Abstract
Yersinia pestis, Y. pseudotuberculosis O:1, and Y. enterocolitica biogroup 1B strains carry a high-pathogenicity island (HPI), which mediates biosynthesis and uptake of the siderophore yersiniabactin and a mouse-lethal phenotype. The HPI of Y. pestis and Y. pseudotuberculosis (Yps HPI) are highly conserved in sequence and organization, while the HPI of Y. enterocolitica (Yen HPI) differs significantly. The 43,393-bp Yen HPI sequence of Y. enterocolitica WA-C, serotype O:8, was completed and compared to that of the Yps HPI of Y. pseudotuberculosis PB1, serotype O:1A. A common GC-rich region (G+C content, 57.5 mol%) of 30.5 kb is conserved between yersinia strains. This region carries genes for yersiniabactin biosynthesis, regulation, and uptake and thus can be considered the functional “core” of the HPI. In contrast, the second part of the HPI is AT rich and completely different in two evolutionary lineages of the HPI, being 12.8 kb in the Yen HPI and 5.6 kb in the Yps HPI. The variable part acquired one IS100 element in the Yps HPI and accumulated four insertion elements, IS1328, IS1329, IS1400, and IS1222, in the Yen HPI. The insertion of a 125-bp ERIC sequence modifies the structure of the promoter of the ybtA yersiniabactin regulator in the Yen HPI. In contrast to the precise excision of the Yps HPI in Y. pseudotuberculosis, the Yen HPI suffers imprecise deletions. The Yen HPI is stably integrated in one of the three asn tRNA copies in Y. enterocolitica biogroup 1B (serotypes O:8, O:13, O:20, and O:21), probably due to inactivation of the putative integrase. The 17-bp duplications of the 3′ end of the asnT RNA are present in both Yersinia spp. The HPI attachment site is unoccupied in nonpathogenic Y. enterocolitica NF-O, biogroup 1A, serotype O:5. The HPI of Yersinia is a composite and widely spread genomic element with a highly conserved yersiniabactin functional “core” and a divergently evolved variable part.
The genus Yersinia consists of 11 species. Strains of Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica are pathogenic for mammals. Pathogenicity determinants have been localized on plasmids and on the chromosome of yersiniae. Pathogenic Yersinia can be divided into a high-pathogenicity group (Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica biogroup [BG] 1B) and a low-pathogenicity group (Y. enterocolitica BG 2 to 4), on the basis of the lethal infectious dose in the mouse model (9). Lethality for mice (50% lethal dose < 1,000 microorganisms) depends on the presence of the yersiniabactin (ybt) locus, which carries genes for biosynthesis, transport, and regulation of the siderophore yersiniabactin (22–24, 28, 31). In Y. pestis, the unstable 102-kb chromosomal fragment is associated with pigmentation (pgm), i.e., the ability of bacterial cells to form pigmented colonies on hemin or Congo red agar plates at 26°C (31). The pgm locus is composed of yersiniabactin (ybt) and hemin storage (hms) loci (15, 25, 27, 29). In contrast, Y. pseudotuberculosis contains nonclustered ybt and hms loci and only the ybt locus is present in Y. enterocolitica (5, 8). The in vitro instability of the pgm locus in Y. pestis has been observed as a complete or partial (hms or ybt locus) deletion of the 102-kb fragment due to the presence of two IS100 flanking sequences (12, 14, 15). The ybt locus comprises 36 to 43 kb (8). Sequencing of genes involved in yersiniabactin synthesis and uptake revealed a G+C content higher than that of the host genome (21, 36). The ybt locus is flanked by an asn tRNA gene at one extremity and carries a gene for a putative integrase (4, 8). These are features typical of pathogenicity islands (20). Therefore, the ybt locus is termed a high-pathogenicity island (HPI) to emphasize its involvement in the mouse-lethal phenotype (8). Interestingly, the ybt cluster has been detected in certain pathotypes of Escherichia coli, suggesting that it originates from a horizontal transfer (40).
Five synthesis genes, irp1 to irp5 (ybtU, ybtT, and ybtE are the orthologs of irp3, irp4, and irp5 in Y. pestis) of the ybt locus are clustered in one large 19-kb operon (2, 28). The genes for the yersiniabactin receptor FyuA (also named Psn in Y. pestis) and an AraC-type yersiniabactin regulator YbtA flank the biosynthetic genes (16, 17, 33). An RS3 repeated sequence and two IS elements were identified downstream of fyuA in Y. enterocolitica (8, 34).
Two distinct variants of the HPI were identified in Y. pseudotuberculosis/Y. pestis (Yps HPI) and in Y. enterocolitica (Yen HPI) (36). The part of the island that contains yersiniabactin synthesis, receptor, and regulator genes is highly conserved in HPIs of both evolutionary lineages (28, 36), while the right end (downstream of fyuA) differs markedly between the Yps HPI and the Yen HPI (5, 8). The Yps HPI is able to occupy any of the three asparagine tRNA genes in Y. pseudotuberculosis, suggesting that the HPI has retained its mobility functions (4). The HPI is flanked by 24-bp (21) or 17-bp (4) direct repeats that are duplications of the 3′ end of asn tRNA.
In this work, we determined the complete molecular genetic structure of the HPI in Y. enterocolitica WA-314 and compared it with that of the Y. pseudotuberculosis HPI to gain insight into the divergent evolution of the HPI and Yersinia in general.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in the study are listed in Table 1. The strains were grown in Luria-Bertani (LB) broth or on LB agar plates (Difco Laboratories, Detroit, Mich.) at 28°C (Yersinia) or 37°C (E. coli). Iron-chelating compounds were screened on a chrome azurol S ferric ion indicator dye (CAS) agar (41). A red-orange halo around bacterial colonies indicated siderophore production (i.e., colonies were CAS agar positive). Y. pestis spontaneous mutants, unable to accumulate the Congo red dye (pgm), were selected on LB medium containing 15 μg of Congo red per ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Y. pestis | ||
| KIM | BG mediavalis | R. R. Brubaker |
| KIM pgm | Spontaneous nonpigmented mutant of KIM | This study |
| Y. pseudotuberculosis | ||
| PB1 | Serotype O:1A | R. R. Brubaker |
| PB1 Nalr | Nalr derivative of PB1 | This study |
| Y. enterocolitica | ||
| WA-314 | Clinical isolate, O:8 BG 1B | 23 |
| WA-C | Plasmidless derivative of WA-314, Nalr | 23 |
| 8081 | Clinical isolate, O:8 BG 1B | 32 |
| Ye1209-79 | Clinical isolate, O:13 BG 1B | 23 |
| Ye1223-75-1 | Clinical isolate, O:20 BG 1B | 23 |
| Ye737 | Clinical isolate, O:21 BG 1B | 23 |
| Y-108 | Clinical isolate, O:3 BG 4 | 23 |
| 108-C | Plasmidless derivative of Y-108 | 23 |
| H567/90 | Clinical isolate, O:5,27 BG 3 | 23 |
| Y-96 | Clinical isolate, O:9 BG 2 | 23 |
| 96-C | Plasmidless derivative of Y-96 | 23 |
| NF-O | Clinical isolate, O:5 BG 1A | 37 |
| E. coli | ||
| Phi | Pesticin sensitive, carrying HPI | R. R. Brubaker |
| K49 | Pesticin sensitive, carrying HPI, ColK, O:156 | 36 |
| K235 | Pesticin sensitive, carrying HPI, ColK, O:1 | 36 |
| D-1041-86 | Enteroaggregative, O:44 | 40 |
| C-4441 | Enteroaggregative, O:128 | 40 |
| 12860 | Enteroinvasive, O:124 | 40 |
| Plasmids | ||
| 12H2 | pLAFR2 cosmid carrying Y. enterocolitica WA-C HPI sequences upstream of the irp2 gene, Tcr | 35 |
| D11 | pLAFR2 cosmid carrying Y. pseudotuberculosis PB1 HPI sequences downstream of the irp1 gene | 36 |
| pRS3 | EcoRI-BamHI fragment of 8081 HPI carrying RS3 | 8 |
| pEBa | BamHI-EcoRI fragment of 8081 between pRS3 and EBg2.4 | 8 |
| EBg2.4 | EcoRI-BglII fragment of 8081 HPI carrying IS1400 | 8 |
| pMOS | Cloning vector, Apr | Amersham |
| pBluescript KSII | Cloning vector, Apr | Stratagene |
DNA manipulations.
Bacterial DNA was isolated by the method of Davis et al. (11). A Y. enterocolitica gene bank was prepared from WA-314 serotype O:8, and cosmid 12H2 was used to determine the 5′ end of the HPI (35). The D11 cosmid of the Y. pseudotuberculosis PB1 serotype O1A gene bank was used to determine the 3′ end of the HPI (36). PCR amplifications were performed in an automated thermal cycler (GeneAmp PCR system 2400; Perkin-Elmer) as described by Saiki et al. (39) with TaqI polymerase and different pairs of oligonucleotides (Roth, Karlsruhe, Germany, and Metabion, Munich, Germany). Self-designed PCR primers used to define the ends of HPI and to amplify specific HPI sequences are listed in Table 2. Southern blot hybridizations were performed with digoxigenin (DIG)-labeled PCR probes, using different primer pairs plus DIG–11-dUTP, as specified by Boeringer Mannheim Biochemica.
TABLE 2.
Primers used in this study
| Primer | Primer sequence (5′ to 3′) | Orientation |
|---|---|---|
| Integrase gene | ||
| int520 | ACATCCTTGCGAATCCTTATC | Forward |
| c15-205 | TACAGGCAGGTTCCCGATGAC | Reverse |
| int1597 | CCTGTGGAGGTGGTGGTAAT | Reverse |
| ybtA promoter | ||
| ybt7524 | AAACAGGGTCGGGAGAGGATT | Forward |
| D7 | GGACATCGATTCAGTATTGGA | Forward |
| ybt1012 | GCCATAGACGCTGTTGTTGAA | Reverse |
| D340 | GGATTGCGTTTGCGGTGACTC | Reverse |
| 3′ end of Yen HPI | ||
| HPI878 | GGGGGCAAGAAAAACTAACC | Forward |
| HPI387 | TCATTAAATAGTCACCCCATAG | Forward |
| Ye9765 | CAAGTGTCACCAGCGTAGGGATTTC | Forward |
| HPI1220 | TTTGTTTTATGGCTTTGGTAG | Reverse |
| HPI283 | TTTCAGTGTTTCAGCGTCCAG | Reverse |
| 3P6057 | CAACTTGAGCGTGATAAACA | Reverse |
| Left junction of Yen HPI | ||
| W250 | TCGGCTCGACGCTTTAGGTAG | Forward |
| W598 | CCAATTTTTCCCGATGCGTAG | Reverse |
| Right junction of HPI | ||
| 3P345 | GAGGCGATCCCAGTCAGAG | Forward |
| Ye262 | TTTTCCCCCGAGAGGCTGAGTAACC | Reverse for Yen HPI |
| W36878 | TGGCTTCCCCGCTTTTTATCGTAT | Reverse for Yps HPI |
| asn tRNA gene | ||
| asn468 | CCGTATGTCACTGGTTCG | Forward |
DNA sequencing and sequence comparison.
Cosmid 12H2 (35) was used to determine sequences located between ybtA and the left end of the island associated with asn tRNA. Four ClaI fragments of the 12H2 cosmid subcloned in pBluescript KSII cloning vector (Stratagene), and the original 12H2 cosmid was used to determine the sequence of the 5′ boundary of the HPI by primer walking.
The sequence between fyuA and the 3′ end of the Yps HPI was determined by a combination of subcloning and primer walking with the Y. pseudotuberculosis O:1A cosmid D11, which contains sequences downstream of fyuA (36). Four EcoRI and three PstI fragments of the D11 cosmid subcloned in pBluescript KS II vector, and the D11 cosmid itself were sequenced to establish the 3′ boundary of the island.
Primers 3P345 (corresponding to the right junction of the island) and 3P6057 (which resides in the region downstream of the HPI that is similar in both evolutionary lineages) amplified a sequence downstream of the pathogenicity island in Y. enterocolitica WA-C.
The sequence between IS1328 and IS1400 was determined in Y. enterocolitica 8081 with the three subclones pRS3 (containing IS1329 and partial IS1222 insertion sequences), pEBa (containing sequences between pRS3 and EBg2.4), and EBg2.4 (carrying the IS1400 mobile element) (8). Plasmids pRS3, pEBa, and EBg2.4 were a kind gift from E. Carniel, Institute Pasteur, Paris, France. Sequencing of the PCR fragments obtained with Y. enterocolitica 8081 chromosomal DNA confirmed the junctions between the subcloned fragments. Bearing in mind the high identity of the Y. enterocolitica WA-C and 8081 sequences (8), the primers designed for the HPI in strain 8081 were used to determine the sequence in strain WA-C. The 5.3-kb DNA fragment between IS1400 and the 3′ end of the Yen HPI was obtained in Y. enterocolitica WA-C by a PCR with the Ye9765 (located downstream of IS1400) and Ye262 (positioned downstream of the right direct repeat DR17 flanking the HPI) primers.
DNA sequencing was performed by the chain termination method with a model ABI 377 DNA sequencer (ABI Prism; Perkin-Elmer). Alignment and sequence comparison were performed with the HIBIO Mac DNASIS (Hitachi Software Engineering Co.) and DNAMAN (Lynnon BioSoft) programs and with the sequence analysis software package of the Genetics Computer Group (University of Wisconsin, Madison, Wis.). The island was analyzed for the presence of open reading frames (ORFs) containing at least 100 codons.
BLAST searches were performed on the NCBI server (27a). HPI sequences were also compared with the Y. pestis CO-92 sequences presented by the Y. pestis Sequencing Group at the Sanger Centre (39a).
Since there is no consensus on uniform nomenclature, we have used the irp (iron-repressed proteins [8]) designation for the genes located on the HPI. The orientation of the genes on the HPI is the same as proposed by Fetherston and Perry (15). We have defined that the intB gene adjacent to the asn tRNA bacterial attachment site is at the 5′ or left extremity of the HPI and that the fyuA gene resides at the 3′ or right extremity of the island.
Nucleotide sequence accession numbers.
The sequences determined in this study were deposited at the EMBL/GenBank database under accession no. AJ132668, AJ132945 and AJ236887.
RESULTS
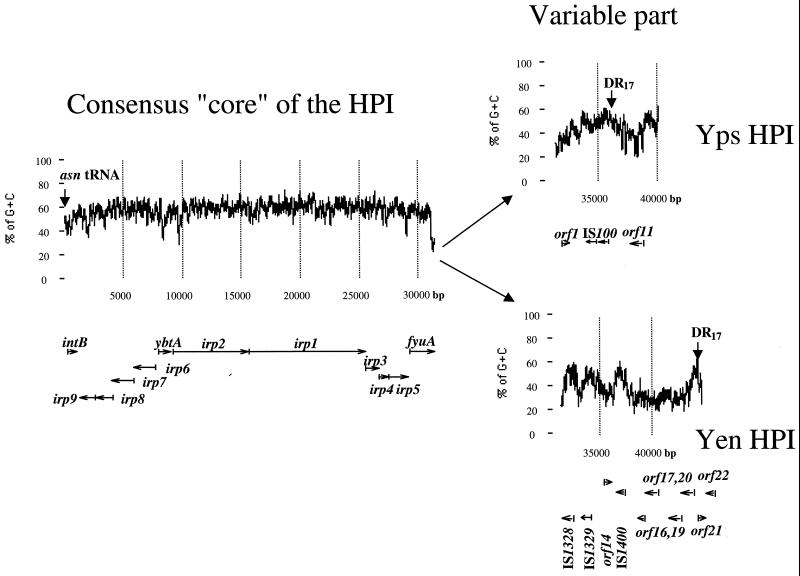
To complete the structure of the Yen HPI in Y. enterocolitica WA-C, serotype O:8, we sequenced both ends of the island (Fig. 1). The borders of the island can be defined by 17-bp direct repeats flanking the HPI. The Yen HPI is 43,393 bp. The left (5′) end, which is associated with the asn tRNA gene, is nearly identical to that of the Yps HPI, while the right (3′) end shows significant differences between the Y. enterocolitica O:8 HPI and the Yps HPI (5, 8). Therefore, the DNA sequence of the 3′ end of the HPI in Y. pseudotuberculosis PB1, O1A (Yps HPI), was also determined and compared to the 3′ end of the Yen HPI. The comparison of Yen and Yps evolutionary lineages of the HPI reveals that the HPI has two distinct parts (Fig. 2). The first part spans the region between asn tRNA and the fyuA stop codon and represents the functional “core” of the island. It is nearly identical (98 to 99% identity) in both evolutionary lineages and has a G+C content significantly higher than that of the Yersinia genome (57.5 mol% versus 46 to 48 mol% for the Yersinia chromosome) (3). A considerably lower G+C content is found downstream of the fyuA gene. This AT-rich variable part differs completely between the HPIs of the two evolutionary lineages.
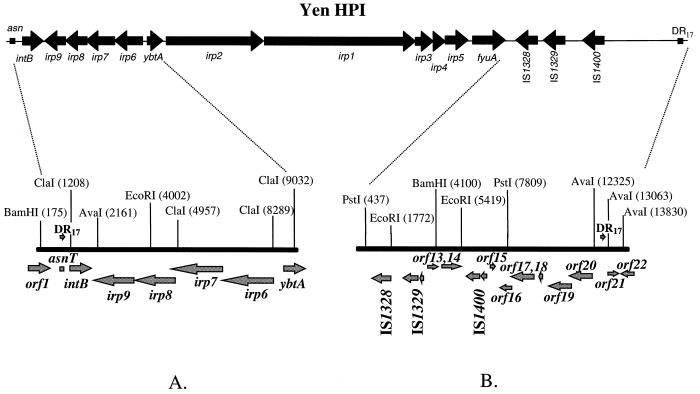
FIG. 1.
Complete structure of the HPI in Y. enterocolitica WA-C. The Left (5′) (A) and right (3′) (B) ends of the island are shown. Arrows show the positions of the ORFs and the direction of transcription. Positions of restriction sites are depicted by vertical lines; numbers above the lines show the distance from the beginning of the sequenced DNA fragment in base pairs.
FIG. 2.
Complete structure and the G+C content of the HPIs in Y. pseudotuberculosis PB1 and Y. enterocolitica WA-C. Arrows below the graph show position of genes and direction of transcription. Vertical arrows show the borders of the HPI, the left DR17 within asn tRNA defines the left border, and the right DR17 defines the right border.
Left (5′) end of the HPI in Y. enterocolitica.
The nucleotide sequence of the 8.1-kb fragment at the 5′ end of the Yen HPI contains five ORFs flanked by ybtA and asn tRNA (Fig. 1A). Four ORFs, irp6 to irp9, have the opposite transcriptional polarity to ybtA. These four genes display 98 to 99% identities to the ybtP, ybtQ, ybtX, and ybtS genes, respectively, recently described in Y. pestis (18, 19). YbtP and YbtQ are thought to be involved in the uptake of the ferric yersiniabactin and YbtS might be involved in biosynthesis of the yersiniabactin in Y. pestis.
Integrase.
The next ORF has an opposite transcriptional polarity to irp6 to irp9 (Fig. 1A). It has high similarity to the genes encoding putative integrase from Y. pseudotuberculosis (4) and to the P4 prophage integrase (6). In contrast to the putative integrase genes in Y. pseudotuberculosis and Y. pestis (4, 21), a TAA stop codon interrupts this ORF, henceforth designated intB, 415 bp downstream of the ATG start codon. The 420-amino-acid (aa) polypeptide that can be predicted from the intB pseudogene sequence shows 98% identity to the putative integrase of Y. pseudotuberculosis and 49.7% identity to the integrase of the prophage P4.
The int520 and int1597 primers amplifying the putative integrase gene were used to generate a probe for Southern hybridization with different isolates of Y. pestis, Y. pseudotuberculosis O1A, Y. enterocolitica, and E. coli. Surprisingly, two reactive bands appeared in both Y. pseudotuberculosis PB1 and Y. pestis KIM, suggesting the presence of two copies of intB (or, alternatively, of a sequence highly identical to the designed probe), in contrast to one copy present in Y. enterocolitica WA-C and E. coli 12860 (data not shown). The DNA of European Y. enterocolitica serotype O:9, O:5,27, and O:3 (BG 2, 3, and 4, respectively) strains did not hybridize with the intB probe. The nonpigmented Y. pestis KIM isolate, which had lost the 102-kb pgm locus, retained one band hybridizing with the intB probe.
intB was sequenced in several HPI-positive isolates. In contrast to Y. enterocolitica WA-C, a T-to-G change turned a stop codon into a GAA triplet in Y. pestis KIM, Y. pseudotuberculosis PB1, and five HPI-positive E. coli isolates (K49, K235, D-1041-86, C-4441, and 12860). Two other Y. enterocolitica 1B representatives (serotypes O:13 and O:20) contain the terminating codon in the same position as in serotype O:8 WA-C and 8081 strains.
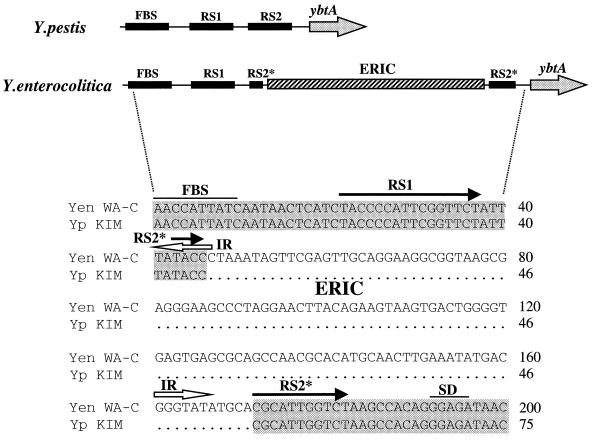
ERIC sequence.
A 127-bp enterobacterial repetitive intergenic consensus (ERIC) sequence (26), also known as the intergenic repeated unit (IRU) (42), was recognized in the promoter of the ybtA yersiniabactin regulator (Fig. 3). D7 and D340 primers, designed to amplify the ybtA promoter, yielded a 229-bp product in Y. pseudotuberculosis PB1, Y. pestis KIM, and HPI-positive E. coli strains (K49, K235, D-1041-86, C-4441, and 12860). In contrast, the same primers amplified a larger (354-bp) product in all Y. enterocolitica 1B isolates (O:8, O:13, O:20, and O:21). Sequencing of the Y. enterocolitica 1B amplicons demonstrated that the 125-bp DNA insertion represents an ERIC sequence.
FIG. 3.
Insertion of a 125-bp ERIC sequence into the ybtA promoter of the Yen HPI. The upper panel shows the structure of the ybtA promoter in Y. pestis and Y. enterocolitica. The lower panel depicts aligned nucleotide sequences of both promoters. Identical bases are boxed in grey. Black arrows show two repeated sequences (RS1 and RS2) of the ybtA promoter. The asterisk in the RS2* sequence depicts an interruption in the repeated sequence. Open arrows show the position and direction of the inverted repeats (IR) of the ERIC. Yen WA-C, Y. enterocolitica WA-C; Yp KIM, Y. pestis KIM; FBS, Fur protein-binding site; RS, repeated sequences, SD, potential ribosome-binding site.
The ERIC element positioned within the ybtA promoter sequence is almost identical to the consensus ERIC sequence (26, 42). Such ERIC motifs are present in multiple copies in intergenic regions or in untranslated regions upstream or downstream of ORFs in various genomes. However, the possible function of the ERIC sequence is still enigmatic.
The ybtA promoter, as well as promoters of the other genes that control yersiniabactin biosynthesis and uptake, contains putative binding sites for the YbtA transcriptional regulator (17). The YbtA-binding site has a palindromic structure with inverted and direct repeats. Six nucleotides, TATACC, of the middle part of the ybtA operator are identical to six of seven nucleotides (TATACCC) representing the inverted repeats of the ERIC consensus (Fig. 3). This sequence seems to be a recognition site for integration of the ERIC element that possibly exploits a site-specific mechanism of integration into a target site. Taken together, these observations suggest that integration of the ERIC sequence modifies the secondary structure of the ybtA promoter in Y. enterocolitica, resulting in modulation of yersiniabactin activity (36a).
Right (3′) end of the HPI.
The size of the 3′ end of the HPI differs between the evolutionary groups. We determined a 14-kb sequence downstream of the fyuA gene in Y. enterocolitica WA-C and compared it to the corresponding sequence of Y. pseudotuberculosis PB1.
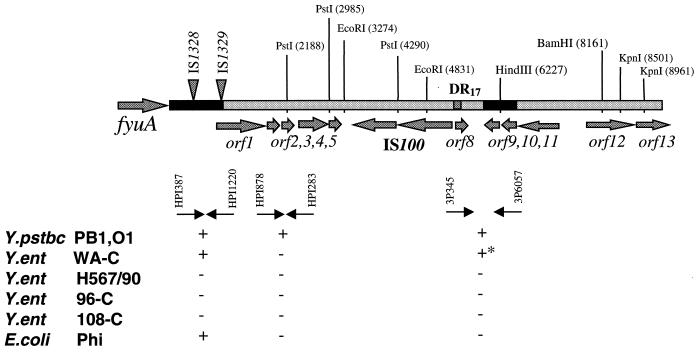
The right end of the Yen HPI contains 13 ORFs (Fig. 1B; Table 3). Six of them are putative transposases of four insertion elements, IS1328, IS1329 (two ORFs), IS1400 (two ORFs) and ORF13, which is a truncated transposase gene of the IS1222 sequence. The ORFs encoding IS1328, IS1329, and IS1400 transposases are transcribed in the same orientation. Comparing the Yen and Yps HPI sequences, we determined the nucleotide sequence of an IS3 family IS1329 mobile element (designated RS3 by Carniel et al. [8]) and defined the precise location of the IS elements on the Yen HPI. Southern hybridization revealed that the 947-bp fragment located downstream of fyuA is common to all HPI-positive isolates, including E. coli, while sequences located downstream of this 947-bp fragment (left black bar in Fig. 4) are present only in Yen HPI. Proteins that might be encoded by the three ORFs, ORF16, ORF17, and ORF19 (Table 3), showed some similarity to the hypothetical proteins YfjK (P52126) and YfjL (P52127) clustered in the alpA-gabD region of the E. coli K-12 chromosome.
TABLE 3.
ORFs defined in the right extremity of the Yen HPI and downstream of the DR17
| Protein | Predicted mass (kDa) | Predicted function | Amino acid identity, protein, organism (accession no.) |
|---|---|---|---|
| Right end of Yen HPI | |||
| Tnp IS1328 | 35.5 | Transposase | 67% to plasmid R751 TnpA (U60777) |
| TnpA IS1329 | 12.9 | Transposase | 42% to IS911 12.7-kDa protein (P39213) |
| TnpB IS1329 | 28.7 | Transposase | 40% to IS911 OrfB (AF074613) |
| Orf13 | 18.7 | Transposase | 98% to IS1222 transposase (B38965) |
| Orf14 | 51.2 | Unknown | |
| TnpA IS1400 | 11.1 | Transposase | 88% to ORFA transposase of Salmonella enteritidis (Z83734) |
| TnpB IS1400 | 35.1 | Transposase | 40% to OrfB of IS3 family (U39501) |
| Orf15 | 5.7 | Unknown | |
| Orf16 | 22.8 | 24% to YfjK of E. coli (P52126) | |
| Orf17 | 44.5 | 35% to YfjK of E. coli (P52126) | |
| Orf18 | 12 | Unknown | |
| Orf19 | 43.1 | 22% to YfjL of E. coli (P52127) | |
| Orf20 | 43.2 | 24% to ComE integral protein of Bacillus subtilis (P39695) | |
| Downstream of the DR17 | |||
| Orf21 | 13.6 | 43% to uracyl hydrolase of E. coli (P39219) | |
| Orf22 | >14.7 | 86% to E. coli nucleoprotein/polynucleotide-associated enzyme (U73857) |
FIG. 4.
Fragment of the Y. pseudotuberculosis PB1 chromosome with the 3′ end of the Yps HPI. The large arrow on the left indicates the position of fyuA. Arrows show positions of the ORFs and their direction of transcription. Black bars within the Y. pseudotuberculosis sequence represent regions of identity with the Y. enterocolitica DNA. Triangles indicate positions of the IS elements within the Yen HPI. PstI, EcoRI, BamHI, and KpnI depict the positions of recognition sites of the corresponding enzymes. Small black arrows under the graph indicate PCR primers used. The DIG–11-dUTP-labelled PCR products obtained with the same primer pairs were used as hybridization probes. + and −, presence or absence, respectively, of hybridization products. +*, larger PCR amplicon in Y. enterocolitica WA-C; Y.ent, Y. enterocolitica; Y.pstbc, Y. pseudotuberculosis.
ORF21 and ORF22 are located outside of the Yen HPI. The ORF21 product has 40 and 43% identity to the uracyl hydrolase of Haemophilus influenzae (accession no. P44782) and E. coli (accession no. P39219), respectively. The ORF22 product has high (86%) identity (113 of 131 aa) to a YaiL nucleotide/polynucleotide-associated enzyme of E. coli (accession no. U73857). Moreover, ORF22 displays 89.9% similarity to ORF9 and ORF10 located downstream of the 17-bp right direct repeat (DR17) of the Yps HPI (Fig. 4; Table 4). Thus, identical chromosomal sequences, ORF22 in Y. enterocolitica WA-C and ORF9 and ORF10 in Y. pseudotuberculosis PB1, are clustered with the 3′ end of the HPI.
TABLE 4.
ORFs defined in the right extremity of the Yps HPI and downstream of the DR17
| Protein, ORF | Predicted mass (kDa) | Predicted function | Amino acid identity, protein, organism (accession no.) |
|---|---|---|---|
| Right end of Yps HPI | |||
| Orf1 | 35.4 | 99% to Orf73, Orf74 of Y. pestis HPI (AL031866) | |
| Orf2 | 7.2 | DNA recognition | 36% to putative DNA-binding protein of phage P4 (P12552) |
| Orf3 | 6.8 | 100% to Orf76 of Y. pestis HPI (AL031866) | |
| Orf4 | 18.8 | 100% to Orf77 of Y. pestis HPI (AL031866) | |
| Orf5 | 8 | DNA recognition | 50% to TraR of F-plasmid (U01159) |
| TnpB IS100 | 29.2 | Transposase | 47% to IstB of IS21 (P15026) |
| TnpA IS100 | 39.8 | Transposase | 32% to IstA of IS21 (P15025) |
| Orf8 | 7.5 | Unknown | |
| Downstream of the DR17 | |||
| Orf9 | 11 | 84% to E. coli nucleoprotein/polynucleotide-associated enzyme (U73857) | |
| Orf10 | 9.1 | 83% to E. coli nucleoprotein/polynucleotide-associated enzyme (U73857) | |
| Orf11 | 26.5 | 31% to soybean transcription factor 5 (S59539) | |
| Orf12 | 34.9 | 75% to conserved hypothetical protein of Helicobacter pylori (AE000549) | |
| Orf13 | >13.2 | Unknown |
The right end of Y. pseudotuberculosis PB1 HPI contains eight ORFs, two of them encoding a putative transposase of the IS100 element (Fig. 4; Table 4). The IS100 insertion sequence is located 3,429 bp downstream of the fyuA stop codon. A 249-bp DNA fragment separates IS100 from the DR17 that is identical to the 3′ end of the asn tRNA. ORF1 has a high A+T content (63.3% A+T) and is located 874 bp downstream of the fyuA stop codon. It is able to encode a 20.8-kDa protein with no obvious homology to any sequence in the database besides its orthologs in the Y. pestis HPI (Table 4).
A small ORF, ORF2, is able to encode a 61-aa protein with 37% similarity to the hypothetical protein 88 of phage φR73 (G42465) and the prophage CP4-57 protein AlpA (P33997) and 36% similarity to a putative DNA-binding protein (the ORF88 product) of the bacteriophage P4 (P12552).
ORF5 may encode a 69-aa small product with possible DNA-binding activity. The ORF5 protein has 50% identity to a zinc finger region of the TraR protein of the F factor (accession no. AF005044). No function has been assigned to the TraR protein. The ORF5 product also has 37% identity to the ORF39 product of Pseudomonas aeruginosa phage φCTX (accession no. AB008550) and shows a similar level of identity to DnaK suppressor proteins of Treponema pallidum (33%), H. influenzae (31%), and E. coli (27%). These small proteins may play a role in interactions with DNA and may be remnants of the HPI association with self-transmissible elements. However, ORF2 and ORF5 are restricted to Y. pestis and Y. pseudotuberculosis, and are not present in the Yen HPI.
ORF9 and ORF10 are located outside the HPI and show 84 and 83% identity to a gene encoding a hypothetical nucleoprotein of E. coli (accession no. U73857). The ORF12 product has 75% identity to a conserved hypothetical protein of Helicobacter pylori (accession no. AE000549).
The ORF22 sequence of the Yen HPI (Fig. 1B) starts 839 bp downstream of the DR17. It has 89.9% similarity to ORF9 and ORF10 of the Yps HPI located 356 bp downstream of the DR17 (Fig. 4). The same region has 98.5% similarity to the Y. pestis CO-92 sequence presented by the Y. pestis Sequencing Group of the Sanger Centre (39a). In contrast to Y. pseudotuberculosis and Y. enterocolitica, this DNA fragment is not contiguous with the HPI in Y. pestis CO-92. In Y. pestis, this highly conserved region is associated with another asn tRNA copy. Thus, the chromosomal location of the HPI is the same in Y. enterocolitica WA-314 and Y. pseudotuberculosis PB1 but differs from that in Y. pestis.
Junctions of the high-pathogenicity island.
The 5′ intB junction of the HPI in Y. enterocolitica 8081, serotype O:8, is associated with the 3′ end of the asnT tRNA (8). asnT tRNA is also used by the HPI in E. coli (40). Integration of pathogenicity islands into highly conserved tRNA copies is a common feature of these islands (20). Four identical copies of the asn tRNA genes (asnT, asnU, asnV, and asnW) have been identified on the chromosome of E. coli K-12 (E. coli MG1655 Genome Project; accession no. AE000289, AE000290, and AE000291) and three have been identified in Yersinia (4). The sequence upstream of the asnT tRNA gene in Y. enterocolitica WA-C O:8 has 84% identity to the sequences upstream of the HPI in Y. pseudotuberculosis O:1 (accession no. AJ009592 [4]). This agrees with the proposal that the Yen HPI in Y. enterocolitica WA-C and the Yps HPI in Y. pseudotuberculosis PB1 occupy the same asn tRNA copy.
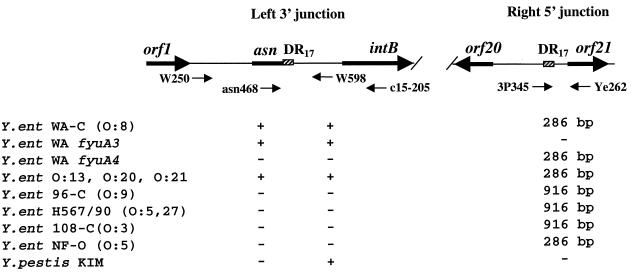
To test whether the HPI is associated with the same asnT tRNA copy in other HPI-positive Yersinia strains, we performed PCR with W250 and W598 primers complementary to the sequences flanking the left junction of the HPI (Fig. 5). Y. enterocolitica BG 1B strains of serotypes O:8, O:13, O:20, and O:21 were positive in this PCR. Y. pestis KIM and Y. enterocolitica Ye H567/90 (O:5,27, BG3), Ye 96-C (O:9, BG2), Ye 108-C (O:3, BG4), and NF-O (O:5, BG 1A) were negative in PCR with these primers. In contrast, PCR with the asn468 (annealing to the conserved part of asn tRNA) and c15-205 (annealing to the middle of intB) primers resulted in a PCR product of the same size in all HPI-positive isolates. This indicates that the HPI is integrated into the same asnT gene in all four serotypes of Y. enterocolitica BG 1B.
FIG. 5.
Left (5′) and right (3′) junctions of the Yen HPI. Arrows in the graph show position and orientation of the genes. Black arrows under the graph show PCR primers used. The + and − indicate the presence or absence of a PCR product amplified with W250 plus W598 and asn468 plus c15-205 primer pairs. Numbers correspond to the size of the PCR products in base pairs. Y.ent, Y. enterocolitica.
A 22-bp stretch of identical DNA, gtCCAGTCAGAGGAGCCAAaTT, can be recognized in the 3′ fyuA junction of both HPIs. This short fragment is a 20-bp imperfect duplication (or a DR17) of the 3′ end of the asn tRNA. Such short duplications are hallmarks of site-specific recombination, whereas the 3′ end of tRNA serves as the core of the bacterial attachment site (att) for integration of temperate phages and conjugative plasmids (7, 38).
The HPI att site is unoccupied in Y. enterocolitica biogroup 1A.
Primers W250 (located upstream of the asnT tRNA) and Ye262 (anneals downstream of DR17) flank the HPI (Fig. 5). As expected, they failed to amplify the whole HPI in Y. enterocolitica WA-C and 8081. However, these primers successfully amplified a 515-bp fragment in avirulent Y. enterocolitica NF-O (O:5, BG 1A). The sequence of this amplicon contains a 16-bp DNA stretch identical to DR17 and overlaps with the sequences flanking the junctions of the HPI in Y. enterocolitica WA-C (Fig. 6).
FIG. 6.
Unoccupied attachment site of the HPI in Y. enterocolitica NF-O. The NF-O sequence was amplified with primers W250 and Ye262 (Fig. 5) and aligned with the 5′ and 3′ boundaries of the Yen HPI. Grey boxes show identical nucleotide sequences and a 16-bp part of the DR17. Numbers on the right show relative positions of nucleotides in base pairs. 5′ end, 5′ boundary of the Yen HPI, asnT RNA; 3′ end, 3′ boundary of the Yen HPI; NF-O, a PCR product amplified by the W250 and Ye262 primers in Y. enterocolitica NF-O.
Y. enterocolitica of other biogroups such as Ye108-C (O:3, BG 4), H597/90 (O:5,27, BG 3), and Ye96-C (O:9, BG 2) were negative in a PCR with the W250/Ye262 primers flanking the HPI. However, these yersiniae yielded a 916-bp PCR product with primers 3P345 (overlaps with the DR17) and Ye262 (Fig. 5). This amplicon was 630 bp larger than the product amplified with the same primers in Y. enterocolitica WA-C. The PCR product reveals identity over 90 bp to the sequence located 134 bp downstream of the right HPI junction in yersiniae lethal for mice. Negative results with the HPI-flanking primers and presence of an additional DNA fragment adjacent to the HPI att site imply that the same recognition site can be used for integration of different DNA in American (BG 1B) and European (BG 2, 3, and 4) Y. enterocolitica. In turn, the att site (asnT RNA) that is used by the Yen HPI in Y. enterocolitica BG 1B strains is unoccupied in avirulent Y. enterocolitica O:5 BG 1A isolates.
Deletions in Y. enterocolitica HPI are imprecise.
The yersiniabactin receptor FyuA has a dual function, since it also serves as a receptor for the Y. pestis bacteriocin pesticin (31, 33). Thus, pesticin sensitivity can be used as a selective marker for the presence of FyuA. Previously we selected several Y. enterocolitica pesticin-resistant spontaneous mutants that failed to produce the yersiniabactin on CAS agar (35). This is indicative that yersiniabactin biosynthetic genes were coinactivated with the yersiniabactin receptor gene. The WA fyuA3 and WA fyuA4 mutants were resistant to the pesticin due to deletions in fyuA. We have analyzed both mutants for the presence of the HPI genes (Table 5). intB through irp4 sequences were detected in the WA fyuA3 mutant but were absent from the WA fyuA4 mutant. Both mutants lost IS1328 and IS1329 insertion sequences and either of the junctions (Fig. 5). However, WA fyuA4 contains the IS1400 element as well as the right junction of the HPI. WA fyuA3 and WA fyuA4 were negative with the W250 and Ye262 primers that amplified the island-free attachment site in Y. enterocolitica NF-O. In summary, WA fyuA4 has lost the HPI sequences upstream of IS1400, whereas WA fyuA3 has lost the right extremity of the island downstream of the irp4 gene. Thus, spontaneous elimination of the Yen HPI, which results in the inactive yersiniabactin iron acquisition system, occurs through imprecise deletions in contrast to the precise excision described for the Yps HPI (4).
TABLE 5.
Presence of the HPI-associated sequences in Y. enterocolitica strains
| Strain | Presence of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| attLa | intB | irp2 | irp1 | irp4a | fyuA | IS1328 | IS1329 | IS1400 | attRa | |
| WA-C | + | + | + | + | + | + | + | + | + | + |
| WA fyuA3 | + | + | + | + | + | − | − | − | − | − |
| WA fyuA4 | − | − | − | − | − | − | − | − | + | + |
The presence of these sequences was checked by a PCR. The presence of the other sequences was proved by Southern hybridization.
DISCUSSION
The HPI of Yersinia spp. is responsible for lethality for mice and for biosynthesis and uptake of the siderophore yersiniabactin. Sequence comparison depicts two evolutionary lineages of the HPI, Y. enterocolitica (Yen HPI) and Y. pestis/Y. pseudotuberculosis (Yps HPI), at the nucleotide level (21, 36) as well as in the genetic organization of the fyuA end of the island (8). We have determined the complete size of the Yen HPI to be 43,393 bp, which is 7.3 kb larger than that of the corresponding HPI in Y. pseudotuberculosis. Such a difference infers the presence of additional genes on the Yen HPI, which might support functions different from yersiniabactin-mediated iron acquisition.
Yersiniabactin-mediated iron acquisition is thought to be the main function of the HPI (19), and genes involved in yersiniabactin biosynthesis, transport, and regulation (irp1 to irp9, ybtA, and fyuA) are clustered on the “core” of the HPI. The P4-like integrase gene intB resides on the 5′ end of the HPI next to the asn tRNA bacterial attachment site and belongs to the “core” of the island as well. The genes of the “core” show extremely high conservation in both evolutionary lineages and are characterized by a higher G+C (57.5 mol%) content than average in yersiniae (48 mol%) (2, 4, 19, 36). In contrast, the second component of the HPI is an AT-rich region that is completely different in the Yen HPI and Yps HPI. Thus, the genes of the HPI variable region are not expected to be involved in yersiniabactin production.
We could identify 13 ORFs within the fyuA end of the Yen HPI (Table 3). Six of them encode putative transposases of the four insertion sequences IS1328, IS1329, IS1400, and IS1222. The fyuA end of the Yps HPI in Y. pseudotuberculosis contains eight ORFs (Table 4). Two of them encode a putative transposase of IS100, while two other ORFs encode products with similarities to putative DNA-binding proteins. These proteins might be the remnants of self-transmissible elements, which were lost during stabilization of the HPI. The ORF2 and ORF5 products might be involved in DNA recognition, but they are not sufficient for the horizontal transfer of the island. Moreover, these proteins are not present in the Y. enterocolitica or E. coli HPI.
Yersinia might have acquired the HPI horizontally from a common progenitor with a high G+C content. The HPI has evolved divergently in the two evolutionary lineages, although the DNA similarity between the yersiniabactin genes of the two groups is about 98%. Y. enterocolitica 1B strains are the only ones that produce a halo on a CAS indicator agar, indicating yersiniabactin production. Actually, the yersiniabactin is the sole endogenous siderophore of Y. enterocolitica. Y. pestis, although containing a complete set of yersiniabactin genes, appears to be CAS negative. The fine-tuning of yersiniabactin genes to specific requirements of the Y. enterocolitica cell might be achieved in a stepwise mode by a single integration of the ERIC element into the promoter of the ybtA yersiniabactin regulator in Y. enterocolitica. The ERIC sequence modifies the structure of the ybtA operator and thus might be responsible for the different expression of the yersiniabactin biosynthetic genes in yersiniae (36a).
The HPI is associated with the asn tRNA genes in Y. enterocolitica (8), Y. pseudotuberculosis (4), Y. pestis (21), and E. coli (40). The Yps HPI can be inserted into any of the three asn tRNA copies of Y. pseudotuberculosis (4) and can use different asn tRNA genes in Y. pestis and Y. pseudotuberculosis (21). The comparison of the HPI integration sites in two Y. pseudotuberculosis strains, described in recent publications (4, 21), demonstrates that the Yps HPI integrates into two different asn tRNA copies in these strains. In contrast to the Yps HPI, the Yen HPI is stably integrated into the same asnT RNA gene in all serotypes of Y. enterocolitica BG 1B strains. Therefore, the HPI seems to be “immobile” in Y. enterocolitica, perhaps due to the inactivated putative integrase. Y. enterocolitica is thus not expected to be the original donor of the HPI that is widely distributed among the members of the Enterobacteriaceae (40). The presence of the Y. pestis-type HPI in the Enterobacteriaceae supports this prediction.
The Yen HPI, like the Yps HPI, is flanked by a 17-bp perfect duplication of the 3′ end of the asn tRNA gene. Such duplications indicate a site-specific recombination event that results in integration of prophages and plasmids (7). The excision of the integrated units is predominantly precise and leads to the reconstruction of the original attachment site. A precise excision seems to be responsible for the HPI disintegration in Y. pseudotuberculosis O:1A (4). In contrast, deletions of the Yen HPI sequences in two pesticin-resistant mutants resulted in different endpoints and extensions. Moreover, the direct repeats of the island do not play a role in these deletion events. Three complete IS elements, which are present in the Yen HPI, might be responsible for the above deletions. Consequently, in contrast to Y. pseudotuberculosis, the precise excision of the Yen HPI is a rare or perhaps even impossible event in Y. enterocolitica.
We have identified an “island-free” bacterial att site for the HPI in apathogenic Y. enterocolitica NF-O (serotype O:5, BG 1A) (Fig. 6). The same site is “occupied” in HPI-negative yersiniae of BG 2, 3, and 4. This indicates that the asn tRNA genes might be used as integration sites for a foreign DNA in human pathogenic yersiniae.
The widespread presence of the HPI in E. coli of different pathotypes (40) implies an efficient mechanism of its transfer. Temperate phages or transmissible plasmids are candidates as HPI vehicles. The presence of a variable AT-rich “additive” to the highly conserved “core” points to a passive HPI transfer by a head-full phage transduction. It is also possible that the mobility genes were already lost by the island as in the locus of enterocyte effacing (LEE) island in enteropathogenic E. coli strains compared to enterohemorrhagic E. coli O157:H7 (30). Alternatively, the LEE island in O157:H7 may acquire the mobility genes. ORF2 and ORF5 with possible DNA-binding ability might be remnants of a mobility fraction of the ancestral HPI.
Different pathotypes of E. coli carry the yersiniabactin “core” of the HPI (40). In addition to the yersiniabactin iron acquisition system, E. coli has the enterochelin system, with higher affinity for iron (10). Reportedly, isolated irp2-positive Y. pseudotuberculosis O:3 strains do not express siderophore activity on the CAS agar and lack the yersiniabactin receptor (4, 36). Therefore, one can envisage alternative functions of the HPI-encoded genes besides production of the yersiniabactin and iron uptake. Modulation of cellular host defense (1, 13) may be a complementary function of the yersiniabactin. Determination of alternative functions and mechanisms of HPI transfer will provide interesting insights into the evolution of genomic islands.
ACKNOWLEDGMENTS
This study was supported by a grant from the Deutsche Forschungsgemeinschaft to J.H. (HE 1297/8-1).
We thank C. Pelludat, W.-D. Hardt, and M. Hensel for critical reading of the manuscript.
REFERENCES
- 1.Autenrieth I B, Bohn E, Ewald J H, Heesemann J. Deferoxamine B but not deferoxamine G1 inhibits cytokine production in murine bone marrow macrophages. J Infect Dis. 1995;172:490–496. doi: 10.1093/infdis/172.2.490. [DOI] [PubMed] [Google Scholar]
- 2.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bercovier H, Mollaret H H. Yersinia. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 498–506. [Google Scholar]
- 4.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 5.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burland V, Plunkett III G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome VI: DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal high pathogenicity island in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter P B. Pathogenecity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis L G, Dibner M D, Battley J F. Basic methods in molecular biology. New York, N.Y: Elsevier Biomedical Press; 1990. [Google Scholar]
- 12.de Almeida A M, Guiyoule A, Leal N C, Carniel E. Survey of the irp2 gene among Yersinia pestis strains isolated during several plague outbreaks in northeast Brazil. Mem Inst Oswaldo Cruz. 1994;89:87–92. doi: 10.1590/s0074-02761994000100015. [DOI] [PubMed] [Google Scholar]
- 13.Ewald J H, Heesemann J, Rudiger H, Autenrieth I B. Interaction of polymorphonuclear leukocytes with Yersinia enterocolitica: role of the Yersinia virulence plasmid and modulation by the iron-chelator desferrioxamine B. J Infect Dis. 1994;170:140–150. doi: 10.1093/infdis/170.1.140. [DOI] [PubMed] [Google Scholar]
- 14.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 15.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 16.Fetherston J D, Lillard J W, Jr, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 18.Fetherston J D, Bertolino V J, Perry R D. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32:289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 19.Gehring A M, DeMoll E, Fetherston J D, Mori I, Mayhew G F, Blattner F R, Walsh C T, Perry R D. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 20.Hacker J, Blum Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 21.Hare J M, Wagner A K, McDonough K A. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol. 1999;31:291–303. doi: 10.1046/j.1365-2958.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 22.Heesemann J, Algermissen B, Laufs R. Genetically manipulated virulence of Yersinia enterocolitica. Infect Immun. 1984;46:105–110. doi: 10.1128/iai.46.1.105-110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heesemann J. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol Lett. 1987;32:229–233. [Google Scholar]
- 24.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 26.Hulton C S, Higgins C F, Sharp P M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5:825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 27.Lillard J W, Jr, Fetherston J D, Pedersen L, Pendrak M L, Perry R D. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 27a.National Center for Biotechnology Information. 21 December 1998, posting date. Sequences. [Online.] http://www.ncbi.nlm.nih.gov/blast/blast.cgi site. [10 August 1999, last date accessed.]
- 28.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pendrak M L, Perry R D. Characterization of a hemin-storage locus of Yersinia pestis. Biol Metals. 1991;4:41–47. doi: 10.1007/BF01135556. [DOI] [PubMed] [Google Scholar]
- 30.Perna N T, Mayhew G F, Postfai G, Elliott S, Donneberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry R D, Fetherston J D. Yersinia pestis—etiological agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portnoy D A, Falkow S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol. 1981;148:877–883. doi: 10.1128/jb.148.3.877-883.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 34.Rakin A, Heesemann J. Virulence-associated fyuA/irp2 gene cluster of Yersinia enterocolitica biotype 1B carries a novel insertion sequence IS1328. FEMS Microbiol Lett. 1995;129:287–292. doi: 10.1111/j.1574-6968.1995.tb07594.x. [DOI] [PubMed] [Google Scholar]
- 35.Rakin A, Heesemann J. Yersiniabactin/pesticin receptor: a component of an iron uptake system of highly pathogenic Yersinia. Contrib Microbiol Immunol. 1995;13:244–247. [PubMed] [Google Scholar]
- 36.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Rakin, A., et al. Unpublished data.
- 37.Ratnam S, Merser E, Picco B, Parsons S, Butler R. A nosocomial outbreak of diarrheal disease due to Yersinia enterocolitica serotype O:5, biotype 1. J Infect Dis. 1982;145:242. doi: 10.1093/infdis/145.2.242. [DOI] [PubMed] [Google Scholar]
- 38.Reiter W D, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 39a.Sanger Centre. Yersinia pestis Genome Project. 3 February 1999, posting date. Yersinia pestis CO92 genomic sequence database. [Online.] http://www.sanger.ac.uk/Projects/Y_pestis/blast_server.shtml site. [10 August 1999, last date accessed.
- 40.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 42.Sharples G J, Lloyd R G. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18:6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]