Abstract
Atopic dermatitis (AD) has been shown to be closely related to gut dysbiosis mediated through the gut–skin axis, and thus the gut microbiome has recently been explored as a potential therapeutic target for the treatment of AD. Contrasting and varying efficacy have been reported since then. In order to investigate the determining factor of probiotics responsiveness in individuals with AD, we initiated the analysis of 41 AD patients with varying disease severity in Hong Kong, whereas the severity was assessed by Eczema Area and Severity Index (EASI) by board certified dermatologist. 16S rRNA sequencing on the fecal samples from AD patients were performed to obtain the metagenomics profile at baseline and after 8 weeks of oral administration of a novel E3 probiotics formula (including prebiotics, probiotics and postbiotics). While EASI of the participants were significantly lower after the probiotics treatment (p < 0.001, paired Wilcoxon signed rank), subjects with mild AD were found to be more likely to respond to the probiotics treatment. Species richness among responders regardless of disease severity were significantly increased (p < 0.001, paired Wilcoxon signed rank). Responders exhibited (1) elevated relative abundance of Clostridium, Fecalibacterium, Lactobacillus, Romboutsia, and Streptococcus, (2) reduced relative abundance of Collinsella, Bifidobacterium, Fusicatenibacter, and Escherichia-Shigella amid orally-intake probiotics identified using the machine learning algorithm and (3) gut microbiome composition and structure resembling healthy subjects after probiotics treatment. Here, we presented the gut microbiome dynamics in AD patients after the administration of the E3 probiotics formula and delineated the unique gut microbiome signatures in individuals with AD who were responding to the probiotics. These findings could guide the future development of probiotics use for AD management.
Keywords: atopic dermatitis, gut microbiome, probiotic, metagenomics, machine learning, Lactobacillus
1. Introduction
Atopic dermatitis (AD) is a complicated chronic immune-mediated skin disorder presenting with remarkable and recurrent pruritic eczematous lesions, which could be provoked by environmental stimulus and skin hyperactivity. It could not only affect infants, children, and adolescents but also be increasingly identified in adults [1,2,3,4,5]. AD contributes as the leading cause of skin disorders globally and is one of the top non-fatal illnesses that may be regarded as an emerging endemic which poses a significant socio-economic burden [6].
Several factors are evidenced to be associated with AD incidence, susceptibility and severity, including but not limited to environmental factors, genetic composition, integrity of skin barrier, and variability in immune response. For example, it has been demonstrated that greater helper T cell Th17/Th22 polarization is observed in Asian populations with a combined manifest of AD and psoriasis [7,8]. Loss-of-function mutation in the filaggrin (FLG) gene is another well-recognized risk factor leading to severe AD with a diverge reported prevalence in different ethnic groups [9,10,11,12,13,14,15]. On top of the regular risk factors, a growing number of studies about the association between intestinal microbiome dysbiosis and AD has emerged owing to the recent advancement in next-generation sequencing (NGS) [13,16,17,18,19]. Decreased intestinal bacterial biodiversity, lower relative abundance of Bifidobacterium, Akkermansia and Fecalibacterium, depletion of Coprococcus eutactus, and enrichment of Clostridia and Fecalibacterium prausnitzii have been observed to be extremely related to the infants with eczema or AD onset early in life [17,20,21,22,23,24,25,26]. This association of gut microbiome with the skin condition is commonly known as the “gut-skin axis”, which was originally postulated in 1930 [27]. The concept and the importance of the axis are becoming increasingly appreciated by the wider dermatologists community nowadays [28].
Hence, the use of probiotics appeals as a possible intervention to augment the standard-of-care treatment of using moisturizing cream for hydration, topical/oral anti-inflammatory drugs for reducing inflammation. Compared with conventional therapies, probiotics have a favourable pharmacological profile and a low production cost, which makes it a more feasible and accessible option. At a cellular and molecular level, it has been demonstrated that probiotics could potentially regulate allergic responses through Th2 suppression and Treg activation [22,29,30,31,32]. There are a number of trials evaluating the clinical efficacy of the use of probiotics prenatally on mothers, infants, and children in preventing and treating AD, but the results remain inconclusive. Recent meta-analyses reckoned the administration of probiotics to significantly reduce SCORAD index in AD patients and might be beneficial in preventing AD onset with a less confident extent [10,16,24,25,29,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50].
In this study, our group aims to evaluate the effectiveness and gut microbiome evolution upon the application of prebiotics, probiotics and postbiotics mixture in southern Chinese AD patients through 16S rRNA sequencing. The findings could help to evaluate, refine, and improve the clinical efficacy of probiotics as an intervention in AD patients.
2. Materials and Methods
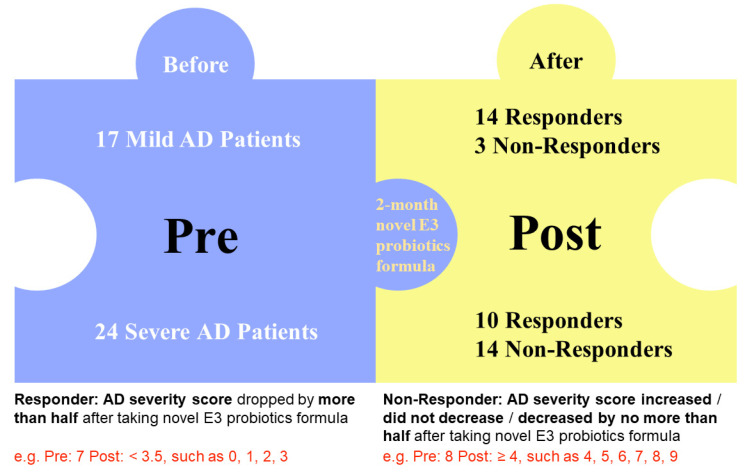
Study design Forty-one adult (18–73 years) AD patients of Chinese ethnicity were recruited from a community trial through a collaboration between The Chinese University of Hong Kong and the BioMed Microbiome Research Centre. All participants (1) with chronic AD that has been present for at least 3 years before the screening visit with any severity; (2) aged above 18; and (3) who provided informed consent were included. Subjects with any one of the following conditions were not recruited or were excluded from the study: (1) history of adverse reaction to probiotics; (2) known overt bacterial infections in the skin; (3) known pregnancy; (4) premorbid medical conditions, such as cardiovascular, liver or renal dysfunction or diabetes mellitus; (5) having used oral corticosteroids, oral antibiotics, other immunosuppressive or any preparation of oral herbal medicines for the treatment of AD in the past one month; (6) having been diagnosed with scabies, allergic contact dermatitis, seborrheic dermatitis or psoriasis; and (7) had taken anti-coagulant or anti-platelet drugs in the past month. All forty-one recruited subjects were included in the subsequent analysis. All patients involved in this study were first diagnosed with AD and evaluated the AD severity by a professional dermatologist according to the EASI scale and fecal samples were collected for downstream sequencing. Then, the patients were orally administered the probiotic mixture for two consecutive months after which AD severity of each recruitment was assessed again and fecal samples were collected for the follow-up studies. Moreover, AD patients were separated into responders and non-responders according to the alterations of EASI score. Those whose AD severity score dropped by more than half before and after taking probiotic mixture were considered as responders, while non-responders were defined as the patients whose AD severity score increased, or did not decrease, or decreased by no more than half after probiotic mixture administration. Informed consent statements were obtained from all recruited subjects in this study. This study received approval from the Research Ethics Committee of Hong Kong Doctors Union. There was no change to the trial protocol after it commenced.
Eczema Area and Severity Index (EASI) EASI score assess the extent (i.e., area) and severity of inflamed areas in AD [51]. It covers 4 body regions, namely head and neck, trunk, upper limbs, and lower limbs. Each body region will be evaluated according to the average intensity of 4 signs including redness, thickness, scratching, and lichenification against a 3-point scale. The severity score of respective body region will be the sum of the average intensity score of the above-mentioned signs. Another component of EASI score involves the percentage of skin affected by AD rated against a 6-point scale. The final EASI score is the sum of severity score multiplied by area score and a multiplier of respective body region. EASI score could therefore range from 0 to 72 with higher score indicating worse severity [52]. Owing to the total number of subjects recruited, subjects were categorized into two subgroups of which subjects with EASI less than 16 were regarded as mild AD, and subjects with EASI larger than or equal to 16 were regarded as severe AD group.
Probiotic mixture All AD patients received daily capsule of a novel E3 probiotics formula developed by BioMed Microbiome Research Centre (BioMed Laboratory Company Limited, Hong Kong) containing a mixture of 7 types of highly effective gastro-resistant probiotics (not less than 2 × 1010 CFU/capsule at the time of production), effective postbiotic HK-LP (heat killed L. plantarum, 10 mg/capsule), and triple prebiotics containing inulin (22 mg/capsule), Galacto-oligosaccharides (GOS) (8.1 mg/capsule), and Fructo-oligosaccharides (FOS) (0.9 mg/capsule) for two months. The product was designed not as a single strain but as a bacteria mixture with Lactobacilli and Bifidobacterium. The probiotic mixture was composed of Lactobacillus rhamnosus GG, Lactobacillus acidophilus GKA7, Lactococcus lactis GKL2, Lactobacillus casei GKC1, Lactobacillus paracasei GKS6, Bifidobacterium bifidum GKB2, and Bifidobacterium lactis GKK2. L. rhamnosus GG formula was evidenced to reduce the occurrence and recurrence risks of allergy and eczema simultaneously and B. lactis was previously proved to strengthen the immunity system and improve symptoms of allergy and eczema [53,54,55,56,57]. Additionally, postbiotics HK-LP involved in this formula was proved to enhance the probiotics functions [58,59]. Moreover, prebiotics act as an energy source for probiotics, which not only enhance the probiotics function but also foster intestinal peristalsis as well as detoxification [60,61,62,63,64].
Library Preparation and 16SrRNA Sequencing All the fecal samples were processed in BioMed Laboratory (BioMed Laboratory Company Limited, Hong Kong) and were first homogenized in PurSafe® DNA and RNA preservative (Puritan, Guilford, ME, USA) and subjected to beating with glass beads (425–600 μm, Sigma-Aldrich, Burlington, MA, USA) for 1 h by following the instructions provided. DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) was used to conduct the isolation of Microbial DNA from fecal samples. The extracted DNA concentration of each sample was quantified using a Qubit™ dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA, USA) with Qubit 3 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Amplicon library was constructed using 515F(5′-GTGCCAGCMGCCGCGG-3′)/907R(5′-CCGTCAATTTCMTTTRAGTTT-3′) primer pair spanning targeting at V4-V5 hypervariable of 16S rRNA genes, together with adapter sequences, multiplex identifier tags, and library keys. 16S rRNA gene sequencing was performed using the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA) following the original Earth Microbiome Project Protocols. In the end, index barcodes and adapters removed pair-end clean reads were obtained for the downstream analysis [65].
Microbiome bioinformatics analysis Microbiome bioinformatics data were analyzed using a plugin-based system, QIIME 2-2021.4, integrating various microbiome analysis algorithms and tools [66]. Demultiplexed reads were firstly subjected to quality control and denoising filter of sequence data with DADA2 [67] using the q2-dada2 plugin to retrieve exact amplicon sequence variants (ASVs) [68]. All ASVs were then aligned with mafft [69] and then a phylogenetic tree was generated using fastree2 [70] via the q2-phylogeny plugin. Taxonomic annotation of the resulting ASV was carried out using the q2-feature-classifier [71] plugin and a pre-trained Naive Bayes classifier which was based on SILVA v138 taxonomic reference database with 99% similarity [72,73,74]. Diversity analyses were performed using the R package microeco (v0.3.2) [75]. We used six metrics to indicate alpha diversity: Observed OTUs, Chao1 Index (Chao1), ACE Index (ACE), Shannon Diversity Index (Shannon), Simpson Index (Simpson), and Faith’s phylogenetic diversity (PD). Furthermore, Beta diversity was calculated based on the Jaccard distance metric, Bray–Curtis distance metric, weighted UniFrac, and unweighted UniFrac distance metrics. The PERMANOVA test on beta diversity (999 permutations) was applied to compare the microbial community dissimilarity across groups using the adonis function in vegan R package to adjust the clinical variables and batch effects [76]. Differential abundance test between the Pre and Post groups was conducted using random forest and non-parametric test.
Statistical analysis All the statistical analysis and visualization of results were conducted in R 4.0.4. Shapiro–Wilk normality test were carried out for normality of all data. Demographic characteristics across groups were compared using Wilcoxon rank-sum tests for continuous variables and Chi-square tests or the Fisher exact test for categorical variables. Paired t-test or paired Wilcoxon signed-rank test was performed to determine the differences in AD severity and alpha diversity before and after probiotic use in the same patient. Statistical significance was set as a p < 0.05.
3. Results
3.1. Study Population
A total of 41 AD patients were recruited in this study, including 17 mild AD patients and 24 severe AD patients. After 2 months of oral administration of probiotic mixture (one capsule daily), severity of AD patients was re-evaluated by a board certified dermatologist (S.K.F.L) with EASI. Significant improvement in AD severity was seen in 24 patients, which was considered as responders, 14 of which was from mild AD group and 10 was from severe AD group. The AD severity of the remaining 17 patients failed to improve, who were recognized as non-responders including 3 mild AD patients and 14 severe AD patients (Figure 1). As detailed in Table 1, the demographic characteristics and presence of comorbidity including sex (p = 0.5737), age (p = 0.8633), BMI (p = 0.3898), allergy (food allergy: p > 0.999 and other p = 0.7417, respectively), GI symptoms (constipation: p > 0.999 and diarrhea p = 0.2118) were similar between mild and severe AD subgroups. No other drugs were administrated during the study period.
Figure 1.
Study design.
Table 1.
Baseline Demographic and Disease Characteristics of Patients.
| Patients (No.) | ||||||
|---|---|---|---|---|---|---|
| Variable | Overall (n = 41) | Mild AD (n = 17) | Severe AD (n = 24) | p Value | ||
| Characteristics | ||||||
| Sex, No. (%) | 0.5737 | |||||
| Male | 16 (39.0) | 8 (47.1) | 8 (33.3) | |||
| Female | 25 (61.0) | 9 (52.9) | 16 (66.6) | |||
| Age, mean (SD) [range], y | 47.0 (15.6) [18–73] | 47.6 (15.5) [26–66] | 46.6 (16.0) [18–73] | 0.8633 | ||
| Weight, mean (SD), kg | 59.9 (11.1) | 62.5 (11.1) | 58.0 (10.9) | 0.1414 | ||
| BMI, mean (SD) † | 22.5 (3.3) | 23.1 (3.3) | 22.1 (3.3) | 0.3898 | ||
| EASI, mean (SD) | 17.7 (7.0) | 10.7 (2.1) | 22.7 (4.5) | <0.001 | ||
| Presence of Comorbidity | ||||||
| Allergy ever, No. (%) | ||||||
| Food allergy | 3 (7.3) | 1 (5.9) | 2 (8.3) | >0.999 | ||
| Others | 14 (34.2) | 5 (29.4) | 9 (37.5) | 0.7417 | ||
| GI, No. (%) | ||||||
| Constipation | 14 (34.2) | 6 (35.3) | 8 (33.3) | >0.999 | ||
| Diarrhea | 6 (14.6) | 4 (23.5) | 2 (8.3) | 0.2118 | ||
BMI, body mass index; EASI, Eczema Area and Severity Index. † BMI between 23.0–25.0 kg/m2 is classified as overweight, while BMI > 25.0 kg/m2 is classified as obese.
3.2. Probiotic Mixture Significantly Ameliorates AD Severity
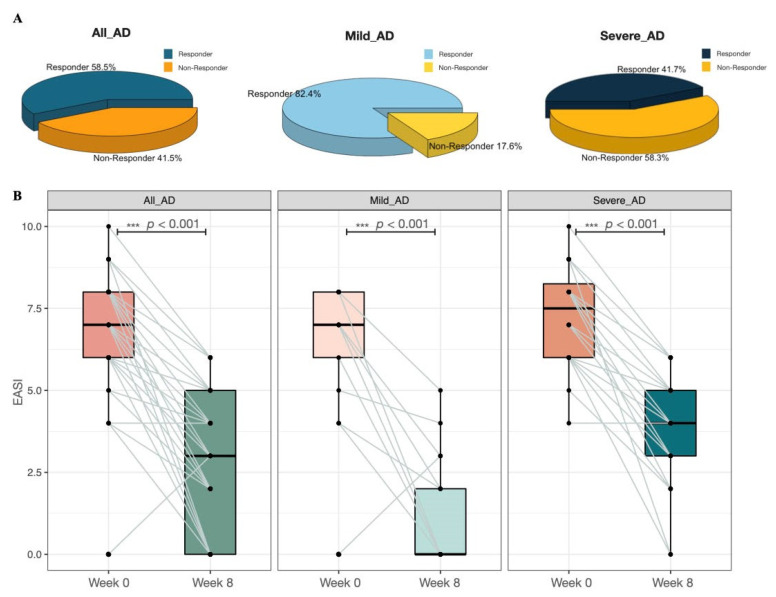
As shown in Figure 2, more mild AD patients significantly improved (p < 0.001) their AD condition after taking novel E3 probiotics formula, compared with the severe AD group. 82.4% of the patients in the mild AD group responded to the probiotics mixture, while only 41.7% of the patients in the severe AD group responded to the probiotic blend (Figure 2A). Our results also illustrated that the EASI of AD patients was significantly reduced (p < 0.001) after oral administration of the probiotic mixture regardless of baseline disease severity (Figure 2B).
Figure 2.
(A) Distribution of responders and non-responders among All_AD, Mild AD, and Severe AD patients. (B) Alteration of the AD severity score before and after novel E3 probiotics mixture administration among all AD, mild AD, and severe AD patients. *** denoted p < 0.001.
3.3. Probiotic Mixture Improves the Diversity of Gut Microbiome in AD Patients
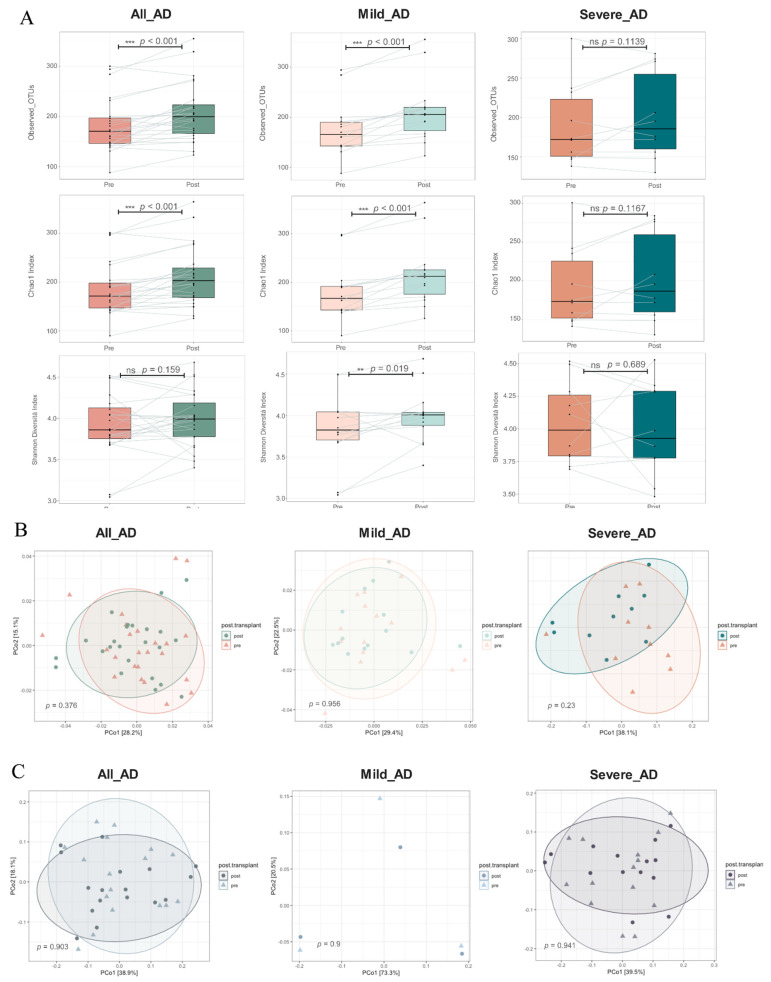
Alpha diversity, also called within-habitat diversity, is usually calculated to describe the richness and evenness of the community within a sample. The richness was measured by the Chao1 index, ACE index, and observed OTUs. Shannon diversity index and InvSimpson diversity index comprehensively consider the richness and uniformity of the community. For responders, significant increase of the species richness was obtained in mild AD patients (p < 0.001), while only a slight increase was identified in the severe AD group (p = 0.1139 for Observed OTUs; p = 0.1167 for Chao1 Index; Table S1, Figure 3A). In addition, for responders, a considerable increase of Shannon diversity index was obtained in mild AD group (p = 0.019) after taking probiotic mixture, while the Shannon diversity index of severe AD patients did not change significantly after the use of probiotics (p = 0.689; Table S1, Figure 3A). For non-responders, after probiotic mixture administration, no significant alteration was identified in alpha diversity of the gut microbiome among AD patients (Table S2, Figure S1). Moreover, in terms of beta diversity analysis, a similar intestinal bacterial community was obtained between pre and post groups in both responders and non-responders based on the Jaccard distance metric, Bray–Curtis distance metric, weighted UniFrac, and unweighted UniFrac distance metrics by PERMANOVA test (Tables S3 and S4, Figure 3B).
Figure 3.
(A) Alteration of species richness, including three metrics, observed OTUs, Chao1 index and Shannon diversity index, before and after probiotic mixture intake among all AD, mild AD, and severe AD patients. (B) PCoA plots based on the weighted UniFrac distance metric across pre and post groups among the responders and (C) non-responders in All AD, Mild AD, and Severe AD patients. p value was calculated by PERMANOVA test with permutation = 999. ** denoted p < 0.05. *** denoted p < 0.001.
3.4. Gut Microbiome Profiling
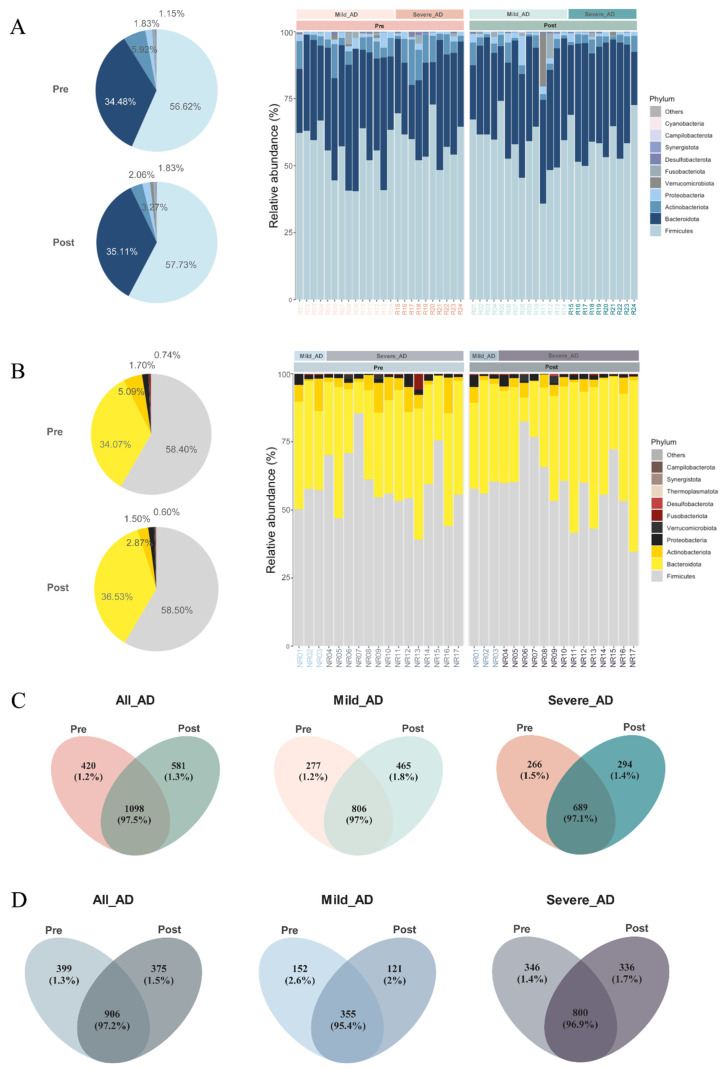
At the phylum level, a total of 15 phyla, including 13 from the kingdom of bacteria and 2 from archaea, were detected in both responders and non-responders, and the top 4 most abundant phyla accounted for over 99% of sequences in the dataset. Before and after probiotic use, Firmicutes was dominant in the gut microbiome of all AD patients, followed by Bacteroidota, Actinobacteriota, and Proteobacteria (Figure 4A,B). At the genus level, the top five genera in the gut microbiome of responders were Bacteroides, Fecalibacterium, Blautia, Bifidobacterium, and Fusicatenibacter. However, in non-responders’ group, the top five genera were Bacteroides, Blautia, Prevotella, Fecalibacterium, and Bifidobacterium (Figures S2 and S3). After probiotics administration, 1098 ASVs were shared and persisted among responders. 420 unique ASVs and 581 unique ASVs were identified in pre and post groups separately (Figure 4C). For non-responders, Venn diagrams illustrated 399 and 375 unique ASVs from pre and post groups separately and a total of 906 shared ASVs (Figure 4D).
Figure 4.
Gut microbiome composition of the pre and post groups at the phylum level for (A) responders and (B) non-responders. Venn diagrams illustrating the unique and shared ASVs of pre and post groups among (C) responders and (D) non-responders in All AD, Mild AD, and Severe AD groups. The integer data is ASV number. The percentage data is the sequence number/total sequence number.
3.5. The Relative Abundance of Lactobacillus Increased Significantly after Oral Administration of Probiotic Mixture
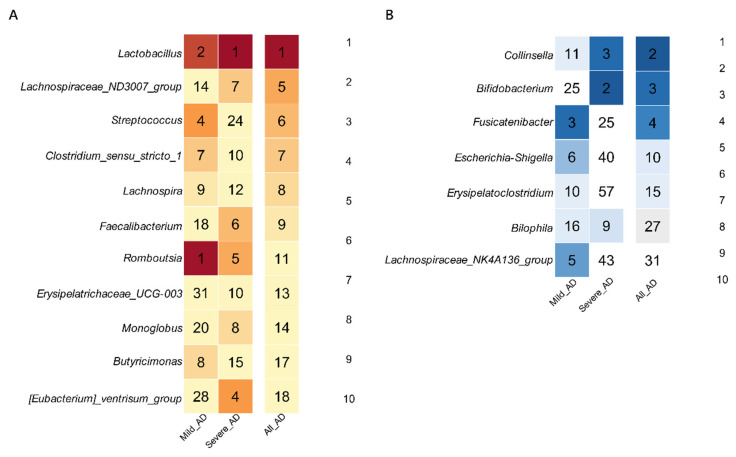
In order to determine the changes in gut microbiota composition following probiotic administration in AD patients, the random forest algorithm was conducted to select the key features affected by probiotics. Mean decrease in Gini coefficient was selected as the indicator value in the random forest analysis. The higher the value of the mean decrease in the Gini coefficient, the higher the importance of the genera responding to the treatment of probiotic mixture [77,78,79]. For responders, a total of 130, 98, and 92 features were identified between pre and post groups among all participants, mild AD and severe AD patients, respectively (Figure S4). We sorted the selected key features from high to low according to the mean decrease in the Gini coefficient and marked the rank of each genus in mild AD and severe AD group. We found that the relative abundance of Lactobacillus, Lachnospiraceae_ND3007_group, Streptococcus, Clostridium_sensu_stricto_1, Lachnospira, Fecalibacterium, Romboutsia, Erysipelatrichaceae_UCG-003, Monoglobus, Butyricimonas, and [Eubacterium]_ventrisum_group increased significantly after oral administration of probiotic mixture, while the relative abundance of Collinsella, Bifidobacterium, Fusicatenibacter, Escherichia-Shigella, Erysipelatoclostridium, Bilophila, and Lachnospiraceae_NK4A136_group decreased considerably (Figure 5). For non-responders, a total of 112, 25, and 99 features were identified between pre and post groups among all participants, Mild AD and Severe AD patients, respectively, and the relative abundance of Lactobacillus increased significantly (Figure S5).
Figure 5.
Significant genera selected across the pre and post groups in responders of (A) increasing and (B) decreasing trends. Only genera appearing in the ten top-ranking features in at least Mild AD or Severe AD groups were reported.
4. Discussion
Despite the on-going debate on whether human gut microbiome dysbiosis is a cause or effect in the development of AD, there is convincing evidence showing that gut dysbiosis has a significant association with AD through the gut-skin axis. [16,47]. Therefore, probiotics have been explored as a therapeutic option for the treatment of AD [10,11,24,33,34,35,36,37,38,40,42,44,47,48,49,50]. In this study, we focus on the effect of probiotics in adult AD patients, the gut microbiome dynamics upon the course of probiotics and the gut microbiome signatures in responders under real world setting.
First of all, AD severity was significantly improved as evidenced by the drop in EASI after 8 weeks of oral probiotics in this cohort, although the minimal clinical important difference (MCID) had not been reached [80]. The effect of probiotics was more apparent in mild AD patients, likely because it would be relatively easier to restore the dysbiosis in mild AD patients than the heavily imbalanced gut flora in severe AD patients by probiotics [23]. Unsurprisingly, there is a surge in species richness among responders and it is consistent with the results reported by other groups [81]. For non-responders, no notable change in both alpha- and beta-diversity was observed. The taxonomic profile was highly comparable as illustrated in Figure 4 at the phylum level and in terms of ASVs across AD severity and time point.
At the genera level, our studies unrevealed the plausible colonization of probiotics in the responders’ gut. The probiotics strain, Lactobacillus, blended in the probiotics may directly colonize the gut microflora [81] or facilitate the expansion of existing beneficial communities. Further experiments would be required to validate the exact mechanism of colonization. Still, we presented concrete evidence that the successful colonization and/or expansion of Lactobacillus could be the key to stimulate response towards probiotics in AD patients. Furthermore, the relative abundances of commonly recognized beneficial bacteria including Clostridium, Fecalibacterium, Romboutsia, and Streptococcus were found to be enriched in AD patients after the course of probiotics, in addition to Lactobacillus. The expansion of beneficial bacteria could exert anti-inflammatory effects by the production of short-chain fatty acids (SCFAs) [20,82], including but not limited to acetate, butyrate, and propionate [83,84,85,86]. Lachnospira and Butyricimonas likely augment the production of SCFAs [87,88]. Lachnospiraceae ND3007 group and [Eubacterium] ventrisum group were also reported as a putative SCFA producer [88,89,90], while Erysipelotrichaceae UCG-003 was reported to be closely related to Fecalibacillus genus [91], namely Fecalibacillus intestinalis and Fecalibacillus faecis. Fecalibacillus intestinalis and Fecalibacillus faecis are recently discovered bacterial species from human clinical samples [92] and is associated with Type II diabetes (T2D), hypertension and ageing [91,93]. Although the definitive role of both Lachnospiraceae ND3007 group, [Eubacterium] ventrisum group, and Erysipelotrichaceae UCG-003 remains unclear, it is anticipated that they function similarly to modulate inflammation activity by SCFAs or other anti-inflammatory metabolites.
On the other hand, responders were characterized by the decline of relative abundance of detrimental bacteria (including Collinsella, Escherichia-Shigella) and other genera without an explicit role (Fusicatenibacter, Erysipelatoclostridium and Bilophila). For Fusicatenibacter, Erysipelatoclostridium and Bilophila, they have been described to correlate with a high fat diet, obesity, T2D, Crohn’s disease, and ulcerative colitis [94,95,96,97]. However, the role and relationship between the genera and AD are largely uncertain; it is anticipated that they would facilitate inflammation mediated by inducing the expression of pro-inflammatory cytokines, such as IL-17A [98]. Thus, lower relative abundance of these bacteria might relive the symptoms in AD patients. Nonetheless, the apparent reduction in the genera may not necessarily reflect the absolute bacteria counts [99]. Instead, the beneficial bacteria and SCFA-rich intestinal environment may outcompete these bacteria and discourage their expansion rather than inhibiting their growth.
Most importantly, the gut microbiome signatures among responders substantially overlapped with the gut microbiome signatures of AD patients previously reported by our group [100]. In particular, depletion of Clostridium_sensu_stricto_1, Romboutsia, and Erysipelatrichaceae UCG-003 were detected in AD patients compared with healthy subjects, and their relative abundance were shown to be inversely correlated with AD severity. In this study, we reported the elevated relative abundance of both Clostridium_sensu_stricto_1, Romboutsia, and Erysipelatrichaceae UCG-003 among responders with improving disease severity as evidenced by lower EASI score. An elevated relative abundance of Erysipelatoclostridium has been noted in AD patients, while the findings of reduced relative abundance of Erysipelatoclostridium in responders discussed herein further resonate the results. In other words, AD patients who responded to probiotics acquired a gut microbiome composition and structure resembling healthy subjects. To the best of our knowledge, this is the first depiction of gut microbiome composition shift from AD status to healthy status.
Interestingly, the relative abundance of Lachnospiraceae NK4A136 group and Bifidobacterium were significantly shrunken in the responders of mild and severe AD patients, respectively. The alterations could be an outcome instead of the causal driver of the responsiveness towards oral probiotics. For example, the Bifidobacterium in the probiotics blend and the pre-existing Bifidobacterium may compete for nutrients with each other and other bacteria in the gut, which Lachnospiraceae NK4A136 group may face comparable challenges in the presence of Lachnospira. Or in the contrary but less probably, the decrease in relative abundance might also reflect decrease shredding into stool following colonization. Nevertheless, it indicated a complicated reciprocity between micro-organisms in the human intestinal environment even though the observation might be counter-intuitive, and the utilization of probiotics to revert the gut microbiome balance from dysbiosis status might not be straightforward [101,102].
Taken all together, lines of evidence about the reshape of gut microbiome composition, especially in Southern Chinese atopic dermatitis patients in this study, has been presented. Although the duration of oral probiotics being taken, the optimal dosage of probiotics intake, and when should probiotics being administrated remain unresolved [101,102] due to the limitation of resources to conduct a more comprehensive longitudinal study, our findings hint at important clues on the effect of probiotics in AD patients and the distinctive microbiome signatures between responders and non-responders to probiotics. Reddel and colleagues conducted a 90-day gut microbiota profile with 3 time points recorded in 18 child AD patients from Italy [81], but further investigation to record the temporal evolution of gut microbiome composition and the persistence of probiotics in human gut upon the course of probiotics could potentially exacerbate the scientific ground for oral administration of probiotics for skin disease symptoms alleviation. Given the disparity of gut microbiome profile between mild and severe AD patients as previously reported [100], the impact of probiotics on the gut flora were expected to be inherently dissimilar, implying the potential of personalized probiotics blend [103,104] to improve efficacy with baseline and following microbiome profiling. Despite the fact that MCID could not be reached in this cohort, likely due to the relatively small sample size, it lays the foundation to incorporate probiotics into the management of AD patients. Of note, the role of fungi and virus in the gut flora of AD patients remains poorly elucidated. Shotgun metagenomics analysis could provide valuable insights in the whole picture down to the species and sub-species levels, with the hope that the complex interplay between numerous micro-organisms and hosts will be delineated in a more accurate and precise manner [105,106]. Last but not least, additional investigations would be required to validate and establish the definitive association of the above-mentioned observations and speculations with the management of AD in a generalizable manner.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10112904/s1, Figure S1 Alteration of Shannon diversity index before and after novel E3 probiotics formula intake among; Figure S2 Heatmap of the gut bacterial community of pre and post groups for responders at the genus level, only top 20 general in relative abundance was reported; Figure S3 Heatmap of the gut bacterial community of pre and post groups for non-responders at the genus level, only top 20 general in relative abundance was reported; Figure S4 Significant genera selected across the pre and post groups in responders of (A) All_AD patients, (B) Mild AD patients, and (C) Severe AD patients using the random forest algorithms with the parameters of tree number = 1000, boots = 30, nresam = 0.667. MeanDecreaseGini was used to indicate the importance of each genus. Only top 30 was displayed. Relative abundance of each genus in pre and post groups demonstrated on the right panel separately; Figure S5 Significant genera selected across the pre and post groups in non-responders of (A) All_AD patients, (B) Mild AD patients, and (C) Severe AD patients using the random forest algorithms with the parameters of tree number = 1000, boots = 30, nresam = 0.667. MeanDecreaseGini was used to indicate the importance of each genera. Only top 30 was displayed. Relative abundance of each genera in pre and post groups demonstrated on the right panel separately. Table S1 Alpha diversities among responders stratified by disease severity. Table S2 Alpha diversities among non-responders stratified by disease severity. Table S3 Beta diversities of responder stratified by disease severity. Table S4 Beta diversities of non-responders stratified by diseases severity.
Author Contributions
Conceptualization, S.K.F.L. and S.K.W.T.; formal analysis, Y.W., Y.L., J.H., L.W.; investigation, J.C.C.T., J.Z., C.H.W., T.K.Y., W.K.T.; data curation, C.T.C., U.K.C. and P.L.K.S.; writing—original draft preparation, Y.W., C.T.C.; visualization, Y.W.; supervision, S.K.F.L. and S.K.W.T.; project administration, C.T.C., U.K.C. and P.L.K.S.; funding acquisition, S.K.F.L. and S.K.W.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Research Ethics Committee of Hong Kong Doctors Union (protocol number HKSGM-2020AD-Study-protocol-vl-20220211).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The raw sequence data are available in NCBI (BioProject PRJNA843860).
Conflicts of Interest
C.T.C., J.C.C.T., J.Z., C.H.W., U.K.C., T.K.Y., P.L.K.S. and W.K.T. are employees of BioMed Laboratory Company Limited but the relationship did not to constitute a conflict in this study. S.K.F.L. and S.K.W.T. are the consultants of the BioMed Laboratory Company Limited but the relationship did not constitute a conflict in this study.
Funding Statement
This project was funded by the General Research Fund from Research Grants Council of Hong Kong (Reference numbers: 14119219 and 14119420), Health and Medical Research Fund from Food and Health Bureau of Hong Kong (Reference numbers: 06171061), and Hong Kong Society of Gut Microbiome (HKSGM).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lopez Carrera Y.I., al Hammadi A., Huang Y.H., Llamado L.J., Mahgoub E., Tallman A.M. Epidemiology, Diagnosis, and Treatment of Atopic Dermatitis in the Developing Countries of Asia, Africa, Latin America, and the Middle East: A Review. Dermatol. Ther. 2019;9:685–705. doi: 10.1007/s13555-019-00332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaufman B.P., Guttman-Yassky E., Alexis A.F. Atopic Dermatitis in Diverse Racial and Ethnic Groups-Variations in Epidemiology, Genetics, Clinical Presentation and Treatment. Exp. Dermatol. 2018;27:340–357. doi: 10.1111/exd.13514. [DOI] [PubMed] [Google Scholar]
- 3.Mei-Yen Yong A., Tay Y.K. Atopic Dermatitis: Racial and Ethnic Differences. Dermatol. Clin. 2017;35:395–402. doi: 10.1016/j.det.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Flohr C., Mann J. New Insights into the Epidemiology of Childhood Atopic Dermatitis. Allergy. 2014;69:3–16. doi: 10.1111/all.12270. [DOI] [PubMed] [Google Scholar]
- 5.Deckers I.A.G., McLean S., Linssen S., Mommers M., van Schayck C.P., Sheikh A. Investigating International Time Trends in the Incidence and Prevalence of Atopic Eczema 1990–2010: A Systematic Review of Epidemiological Studies. PLoS ONE. 2012;7:e39803. doi: 10.1371/journal.pone.0039803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verboom P., Hakkaart-Van Roijen L., Sturkenboom M., de Zeeuw R., Menke H., Rutten F. The Cost of Atopic Dermatitis in the Netherlands: An International Comparison. Br. J. Dermatol. 2002;147:716–724. doi: 10.1046/j.1365-2133.2002.04964.x. [DOI] [PubMed] [Google Scholar]
- 7.Noda S., Suárez-Fariñas M., Ungar B., Kim S.J., de Guzman Strong C., Xu H., Peng X., Estrada Y.D., Nakajima S., Honda T., et al. The Asian Atopic Dermatitis Phenotype Combines Features of Atopic Dermatitis and Psoriasis with Increased TH17 Polarization. J. Allergy Clin. Immunol. 2015;136:1254–1264. doi: 10.1016/j.jaci.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 8.David Boothe W., Tarbox J.A., Tarbox M.B. Atopic Dermatitis: Pathophysiology. Adv. Exp. Med. Biol. 2017;1027:21–37. doi: 10.1007/978-3-319-64804-0_3. [DOI] [PubMed] [Google Scholar]
- 9.Barnes E.M., Carter E.L., Lewis J.D. Predicting Microbiome Function Across Space Is Confounded by Strain-Level Differences and Functional Redundancy Across Taxa. Front. Microbiol. 2020;11:101. doi: 10.3389/fmicb.2020.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S., Chang G., Zhang L. The Prevention Effect of Probiotics against Eczema in Children: An Update Systematic Review and Meta-Analysis. J. Dermatol. Treat. 2022;33:1844–1854. doi: 10.1080/09546634.2021.1925077. [DOI] [PubMed] [Google Scholar]
- 11.Ambrożej D., Kunkiel K., Dumycz K., Feleszko W. The Use of Probiotics and Bacteria-Derived Preparations in Topical Treatment of Atopic Dermatitis-A Systematic Review. J. Allergy Clin. Immunol. Pract. 2021;9:570–575.e2. doi: 10.1016/j.jaip.2020.07.051. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds G., Vegh P., Fletcher J., Poyner E.F.M., Stephenson E., Goh I., Botting R.A., Huang N., Olabi B., Dubois A., et al. Developmental Cell Programs Are Co-Opted in Inflammatory Skin Disease. Science. 2021;371:eaba6500. doi: 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams H.C., Grindlay D.J.C. What’s New in Atopic Eczema? An Analysis of Systematic Reviews Published in 2007 and 2008. Part 1. Definitions, Causes and Consequences of Eczema. Clin. Exp. Dermatol. 2010;35:12–15. doi: 10.1111/j.1365-2230.2009.03733.x. [DOI] [PubMed] [Google Scholar]
- 14.On H.R., Lee S.E., Kim S.E., Hong W.J., Kim H.J., Nomura T., Suzuki S., Shimizu H., Kim S.C. Filaggrin Mutation in Korean Patients with Atopic Dermatitis. Yonsei Med. J. 2017;58:395–400. doi: 10.3349/ymj.2017.58.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassani B., Isaian A., Shariat M., Mollanoori H., Sotoudeh S., Babaei V., Ziaali A., Teimourian S. Filaggrin Gene Polymorphisms in Iranian Ichthyosis Vulgaris and Atopic Dermatitis Patients. Int. J. Dermatol. 2018;57:1485–1491. doi: 10.1111/ijd.14213. [DOI] [PubMed] [Google Scholar]
- 16.Rather I.A., Bajpai V.K., Kumar S., Lim J., Paek W.K., Park Y.H. Probiotics and Atopic Dermatitis: An Overview. Front. Microbiol. 2016;7:507. doi: 10.3389/fmicb.2016.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas C.L., Fernández-Peñas P. The Microbiome and Atopic Eczema: More than Skin Deep. Australas. J. Dermatol. 2017;58:18–24. doi: 10.1111/ajd.12435. [DOI] [PubMed] [Google Scholar]
- 18.Sugita K., Akdis C.A. Recent Developments and Advances in Atopic Dermatitis and Food Allergy. Allergol. Int. 2020;69:204–214. doi: 10.1016/j.alit.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Shen X., Wang M., Zhang X., He M., Li M., Cheng G., Wan C., He F. Dynamic Construction of Gut Microbiota May Influence Allergic Diseases of Infants in Southwest China. BMC Microbiol. 2019;19:1–13. doi: 10.1186/s12866-019-1489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song H., Yoo Y., Hwang J., Na Y.C., Kim H.S. Faecalibacterium Prausnitzii Subspecies-Level Dysbiosis in the Human Gut Microbiome Underlying Atopic Dermatitis. J. Allergy Clin. Immunol. 2016;137:852–860. doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Lee E., Lee S.Y., Kang M.J., Kim K., Won S., Kim B.J., Choi K.Y., Kim B.S., Cho H.J., Kim Y., et al. Clostridia in the Gut and Onset of Atopic Dermatitis via Eosinophilic Inflammation. Ann. Allergy Asthma Immunol. 2016;117:91–92.e1. doi: 10.1016/j.anai.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Fujimura K.E., Sitarik A.R., Havstad S., Lin D.L., Levan S., Fadrosh D., Panzer A.R., Lamere B., Rackaityte E., Lukacs N.W., et al. Neonatal Gut Microbiota Associates with Childhood Multisensitized Atopy and T Cell Differentiation. Nat. Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nylund L., Nermes M., Isolauri E., Salminen S., de Vos W.M., Satokari R. Severity of Atopic Disease Inversely Correlates with Intestinal Microbiota Diversity and Butyrate-Producing Bacteria. Allergy. 2015;70:241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 24.Bertelsen R.J., Brantsæter A.L., Magnus M.C., Haugen M., Myhre R., Jacobsson B., Longnecker M.P., Meltzer H.M., London S.J. Probiotic Milk Consumption in Pregnancy and Infancy and Subsequent Childhood Allergic Diseases. J. Allergy Clin. Immunol. 2014;133:165–171. doi: 10.1016/j.jaci.2013.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penders J., Gerhold K., Stobberingh E.E., Thijs C., Zimmermann K., Lau S., Hamelmann E. Establishment of the Intestinal Microbiota and Its Role for Atopic Dermatitis in Early Childhood. J. Allergy Clin. Immunol. 2013;132:601–607. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 26.Han P., Gu J.Q., Li L.S., Wang X.Y., Wang H.T., Wang Y., Chang C., Sun J.L. The Association Between Intestinal Bacteria and Allergic Diseases-Cause or Consequence? Front. Cell. Infect. Microbiol. 2021;11:284. doi: 10.3389/fcimb.2021.650893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stokes J.H., Pillsbury D.M. The effect on the skin of emotional and nervous states: iii. theoretical and practical consideration of a gastro-intestinal mechanism. Arch. Derm. Syphilol. 1930;22:962–993. doi: 10.1001/archderm.1930.01440180008002. [DOI] [Google Scholar]
- 28.Bowe W.P., Logan A.C. Acne Vulgaris, Probiotics and the Gut-Brain-Skin Axis—Back to the Future? Gut Pathog. 2011;3:1. doi: 10.1186/1757-4749-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feleszko W., Jaworska J., Rha R.D., Steinhausen S., Avagyan A., Jaudszus A., Ahrens B., Groneberg D.A., Wahn U., Hamelmann E. Probiotic-Induced Suppression of Allergic Sensitization and Airway Inflammation Is Associated with an Increase of T Regulatory-Dependent Mechanisms in a Murine Model of Asthma. Clin. Exp. Allergy. 2007;37:498–505. doi: 10.1111/j.1365-2222.2006.02629.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim H.J., Kim Y.J., Kang M.J., Seo J.H., Kim H.Y., Jeong S.K., Lee S.H., Kim J.M., Hong S.J. A Novel Mouse Model of Atopic Dermatitis with Epicutaneous Allergen Sensitization and the Effect of Lactobacillus Rhamnosus. Exp. Dermatol. 2012;21:672–675. doi: 10.1111/j.1600-0625.2012.01539.x. [DOI] [PubMed] [Google Scholar]
- 31.Kim J.Y., Park B.K., Park H.J., Park Y.H., Kim B.O., Pyo S. Atopic Dermatitis-Mitigating Effects of New Lactobacillus Strain, Lactobacillus Sakei Probio 65 Isolated from Kimchi. J. Appl. Microbiol. 2013;115:517–526. doi: 10.1111/jam.12229. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto M., Aranami A., Ishige A., Watanabe K., Benno Y. LKM512 Yogurt Consumption Improves the Intestinal Environment and Induces the T-Helper Type 1 Cytokine in Adult Patients with Intractable Atopic Dermatitis. Clin. Exp. Allergy. 2007;37:358–370. doi: 10.1111/j.1365-2222.2007.02642.x. [DOI] [PubMed] [Google Scholar]
- 33.Weston S., Halbert A., Richmond P., Prescott S.L. Effects of Probiotics on Atopic Dermatitis: A Randomised Controlled Trial. Arch. Dis. Child. 2005;90:892–897. doi: 10.1136/adc.2004.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalliomäki M., Salminen S., Arvilommi H., Kero P., Koskinen P., Isolauri E. Probiotics in Primary Prevention of Atopic Disease: A Randomised Placebo-Controlled Trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 35.Kukkonen K., Savilahti E., Haahtela T., Juntunen-Backman K., Korpela R., Poussa T., Tuure T., Kuitunen M. Probiotics and Prebiotic Galacto-Oligosaccharides in the Prevention of Allergic Diseases: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Allergy Clin. Immunol. 2007;119:192–198. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Lee J., Seto D., Bielory L. Meta-Analysis of Clinical Trials of Probiotics for Prevention and Treatment of Pediatric Atopic Dermatitis. J. Allergy Clin. Immunol. 2008;121:116–121. doi: 10.1016/j.jaci.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 37.Michail S.K., Stolfi A., Johnson T., Onady G.M. Efficacy of Probiotics in the Treatment of Pediatric Atopic Dermatitis: A Meta-Analysis of Randomized Controlled Trials. Ann. Allergy Asthma Immunol. 2008;101:508–516. doi: 10.1016/S1081-1206(10)60290-6. [DOI] [PubMed] [Google Scholar]
- 38.Wickens K., Black P.N., Stanley T.V., Mitchell E., Fitzharris P., Tannock G.W., Purdie G., Crane J. A Differential Effect of 2 Probiotics in the Prevention of Eczema and Atopy: A Double-Blind, Randomized, Placebo-Controlled Trial. J. Allergy Clin. Immunol. 2008;122:788–794. doi: 10.1016/j.jaci.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Woo S.I., Kim J.Y., Lee Y.J., Kim N.S., Hahn Y.S. Effect of Lactobacillus Sakei Supplementation in Children with Atopic Eczema-Dermatitis Syndrome. Ann. Allergy Asthma Immunol. 2010;104:343–348. doi: 10.1016/j.anai.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 40.Betsi G.I., Papadavid E., Falagas M.E. Probiotics for the Treatment or Prevention of Atopic Dermatitis: A Review of the Evidence from Randomized Controlled Trials. Am. J. Clin. Dermatol. 2008;9:93–103. doi: 10.2165/00128071-200809020-00002. [DOI] [PubMed] [Google Scholar]
- 41.Han Y., Kim B., Ban J., Lee J., Kim B.J., Choi B.S., Hwang S., Ahn K., Kim J. A Randomized Trial of Lactobacillus Plantarum CJLP133 for the Treatment of Atopic Dermatitis. Pediatr. Allergy Immunol. 2012;23:667–673. doi: 10.1111/pai.12010. [DOI] [PubMed] [Google Scholar]
- 42.Rautava S., Kainonen E., Salminen S., Isolauri E. Maternal Probiotic Supplementation during Pregnancy and Breast-Feeding Reduces the Risk of Eczema in the Infant. J. Allergy Clin. Immunol. 2012;130:1355–1360. doi: 10.1016/j.jaci.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Wickens K., Black P., Stanley T.V., Mitchell E., Barthow C., Fitzharris P., Purdie G., Crane J. A Protective Effect of Lactobacillus Rhamnosus HN001 against Eczema in the First 2 Years of Life Persists to Age 4 Years. Clin. Exp. Allergy. 2012;42:1071–1079. doi: 10.1111/j.1365-2222.2012.03975.x. [DOI] [PubMed] [Google Scholar]
- 44.Kim S.O., Ah Y.M., Yu Y.M., Choi K.H., Shin W.G., Lee J.Y. Effects of Probiotics for the Treatment of Atopic Dermatitis: A Meta-Analysis of Randomized Controlled Trials. Ann. Allergy Asthma Immunol. 2014;113:217–226. doi: 10.1016/j.anai.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Enomoto T., Sowa M., Nishimori K., Shimazu S., Yoshida A., Yamada K., Furukawa F., Nakagawa T., Yanagisawa N., Iwabuchi N., et al. Effects of Bifidobacterial Supplementation to Pregnant Women and Infants in the Prevention of Allergy Development in Infants and on Fecal Microbiota. Allergol. Int. 2014;63:575–585. doi: 10.2332/allergolint.13-OA-0683. [DOI] [PubMed] [Google Scholar]
- 46.Foolad N., Armstrong A.W. Prebiotics and Probiotics: The Prevention and Reduction in Severity of Atopic Dermatitis in Children. Benef. Microbes. 2014;5:151–160. doi: 10.3920/BM2013.0034. [DOI] [PubMed] [Google Scholar]
- 47.Makrgeorgou A., Leonardi-Bee J., Bath-Hextall F.J., Murrell D.F., Tang M.L.K., Roberts A., Boyle R.J. Probiotics for Treating Eczema. Cochrane Database Syst. Rev. 2018;11 doi: 10.1002/14651858.CD006135.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan-Lim C.S.C., Esteban-Ipac N.A.R., Recto M.S.T., Castor M.A.R., Casis-Hao R.J., Nano A.L.M. Comparative Effectiveness of Probiotic Strains on the Prevention of Pediatric Atopic Dermatitis: A Systematic Review and Network Meta-Analysis. Pediatr. Allergy Immunol. 2021;32:1255–1270. doi: 10.1111/pai.13514. [DOI] [PubMed] [Google Scholar]
- 49.Pachacama López A.F., Tapia Portilla M.F., Moreno-Piedrahíta Hernández F., Palacios-Álvarez S. Probiotics to Reduce the Severity of Atopic Dermatitis in Pediatric Patients: A Systematic Review and Meta-Analysis. Actas Dermo Sifiliográficas. 2021;112:881–890. doi: 10.1016/j.ad.2021.06.006. [DOI] [Google Scholar]
- 50.Rosenfeldt V., Benfeldt E., Nielsen S.D., Michaelsen K.F., Jeppesen D.L., Valerius N.H., Paerregaard A. Effect of Probiotic Lactobacillus Strains in Children with Atopic Dermatitis. J. Allergy Clin. Immunol. 2003;111:389–395. doi: 10.1067/mai.2003.389. [DOI] [PubMed] [Google Scholar]
- 51.Leshem Y.A., Hajar T., Hanifin J.M., Simpson E.L. What the Eczema Area and Severity Index Score Tells Us about the Severity of Atopic Dermatitis: An Interpretability Study. Br. J. Dermatol. 2015;172:1353–1357. doi: 10.1111/bjd.13662. [DOI] [PubMed] [Google Scholar]
- 52.Simpson E.L., Bieber T., Guttman-Yassky E., Beck L.A., Blauvelt A., Cork M.J., Silverberg J.I., Deleuran M., Kataoka Y., Lacour J.-P., et al. Two Phase 3 Trials of Dupilumab versus Placebo in Atopic Dermatitis. N. Engl. J. Med. 2016;375:2335–2348. doi: 10.1056/NEJMoa1610020. [DOI] [PubMed] [Google Scholar]
- 53.Szajewska H., Horvath A. Lactobacillus Rhamnosus GG in the Primary Prevention of Eczema in Children: A Systematic Review and Meta-Analysis. Nutrients. 2018;10:1319. doi: 10.3390/nu10091319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan W., Zhou Z., Li W., Lu H., Qiu Z. Lactobacillus Rhamnosus GG for Cow’s Milk Allergy in Children: A Systematic Review and Meta-Analysis. Front. Pediatr. 2021;9:1127. doi: 10.3389/fped.2021.727127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyle R.J., Ismail I.H., Kivivuori S., Licciardi P.V., Robins-Browne R.M., Mah L.J., Axelrad C., Moore S., Donath S., Carlin J.B., et al. Lactobacillus GG Treatment during Pregnancy for the Prevention of Eczema: A Randomized Controlled Trial. Allergy. 2011;66:509–516. doi: 10.1111/j.1398-9995.2010.02507.x. [DOI] [PubMed] [Google Scholar]
- 56.Viljanen M., Savilahti E., Haahtela T., Juntunen-Backman K., Korpela R., Poussa T., Tuure T., Kuitunen M. Probiotics in the Treatment of Atopic Eczema/Dermatitis Syndrome in Infants: A Double-Blind Placebo-Controlled Trial. Allergy. 2005;60:494–500. doi: 10.1111/j.1398-9995.2004.00514.x. [DOI] [PubMed] [Google Scholar]
- 57.Viljanen M., Kuitunen M., Haahtela T., Juntunen-Backman K., Korpela R., Savilahti E. Probiotic Effects on Faecal Inflammatory Markers and on Faecal IgA in Food Allergic Atopic Eczema/Dermatitis Syndrome Infants. Pediatr. Allergy Immunol. 2005;16:65–71. doi: 10.1111/j.1399-3038.2005.00224.x. [DOI] [PubMed] [Google Scholar]
- 58.Hirose Y., Yamamoto Y., Yoshikai Y., Murosaki S. Oral Intake of Heat-Killed Lactobacillus Plantarum L-137 Decreases the Incidence of Upper Respiratory Tract Infection in Healthy Subjects with High Levels of Psychological Stress. J. Nutr. Sci. 2013;2:1–8. doi: 10.1017/jns.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirose Y., Murosaki S., Yamamoto Y., Yoshikai Y., Tsuru T. Daily Intake of Heat-Killed Lactobacillus Plantarum L-137 Augments Acquired Immunity in Healthy Adults. J. Nutr. 2006;136:3069–3073. doi: 10.1093/jn/136.12.3069. [DOI] [PubMed] [Google Scholar]
- 60.Cuello-Garcia C.A., Fiocchi A., Pawankar R., Yepes-Nuñez J.J., Morgano G.P., Zhang Y., Ahn K., Al-Hammadi S., Agarwal A., Gandhi S., et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Prebiotics. World Allergy Organ. J. 2016;9:10. doi: 10.1186/s40413-016-0102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osborn D.A., Sinn J.K. Prebiotics in Infants for Prevention of Allergy. Cochrane Database Syst. Rev. 2013;2013 doi: 10.1002/14651858.CD006474.pub3. [DOI] [PubMed] [Google Scholar]
- 62.Closa-Monasterolo R., Gispert-Llaurado M., Luque V., Ferre N., Rubio-Torrents C., Zaragoza-Jordana M., Escribano J. Safety and Efficacy of Inulin and Oligofructose Supplementation in Infant Formula: Results from a Randomized Clinical Trial. Clin. Nutr. 2013;32:918–927. doi: 10.1016/j.clnu.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Costalos C., Kapiki A., Apostolou M., Papathoma E. The Effect of a Prebiotic Supplemented Formula on Growth and Stool Microbiology of Term Infants. Early Hum. Dev. 2008;84:45–49. doi: 10.1016/j.earlhumdev.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Bruzzese E., Volpicelli M., Squeglia V., Bruzzese D., Salvini F., Bisceglia M., Lionetti P., Cinquetti M., Iacono G., Amarri S., et al. A Formula Containing Galacto- and Fructo-Oligosaccharides Prevents Intestinal and Extra-Intestinal Infections: An Observational Study. Clin. Nutr. 2009;28:156–161. doi: 10.1016/j.clnu.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 65.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Callahan B.J., McMurdie P.J., Holmes S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K., Misawa K., Kuma K.I., Miyata T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price M.N., Dehal P.S., Arkin A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glöckner F.O., Yilmaz P., Quast C., Gerken J., Beccati A., Ciuprina A., Bruns G., Yarza P., Peplies J., Westram R., et al. 25 Years of Serving the Community with Ribosomal RNA Gene Reference Databases and Tools. J. Biotechnol. 2017;261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 75.Liu C., Cui Y., Li X., Yao M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021;97:fiaa255. doi: 10.1093/femsec/fiaa255. [DOI] [PubMed] [Google Scholar]
- 76.Anderson M.J. Permutational Multivariate Analysis of Variance (PERMANOVA) Wiley StatsRef Stat. Ref. Online. 2017:1–15. doi: 10.1002/9781118445112.STAT07841. [DOI] [Google Scholar]
- 77.Beck D., Foster J.A. Machine Learning Techniques Accurately Classify Microbial Communities by Bacterial Vaginosis Characteristics. PLoS ONE. 2014;9:e87830. doi: 10.1371/journal.pone.0087830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., et al. Human Gut Microbiome Viewed across Age and Geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinez-Taboada F., Redondo J.I. The SIESTA (SEAAV Integrated Evaluation Sedation Tool for Anaesthesia) Project: Initial Development of a Multifactorial Sedation Assessment Tool for Dogs. PLoS ONE. 2020;15:e0230799. doi: 10.1371/journal.pone.0230799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schram M.E., Spuls P.I., Leeflang M.M.G., Lindeboom R., Bos J.D., Schmitt J. EASI, (Objective) SCORAD and POEM for Atopic Eczema: Responsiveness and Minimal Clinically Important Difference. Allergy. 2012;67:99–106. doi: 10.1111/j.1398-9995.2011.02719.x. [DOI] [PubMed] [Google Scholar]
- 81.Reddel S., del Chierico F., Quagliariello A., Giancristoforo S., Vernocchi P., Russo A., Fiocchi A., Rossi P., Putignani L., el Hachem M. Gut Microbiota Profile in Children Affected by Atopic Dermatitis and Evaluation of Intestinal Persistence of a Probiotic Mixture. Sci. Rep. 2019;9:4996. doi: 10.1038/s41598-019-41149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miquel S., Martín R., Rossi O., Bermúdez-Humarán L.G., Chatel J.M., Sokol H., Thomas M., Wells J.M., Langella P. Faecalibacterium Prausnitzii and Human Intestinal Health. Curr. Opin. Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 83.Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam Leitch C., Scott K.P., Flint H.J., Louis P. Phylogenetic Distribution of Three Pathways for Propionate Production within the Human Gut Microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shu M., Wang Y., Yu J., Kuo S., Coda A., Jiang Y., Gallo R.L., Huang C.M. Fermentation of Propionibacterium Acnes, a Commensal Bacterium in the Human Skin Microbiome, as Skin Probiotics against Methicillin-Resistant Staphylococcus Aureus. PLoS ONE. 2013;8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Benedetto A., Rafaels N.M., McGirt L.Y., Ivanov A.I., Georas S.N., Cheadle C., Berger A.E., Zhang K., Vidyasagar S., Yoshida T., et al. Tight Junction Defects in Patients with Atopic Dermatitis. J. Allergy Clin. Immunol. 2011;127:773–786.e7. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Macia L., Thorburn A.N., Binge L.C., Marino E., Rogers K.E., Maslowski K.M., Vieira A.T., Kranich J., Mackay C.R. Microbial Influences on Epithelial Integrity and Immune Function as a Basis for Inflammatory Diseases. Immunol. Rev. 2012;245:164–176. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 87.Paliy O., Rajakaruna S. Development of Microbiota—Is the Process Continuing Through Adolescence? Compr. Gut Microb. 2022:59–68. doi: 10.1016/B978-0-12-819265-8.00022-X. [DOI] [Google Scholar]
- 88.Holmes Z.C., Silverman J.D., Dressman H.K., Wei Z., Dallow E.P., Armstrong S.C., Seed P.C., Rawls J.F., David L.A. Short-Chain Fatty Acid Production by Gut Microbiota from Children with Obesity Differs According to Prebiotic Choice and Bacterial Community Composition. mBio. 2020;11:1–15. doi: 10.1128/mBio.00914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou W., Zhang D., Li Z., Jiang H., Li J., Ren R., Gao X., Li J., Wang X., Wang W., et al. The Fecal Microbiota of Patients with Pancreatic Ductal Adenocarcinoma and Autoimmune Pancreatitis Characterized by Metagenomic Sequencing. J. Transl. Med. 2021;19:215. doi: 10.1186/s12967-021-02882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Han Y., Gong Z., Sun G., Xu J., Qi C., Sun W., Jiang H., Cao P., Ju H. Dysbiosis of Gut Microbiota in Patients With Acute Myocardial Infarction. Front. Microbiol. 2021;12:1489. doi: 10.3389/fmicb.2021.680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Louca P., Nogal A., Wells P.M., Asnicar F., Wolf J., Steves C.J., Spector T.D., Segata N., Berry S.E., Valdes A.M., et al. Gut Microbiome Diversity and Composition Is Associated with Hypertension in Women. J. Hypertens. 2021;39:1810–1816. doi: 10.1097/HJH.0000000000002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seo B., Jeon K., Baek I., Lee Y.M., Baek K., Ko G.P. Faecalibacillus Intestinalis Gen. Nov., Sp. Nov. and Faecalibacillus Faecis Sp. Nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2019;69:2120–2128. doi: 10.1099/ijsem.0.003443. [DOI] [PubMed] [Google Scholar]
- 93.Singh H., Torralba M.G., Moncera K.J., DiLello L., Petrini J., Nelson K.E., Pieper R. Gastro-Intestinal and Oral Microbiome Signatures Associated with Healthy Aging. Geroscience. 2019;41:907–921. doi: 10.1007/s11357-019-00098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.JDDW 2014 Abstracts: The Relationship between Fusicatenibacter Saccharivorans and Inflammatory Bowel Disease. [(accessed on 3 November 2022)]. Available online: https://www.jddw.jp/jddw2014/abstracts-eng/abst/60011.html.
- 95.Plaut A.G., Qiu J. IgA-Specific Metalloendopeptidase. Handb. Proteolytic Enzymes. 2013;1:1243–1248. doi: 10.1016/B978-0-12-382219-2.00279-9. [DOI] [Google Scholar]
- 96.Natividad J.M., Lamas B., Pham H.P., Michel M.L., Rainteau D., Bridonneau C., da Costa G., van Hylckama Vlieg J., Sovran B., Chamignon C., et al. Bilophila Wadsworthia Aggravates High Fat Diet Induced Metabolic Dysfunctions in Mice. Nat. Commun. 2018;9:2802. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li R., Sun X., Liu X., Yang Y., Li Z. Autoimmune Diseases in China. Adv. Immunol. 2019;144:173–216. doi: 10.1016/BS.AI.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 98.Douzandeh-Mobarrez B., Kariminik A. Gut Microbiota and IL-17A: Physiological and Pathological Responses. Probiotics Antimicrob. Proteins. 2019;11:1–10. doi: 10.1007/s12602-017-9329-z. [DOI] [PubMed] [Google Scholar]
- 99.Greenacre M., Martínez-Álvaro M., Blasco A. Compositional Data Analysis of Microbiome and Any-Omics Datasets: A Validation of the Additive Logratio Transformation. Front. Microbiol. 2021;12:727398. doi: 10.3389/fmicb.2021.727398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y., Hou J., Chi-Ching Tsui J., Wang L., Zhou J., Kei Chan U., Jun Yi Lo C., Ling Kella Siu P., King Fan Loo S., Kwok Wing Tsui S., et al. Unique Gut Microbiome Signatures among Adult Patients with Moderate to Severe Atopic Dermatitis in Southern Chinese. bioRxiv. 2022 doi: 10.1101/2022.05.14.491964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gagliardi A., Totino V., Cacciotti F., Iebba V., Neroni B., Bonfiglio G., Trancassini M., Passariello C., Pantanella F., Schippa S. Rebuilding the Gut Microbiota Ecosystem. Int. J. Environ. Res. Public Health. 2018;15:1679. doi: 10.3390/ijerph15081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McFarland L.V. Use of Probiotics to Correct Dysbiosis of Normal Microbiota Following Disease or Disruptive Events: A Systematic Review. BMJ Open. 2014;4:e005047. doi: 10.1136/bmjopen-2014-005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh T.P., Natraj B.H. Next-Generation Probiotics: A Promising Approach towards Designing Personalized Medicine. Crit Rev. Microbiol. 2021;47:479–498. doi: 10.1080/1040841X.2021.1902940. [DOI] [PubMed] [Google Scholar]
- 104.Zmora N., Zilberman-Schapira G., Suez J., Mor U., Dori-Bachash M., Bashiardes S., Kotler E., Zur M., Regev-Lehavi D., Brik R.B.Z., et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 105.Vemuri R., Shankar E.M., Chieppa M., Eri R., Kavanagh K. Beyond Just Bacteria: Functional Biomes in the Gut Ecosystem Including Virome, Mycobiome, Archaeome and Helminths. Microorganisms. 2020;8:483. doi: 10.3390/microorganisms8040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guzzo G.L., Andrews J.M., Weyrich L.S. The Neglected Gut Microbiome: Fungi, Protozoa, and Bacteriophages in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2022;28:1112–1122. doi: 10.1093/ibd/izab343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data are available in NCBI (BioProject PRJNA843860).