Abstract
In Aeromonas hydrophila, the ahyI gene encodes a protein responsible for the synthesis of the quorum sensing signal N-butanoyl-l-homoserine lactone (C4-HSL). Inactivation of the ahyI gene on the A. hydrophila chromosome abolishes C4-HSL production. The exoprotease activity of A. hydrophila consists of both serine protease and metalloprotease activities; in the ahyI-negative strain, both are substantially reduced but can be restored by the addition of exogenous C4-HSL. In contrast, mutation of the LuxR homolog AhyR results in the loss of both exoprotease activities, which cannot be restored by exogenous C4-HSL. Furthermore, a substantial reduction in the production of exoprotease by the ahyI+ parent strain is obtained by the addition of N-acylhomoserine lactone analogs that have acyl side chains of 10, 12, or 14 carbons. The inclusion of N-(3-oxododecanoyl)-l-homoserine lactone or N-(3-oxotetradecanoyl)-l-homoserine lactone at 10 μM in overnight cultures of A. hydrophila abolishes exoprotease production in azocasein assays and reduces the activity of all the exoprotease species seen in zymograms.
Aeromonas species are pathogens of humans and fish. Aeromonas salmonicida is the causative agent of furunculosis in salmonid fish, whereas Aeromonas hydrophila is responsible for motile aeromonad septicemia; both are a significant problem in aquaculture (11). Importantly, interest in the pathogenesis of Aeromonas now extends beyond the economic consequences to the fish farming industry, as members of this genus are increasingly implicated in intestinal and extraintestinal infections in humans (54).
The virulence of Aeromonas spp. is multifactorial. Surface-associated factors include adhesins (e.g., pili), the S-layer, and lipopolysaccharide. Extracellular factors include siderophores for iron acquisition and an array of exoenzymes and exotoxins, i.e., enterotoxins, glycero-phospholipid-cholesterol acetyltransferase (GCAT), hemolysins, lipases, and proteases (30, 33, 38, 54). Many of the proteins involved in pathogenicity are reliant on the general secretory pathway for export (16, 38).
The regulation of virulence determinants by pathogenic bacteria, such as Aeromonas, throughout the infection and transmission cycle is an important consideration for the etiology of disease. A major objective of an infecting bacterium is the evasion of host defenses. Hence the premature elaboration of an aggressive phenotype, which could be recognized by the host as the signal to elicit the induction of immune defenses, would constitute a poor strategy for a pathogen. Where the bacterium is able to evade host defenses and find a suitable niche, it can then proliferate to a level where the combined aggressive phenotype of the population is capable of overwhelming host defenses. In this respect, the regulation of gene expression by the process termed quorum sensing (12) can be used for a concerted activation of a modulon of genes coding for the components of an aggressive phenotype only when a bacterial population sufficient to make the phenotype effective is present. Quorum sensing relies on the release of a low-molecular-mass signalling molecule into the extracellular milieu (for reviews, see references 12, 13, 46, and 52). Accumulation of the signal (often an N-acylhomoserine lactone [AHL]) above a threshold concentration, indicative of a critical cell population density, activates the relevant gene expression. The system has been shown to regulate virulence and secondary metabolism in a number of gram-negative bacteria (for reviews, see references 12, 13, 46, and 52), where the AHL is produced by members of the LuxI family of synthases and recognized by the LuxR family of response regulators.
The discovery of AHL-based quorum sensing in Aeromonas (51) has placed our focus on this genus for the elucidation of its role in pathogenesis. A number of investigations of different strains of Aeromonas have demonstrated that exoprotease activity correlates with the establishment of infection (8, 9, 25, 44). There is, however, good evidence for A. salmonicida that protease is not an absolute requirement for pathogenicity. Vipond et al. (58) demonstrated that a defined protease mutant of a highly pathogenic strain of A. salmonicida exhibited no significant change in virulence. This study was of further interest because the protease-dependent activation of GCAT was also abolished (58).
The regulation of exoprotease activity by Aeromonas may be important because if it is elaborated too early, host defenses will be alerted and the bacterial infection may well be contained. Exoprotease production is therefore a likely candidate for quorum sensing-dependent regulation. A number of factors contribute data to this concept. Proteolytic activity is observed in the culture supernatant when cells are at high population density in the stationary phase of growth (6, 45), a phenomenon closely associated with quorum sensing control. In Pseudomonas aeruginosa (15, 22) and Erwinia carotovora (22, 39), protease expression is positively regulated by quorum sensing.
To explore the role of quorum sensing in regulating exoproteases in A. hydrophila, we mutagenized ahyI, encoding the N-butanoyl-l-homoserine lactone (C4-HSL) synthase of A. hydrophila, and ahyR, encoding a LuxR-type response regulator. In this study, we show that quorum sensing regulates both serine protease and metalloprotease activities and demonstrate that exoprotease production can be blocked by C4-HSL analogs.
MATERIALS AND METHODS
Strains.
A. hydrophila AH-1N is a spontaneous mutant of A. hydrophila AH-1 that lacks an S-layer and the O-11 antigen while retaining other surface characteristics (57). Escherichia coli JM109 [recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) mcrA / F′ traD36 proAB lacIq lacZΔM15 (60)] was used as the host for plasmids not requiring the λpir protein for replication. E. coli CC118 λpir [λpir lysogen of CC118 (Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 (17)] was used as a permissive host for suicide plasmids requiring the λpir protein, and E. coli S17/1 λpir [λpir lysogen of S17-1 (thi pro hsdR− hsdM+ recA RP4 2-Tc::Mu-Km::Tn7(Tpr Smr)] (49)) was used as a permissive host able to transfer suicide plasmids requiring the λpir protein by conjugation to A. hydrophila. Chromobacterium violaceum CV026 (double mini-Tn5 mutant derived from C. violaceum ATCC31532, Hgr cviI::Tn5 xylE Kmr, plus spontaneous Smr) was used as an AHL biosensor (28). Unless otherwise stated, Aeromonas and Chromobacterium cultures were grown at 30°C and E. coli cultures were grown at 37°C. In cases of mixed cultures, e.g., conjugations, incubations were at 30°C.
Plasmids.
pAHP11 contains an active ahyI gene, confers ampicillin resistance, and has the pUC origin of replication (51). pAHH2 is analogous to pAHH1 (51), containing the ahyRI region from A. hydrophila AH-1 as a HindIII fragment cloned into pUC18. pBScam contains the chloramphenicol resistance (cat) cassette from pACYC184 (HincII/XmnI fragment) cloned into the EcoRV site of pBluescript SKII+ (Stratagene, La Jolla, Calif.). For an AhyR expression vector, ahyR was PCR amplified from A. hydrophila AH-1N, using the primer pair RMBP1 5′ AGGGGGGCCAGCTGATGAAA plus AHYRB 5′ TCACTCTGCAGCGAGAATCATCGGGTT, and T-cloned into pUC57/T. The ahyR gene was subcloned into pRK415 (23a), using BamHI and XbaI restriction endonuclease sites in the pUC57/T multicloning region, to give pDK42. The expression of ahyR on pDK42 is driven by Plac. pKNG101 (23) and pDM4 (31a) are suicide vectors able to replicate only in the presence of the λpir protein and which contain the sacBR genes for sucrose sensitivity. pKNG101 confers resistance to streptomycin and pDM4 resistance to chloramphenicol.
Media.
Unless otherwise stated, growth was in L-broth, Lennox (Difco, Detroit, Mich.) (LB) medium with agar (No. 1; Oxoid, Basingstoke, United Kingdom) (1.5% [wt/vol]) and antibiotics added appropriately. Biochemical tests were performed with the API20E system (BioMerieux UK, Basingstoke, United Kingdom) according to the manufacturer’s instructions. Selection of A. hydrophila over E. coli S17/1 λpir was accomplished by using either Aeromonas selective medium (Difco) or modified Griffin’s liquid medium (MGLM) (35). Selection against the presence of the sacB gene was performed by selection for resistance to sucrose in LB-sucrose medium (10 g of tryptone and 5 g of yeast extract per liter, sucrose at 10% [wt/vol]).
DNA manipulations.
Genomic DNA was purified as described by Swift et al. (53); plasmid DNA was isolated by alkaline lysis (47) and further purified by using Qiagen plasmid preparation columns (Qiagen Ltd., Crawley, United Kingdom). Restriction enzyme digests and DNA ligations were performed as instructed by the manufacturer (Promega UK, Southampton, United Kingdom). Southern hybridizations were performed as described previously (55). DNA sequencing was performed by the University of Nottingham Sequencing Laboratory. Oligonucleotides were synthesized by the Biopolymer Synthesis and Analysis Unit, University of Nottingham. PCR amplifications were performed according to a standard protocol (43). Long-range PCRs were performed with the Expand Long Template PCR system (Boehringer Mannheim UK, Lewes, United Kingdom). Ligation of PCR products to blunt-ended DNA fragments was accomplished by the method of Throup and Francis (56). Ligation of PCR products by T-cloning used the vector pUC57/T (Immunogen International, Sunderland, United Kingdom) at the vector/insert ratio recommended by the supplier.
Construction of A. hydrophila ahyI mutant strains.
Long-range PCR was performed on pAHP11 with primers 5′-TTACATGCTGCCCAGCATC and 5′-AAGACGCGATTGCGAAAGCG, which are divergent and lie within ahyI. The PCR product was ligated to a blunt-ended (SmaI-HincII) cat cassette from pBScam, and an ampicillin- and chloramphenicol-resistant, AHL-negative derivative of pAHP11 was selected. The cat insertion runs in the same direction as ahyI. A BamHI fragment containing cat and a 188-bp central portion of ahyI (ahyI nucleotides 223 to 411, of a total 623) was taken from this plasmid and ligated into the suicide vector pKNG101. Transformants of E. coli CC118 λpir were analyzed, and two plasmids, pIcam1 and pIcam4, were selected. In pIcam1, cat and strAB run in opposite directions; in pIcam4, cat and strAB run in the same direction. Chloramphenicol- and streptomycin-resistant, ahyI-negative (as determined by C. violaceum CV026 T-streaks [28]) A. hydrophila mutants were selected from independent matings of either E. coli S17/1 λpir(pIcam1) or E. coli S17/1 λpir(pIcam4) and A. hydrophila AH-1N plated onto either MGLM or Aeromonas selective agar containing 30 mg of chloramphenicol and 30 mg of ampicillin per liter. Six independent isolates, termed AH-1NahyI-1 (pIcam1 derived), AH-1NahyI-2 (pIcam1 derived), AH-1NahyI-3 (pIcam1 derived), AH-1NahyI-4 (pIcam4 derived), AH-1NahyI-5 (pIcam4 derived), and AH-1NahyI-6 (pIcam4 derived), were selected for further study. Southern blot analysis demonstrated that in each case, a single-crossover event creating two truncated copies the ahyI gene, linked to cat and the pKNG101 plasmid backbone, had taken place (data not shown).
Construction of an A. hydrophila ahyR mutant strain.
Overlap extension PCR (17a) was used to generate an in-frame deletion of the ahyR gene on the A. hydrophila AH-1N chromosome. Two PCR fragments were generated from the template pAHP2 with the primer pairs AHYR-1 (5′ GAGTACCTGAGCATTTCACTTCGG) plus AHYR-2 (5′ GTACTTGGACATCCAGGCAAGACTGCCCTCTTGCAG) and AHYR-3 (5′ CCTGGATGTCCAACTACATCTTCGAGGCGGCG) plus AHYR-4 (5′ GGGGAAGTTGGTGACCACGACCTGC). The resulting products contained a 315-bp fragment containing the 5′ end of ahyR and a 302-bp fragment containing the 3′ of ahyR, respectively. A 17-bp overlap in their sequences (underlined) permitted amplification of a 600-bp product during a second PCR with primers AHYR-1 and AHYR-4. The resultant product contained a deletion from nucleotides 320 to 466 of ahyR (GenBank accession no. X89469) corresponding to AhyR amino acid residues 107 to 156 and was T-cloned into pUC57/T. DNA sequencing was used to confirm that the cloned overlap extension PCR product was correct. The T-cloned PCR product was transferred to pDM4 as a XbaI/SalI fragment, using the restriction endonuclease sites in the pUC57/T and pDM4 multicloning regions, to give plasmid pDM600. Conjugation from E. coli S17/1 λpir was used to introduce pDM600 into A. hydrophila AH-1N, and A. hydrophila cells containing single-crossover events were isolated on Aeromonas selective agar containing chloramphenicol at 30 mg/liter. Double-crossover events were selected on LB-sucrose agar, and chloramphenicol-sensitive colonies were screened by PCR with primers AHYR-1 and AHYR-4. In putative ahyR deletion mutants, a 600-bp PCR product was obtained, compared with a 747-bp product from the parent strain. Southern hybridization using a probe comprising the ahyR region amplified from pAHH2 with primers AHYR-1 and AHYR-4 confirmed the chromosomal deletion. In a PstI/XhoI digestion of chromosomal DNA from the parent strain, a single band corresponding to the predicted size of 1,381 bp hybridized to the probe. In the ahyR deletion mutant this band was 1,234 bp. In a BglI digestion of chromosomal DNA from the parent strain two bands corresponding to the predicted sizes of 222 and 722 bp hybridize to the probe. The central BglI site is in the deleted region of ahyR, and in BglI-digested DNA from the ahyR mutant, hybridization to a single band of 847 bp is seen.
Polyacrylamide gel electrophoresis (PAGE).
Extracellular protein samples for Nu-PAGE (Novex, San Diego, Calif.) were precipitated with trichloroacetic acid (TCA); 150 μl of 50% (wt/vol) TCA was added to 1.35 ml of culture supernatant, and proteins were precipitated by centrifugation at 15,000 × g at 4°C for 20 min after 1 h on ice. Pellets were drained, rinsed with 0.5 ml of ice-cold acetone, air dried, and resuspended in 65 μl of H2O plus 25 μl of 4× lithium dodecyl sulfate gel loading buffer (Novex) and 10 μl of reducing agent (Novex). Proteins were denatured at 90°C for 10 min and placed on ice; 15 μl was loaded to individual lanes, and electrophoresis was performed according to the manufacturer’s instructions in morphonlinepropanesulfonic acid buffer. Extracellular proteins for native gels were concentrated 100 times in Microcon 10 microconcentrators (Amicon, Inc., Beverly, Mass). Ten microliters of protein sample was mixed with 10 μl of 2× Tris-glycine sodium dodecyl sulfate (SDS) sample buffer (Novex), incubated for 10 min at room temperature, and loaded onto a discontinuous Tris-HCl–10% polyacrylamide gel (24) lacking SDS. Proteins were stained with Coomassie brilliant blue R250 (0.25% [wt/vol] in 25% [vol/vol] propan-2-ol–10% [vol/vol] glacial acetic acid), and the gel was destained in 10% (vol/vol) propan-2-ol–10% (vol/vol) glacial acetic acid. Protease zymography was performed with 10% polyacrylamide gelatin gels (Novex) or 12% polyacrylamide casein gels (Novex) according to the manufacturer’s protocol. The equivalent of 0.8 μl of supernatant was loaded onto each lane.
Exoenzyme assays.
Qualitative assays of exoprotease activity were performed on LB agar containing 2% (wt/vol) skimmed milk (SMLB; Oxoid). Similarly, hemolysin activity was assayed on LB agar containing 5% (vol/vol) sheep blood, amylase activity was assessed on starch agar (Difco), and nuclease activity was assayed on DNase agar (Oxoid).
For quantitative exoprotease assays, A. hydrophila was grown overnight at the given temperature in L-broth containing AHLs where appropriate. Cells were removed from the medium by centrifugation, and 50-μl aliquots of supernatant were taken for assay; 500 μl of 0.25% (wt/vol) azocasein (Sigma-Aldrich Ltd., Poole, United Kingdom) in 0.1 M sodium citrate (pH 6) was added to each supernatant aliquot to be tested and incubated at 37°C for 2 h. The protease reaction was stopped, and protein was precipitated, by the addition of 550 μl of ice-cold 10% (wt/vol) TCA followed by incubation on ice for 15 min. Azodye released by the action of proteases in supernatant aliquots was determined at A366 after the removal of precipitated protein by centrifugation. The serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) was included at 1 mM, and the metalloprotease inhibitor EDTA was included at 10 mM (25). For quantitative hemolysin assay, doubling dilutions of sterile filtered culture supernatant were prepared in a U-bottomed microplate with phosphate-buffered saline, pH 7.2 (70 mM phosphate, 150 mM sodium chloride), to leave a 50-μl volume. Then 100 μl of 3% (vol/vol) sheep blood in phosphate-buffered saline was added to each and incubated at 37°C for 1 to 2 h, and the lowest dilution at which hemolysis occurred was recorded.
AHLs.
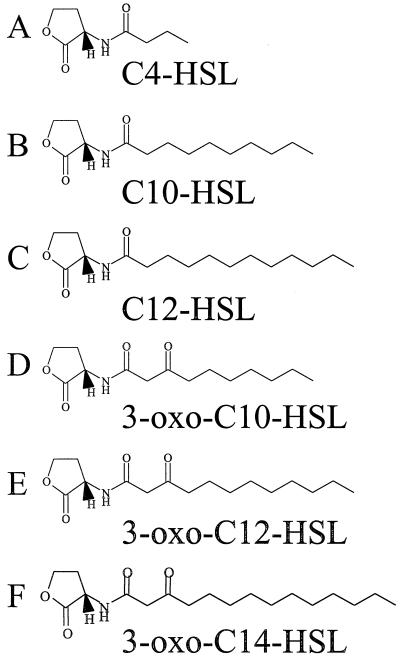
The AHLs used in this study, C4-HSL, N-(hexanoyl)-l-homoserine lactone (C6-HSL), N-(octanoyl)-l-homoserine lactone (C8-HSL), N-(decanoyl)-l-homoserine lactone (C10-HSL), N-(dodecanoyl)-l-homoserine lactone (C12-HSL), N-(3-oxobutanoyl)-l-homoserine lactone (3-oxo-C4-HSL), N-(3-oxohexanoyl)-l-homoserine lactone (3-oxo-C6-HSL), N-(3-oxooctanoyl)-l-homoserine lactone (3-oxo-C8-HSL), N-(3-oxodecanoyl)-l-homoserine lactone (3-oxo-C10-HSL), N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), and N-(3-oxotetradecanoyl)-l-homoserine lactone (3-oxo-C14-HSL), were synthesized as described by Chhabra et al. (3). Figure 1 shows the structures of the major A. hydrophila AHL (C4-HSL) and its long acyl chain antagonists. Stock solutions at 10 mM in acetonitrile (far-UV grade) were diluted into the growth medium to the stated concentration. Control assays of protease activity with acetonitrile alone demonstrated that any effects were due to the presence of the AHL. The AHL biosynthesis activity of individual strains was assayed in T-streaks by using the biosensor C. violaceum CV026 (28), where the induction of the purple pigment violacein indicates the production of AHLs with short acyl chains (i.e., C4-HSL, C6-HSL, C8-HSL, 3-oxo-C4-HSL, 3-oxo-C6-HSL, and 3-oxo-C8-HSL).
FIG. 1.
Structures of AHL molecules used in this study. See text for definitions.
RESULTS
Exoprotease activity is abolished in A. hydrophila quorum sensing mutants.
Agar plate assays were used to qualitatively screen candidate phenotypes regulated by quorum sensing in the ahyI mutant strains AH-1NahyI-1 to -6. All behaved the same, and AH-1NahyI-6 was taken as a representative for use in further assays. In this mutant, exoprotease activity on SMLB was substantially down-regulated, but amylase, nuclease, and lipase activities were unaffected and β-hemolysin activity was increased. Microplate assay of β-hemolysin demonstrated a twofold increase in activity in the mutant strain. Analysis of the API-20E profile of mutants AH-1NahyI-1 to -6 and parent showed that each mutant was unable to liquefy gelatin (a function of protease activity). The inclusion of the major AHL product of AhyI, C4-HSL, at 1 μM in SMLB and in API-20E assays restored the exoprotease activity and consequently the ability to both digest casein and liquefy gelatin.
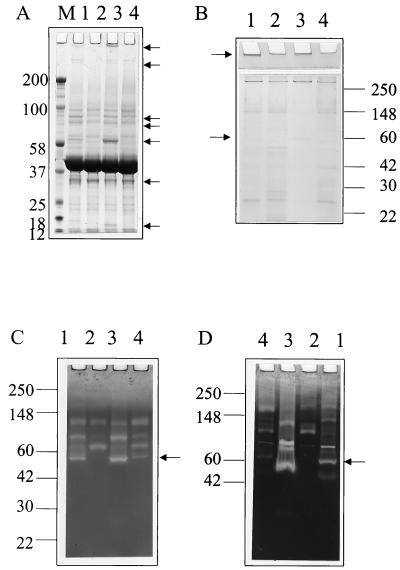
The analysis of supernatant proteins on denaturing PAGE highlights a number of quorum sensing-dependent proteins (arrowed in Fig. 2A). The largest of these is also observable on nondenaturing PAGE (Fig. 2B) and is possibly multimerized aerolysin, formed in a process reliant upon protease activity (7, 18). The reduction in the number of protein species in the lanes where concentrated supernatants were loaded is likely due simply to proteolytic degradation. The resistance to protease of the large protein present in the stacking gel (Fig. 2B, lane 3) is further evidence for this protein being a multimerized form of aerolysin, as this has been reported to be resistant to proteolytic digestion (7, 18).
FIG. 2.
PAGE analysis of A. hydrophila exoproteins. (A) Denaturing PAGE of TCA-precipitated proteins present in the supernatant of an overnight culture. C4-HSL-dependent proteins are arrowed. (B) Nondenaturing PAGE of Microcon 10-concentrated supernatant proteins. Proteins predicted to be multimerized aerolysin and serine protease are arrowed. (C) Casein zymogram of supernatant proteins. Activity predicted to be due to the serine protease is arrowed. (D) Gelatin zymogram of supernatant proteins. Activity predicted to be due to the serine protease is arrowed. In each case, lane 1 is a sample derived from A. hydrophila AH-1N, lane 2 is from A. hydrophila AH-1NahyI-6, lane 3 is from A. hydrophila AH-1NahyI-6 cultured with 5 μM C4-HSL, and lane 4 is from A. hydrophila AH-1N cultured with 10 μM 3-oxo-C12-HSL. M denotes standards, with molecular masses in kilodaltons indicated.
Analysis of supernatants on casein and gelatin zymograms (Fig. 2C and D) revealed a reduction in the number of bands exhibiting protease activity in the ahyI mutant. The analysis of protease activity in Aeromonas supernatants is complicated by the presence of the number of protease bands. Nieto and Ellis (32) suggested that these could represent both additional cryptic proteases and different forms of the same protease after casein overlay analysis of extracellular proteins separated by isoelectric focusing (32). After PAGE performed under nondenaturing conditions equivalent to those used in the zymography (Fig. 2B), in concentrated supernatants from the ahyI mutant strain cultured with C4-HSL we see a protein band migrating with the 60-kDa marker that is most probably the A. hydrophila serine protease. Denatured, prestained markers were run with a gap of at least one lane to sample proteins to minimize any effect of reducing agents present in the markers. Protease bands at 60 kDa are also associated with A. hydrophila AH-1N and the A. hydrophila AH-1N ahyI mutant cultured with C4-HSL in both casein and gelatin zymograms (Fig. 2C and D) but are absent in the A. hydrophila AH-1N ahyI mutant and less intense in A. hydrophila AH-1N cultured with 3-oxo-C12-HSL.
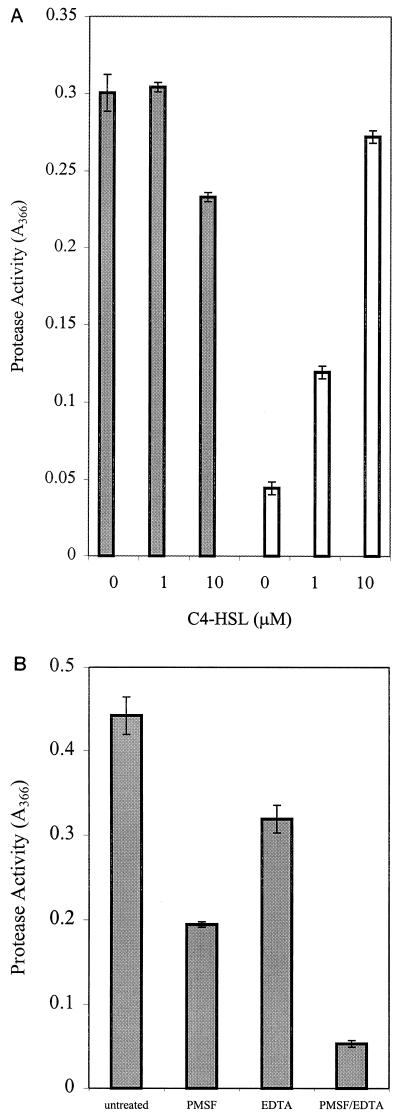
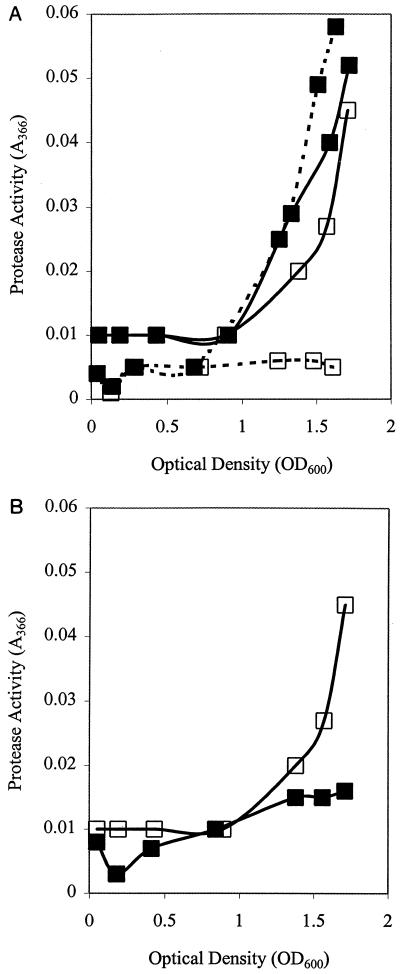
For quantitative assay of ahyI-regulated exoprotease activity in A. hydrophila, assays using culture supernatants from AH-1NahyI-6 and the isogenic parent were performed after overnight growth in LB at 30°C in the presence of 0, 1, and 10 μM C4-HSL. Although little effect is seen on the parent, a dose response-dependent restoration of exoprotease activity is seen in the ahyI mutant (Fig. 3A). As exoprotease production in A. hydrophila is population density dependent, we also assayed exoprotease activity in the supernatant throughout growth to the stationary phase. The exoprotease induction profiles from the parent and from the mutant grown in the presence of 1 μM C4-HSL are similar, but little activity is detected in the ahyI mutant in the absence of C4-HSL (Fig. 4A). Furthermore, this experiment shows that the exogenous provision of the quorum sensing signalling molecule does not immediately induce any detectable increase in exoprotease production.
FIG. 3.
Exoprotease production by A. hydrophila AH-1N and AH-1NahyI-6. (A) Response to C4-HSL (added to give 0, 1, or 10 μM) in the culture medium of A. hydrophila AH-1N (solid bars) and AH-1NahyI-6 (open bars); (B) ratio of serine protease and metalloprotease activities. The exoprotease activity induced by 1 μM C4-HSL in A. hydrophila AH-1NahyI-6 supernatant and that remaining after inhibition with 1 mM PMSF (due to metalloprotease), 10 mM EDTA (due to serine protease), or 1 mM PMSF and 10 mM EDTA are shown in panel B. In both panels, n = 3 and error bars represent 1 standard deviation.
FIG. 4.
Exoprotease production by A. hydrophila AH-1N and AH-1NahyI-6 as a function of cell density. (A) A. hydrophila AH-1N (solid line) and A. hydrophila AH-1NahyI-6 (broken line) in the presence (■) or absence (□) of 1 μM C4-HSL in the culture medium; (B) inhibition of exoprotease activity by A. hydrophila in the presence (■) or absence (□) of 10 μM 3-oxo-C12-HSL in the culture medium. The data are representative of three experiments.
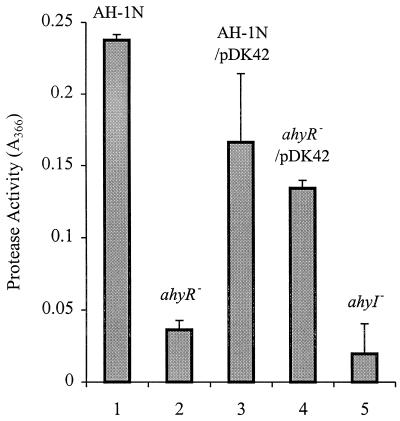
The exoproduct profile of the A. hydrophila ahyR mutant matches that of the ahyI mutant strain in agar plate assays for exoprotease, lipase, hemolysin, nuclease, and amylase activities. Analysis of exoprotease activity from supernatants of overnight A. hydrophila cultures in azocasein assays (Fig. 5) demonstrates that the A. hydrophila AH-1N ahyR mutant is substantially reduced in its ability to produce exoprotease activity. Complementation of the ahyR mutation with a plasmid encoding the ahyR gene restores approximately 50% of protease production (Fig. 5). We cannot explain why full complementation is not obtained but believe that this may be an effect of the introduction of multiple copies of ahyR uncoupled from its normal regulation into A. hydrophila.
FIG. 5.
Exoprotease production in supernatants of A. hydrophila AH-1N and the AH-1N ahyR mutant (ahyR−). ahyI−, A. hydrophila AH-1NahyI-6; n = 3 and error bars represent 1 standard deviation.
The effect of the ahyR mutation on protease production does not appear to be due to an effect on AHL production. C. violaceum CV026 T-streaks show that the A. hydrophila AH-1N ahyR mutant and A. hydrophila AH-1N produce similar levels of AHL after overnight incubation (data not shown) and that the exogenous addition of 5 μM C4-HSL to the A. hydrophila AH-1N ahyR mutant does not restore any protease production (data not shown).
Metalloprotease and serine protease activities are under quorum sensing control.
Previous studies have demonstrated that A. hydrophila possesses both serine protease and metalloprotease activities (25). To investigate the contribution of quorum sensing to the activation of both activities in A. hydrophila, exoprotease activity was assayed in the supernatant of overnight cultures of the ahyI-negative mutant AH-1NahyI-6 induced with 1 μM C4-HSL. In the presence of 10 mM EDTA (metalloprotease inhibitor), 1 mM PMSF (serine protease inhibitor), or 10 mM EDTA plus 1 mM PMSF, it was demonstrated that both activities were activated by C4-HSL (Fig. 3B). Serine protease accounted for approximately 60% of the induced activity, while metalloprotease accounted for approximately 30%. The residual 10% activity is presumably attributable to the azocaseinolytic activity of one or more of the additional exoproteases observed in zymography (Fig. 2C and D). Further evidence for the control of both protease activities was obtained from SDS-PAGE analysis of the TCA-precipitable exoproducts. A. hydrophila AH-1N ahyI-6 lacks proteins at approximately 35 kDa and approximately 65 kDa that are present in the wild type and in the mutant cultured with 1 μM C4-HSL (Fig. 2A). The molecular masses of these proteins correspond to the published sizes of the serine protease (70 kDa [25, 42]) and metalloprotease (35 kDa [25, 41]) of A. hydrophila.
Under suitable culture conditions, protease production by A. hydrophila at a high population density occurs at 22 and 30°C; however, a growth temperature of 37°C inhibits exoprotease production (27, 36). A. hydrophila AH-1N and the ahyI mutant AH-1NahyI-6 were incubated in LB overnight at 22, 30, and 37°C. In line with previous studies (27, 36), no protease expression was seen at 37°C, and the addition of 1 μM C4-HSL to either parent or mutant failed to stimulate activity (data not shown). Nevertheless, C. violaceum CV026 T-streak experiments showed that A. hydrophila AH-1N does produce C4-HSL at 37°C, and the serine protease and metalloprotease are active in assays at 37°C, demonstrating that protease production is inhibited at 37°C in a quorum sensing-independent manner.
Inhibition of exoprotease activity by quorum sensing blocking.
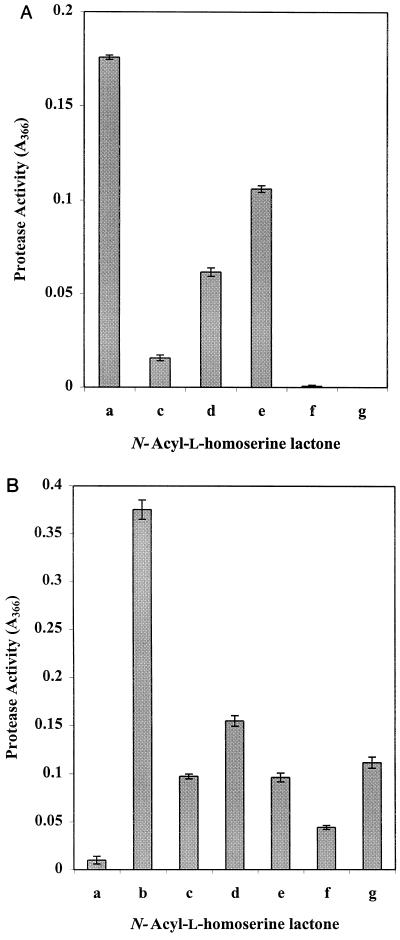
Empirical studies with AHL analogs of the natural ligands for LuxR, LasR, and CarR have shown certain compounds to be antagonistic (3, 37, 48, 61). The application of this antagonism to the induction of pigment by C. violaceum CV026 by 3-oxo-C6-HSL has been used as an assay for long-chain (C>8) AHLs (28) and in the characterization of 3-oxo-C10-HSL, the AHL produced via VanI in Vibrio anguillarum (31). In A. salmonicida, 3-oxo-C10-HSL has been shown to antagonize both the time of induction and final level of exoprotease (51). In this study, we demonstrate that 3-oxo-C10-HSL has the same inhibitory effect on exoprotease production by A. hydrophila (Fig. 4B). To further investigate this phenomenon, we analyzed the antagonistic effects of a range of C4-HSL analogs toward exoprotease production by A. hydrophila (Fig. 6A) and the ahyI mutant AH-1NahyI-6 (Fig. 6B). Exoprotease activity in the supernatant was assayed against azocasein after overnight incubation at 30°C in the presence of AHLs. Results consistently showed that AHLs with an acyl chain of C10, C12, or C14 at 10 μM antagonized protease production and that for the parent strain, 3-oxo-C12-HSL and 3-oxo-C14-HSL almost totally inhibited protease production (Fig. 6A). AHLs with acyl chains of C6 and C8 had little or no antagonistic activity in equivalent experiments (data not shown). Denaturing and nondenaturing PAGE and zymography also demonstrate the activity of 3-oxo-C12-HSL upon the expression of Aeromonas exoproteases (Fig. 2).
FIG. 6.
Influence of long-chain AHLs on exoprotease production by A. hydrophila. (A) A. hydrophila AH-1N exoprotease activity in the presence of no treatment (a), 10 μM C10-HSL (c), 10 μM C12-HSL (d), 10 μM 3-oxo-C10-HSL (e), 10 μM 3-oxo-C12-HSL (f), and 10 μM 3-oxo-C14-HSL (g) in the culture medium. (B) A. hydrophila AH-1NahyI-6 exoprotease activity in the presence of no treatment (a), 1 μM C4-HSL (b), 1 μM C4-HSL plus 10 μM C10-HSL (c), 1 μM C4-HSL plus 10 μM C12-HSL (d), 1 μM C4-HSL plus 10 μM 3-oxo-C10-HSL (e), 1 μM C4-HSL plus 10 μM 3-oxo-C12-HSL (f), and 1 μM C4-HSL plus 10 μM 3-oxo-C14-HSL (g) in the culture medium. In both panels, n = 3 and error bars represent 1 standard deviation.
DISCUSSION
Mutagenesis of the ahyI and the ahyR genes abolishes the ability of A. hydrophila to produce C4-HSL, which in turn substantially reduces exoprotease production. Addition of exogenous C4-HSL restored exoprotease production to the ahyI mutant but not to the ahyR mutant. The serine protease(s) A. hydrophila and A. salmonicida secrete is via the general secretory pathway (16, 38). We have previously shown that unlike the case of P. aeruginosa (2), the general secretory pathway of A. salmonicida (exe) is not under the control of quorum sensing (51). In this study we show that apart from exoproteases, the secretion of proteins using the Exe system, e.g., aerolysin, amylase, and lipase (16), is unaffected by the ahyI mutation, implying that the regulation of protease production by quorum sensing is direct. Further work in A. hydrophila is required to prove this assumption, as the ahyRI system may control expression of a regulatory protein that directly activates transcription of the genes encoding the exoprotease activities.
This work shows that both serine protease and metalloprotease activities are under quorum sensing control and that the inhibition of protease production at low cell population densities and at 37°C (in both ahyI mutant and parent strain) cannot be recovered simply by the addition of C4-HSL. PAGE analysis of A. hydrophila and A. hydrophila ahyI mutant culture supernatant highlights a number of quorum sensing-dependent proteins (Fig. 2A). Identification of some of these requires further work; however, the high-molecular-mass band that barely migrates into the gel is likely to be a multimerized form of the pore-forming toxin aerolysin, absent from the ahyI mutant because of the requirement for proteolytic processing during multimerization (7, 18). No other regulated traits were observed, although we detected an increase in the activity of β-hemolysin, as has been noted previously for protease mutants of A. hydrophila (1). The molecular basis of this phenomenon is still to be elucidated (1).
There is considerable evidence implicating exoprotease activity as a factor in the virulence of Aeromonas. Histopathology studies have revealed tissue damage associated with proteolytic activity (8, 44), and the injection of exoprotease can re-create certain aspects of the pathology of an Aeromonas infection (44). In experimental animal models, protease-null mutants of both A. hydrophila and A. salmonicida exhibit reduced virulence (25, 44), although recent work with defined mutants of A. salmonicida contradicts this finding (58). Nevertheless, the evidence suggests a role for exoprotease in the establishment of infection. This role is consistent with the correlation between the increasing levels of exoprotease inhibitors in fish serum and the decreasing susceptibility to furunculosis seen in comparisons of rainbow trout (Onchorhynchus mykiss), atlantic salmon (Salmo salar), and brown trout (Salmo trutta) (9). Furthermore, a comparison of the furunculosis-resistant rainbow trout with the furunculosis-sensitive brook trout (Salveninus fontinalis) demonstrated a 10-fold reduction in the levels of α2-macroglobulin (10). The exoprotease of A. salmonicida has been proposed as a candidate vaccine target, and its effective use in fish vaccination trials provides practical evidence for a key role in pathogenicity (4).
The regulation of exoprotease activity is therefore important, for if it is expressed too early, effective host defenses will be induced and the infection will most probably be contained. Indeed, null mutations in the S-layer (33), protease (58), hemolysin (59), and GCAT (58) suggest that while Aeromonas spp. can be effective pathogens when lacking one or more secreted virulence factors, a significant reduction in pathogenicity occurs when the ability of the bacterium to evade host defenses is compromised. The tight regulation of virulence gene activation by quorum sensing is seen, for example, in Agrobacterium (20, 34, 62) and Erwinia (5), where it can be proposed to prevent host alert through prevention of exoenzyme production at low cell numbers. A similar situation may exist in Aeromonas (Fig. 4A), and the role of quorum sensing may simply be to rapidly induce the expression of certain virulence factors when a significant population density has been achieved.
Mechanisms have been described whereby information about, for example, ambient temperature (19, 29) and oxygen tension (21, 50) can be transduced and effect changes in gene expression. Prevailing growth environment with both nutrient deprivation and growth rate influencing expression (36) regulates the synthesis of protease. Exoprotease production is therefore dependent on the particular environmental stimuli such as iron, nitrogen availability, temperature, pH, oxygen concentration but in general does not occur until a high cell population density is achieved (1, 6, 14, 26, 27, 36, 40). The integration of these regulatory networks at the corresponding protease promoters with quorum sensing signals is, therefore, an important question for the future.
The potential of blocking quorum sensing to control virulence and hence prevent infection by A. hydrophila was examined in empirical studies where C4-HSL analogs were added to cultures of A. hydrophila. The inhibition of protease activity by the AHLs 3-oxo-C12-HSL and 3-oxo-C14-HSL is an indication of the potential value of the blockade of quorum sensing. Interestingly, a second fish pathogen, V. anguillarum, produces 3-oxo-C10-HSL, which we have shown has antagonistic activity toward the protease activities of both A. hydrophila (Fig. 4B and 6) and A. salmonicida (51). Therefore, a role for this molecule in nature might be to antagonize quorum sensing-dependent virulence in Aeromonas and perhaps provide V. anguillarum with a competitive edge.
ACKNOWLEDGMENTS
This work was supported by grants from the United Kingdom Biotechnology and Biological Sciences Research Council (A01755) to P.W. and G.S.A.B.S., by a Wellcome Trust Prize Studentship to L.F., and by grants from DGICYT and Plan Nacional de I+D (Ministerio de Educación y Cultura, Spain) to J.M.T.
REFERENCES
- 1.Allan B J, Stevenson R M. Extracellular virulence factors of Aeromonas hydrophila in fish infections. Can J Microbiol. 1981;27:1114–1122. doi: 10.1139/m81-174. [DOI] [PubMed] [Google Scholar]
- 2.Chapon-Hervé V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of xcp secretion pathway by multiple quorum sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 3.Chhabra S R, Stead P, Bainton N J, Salmond G P C, Stewart G S A B, Williams P, Bycroft B W. Autoregulation of carbapenem biosynthesis in Erwinia carotovora ATCC 39048 by analogues of N-(3-oxohexanoyl)-l-homoserine lactone. J Antibiot. 1993;46:441–454. doi: 10.7164/antibiotics.46.441. [DOI] [PubMed] [Google Scholar]
- 4.Coleman G, Bennett A J, Whitby P W, Bricknell I R. A 70 kDa Aeromonas salmonicida serine protease-β-galactosidase hybrid protein as an antigen and its protective effect on Atlantic salmon (Salmo salar L.) against a virulent A. salmonicida challenge. Biochem Soc Trans. 1993;21:49S. doi: 10.1042/bst021049s. [DOI] [PubMed] [Google Scholar]
- 5.Cui Y, Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol. 1995;177:5108–5115. doi: 10.1128/jb.177.17.5108-5115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis F, Veillet-Poncet L. Essai de mise en evidence d’une activité proteolytique d’origine intracellulaire chez Aeromonas hydrophila LP50. Can J Microbiol. 1984;30:1190–1192. [PubMed] [Google Scholar]
- 7.Diep D B, Lawrence T S, Ausio J, Howard S P, Buckley J T. Secretion and properties of the large and small lobes of the channel-forming toxin aerolysin. Mol Microbiol. 1998;30:341–352. doi: 10.1046/j.1365-2958.1998.01068.x. [DOI] [PubMed] [Google Scholar]
- 8.Ellis A E, Hastings T S, Munro A L S. The role of Aeromonas salmonicida extracellular products in the pathology of furunculosis. J Fish Dis. 1981;4:41–51. [Google Scholar]
- 9.Ellis A E, Stapleton K J. Differential susceptibility of salmonid fishes to furunculosis correlates with differential serum enhancement of Aeromonas salmonicida extracellular protease activity. Microb Pathog. 1988;4:299–304. doi: 10.1016/0882-4010(88)90090-3. [DOI] [PubMed] [Google Scholar]
- 10.Freedman S J. The role of alpha 2-macroglobulin in furunculosis: a comparison of rainbow trout and brook trout. Comp Biochem Physiol B. 1991;98:549–553. doi: 10.1016/0305-0491(91)90252-9. [DOI] [PubMed] [Google Scholar]
- 11.Fryer J L, Bartholomew J L. Established and emerging infectious diseases of fish. ASM News. 1996;62:592–594. [Google Scholar]
- 12.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 14.Fyfe L, Coleman G, Munro A L. A comparison of the distribution of extracellular proteins produced by the protease-secreting organism Aeromonas salmonicida during aerobic and anaerobic growth. Ann Inst Pasteur Microbiol. 1986;137A:117–123. doi: 10.1016/s0769-2609(86)80016-3. [DOI] [PubMed] [Google Scholar]
- 15.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie K R, Schulze A, Parker M W, Buckley J T. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bind formation. Mol Microbiol. 1995;17:1035–1044. doi: 10.1111/j.1365-2958.1995.mmi_17061035.x. [DOI] [PubMed] [Google Scholar]
- 17.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Howard S P, Buckley J T. Activation of the hole-forming toxin aerolysin by extracellular processing. J Bacteriol. 1985;163:336–340. doi: 10.1128/jb.163.1.336-340.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurme R, Rhen M. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol Microbiol. 1998;30:1–6. doi: 10.1046/j.1365-2958.1998.01049.x. [DOI] [PubMed] [Google Scholar]
- 20.Hwang I, Cook D M, Farrand S K. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J Bacteriol. 1995;177:449–458. doi: 10.1128/jb.177.2.449-458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iuchi S, Lin E C C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J, Golby P, Reeves P J, Stephens S, Winson M K, Salmond G P C, Stewart G S A B, Williams P. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 23a.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Leung K Y, Stevenson R M. Tn5-induced protease-deficient strains of Aeromonas hydrophila with reduced virulence for fish. Infect Immun. 1988;56:2639–2644. doi: 10.1128/iai.56.10.2639-2644.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P V, Hsieh H C. Inhibition of protease production of various bacteria by ammonium salts: its effect on toxin production and virulence. J Bacteriol. 1969;99:406–413. doi: 10.1128/jb.99.2.406-413.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateos D, Anguita J, Naharro G, Paniagua C. Influence of growth temperature on the production of extracellular virulence factors and pathogenicity of environmental and human strains of Aeromonas hydrophila. J Appl Bacteriol. 1993;74:111–118. doi: 10.1111/j.1365-2672.1993.tb03003.x. [DOI] [PubMed] [Google Scholar]
- 28.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation and violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 29.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merino S, Rubires X, Knøchel S, Tomas J M. Emerging pathogens: Aeromonas spp. Int J Food Microbiol. 1995;28:157–168. doi: 10.1016/0168-1605(95)00054-2. [DOI] [PubMed] [Google Scholar]
- 31.Milton D L, Hardman A, Camara M, Chhabra S R, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing in Vibrio anguillarum: characterization of the vanI/R locus and identification of the autoinducer N-(3-oxododecanoyl)-l-homoserine lactone. J Bacteriol. 1996;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Milton D L, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieto T P, Ellis A E. Characterization of extracellular mettalo- and serine-proteases of Aeromonas hydrophila strain B51. J Gen Microbiol. 1986;132:1975–1979. doi: 10.1099/00221287-132-7-1975. [DOI] [PubMed] [Google Scholar]
- 33.Noonan B, Trust T J. The synthesis, secretion and role in virulence of the paracrystalline surface protein layers of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol Lett. 1997;154:1–7. doi: 10.1111/j.1574-6968.1997.tb12616.x. [DOI] [PubMed] [Google Scholar]
- 34.Oger P, Kim K-S, Sackett R L, Piper K R, Farrand S K. Octopine-type Ti plasmids code mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol Microbiol. 1998;27:277–288. doi: 10.1046/j.1365-2958.1998.00671.x. [DOI] [PubMed] [Google Scholar]
- 35.O’Leary W M, Panos C, Helz G E. Studies on the nutrition of Bacterium salmonicida. J Bacteriol. 1956;72:673–676. doi: 10.1128/jb.72.5.673-676.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Reilly T, Day D F. Effects of cultural conditions on protease production by Aeromonas hydrophila. Appl Environ Microbiol. 1983;45:1132–1135. doi: 10.1128/aem.45.3.1132-1135.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Passador L, Tucker K D, Guertin K R, Journet M P, Kende A S, Iglewski B H. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pemberton J M, Kidd S P, Schmidt R. Secreted enzymes of Aeromonas. FEMS Microbiol Lett. 1997;152:1–10. doi: 10.1111/j.1574-6968.1997.tb10401.x. [DOI] [PubMed] [Google Scholar]
- 39.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riddle L M, Graham T E, Amborski R L. Medium for the accumulation of extracellular hemolysin and protease by Aeromonas hydrophila. Infect Immun. 1981;33:728–733. doi: 10.1128/iai.33.3.728-733.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivero O, Anguita J, Paniagua C, Naharro G. Molecular cloning and characterization of an extracellular protease gene from Aeromonas hydrophila. J Bacteriol. 1990;172:3905–3908. doi: 10.1128/jb.172.7.3905-3908.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivero O, Anguita J, Mateos D, Paniagua C, Naharro G. Cloning and characterization of an extracellular temperature-labile serine protease gene from Aeromonas hydrophila. FEMS Microbiol Lett. 1991;65:1–7. doi: 10.1016/0378-1097(91)90461-i. [DOI] [PubMed] [Google Scholar]
- 43.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 44.Sakai D K. Loss of virulence in a protease-deficient mutant of Aeromonas salmonicida. Infect Immun. 1985;48:146–152. doi: 10.1128/iai.48.1.146-152.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai D K. Significance of extracellular protease for growth of a heterotrophic bacterium, Aeromonas salmonicida. Appl Environ Microbiol. 1985;50:1031–1037. doi: 10.1128/aem.50.4.1031-1037.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial enigma: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Schaefer A L, Hanzelka B L, Eberhard A, Greenberg E P. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 50.Spiro S, Guest J R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;6:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 51.Swift S, Karlyshev A V, Durant E L, Winson M K, Chhabra S R, Williams P, Macintyre S, Stewart G S A B. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologues AhyRI and AsaRI and their cognate signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swift S, Throup J P, Salmond G P C, Williams P, Stewart G S A B. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 53.Swift S, Winson M K, Chan P F, Bainton N J, Birdsall M, Reeves P J, Rees C E D, Chhabra S R, Hill P J, Throup J P, Bycroft B W, Salmond G P C, Williams P, Stewart G S A B. A novel strategy for the isolation of luxI homologues: evidence for the widespread distribution of a LuxR:LuxI superfamily in enteric bacteria. Mol Microbiol. 1993;10:511–520. doi: 10.1111/j.1365-2958.1993.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 54.Thornley J P, Shaw J G, Gryllos I A, Eley A. Virulence properties of clinically significant Aeromonas species: evidence for pathogenicity. Rev Med Microbiol. 1997;8:61–72. [Google Scholar]
- 55.Throup J P, Camara M, Briggs G S, Winson M K, Chhabra S R, Bycroft B W, Williams P, Stewart G S A B. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acyl-homoserine lactone signal molecules. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]
- 56.Throup J P, Francis K P. Subscriber’s notebook. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology, suppl. 33. The Red Book Bulletin. New York, N.Y: John Wiley & Sons; 1996. p. 1. [Google Scholar]
- 57.Tomas, J. M. Unpublished data.
- 58.Vipond R, Bricknell I R, Durant E, Bowden T J, Ellis A E, Smith M, MacIntyre S. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect Immun. 1998;66:1990–1998. doi: 10.1128/iai.66.5.1990-1998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong C Y F, Heuzenroeder M W, Flower R L P. Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology. 1998;144:291–298. doi: 10.1099/00221287-144-2-291. [DOI] [PubMed] [Google Scholar]
- 60.Yanisch-Peron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 61.Zhu J, Beaber J W, Moré M I, Fuqua C, Eberhard A, Winans S C. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu J, Winans S C. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol Microbiol. 1998;27:289–297. doi: 10.1046/j.1365-2958.1998.00672.x. [DOI] [PubMed] [Google Scholar]