Abstract
Simple Summary
Ameloblastoma is a benign odontogenic tumour, and the patient always presents at a later stage when the tumour is already in an aggressive state. The finding of high mutation of BRAF V600E indicates the need to explore the molecular pathogenesis of ameloblastoma. However, there is inconsistent evidence regarding this mutation occurrence and its association with clinical information. This systematic review and meta-analysis aim to pool the overall mutation prevalence of BRAF V600E in reported ameloblastoma cases and to determine its association with patient demographic and clinicopathological features. This meta-analysis shows that BRAF V600E mutation has a high pooled prevalence of 70.49% in ameloblastoma. Furthermore, there was a significant meta-analysis association for those younger than 54 years old and in the mandible. Researchers could utilise these findings to improve the treatment option and find a possible new biomarker for the early detection of ameloblastoma.
Abstract
The discovery that ameloblastoma has a high mutation incidence of BRAF V600E may enable a better investigation of pathophysiology. However, there is inconsistent evidence regarding this mutation occurrence and its association with clinical information. This systematic review and meta-analysis aim to pool the overall mutation prevalence of BRAF V600E in reported ameloblastoma cases and to determine its association with patient demographic and clinicopathological features. Following the PRISMA guidelines, a comprehensive article search was conducted through four databases (Scopus, Google Scholar, PubMed, and Web of Science). Seventeen articles between 2014 and 2022 met the inclusion criteria with 833 ameloblastoma cases. For each included study, the significance of BRAF V600E on the outcome parameters was determined using odd ratios and 95% confidence intervals. Meta-analysis prevalence of BRAF V600E in ameloblastoma was 70.49%, and a significant meta-analysis association was reported for those younger than 54 years old and in the mandible. On the contrary, other factors, such as sex, histological variants, and recurrence, were insignificant. As a result of the significant outcome of BRAF V600E mutation in ameloblastoma pathogenesis, targeted therapy formulation can be developed with this handful of evidence.
Keywords: ameloblastoma, odontogenic tumour, proto-oncogene proteins B-Raf, BRAF V600E, clinicopathological features
1. Introduction
Ameloblastoma is a benign, slow-growing epithelial odontogenic tumour. It is the second most common, constituting about 10% of all jaw neoplasms, and the annual pooled incidence rate of ameloblastoma was 0.92 cases per million [1,2,3]. Ameloblastoma affects both the maxilla and mandible. Due to the slow-growing nature of the tumour, it is usually neglected unknowingly at the early stage [1,4]. At a later stage, patients present with significant swelling and other accompanying signs and symptoms such as facial asymmetry, dental malocclusion, pain, and paraesthesia. In exceptional cases, it metastasises despite having a benign histologic appearance [4,5]. The mainstay treatment for ameloblastoma relies on surgical treatment; nonetheless, conservative treatments such as enucleation or curettage risk potential recurrence, whereas extensive surgical resection for massive ameloblastoma results in high morbidity and postoperative deformity [1,4].
For the past decade, the pathogenesis underlying ameloblastoma has unfolded. Ameloblastoma can be defined by uncontrolled cell proliferation, primarily driven by the mitogen-activated protein kinase (MAPK) signalling pathway, one of the main molecular pathways [5,6,7]. In this pathway, the most significant molecular event is the mutated BRAF gene, resulting in the substitution of amino acid valine (V) by glutamic acid (E) at position 600 (mutated BRAF V600E). Mutated BRAF V600E in this MAPK pathway enables the cells to proliferate excessively, leading to neoplasm formation [8]. It has been shown that this mutated BRAF V600E is commonly found in ameloblastoma of the mandible [9,10,11]. In contrast, for ameloblastoma developed in the maxilla, mutations of the protein Smoothened (SMO) of the Hedgehog pathway, a non-MAPK pathway, is involved [5].
Given the evolving molecular discoveries in ameloblastoma, this work presented a systematic review and meta-analysis to pool the mutation prevalence of BRAF V600E and to seek any association between BRAF V600E mutation and demographic profiles (age and sex) as well as clinicopathological features (site, histological variants, and recurrence) in ameloblastoma.
2. Materials and Methods
2.1. Research Questions
In this study, the following research questions were formulated: (1) What is the role of BRAF V600E mutation in ameloblastoma regarding its pooled prevalence, and (2) how does BRAF V600E mutation in ameloblastoma associate with sociodemographic profiles and clinicopathological features?
2.2. Protocol and Eligibility Criteria
The report presentation followed preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines from the screening protocol to the final analysis [12]. The protocol has been registered in the PROSPERO database (CRD42022328296).
The inclusion criteria for the studies to be considered in this systematic review and meta-analysis were as follows: (1) studies related to BRAF V600E mutation in ameloblastoma; (2) the studies with adequate clinical information on at least three of the following features: age, sex, site, histological variants, and recurrence; and (3) English-language articles.
The exclusion criteria were as follows: (1) the studies reported as review papers, books, practice guidelines, letters, editorials, commentaries, case reports, and pilot studies; (2) articles on metastatic ameloblastoma and ameloblastic carcinomas; and (3) case studies with less than 10 patients.
2.3. Information Sources and Search Criteria
The search was conducted in Google Scholar, PubMed, Web of Science, and Scopus databases. The main fraction keywords according to the PICO tool of the article [13] were selected as follows: ameloblastoma (Population), BRAF (Indicator), clinicopathological features (Comparison), and recurrence (Outcome). The search strategy involved combinations of keyword concepts by medical subject heading (MeSH) terminology. The article search was done by 30 April 2022 using the keywords of ‘Ameloblastoma’, ‘B-Raf protein’, ‘Proto-Oncogene Protein B-Raf’, ‘BRAF’, and ‘BRAF V600E’.
2.4. Study Selection
Preliminarily, the selected articles were screened for validity and relevance with the inclusion and exclusion criteria regarding the information, selection bias, and quality of data analysis. Then, we proceeded with the article’s title and abstract reading to verify the content. Next, the screening process was done by reading the full-text articles to finalize which articles were eligible based on the study’s aims. The articles that did not fulfil the criteria and were out of scope were removed for each step. The final included articles have proceeded with the risk of bias assessment and quantitative analysis.
2.5. Data Collection Process and Data Items
Data were extracted by two authors independently (M.N.M.@Y. and N.R.A.R), and the third author, E.S.C., participated if any discrepancy was raised for analysis starting from the initial screening till the assessment of the bias. Relevant information was listed in a table as follows: authors and year of publication, demographic aspects of sex and age, number of cases, percentage of BRAF V600E positive mutation, tumour location, histological variants, and number of recurrences. Eligible, open-access or restricted-access articles were retrieved by Universiti Sains Malaysia (USM) library support. Research papers from those sources were uploaded into Mendeley reference manager software, and duplicate articles were removed.
2.6. Assessment of Risk of Bias in Individual Studies
The risks of bias in the selected studies were assessed using the Agency for Healthcare Research and Quality (AHRQ) modified scale for observational studies [14,15]. This scale assessment tool consists of nine main evaluation components with sub-elements. The evaluation was assessed for each study to obtain the overall score, which is a score for each component, with ‘adequate’ (A) when the criteria were fulfilled, ‘inadequate’ (I) when the criteria were not fulfilled ‘not reported’ (N) when the study failed to provide the required information, and ‘no information’ (-) when the criteria do not apply to the study design [15].
2.7. Statistical Methods
The mutation pooled prevalence of BRAF V600E among ameloblastoma patients was analysed using Stata software (version 17, College Station, TX, USA) with a 95% confidence interval (CI) [16]. Cochrane Q of heterogeneity test is significant when the I-squared (I2) statistic value is more than 50% with the p-value less than 0.05. The value of meta-analysis was used depending on the fixed effect models (FEM) if the heterogeneity was not significant, and random or the quality effect models (QEM) were used if heterogeneity was significant [17].
A funnel plot of included studies was extracted from Stata software (version 17) and used to evaluate the risk of publication bias. Plotted graph with symmetrical distribution of inverted funnel shape and without outliers indicates a low risk of bias.
A meta-analysis of associations between BRAF V600E mutation in ameloblastoma and clinicopathological features were analysed using Review Manager software (RevMan version 5.4, London, UK). Heterogeneity test data from RevMan version 5.4 was evaluated depending on the chi-square (χ2) test and determined by the I2 statistic and statistical significance with a p-value of less than 0.05. Each association study was presented in forest plots to see the outcomes of individual studies’ effects and to conclude with overall pooled studies.
Age was divided into three age groups (young, adult, and older). First, age groups were determined by calculating the area under the curve of normal distribution using the IBM SPSS version 24. Then, the area under the curve was divided into four quarters (Q1, Q2, Q3, and Q4). The cut-off point for the young age group was Q1 and below, the older age group was Q4 and above, and the adult age group was a combination of Q2 and Q3.
3. Results
3.1. Search Sequence and Quality Assessment of Selected Publications
In total, 782 abstracts and titles were obtained through electronic database searches, and 175 articles were excluded due to duplicate articles (Figure 1). Then, the remaining 697 articles proceeded with screening by reading the titles and abstracts. Subsequently, 521 articles were excluded as they did not fulfil the criteria. Lastly, the relevance of 86 full-text articles was screened in detail. A total of 69 articles were excluded in the final step as did not meet the study aims, and reasons for excluded full-text articles were listed (Table 1). The remaining 17 studies were all evaluated for risk of bias according to AHRQ (Table 2). Elements which did not apply to the study design (-) were excluded from the domain summary. Only the minimum 50% of the elements in each domain were accounted for as an A score. Six studies were evaluated with an A score for nine domains [6,9,18,19,20,21], six studies with an A score for eight out of nine domains [22,23,24,25,26,27], three studies with an A score for seven out of nine domains [5,11,28], and two studies with an A score for six out of nine domains [29,30].
Figure 1.
PRISMA flow diagram of study selection and screening [12].
Table 1.
Full-text articles excluded with reason.
| Articles Excluded | Reason for Exclusion | No. of Articles |
|---|---|---|
| Abe et al., 2018 [31] | Letter/correspondence/commentary/ response/communication |
13 |
| Abe et al., 2018 [32] | ||
| Brunnet et al., 2015 [33] | ||
| De Sousa et al., 2016 [34] | ||
| Coura et al., 2021 [35] | ||
| Faden and Algazi, 2017 [36] | ||
| Gomes et al., 2014 [37] | ||
| Kaye et al., 2015 [38] | ||
| Kaye et al., 2017 [39] | ||
| Magliocca et al., 2016 [40] | ||
| Mota santana et al., 2020 [41] | ||
| Saffari et al., 2019 [42] | ||
| Waqa et al., 2020 [43] | ||
| Effiom et al., 2018 [2] | Review paper | 17 |
| McClary et al., 2016 [4] | ||
| Brown and Betz, 2015 [7] | ||
| Diniz et al., 2017 [44] | ||
| Daws et al., 2021 [45] | ||
| do Canto et al., 2016 [46] | ||
| Fuchigami et al., 2021 [47] | ||
| Heikinheimo et al., 2015 [48] | ||
| Jhamb and Kramer, 2014 [49] | ||
| Khalele and Al-Shiaty, 2016 [50] | ||
| Kreppel and Zöller, 2018 [51] | ||
| Marín et al., 2021 [52] | ||
| Martins-de-Barros et al., 2022 [53] | ||
| Ngan et al., 2022 [54] | ||
| Ritterhouse and Barletta, 2015 [55] | ||
| Shi et al., 2021 [56] | ||
| You et al., 2019 [57] | ||
| Abramson et al., 2022 [58] | Case report | 10 |
| Bernaola-Paredes et al., 2021 [59] | ||
| Broudic-Guibert et al., 2019 [60] | ||
| Brunet et al., 2019 [61] | ||
| Fernandes et al., 2018 [62] | ||
| Hirschhorn et al., 2021 [63] | ||
| Roque and Yazmin, 2017 [64] | ||
| Rotellini et al., 2016 [65] | ||
| Suzuki et al., 2020 [66] | ||
| Tan et al., 2016 [67] | ||
| Bartels et al., 2018 [68] | Number of cases less than 10 | 9 |
| Diniz et al., 2017 [69] | ||
| Kennedy et al., 2016 [70] | ||
| Kondo et al., 2020 [71] | ||
| Pereira et al., 2016 [72] | ||
| Sant’Ana et al., 2021 [73] | ||
| Shi et al., 2021 [74] | ||
| Shimura et al., 2020 [75] | ||
| You et al., 2019 [76] | ||
| Bologna-Molina et al., 2019 [77] | Unrelated to BRAF studies, failure to provide clinical information or inability to present data clearly |
17 |
| Bonacina et al., 2022 [78] | ||
| Coura et al., 2020 [79] | ||
| Duarte-Andrade et al., 2019 [80] | ||
| Fujii et al., 2022 [81] | ||
| Guan et al., 2019 [82] | ||
| Kokubun et al., 2022 [83] | ||
| Lapthanasupkul et al., 2021 [84] | ||
| Oh et al., 2021 [85] | ||
| Oh et al., 2022 [86] | ||
| Owosho et al., 2021 [87] | ||
| Peralta et al., 2019 [88] | ||
| Salama et al., 2020 [89] | ||
| Sharp et al., 2019 [90] | ||
| Soltani et al., 2018 [91] | ||
| Tseng et al., 2022 [92] | ||
| Zhang et al., 2020 [93] | ||
| Diniz et al., 2017 [94] | Samples were taken from the same Hospital as Diniz et al., 2015 [29] |
1 |
Table 2.
Risk of bias among studies analysed.
| Domain | Elements | Sweeney 2014 [5] | Brown 2014 [6] | Kurppa 2014 [9] | Derakhshan 2020 [11] | Fregnani 2016 [18] | Gültekin 2018 [19] | do Canto 2019 [20] | Heikinheimo 2019 [21] | Oh 2019 [22] | Kelppe 2019 [23] | Seki-Soda 2020 [24] | da Silva Marcelino 2021 [25] | Kunmongkolwut 2022 [26] | Santana 2021 [27] | Shirsat 2018 [28] | Diniz 2015 [29] | Yukimori 2017 [30] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study question | Clearly focused and appropriate question | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| Study population | Description of study population | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| Sample size justification | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |
| Comparability of subjects | Specific inclusion/exclusion criteria | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | N | A |
| Criteria applied equally to all groups | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | N | A | |
| Comparability of groups at baseline with regard to disease status and prognostic factors | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Study groups comparable to non-participants with regard to confounding factors | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Use of concurrent controls | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Comparability of follow-up among groups | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Exposure or intervention | Clear definition of exposure | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| Measurement method standard, valid, and reliable | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |
| Exposure measured equally in all study groups | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |
| Outcome measurement | Primary/secondary outcomes clearly defined | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A |
| Outcomes assessed blind to exposure or intervention status | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Method of outcome assessment standard, valid, and reliable | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |
| Length of follow-up adequate for question | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Statistical analysis | Statistical tests appropriate | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | N | N |
| Multiple comparisons taken into consideration | N | A | A | N | A | A | A | A | A | A | A | A | A | A | N | A | A | |
| Modelling and multivariate techniques appropriate | N | A | N | N | A | N | A | A | N | N | N | A | A | N | N | N | N | |
| Power calculation provided | N | A | A | N | A | A | I | A | A | A | A | A | A | A | N | N | N | |
| Assessment of confounding variables | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Dose–response assessment, if appropriate | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Results | Measure of effect for outcomes and appropriate measure of precision | A | A | A | I | A | A | A | A | A | A | A | A | A | A | A | I | A |
| Adequacy of follow-up for each study group | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | _ | |
| Discussion | Conclusions supported by results with biases and limitations taken into consideration | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | N |
| Funding or sponsorship | Type and sources of support for study | N | A | A | A | A | A | A | A | I | N | N | N | I | A | I | A | N |
Abbreviations: A, adequate; I, inadequate; N, not reported; -, not applicable to the study design.
3.2. Study Characteristics
The final 17 studies were included for qualitative and quantitative analysis with a total of 833 patients. The research by Sweeney et al. in 2014 was the earliest study, and the latest publication was in 2022, by Kunmongkolwut and colleagues [26]. The most extensive study was by da Silva Marcelino et al. [25] and included 128 patients; the lowest number of samples was by Yukimori et al. [30], with 14 patients. A summary of the 17 selected studies and association variables with BRAF V600E mutation in ameloblastoma are summarised in Table 3.
Table 3.
The summary of 17 included ameloblastoma studies with BRAF V600E, demographic and clinicopathological features profile.
| Author/ Year |
Country | No. of Cases (n) | BRAF+, Detection Method (n) | No. of BRAF+, n (%) a |
Demographic (BRAF+/Total Case) (n) |
Clinicopathological Features (BRAF+/Total Case) (n) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| PCR | IHC | Sex | Age Group b | Location | Histological Variant | Recurrence | ||||
| Brown et al., 2014 [6] | USA | 83 | Pos = 30 Neg = 19 NA = 34 |
Pos = 53 Neg = 30 |
53(63.9) | M = 31/47 F = 22/36 |
Young = 18/21 Adult = 29/39 Older = 6/25 |
Mn = 50/67 Mx = 3/16 |
CA = 45/71 UA = 5/5 DA = 3/6 PA = 0/1 |
Yes = 7/14 No = 46/69 |
| da Silva Marcelino et al., 2021 [25] | Brazil | 128 | NA | Pos = 104 Neg = 24 |
104(81.2) | M = 60/71 F = 44/57 |
NA | Mn = 104/128 | CA = 89/110 UA = 15/18 |
Yes = 10/11 No = 94/117 |
| Derakhshan et al., 2020 [11] | Iran | 50 | Pos = 46 Neg = 4 |
Pos = 39 Neg = 11 |
46(92.0) | M = 27/29 F = 19/21 |
Young = 6/6 Adult = 32/35 Older = 8/9 |
Mn = 40/44 Mx = 5/5 Both = 1/1 |
NA | Yes = 12/13 No = 34/37 |
| Diniz et al., 2015 [29] | Brazil | 17 | Pos = 14 Neg = 3 |
NA | 14(82.4) | M = 7/9 F = 7/8 |
Young = 4/6 Adult = 7/8 Older = 1/1 NA = 2 |
Mn = 11/13 Mx = ¾ |
CA = 7/9 UA = 5/6 DA = 2/2 |
NA |
| do Canto et al., 2016 [20] | Brazil | 84 | NA | Pos = 66 Neg = 18 |
66(78.6) | M = 35/44 F = 31/40 |
NA | Mn = 66/84 | CA = 58/73 UA = 8/11 |
Yes = 4/7 No = 62/77 |
| Fregnani et al., 2017 [18] | Brazil | 73 | NA | Pos = 34 Neg = 39 |
34(46.6) | M = 17/35 F = 17/38 |
NA | Mn = 32/63 Mx = 2/10 |
NA | Yes = 13/15 No = 21/58 |
| Gültekin et al., 2018 [19] | Germany | 62 | Pos = 34 Neg = 28 |
NA | 34(54.8) | M = 21/42 F = 13/20 |
NA | Mn = 33/46 Mx = 1/16 |
CA = 21/45 UA = 8/11 PA = 5/6 |
Yes = 6/12 No = 14/18 NA = 32 |
| Heikinheimo et al., 2019 [21] | Finland | 49 | Pos = 42 Neg = 7 NA = 5 |
Pos = 39 Neg = 11 NA = 4 |
42(85.7) | M = 18/22 F = 24/27 |
Young = 17/21 Adult = 15/16 Older = 10/12 |
Mn = 41/45 Mx = 1/4 |
CA = 12/15 UA = 30/34 |
Yes = 15/18 No = 27/31 |
| Kelppe et al., 2019 [23] | Finland | 36 | NA | Pos = 26 Neg = 10 |
26(72.2) | M = 12/20 F = 14/16 |
NA | Mn = 26/29 Mx = 0/7 |
CA = 18/27 UA = 7/7 PA = 1/2 |
Yes = 9/14 No = 17/22 |
| Kunmongkol-wut et al., 2022 [26] | Thai | 74 | NA | Pos = 50 Neg = 24 |
50(67.6) | M = 30/40 F = 20/34 |
NA | Mn = 45/67 Mx = 5/7 |
NA | Yes = 9/16 No = 22/29 NA = 29 |
| Kurppa et al., 2014 [9] | Finland | 24 | Pos = 15 Neg = 9 |
Pos = 11 Neg = 9 NA = 4 |
15(62.5) | M = 7/15 F = 8/9 |
Young = 4/4 Adult = 9/12 Older = 2/8 |
Mn = 15/24 | CA = 15/24 | Yes = 5/7 No = 10/17 |
| Oh et al., 2019 [22] | Korea | 30 | Pos = 27 Neg = 3 |
Pos = 17 Neg = 10 NA = 3 |
27(90.0) | M = 18/19 F = 9/11 |
Young = 4/5 Adult = 17/19 Older = 6/6 |
Mn = 25/28 Mx = 2/2 |
CA = 25/27 UA = 2/3 |
Yes = 12/15 No = 15/15 |
| Santana et al., 2021 [27] | Brazil | 30 | NA | Pos = 20 Neg = 10 |
20(66.7) | M = 13/16 F = 7/14 |
Young = 6/10 Adult = 10/15 Older = 4/5 |
Mn = 17/26 Mx = 3/4 |
CA = 17/27 UA = 3/3 |
Yes = 7/10 No = 13/20 |
| Seki-Soda et al., 2020 [24] | Japan | 21 | Pos = 16 Neg = 5 |
Pos = 20 Neg = 1 |
16(76.2) | M = 12/15 F = 4/6 |
NA | Mn = 16/21 | CA = 14/17 UA = 2/4 |
Yes = 0/2 No = 16/19 |
| Shirsat et al., 2018 [28] | India | 30 | NA | Pos = 10 Neg = 20 |
10(33.3) | M = 6/18 F = 4/12 |
Young = 5/12 Adult = 4/12 Older = 1/6 |
Mn = 10/30 | NA | Yes = 4/6 No = 6/24 |
| Sweeney et al., 2014 [5] | USA | 28 | Pos = 12 Neg = 16 |
NA | 12(42.9) | M = 4/13 F = 0/5 NA = 8/10 |
Young = 0/0 Adult = 1/4 Older = 3/14 NA = 8 |
Mn = 9/14 Mx = 0/11 Other = 3/3 |
NA | Yes = 7/15 No = 3/9 NA = 4 |
| Yukimori et al., 2017 [30] | Japan | 14 | Pos = 12 Neg = 2 |
Pos = 12 Neg = 2 |
12(85.7) | M = 10/11 F = 2/3 |
Young = 2/2 Adult = 4/4 Older = 6/8 |
Mn = 7/8 Mx = 5/6 |
CA = 10/11 PA = 2/3 |
NA |
Abbreviations: a, total no. of BRAF mutations used for prevalence analysis; b, age group, young (≤24), adult (24 < x < 54), older (≥54); NA, data not available/unknown status; Pos, positive; Neg, negative; M, male; F, female; Mn, mandibular; Mx, maxilla; CA, conventional ameloblastoma; UA, unicystic ameloblastoma; DA, desmoplastic ameloblastoma; PA, peripheral ameloblastoma.
3.3. Quantitative Synthesis
3.3.1. Prevalence of BRAF V600E Mutation
The total number of BRAF mutations for further analysis includes the number of positive mutations detected by polymerase chain reaction (PCR) [5,9,11,19,21,22,24,29,30], and data from immunohistochemistry (IHC) results whereby IHC was the sole method available in seven individual studies [18,20,23,25,26,27,28] (Table 3). For Brown et al.’s [6] study, IHC data were used because not all cases had PCR data; moreover, for cases with both IHC and PCR data, 100% concordance was recorded for both methods in this individual study.
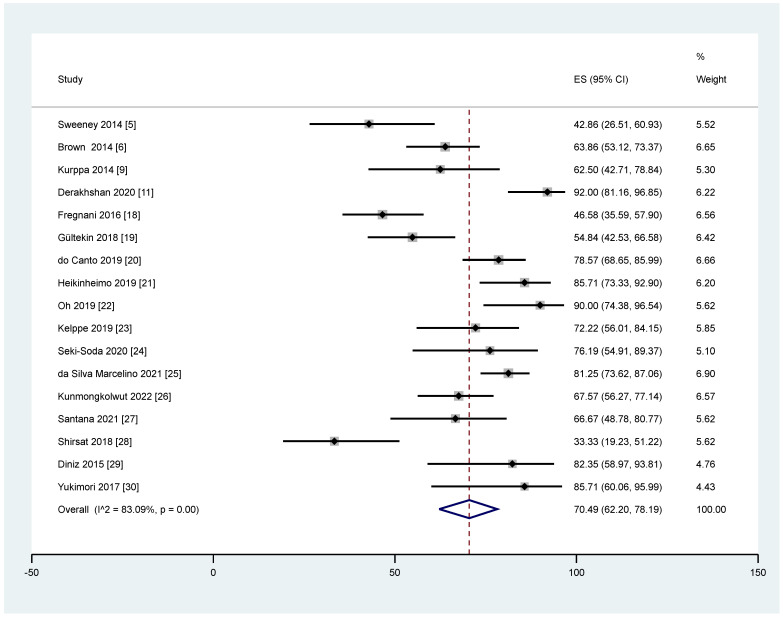
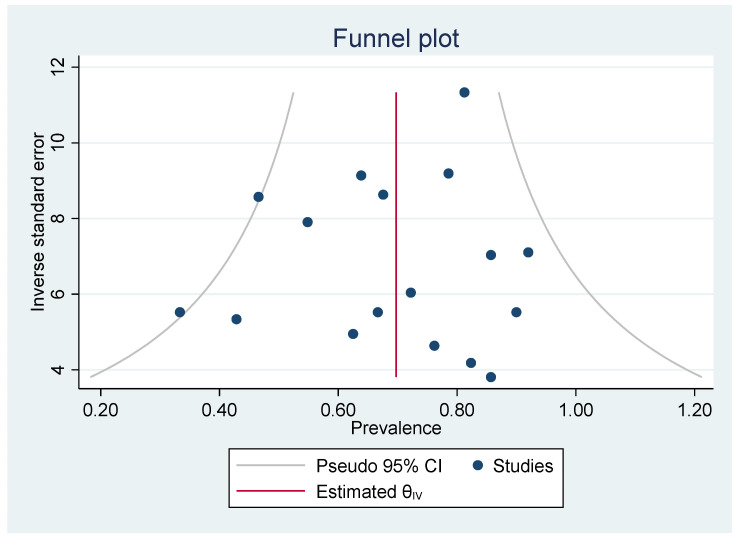
Heterogeneity was significant for the pooled prevalence of BRAF mutation among ameloblastoma, which had a p < 0.05 in Cochrane Q statistics, and I2 statistics values of 83.09%. From 17 studies that reported total BRAF mutation cases, the overall pooled prevalence among ameloblastoma based on QEM was 70.49% (95% CI = 62.20–78.19%; p < 0.05) (Figure 2). Publication bias of this pooled prevalence was also evaluated using a funnel plot, showing a symmetrical plot, indicating a low potential risk of publication bias (Figure 3).
Figure 2.
The pooled prevalence of BRAF V600E mutation in ameloblastoma cases.
Figure 3.
Funnel plot for publication bias evaluation.
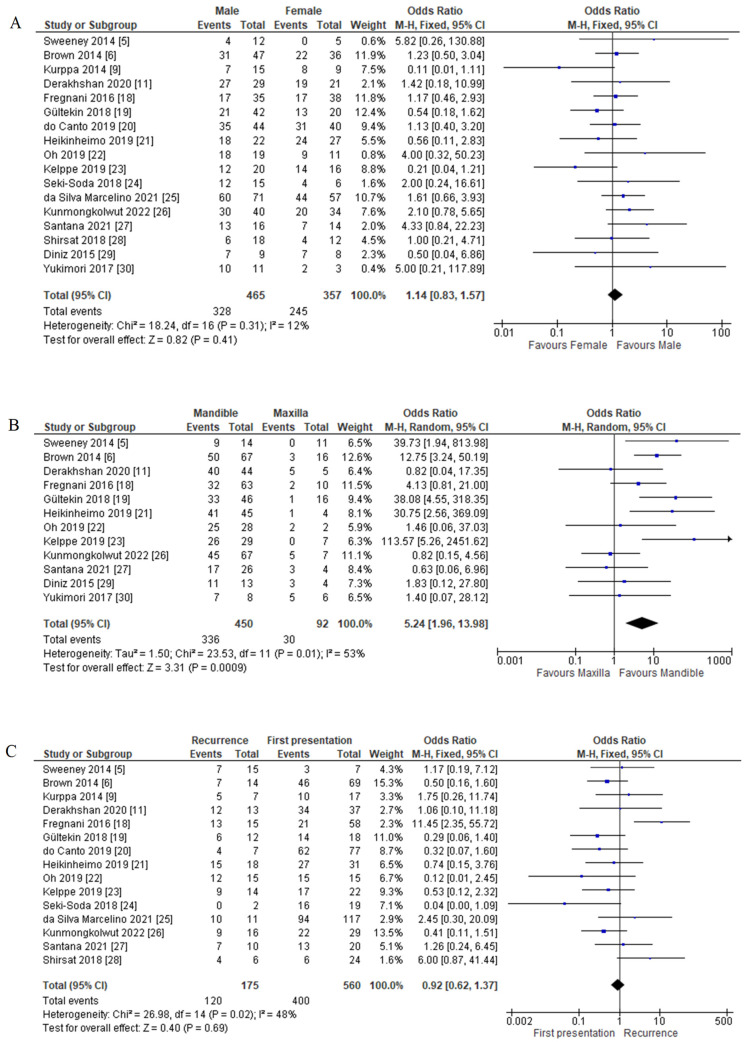
3.3.2. BRAF V600E Mutation and Demographic Profiles
Age with BRAF V600E Mutation
Based on available data, a sensitivity study for the association between age and BRAF V600E mutation was conducted for 10 out of 17 studies. First, histogram and normal age distribution for total cases of ameloblastoma were plotted. From the quartile analysis, the age was then grouped into three: young (less and equal to 24 years old), adult (more than 24 years old and less than 54 years old), and older (more and equal to 54 years old) (Supplementary Figure S1). Finally, the intergroup comparison was made of young versus adult, young versus older, and adult versus older.
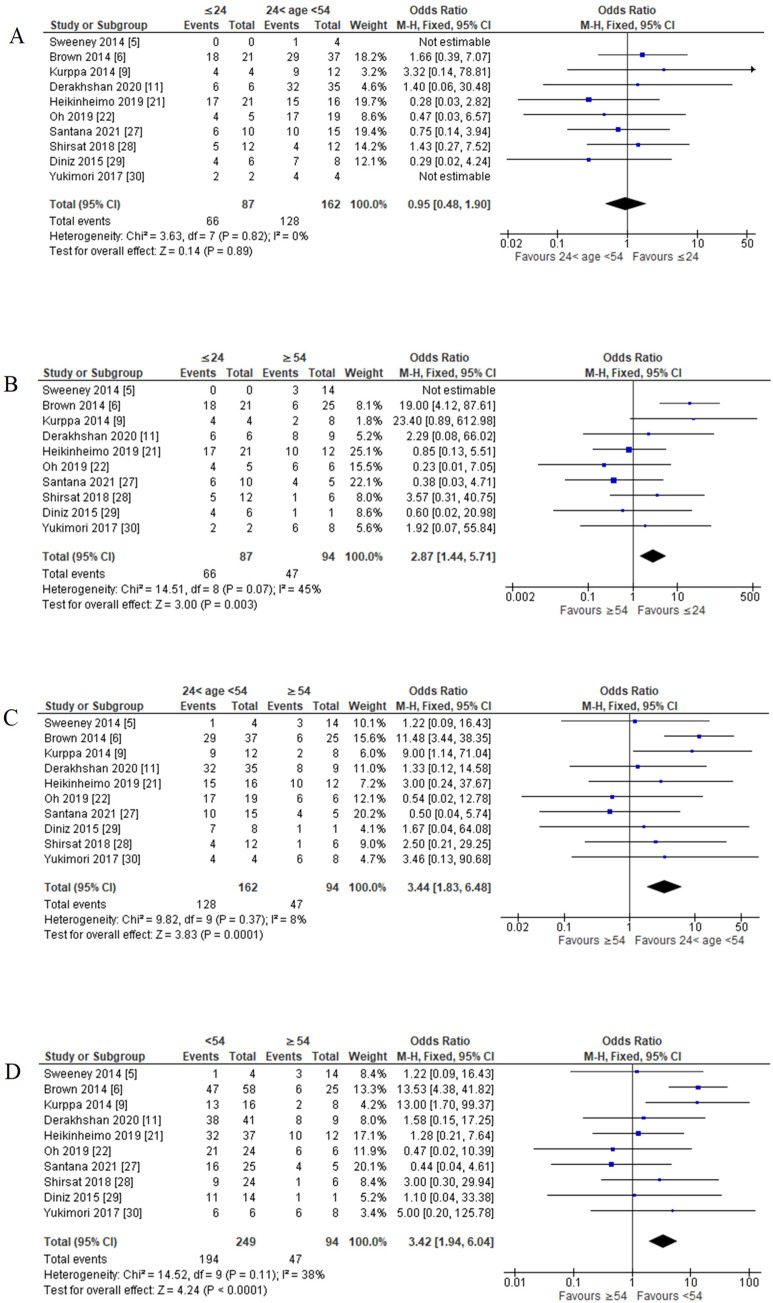
For young versus adult comparison, young patients recorded 75.86% (66 out of 87 cases had a mutation of BRAF V600E), and adult patients recorded 79.01% (128 out of 162 cases had a mutation of BRAF V600E). FEM was used as there was no significant amount of heterogeneity (p = 0.82; I2 = 0%). The pooled analysis showed no significant association of BRAF V600E mutation in the young age group compared to the adult age group (OR = 0.95; 95% CI = 0.48–1.90; p = 0.89) (Figure 4A).
Figure 4.
Forest plot of age groups with BRAF V600E mutation in ameloblastoma cases. (A) Young age group versus adult age group; (B) young age group versus older age group; (C) adult age group versus older age group; (D) young and adult age group versus older age group.
For the young versus older comparison, young patients recorded 75.86%, and older patients recorded 50.00% (47 out of 94 cases had a mutation of BRAF V600E). FEM was used as there was no significant amount of heterogeneity (p = 0.07; I2 = 45%). The pooled analysis showed a significant association of BRAF V600E mutation in the young age group compared to the older age group (OR = 2.87; 95% CI = 1.44–5.71; p = 0.003) (Figure 4B).
For adult versus older comparison, 79.01% of adult patients recorded a mutation of BRAF V600E, and 50.00% of older patients. FEM was used as there was no significant heterogeneity (p = 0.37; I2 = 8%). The pooled analysis showed a significant association of BRAF V600E mutation in the adult age group compared with the older age group (OR = 3.44; 95% CI = 1.83–6.48; p = 0.0001) (Figure 4C).
There was no significant association between BRAF V600E mutation for the young and adult groups. However, it was significant in the young age group compared with the older age group and the adult age group compared with the older age group. Therefore, BRAF V600E mutation was significantly associated with age less than 54 years old among ameloblastoma patients, as shown in the pooled analysis (OR = 3.42; 95% CI = 1.94–6.04; p < 0.0001) based on FEM, as the heterogeneity was not significant (p = 0.11; I2 = 38%). (Figure 4D).
Sex with BRAF V600E Mutation
The association analysis between BRAF V600E mutation and sex proceeded with 17 studies of 590 patients. Among 465 male patients, 70.54% of patients were BRAF positive. Females showed a slightly higher percentage than males (68.63%), consisting of 245 out of 357 female patients. FEM data used as the heterogeneity test was not significant (p = 0.31; I2 = 12%). There was no association between BRAF V600E mutation and sex, as the statistical analysis was not significant (OR = 1.14; 95% CI = 0.83–1.57: p = 0.41) (Figure 5A).
Figure 5.
Forest plot of clinicopathological features association with BRAF V600E mutation in ameloblastoma cases. (A) Sex with BRAF V600E mutation; (B) tumour location with BRAF V600E mutation; (C) recurrence with BRAF V600E mutation.
3.3.3. BRAF V600E Mutation and Clinicopathological Features Association
Tumour Location with BRAF V600E Mutation
A sensitivity study was conducted for an association between tumour location (mandible versus maxilla) and BRAF V600E mutation for 12 out of 17 studies based on available data. 74.67% of the mandible (336 out of 450 patients) and 30.00% of the maxilla (30 out of 100 patients) had BRAF V600E mutation. QEM data used as the heterogeneity test was significant (p = 0.01; I2 = 53%). There was an association between the mandible and BRAF V600E mutation, as the statistical analysis proved significant (OR = 5.24; 95% CI = 1.96–13.98; p = 0.0009) (Figure 5B).
Recurrence with BRAF V600E Mutation
Based on available data, a sensitivity study for the association between recurrence and BRAF V600E mutation was conducted for 15 out of 17 studies. Of 175 recurrence cases, 120 (68.57%) were BRAF V600E positive. First presentation or primary cases reported mutations in 400 out of 560 patients (71.43%). FEM data used as the heterogeneity test was not significant (p = 0.02; I2 = 48%). There was no association between recurrence and BRAF V600E mutation (OR = 0.92; 95% CI = 0.62–1.1.37; p = 0.69) (Figure 5C).
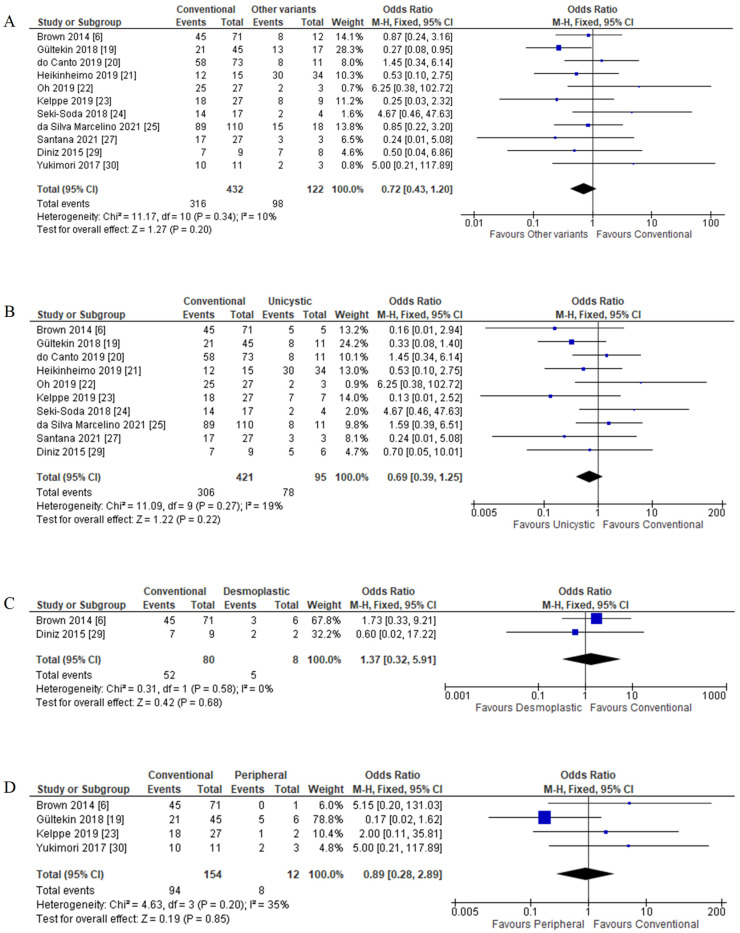
Histological Variants with BRAF V600E Mutation
A sensitivity study for the association between histological variants and BRAF V600E mutation was conducted for 11 of 17 studies based on available data. Out of 432 conventional ameloblastoma cases, 316 (73.15%) had BRAF mutation, while other variants (including unicystic, desmoplastic, and peripheral) reported that 80.33% (98 out of 122 patients) also had the mutation. FEM was used as there was no significant amount of heterogeneity (p = 0.30; I2 = 10%). There was no significant association between histological variants and BRAF V600E mutation (OR = 0.72; 95% CI = 0.43–1.20; p = 0.20) (Figure 6A).
Figure 6.
Forest plot of histological variants association with BRAF V600E mutation in ameloblastoma cases. (A) Conventional versus other variants; (B) conventional versus unicystic; (C) conventional versus desmoplastic; (D) conventional versus peripheral.
Other histological variants were also reported with the same range of BRAF V600E mutation with conventional ameloblastoma: 82.11% mutation in unicystic, 62.50% in desmoplastic, and 66.67% mutation in peripheral ameloblastoma. Further subgroup analysis of histological variants of conventional with unicystic, desmoplastic, and peripheral ameloblastoma also showed no significant association with BRAF V600E mutation (Figure 6B–D).
4. Discussion
Ameloblastoma is an aggressive benign odontogenic tumour of the jaws. Brown et al. [6] have found the mutation in the MAPK pathways, remarkably high in the BRAF gene. More researchers then began to explore and refine this discovery of oncogenic mutation, correlating it to the clinical implication for the improvement in disease control and treatments [29]. This systematic review and meta-analysis were conducted to identify the pooled prevalence of BRAF mutation and associate it with the sociodemographic and clinicopathological features.
This study reported a 70.49 % mutation prevalence with the BRAF V600E gene among ameloblastoma patients. Among individual studies, the lowest prevalence of BRAF V600E mutation was reported by Shirsat et al. [28] (33.33%), and the highest prevalence (92.00%) was reported by Derakhshan et al. [11]. This meta-analysis outcome of high prevalence has shown that BRAF V600E mutation has played a role in molecular pathogenesis in ameloblastoma incidence. Even though the status and exact mechanisms are still unclear, a high prevalence status may indicate a new treatment area exploration concerning this specific mutation. Previously, the BRAF V600E mutation focused on malignant tumours, where the prevalence was approximately 74.6% for papillary thyroid carcinoma [95], 7.4% for colorectal cancer [96], and 60% for melanomas [97]. According to previous studies, several gene mutations have been identified in the background of BRAF V600E positive mutation in ameloblastoma, including somatic mutation in cyclin-dependent kinase inhibitor 2A (CDKN2A), catenin beta 1 (CTNNB1), fibroblast growth factor receptors (FGFR), Kirsten rat sarcoma virus (KRAS), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and phosphatase and tensin homolog (PTEN) [19,98].
There was also a case report which was ameloblastoma initially but later presented with ameloblastic carcinoma; positive BRAF V600E mutation was subsequently detected [99]. This malignant counterpart also expresses BRAF V600E mutation (about 25 to 33%) [100,101]. Hence, there is a possibility that BRAF V600E mutation may play a role in the malignant transformation of ameloblastoma, as this rare odontogenic malignancy has close features that combine the histologic features of ameloblastoma with cytologic atypia [100,101,102].
Having a mutation of BRAF V600E will alter the MAPK pathway, and Brown et al. [7] suggested this alteration may be crucial in the early stage of ameloblastoma pathogenesis. Other than the BRAF gene in the MAPK pathway, FGFR2 and RAS mutations were also part of the pathogenesis of most ameloblastoma cases [6]. On the other hand, the Hedgehog pathway has also been reported as the secondary mutation, specifically the somatic mutation of Smoothened (SMO) gene [5,7].
The BRAF V600E mutation remains the most critical molecular marker studied in ameloblastoma pathogenesis [9,33,37,56]. Hence, a valid method for detecting this mutation is crucial in accurately evaluating mutation occurrence, either by IHC or molecular assay, usually by PCR. PCR is the gold standard for detecting any gene mutation. However, due to its costliness and the limited number of facilities that offers this technique, BRAF V600E mutation was rarely assessed. Later, establishing a mouse monoclonal antibody (VE-1) allowed for the analysis of IHC for the BRAF V600E mutation in numerous tumours [103,104]. Therefore, IHC is a preferred technique as it is easy, reliable, affordable, and widely used as a standard diagnostic histopathological procedure [55,72].
These two methods reported some discrepancies, mainly false-positive and false-negative results. For example, colorectal carcinomas [105] and melanomas [9,106] had false-positive results from the IHC assessment but were negative in the molecular assay. Even in this meta-analysis, some studies reported a few differences in the BRAF V600E mutation detection by IHC compared with PCR [9,11,21]. In those mentioned studies, when there were false-positive or false-negative cases, further molecular assays such as DNA sequencing were performed to confirm the mutation status [9,11,21].
IHC false positives are attributed to sampling contamination with other tissues with positive immunoreactivity [104,107]. In addition, IHC false negatives are possible due to decalcified old histological samples. Therefore, it is suggested that tissues be preserved and processed thoroughly within two hours following collection [103]. Other likely explanations of IHC false negatives include loss of expression of the altered antigen, such as in necrotic tumour regions, and additional mutations that prevent the mutated messenger RNA from being translocated into a functional protein [104,108].
Although they possess some pitfalls, several studies have proven a strong concordance between IHC and molecular assays [103,109,110]. Thus, IHC staining for BRAF V600E detection is a reliable technique compared to PCR for identifying the BRAF V600E mutation in various tumours, and it has the benefit of substantial cost and labour savings. In addition, the identification of BRAF V600E mutation using the IHC method should be interpreted carefully by expert pathologists [72].
This meta-analysis reported that the percentage of BRAF V600E mutation is higher in patients less than 54 years old—the age group for the young and adults. However, most individual studies reported no association between mutation status and age [9,29,91]. Thus, an objective comparison was made in this study by plotting a normal distribution graph of total ameloblastoma cases in all eligible studies. As a result, the meta-analysis revealed a significant association between BRAF V600E mutation and young and adult age groups among ameloblastoma patients.
The association between age and disease risk has always been one of the primary criteria for epidemiology [111,112]. Our findings showed that the younger generation (less than 54 years old) was associated with BRAF V600E mutation in ameloblastoma patients. Based on this result, we suggest a correlation between odontogenesis and BRAF V600E mutation, which led to the ameloblastoma incidence. Fibroblast growth factor (FGF) is one of the signalling molecules in mammalian tooth development, which initiates signalling through multiple downstream intracellular pathways and later activates Ras signals, including RAF/MEK/ERK [113]. The BRAF V600E mutation essentially activates MEK/ERK signalling, leading to tumour formation [8] and, in this case, ameloblastoma formation. This finding may reflect why the young age group has a significant association, as odontogenesis is more prevalent. In resource-limited settings, BRAF V600E mutation screening should be prioritised for patients below the age of 54, as proven by our findings. Besides, they also have a longer lifespan and thus have a higher risk of recurrence. Testing in advance will offer an option for better treatment with targeted therapies against BRAF V600E mutation that could prevent the tumour from recurring.
Sex can sometimes significantly influence disease formation [114]. Hence, health professionals investigate the influence of sex in many ways in terms of aetiology, diagnosis, progression, prevention, treatment, health outcomes of disease, and exposure to risk [114]. In the current study, the statistical analysis failed to prove association between sex and mutation occurrence. This finding supported previous research showing that mutation of BRAF V600E is not affected by sex in ameloblastoma patients.
The studies of tumour site-specific mutations in ameloblastoma were first reported in 2014 and were proposed as a new paradigm [5,6]. Many studies have reported a higher frequency of association between BRAF V600E mutation and the mandible and showed a statistically significant result compared with the maxilla [5,23,32]. The result is in line with the current report. On the other hand, five studies revealed no significant differences between the mandible or maxilla tumour locations [11,18,22,29,30]. The effects of sample sizes can explain the differences in the results. A meta-analysis helps pool the included studies; hence, a better sample size calculation can be generated and more representative. According to Sweeney et al. [5], the SMO gene mutation substantially affects the maxilla rather than the mandible, which may explain the independent odontogenic pathways in the jaw location [115]. On a broader spectrum, this finding emphasised the understanding of the anatomical specificity in mutation-driven pathogenesis, reflecting the distinctive developmental pathways of the jaws [5]. Significant mutation of BRAF V600E in mandibular ameloblastoma may allow for better risk assessment and the possibility of personalized adjunctive therapy to sustain jaw functionality.
Furthermore, there was no significant difference between the mutation of BRAF V600E and ameloblastoma histological variants via meta-analysis. Only a study by Gültekin et al. [19] showed a significant association with conventional ameloblastoma. It investigates whether BRAF mutation occurs in different variants at a similar proportion [29]. They concluded that all histological variants of ameloblastoma underwent similar molecular alterations for this benign odontogenic neoplasm [29].
Our meta-analysis revealed that the mutation of BRAF V600E was not significantly associated either with the first presentation or the recurrence cases. It was consistent with most studies, except one study had an association of BRAF V600E positivity with the recurrence cases with an odds ratio of 11.45 [18]. On the other hand, BRAF wild type was found to have an earlier recurrence, especially in those treated with surgical enucleation rather than surgical resection [2,116]. Maxilla has a higher recurrence rate, most probably due to the anatomy, causing limited treatment options and difficulty achieving a safe and clear surgical margin [11,116]. However, current studies on the relationship between BRAF V600E and the recurrences are still unclear. Further studies that provide a definite conclusion on this matter can improve the clinical management of ameloblastoma [2,117].
BRAF V600E mutation has a significant association with a broad range of neoplasms [105,106,118,119], and in ameloblastoma specifically [5,6,18,19,21,22]. Furthermore, once the mutation occurs, the complicated MAPK pathway, which involves the activation of downstream RAS, RAF, MEK, and ERK, becomes activated in tumorigenesis [120]. Hence, each MAPK component that undergoes mutation needs to be understood to formulate the best treatment regimen for ameloblastoma patients [5,6,7,37].
Due to its benign and locally aggressive behaviour, current ameloblastoma management is by surgical intervention. However, these standard surgical treatments are either by resection or conservative (curettage or enucleation), with the latter possessing a higher recurrence rate [121]. The meta-analysis result has significantly upheld the correlation of the BRAF gene to ameloblastoma occurrence. Hence, taking advantage of this molecular pathway, a BRAF inhibitor treatment may be used to avoid wide surgical resection or multiple surgical procedures due to recurrence [7].
BRAF inhibitor studies actively explored their effectiveness, where the data collected shows promising results. A study on ameloblastoma cell lines has reported an in vitro sensitivity of BRAF inhibitor (vemurafenib) to hinder V600E mutation [5,6]. A case report of a 29-year-old woman with a recurrence of ameloblastoma with BRAF V600E mutation, who received vemurafenib, was symptomless and had tumour shrinkage after 11 months of therapy [62]. Vemurafenib is a well-known BRAF inhibitor that the FDA has authorised for treating metastatic melanoma with BRAF V600E [122]. Another inhibitor, dabrafenib, is also used to control the MAPK pathway with different neoplasms, including ameloblastoma [123,124]. Faden and Algazi [36], and Tan et al. [67] have reported using dabrafenib in recurrence cases of ameloblastoma, which also showed promising results. This limited clinical data reflected that this inhibitor therapy seems to be an effective treatment modality. However, there were downsides to this therapy, such as the development of resistance and acquiring skin tumours, thus proposing a dual-agent therapy instead of single-agent therapy [56].
The current study profiled the relationship between the mutation of BRAF V600E and the incidence of ameloblastoma. We have found significant findings related to the age groups and tumour location. However, some limitations might influence the interpretation. Firstly, we could not do the meta-analysis on each method (IHC versus molecular assay) in detecting BRAF V600E due to a lack of data from the search strategy criteria. Therefore, we suggest that future research on a meta-analysis related to comparing the validated methods to detect BRAF V600E mutation in ameloblastoma. For example, this has been done in the study of papillary thyroid carcinoma, a type of cancer with significant BRAF V600E mutation [125]. Secondly, the definition of recurrences in the studies was not clear. Most studies reported the cases as primary or recurrence cases without specifying whether it was a true recurrence or a residual tumour. Therefore, despite no significant finding in the current study, the chance of BRAF V600E mutation occurring in recurrence cases versus primary presentation is still debatable. Thus, it is recommended that clinicians and researchers record those details when documenting a case report, which is essential for further analysis and later can be translated into clinical management. Finally, data on treatment options for ameloblastoma has been excluded in this review due to the limited information available in the articles. It may be because BRAF inhibitor is not widely used in treating ameloblastoma. Therefore, further clinical trials are recommended to determine the effectiveness of this drug in ameloblastoma management. Once the data is more widely available, future studies on this aspect should be explored by meta-analysis. This may help improve the clinical outcome of ameloblastoma patients.
5. Conclusions
The present systematic review and meta-analysis show that BRAF V600E mutation has a high pooled prevalence of 70.49% in ameloblastoma. Furthermore, a significant meta-analysis association was reported for those younger than 54 years old, and in the mandible. On the contrary, other factors, such as sex, histological variants, and recurrence, were insignificant among ameloblastoma cases with BRAF V600E mutation. Researchers could utilise these findings to improve the treatment option and find a possible new biomarker for the early detection of ameloblastoma. This evidence-based medicine information is essential in targeted therapy development. However, further well-designed cohort studies are needed to verify the association of BRAF V600E mutation in ameloblastoma before applying new medical interventions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14225593/s1, Supplementary Figure S1: Histogram of age group distribution and quartiles.
Author Contributions
Conceptualization, M.N.M.@.Y., E.S.C. and N.R.A.R.; methodology, M.N.M.@.Y., E.S.C. and N.R.A.R.; software, M.N.M.@.Y. and E.S.C.; validation, N.R.A.R. and E.S.C.; formal analysis, M.N.M.@.Y.; investigation, M.N.M.@.Y., E.S.C. and N.R.A.R.; resources, N.R.A.R.; data curation, M.N.M.@.Y.; writing—original draft preparation, M.N.M.@.Y.; writing—review and editing, M.N.M.@.Y., E.S.C. and N.R.A.R.; visualization, M.N.M.@.Y.; supervision, N.R.A.R. and E.S.C.; project administration, N.R.A.R. and E.S.C.; funding acquisition, N.R.A.R. and E.S.C. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received funding from Universiti Sains Malaysia Short Term Grant, 304/CIPPT/6315238.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Becelli R., Carboni A., Cerulli G., Perugini M., Iannetti G. Mandibular ameloblastoma: Analysis of surgical treatment carried out in 60 patients between 1977 and 1998. J. Craniofac. Surg. 2002;13:395–400. doi: 10.1097/00001665-200205000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Effiom O.A., Ogundana O.M., Akinshipo A.O., Akintoye S.O. Ameloblastoma: Current etiopathological concepts and management. Oral Dis. 2018;24:307–316. doi: 10.1111/odi.12646. [DOI] [PubMed] [Google Scholar]
- 3.Hendra F.N., Van Cann E.M., Helder M.N., Ruslin M., De Visscher J.G., Forouzanfar T., De Vet H.C. Global incidence and profile of ameloblastoma: A systematic review and meta-analysis. Oral Dis. 2020;26:12–21. doi: 10.1111/odi.13031. [DOI] [PubMed] [Google Scholar]
- 4.McClary A.C., West R.B., McClary A.C., Pollack J.R., Fischbein N.J., Holsinger C.F., Sunwoo J., Colevas A.D., Sirjani D. Ameloblastoma: A clinical review and trends in management. Eur. Arch. Oto-Rhino-Laryngol. 2016;273:1649–1661. doi: 10.1007/s00405-015-3631-8. [DOI] [PubMed] [Google Scholar]
- 5.Sweeney R.T., McClary A.C., Myers B.R., Biscocho J., Neahring L., Kwei K.A., Qu K., Gong X., Ng T., Jones C.D., et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat. Genet. 2014;46:722–725. doi: 10.1038/ng.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown N.A., Rolland D., McHugh J.B., Weigelin H.C., Zhao L., Lim M.S., Elenitoba-Johnson K.S., Betz B.L. Activating FGFR2—RAS—BRAF mutations in ameloblastoma. Clin. Cancer Res. 2014;20:5517–5526. doi: 10.1158/1078-0432.CCR-14-1069. [DOI] [PubMed] [Google Scholar]
- 7.Brown N.A., Betz B.L. Ameloblastoma: A review of recent molecular pathogenetic discoveries. Biomark. Cancer. 2015;7:19–24. doi: 10.4137/BIC.S29329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantwell-Dorris E.R., O’Leary J.J., Sheils O.M. BRAFV600E: Implications for carcinogenesis and molecular therapy. Mol. Cancer Ther. 2011;10:385–394. doi: 10.1158/1535-7163.MCT-10-0799. [DOI] [PubMed] [Google Scholar]
- 9.Kurppa K.J., Catón J., Morgan P.R., Ristimäki A., Ruhin B., Kellokoski J., Elenius K., Heikinheimo K. High frequency of BRAF V600E mutations in ameloblastoma. J. Pathol. 2014;232:492–498. doi: 10.1002/path.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright J.M., Vered M. Update from the 4th Edition of the World Health Organization Classification of Head and Neck Tumours: Odontogenic and Maxillofacial Bone Tumors. Head Neck Pathol. 2017;11:68–77. doi: 10.1007/s12105-017-0794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derakhshan S., Aminishakib P., Karimi A., Saffar H., Abdollahi A., Mohammadpour H., Fard M.J.K., Memarha A. High frequency of BRAF V600E mutation in Iranian population ameloblastomas. Med. Oral Patol. Oral Cir. Bucal. 2020;25:e502–e507. doi: 10.4317/medoral.23519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ. 2009;339:332–336. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Methley A.M., Campbell S., Chew-Graham C., McNally R., Cheraghi-Sohi S. PICO, PICOS and SPIDER: A comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv. Res. 2014;14:579. doi: 10.1186/s12913-014-0579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West S., King V., Carey T.S., Lohr K.N., McKoy N., Sutton S.F., Lux L. Systems to rate the strength of scientific evidence. Evid. Rep. Technol. Assess (Summ.) 2002;47:1–11. [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida R.D.A.C., Andrade E.S.D.S., Barbalho J.C., Vajgel A., Vasconcelos B.C.D.E. Recurrence rate following treatment for primary multicystic ameloblastoma: Systematic review and meta-analysis. Int. J. Oral Maxillofac. Surg. 2016;45:359–367. doi: 10.1016/j.ijom.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Higgins J.P.T., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. Br. Med. J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi S.A.R., Thalib L. A quality-effects model for meta-analysis. Epidemiology. 2008;19:94–100. doi: 10.1097/EDE.0b013e31815c24e7. [DOI] [PubMed] [Google Scholar]
- 18.Fregnani E.R., Perez D.E.d.C., de Almeida O., Fonseca F.P., Soares F.A., Castro-Junior G., Alves F.A. BRAF-V600E expression correlates with ameloblastoma aggressiveness. Histopathology. 2017;70:473–484. doi: 10.1111/his.13095. [DOI] [PubMed] [Google Scholar]
- 19.Gültekin S.E., Aziz R., Heydt C., Sengüven B., Zöller J., Safi A.F., Kreppel M., Buettner R. The landscape of genetic alterations in ameloblastomas relates to clinical features. Virchows Arch. 2018;472:807–814. doi: 10.1111/his.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.do Canto A.M., da Silva Marcelino B.M.R., Schussel J.L., Wastner B.F., Sassi L.M., Corrêa L., de Freitas R.R., Hasséus B., Kjeller G., Junior C.A.L., et al. Immunohistochemical analysis of BRAF V600E mutation in ameloblastomas. Clin. Oral Investig. 2019;23:779–784. doi: 10.1007/s00784-018-2494-y. [DOI] [PubMed] [Google Scholar]
- 21.Heikinheimo K., Huhtala J.-M., Thiel A., Kurppa K., Kovac M., Kragelund C., Warfvinge G., Dawson H., Elenius K., Ristimäki A., et al. The Mutational Profile of Unicystic Ameloblastoma. J. Dent. Res. 2019;98:54–60. doi: 10.1177/0022034518798810. [DOI] [PubMed] [Google Scholar]
- 22.Oh K.-Y., Cho S.-D., Yoon H.-J., Lee J.-I., Ahn S.-H., Hong S.-D. High prevalence of BRAF V600E mutations in Korean patients with ameloblastoma: Clinicopathological significance and correlation with epithelial-mesenchymal transition. J. Oral Pathol. Med. 2019;48:413–420. doi: 10.1111/jop.12851. [DOI] [PubMed] [Google Scholar]
- 23.Kelppe J., Thorén H., Ristimäki A., Haglund C., Sorsa T., Hagström J. BRAF V600E expression in ameloblastomas—A 36-patient cohort from Helsinki University Hospital. Oral Dis. 2019;25:1169–1174. doi: 10.1111/odi.13072. [DOI] [PubMed] [Google Scholar]
- 24.Seki-Soda M., Sano T., Ito K., Yokoo S., Oyama T. An immunohistochemical and genetic study of BRAFV600E mutation in Japanese patients with ameloblastoma. Pathol. Int. 2020;70:224–230. doi: 10.1111/pin.12899. [DOI] [PubMed] [Google Scholar]
- 25.da Silva Marcelino B.M.R., Parise G.K., do Canto A.M., Sassi L.M., Sarmento D.J.S., Costa A.L.F., Hasséus B., Kjeller G., Schussel J.L., Braz-Silva P.H. Comparison of immunohistochemistry and DNA sequencing for BRAF V600E mutation detection in mandibular ameloblastomas. Appl. Immunohistochem. Mol. Morphol. 2021;29:390–393. doi: 10.1097/PAI.0000000000000904. [DOI] [PubMed] [Google Scholar]
- 26.Kunmongkolwut S., Chaisuparat R. Analysis of BRAF V600E expression and disease-free survival in patients with ameloblastoma. Int. J. Oral Maxillofac. Surg. 2022;51:1034–1042. doi: 10.1016/j.ijom.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Santana L.A.D.M., Santana E.M.R., de Albuquerque-Júnior R.L.C., Takeshita W.M., Pereira N.B., Gomez R.S., Gomes C.C., de Sousa S.F. Ameloblastoma shows nuclear BAP1 immunoexpression, independently of the BRAF V600E status. Oral Dis. 2021;27:1238–1242. doi: 10.1111/odi.13644. [DOI] [PubMed] [Google Scholar]
- 28.Shirsat P.M., Bansal S., Prasad P., Desai R.S. Low frequency of BRAF V600E immunoexpression in mandibular ameloblastomas: An institutional study. J. Oral Maxillofac. Pathol. 2018;22:353–359. doi: 10.4103/jomfp.jomfp_174_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diniz M.G., Gomes C., Guimarães B.V.A., Castro W.H., Lacerda J.C.T., Cardoso S.V., de Faria P.R., Dias F.L., Eisenberg A.L.A., Loyola A.M., et al. Assessment of BRAFV600E and SMOF412E mutations in epithelial odontogenic tumours. Tumor Biol. 2015;36:5649–5653. doi: 10.1007/s13277-015-3238-0. [DOI] [PubMed] [Google Scholar]
- 30.Yukimori A., Oikawa Y., Morita K.-I., Nguyen C.T.K., Harada H., Yamaguchi S., Kayamori K., Yamaguchi A., Ikeda T., Sakamoto K. Genetic basis of calcifying cystic odontogenic tumors. PLoS ONE. 2017;12:e0180224. doi: 10.1371/journal.pone.0180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe M., Zong L., Abe T., Hoshi K. A turning point in therapy for ameloblastomas. Oral Oncol. 2018;80:95–96. doi: 10.1016/j.oraloncology.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Abe M., Zong L., Abe T., Takeshima H., Ji J., Ushijima T., Hoshi K. BRAF inhibitor: A novel therapy for ameloblastoma in mandible. Chin. J. Cancer Res. 2018;30:677–678. doi: 10.21147/j.issn.1000-9604.2018.06.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunner P., Bihl M., Jundt G., Baumhoer D., Hoeller S. BRAF p.V600E mutations are not unique to ameloblastoma and are shared by other odontogenic tumors with ameloblastic morphology. Oral Oncol. 2015;51:e77–e78. doi: 10.1016/j.oraloncology.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 34.De Sousa S.F., Moreira R.G., Gomez R.S., Gomes C.C. Interrogation of cancer hotspot mutations in 50 tumour suppressor genes and oncogenes in calcifying cystic odontogenic tumour. Oral Oncol. 2016;57:e1–e3. doi: 10.1016/j.oraloncology.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Coura B.P., dos Santos J.N., Fonseca F.P., Bernardes V.F., de Aquino S.N., Júnior J.J., Vargas P.A., Romañach M.J., de Andrade B.A.B., Gomez R.S., et al. Adenoid ameloblastoma with dentinoid is molecularly different from ameloblastomas and adenomatoid odontogenic tumors. J. Oral Pathol. Med. 2021;50:1067–1071. doi: 10.1111/jop.13243. [DOI] [PubMed] [Google Scholar]
- 36.Faden D.L., Algazi A. Durable treatment of ameloblastoma with single agent BRAFi Re: Clinical and radiographic response with combined BRAF-targeted therapy in stage 4 ameloblastoma. J. Natl. Cancer Inst. 2017;109:djw190. doi: 10.1093/jnci/djw190. [DOI] [PubMed] [Google Scholar]
- 37.Gomes C.C., Diniz M.G., Gomez R.S. Progress towards personalized medicine for ameloblastoma. J. Pathol. 2014;232:488–491. doi: 10.1002/path.4331. [DOI] [PubMed] [Google Scholar]
- 38.Kaye F.J., Ivey A.M., Drane W.E., Mendenhall W.M., Allan R.W. Clinical and radiographic response with combined BRAF-targeted therapy in stage 4 ameloblastoma. J. Natl. Cancer Inst. 2015;107:2014–2016. doi: 10.1093/jnci/dju378. [DOI] [PubMed] [Google Scholar]
- 39.Kaye F.J., Ivey A.M., Drane W.E., Mendenhall W.M., Allan R.W. Response. J. Natl. Cancer Inst. 2017;109:2017. doi: 10.1093/jnci/djw191. [DOI] [PubMed] [Google Scholar]
- 40.Magliocca K.R., Steuer C.E., Hudgins P.A., Bouloux G.F. Regarding BRAF-inhibitor therapy of primary ameloblastoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016;122:517–518. doi: 10.1016/j.oooo.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 41.da Mota Santana L.A., Cunha J.L.S., Oliveira E.M., Sabey M.J.S., Rezende-Silva E., Almeida Souza L.M., Trento C.L. BRAF V600E expression in ameloblastomas: Report of two cases. Oral Surg. 2020;15:467–470. doi: 10.1111/ors.12586. [DOI] [Google Scholar]
- 42.Saffari P.S., Vapniarsky N., Pollack A.S., Gong X., Vennam S., Pollack A.J., Verstraete F.J.M., West R.B., Arzi B., Pollack J.R. Most canine ameloblastomas harbor HRAS mutations, providing a novel large-animal model of RAS-driven cancer. Oncogenesis. 2019;8:11. doi: 10.1038/s41389-019-0119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waqa O., Rich A., Seo B., Tong D., DeSilva R. BRAF mutations in ameloblastoma: Correlation with clinical and histopathological features. Br. J. Oral Maxillofac. Surg. 2020;58:e206. doi: 10.1016/j.bjoms.2020.10.213. [DOI] [Google Scholar]
- 44.Diniz M.G., Gomes C.C., de Sousa S.F., Xavier G.M., Gomez R.S. Oncogenic signalling pathways in benign odontogenic cysts and tumours. Oral Oncol. 2017;72:165–173. doi: 10.1016/j.oraloncology.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Daws S., Chaiyasate K., Lehal A. Treatment of a BRAF V600E Positive Ameloblastoma in a Pediatric Patient with MEK Inhibitor Monotherapy. FACE. 2021;2:179–182. doi: 10.1177/27325016211005126. [DOI] [Google Scholar]
- 46.do Canto A.M., Rozatto J.R., Schussel J.L., de Freitas R.R., Hasséus B., Braz-Silva P.H. Immunohistochemical biomarkers in ameloblastomas. Acta Odontol. Scand. 2016;74:585–590. doi: 10.1080/00016357.2016.1224918. [DOI] [PubMed] [Google Scholar]
- 47.Fuchigami T., Ono Y., Kishida S., Nakamura N. Molecular biological findings of ameloblastoma. Jpn. Dent. Sci. Rev. 2021;57:27–32. doi: 10.1016/j.jdsr.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heikinheimo K., Kurppa K.J., Elenius K. Novel targets for the treatment of ameloblastoma. J. Dent. Res. 2015;94:237–240. doi: 10.1177/0022034514560373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jhamb T., Kramer J.M. Molecular concepts in the pathogenesis of ameloblastoma: Implications for therapeutics. Exp. Mol. Pathol. 2014;97:345–353. doi: 10.1016/j.yexmp.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Khalele B.A.E.O., Al-Shiaty R.A. A novel marker of ameloblastoma and systematic review of immunohistochemical findings. Ann. Diagn. Pathol. 2016;22:18–24. doi: 10.1016/j.anndiagpath.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Kreppel M., Zöller J. Ameloblastoma—Clinical, radiological, and therapeutic findings. Oral Dis. 2018;24:63–66. doi: 10.1111/odi.12702. [DOI] [PubMed] [Google Scholar]
- 52.Marín C., Niklander S.E., Martínez-Flores R. Genetic Profile of Adenomatoid Odontogenic Tumor and Ameloblastoma. A Systematic Review. Front. Oral Health. 2021;2:767474. doi: 10.3389/froh.2021.767474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins-De-Barros A.V., dos Anjos R.S., Silva C.C.G., Silva E.D.D.O.E., Araújo F.A.D.C., Carvalho M.D.V. Diagnostic accuracy of immunohistochemistry compared with molecular tests for detection of BRAF V600E mutation in ameloblastomas: Systematic review and meta-analysis. J. Oral Pathol. Med. 2022;51:223–230. doi: 10.1111/jop.13278. [DOI] [PubMed] [Google Scholar]
- 54.Ngan H.L., Law C.H., Choi Y.C.Y., Chan J.Y.S., Lui V.W.Y. Precision drugging of the MAPK pathway in head and neck cancer. NPJ Genom. Med. 2022;7:20. doi: 10.1038/s41525-022-00293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritterhouse L.L., Barletta J.A. BRAF V600E mutation-specific antibody: A review. Semin. Diagn. Pathol. 2015;32:400–408. doi: 10.1053/j.semdp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 56.Shi H.A., Ng C.W.B., Kwa C.T., Sim Q.X.C. Ameloblastoma: A succinct review of the classification, genetic understanding and novel molecular targeted therapies. Surgeon. 2021;19:238–243. doi: 10.1016/j.surge.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 57.You Z., Liu S.P., Du J., Wu Y.H., Zhang S.Z. Advancements in MAPK signaling pathways and MAPK-targeted therapies for ameloblastoma: A review. J. Oral Pathol. Med. 2019;48:201–205. doi: 10.1111/jop.12807. [DOI] [PubMed] [Google Scholar]
- 58.Abramson Z., Dayton O.L., Drane W.E., Mendenhall W.M., Kaye F.J. Managing stage 4 ameloblastoma with dual BRAF/MEK inhibition: A case report with 8-year clinical follow-up. Oral Oncol. 2022;128:105854. doi: 10.1016/j.oraloncology.2022.105854. [DOI] [PubMed] [Google Scholar]
- 59.Bernaola-Paredes W.E., Albuja-Rivadeneira E., Novelli F., Dalcin J.F., Vartanian J.G., Kohler H.F., Pellizzon A.C.A. Refractory Ameloblastoma in the Maxilla: Clinical, Imaging, Histological, Surgical and Mutational Characterization: A Case Series. Craniomaxillofac. Trauma Reconstr. Open. 2021;6:247275122110337. doi: 10.1177/24727512211033724. [DOI] [Google Scholar]
- 60.Broudic-Guibert M., Blay J.-Y., Vazquez L., Evrard A., Karanian M., Taïeb S., Hoog-Labouret N., Oukhatar C.M.A., Boustany-Grenier R., Arnaud A. Persistent response to vemurafenib in metastatic ameloblastoma with BRAF mutation: A case report. J. Med. Case Rep. 2019;13:245. doi: 10.1186/s13256-019-2140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunet M., Khalifa E., Italiano A. Enabling Precision Medicine for Rare Head and Neck Tumors: The Example of BRAF/MEK Targeting in Patients with Metastatic Ameloblastoma. Front Oncol. 2019;9:1204. doi: 10.3389/fonc.2019.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandes G.S., Girardi D.M., Bernardes J.P.G., Fonseca F.P., Fregnani E.R. Clinical benefit and radiological response with BRAF inhibitor in a patient with recurrent ameloblastoma harboring V600E mutation. BMC Cancer. 2018;18:887. doi: 10.1186/s12885-018-4802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirschhorn A., Campino G.A., Vered M., Greenberg G., Yacobi R., Yahalom R., Barshack I., Toren A., Amariglio N., Rechavi G. Upfront rational therapy in BRAF V600E mutated pediatric ameloblastoma promotes ad integrum mandibular regeneration. J. Tissue Eng. Regen. Med. 2021;15:1155–1161. doi: 10.1002/term.3254. [DOI] [PubMed] [Google Scholar]
- 64.Roque A., Odia Y. BRAF-V600E mutant papillary craniopharyngioma dramatically responds to combination BRAF and MEK inhibitors. CNS Oncol. 2017;6:95–99. doi: 10.2217/cns-2016-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rotellini M., Maggiore G., Trovati M., Saraceno M.S., Franchi A. Metastasizing maxillary ameloblastoma: Report of a case with molecular characterization. J. Oral Maxillofac. Res. 2016;7:e5. doi: 10.5037/jomr.2016.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki H., Sasaki E., Nakamura R., Sawabe M., Hagiwara S., Hyodo I., Hanai N. Recurrent ameloblastoma with both hypercalcemia and BRAF mutation: A case report. Clin. Case Rep. 2020;8:3462–3466. doi: 10.1002/ccr3.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan S., Pollack J.R., Kaplan M.J., Colevas A.D., West R.B. BRAF inhibitor treatment of primary BRAF-mutant ameloblastoma with pathologic assessment of response. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016;122:e5–e7. doi: 10.1016/j.oooo.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 68.Bartels S., Adisa A., Aladelusi T., Lemound J., Stucki-Koch A., Hussein S., Kreipe H., Hartmann C., Lehmann U., Hussein K. Molecular defects in BRAF wild-type ameloblastomas and craniopharyngiomas—Differences in mutation profiles in epithelial-derived oropharyngeal neoplasms. Virchows Arch. 2018;472:1055–1059. doi: 10.1007/s00428-018-2323-3. [DOI] [PubMed] [Google Scholar]
- 69.Diniz M.G., Duarte A.P., Villacis R.A., Guimarães B.V.A., Duarte L.C.P., Rogatto S.R., Gomez R.S., Gomes C.C. Rare copy number alterations and copy-neutral loss of heterozygosity revealed in ameloblastomas by high-density whole-genome microarray analysis. J. Oral Pathol. Med. 2017;46:371–376. doi: 10.1111/jop.12505. [DOI] [PubMed] [Google Scholar]
- 70.Kennedy W.R., Werning J.W., Kaye F.J., Mendenhall W.M. Treatment of ameloblastoma and ameloblastic carcinoma with radiotherapy. Eur. Arch. Oto-Rhino-Laryngol. 2016;273:3293–3297. doi: 10.1007/s00405-016-3899-3. [DOI] [PubMed] [Google Scholar]
- 71.Kondo S., Ota A., Ono T., Karnan S., Wahiduzzaman M., Hyodo T., Rahman L., Ito K., Furuhashi A., Hayashi T., et al. Discovery of novel molecular characteristics and cellular biological properties in ameloblastoma. Cancer Med. 2020;9:2904–2917. doi: 10.1002/cam4.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pereira N.B., Pereira K.M.A., Coura B.P., Diniz M.G., de Castro W.H., Gomes C.C., Gomez R.S. BRAFV600E mutation in the diagnosis of unicystic ameloblastoma. J. Oral Pathol. Med. 2016;45:780–785. doi: 10.1111/jop.12443. [DOI] [PubMed] [Google Scholar]
- 73.Sant’Ana M.S.P., dos Santos Costa S.F., da Silva M.P., Martins-Chaves R.R., Pereira T.d.S.F., de Oliveira E.M., Pedraza R.M., de Castro W.H., Gomes C.C., Gomes R.S., et al. BRAF p.V600E status in epithelial areas of ameloblastoma with different histological aspects: Implications to the clinical practice. J. Oral Pathol. Med. 2021;50:478–484. doi: 10.1111/jop.13155. [DOI] [PubMed] [Google Scholar]
- 74.Shi Y., Li M., Yu Y., Zhou Y., Wang S. Whole exome sequencing and system biology analysis support the “two-hit” mechanism in the onset of ameloblastoma. Med. Oral Patol. Oral Cir. Bucal. 2021;26:e510–e517. doi: 10.4317/medoral.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimura M., Nakashiro K.-I., Sawatani Y., Hasegawa T., Kamimura R., Izumi S., Komiyama Y., Fukumoto C., Yagisawa S., Yaguchi E., et al. Whole Exome Sequencing of SMO, BRAF, PTCH1 and GNAS in Odontogenic Diseases. In Vivo. 2020;34:3233–3240. doi: 10.21873/invivo.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.You Z., Sun L., Yan X., Zhang J., Du J., Li T., Zhao H. Clinicopathologic study on a rare variant of ameloblastoma with basal cell features. Oral Dis. 2019;25:788–795. doi: 10.1111/odi.13018. [DOI] [PubMed] [Google Scholar]
- 77.Bologna-Molina R., Ogawa I., Mosqueda-Taylor A., Takata T., Sánchez-Romero C., Villarroel-Dorrego M., Takeda Y., Mikami T. Detection of MAPK/ERK pathway proteins and KRAS mutations in adenomatoid odontogenic tumors. Oral Dis. 2019;25:481–487. doi: 10.1111/odi.12989. [DOI] [PubMed] [Google Scholar]
- 78.Bonacina R., Indini A., Massazza G., Rulli E., Gianatti A., Mandalà M. Correlation of BRAF mutational status with clinical characteristics and survival outcomes of patients with ameloblastoma: The experience of 11 Italian centres. J. Clin. Pathol. 2022;75:555–559. doi: 10.1136/jclinpath-2021-207527. [DOI] [PubMed] [Google Scholar]
- 79.Coura B.P., de Resende T.A.C., de Menezes V.C.B., Bernardes V.F., de Sousa S.F., Diniz M.G., Gomez R.S., Gomes C.C. Assessing pathogenic mutations in dental follicles as an attempt to identify early events in odontogenic tumours tumourigenesis. Arch. Oral Biol. 2020;113:104523. doi: 10.1016/j.archoralbio.2019.104523. [DOI] [PubMed] [Google Scholar]
- 80.Duarte-Andrade F., Silva A.M.B., Vitório J., Canuto G.A.B., Costa S.F.S., Diniz M.G., Fernandes A.P., De Toledo J.S., André L.C., Gomes C.C., et al. The importance of BRAF-V600E mutation to ameloblastoma metabolism. J. Oral Pathol. Med. 2019;48:307–314. doi: 10.1111/jop.12839. [DOI] [PubMed] [Google Scholar]
- 81.Fujii S., Ishibashi T., Kokura M., Fujimoto T., Matsumoto S., Shidara S., Kurppa K.J., Pape J., Caton J., Morgan P.R., et al. RAF1–MEK/ERK pathway-dependent ARL4C expression promotes ameloblastoma cell proliferation and osteoclast formation. J. Pathol. 2022;256:119–133. doi: 10.1002/path.5814. [DOI] [PubMed] [Google Scholar]
- 82.Guan P., Wong S., Lim J.Q., Ng C., Soong P., Sim C., Ong C., Rajasegaran V., Myint S., Lee J., et al. Mutational Signatures in Mandibular Ameloblastoma Correlate with Smoking. J. Dent. Res. 2019;98:652–658. doi: 10.1177/0022034519837248. [DOI] [PubMed] [Google Scholar]
- 83.Kokubun K., Yamamoto K., Akashi Y., Chujo T., Nakajima K., Matsuzaka K. Genetic Study of BRAF V600E and SMO L412F Mutations in Japanese Patients with Ameloblastoma. Int. J. Surg. Pathol. 2022;30:378–384. doi: 10.1177/10668969211064203. [DOI] [PubMed] [Google Scholar]
- 84.Lapthanasupkul P., Laosuk T., Ruangvejvorachai P., Aittiwarapoj A., Kitkumthorn N. Frequency of BRAF V600E mutation in a group of Thai patients with ameloblastomas. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2021;132:e180–e185. doi: 10.1016/j.oooo.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 85.Oh K.Y., Kim J.H., Cho S.D., Yoon H.J., Lee J.I., Hong S.D. BRAF V600E and previously unidentified KRAS G12C mutations in odontogenic tumors may affect MAPK activation differently depending on tumor type. Genes Chromosom. Cancer. 2022;61:481–490. doi: 10.1002/gcc.23040. [DOI] [PubMed] [Google Scholar]
- 86.Oh K.Y., Cho S.D., Yoon H.J., Lee J.I., Hong S.D. Discrepancy between immunohistochemistry and sequencing for BRAF V600E in odontogenic tumours: Comparative analysis of two VE1 antibodies. J. Oral Pathol. Med. 2021;50:85–91. doi: 10.1111/jop.13108. [DOI] [PubMed] [Google Scholar]
- 87.Owosho A.A., Ladeji A.M., Adebiyi K.E., Olajide M.A., Okoye I.S.I., Kehinde T., Nwizu N.N., Summersgill K.F. BRAF V600E mutation-specific immunohistochemical analysis in ameloblastomas: A 44-patient cohort study from a single institution. Eur. Arch. Oto-Rhino-Laryngol. 2021;278:3065–3071. doi: 10.1007/s00405-020-06491-w. [DOI] [PubMed] [Google Scholar]
- 88.Peralta S., McCleary-Wheeler A.L., Duhamel G.E., Heikinheimo K., Grenier J.K. Ultra-frequent HRAS p. Q61R somatic mutation in canine acanthomatous ameloblastoma reveals pathogenic similarities with human ameloblastoma. Vet. Comp. Oncol. 2019;17:439–445. doi: 10.1111/vco.12487. [DOI] [PubMed] [Google Scholar]
- 89.Salama A.K.S., Li S., Macrae E.R., Park J.-I., Mitchell E.P., Zwiebel J.A., Chen H.X., Gray R.J., McShane L.M., Rubinstein L.V., et al. Dabrafenib and trametinib in patients with tumors with BRAFV600E mutations: Results of the NCI-MATCH trial subprotocol H. J. Clin. Oncol. 2020;38:3895–3904. doi: 10.1200/JCO.20.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharp R.C., Effiom O.A., Dhingra A., Odukoya O., Olawuyi A., Arotiba G.T., Boesze-Battaglia K., Akintoye S.O. Enhanced basal autophagy supports ameloblastoma-derived cell survival and reactivation. Arch. Oral Biol. 2019;98:61–67. doi: 10.1016/j.archoralbio.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soltani M., Tabatabaiefar M.A., Mohsenifar Z., Pourreza M.R., Moridnia A., Shariati L., Razavi S.M. Genetic study of the BRAF gene reveals new variants and high frequency of the V600E mutation among Iranian ameloblastoma patients. J. Oral Pathol. Med. 2018;47:86–90. doi: 10.1111/jop.12610. [DOI] [PubMed] [Google Scholar]
- 92.Tseng C.H., Lu P.H., Wang Y.P., Chang J.Y.F. Enrichment of SOX2-Positive Cells in BRAF V600E Mutated and Recurrent Ameloblastoma. J. Pers. Med. 2022;12:77. doi: 10.3390/jpm12010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang R., Yang Q., Qu J., Hong Y., Liu P., Li T. The BRAF p.V600E mutation is a common event in ameloblastomas but is absent in odontogenic keratocysts. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020;129:229–235. doi: 10.1016/j.oooo.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 94.Diniz M.G., Guimarães B.V.A., Pereira N.B., de Menezes G.H.F., Gomes C.C., Gomez R.S. DNA damage response acti-vation and cell cycle dysregulation in infiltrative ameloblastomas: A proposed model for ameloblastoma tumor evolution. Exp Mol Pathol. 2017;102:391–395. doi: 10.1016/j.yexmp.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 95.Wei X., Wang X., Xiong J., Li C., Liao Y., Zhu Y., Mao J. Risk and Prognostic Factors for BRAFV600E Mutations in Papillary Thyroid Carcinoma. Biomed Res. Int. 2022;2022:9959649. doi: 10.1155/2022/9959649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ni Nyoman A.D., Suksmarini N.M.P.W., Pranata A.A.N.S., Rompis A.Y., Sumadi I.W.J. The prevalence of KRAS and BRAF mutation in colorectal cancer patients in Bali. Indones. J. Biotechnol. 2022;27:29. doi: 10.22146/ijbiotech.67506. [DOI] [Google Scholar]
- 97.Porumb-Andrese E., Ursu R.G., Ivanov I., Caruntu I.-D., Porumb V., Ferariu D., Damian C., Ciobanu D., Terinte C., Iancu L.S. The braf v600e mutation detection by quasa sensitive real-time pcr assay in northeast romania melanoma patients. Appl. Sci. 2021;11:9511. doi: 10.3390/app11209511. [DOI] [Google Scholar]
- 98.Guimarães L.M., Coura B.P., Gomez R.S., Gomes C.C. The Molecular Pathology of Odontogenic Tumors: Expanding the Spectrum of MAPK Pathway Driven Tumors. Front. Oral Health. 2021;2:740788. doi: 10.3389/froh.2021.740788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brukas M., Pedersen T.Ø., Lybak S., Skarstein K., Løes S. Ameloblastic carcinoma of the mandible: A case report and literature review. Oral Maxillofac. Surg. Cases. 2020;6:100183. doi: 10.1016/j.omsc.2020.100183. [DOI] [Google Scholar]
- 100.Niu Z., Li Y., Chen W., Zhao J., Zheng H., Deng Q., Zha Z., Zhu H., Sun Q., Su L. Study on clinical and biological characteristics of ameloblastic carcinoma. Orphanet J. Rare Dis. 2020;15:316. doi: 10.1186/s13023-020-01603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marin C., Dave M., Hunter K.D. Malignant Odontogenic Tumours: A Systematic Review of Cases Reported in Literature. Front. Oral Health. 2021;2:82. doi: 10.3389/froh.2021.775707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Togni L., Zizzi A., Mazzucchelli R., Santarelli A., Rubini C., Mascitti M. Identification of BRAF V600E mutation in odontogenic tumors by high-performance MALDI-TOF analysis. Int. J. Oral Sci. 2022;14:22. doi: 10.1038/s41368-022-00170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dvorak K., Aggeler B., Palting J., McKelvie P., Ruszkiewicz A., Waring P. Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: Impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma. Pathology. 2014;46:509–517. doi: 10.1097/PAT.0000000000000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paja Fano M., Ugalde Olano A., Fuertes Thomas E., Oleaga Alday A. Immunohistochemical detection of the BRAF V600E mutation in papillary thyroid carcinoma. Evaluation against real-time polymerase chain reaction. Endocrinol. Diabetes Nutr. 2017;64:75–81. doi: 10.1016/j.endinu.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 105.Estrella J.S., Tetzlaff M.T., Roland L.B., Jr., Patel K.P., Williams M.D., Curry J.L., Rashid A., Hamilton S.R., Broaddus R.R. Assessment of BRAF V600E status in colorectal carcinoma: Tissue-specific discordances between immunohistochemistry and sequencing. Mol. Cancer Ther. 2015;14:2887–2895. doi: 10.1158/1535-7163.MCT-15-0615. [DOI] [PubMed] [Google Scholar]
- 106.Thiel A., Moza M., Kytölä S., Orpana A., Jahkola T., Hernberg M., Virolainen S., Ristimäki A. Prospective immunohistochemical analysis of BRAF V600E mutation in melanoma. Hum. Pathol. 2015;46:169–175. doi: 10.1016/j.humpath.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 107.Jones R.T., Abedalthagafi M.S., Brahmandam M., Greenfield E.A., Hoang M.P., Louis D.N., Hornick J., Santagata S. Cross-reactivity of the BRAF VE1 antibody with epitopes in axonemal dyneins leads to staining of cilia. Mod. Pathol. 2015;28:596–606. doi: 10.1038/modpathol.2014.150. [DOI] [PubMed] [Google Scholar]
- 108.Capper D., Berghoff A.S., Magerle M., Ilhan A., Wöhrer A., Hackl M., Pichler J., Pusch S., Meyer J., Habel A., et al. Immunohistochemical testing of BRAF V600E status in 1120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012;123:223–233. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 109.Jung Y.Y., Yoo J.H., Park E.S., Kim M.K., Lee T.J., Cho B.Y., Chung Y.J., Kang K.H., Ahn H.Y., Kim H.S. Clinicopathologic correlations of the BRAFV600E mutation, BRAF V600E immunohistochemistry, and BRAF RNA in situ hybridization in papillary thyroid carcinoma. Pathol. Res. Pract. 2015;211:162–170. doi: 10.1016/j.prp.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 110.Sun J., Zhang J., Lu J., Gao J., Lu T., Ren X., Duan H., Liang Z. Immunohistochemistry is highly sensitive and specific for detecting the BRAF V600E mutation in papillary thyroid carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:15072–15078. [PMC free article] [PubMed] [Google Scholar]
- 111.Deeks S.G. HIV Infection, Inflammation, Immunosenescence, and Aging. Annu. Rev. Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.White M.C., Holman D.M., Boehm J.E., Peipins L.A., Grossman M., Henley S.J. Age and cancer risk: A potentially modifiable relationship. Am. J. Prev. Med. 2014;46:S7–S15. doi: 10.1016/j.amepre.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li C.Y., Prochazka J., Goodwin A.F., Klein O.D. Fibroblast growth factor signaling in mammalian tooth development. Odontology. 2014;102:1–13. doi: 10.1007/s10266-013-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buvini M., Medici A., Fernández E., Torres A.C. Disease Control Priorities in Developing Countries. 2nd ed. Oxford University Press; Oxford, UK: 2006. Chapter 10. Gender Differentials in Health; pp. 195–210. [DOI] [Google Scholar]
- 115.Tucker A., Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 116.Dandriyal R., Pant S., Gupta A., Baweja H. Surgical management of ameloblastoma: Conservative or radical approach. Natl. J. Maxillofac. Surg. 2011;2:22. doi: 10.4103/0975-5950.85849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bauer J., Büttner P., Murali R., Okamoto I., Kolaitis N.A., Landi M.T., Scolyer R.A., Bastian B.C. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma. Res. 2011;24:345–351. doi: 10.1111/j.1755-148X.2011.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Capper D., Preusser M., Habel A., Sahm F., Ackermann U., Schindler G., Pusch S., Mechtersheimer G., Zentgraf H., von Deimling A. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 119.Sasaki H., Shimizu S., Tani Y., Shitara M., Okuda K., Hikosaka Y., Moriyama S., Yano M., Fujii Y. Usefulness of immunohistochemistry for the detection of the BRAF V600E mutation in Japanese lung adenocarcinoma. Lung Cancer. 2013;82:51–54. doi: 10.1016/j.lungcan.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 120.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 121.Mendenhall W.M., Werning J.W., Fernandes R., Malyapa R.S., Mendenhall N.P. Ameloblastoma. Am. J. Clin. Oncol. Cancer Clin. Trials. 2007;30:645–648. doi: 10.1097/COC.0b013e3181573e59. [DOI] [PubMed] [Google Scholar]
- 122.Kim G., McKee A.E., Ning Y.-M., Hazarika M., Theoret M., Johnson J.R., Xu Q.C., Tang S., Sridhara R., Jiang X., et al. FDA approval summary: Vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation mutation. Clin. Cancer Res. 2014;20:4994–5000. doi: 10.1158/1078-0432.CCR-14-0776. [DOI] [PubMed] [Google Scholar]
- 123.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hauschild A., Grob J.-J., Demidov L.V., Jouary T., Gutzmer R., Millward M., Rutkowski P., Blank C.U., Miller W.H., Jr., Kaempgen E., et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 125.Parker K.G., White M.G., Cipriani N.A. Comparison of Molecular Methods and BRAF Immunohistochemistry (VE1 Clone) for the Detection of BRAF V600E Mutation in Papillary Thyroid Carcinoma: A Meta-Analysis. Head Neck Pathol. 2020;14:1067–1079. doi: 10.1007/s12105-020-01166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.