Abstract
To investigate the kinetics and mechanisms of extracellular protein release by Helicobacter pylori, we analyzed the entry of metabolically radiolabeled bacterial proteins into broth culture supernatant. At early time points, vacuolating cytotoxin (VacA) constituted a major extracellular protein. Subsequently, culture supernatants accumulated many proteins that were components of intact bacterial cells. This nonselective release of proteins was associated with a decreasing turbidity of cultures and loss of bacterial viability, indicative of an autolytic process. The rates of VacA secretion and autolysis were each influenced by medium composition, and therefore these may be regulated phenomena. Extracellular release of proteins by H. pylori may be an important adaptation that facilitates the persistence of H. pylori in the human gastric mucus layer. Moreover, entry of proinflammatory proteins into the gastric mucosa may contribute to the induction of a mucosal inflammatory response.
Helicobacter pylori are noninvasive gram-negative bacteria that colonize the gastric mucosae of more than half of the world’s human population. Gastric colonization by H. pylori is consistently associated with a mucosal inflammatory response and is a risk factor for peptic ulcer disease and gastric malignancies (6, 10, 18, 22). The mechanisms by which H. pylori colonization leads to gastric mucosal inflammation are not yet well understood, but entry of proinflammatory bacterial components into the mucosa may be contributory. One such bacterial component, urease, has been directly localized within the lamina propria by immunohistochemical staining (15, 20, 21).
H. pylori proteins that are secreted or released into the extracellular space may play important roles in enabling H. pylori to persistently colonize the gastric mucosa and may be relevant to the pathogenesis of H. pylori-associated diseases. Several previous studies have analyzed the release of H. pylori proteins into broth culture supernatants during bacterial growth in vitro (3, 5, 7, 29, 35). One of the proteins found in broth culture supernatants is a vacuolating cytotoxin (VacA) that alters the morphology and function of gastric epithelial cells (4, 23). H. pylori broth culture supernatants also contain an assortment of additional proteins, including urease and heat shock proteins (HspA and HspB), which in most bacterial species are localized exclusively in the cytoplasm (3, 5, 7, 29, 35). The pathways by which these proteins enter the extracellular space are not yet completely understood. Therefore, in this study we sought to investigate the kinetics and mechanisms of extracellular protein release by H. pylori.
MATERIALS AND METHODS
Bacterial culture conditions.
H. pylori 60190 (ATCC 49503) is a cag+ strain containing a type s1a/m1 vacA allele (8). This strain was routinely grown at 37°C on blood agar plates (Trypticase soy agar containing 5% sheep blood) in room air supplemented with 8% CO2. Broth cultures were inoculated by scraping bacteria from plates into sterile 0.9% saline solution and immediately adding this suspension to liquid media. The initial optical density at 600 nm (OD600) of all cultures was about 0.13. Broth cultures were agitated with a rotary shaker and incubated at 37°C in room air containing 8% CO2. Bacterial growth was monitored by measuring the OD600 of cultures at serial time points, using uninoculated culture media as blanks. Concentrations of viable bacteria in broth cultures were quantified by plating 10-μl aliquots of appropriate 10-fold dilutions on blood agar plates. Plates were incubated at 37°C for 3 days, colonies were counted, and results were expressed as CFU/ml.
Metabolic labeling of bacterial proteins.
H. pylori were serially passaged at 24-h intervals on blood agar plates and then inoculated into sulfite-free brucella broth (SFBB) (16) supplemented with 20 to 50 μCi of [35S]cysteine and [35S]methionine per ml (Express protein labeling mix; NEN Dupont), 0.025% (wt/vol) finely granulated activated charcoal, and 10 μg of vancomycin per ml. Activated charcoal was placed adjacent to culture flasks to absorb volatilized radioactivity. At varying times, 15-ml aliquots of the culture were removed and centrifuged at 17,000 × g for 20 min at 4°C to remove bacteria. Proteins in the broth culture supernatant were precipitated by the addition of 70% (wt/vol) solid ammonium sulfate. H. pylori proteins also were radiolabeled in methionine-and-cysteine-free RPMI 1640 medium (GIBCO BRL) containing 10 μg of vancomycin per ml.
Analysis of radiolabeled H. pylori proteins.
Radioactivity in preparations of bacterial cells and preparations of concentrated supernatant proteins was quantified by liquid scintillation counting (5 to 10 μl per 4 ml of Envirosafe) (ANOROC Scientific, Hackensack, N.J.). Equal amounts of radioactivity (0.5 × 105 to 1 × 105 cpm) were loaded onto 12% polyacrylamide gels, and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with the method of Laemmli (19). Following SDS-PAGE, 35S-containing gels were soaked in Entensify (NEN Dupont) according to the manufacturer’s instructions, were vacuum-dried at 80°C for 1 h, and were exposed to X-ray film (full-speed blue film; Kodak) with an intensifier screen at −70°C.
Analysis of extracellular membrane blebs.
To identify proteins that may be components of extracellular membrane blebs, H. pylori cultures were centrifuged at 17,000 × g to remove intact bacteria, and the culture supernatant was recentrifuged at 100,000 × g for 60 min at 4°C. Putative membrane bleb preparations and soluble proteins were analyzed by SDS-PAGE and silver staining.
N-terminal sequence analysis.
H. pylori proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) paper (Bio-Rad) by electroblotting for 30 min at 50 V in 10 mM CAPS (3-cyclohexylamino-1-propanesulfonic acid) buffer (pH 11). After Coomassie blue staining, the bands of interest were excised, and N-terminal sequence analysis was performed with a PE Biosystems Procise 492 protein sequencing apparatus in the Protein Chemistry Core Laboratory of Vanderbilt University School of Medicine. N-terminal amino acid sequences were identified by searching for matches in the known genome sequence of H. pylori 26695 (34).
RESULTS
Time-dependent extracellular release of radiolabeled H. pylori proteins.
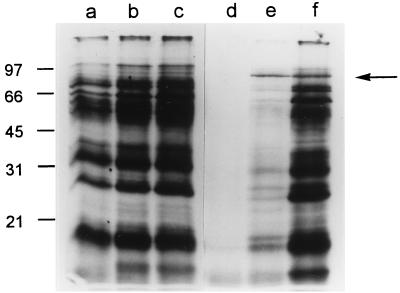
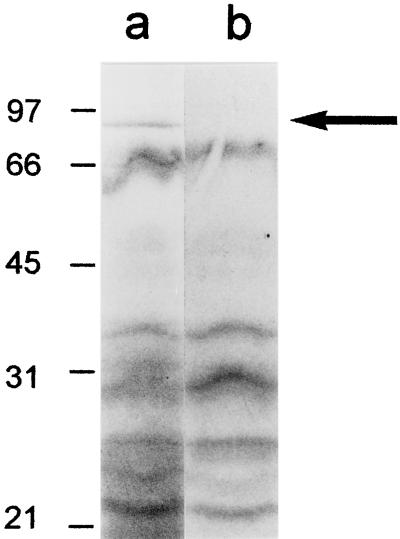
To investigate the kinetics and mechanisms by which H. pylori proteins are released into the extracellular space, we metabolically labeled H. pylori proteins during growth in liquid media and analyzed the entry of radiolabeled proteins into the extracellular space at serial time points. During growth of H. pylori 60190 in SFBB medium, bacterial proteins were effectively radiolabeled by the 8-h time point, but radiolabeled proteins were not detected in any substantial quantity in the culture supernatant at this time point (Fig. 1). At the 24-h time point, the most prominent radiolabeled supernatant protein migrated as an 87-kDa band, which was detected in only trace quantities in the bacterial cell pellet. Most other supernatant bands detected at the 24-h time point corresponded to major bands in the bacterial cells. At the 48-h time point, the supernatant contained numerous dense bands that were also present in intact bacterial cells. The size of the 87-kDa band detected in the culture supernatant at early time points corresponds to the known molecular mass of VacA (4, 5). To determine whether the radiolabeled 87-kDa band indeed represented VacA, wild-type H. pylori 60190 and an isogenic strain containing an insertion mutation in vacA (8) were each metabolically labeled, and culture supernatants were analyzed. A radiolabeled 87-kDa band was detected in supernatant from the wild-type strain but not from the mutant (Fig. 2).
FIG. 1.
Metabolic labeling of H. pylori during growth in broth culture. H. pylori 60190 was inoculated into 50 ml of SFBB containing 50 μCi of [35S]cysteine and [35S]methionine per ml (initial OD600, 0.13), and aliquots were removed after 8, 24, and 48 h of growth at 37°C. Turbidities (judged by OD600) of the culture at these time points were 0.256, 0.872, and 0.726, respectively. Bacterial cell pellets and supernatant proteins were prepared and analyzed by SDS-PAGE and autoradiography, as described in Materials and Methods. Lanes represent bacterial cells from 8- (a), 24- (b), and 48-h (c) time points and supernatant proteins from 8- (d), 24- (e), and 48-h (f) time points. An 87-kDa band constituted the major extracellular protein at the 24-h time point (arrow). Molecular mass in kilodaltons is shown on the left.
FIG. 2.
Secretion of H. pylori VacA. H. pylori 60190 (wild-type strain) and an isogenic vacA-null mutant (60190-v2) were metabolically labeled with 50 μCi of [35S]cysteine and [35S]methionine per ml of SFBB medium for 30 h. Bacterial cell pellets and supernatant proteins were prepared and analyzed by SDS-PAGE and autoradiography. Lane a, H. pylori 60190 supernatant proteins; lane b, H. pylori 60190-v2 supernatant proteins. An 87-kDa band corresponding to VacA is present in lane a (arrow) but not in lane b. Molecular mass in kilodaltons is shown on the left.
These data indicate that VacA enters the extracellular space at relatively early time points via a selective secretory process, whereas most other H. pylori proteins enter the extracellular space at later time points via a less selective process. Scintillation counting indicated that culture supernatant proteins from a 72-h broth culture contained four- to fivefold higher levels of radioactivity (in counts per minute) than supernatant proteins from a 24-h culture, whereas there was no substantial increase during this time period in the radioactivity of bacterial cells (Table 1). In addition, the turbidity (judged by OD600) of 72-h cultures was about 30% less than the maximum turbidity measured at earlier time points (Table 1). These data suggest that H. pylori releases proteins into the extracellular space as a result of autolysis.
TABLE 1.
Incorporation of 35S into H. pylori cells and release of 35S-labeled proteins into the extracellular spacea
| Time (h) | Radioactivity (cpm/μl)
|
Turbidity (OD600) | |
|---|---|---|---|
| Bacterial cells | Supernatant proteins | ||
| 0 | NAb | NA | 0.120 |
| 24 | 12,819 | 1,499 | 0.387 |
| 48 | 15,742 | 3,173 | 0.605 |
| 72 | 14,985 | 6,928 | 0.428 |
H. pylori 60190 was metabolically labeled during growth in SFBB medium, and aliquots of the culture were removed at serial time points. Lysates of bacterial cells (500 μl) and concentrated supernatant proteins (700 μl) were prepared from each culture aliquot as described in Materials and Methods, and radioactivity was quantified by liquid scintillation counting.
NA, not applicable.
Effect of culture medium composition on VacA secretion and autolysis.
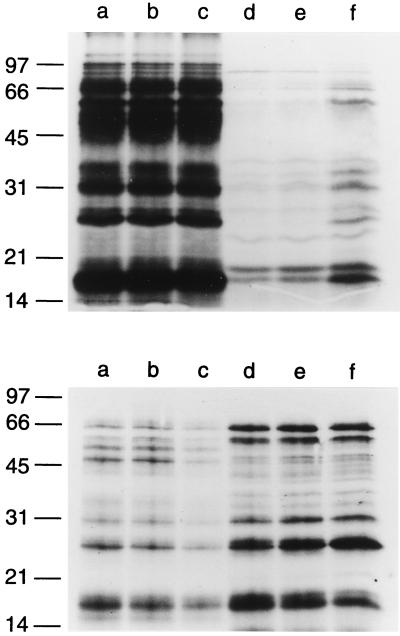
We next sought to determine whether rates of VacA secretion and autolysis were dependent upon the composition of the liquid culture medium. Two different media (SFBB as described above and methionine-and-cysteine-free RPMI 1640 medium [GIBCO BRL]) were inoculated simultaneously with H. pylori 60190, and proteins were metabolically radiolabeled. The efficiency of metabolic labeling of bacterial cells was about sevenfold greater in RPMI medium than in SFBB medium (data not shown). Diminished incorporation of [35S]methionine and [35S]cysteine into proteins of bacteria grown in SFBB medium may be attributable to the competitive effects of nonradioactive methionine and cysteine in the SFBB medium. Analysis of radiolabeled proteins in supernatant from SFBB-based cultures showed results similar to those in the first experiment, i.e., evidence of VacA secretion within 16 h and increasing autolysis at the 48-h time point (Fig. 3, top panel). In contrast, VacA did not constitute a prominent component of the supernatant proteins from RPMI medium-based cultures (Fig. 3, lower panel). A comparison of supernatant protein profiles with bacterial cell protein profiles indicates that substantial autolysis occurred within 16 h of inoculation of H. pylori into RPMI medium. Additional experiments indicated that radiolabeled proteins were released and detectable by autoradiography within 1 h of the addition of H. pylori to RPMI medium (data not shown).
FIG. 3.
Effect of culture medium composition on VacA secretion and autolysis. H. pylori 60190 was inoculated simultaneously into 50 ml of SFBB and 50 ml of methionine-and-cysteine-free RPMI medium and metabolically labeled with [35S]cysteine and [35S]methionine. Aliquots were removed after 16, 24, and 48 h at 37°C. Turbidities (judged by OD600) of the cultures at 0, 16, 24, and 48 h were as follows: SFBB, 0.130, 0.585, 0.736, and 0.532; RPMI, 0.130, 0.045, 0.047, and 0.034. Bacterial cell pellets and broth culture supernatant proteins were prepared and analyzed by SDS-PAGE and autoradiography, as described in Materials and Methods. Upper panel, culture from SFBB; lower panel, culture from methionine-and-cysteine-free RPMI medium. Lanes represent bacterial cells from 16- (a), 24- (b), and 48-h (c) time points and culture supernatant proteins from 16- (d), 24- (e), and 48-h (f) time points. Molecular mass in kilodaltons is shown on the left.
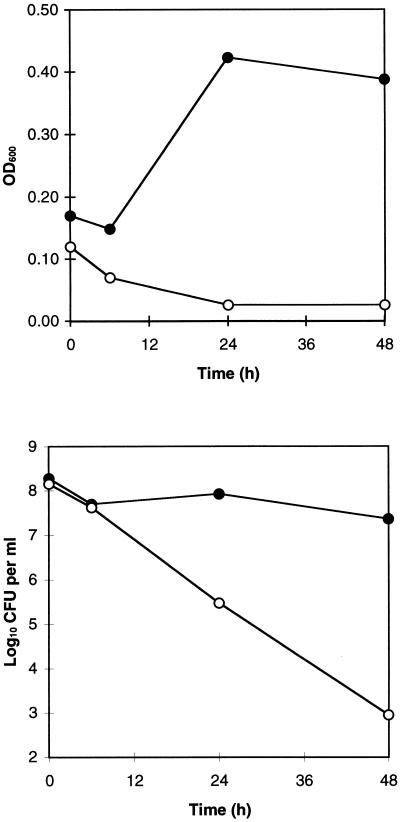
Further studies were carried out to investigate and compare the physiological condition of the bacteria in the two different media. The turbidity (judged by OD600) of H. pylori cultures in SFBB medium increased over the first 24 h, whereas there was a progressive decrease in the OD of the culture in RPMI medium (Fig. 4, top panel). In addition, the number of CFU remained stable in SFBB, but steadily decreased in RPMI medium (Fig. 4, bottom panel). Thus, during culture in SFBB medium, bacterial growth seems to be balanced by a loss of bacterial viability, whereas during culture in RPMI medium, no bacterial growth is detectable. However, the capacity of H. pylori to incorporate [35S]methionine and [35S]cysteine into newly synthesized proteins indicates that the bacteria are metabolically active in both types of media.
FIG. 4.
Effect of culture medium composition on H. pylori growth and viability. H. pylori 60190 was inoculated simultaneously into 50 ml of SFBB and 50 ml of methionine-and-cysteine-free RPMI medium. Aliquots were removed at 0, 6, 24, and 48 h for measurement of turbidity (judged by OD600) (top panel) and viable bacterial density (CFU per ml) (bottom panel). Measurements of CFU per ml represent the means of quadruplicate determinations; standard deviations were smaller than the symbol size at all time points. Closed symbols, SFBB; open symbols, RPMI medium.
Extracellular release of H. pylori superoxide dismutase.
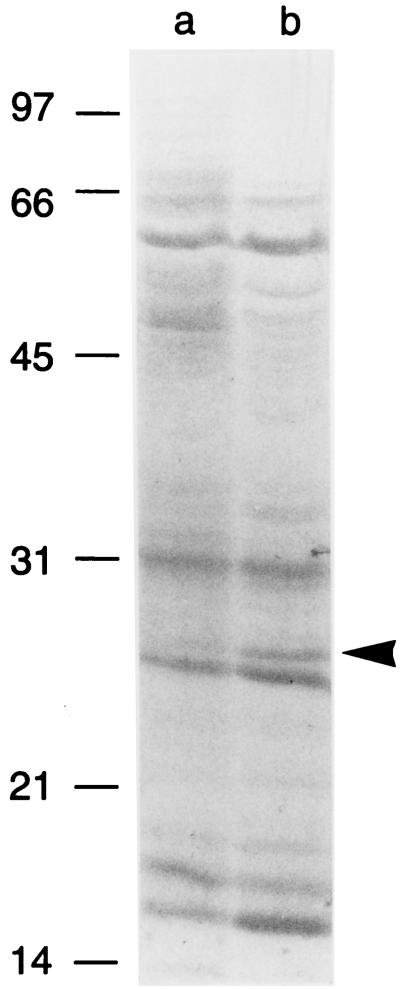
In an effort to identify novel proteins other than VacA that might be specifically secreted into the extracellular space, we carefully compared the SDS-PAGE protein profiles of H. pylori cells and extracellular proteins. A 25-kDa band was identified, which appeared to be overrepresented in supernatant compared to intact bacterial cells from RPMI-based cultures (Fig. 5). N-terminal sequence analysis of this band yielded the sequence MFTLRELPFA, which coincides with the sequence of the N terminus of H. pylori superoxide dismutase (SodB) (25, 31).
FIG. 5.
Analysis of proteins released into the extracellular space during incubation of H. pylori in RPMI medium. H. pylori 60190 was grown for 24 h in 500 ml of SFBB, and the bacterial cells were then pelleted, washed in RPMI medium, and inoculated into 50 ml of methionine-and-cysteine-free RPMI medium. After incubation at 37°C for 24 h, the culture was centrifuged, and both cells and concentrated supernatant proteins (10 μg of each protein) were separated by SDS-PAGE, electroblotted to PVDF paper, and stained with Coomassie blue. The indicated 25-kDa band in the supernatant (arrowhead) was excised, and N-terminal sequence analysis yielded a sequence (MFTLRELPFA) that coincides with the sequence of the N terminus of superoxide dismutase (25, 31). Lane a, H. pylori cells; lane b, H. pylori supernatant proteins. Molecular mass in kilodaltons is shown on the left.
Role of membrane vesicles in extracellular protein release.
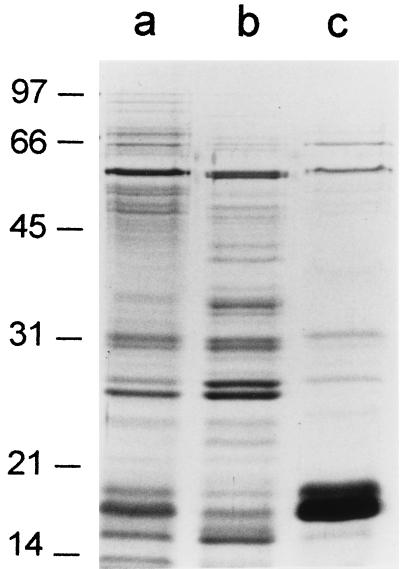
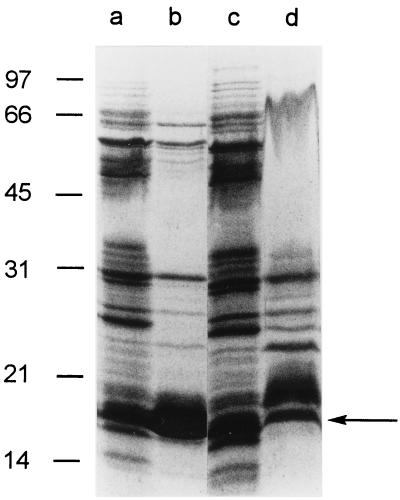
In addition to undergoing bacterial autolysis, H. pylori may release proteins in the form of membrane vesicles or blebs (12, 17, 30). To identify proteins that may be components of such structures, H. pylori was incubated in methionine-and-cysteine-free RPMI medium for 24 h as described above, and then the culture was centrifuged at 17,000 × g to remove intact bacteria. To pellet membranous material, the culture supernatant then was recentrifuged at 100,000 × g for 60 min. As shown in Fig. 6, several proteins were pelleted by ultracentrifugation (lane c), but most supernatant proteins remained soluble (lane b). This indicates that most H. pylori proteins released into the extracellular space are not components of membrane vesicles or blebs. The protein compositions of putative membrane vesicle preparations from RPMI and SFBB media were similar (Fig. 7). One of the most prominent extracellular proteins pelleted by centrifugation at 100,000 × g migrated at 19 kDa (Fig. 6 and 7). N-terminal sequence analysis of 19-kDa bands derived from RPMI- and SFBB-based culture supernatants yielded the same sequence (MLSKDIIKLL), which coincides with the sequence of the N terminus of non-heme iron-containing ferritin (Pfr) (9, 13).
FIG. 6.
Analysis of potential membrane bleb-associated proteins in H. pylori supernatant. H. pylori 60190 was grown for 24 h in 100 ml of SFBB, pelleted, washed, and then resuspended and incubated in 25 ml of methionine-and-cysteine-free RPMI medium for 24 h at 37°C. The culture was centrifuged at 17,000 × g for 20 min to remove intact bacteria, and the supernatant was then recentrifuged at 100,000 × g for 1 h. Proteins (5 μg/lane) were separated by SDS-PAGE followed by visualization with silver staining. Lane a, bacterial cells pelleted by 17,000 × g centrifugation; lane b, proteins remaining in supernatant after centrifugation at 100,000 × g; and lane c, proteins present in supernatant after 17,000 × g centrifugation and subsequently pelletted by centrifugation at 100,000 × g. Molecular mass in kilodaltons is shown on the left.
FIG. 7.
Analysis of proteins in putative membrane bleb fractions isolated from H. pylori broth culture supernatants. H. pylori 60190 was incubated in methionine-and-cysteine-free RPMI medium or in SFBB medium for 24 h at 37°C. Cultures were centrifuged at 17,000 × g for 20 min to remove intact bacteria, and supernatants were then recentrifuged at 100,000 × g for 1 h. Proteins in intact bacterial cells and putative membrane bleb fractions then were separated by SDS-PAGE, electroblotted to PVDF paper, and stained with Coomassie blue. Lane a, bacterial cells from RPMI medium-based culture; lane b, putative membrane bleb components from RPMI medium-based culture; lane c, bacterial cells from SFBB-based culture; and lane d, putative membrane bleb components from SFBB-based culture. N-terminal sequence analysis of a 19-kDa band (arrow) yielded a sequence (MLSKDIIKLL) that coincides with the sequence of the N terminus of non-heme iron-containing ferritin (Pfr) (9, 13). Molecular mass in kilodaltons is shown on the left.
DISCUSSION
In summary, these results indicate that H. pylori proteins can enter the extracellular space via at least three different mechanisms. (i) An early, selective appearance of radiolabeled VacA in culture supernatant relative to other proteins, combined with its preferential localization in supernatant rather than the cell pellet, provides convincing evidence that VacA is specifically secreted. This conclusion is consistent with the structural organization of VacA, which is similar to that of the immunoglobulin A protease family of secreted proteins (8, 28, 33). (ii) Nonselective entry of H. pylori proteins into culture supernatant, concomitant with a gradual decrease in the OD of broth cultures and loss of bacterial viability, is attributed to bacterial autolysis. The occurrence of autolysis in H. pylori has been proposed previously based on ultrastructural evidence of bacterial membrane fragmentation (26) and may account for the surface localization of proteins such as urease and HspB (11, 26). (iii) Several previous studies have reported that H. pylori releases membrane vesicles or blebs (12, 17, 30). Many extracellular proteins that are sedimented by ultracentrifugation are likely to be components of such membranous structures (17). However, the 19-kDa protein (Pfr) is primarily a cytoplasmic protein (9, 13), and therefore, we speculate that it is pelleted in the form of paracrystalline inclusions (9, 13) released from autolysed bacteria rather than as a component of membranous blebs.
An unexpected finding was that a 25-kDa protein corresponding to superoxide dismutase seemed to be selectively released into the extracellular space. In accordance with this observation, Spiegelhalder et al. demonstrated that superoxide dismutase is localized on the surface of H. pylori (31). H. pylori superoxide dismutase does not contain an N-terminal signal sequence, and there are no contiguous genes in the genome of H. pylori 26695 (34) that encode putative proteins involved in secretory pathways. Thus, at present, there is no obvious explanation for the selective release of H. pylori superoxide dismutase into broth culture supernatant, but a specific secretion pathway must be considered. Similarly, a recent study concluded that H. pylori urease, HspA, and HspB are selectively released into the extracellular space, despite the absence of any known specific secretion pathways for these proteins (35).
In this study, we noted that the rate of H. pylori autolysis is increased when organisms are incubated in RPMI medium compared to SFBB. Conversely, VacA secretion was more prominent in SFBB. SFBB is a rich culture medium prepared from animal tissue, yeast extract, and casein, whereas RPMI medium is comparatively sparse in nutrient content. In particular, the RPMI medium used in this study was free of alanine and methionine, both of which are thought to be essential amino acids for H. pylori growth (24, 27). Thus, both autolysis and VacA secretion may be regulated phenomena that are influenced either by the composition of the bacterial culture medium or by the growth phase of the organism.
Autolysis of H. pylori potentially may serve several useful functions. One model suggests that the autolytic release of H. pylori proteins is a mechanism for evading host defenses, either by modifying surface properties of viable organisms or by overwhelming host defenses with decoy antigens (26). Another model suggests that released proinflammatory H. pylori proteins enter the gastric mucosa, stimulate an inflammatory response, and thereby provide H. pylori with an enhanced supply of nutrients (2). Finally, autolysis is an effective mechanism for releasing bacterial DNA, which can be taken up by viable, naturally competent organisms and used to generate recombinant alleles. Genetic recombination seems to have played an important role in the evolution of H. pylori (14, 32) and presumably provides a mechanism for generating diversity that is more rapid than mutation alone (1). Thus, autolysis may be an important adaptation that facilitates persistence of H. pylori in the gastric mucus layer.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health (AI 39657) and the Medical Research Service of the Department of Veterans Affairs. Protein sequencing facilities are supported by funding to the Vanderbilt Cancer Center (P30 CA68485).
We thank M. H. Forsyth for helpful discussions.
REFERENCES
- 1.Berg D E, Hoffman P S, Appelmelk B J, Kusters J G. The Helicobacter pylori genome sequence: genetic factors for long life in the gastric mucosa. Trends Microbiol. 1997;5:468–474. doi: 10.1016/s0966-842x(97)01164-5. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M J. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology. 1992;102:720–727. doi: 10.1016/0016-5085(92)90126-j. [DOI] [PubMed] [Google Scholar]
- 3.Cao P, McClain M S, Forsyth M H, Cover T L. Extracellular release of antigenic proteins by Helicobacter pylori. Infect Immun. 1998;66:2984–2986. doi: 10.1128/iai.66.6.2984-2986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 5.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 6.Cover T L, Blaser M J. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996;41:85–117. [PubMed] [Google Scholar]
- 7.Cover T L, Hanson P I, Heuser J E. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin reveals its pattern of assembly. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 9.Doig P, Austin J W, Trust T J. The Helicobacter pylori 19.6-kilodalton protein is an iron-containing protein resembling ferritin. J Bacteriol. 1993;175:557–560. doi: 10.1128/jb.175.2.557-560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn B E, Vakil N B, Schneider B G, Miller M M, Zitzer J B, Peutz T, Phadnis S H. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect Immun. 1997;65:1181–1188. doi: 10.1128/iai.65.4.1181-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover T L, Solcia E. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J Pathol. 1999;188:220–226. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Frazier B A, Pfeifer J D, Russell D G, Falk P, Olsen A N, Hammar M, Westblom T U, Normark S J. Paracrystalline inclusions of a novel ferritin containing nonheme iron, produced by the human gastric pathogen Helicobacter pylori: evidence for a third class of ferritins. J Bacteriol. 1993;175:966–972. doi: 10.1128/jb.175.4.966-972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Go M F, Kapur V, Graham D Y, Musser J M. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris P R, Mobley H L T, Perez-Perez G I, Blaser M J, Smith P D. Helicobacter pylori urease is a potent stimulus of mononuclear phagocyte activation and inflammatory cytokine production. Gastroenterology. 1996;111:419–425. doi: 10.1053/gast.1996.v111.pm8690207. [DOI] [PubMed] [Google Scholar]
- 16.Hawrylik S J, Wasilko D J, Haskell S L, Gootz T D, Lee S E. Bisulfite or sulfite inhibits growth of Helicobacter pylori. J Clin Microbiol. 1994;32:790–792. doi: 10.1128/jcm.32.3.790-792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keenan J I, Allardyce R A, Bagshaw P F. Lack of protection following immunisation with H. pylori outer membrane vesicles highlights antigenic differences between H. felis and H. pylori. FEMS Microbiol Lett. 1998;161:21–27. doi: 10.1111/j.1574-6968.1998.tb12924.x. [DOI] [PubMed] [Google Scholar]
- 18.Labigne A, deReuse H. Determinants of Helicobacter pylori pathogenicity. Infect Agents Dis. 1996;5:191–202. [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Mai U E H, Perez-Perez G I, Allen J B, Wahl S M, Blaser M J, Smith P D. Surface proteins from Helicobacter pylori exhibit chemotactic activity for human leukocytes and are present in gastric mucosa. J Exp Med. 1992;175:517–525. doi: 10.1084/jem.175.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mai U E H, Perez-Perez G I, Wahl L M, Wahl S M, Blaser M J, Smith P D. Soluble surface proteins from Helicobacter pylori activate monocytes/macrophages by lipopolysaccharide-independent mechanism. J Clin Investig. 1991;87:894–900. doi: 10.1172/JCI115095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobley H L. Helicobacter pylori factors associated with disease development. Gastroenterology. 1997;113(Suppl. 6):S21–S28. doi: 10.1016/s0016-5085(97)80006-6. [DOI] [PubMed] [Google Scholar]
- 23.Montecucco C. Protein toxins and membrane transport. Curr Opin Cell Biol. 1998;10:530–536. doi: 10.1016/s0955-0674(98)80069-0. [DOI] [PubMed] [Google Scholar]
- 24.Nedenskov P. Nutritional requirements for growth of Helicobacter pylori. Appl Environ Microbiol. 1994;60:3450–3453. doi: 10.1128/aem.60.9.3450-3453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pesci E C, Pickett C L. Genetic organization and enzymatic activity of a superoxide dismutase from the microaerophilic human pathogen, Helicobacter pylori. Gene. 1994;143:111–116. doi: 10.1016/0378-1119(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 26.Phadnis S H, Parlow M H, Levy M, Ilver D, Caulkins C M, Connors J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds D J, Penn C W. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology. 1994;140:2649–2656. doi: 10.1099/00221287-140-10-2649. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 29.Scott D R, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 30.Sommi P, Ricci V, Fiocca R, Necchi V, Romano M, Telford J L, Solcia E, Ventura U. Persistence of Helicobacter pylori VacA toxin and vacuolating potential in cultured gastric epithelial cells. Am J Physiol. 1998;275:G681–G688. doi: 10.1152/ajpgi.1998.275.4.G681. [DOI] [PubMed] [Google Scholar]
- 31.Spiegelhalder C, Gerstenecker B, Kersten A, Schiltz E, Kist M. Purification of Helicobacter pylori superoxide dismutase and cloning and sequencing of the gene. Infect Immun. 1993;61:5315–5325. doi: 10.1128/iai.61.12.5315-5325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suerbaum S, Maynard Smith J, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telford, J. L., P. Ghiara, M. Dell’Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, Z. Xiang, E. Papini, C. Montecucco, L. Parente, and R. Rappuoli. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. 179:1653–1658. [DOI] [PMC free article] [PubMed]
- 34.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 35.Vanet A, Labigne A. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect Immun. 1998;66:1023–1027. doi: 10.1128/iai.66.3.1023-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]