiency for plasmid-based CRISPR knock-in in C. elegans. microPubl. Biol. 2021, 10.17912/micropub.biology.000499. 10.17912/micropub.biology.000499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano, B., Luo, E.-C., Haggerty, C., Aigner, S., Charlton, J., Brumbaugh, J., Ji, F., Rabano Jiménez, I., Clowers, K. J., Huebner, A. J.et al. (2019). The RNA helicase DDX6 controls cellular plasticity by modulating P-body homeostasis. Cell Stem Cell 25, 622-638.e13. 10.1016/j.stem.2019.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson, A. E. and Kennedy, S. (2019). Germ granules coordinate RNA-based epigenetic inheritance pathways. Dev. Cell 50, 704-715.e4. 10.1016/j.devcel.2019.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann, C. R., Kraemer, B., Wickens, M. and Kimble, J. (2002). GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3, 697-710. 10.1016/S1534-5807(02)00322-2 [DOI] [PubMed] [Google Scholar]
- Eichler, C. E., Hakes, A. C., Hull, B. and Gavis, E. R. (2020). Compartmentalized oskar degradation in the germ plasm safeguards germline development. eLife 9, e49988. 10.7554/eLife.49988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elewa, A., Shirayama, M., Kaymak, E., Harrison, P. F., Powell, D. R., Du, Z., Chute, C. D., Woolf, H., Yi, D., Ishidate, T.et al. (2015). POS-1 promotes endo-mesoderm development by inhibiting the cytoplasmic polyadenylation of neg-1 mRNA. Dev. Cell 34, 108-118. 10.1016/j.devcel.2015.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley, B. M., Pagano, J. M. and Ryder, S. P. (2008). RNA target specificity of the embryonic cell fate determinant POS-1. RNA 14, 2685-2697. 10.1261/rna.1256708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann, A. W., Putnam, A., Lee, C. F. and Seydoux, G. (2021). Regulation of biomolecular condensates by interfacial protein clusters. Science 373, 1218-1224. 10.1126/science.abg7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, R., Barton, M. K., Kimble, J. and Schedl, T. (1995). gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139, 579-606. 10.1093/genetics/139.2.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo, C. M., Munro, E., Rasoloson, D., Merritt, C. and Seydoux, G. (2008). Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 323, 76-87. 10.1016/j.ydbio.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Gallo, C. M., Wang, J. T., Motegi, F. and Seydoux, G. (2010). Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 330, 1685-1689. 10.1126/science.1193697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, B., Antkowiak, K. R., Fan, X., Rutigliano, M., Ryder, S. P. and Griffin, E. E. (2018). Polo-like kinase couples cytoplasmic protein gradients in the C. elegans zygote. Curr. Biol. 28, 60-69.e8. 10.1016/j.cub.2017.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa, M., Yonetani, M. and Sugimoto, A. (2011). PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 192, 929-937. 10.1083/jcb.201010106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, P., Kedersha, N. and Anderson, P. (2019). Stress granules and processing bodies in translational control. Cold Spring Harbor Perspect. Biol. 11, a032813. 10.1101/cshperspect.a032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav, S., Rana, M. and Subramaniam, K. (2008). Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803-1812. 10.1242/dev.013656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, L., Venkei, Z. G., Watase, G. J., Bisai, B., Pletcher, S., Lee, C.-Y. and Yamashita, Y. M. (2021). me31B regulates stem cell homeostasis by preventing excess dedifferentiation in the Drosophila male germline. J. Cell Sci. 134, jcs258757. 10.1242/jcs.258757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. R., Francis, R. and Schedl, T. (1996). GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180, 165-183. 10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Kapelle, W. S. and Reinke, V. (2011). C. elegans meg-1 and meg-2 differentially interact with nanos family members to either promote or inhibit germ cell proliferation and survival. Genesis 49, 380-391. 10.1002/dvg.20726 [DOI] [PubMed] [Google Scholar]
- Kemph, A. and Lynch, J. A. (2022). Evolution of germ plasm assembly and function among the insects. Curr. Opin. Insect Sci. 50, 100883. 10.1016/j.cois.2022.100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Langmead, B. and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos, E. T. (2005). Ubiquitin-mediated pathways in C. elegans. WormBook (ed. The C. elegans Research Community). doi/10.1895/wormbook.1.36.1
- Kulkarni, A. and Extavour, C. G. (2017). Convergent evolution of germ granule nucleators: a hypothesis. Stem Cell Research 24, 188-194. 10.1016/j.scr.2017.07.018 [DOI] [PubMed] [Google Scholar]
- Leacock, S. W. and Reinke, V. (2008). MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics 178, 295-306. 10.1534/genetics.107.080218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. Y. S., Lu, T. and Seydoux, G. (2017). Nanos promotes epigenetic reprogramming of the germline by down-regulation of the THAP transcription factor LIN-15B. eLife 6, e30201. 10.7554/eLife.30201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.-Y. S., Putnam, A., Lu, T., He, S. X., Ouyang, J. P. T. and Seydoux, G. (2020). Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife 9, e52896. 10.7554/eLife.52896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainpal, R., Nance, J. and Yanowitz, J. L. (2015). A germ cell determinant reveals parallel pathways for germ line development in Caenorhabditis elegans. Development 142, 3571-3582. 10.1242/dev.125732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCambridge, A., Solanki, D., Olchawa, N., Govani, N., Trinidad, J. C. and Gao, M. (2020). Comparative proteomics reveal Me31B's interactome dynamics, expression regulation, and assembly mechanism into germ granules during Drosophila germline development. Sci. Rep. 10, 564. 10.1038/s41598-020-57492-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil, C. R., Jeschonek, S. P., Cabral, S. E., O'Connell, L. C., Powrie, E. A., Otis, J. P., Wood, T. R. and Mowry, K. L. (2021). L-bodies are RNA-protein condensates driving RNA localization in Xenopus oocytes. Mol. Biol. Cell 32, ar37. 10.1091/mbc.E21-03-0146-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, S., Okawa, S., Hillje, A.-L., González-Cano, L., del Sol, A. and Schwamborn, J. C. (2015). The RNA helicase DDX6 regulates cell-fate specification in neural stem cells via miRNAs. Nucleic Acids Res. 43, 2638-2654. 10.1093/nar/gkv138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, S. L., Allen, B. L., Goh, L. K., Nordick, K. and Evans, T. C. (2008). Maternal mRNAs are regulated by diverse P-body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559-572. 10.1083/jcb.200802128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, J. P. T., Folkmann, A., Bernard, L., Lee, C. Y., Seroussi, U., Charlesworth, A. G., Claycomb, J. M. and Seydoux, G. (2019). P granules protect RNA interference genes from silencing by piRNAs. Dev. Cell 50, 716-728.e6. 10.1016/j.devcel.2019.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix, A., Wang, Y., Smith, H. E., Lee, C.-Y. S., Calidas, D., Lu, T., Smith, J., Schmidt, H., Krause, M. W. and Seydoux, G. (2014). Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198, 1347-1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix, A., Folkmann, A. and Seydoux, G. (2017). Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121-122, 86-93. 10.1016/j.ymeth.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, D. M., Winkenbach, L. P., Boyson, S., Saxton, M. N., Daidone, C., Al-Mazaydeh, Z. A., Nishimura, M. T., Mueller, F. and Nishimura, E. O. (2020). mRNA localization is linked to translation regulation in the Caenorhabditis elegans germ lineage. Development 147, dev186817. 10.1242/dev.186817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C. M. and Updike, D. L. (2022). Germ granules and gene regulation in the Caenorhabditis elegans germline. Genetics 220, iyab195. 10.1093/genetics/iyab195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam, A., Cassani, M., Smith, J. and Seydoux, G. (2019). A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 26, 220-226. 10.1038/s41594-019-0193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers, E. F., Kaaij, L. J. T., Redl, S., Bronkhorst, A. W., Wiebrands, K., de Jesus Domingues, A. M., Huang, H.-Y., Han, C.-T., Riemer, S., Dosch, R.et al. (2018). Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Dev. Cell 46, 285-301. 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., Weber, C. A., Nousch, M., Adame-Arana, O., Hoege, C., Hein, M. Y., Osborne-Nishimura, E., Mahamid, J., Jahnel, M., Jawerth, L.et al. (2016). Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572-1584.e16. 10.1016/j.cell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckel, C., Gaidatzis, D., Wright, J. E. and Ciosk, R. (2012). Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS Genet. 8, e1002742. 10.1371/journal.pgen.1002742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H., Putnam, A., Rasoloson, D. and Seydoux, G. (2021). Protein-based condensation mechanisms drive the assembly of RNA-rich P granules. eLife 10, e63698. 10.7554/eLife.63698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux, G. and Fire, A. (1994). Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 120, 2823-2834. 10.1242/dev.120.10.2823 [DOI] [PubMed] [Google Scholar]
- Shimada, R., Kiso, M. and Saga, Y. (2019). ES-mediated chimera analysis revealed requirement of DDX6 for NANOS2 localization and function in mouse germ cells. Sci. Rep. 9, 515. 10.1038/s41598-018-36502-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J., Calidas, D., Schmidt, H., Lu, T., Rasoloson, D. and Seydoux, G. (2016). Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife 5, e21337. 10.7554/eLife.21337.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S. and Wood, W. B. (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 79, 1558-1562. 10.1073/pnas.79.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S., Martin, P., Schierenberg, E. and Paulsen, J. (1995). Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development 121, 2961-2972. 10.1242/dev.121.9.2961 [DOI] [PubMed] [Google Scholar]
- Subramaniam, K. and Seydoux, G. (1999). nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861-4871. 10.1242/dev.126.21.4861 [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Igarashi, K., Aisaki, K.-I., Kanno, J. and Saga, Y. (2010). NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc. Natl. Acad. Sci. USA 107, 3594-3599. 10.1073/pnas.0908664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Hill, R. J., Mello, C. C., Priess, J. R. and Kohara, Y. (1999). pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1-11. 10.1242/dev.126.1.1 [DOI] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. (1998). Specific interference by ingested dsRNA. Nature 395, 854. 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Tintori, S. C., Osborne Nishimura, E., Golden, P., Lieb, J. D. and Goldstein, B. (2016). A transcriptional lineage of the early C. elegans embryo. Dev. Cell 38, 430-444. 10.1016/j.devcel.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova, S., Temu, T. and Cox, J. (2016a). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301-2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., Mann, M. and Cox, J. (2016b). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731-740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Uebel, C. J., Agbede, D., Wallis, D. C. and Phillips, C. M. (2020). Mutator foci are regulated by developmental stage, RNA, and the germline cell cycle in Caenorhabditis elegans. G3 10, 3719-3728. 10.1534/g3.120.401514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebel, C. J., Manage, K. I. and Phillips, C. M. (2021). SIMR foci are found in the progenitor germ cells of C. elegans embryos. MicroPubl. Biol. 2021, 10.17912/micropub.biology.000374. 10.17912/micropub.biology.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike, D. and Strome, S. (2010). P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 31, 53-60. 10.2164/jandrol.109.008292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike, D. L., Hachey, S. J., Kreher, J. and Strome, S. (2011). P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192, 939-948. 10.1083/jcb.201010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, H. D. L., Wahiduzzaman, Tindell, S. J., Zheng, J., Gao, M. and Arkov, A. L. (2019). Protein components of ribonucleoprotein granules from Drosophila germ cells oligomerize and show distinct spatial organization during germline development. Sci. Rep. 9, 19190. 10.1038/s41598-019-55747-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, C. R., Kuervers, L., Baillie, D. L. and Yanowitz, J. L. (2010). xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467, 839-843. 10.1038/nature09429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, G., Fields, B. D., Spracklin, G., Shukla, A., Phillips, C. M. and Kennedy, S. (2018). Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679-683. 10.1038/s41586-018-0132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Eckmann, C. R., Kadyk, L. C., Wickens, M. and Kimble, J. (2002). A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312-316. 10.1038/nature01039 [DOI] [PubMed] [Google Scholar]
- Wang, J. T., Smith, J., Chen, B.-C., Schmidt, H., Rasoloson, D., Paix, A., Lambrus, B. G., Calidas, D. and Seydoux, G. (2014). Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591. 10.7554/eLife.04591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Wright, D., Kiso, M. and Saga, Y. (2021). Genetic and structural analysis of the in vivo functional redundancy between murine NANOS2 and NANOS3. Development 148, dev191916. 10.1242/dev.191916 [DOI] [PubMed] [Google Scholar]
- Wu, E., Vashisht, A. A., Chapat, C., Flamand, M. N., Cohen, E., Sarov, M., Tabach, Y., Sonenberg, N., Wohlschlegel, J. and Duchaine, T. F. (2017). A continuum of mRNP complexes in embryonic microRNA-mediated silencing. Nucleic Acids Res. 45, 2081-2098. 10.1093/nar/gkw872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., Dominique, G. M., Champion, M. M. and Huber, P. W. (2022). Remnants of the Balbiani body are required for formation of RNA transport granules in Xenopus oocytes. IScience 25, 103878. 10.1016/j.isci.2022.103878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavortink, M., Rutt, L. N., Dzitoyeva, S., Henriksen, J. C., Barrington, C., Bilodeau, D. Y., Wang, M., Chen, X. X. L. and Rissland, O. S. (2020). The E2 Marie Kondo and the CTLH E3 ligase clear deposited RNA binding proteins during the maternal-to-zygotic transition. eLife 9, e53889. 10.7554/eLife.53889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.