ABSTRACT
Num1 is a multifunctional protein that both tethers mitochondria to the plasma membrane and anchors dynein to the cell cortex during nuclear inheritance. Previous work has examined the impact loss of Num1-based mitochondrial tethering has on dynein function in Saccharomyces cerevisiae; here, we elucidate its impact on mitochondrial function. We find that like mitochondria, Num1 is regulated by changes in metabolic state, with the protein levels and cortical distribution of Num1 differing between fermentative and respiratory growth conditions. In cells lacking Num1, we observe a reproducible respiratory growth defect, suggesting a role for Num1 in not only maintaining mitochondrial morphology, but also function. A structure–function approach revealed that, unexpectedly, Num1-mediated cortical dynein anchoring is important for normal growth under respiratory conditions. The severe respiratory growth defect in Δnum1 cells is not specifically due to the canonical functions of dynein in nuclear migration but is dependent on the presence of dynein, as deletion of DYN1 in Δnum1 cells partially rescues respiratory growth. We hypothesize that misregulated dynein present in cells that lack Num1 negatively impacts mitochondrial function resulting in defects in respiratory growth.
Keywords: Membrane contact sites, Mitochondria, Organelle positioning
Highlighted Article: The impact of the S. cerevisiae multifunctional mitochondria and dynein anchor protein Num1 on mitochondrial function extends beyond its role in mitochondria–PM tethering to its role in anchoring dynein at the cell cortex.
INTRODUCTION
Eukaryotic cells are defined by the ability to compartmentalize distinct biochemical processes into membrane-bound organelles. However, it has become well established that these organelles do not act in isolation. Rather, organelles engage in non-vesicular interorganelle communication through membrane contact sites (MCSs) in which proteins bring organelle membranes into close proximity without fusion (Scorrano et al., 2019). For example, the multi-protein endoplasmic reticulum (ER) mitochondria encounter structure (ERMES), forms a MCS between the ER and mitochondria, and different sets of proteins, called vCLAMPs, drive the formation of vacuole–mitochondria patches, which tether mitochondria and the vacuole (Elbaz-Alon et al., 2014; Hönscher et al., 2014; Kornmann et al., 2009). Research has demonstrated that MCSs and the proteins that compose them have a myriad of functions within the cell. Several MCS proteins serve solely to tether organelles, and this maintains the spatial distribution of the organelles relative to each other and the overall cell. Other MCS proteins recruit additional factors to tethering sites that facilitate processes like organelle division and inheritance, whereas others facilitate the transport of lipids and metabolites between organelles to maintain proper organelle function and, ultimately, cellular fitness (Eisenberg-Bord et al., 2016; Prinz et al., 2020; Scorrano et al., 2019). More recently, it has come to be appreciated that many of the proteins that function within MCSs have dual functions in the cell, serving roles within and outside of a contact site, functioning at multiple contact sites, or serving as a hub that coordinates multiple processes at a contact site (reviewed in Harper et al., 2020; Lackner, 2019). For example, the Saccharomyces cerevisiae ERMES component Mdm10 functions in the biogenesis of mitochondrial outer membrane β-barrel proteins (Wiedemann and Pfanner, 2017), and the vCLAMP component Vps39 functions in endolysosomal trafficking as a member of the homotypic fusion and vacuole sorting (HOPS) complex (Bröcker et al., 2012). Thus, the regulation of a single multifunctional MCS protein and the interactions it makes within and outside of a contact site can be used to functionally link processes within the cell.
Num1 is another example of a multifunctional MCS protein in S. cerevisiae. Num1 is the core component of the multi-subunit tethering complex called the mitochondria-ER-cortex-anchor (MECA), which brings three membranes into close proximity – the mitochondrion, plasma membrane (PM) and ER (Cerveny et al., 2007; Klecker et al., 2013; Lackner et al., 2013; Tang et al., 2012). Num1 interacts with the mitochondrial outer membrane through its N-terminal coiled-coil (CC) domain (Lackner et al., 2013; Ping et al., 2016; Tang et al., 2012), and with the PM through its C-terminal pleckstrin homology (PH) domain, which interacts with the PM-enriched phosphoinositide phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (Tang et al., 2009; Yu et al., 2004). Unlike the direct interactions Num1 makes with the mitochondrial membrane and PM, the association of Num1 with the ER is likely mediated through an interaction with the integral ER protein Scs2 (Chao et al., 2014; Omer et al., 2018). Num1 also directly interacts with Mdm36 through its CC domain, and this interaction serves to enhance Num1 assembly into clusters that robustly tether mitochondria to the PM and cortical ER (Hammermeister et al., 2010; Lackner et al., 2013; Ping et al., 2016; Won et al., 2021). Num1-mediated mitochondrial tethering ensures the proper positioning and distribution of mitochondria within the cell, and ensures the mother cell retains mitochondria during cell division (Cerveny et al., 2007; Klecker et al., 2013; Lackner et al., 2013). However, in addition to mitochondrial tethering, Num1 has historically been known for its role in dynein-mediated nuclear inheritance. Specifically, dynein is trafficked to the plus ends of astral microtubules emanating from the mitotic spindle and is offloaded onto Num1 clusters at the cortex and subsequently activated. Dynein then captures and walks along astral microtubules, creating the force required for spindle migration and nuclear inheritance (Adames and Cooper, 2000; Carminati and Stearns, 1997; Eshel et al., 1993; Farkasovsky and Küntzel, 2001; Heil-Chapdelaine et al., 2000; Lammers and Markus, 2015; Lee et al., 2005). Cortical anchoring of dynein requires the Num1 CC domain and the multi-subunit dynein co-factor dynactin (Lammers and Markus, 2015; Lee et al., 2003; Moore et al., 2008; Sheeman et al., 2003; Tang et al., 2012).
Our previous work has demonstrated that the mitochondrial-tethering and dynein-anchoring activities of Num1 are coordinated. Specifically, we have demonstrated that MECA contact sites form in the presence of mitochondria. These mitochondria-assembled MECA contact sites then serve as cortical anchoring sites for dynein (Kraft and Lackner, 2017). In further support of a role for mitochondria in dynein anchoring, we have demonstrated that by artificially driving Num1 cluster formation at the PM in a mitochondria-independent manner, two populations of Num1 clusters are created – those that are associated with mitochondria and those that are not – and dynein is preferentially anchored to clusters that are associated with mitochondria (Schmit et al., 2018). Importantly, when mitochondrial-driven assembly of Num1 clusters in buds is inhibited, defects in dynein-mediated nuclear migration are observed (Kraft and Lackner, 2017; Schmit et al., 2018). Together, these data support a role for Num1 as a cortical hub that promotes spatial organization within the cell by coupling the mitochondrial and nuclear positioning pathways. In certain mutant conditions, such as upon the disruption of the Num1 EF hand-like motif (Anderson et al., 2022), overexpression of Mdm36 (Omer et al., 2020) or deletion of Scs2 (Omer et al., 2018), the non-mitochondria associated pool of Num1 can anchor dynein and support dynein function. However, in wild-type situations, our work demonstrates that mitochondrial inheritance positively impacts Num1 cluster formation in buds and dynein function in spindle positioning (Kraft and Lackner, 2017; Schmit et al., 2018). Although we have gained insight into how disrupting Num1-mediated mitochondrial tethering impacts dynein function, it is not clear how the loss of mitochondrial tethering and consequent loss of dynein anchoring impact mitochondrial function.
Here, we address the role of the multifunctional MCS protein Num1 in the maintenance of mitochondrial function. We find that the levels and cortical distribution of Num1 differ between S. cerevisiae cells in fermentative and respiratory growth conditions, suggesting that, like mitochondria, Num1 is regulated by changes in the metabolic state of the cell. In addition to a dramatic decrease in mitochondrial contact with the cell cortex, cells lacking Num1 also exhibit respiratory growth and oxygen consumption defects. Thus, the loss of Num1 negatively impacts mitochondrial function in addition to mitochondrial distribution. Surprisingly, we find that the respiratory growth defect observed in the absence of Num1 extends beyond the loss of the mitochondrial-tethering function of Num1. We find that the respiratory growth defect is due in part to the loss of Num1-mediated dynein anchoring at the cortex. Interestingly, the severe respiratory growth defect observed in cells lacking Num1 is not linked to the canonical function of dynein in nuclear migration but is dependent on the presence of dynein. We hypothesize that it is the misregulation of dynein activity in cells lacking Num1 that leads to a defect in respiratory growth by a mechanism that has yet to be described.
RESULTS
Cells lacking Num1 exhibit defects in respiratory growth and mitochondrial function
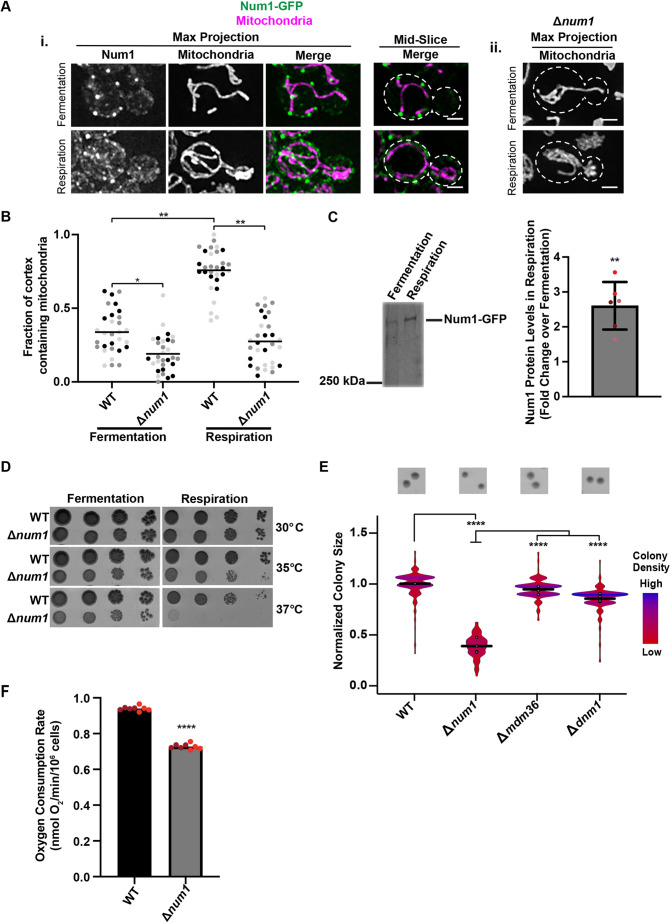
It has been well documented that mitochondrial morphology is impacted by changes in the cellular environment (Stevens, 1981; Visser et al., 1995). In conditions where cells engage predominantly in fermentative growth, mitochondria form simple tubular networks that associate with the PM at discrete attachment points (Fig. 1Ai,B). However, when cells are forced to respire, which requires the process of oxidative phosphorylation, which occurs in mitochondria, mitochondrial mass increases resulting in the formation of an elaborate mitochondrial network that makes extensive contact with the PM in a Num1-dependent manner (Fig. 1Ai,ii,B; Egner et al., 2002; Hoffmann and Avers, 1973; Stevens, 1981). Given these dramatic alterations in mitochondrial morphology and contact with the PM, we expected that Num1 localization would also be altered in respiratory growth conditions. Thus, we tagged Num1 with yeast enhanced GFP (annotated as GFP) at the endogenous locus and visualized this construct in the presence of a mitochondrial marker, mitochondrial matrix-targeted dsRED (mito-dsRED). Concomitant with increased contact between mitochondria and the PM, we also observed changes in the distribution and steady-state levels of Num1 in respiratory conditions (Fig. 1A–C). Compared to fermenting cells (grown in glucose), in which a few discrete Num1 clusters were observed, Num1 assemblies covered a larger portion of the PM in respiring cells (grown in ethanol/glycerol) (Fig. 1Ai). Consistent with this, western blot analysis of Num1–GFP demonstrated a ∼2.6-fold increase in steady-state Num1 protein levels in respiratory growth conditions compared to those seen in fermentative growth conditions (Fig. 1C). Together, these data suggest that, like mitochondria, Num1 is regulated by changes in the metabolic state of cells.
Fig. 1.
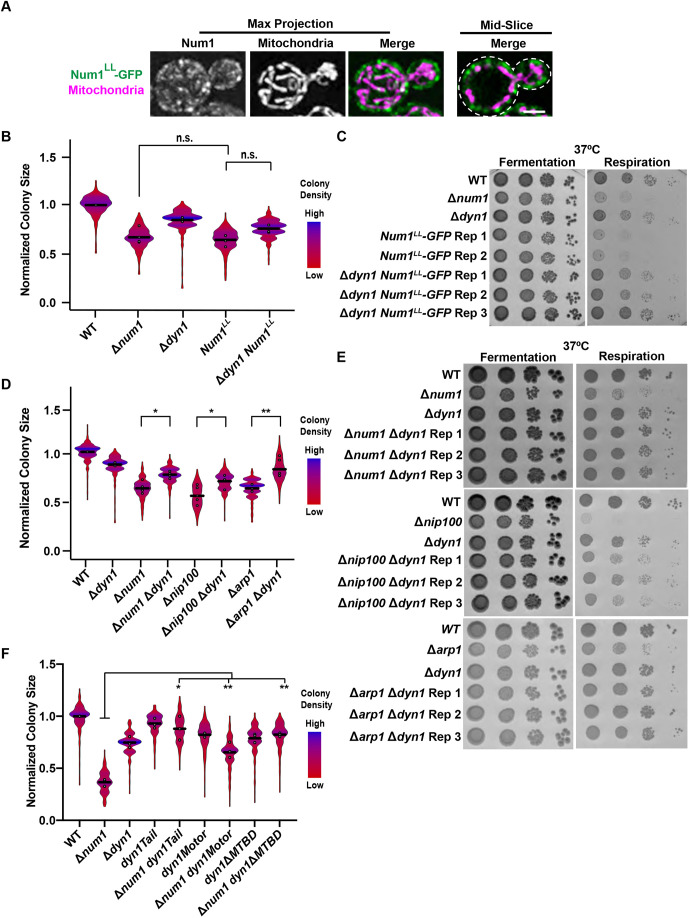
Cells lacking Num1 exhibit a respiratory growth defect and mitochondrial dysfunction. (Ai,ii) WT cells expressing mito-dsRED and Num1–GFP (i) or Δnum1 cells expressing mito-dsRED (ii) were grown in either a fermentative (SCDex) or respiratory (SCEG) growth condition, as indicated, and analyzed by fluorescence microscopy. Whole-cell maximum intensity projections and single focal planes of the middle of each cell, as indicated, are shown. Dashed white lines denote the outline of the cell. Scale bars: 2 µm. (B) The graph represents a quantification of the fraction of the cell cortex at mid-cell that is occupied by mitochondria for WT cells expressing mito-dsRED and Num1–GFP or Δnum1 cells expressing mito-dsRED grown in either a fermentative or respiratory growth condition, as indicated. n=30 cells per strain from three independent experiments, each shown in a different shade, and line representing the grand mean. (C) Whole-cell extracts from strains expressing Num1–GFP grown either in a fermentative or respiratory growth conditions, as indicated, were analyzed by SDS-PAGE and western blotting using an anti-GFP antibody. The graph represents a quantification of the fold change in the levels of the Num1 protein in the respiratory growth condition compared to the fermentative growth condition; n=3 independent experiments each consisting of two technical replicates. The mean±s.d. is shown. The circles denote each technical replicate; and different colors are used to represent the biological replicates. Total protein stain was used as a loading control and normalization standard as described in the Materials and Methods (see Fig. S3A). (D) Serial dilutions of WT or Δnum1 cells onto YPD (fermentative growth condition) or YPEG (respiratory growth condition) agar plates grown at 30°C, 35°C or 37°C, as indicated. Image shown representative of three repeats. (E) Quantification of WT, Δnum1, Δmdm36, and Δdnm1 cell growth at 35°C in respiratory growth conditions. The graph is a violin plot of the radius (in pixels) of WT, Δnum1, Δmdm36, and Δdnm1 colonies as a measure of colony size normalized to the mean radius (in pixels) of WT colonies. The black line denotes the grand mean of at least four independent experiments and circles depict the mean of each independent experiment; n≥344 colonies per strain. Inset images above the graph are representative images of quantified colonies for each strain. (F) Basal oxygen consumption rate of WT or Δnum1 cells, as indicated, grown in YP medium with galactose; n=2 independent experiments with four technical replicates each. The circles denote each technical replicate; and different colors are used to represent the biological replicates. ****P≤0.0001, **P≤0.01, *P≤0.05 (two-tailed unpaired t-test).
Given the corresponding changes in mitochondrial morphology and Num1 localization in response to respiratory growth, we sought to determine whether loss of Num1 impacts mitochondrial function. As one of the crucial functions of mitochondria is the production of ATP through cellular respiration, we examined cell growth on agar plates containing medium that force yeast cells to respire. In this condition, cells that lack NUM1 (Δnum1) exhibited a subtle growth defect at 30°C that increased in severity under temperature stress at 35°C and 37°C (Fig. 1D). To quantify the growth defect at elevated temperatures, we measured colony size for cells grown at 35°C, which allowed us to reproducibly detect and measure the size of individual colonies. In fermentative conditions, the colonies formed by Δnum1 cells were similar in size to those formed by wild-type (WT) cells (Fig. S1A). However, in respiratory growth conditions, a notable and reproducible growth defect was observed for Δnum1 cells (Fig. 1E).
Given that Num1 is the predominant mitochondria–PM tether in yeast, loss of Num1 results in severe defects in mitochondrial morphology and positioning within cells. In both fermentative and respiratory growth conditions, the mitochondrial network in Δnum1 cells collapses and contact with the PM is reduced (Fig. 1Aii,B; Cerveny et al., 2007; Hammermeister et al., 2010; Klecker et al., 2013; Lackner et al., 2013). Thus, we questioned whether the loss of proper mitochondrial morphology and positioning accounts for the defects in respiratory growth observed in Δnum1 cells. To test this idea, we examined how the loss of two proteins known to associate with MECA and to impact mitochondrial morphology, Mdm36 and Dnm1, affect respiratory growth. Mdm36 functions as an accessory component of MECA that bridges Num1–Num1 interactions to enhance Num1 clustering. In the absence of Mdm36, there is a reduction in Num1 tether points and the mitochondrial network is disorganized and collapsed (Fig. S2A; Hammermeister et al., 2010; Lackner et al., 2013; Ping et al., 2016). Dnm1 is a core component of the mitochondrial division machinery and also associates with MECA (Bleazard et al., 1999; Cerveny et al., 2007; Hammermeister et al., 2010; Lackner et al., 2013). In the absence of Dnm1, mitochondria form well-characterized net-like structures that often collapse to one side of the cell (Fig. S2A). Using our colony size assay, we quantified the growth of Δmdm36 and Δdnm1 cells in both fermentative and respiratory growth conditions and found that neither of these cells exhibited a notable growth defect in either growth condition (Fig. S1A; Fig. 1E). Even at 37°C, the respiratory growth of Δmdm36 cells was similar to WT (Fig. S2B). These results suggest that the defect in respiratory growth observed in Δnum1 cells is not a direct result of defects in mitochondrial morphology, but rather is specific to Num1.
Mitochondrial respiration requires the consumption of oxygen; therefore, the rate of oxygen consumption can be used as a readout of mitochondrial function. Thus, to test mitochondrial function more directly in Δnum1 cells, we compared the oxygen consumption rate of Δnum1 cells to WT cells. In agreement with the respiratory growth defect observed for Δnum1 cells, the basal oxygen consumption rate of Δnum1 cells, as measured with a Clark oxygen electrode, was notably decreased in comparison to WT cells (Fig. 1F). Interestingly, the defect in mitochondrial function observed in Δnum1 cells was not a result of defects in individual respiratory chain complex biogenesis and supercomplex assembly (Fig. S3B,C), which also suggests that mitochondrial DNA is not compromised in Δnum1 cells. Together, these data suggest that Num1 impacts mitochondrial function in addition to mitochondrial distribution.
Synthetically clustering the CC domain of Num1 at the PM rescues respiratory growth
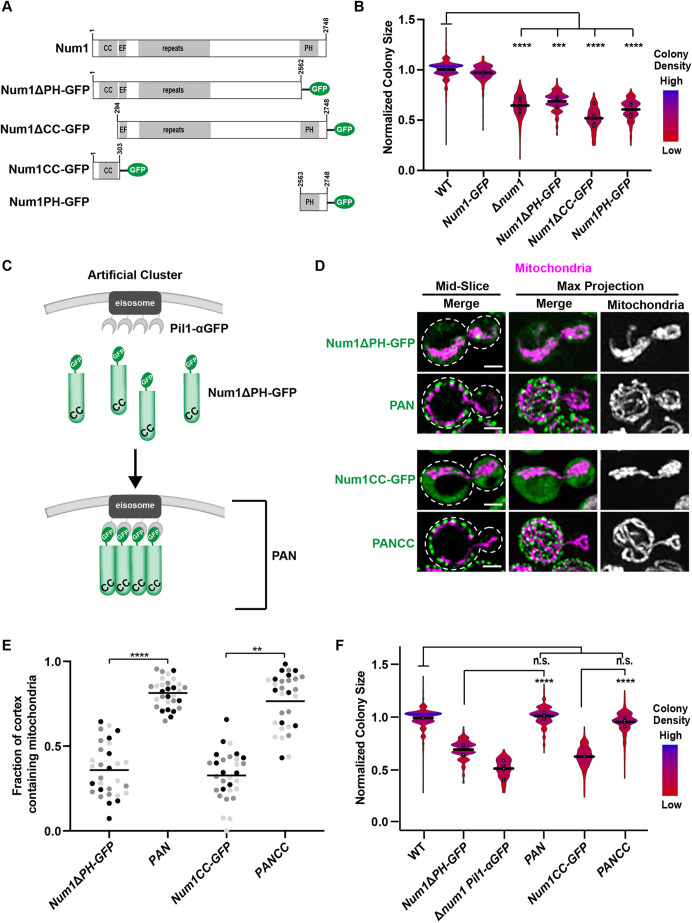
Although mitochondrial morphology is drastically altered by deletion of both MDM36 and DNM1, these cells still have Num1-mediated contacts between mitochondria and the PM (Bleazard et al., 1999; Cerveny et al., 2007; Hammermeister et al., 2010; Lackner et al., 2013; Ping et al., 2016). This raises the possibility that Num1-mediated mitochondria–PM contacts present in Δmdm36 and Δdnm1 cells are able to support respiratory growth and that the respiratory growth defect of Δnum1 cells is a result of the complete loss of these contacts. To test this idea, we sought to determine whether deleting the domains of Num1 that are responsible for Num1-mediated tethering would recapitulate the growth defect of Δnum1 cells. Specifically, we expressed Num1 (Num1–GFP), Num1 lacking its PH domain (Num1ΔPH–GFP), Num1 lacking its CC domain (Num1ΔCC–GFP), or the Num1 PH domain alone (PH–GFP) from the endogenous NUM1 locus as a GFP fusion (Fig. 2A). The PH domain is necessary for Num1 to localize to the PM (Tang et al., 2009; Yu et al., 2004), whereas the CC domain is necessary for the association of Num1 with mitochondria (Lackner et al., 2013; Ping et al., 2016; Tang et al., 2012). Using the colony size assay to quantify growth, we found that constructs that lacked either one of the Num1 domains necessary for tethering had respiratory growth defects similar to Δnum1 cells (Fig. 2B). Similar to Δnum1 cells, none of the Num1 mutants exhibited a notable growth defect in fermentative conditions (Fig. S1B). Together, these data suggest a role for Num1-mediated mitochondria–PM tethering in the maintenance of mitochondrial function and respiratory growth.
Fig. 2.
Anchoring the CC domain of Num1 to the cell cortex rescues the respiratory growth defect of Δnum1 cells. (A) Schematic of Num1 constructs that are used in this study. CC, coiled-coil domain; EF, EF hand-like motif; PH, pleckstrin homology domain. (B) Quantification, presented as a violin plot, of WT, Num1-GFP, Δnum1, Num1ΔPH-GFP, Num1ΔCC-GFP, and Num1PH-GFP cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. The black line denotes the grand mean of at least three independent experiments and the circles depict the mean of each independent experiment; n≥352 colonies per strain. (C) Schematic depicting the GFP–αGFP nanobody artificial Num1 clustering system used in this study. (D,E) Cells expressing mito-dsRED and Num1ΔPH-GFP, PAN, Num1CC–GFP or PANCC were grown in respiratory growth conditions and analyzed by fluorescence microscopy. Whole-cell maximum intensity projections and single focal planes of the middle of each cell are shown (D). Scale bars: 2 µm. Dashed white lines denote the outline of the cell. The graph (E) represents a quantification of the fraction of the cell cortex at mid-cell that is occupied by mitochondria; n=30 cells per strain from three independent experiments, each shown in a different color. The black line denotes the grand mean. (F) Quantification of WT, Num1ΔPH-GFP, Δnum1 Pil1-αGFP, PAN, Num1CC-GFP, and PANCC growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least four independent experiments and the circles depict the mean of each independent experiment; n≥401 colonies per strain. Num1ΔPH-GFP data are recapitulated from Fig. 2B for comparison. ****P≤0.0001, ***P≤0.001; **P≤0.01; n.s., not significant (two-tailed unpaired t-test).
Given that the deletion of the Num1 domains required for tethering reproduces the growth defect seen in Δnum1 cells, we next asked whether the respiratory growth defect could be rescued by artificially tethering mitochondria to the PM. To this end, we artificially clustered Num1 at the PM using the GFP–αGFP nanobody targeting system developed previously in our laboratory (Fridy et al., 2014; Muyldermans, 2013; Schmit et al., 2018). In this system, we appended an αGFP nanobody to the eisosome components Pil1 or Seg1, expressed from their endogenous loci (Pil1–αGFP or Seg1–αGFP). Eisosomes are multi-protein assemblies that form discrete stable puncta at the PM (Moreira et al., 2012; Walther et al., 2006). We then expressed Num1ΔPH–GFP from its endogenous locus in the presence or absence of an αGFP-tagged eisosome component. In the absence of Pil1–αGFP, Num1ΔPH–GFP is diffusely localized in the cytosol with up to two non-cortical, mitochondria-associated puncta per cell and the mitochondrial network is collapsed (Fig. 2D; Schmit et al., 2018). However, in the presence of Pil1–αGFP, Num1ΔPH–GFP is recruited to eisosomes, forming discrete foci on the PM that we refer to as Pil1-associated Num1 (PAN) clusters (Fig. 2C,D; Schmit et al., 2018). We have previously shown that PAN clusters are able to tether mitochondria to the PM and restore the morphology of the mitochondrial network in fermentative growth conditions (Anderson et al., 2022; Schmit et al., 2018). Here, we found that PAN clusters are also able to tether mitochondria to the PM and restore the morphology of the mitochondrial network in respiratory growth conditions (Fig. 2D,E). Using colony size as a quantitative readout of respiratory growth, we found that cells expressing PAN grew similarly to WT cells, whereas cells expressing either Num1ΔPH–GFP or Pil1–αGFP alone exhibited a respiratory growth defect (Fig. 2F). No notable growth defect was observed for any of the strains in fermentative growth conditions (Fig. S1C). These results suggest that the artificial clustering system can bypass the requirement for the Num1 PH domain for full respiratory growth.
The CC domain of Num1 is necessary and sufficient for the interaction of Num1 with mitochondria (Lackner et al., 2013; Ping et al., 2016; Tang et al., 2012). Thus, we sought to determine whether this domain is sufficient to rescue respiratory growth in our artificial clustering system. We expressed the CC domain (amino acids 1–303) of Num1 (Num1CC–GFP) in cells lacking a nanobody fusion or in cells expressing Pil1–αGFP, the latter of which we refer to as PANCC cells (Fig. 2A; Anderson et al., 2022). In cells expressing Num1CC–GFP in the absence of Pil1–αGFP, defects in mitochondrial morphology and respiratory growth similar to those seen in Δnum1 cells were observed (Fig. 2D–F). In contrast, the morphology of mitochondria and respiratory growth of PANCC cells were similar to WT (Fig. 2D–F). No notable growth defect was observed for any of the strains in fermentative growth conditions (Fig. S1C). Thus, synthetically clustering the CC domain of Num1 at the PM is sufficient to rescue respiratory growth.
Restoring mitochondria–PM tethering in the absence Num1 is not sufficient to rescue respiratory growth
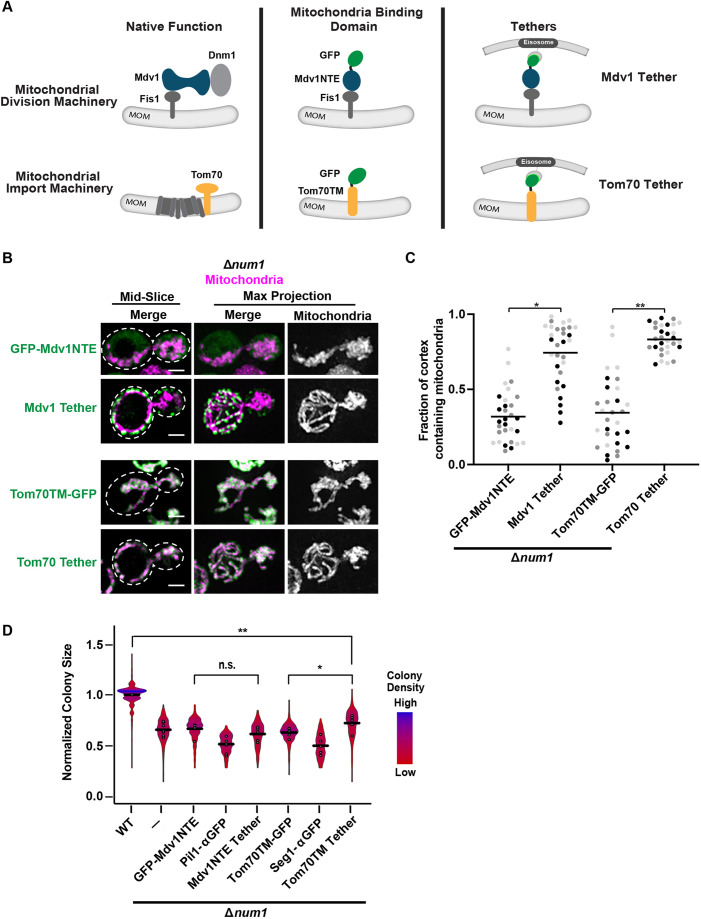
We reasoned that the ability of PANCC to rescue the Δnum1 respiratory growth defect seen above could be occurring in one of two ways – (1) by restoring contact or proximity between mitochondria and the PM, or (2) by providing an activity or function specific to the Num1CC domain. To distinguish between these two possibilities, we constructed artificial mitochondria–PM tethers that lack Num1 domains. To this end, we expressed two mitochondria-binding domains as GFP fusions in our artificial clustering system – the N-terminal extension domain of Mdv1 (amino acids 1–241; GFP–Mdv1NTE) and the transmembrane domain of Tom70 (amino acids 1–30; Tom70TM–GFP). The mitochondria-binding domain–GFP fusions were expressed from an exogenous locus so the native MDV1 and TOM70 genes were not disrupted. These artificial tethering constructs are referred to as Mdv1 Tether and Tom70 Tether (Fig. 3A). Mdv1 is a core component of the mitochondrial fission machinery that associates with Fis1 on the mitochondrial membrane and recruits Dnm1 for mitochondrial fission (Cerveny and Jensen, 2003; Cerveny et al., 2001; Tieu and Nunnari, 2000; Tieu et al., 2002). Previous work has identified the Mdv1 N-terminal extension (NTE) as the domain that directly interacts with Fis1 (Cerveny and Jensen, 2003; Tieu et al., 2002; Zhang and Chan, 2007). The transmembrane (TM) domain of Tom70, a component of the mitochondrial import machinery, has been used successfully in many artificial mitochondria tethers (Kornmann et al., 2009; Lackner et al., 2013).
Fig. 3.
Anchoring mitochondria to the cell cortex in the absence of Num1 does not rescue the Δnum1 phenotype. (A) Schematic of Mdv1 and Tom70 in their native complexes and the mitochondrial-binding domains of each protein used in the modified GFP–αGFP nanobody mitochondria-tethering system. MOM, mitochondrial outer membrane. (B,C) Cells expressing mito-dsRED and GFP–Mdv1NTE, Mdv1 Tether, Tom70TM–GFP or Tom70 Tether in the absence of NUM1 were grown in respiratory growth conditions and analyzed by fluorescence microscopy. Whole-cell maximum intensity projections and single focal planes of the middle of each cell are shown (B). Dashed white lines denote the outline of the cell. Scale bars: 2 µm. The graph (C) represents a quantification of the fraction of the cell cortex at mid-cell that is occupied by mitochondria; n=30 cells per strain from three independent experiments, each shown in a different color. Black line denotes the grand mean. (D) Quantification, presented as a violin plot, of WT, Δnum1, Δnum1 GFP-Mdv1NTE, Δnum1 Pil1-αGFP, Δnum1 Mdv1 Tether, Δnum1 Tom70TM-GFP, Δnum1 Seg1-αGFP and Δnum1 Tom70 Tether cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least five independent experiments and the circles depict the mean of each independent experiment; n≥316 colonies per strain. Δnum1 and Δnum1 Pil1-αGFP data are recapitulated from Fig. 2B and F, respectively, for comparison. **P≤0.01, *P≤0.05, n.s., not significant (two-tailed unpaired t-test).
When GFP–Mdv1NTE or Tom70TM–GFP were expressed in Δnum1 cells in the absence of αGFP-tagged eisosome components, mitochondrial morphology was collapsed (Fig. 3A–C). However, when the GFP fusions were expressed in Δnum1 cells in the presence of the αGFP-tagged eisosome components, which we refer to as Mdv1 Tether and Tom70 Tether, GFP–Mdv1NTE and Tom70TM–GFP were recruited to the PM and mitochondrial morphology was restored with several contact points between mitochondria and the PM (Fig. 3A-C). Interestingly, although these artificial tethering constructs were able to recruit mitochondria to the PM, neither was able to restore respiratory growth to WT levels in cells lacking Num1 (Fig. 3D), which differs from what was observed for PAN and PANCC (Fig. 2F). Neither the Mdv1 Tether nor Tom70 Tether notably affected growth when expressed in either WT or Δnum1 cells grown in fermentative conditions, suggesting the artificial tethers themselves do not have a negative effect on cell growth (Figs S1D and S4A). Similar results were obtained when a fusion between the PH domain of Num1 and the αGFP nanobody (PH–αGFP) was used, instead of an αGFP-tagged eisosome component, to recruit GFP–Mdv1NTE or Tom70TM–GFP to the PM and restore mitochondria–PM tethering. Specifically, respiratory growth was not rescued in Δnum1 cells expressing PH–αGFP with either GFP–Mdv1NTE or Tom70TM–GFP, but was rescued in cells expressing PH–αGFP and Num1ΔPH–GFP (Fig. S4B,C). Thus, these results suggest that it is not simply the loss of mitochondria–PM contact that results in the respiratory growth defect observed in Δnum1 cells, and that the role of Num1 in maintaining mitochondrial function extends beyond its role in tethering mitochondria to the PM.
The Δnum1 respiratory growth defect is not due to defects in dynein-mediated nuclear migration
As mentioned above, Num1 forms a multifunctional MCS. Therefore, we sought to investigate what other Num1 functions could contribute to the maintenance of mitochondrial function. In addition to its role in mitochondrial positioning and tethering, Num1 serves as a cortical anchor for the motor protein, dynein. Once anchored, dynein is then able to capture and walk along astral microtubules to orient the spindle for nuclear inheritance during cell division (Farkasovsky and Küntzel, 2001; Heil-Chapdelaine et al., 2000; Lee et al., 2005). Our previous work demonstrates that in addition to rescuing mitochondrial morphology, PAN and PANCC also support dynein function in nuclear inheritance (Anderson et al., 2022; Schmit et al., 2018). The ability of PAN and PANCC to fully rescue both mitochondrial function and dynein function differs from that of the artificial mitochondria–PM tethers Mdv1 Tether and Tom70 Tether. We found the artificial tethers do not fully restore mitochondrial function (Fig. 3D) nor rescue dynein function, which was evidenced by their inability to support growth in the absence of Kar9, a component of a partially redundant nuclear inheritance pathway (Fig. S4D). Loss of both the dynein and Kar9 pathways for nuclear inheritance results in a severe growth defect (Miller and Rose, 1998), such as that observed for Δkar9 Δnum1 cells and the Δkar9 Δnum1 cells expressing the artificial tethers (Fig. S4D). Given these functional differences between PAN and the artificial tethers, we sought to determine whether the loss of correct dynein function could result in the respiratory growth defect observed in Δnum1 cells.
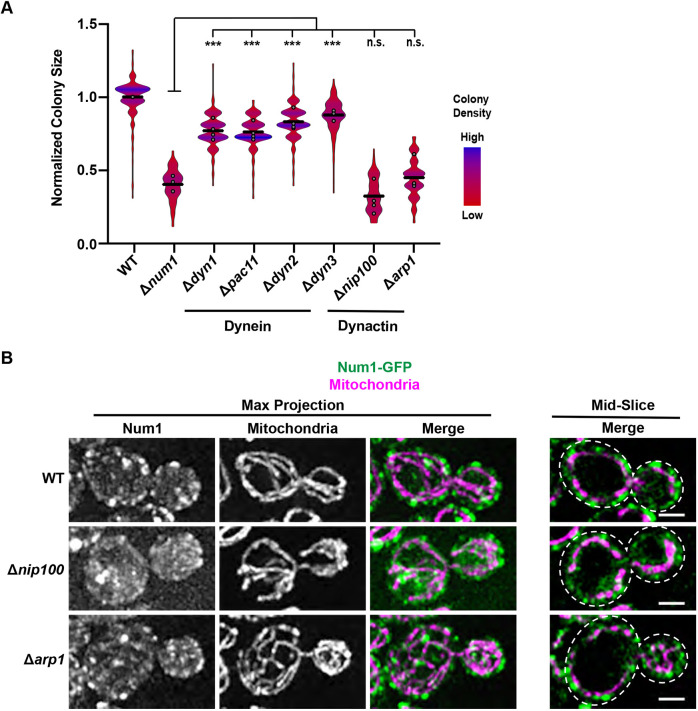
The dynein complex is composed of four components – Dyn1, Pac11, Dyn2 and Dyn3. Cells lacking any one of these components have defects in nuclear inheritance (Carminati and Stearns, 1997; Eshel et al., 1993; Lee et al., 2005; Stuchell-Brereton et al., 2011). Thus, to investigate whether the loss of the dynein-mediated nuclear inheritance role of Num1 could result in the respiratory growth defect observed in Δnum1 cells, we deleted DYN1, PAC11, DYN2 or DYN3 in otherwise WT cells and quantified the ability of these cells to grow when forced to respire. Δdyn1, Δpac11, Δdyn2 and Δdyn3 cells all exhibited a slight growth defect compared to WT cells when forced to respire (Fig. 4A). However, this respiratory growth defect was distinct from that observed in Δnum1 cells. Specifically, the respiratory growth defect observed for Δnum1 cells was reproducibly more severe than that observed in cells lacking any one of the dynein components, despite Δnum1 cells having similar growth to dynein mutants in fermentative conditions (Fig. 4A; Fig. S1E). These results suggest that the more severe respiratory growth defect observed in Δnum1 cells cannot be solely attributed to loss of dynein function in nuclear inheritance.
Fig. 4.
Loss of dynactin function recapitulates the Δnum1 respiratory growth defect. (A) Quantification, presented as a violin plot, of WT, Δnum1, Δdyn1, Δdyn2, Δpac11, Δdyn3, Δnip100, and Δarp1 cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least three independent experiments and the circles depict the mean of each independent experiment; n≥245 colonies per strain. (B) Cells expressing mito-dsRED and Num1–GFP in WT or in the absence of either Nip100 or Arp1, as indicated, were grown in respiratory growth conditions and analyzed by fluorescence microscopy. Whole-cell maximum intensity projections and single focal planes from the middle of each cell are shown. Dashed white lines denote the outline of the cell. Images shown representative of two repeats. Scale bars: 2 µm. ***P≤0.001, n.s., not significant (two-tailed unpaired t-test).
To confirm that the defect in respiratory growth observed in Δnum1 cells is distinct from the specific role of Num1 in dynein-mediated nuclear migration, we deleted components of dynactin, a co-factor for dynein, and assessed the respiratory growth of these strains. Dynactin is required for dynein offloading onto Num1 as well as dynein activation (Kardon et al., 2009; Lee et al., 2003; Moore et al., 2008; Sheeman et al., 2003). The dynactin complex is composed of five subunits – Nip100, Arp1, Jnm1, Ldb18 and Arp10 (Moore et al., 2008; Schroer, 2004). Deletion of any one of the genes that encode components of dynactin, with the exception of ARP10, results in nuclear inheritance defects that phenocopy loss of dynein function including lethality in the absence of Kar9 (Clark and Rose, 2006; Kahana et al., 1998; McMillan and Tatchell, 1994; Moore et al., 2008; Muhua et al., 1994). Thus, to further investigate whether the defects observed in Δnum1 cells are related to defects in dynein-mediated nuclear migration, we deleted ARP1 and NIP100, which encode the two major core components of dynactin, and quantified growth when cells were forced to respire. Interestingly, both Δarp1 and Δnip100 cells exhibited a respiratory growth defect that was similar to that of Δnum1 cells (Fig. 4A). In fermentative growth conditions, no notable defects in growth were observed in Δarp1 and Δnip100 cells (Fig. S1E). Consistent with our results, respiratory growth defects have been observed for cells lacking Num1, Arp1 and Nip100, but not Dyn1, in large-scale screens designed to identify proteins important for mitochondrial function (Dimmer et al., 2002; Steinmetz et al., 2002; Tigano et al., 2015). The respiratory growth defect observed for Δarp1 and Δnip100 cells was not due to defects in Num1-mediated mitochondrial tethering or in mitochondrial morphology (Fig. 4B). The difference in the respiratory growth between the dynein and dynactin mutants is surprising, as dynactin mutants often phenocopy dynein mutants (Schroer, 2004), and provides further support to our notion that the respiratory growth defect observed in cells lacking Num1 and dynactin cannot be solely attributed to loss of dynein function in nuclear inheritance.
The respiratory growth defect of Num1 and dynactin mutants correlates with the inability to anchor dynein at the cell cortex
A shared feature of cells that lack either Num1 or dynactin is the loss of cortically anchored dynein. Specifically, in the absence of Num1 or dynactin components, dynein is not offloaded at the cell cortex and instead remains localized to the plus end of microtubules (Lee et al., 2003; Moore et al., 2008; Sheeman et al., 2003). Given this shared feature, we hypothesized that the respiratory growth defect observed for cells that lack Num1 or components of dynactin is a result of the loss of proper dynein localization and potentially misregulated dynein function. To test this hypothesis, we constructed a strain in which dynein offloading at cortical Num1 clusters is disrupted even when Num1 and the full dynactin complex are present, and determined whether this strain exhibited the same defects in respiratory growth. We made use of a well-characterized Num1 mutant, Num1LL, which harbors two mutations (L167E L170E) that have been shown to disrupt the Num1–dynein association. The Num1LL mutant is able to form clusters that tether mitochondria to the PM; however, the mutant is not able to anchor dynein to the PM (Tang et al., 2012). We confirmed that Num1LL–GFP forms WT-like Num1 clusters at the PM that are able to tether mitochondria (Fig. 5A). We also verified that Num1LL is not able to support dynein function by deleting KAR9 in a strain that expresses Num1LL-GFP from the endogenous NUM1 locus (Δkar9 Num1LL-GFP). Δkar9 Num1LL-GFP cells exhibited a severe growth defect, suggesting that dynein function in nuclear inheritance is disrupted (Fig. S5A).
Fig. 5.
The role of Num1 in anchoring dynein, and not its role in mitochondrial tethering, contributes to the Δnum1 respiratory growth defect. (A) Cells expressing mito-dsRED and Num1LL–GFP were grown in respiratory growth conditions and analyzed by fluorescence microscopy. Whole-cell maximum intensity projections and a single focal plane from the middle of the cell are shown. Dashed white lines denote the outline of the cell. Scale bar: 2 µm. (B) Quantification, presented as a violin plot, of WT, Δnum1, Δdyn1, Num1LL-GFP, and Δdyn1 Num1LL-GFP cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least three independent experiments and the circles depict the mean of each independent experiment; n≥328 colonies per strain. (C) To supplement the colony size assays shown in Fig. 5B, serial dilutions of the indicated strains were spotted onto YPD (fermentative growth condition) or YPEG (respiratory growth condition) agar plates and grown at 37°C. (D) Quantification, presented as a violin plot, of WT, Δnum1, Δdyn1, Δnum1 Δdyn1, Δnip100, Δdyn1 Δnip100, Δarp1 and Δdyn1 Δarp1 growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least three independent experiments and the circles depict the mean of each independent experiment; n≥312 colonies per strain. The fold change in colony size between a double mutant and its respective single mutant was 1.3x for all genotypes examined. (E) To supplement the colony size assays shown in Fig. 5D, serial dilutions of the indicated strains were spotted onto YPD (fermentative growth condition) or YPEG (respiratory growth condition) agar plates and grown at 37°C. Rep 1, 2 and 3 denote three independent isolates of the indicated genotype. Δdyn1 data are recapitulated from Fig. 5B. (F) Quantification, presented as a violin plot, of WT, Δnum1, Δdyn1, dyn1Tail, Δnum1 dyn1Tail, dyn1Motor, Δnum1 dyn1Motor, dyn1ΔMTBD and Δnum1 dyn1ΔMTBD cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. The black line denotes the grand mean of at least three independent experiments and the circles depict the mean of each independent experiment; n≥291 colonies per strain. Images shown in A, C and E are representative of at least two repeats. **P≤0.01, *P≤0.05, n.s., not significant (two-tailed unpaired t-test).
We next examined the ability of Num1LL cells to grow when forced to respire. Similar to what was seen with Δnum1 cells, cells containing the Num1LL-GFP mutation exhibited a severe growth defect when forced to respire at elevated temperatures (Fig. 5B). As this mutant specifically disrupts the association of dynein with Num1 (Tang et al., 2012), these results support the hypothesis that loss of proper dynein localization contributes to the respiratory growth defect observed in cells lacking Num1 or dynactin. We next sought to determine whether deleting dynein in Num1LL-GFP cells could rescue the respiratory growth defect. We found that Num1LL-GFP Δdyn1 cells exhibited a notable and reproducible increase in the ability to grow in respiratory growth conditions in comparison to Num1LL-GFP cells (Fig. 5B), and this increase in respiratory growth was also evident at 37°C (Fig. 5C). In fermentative growth conditions, no notable defects in growth were observed in either Num1LL-GFP or Num1LL-GFP Δdyn1 cells (Fig. S1F; Fig. 5C). These data support the idea that the respiratory growth defect seen in Num1LL-GFP cells is due in part to mislocalized dynein.
Given these results, we sought to determine whether the absence of Dyn1 could alleviate the respiratory growth defect of cells lacking Num1 or components of dynactin. Thus, we deleted DYN1 in strains lacking NUM1, NIP100 or ARP1, and quantified respiratory growth for each double deletion. All of the double-deletion strains, Δnum1 Δdyn1, Δnip100 Δdyn1, and Δarp1 Δdyn1, exhibited a notable increase in the ability to grow in respiratory growth conditions compared to their respective single deletion strains, Δnum1, Δnip100, or Δarp1 (Fig. 5D). The fold change in colony size between a double mutant and its respective single mutant was similar across the strains examined. This increase in respiratory growth in the double mutants was also evident at 37°C (Fig. 5E). Consistent with this, Δnum1 cells lacking the other dynein components, PAC11, DYN2 or DYN3, also exhibited an increase in the ability to grow in respiratory growth conditions at elevated temperatures in comparison to Δnum1 cells (Fig. S5B–D), indicating that the complete dynein complex is required for the respiratory growth defect observed in cells lacking NUM1. In fermentative growth conditions, no notable differences in growth were observed (Figs S1G, S5B–D). The rescue of the respiratory growth defect in the double deletions was not due to the rescue of mitochondrial morphology, as visualizing mitochondria in Δnum1 Δdyn1 cells showed a collapsed mitochondrial network comparable to what was seen in Δnum1 cells (Fig. S5E). Providing further support that the rescue of the respiratory growth defect is independent of rescued mitochondrial morphology, the respiratory growth defect of Δnum1 cells expressing either the Mdv1 Tether or Tom70 Tether, in which cortical tethering of mitochondria is restored, was also rescued by the deletion of DYN1 (Fig. S4E,F). These results further support the idea that the inability to offload and anchor dynein at the cell cortex, and the consequent altered localization of dynein, impact respiratory growth and mitochondrial function.
To examine the properties of dynein that are required for the respiratory growth defect observed, we expressed the following dynein mutants from the endogenous DYN1 locus in Δnum1 cells – (1) Dyn1Tail, which lacks the motor domain and does not depend on plus-end targeting for association with Num1 at the cell cortex (Markus et al., 2009); (2) Dyn1Motor, which is a non-motile form of the motor that localizes to the spindle pole body and plus ends of microtubules but is absent from the cell cortex (Markus et al., 2009); and (3) Dyn1ΔMTBD, which lacks the microtubule-binding domain (MTBD) and has effects on astral microtubule length and dynamics that are equivalent to those seen in the absence of Dyn1 (Lammers and Markus, 2015; Estrem et al., 2017). It is important to note that although each Dyn1 mutant has different effects on the properties of dynein, none of the mutants form a fully functional dynein complex capable of supporting dynein-mediated nuclear migration. Δnum1 cells expressing Dyn1Tail, Dyn1Motor or Dyn1ΔMTBD as the sole source of Dyn1 exhibited respiratory growth similar to Δnum1 Δdyn1 cells (Fig. 5F). In fermentative growth conditions, no notable differences in growth were observed (Fig. S1H). These results indicate that a functional dynein complex and its ability to directly interact with microtubules is required for the respiratory growth defect observed in Δnum1 cells.
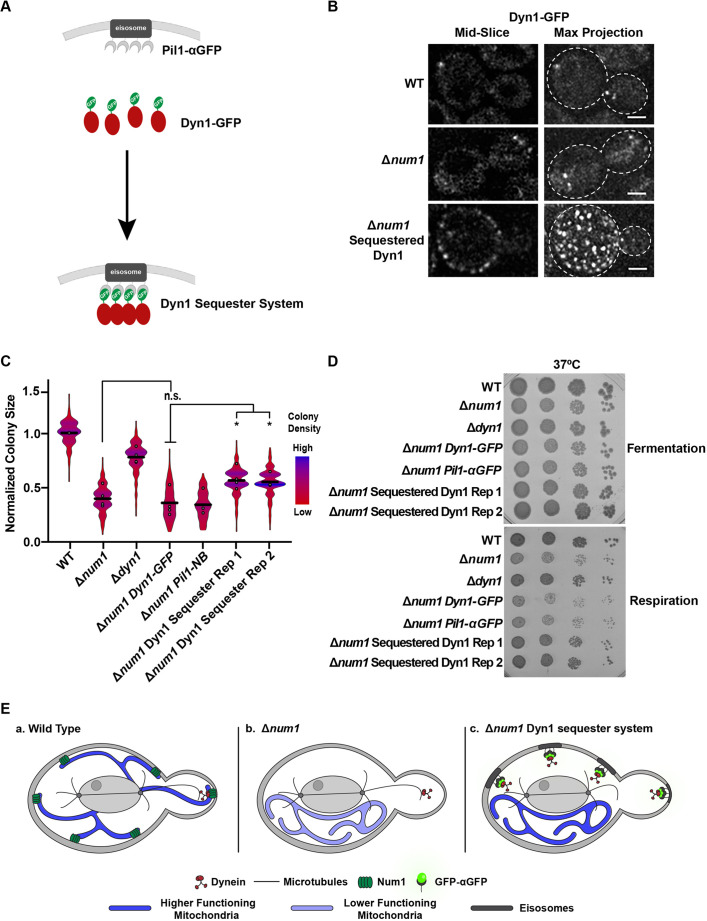
To further test the idea that mitochondrial dysfunction in Δnum1 cells is a consequence of unanchored dynein, we sought to artificially sequester the dynein complex at the PM in Δnum1 cells. To this end, we leveraged the GFP–αGFP nanobody targeting system to sequester Dyn1–GFP at eisosomes in Δnum1 cells expressing Pil1–αGFP (Fig. 6A,B). Consistent with our idea, we found that the respiratory growth defect of Δnum1 DYN1-GFP cells was partially rescued by the expression of Pil1–αGFP (Fig. 6C), and this rescue in respiratory growth was also evident at 37°C (Fig. 6D). In fermentative growth conditions, no notable differences in growth were observed (Fig. S1I). The rescue in respiratory growth was independent of dynein function in nuclear migration as eisosome-sequestered Dyn1–GFP was not functional in nuclear migration. Specifically, the expression of Pil1–αGFP was not able to rescue the growth of Δkar9 Δnum1 DYN1-GFP cells (Fig. S5F). These results provide further evidence that the respiratory growth defect observed in cells lacking Num1 extends beyond the role of Num1 in mitochondrial tethering and the canonical function of dynein in nuclear migration and is dependent on the presence of unanchored dynein (Fig. 6E).
Fig. 6.
Artificially sequestering dynein at the cell cortex rescues the respiratory growth defect of Δnum1 cells. (A) Schematic depicting the GFP–αGFP nanobody system used to artificially sequester Dyn1–GFP at the PM. (B) WT, Δnum1 and Δnum1 Pil1-αGFP cells expressing Dyn1–GFP were grown in fermentative growth conditions and analyzed by fluorescence microscopy. Whole-cell, maximum intensity projections and a single focal plane from the middle of the cell are shown. Dashed white lines denote the outline of the cell. Scale bars: 2 µm. (C) Quantification, presented as a violin plot, of WT, Δnum1, Δdyn1, Δnum1 DYN1-GFP, Δnum1 Pil1-NB and Δnum1 Dyn1 Sequester cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least four independent experiments and the circles depict the mean of each independent experiment; n≥399 colonies per strain. Rep 1 and Rep 2 denote two independent isolates of Δnum1 Sequestered Dyn1. The fold change in colony size between a Δnum1 DYN1-GFP and Dyn1 Sequester Rep 1 and Rep 2 is 1.3 and 1.2, respectively, which is similar to rescue observed in Fig. 5C. (D) To supplement the colony size assays shown in Fig. 6C, serial dilutions of the indicated strains were spotted onto YPD (fermentative growth condition) or YPEG (respiratory growth condition) agar plates and grown at 37°C. Images shown in B and D are representative of two and three repeats, respectively. (E) In WT cells (a), Num1 serves as a cortical anchor for dynein. The loss of Num1-mediated dynein anchoring at the cortex results (b) in reduced mitochondrial function by a yet-to-be-described mechanism that depends on the ability of dynein to directly associate with microtubules. Artificially sequestering the dynein complex at the PM in Δnum1 cells restores mitochondrial function (c), supporting the idea that the reduced mitochondrial function in Δnum1 cells is a consequence of unanchored dynein. *P≤0.05; n.s., not significant (two-tailed unpaired t-test).
DISCUSSION
Many MCSs have been described; however, elucidating the biological functions of these MCSs has been challenging. The challenge, in part, stems from the multifunctional nature of MCS proteins and the sites they create (Eisenberg-Bord et al., 2016; Harper et al., 2020; Prinz et al., 2020; Scorrano et al., 2019). To overcome this challenge, we leveraged synthetic tethering systems and separation of function mutants that allowed us to examine how distinct features and activities of the MCS protein Num1 contribute to the biological functions of MECA contact sites.
Surprisingly, we found that Num1-mediated cortical dynein anchoring is important for normal growth under respiratory conditions (Fig. 6E). Thus, the most straightforward hypothesis to explain the respiratory phenotype observed upon disruption of MECA was incorrect. This unexpected result highlights the complexity of dissecting the functions of MCSs and their resident proteins, and is reminiscent of studies aimed at dissecting the function of Vps39-mediated vCLAMPs. In addition to its role in the formation of Vps39-mediated vCLAMPs (Elbaz-Alon et al., 2014; Hönscher et al., 2014), Vps39 also functions as a component of the HOPS complex that is involved in endosome–vacuole tethering and fusion (Bröcker et al., 2012). Vps39 function in vCLAMPs is independent of its association with other HOPS components, and vCLAMPs and HOPS compete for the pool of Vps39 present in cells (González Montoro et al., 2021). Thus, Vps39 is an excellent example of how a single multifunctional MCS protein can be used to functionally link processes within the cell. Interestingly, Vps39 function is critical in the absence of the ERMES ER–mitochondrial tether, and the loss of Vps39-mediated vacuole-mitochondria tethering was thought to underlie the synthetic interaction observed (Elbaz-Alon et al., 2014; Hönscher et al., 2014; Kornmann et al., 2009). However, a series of Vps39 separation of function mutants was used to demonstrate that the role of Vps39 in the HOPS complex rather than its role in vCLAMPs becomes critical in the absence of ERMES (González Montoro et al., 2018). Thus, similar to our findings, the most straightforward hypothesis was incorrect. Although the exact functions of vCLAMPs are unclear, artificial mitochondria–vacuole tethers cannot rescue the growth defects observed in cells lacking Vps39-mediated vCLAMPs, suggesting that Vps39-mediated vCLAMPs have functions beyond organelle tethering (González Montoro et al., 2021), similar to what we observe for Num1. Thus, the study presented here along with the studies that have dissected the role of Vps39 in vCLAMPs clearly demonstrate the importance of separation of function mutants and synthetic tethering strategies. Such mutants and tools are critical for understanding the unique and specific contributions of a multifunctional MCS protein to MCS function.
To gain a complete picture of MCS function, it is also critical to understand how MCSs and their proteins respond to changes in the cellular environment. Given the dramatic differences in the morphology, proteome and function of mitochondria in cells grown in fermentative versus respiratory growth conditions (Egner et al., 2002; Hoffmann and Avers, 1973; Morgenstern et al., 2017; Stevens, 1981), mitochondrial MCSs are likely altered to help facilitate and/or respond to these changes (Bohnert, 2020). Indeed, this has been observed in this study, and the few other studies that have examined mitochondrial MCSs in both fermentative and respiratory conditions. Compared to fermenting cells, the steady state levels of Num1 are increased and Num1 assemblies cover a larger portion of the PM in respiring cells. ERMES contact sites have also been shown to be upregulated in respiring cells, whereas Vsp39-mediated vCLAMPs are downregulated (Hönscher et al., 2014). Thus, individual mitochondrial MCSs are differentially regulated by changes in the cellular environment. These findings highlight the importance of studying MCSs in a wide range of biological contexts, which will be critical to understanding how the complex, dynamic and interdependent organelle interaction network that exists in cells is integrated and coordinated to respond to cellular needs.
The finding that null mutants in dynein and dynactin did not phenocopy each other with respect to respiratory growth was also unexpected. In yeast, the primary function of the large, multiprotein dynein and dynactin complexes is nuclear migration, and null mutations in dynein and dynactin have similar nuclear migration phenotypes (Carminati and Stearns, 1997; Clark and Rose, 2006; Eshel et al., 1993; Kahana et al., 1998; McMillan and Tatchell, 1994; Moore et al., 2008; Muhua et al., 1994). However, not all phenotypes are shared between dynein and dynactin mutants. For example, dynein and dynactin have opposite effects on microtubule stability; dynein has been shown to destabilize microtubules, whereas dynactin has been shown to stabilize microtubules (Estrem et al., 2017; Laan et al., 2012). In addition, dynactin, but not dynein, has been implicated in a cell wall integrity checkpoint. In response to defects in cell wall synthesis, yeast cells arrest in G2 and this arrest is dependent on components of dynactin but not dynein (Clark and Rose, 2006; Igarashi et al., 2005; Suzuki et al., 2004). Here, we add an additional phenotypic difference between null mutants of dynein and dynactin. We find that cells lacking components of dynactin, but not dynein, have severe respiratory growth defects at elevated temperatures, consistent with results from large-scale screens that observed respiratory growth defects for dynactin mutants, but not dynein mutants (Dimmer et al., 2002; Steinmetz et al., 2002; Tigano et al., 2015). The phenotypic differences observed indicate that dynein and dynactin might each have unique functions outside of nuclear migration or the misregulated activity of one complex as a consequence of the loss of the other complex causes the phenotype observed. For the phenotypic difference in respiratory growth, our data suggest it is the latter. Specifically, the presence of unanchored dynein is the major contributing factor to the respiratory growth defect observed in dynactin mutants.
Why does the inability to anchor dynein at MECA contact sites result in mitochondrial dysfunction? Although we are not yet able to clearly answer this question, there are multiple non-mutually exclusive possibilities to be considered. When the cortical anchoring of dynein is disrupted, dynein accumulates on the plus end of microtubules (Lee et al., 2003; Moore et al., 2008; Sheeman et al., 2003). However, due to the difficulties in imaging dynein, it is unclear if a fraction of the protein is mislocalized to another cellular location and subsequently causes a disruption at that location. In addition, our data suggest that the ability of dynein to directly associate with microtubules is required for the negative effects of dynein on respiratory growth. The dynein–microtubule interaction affects microtubule stability (Estrem et al., 2017; Laan et al., 2012), which might indirectly impact mitochondrial function. It is worth noting that, in mammalian cells, dimeric tubulin has been suggested to interact with and regulate permeability of the mitochondrial outer membrane channel VDAC and consequently impacts respiration (Rostovtseva et al., 2008). Additionally, in yeast, microtubules have been suggested to play a role in the carbon source-dependent regulation of the vacuolar H+-ATPase (Xu and Forgac, 2001), and vacuolar acidification is linked to mitochondrial function and related iron homeostasis (Hughes and Gottschling, 2012; Hughes et al., 2020). Although our data clearly indicate that the inability to anchor dynein at the cortex results in respiratory growth defects that can be alleviated by the deletion of dynein components, we are still working to understand the mechanism underlying this unexpected phenotype.
Considering the only known function of dynein in budding yeast is nuclear migration, the recent connections made between mitochondria and dynein are interesting and unexpected. We have previously shown dynein is anchored by mitochondrial assembled Num1 clusters, and when mitochondrial-driven assembly of Num1 clusters in the bud is inhibited, defects in dynein-mediated nuclear migration are observed (Kraft and Lackner, 2017; Schmit et al., 2018). Here, we extend the functional connection between mitochondria and dynein, and find that the inability to anchor dynein at MECA sites negatively impacts mitochondrial function. Thus, we provide further support for the role of the multifunctional MCS, MECA, as a hub that impacts and integrates the spatial organization and function of organelles within the cell.
MATERIALS AND METHODS
Yeast strains and plasmids
Tables S1 and S2 list all S. cerevisiae strains and primers, respectively, used in this study. All strains and plasmids will be made available upon request. Plasmids used in this study that were previously described are pFA6-link-yEGFP-kanMX6 and pFA6a-link-yEGFP::SpHIS5 (pKT127/pLL55 and pKT128/pLL54; Sheff and Thorn, 2004), pFA6a-His3MX6 and pFA6a-kanMX6 (pLL29 and pLL27; Longtine et al., 1998), and pFA6-NAT-MX3 (pLL25; Goldstein and McCusker, 1999).
The following W303 deletion strains were constructed by replacing the complete open-reading frame (ORF) using PCR-based homologous recombination: Δarp1::HIS (pLL29 template with primers 156/157), Δnip100::HIS (pLL29 template with primers 163/164), Δdyn2::NAT (pLL25 template with 1838/1839 primers), Δpac11::HIS (pLL29 template with primers 1459/1460), and Δdyn3::KAN (pLL27 template with primers 1778/1779). W303 DYN1-yEGFP::KAN was constructed by PCR-based homologous recombination using pLL55 as template with primers 270/271. W303 NUM1LL-yEGFP was constructed by PCR-based homologous recombination of three PCR products (Num1CC with L167E and L170E mutations gblock gene fragment template with primers 513/435, Num1 sequence as template with primers 435/514, and pLL54 template with primers 177/178) into W303 Δnum1::KAN cells. W303 NUM1ΔCC-yEGFP was constructed by PCR-based homologous recombination of two PCR products (Num1 sequence as template with primers 742/514 and pLL54 template with primers 177/178) into W303 cells. W303 Num1PH-yEGFP was constructed by PCR-based homologous recombination of two PCR products (pLL822 with primers 1913/514 and pLL54 template with primers 177/178) into W303 Δnum1::KAN cells. All Num1 mutants were verified by sequencing, with the exception of Num1PH-yEGFP, which was verified by colony PCR and imaging. All W303 strains including all double deletions used as well as Δnum1::HIS Pil1-LaG16::CaURA3, Δnum1::HIS Seg1-LaG16::CaURA3, Seg1-LaG16::CaURA3, Δkar9::HIS NUM1LL-GFP::HIS, Δnip100::HIS NUM1-yEGFP::KAN, Δarp1::HIS NUM1-yEGFP::KAN, Δdyn1::NAT NUM1LL-yEGFP::HIS, Δnum1::HIS DYN1-yEGFP::KAN and Δnum1::HIS DYN1-yEGFP::KAN Pil1-LaG16::CaURA3 were constructed by crossing, sporulation and tetrad analysis.
W303 dyn1Tail-3XFLAG was constructed by PCR-based homologous recombination using 3X-FLAG::HIS (pLL 770) as a template and with primers 271 and 1723. dyn1Motor-3XFLAG was constructed by PCR-based homologous recombination of two PCR products (Dyn1 sequence from genomic prep as template with primers 1780/617 and pLL770 template with primers 270/271). W303 dyn1ΔMTBD-3XFLAG was constructed by PCR-based homologous recombination of three PCR products (dyn1ΔMTBD sequence from genomic prep of a strain yJM1494 given by the Moore Lab was used as a template with primers 614/1449 and 1450/615 as well as pLL770 as a template with primers 270/271. All Dyn1 mutants were verified through sequencing or colony PCR. All other Dyn1 mutant strains were constructed including Δnum1::KAN dyn1Tail-3XFLAG::HIS, Δnum1::KAN dyn1Motor-3XFLAG::HIS and Δnum1::KAN dyn1ΔMTBD-3XFLAG::HIS were constructed by crossing, sporulation and tetrad analysis.
To construct the synthetic Tom70TM mitochondria–PM tether, the GPD::Tom70TM-GFP construct was excised from plasmid p414-GPD::Tom70mito-GFP (pLL357; Lackner et al., 2013) and inserted into plasmid pRS304-TRP1 (pLL181) using SacI/KpnI restriction enzymes to form plasmid pRS304::GPD::Tom70TM-GFP::TRP1 (pLL666). The GPD promoter was replaced with the CYC1 promoter using SacI/SpeI to form plasmid pRS304::CYC1::Tom70TM-GFP::TRP1 (pLL708) used in this study. pLL708 was digested with EcoRV before transformation into Δnum1::KAN cells. Δnum1::KAN Tom70TM Tether strain was constructed by crossing Δnum1::KAN pRS304::CYC1::Tom70TM-GFP::TRP1 cells to Δnum1::KAN Seg1-LaG16::CaURA3 cells, sporulation and tetrad analysis.
To construct the mitochondria–PM tether, Mdv1NTE Tether, yEGFP-Mdv1(1-241) was amplified from MDV1 sequence using primers 1103/1104. Tom70TM-GFP was replaced in plasmid pRS304::ADH1::Tom70TM-GFP::TRP1 (pLL688) with yEGFP-Mdv1(1-241). Specifically, yEGFP-Mdv1(1-241) PCR product was digested with BglII/XhoI and pRS304::ADH1::Tom70TM-GFP::TRP1 was digested with BamHI/XhoI to form pRS304::ADH1::yEGFP-Mdv1(1-241)::TRP1 (pLL698). ADH1 promoter was replaced with CYC1 promoter using SpeI/KpnI to form pRS304::CYC1::yEGFP-Mdv1(1-241)::TRP1 (pLL712) used in this study. pLL712 was digested with EcoRV before transformation into Δnum1::KAN and Δnum1::HIS Pil1-LaG16::CaURA3 cells.
To construct the mitochondria–PM tether using αGFP–Num1PH, the Num1PH construct was inserted downstream of LaG16 using XhoI/BamHI restriction enzymes to form plasmid pR306::GPD::LaG16-NumPH::CaURA3 (pLL822). pLL822 was digested with EcoRV before transformation into Δnum1::KAN, Num1ΔPH-GFP::HIS, Num1CC-GFP::HIS, CYC1::Tom70TM-yEGFP::TRP1, and CYC1::yEGFP-Mdv1NTE::TRP1 to form Num1ΔPH-GFP::HIS LaG16::CaURA3-Num1PH, Num1CC-GFP::HIS LaG16::CaURA3-Num1PH, Δnum1::KAN LaG16::CaURA3-Num1PH, CYC1::Tom70TM-yEGFP::TRP1 LaG16::CaURA3-Num1PH and CYC1::yEGFP-Mdv1NTE::TRP1 LaG16::CaURA3-Num1PH.
For all strains expressing mito-dsRED, plasmid pRS305::GPD::mito-dsRED::LEU/NAT (pLL19; Abrisch et al., 2020) was digested with EcoRI before transformation into the indicated strain.
Imaging
Visualization
All cells were grown to mid-log at 30°C in synthetic complete medium (United States Biological) plus 2% (w/v) dextrose (SCDex) with 2× adenine at pH 6.4 or synthetic complete medium plus 3% ethanol and 3% glycerol (SCEG) with 2× adenine at pH 6.4. Cells were concentrated by centrifugation and mounted on a 4% (w/v) agarose pad (agar was dissolved in SCDex or SCEG medium with 2× adenine at pH 6.4 based on the media cells were grown in). All imaging was performed at room temperature. Images were captured on a spinning disk confocal system (Leica) fit with a spinning disk head (CSU-X1; Yokogawa), a PLAN APO 100×1.44 NA objective (Leica), and an electron-multiplying charge-coupled device camera (Evolve 512 Delta; Photometrics). Image capturing was undertaken using Metamorph software (Molecular Devices). A Z-series of cells using a 0.4-µm step size were taken for a single time point or overtime. Images were deconvolved using the AutoQuantX3 (Media Cybernetics) iterative, constrained 3D deconvolution method. Fiji software (National Institutes of Health) was used to make linear adjustments to brightness and contrast. Deconvolved images are shown.
Image quantification
For quantifying the cortical localization of mitochondria, the proportion of the cell cortex in which mitochondrial signal (red channel) was present was quantified similar to in Kraft and Lackner (2019). Specifically, a five-pixel wide line was drawn around the circumference of the cell in Fiji software. The intensity values of the red channel were collected along the drawn line. Background intensity values were determined by drawing three 5-µm long, five-pixel wide lines at different points near the cells for each field of view used. Intensity values were exported to Excel. The number of data points above the average background intensity value specified for the corresponding field of view divided by the total number of data points was plotted. A total of 30 cells per strain from three independent experiments were quantified. Individual data points from independent experiments are depicted using different colors; no statistical difference was observed between independent experiments for a given strain with the exception of the Mdv1 Tether quantification shown in Fig. 3C. Statistical analyses were performed in GraphPad using a nested two-tailed unpaired t-test with a 95% confidence interval. Designations of significance are indicated in the figure legends.
Cell extracts and western blots
Cells were grown to mid-log in either yeast extract/peptone (YP; BD DIFCO) plus 2% (w/v) dextrose (YPD) medium or YP plus 3% ethanol and 3% glycerol (YPEG) medium. Cells (1.0 OD600) were harvested by centrifugation (3000 g for 1 min) and whole-cell extracts were prepared by a NaOH lysis and trichloroacetic acid (TCA) precipitation procedure. TCA pellets were resuspended in 50 μl of MURB (100 mM MES, pH 7, 1% SDS, and 3 M urea). Whole-cell extracts were analyzed by SDS-PAGE followed by western analysis using Revert Total Protein Stain (LI-COR Biosciences) as loading control as well as anti-GFP [Abclonal Rabbit anti GFP-Tag pAb (AE011), 1:2000] as primary antibody and goat anti-rabbit IgG DyLight 800 (Thermo Fisher Scientific) as secondary antibody (see Fig. 1C and Fig. S3A as an example western blot and corresponding extended western blot and Revert-stained membrane). The total protein stain and immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences). Western blots were quantified using ImageStudio (LI-COR Biosciences). For protein quantification, normalization to total protein was performed as described previously (Pillai-Kastoori et al., 2020). Specifically, the Num1 protein signal (anti-GFP) intensity in each lane was normalized to the total protein stain signal intensity for that lane. Statistical analyses were performed in GraphPad using a two-tailed unpaired t-test with a 95% confidence interval.
Growth assays
To analyze growth by serial dilution, cells were grown in YPD medium overnight at 24°C. Cells were diluted and allowed to grow to mid-log at 30°C, 0.2 OD600 of cells were pelleted (3000 g for 1 min) and resuspended in water to a final OD600 of 0.5. Fivefold serial dilutions were conducted, spotted onto YPD or YPEG agar plates, and grown at 30°C, 35°C or 37°C as indicated.
To analyze growth quantitatively by colony size, cells were grown as described above to achieve mid-log growth. Two 1:100 serial dilutions were performed to achieve cell concentrations of ∼103 cells/ml. Based on the OD600 reading, between 125 and 200 μl of the diluted cell culture were plated onto both YPD and YPEG plates to achieve roughly 100 individual colonies per plate and left to grow at 35°C. YPD plates were removed from the incubator after roughly 48 h of growth, and YPEG plates were removed from the incubator after ∼120 h of growth. For analysis, plates were scanned and images were analyzed using OpenCFU software to identify the radius (in pixels) of individual colonies as a measure of colony size (Geissmann, 2013). In the analysis, colonies were excluded if: (1) the box that defined the area measured for a particular colony encompassed more than one colony; (2) the box that defined the area measured did not completely encompass the colony being measured; or (3) if colony morphology was significantly impacted by touching the side of the plate or other colonies, such that area being measured was not accurate. All strains were normalized to the average colony size of the WT sample grown in the same condition on the same day. Specifically, for each round of the assay, the average radius size of WT colonies was determined for each condition. Colony size for all of the strains from the same experimental round and condition was divided by the average radius size of WT colonies. For graphs that include data collated from experiments that were each normalized to their own WT data set, the WT data sets from all experiments are included in the graph. In addition to showing the grand (i.e. overall pooled) mean of the single colony data, the mean of each independent experiment is shown using circles. Statistical analyses were performed with R programs using a two-tailed unpaired t-test with a 95% confidence interval. The independent experimental means for each genotype were used for the statistical comparisons as described in Lord et al. (2020). Designations of significance are indicated in the figure legends.
Respirometry and biochemical assessment of mitochondrial respiratory chain
Oxygen consumption rates of synchronized WT and Δnum1 cells grown in liquid YP plus 2% (w/v) galactose medium (YP-galactose) at 30°C were assessed polarographically using an Oxygraph system (Hansatech Instruments) according to previously published procedures (Barrientos, 2002; Barrientos et al., 2009). The rates were calculated per manufacturer's instructions from linear responses.
Blue native (BN)-PAGE analyses of respiratory chain complexes II, III, IV, and V solubilized with 1% digitonin were carried out essentially as described before (Swenson et al., 2016; Wittig et al., 2006) using 3–12% gradient polyacrylamide gels (Life Technologies). BN-PAGE- or SDS-PAGE-separated proteins were transferred onto polyvinylidene difluoride or nitrocellulose membranes, respectively, and analyzed by immunoblotting. Protein bands of interest were detected using the following antibodies: anti-porin (1:7500, 459500, Invitrogen); anti-Cox1 (1:2000, ab110270, Abcam); anti-Cox2 (1:2000, ab110271, Abcam); anti-Cox3 (1:2000, ab110259, Abcam); anti-Rip1, anti-Sdh2, and anti-Cyt1 (1:2000, 1:5000, and 1:3000 respectively; kindly provided by Dr Dennis Winge, University of Utah, Salt Lake City, Utah, USA); and anti-F1β (1:5000, a gift from Dr Alexander Tzagoloff, Columbia University, New York, USA). The secondary HRP-coupled antibodies were from Jackson ImmunoResearch Laboratories (anti-mouse IgG; 115-035-068) and Cell Signaling Technology (anti-rabbit IgG; 7074S). All antibodies were validated to ensure specificity and reliability in detection of the proteins of interest.
Supplementary Material
Acknowledgements
We thank members of the Lackner laboratory for suggestions and critical scientific discussions. We also thank Northwestern's Cell Biology Supergroup and the WiLa ICB for constructive feedback on the project. We are extremely grateful to Dennis Winge and Alexander Tzagoloff for providing reagents, Antoni Barrientos (U. Miami) for his help and guidance with the respirometry/polarography measurements, Jeff Moore (CU Anschutz) for the Dyn1ΔMTBD strain, and Jessica Hornick for help and advice on imaging. All microscopy was performed at the Biological Imaging Facility at Northwestern University (RRID:SCR_017767), supported by the Chemistry for Life Processes Institute, the NU Office for Research, the Department of Molecular Biosciences, and the Rice Foundation.
Footnotes
Author contributions
Conceptualization: A.J.W., C.S.H., O.K., L.L.L.; Methodology: A.J.W.; Validation: A.J.W., C.S.H., E.M.R., O.K., L.L.L.; Formal analysis: A.J.W., C.S.H., E.M.R.; Investigation: A.J.W., C.S.H., E.M.R., J.V.D., H.G.A., J.L.F., L.L.L.; Writing - original draft: A.J.W., L.L.L.; Writing - review & editing: C.S.H., E.M.R., J.V.D., J.L.F., O.K.; Visualization: A.J.W., C.S.H., E.M.R., J.L.F., O.K., L.L.L.; Supervision: J.L.F., O.K., L.L.L.; Funding acquisition: O.K., L.L.L.
Funding
This work was supported in part by the National Institutes of Health grants R01 GM120303 (L.L.L.), T32 GM008382 (A.J.W.), T32 GM008449 (C.S.H. and E.M.R.), R35 GM131701-01 (O.K.), P20 GM103499-20, and T32 GM107001-01A1 (J.V.D.) and the National Science Foundation Graduate Research Fellowship GRF DGE-1842165 (C.S.H.). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/lookup/doi/10.1242/jcs.259980.reviewer-comments.pdf.
References
- Abrisch, R. G., Gumbin, S. C., Wisniewski, B. T., Lackner, L. L. and Voeltz, G. K. (2020). Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J. Cell Biol. 219, e201911122. 10.1083/jcb.201911122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adames, N. R. and Cooper, J. A. (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149, 863-874. 10.1083/jcb.149.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, H. L., Casler, J. C. and Lackner, L. L. (2022). Hierarchical integration of mitochondrial and nuclear positioning pathways by the Num1 EF hand. Mol. Biol. Cell 33, mbcE21120610T. 10.1091/mbc.E21-12-0610-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos, A. (2002). In vivo and in organello assessment of OXPHOS activities. Methods 26, 307-316. 10.1016/S1046-2023(02)00036-1 [DOI] [PubMed] [Google Scholar]
- Barrientos, A., Fontanesi, F. and Díaz, F. (2009). Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet. Chapter 19, Unit19.3. 10.1002/0471142905.hg1903s63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J. M., King, E. J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J. and Shaw, J. M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298-304. 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert, M. (2020). Tether me, tether me not-dynamic organelle contact sites in metabolic rewiring. Dev. Cell 54, 212-225. 10.1016/j.devcel.2020.06.026 [DOI] [PubMed] [Google Scholar]
- Bröcker, C., Kuhlee, A., Gatsogiannis, C., Balderhaar, H. J. K., Hönscher, C., Engelbrecht-Vandré, S., Ungermann, C. and Raunser, S. (2012). Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA 109, 1991-1996. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, J. L. and Stearns, T. (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138, 629-641. 10.1083/jcb.138.3.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K. L. and Jensen, R. E. (2003). The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 14, 4126-4139. 10.1091/mbc.e03-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K. L., McCaffery, J. M. and Jensen, R. E. (2001). Division of mitochondria requires a novel DNM1-interacting protein, Net2p. Mol. Biol. Cell 12, 309-321. 10.1091/mbc.12.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K. L., Studer, S. L., Jensen, R. E. and Sesaki, H. (2007). Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell 12, 363-375. 10.1016/j.devcel.2007.01.017 [DOI] [PubMed] [Google Scholar]
- Chao, J. T., Wong, A. K. O., Tavassoli, S., Young, B. P., Chruscicki, A., Fang, N. N., Howe, L. A. J., Mayor, T., Foster, L. J. and Loewen, C. J. R. (2014). Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158, 620-632. 10.1016/j.cell.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Clark, S. W. and Rose, M. D. (2006). Arp10p is a pointed-end-associated component of yeast dynactin. Mol. Biol. Cell 17, 738-748. 10.1091/mbc.e05-05-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer, K. S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W. and Westermann, B. (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847-853. 10.1091/mbc.01-12-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner, A., Jakobs, S. and Hell, S. W. (2002). Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. USA 99, 3370-3375. 10.1073/pnas.052545099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord, M., Shai, N., Schuldiner, M. and Bohnert, M. (2016). A tether is a tether is a tether: tethering at membrane contact sites. Dev. Cell 39, 395-409. 10.1016/j.devcel.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon, Y., Rosenfeld-Gur, E., Shinder, V., Futerman, A. H., Geiger, T. and Schuldiner, M. (2014). A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 30, 95-102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Eshel, D., Urrestarazu, L. A., Vissers, S., Jauniaux, J. C., van Vliet-Reedijk, J. C., Planta, R. J. and Gibbons, I. R. (1993). Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA 90, 11172-11176. 10.1073/pnas.90.23.11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem, C., Fees, C. P. and Moore, J. K. (2017). Dynein is regulated by the stability of its microtubule track. J. Cell Biol. 216, 2047-2058. 10.1083/jcb.201611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky, M. and Küntzel, H. (2001). Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 152, 251-262. 10.1083/jcb.152.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy, P. C., Li, Y., Keegan, S., Thompson, M. K., Nudelman, I., Scheid, J. F., Oeffinger, M., Nussenzweig, M. C., Fenyö, D., Chait, B. T.et al. (2014). A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253-1260. 10.1038/nmeth.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann, Q. (2013). OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 8, e54072. 10.1371/journal.pone.0054072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L. and McCusker, J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553. [DOI] [PubMed] [Google Scholar]
- González Montoro, A., Auffarth, K., Hönscher, C., Bohnert, M., Becker, T., Warscheid, B., Reggiori, F., van der Laan, M., Fröhlich, F. and Ungermann, C. (2018). Vps39 interacts with Tom40 to establish one of two functionally distinct vacuole-mitochondria contact sites. Dev. Cell 45, 621-636.e7. 10.1016/j.devcel.2018.05.011 [DOI] [PubMed] [Google Scholar]
- González Montoro, A., Vargas Duarte, P., Auffarth, K., Walter, S., Fröhlich, F. and Ungermann, C. (2021). Subunit exchange among endolysosomal tethering complexes is linked to contact site formation at the vacuole. Mol. Biol. Cell 32, br14. 10.1091/mbc.E21-05-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammermeister, M., Schödel, K. and Westermann, B. (2010). Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2443-2452. 10.1091/mbc.e10-02-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, C. S., White, A. J. and Lackner, L. L. (2020). The multifunctional nature of mitochondrial contact site proteins. Curr. Opin. Cell Biol. 65, 58-65. 10.1016/j.ceb.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R. A., Oberle, J. R. and Cooper, J. A. (2000). The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151, 1337-1344. 10.1083/jcb.151.6.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, H.-P. and Avers, C. J. (1973). Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science 181, 749-751. 10.1126/science.181.4101.749 [DOI] [PubMed] [Google Scholar]
- Hönscher, C., Mari, M., Auffarth, K., Bohnert, M., Griffith, J., Geerts, W., van der Laan, M., Cabrera, M., Reggiori, F. and Ungermann, C. (2014). Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30, 86-94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Hughes, A. L. and Gottschling, D. E. (2012). An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261-265. 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, C. E., Coody, T. K., Jeong, M.-Y., Berg, J. A., Winge, D. R. and Hughes, A. L. (2020). Cysteine toxicity drives age-related mitochondrial decline by altering iron homeostasis. Cell 180, 296-310.e18. 10.1016/j.cell.2019.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi, R., Suzuki, M., Nogami, S. and Ohya, Y. (2005). Molecular dissection of ARP1 regions required for nuclear migration and cell wall integrity checkpoint functions in Saccharomyces cerevisiae. Cell Struct. Funct. 30, 57-67. 10.1247/csf.30.57 [DOI] [PubMed] [Google Scholar]
- Kahana, J. A., Schlenstedt, G., Evanchuk, D. M., Geiser, J. R., Hoyt, M. A. and Silver, P. A. (1998). The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell 9, 1741-1756. 10.1091/mbc.9.7.1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon, J. R., Reck-Peterson, S. L. and Vale, R. D. (2009). Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc. Natl. Acad. Sci. USA 106, 5669-5674. 10.1073/pnas.0900976106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker, T., Scholz, D., Förtsch, J. and Westermann, B. (2013). The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J. Cell Sci. 126, 2924-2930. 10.1242/jcs.126045 [DOI] [PubMed] [Google Scholar]
- Kornmann, B., Currie, E., Collins, S. R., Schuldiner, M., Nunnari, J., Weissman, J. S. and Walter, P. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477-481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, L. M. and Lackner, L. L. (2017). Mitochondria-driven assembly of a cortical anchor for mitochondria and dynein. J. Cell Biol. 216, 3061-3071. 10.1083/jcb.201702022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, L. M. and Lackner, L. L. (2019). A conserved mechanism for mitochondria-dependent dynein anchoring. Mol. Biol. Cell 30, 691-702. 10.1091/mbc.E18-07-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan, L., Pavin, N., Husson, J., Romet-Lemonne, G., van Duijn, M., López, M. P., Vale, R. D., Jülicher, F., Reck-Peterson, S. L. and Dogterom, M. (2012). Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 148, 502-514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, L. L. (2019). The expanding and unexpected functions of mitochondria contact sites. Trends Cell Biol. 29, 580-590. 10.1016/j.tcb.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, L. L., Ping, H., Graef, M., Murley, A. and Nunnari, J. (2013). Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA 110, E458-E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers, L. G. and Markus, S. M. (2015). The dynein cortical anchor Num1 activates dynein motility by relieving Pac1/LIS1-mediated inhibition. J. Cell Biol. 211, 309-322. 10.1083/jcb.201506119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.-L., Oberle, J. R. and Cooper, J. A. (2003). The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160, 355-364. 10.1083/jcb.200209022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.-L., Kaiser, M. A. and Cooper, J. A. (2005). The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 168, 201-207. 10.1083/jcb.200407036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Lord, S. J., Velle, K. B., Mullins, R. D. and Fritz-Laylin, L. K. (2020). SuperPlots: communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064. 10.1083/jcb.202001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus, S. M., Punch, J. J. and Lee, W.-L. (2009). Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19, 196-205. 10.1016/j.cub.2008.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J. N. and Tatchell, K. (1994). The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J. Cell Biol. 125, 143-158. 10.1083/jcb.125.1.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K. and Rose, M. D. (1998). Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140, 377-390. 10.1083/jcb.140.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., Li, J. and Cooper, J. A. (2008). Dynactin function in mitotic spindle positioning. Traffic 9, 510-527. 10.1111/j.1600-0854.2008.00710.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, K. E., Schuck, S., Schrul, B., Fröhlich, F., Moseley, J. B., Walther, T. C. and Walter, P. (2012). Seg1 controls eisosome assembly and shape. J. Cell Biol. 198, 405-420. 10.1083/jcb.201202097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, M., Stiller, S. B., Lübbert, P., Peikert, C. D., Dannenmaier, S., Drepper, F., Weill, U., Höß, P., Feuerstein, R., Gebert, M.et al. (2017). Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 19, 2836-2852. 10.1016/j.celrep.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua, L., Karpova, T. S. and Cooper, J. A. (1994). A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell 78, 669-679. 10.1016/0092-8674(94)90531-2 [DOI] [PubMed] [Google Scholar]
- Muyldermans, S. (2013). Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775-797. 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- Omer, S., Greenberg, S. R. and Lee, W.-L. (2018). Cortical dynein pulling mechanism is regulated by differentially targeted attachment molecule Num1. eLife 7, e36745. 10.7554/eLife.36745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, S., Brock, K., Beckford, J. and Lee, W. L. (2020). Overexpression of Mdm36 reveals Num1 foci that mediate dynein-dependent microtubule sliding in budding yeast. J. Cell Sci. 133, jcs246363. 10.1242/jcs.246363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai-Kastoori, L., Schutz-Geschwender, A. R. and Harford, J. A. (2020). A systematic approach to quantitative Western blot analysis. Anal. Biochem. 593, 113608. 10.1016/j.ab.2020.113608 [DOI] [PubMed] [Google Scholar]
- Ping, H. A., Kraft, L. M., Chen, W. T., Nilles, A. E. and Lackner, L. L. (2016). Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J. Cell Biol. 213, 513-524. 10.1083/jcb.201511021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, W. A., Toulmay, A. and Balla, T. (2020). The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7-24. 10.1038/s41580-019-0180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtseva, T. K., Sheldon, K. L., Hassanzadeh, E., Monge, C., Saks, V., Bezrukov, S. M. and Sackett, D. L. (2008). Tubulin binding blocks mitochondrial voltage-dependent anion channel and regulates respiration. Proc. Natl. Acad. Sci. USA 105, 18746-18751. 10.1073/pnas.0806303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit, H. L., Kraft, L. M., Lee-Smith, C. F. and Lackner, L. L. (2018). The role of mitochondria in anchoring dynein to the cell cortex extends beyond clustering the anchor protein. Cell Cycle 17, 1345-1357. 10.1080/15384101.2018.1480226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer, T. A. (2004). Dynactin. Annu. Rev. Cell Dev. Biol. 20, 759-779. 10.1146/annurev.cellbio.20.012103.094623 [DOI] [PubMed] [Google Scholar]
- Scorrano, L., De Matteis, M. A., Emr, S., Giordano, F., Hajnóczky, G., Kornmann, B., Lackner, L. L., Levine, T. P., Pellegrini, L., Reinisch, K.et al. (2019). Coming together to define membrane contact sites. Nat. Commun. 10, 1287. 10.1038/s41467-019-09253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman, B., Carvalho, P., Sagot, I., Geiser, J., Kho, D., Hoyt, M. A. and Pellman, D. (2003). Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13, 364-372. 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Sheff, M. A. and Thorn, K. S. (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661-670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Steinmetz, L. M., Scharfe, C., Deutschbauer, A. M., Mokranjac, D., Herman, Z. S., Jones, T., Chu, A. M., Giaever, G., Prokisch, H., Oefner, P. J.et al. (2002). Systematic screen for human disease genes in yeast. Nat. Genet. 31, 400-404. 10.1038/ng929 [DOI] [PubMed] [Google Scholar]
- Stevens, B. (1981). Mitochondrial Structure. Cold Spring Harb. Monograph Archive 11, 471-504. [Google Scholar]
- Stuchell-Brereton, M. D., Siglin, A., Li, J., Moore, J. K., Ahmed, S., Williams, J. C. and Cooper, J. A. (2011). Functional interaction between dynein light chain and intermediate chain is required for mitotic spindle positioning. Mol. Biol. Cell 22, 2690-2701. 10.1091/mbc.e11-01-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Igarashi, R., Sekiya, M., Utsugi, T., Morishita, S., Yukawa, M. and Ohya, Y. (2004). Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 6, 861-871. 10.1038/ncb1162 [DOI] [PubMed] [Google Scholar]
- Swenson, S., Cannon, A., Harris, N. J., Taylor, N. G., Fox, J. L. and Khalimonchuk, O. (2016). Analysis of oligomerization properties of heme a synthase provides insights into its function in eukaryotes. J. Biol. Chem. 291, 10411-10425. 10.1074/jbc.M115.707539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Punch, J. J. and Lee, W.-L. (2009). A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle 8, 3182-3190. 10.4161/cc.8.19.9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Germain, B. S. and Lee, W.-L. (2012). A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J. Cell Biol. 196, 743-756. 10.1083/jcb.201112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q. and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353-366. 10.1083/jcb.151.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K. and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445-452. 10.1083/jcb.200205031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano, M., Ruotolo, R., Dallabona, C., Fontanesi, F., Barrientos, A., Donnini, C. and Ottonello, S. (2015). Elongator-dependent modification of cytoplasmic tRNALysUUU is required for mitochondrial function under stress conditions. Nucleic Acids Res. 43, 8368-8380. 10.1093/nar/gkv765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, W., van Spronsen, E. A., Nanninga, N., Pronk, J. T., Gijs Kuenen, J. and van Dijken, J. P. (1995). Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Leeuwenhoek 67, 243-253. 10.1007/BF00873688 [DOI] [PubMed] [Google Scholar]
- Walther, T. C., Brickner, J. H., Aguilar, P. S., Bernales, S., Pantoja, C. and Walter, P. (2006). Eisosomes mark static sites of endocytosis. Nature 439, 998-1003. 10.1038/nature04472 [DOI] [PubMed] [Google Scholar]
- Wiedemann, N. and Pfanner, N. (2017). Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86, 685-714. 10.1146/annurev-biochem-060815-014352 [DOI] [PubMed] [Google Scholar]
- Wittig, I., Braun, H.-P. and Schägger, H. (2006). Blue native PAGE. Nat. Protoc. 1, 418-428. 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- Won, J., Choi, Y., Yun, Y. and Lee, H. H. (2021). Biochemical characterization of the Num1-Mdm36 complex at the mitochondria-plasma membrane contact site. Mol. Cells 44, 207-213. 10.14348/molcells.2021.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T. and Forgac, M. (2001). Microtubules are involved in glucose-dependent dissociation of the yeast vacuolar [H+]-ATPase in vivo. J. Biol. Chem. 276, 24855-24861. 10.1074/jbc.M100637200 [DOI] [PubMed] [Google Scholar]
- Yu, J. W., Mendrola, J. M., Audhya, A., Singh, S., Keleti, D., DeWald, D. B., Murray, D., Emr, S. D. and Lemmon, M. A. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13, 677-688. 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. and Chan, D. C. (2007). Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. USA 104, 18526-18530. 10.1073/pnas.0706441104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.