Abstract
The lipopolysaccharide (LPS) of Helicobacter pylori expresses the Lewis x (Lex) and/or Ley antigen. We have shown previously that H. pylori LPS displays phase variation whereby an Lex-positive strain yields variants with different LPS serotypes, for example, Lex plus Ley or nonfucosylated polylactosamine. H. pylori has two α3-fucosyltransferase genes that both contain poly(C) tracts. We now demonstrate that these tracts can shorten or lengthen randomly, which results in reversible frameshifting and inactivation of the gene products. We provide genetic and serological evidence that this mechanism causes H. pylori LPS phase variation and demonstrate that the on or off status of α3-fucosyltransferase genes determines the LPS serotypes of phase variants and clinical isolates. The role of the α3-fucosyltransferase gene products in determining the LPS serotype was confirmed by structural-chemical analysis of α3-fucosyltransferase knockout mutants. The data also show that the two α3-fucosyltransferase genes code for enzymes with different fine specificities, and we propose the names futA and futB to designate the orthologs of the H. pylori 26695 α3-fucosyltransferase genes HP0379 and HP0651, respectively. The data also show that the α3-fucosylation in H. pylori precedes α3-fucosyltransferase, an order of events opposite to that which prevails in mammals. Finally, the data provide an understanding at the molecular level of the mechanisms underlying LPS diversity in H. pylori, which may play an important role in adaptation to the host.
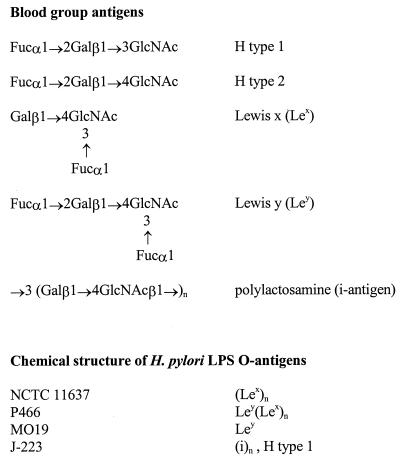
Helicobacter pylori causes lifelong infection in humans and is involved in diverse diseases: gastritis, gastric and duodenal ulcer, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma (16). Through which mechanism(s) H. pylori is able to persist chronically is not known, but possibly molecular mimicry plays a role (2, 3). In mimicry of the host, H. pylori lipopolysaccharide (LPS) expresses Lewis blood group antigens (Fig. 1). Polymeric Lewis x (Lex), Ley, or both (5, 6) are expressed most often, but Lea, H type 1, and the i antigen can also be present (21). The expression of Lewis antigens appears to be a highly conserved feature, and only a few strains lack these epitopes (29); this is striking because genetically H. pylori is very diverse (17). This conservation might be related to the restricted ecological niche of H. pylori: the human stomach. Gastric mucosal epithelial cells also express Lewis antigens. Molecular mimicry (2, 3) might mediate evasion by the microorganism of host immune attack and allow colonization to persist. A similar mimicry is seen in the ferret, where both Helicobacter mustelae and the host express blood group A (22, 25). Thus, Helicobacter seems capable of expressing an LPS serotype similar and adapted to that of the host. Data supporting this concept were obtained from both human studies (38) and experimental infection studies where, depending on the Lewis phenotype of the host, the infecting H. pylori strain expressed mainly Lex or mainly Ley (39). These data suggest that H. pylori LPS Lewis antigen expression may change, depending on the host. The mechanisms responsible for these phenotypical changes were the subject of this study.
FIG. 1.
Structures of Lewis blood group antigens and H. pylori LPS. Gal, d-galactose; Fuc, l-fucose; GlcNAc, N-acetyl-d-glucosamine. The general structure of H. pylori LPS is O-antigen–core–lipid A.
Previously we have shown that H. pylori LPS displays phase variation (4). This is the occurrence of spontaneous, high-frequency (up to 0.5%), reversible on-off switching of LPS epitopes. Bacterial cells of the parent strain NCTC 11637 that express Lex can yield phase variants (variant K4.1) that express the nonfucosylated i antigen; back switches from K4.1 to the α3-fucosylated parent phenotype are also observed. Other variants strongly express both Lex and Ley (variant 1c) or related epitopes (4).
Phase variation in the LPSs of Neisseria spp. (40) and Haemophilus influenzae (27) is well documented and is caused by reversible on-off switching of LPS biosynthesis genes. On-off switching occurs during replication due to a strand slip mechanism which changes the length of polynucleotide repeats, for example, of G tracts present in certain glycosyltransferase genes of Neisseria spp. (40). Changes in these polynucleotide tracts introduce translational frameshifts, leading to the production of inactive truncated gene products, i.e., the gene is switched off. Subsequent changes during replication may switch the gene back on by restoring the reading frame and restoring production of an active gene product. The consequence is a variable LPS phenotype. There is evidence which suggests that the LPS phase variation in Neisseria spp. plays an adaptive role and generates microorganisms that either adhere better to host cells or are more resistant to being killed by complement (34). Phase variation in H. pylori LPS causes considerable changes in Lewis antigen expression and might be responsible for the changes in Lewis antigen expression observed in vivo (39). The molecular mechanisms of H. pylori LPS phase variation are unknown.
H. pylori requires a series of enzymes to synthesize LPS O antigen containing an Lex polymer plus an Ley terminus: α3- and α2-fucosyltransferases that link fucose to C-3 of N-acetylglucosamine (GlcNAc) and C-2 of galactose (Gal), respectively; GlcNAc transferases (GlcNAcT) and Gal transferases (GalT) that form the main polylactosamine O chain are also required. Two H. pylori α3-fucosyltransferase genes (HP0379 and HP0651 in strain 26695; JHP 1002 and 596 in strain J99) have been identified, cloned, and expressed (1, 13, 18, 31). An α2-fucosyltransferase gene (HP0093/94 in strain 26695; JHP 86 in strain J99) was also identified and characterized (7, 28, 35). These genes contain poly(C) tracts: in strain 26695, both α3-fucosyltransferase genes contain C13 tracts, while the α2-fucosyltransferase gene contains a C14 tract (31). C tracts are also found in the homologous genes in strain J99 (1). Another feature of α3-fucosyltransferase genes is the presence of oligonucleotide repeats at the 3′ end. We hypothesized that the LPS phase variation in H. pylori is caused by (reversible) inactivation of glycosyltransferases through translational frameshifts due to the presence of these C tracts.
In the present paper, we provide evidence that length changes in the poly(C) tracts of α3-fucosyltransferase genes indeed lead to phase variation in the LPS of H. pylori. The on or off status of the two α3-fucosyltransferase genes determined the LPS serotypes of selected phase variants and clinical isolates. The data show that the two α3-fucosyltransferase gene products have different specificities and, by analogizing with the nomenclature for eukaryotic fucosyltransferases (8a), we propose the names futA and futB to designate the orthologs of the H. pylori 26695 α3-fucosyltransferase genes HP0379 and HP0651, respectively. The role of futA and futB gene products was confirmed through the structural-chemical and serological analysis of mutant strains in which one or both α3-fucosyltransferase genes were inactivated. Our results provide a molecular basis for an understanding of how H. pylori might adapt to the host.
MATERIALS AND METHODS
Bacterial strains.
Strain NCTC 11637 and the LPS phase variants 2b, K4.1, K5.1, and 1c have been described before (4, 5). NCTC 11637 and variant 2b express mainly Lex. Variant K4.1 expresses the i antigen. Variant K5.1, derived from strain K4.1, expresses mainly Lex and represents a back switch to the serotype of the parent strain. Variant 1c expresses Lex and Ley. Strain P466 (6) was obtained from T. Boren; strain 26695 (31) was obtained from S. Krakowka; strain 4187E was described before (19); strain J223 was obtained from H. P. Wirth (21); strain N6 was obtained from A. Labigne; strain J99 was obtained from R. Alm (1); and strain SS-1 was obtained from A. Lee. Bacteria were grown in brucella broth supplemented with 10% newborn-calf serum as described before (4).
Monoclonal antibodies and ELISA.
The monoclonal antibodies (MAbs) used in this study and their specificities are shown in Table 1. For enzyme-linked immunosorbent assays (ELISAs), polystyrene 96-well microtiter plates were coated at 7.5 × 106 CFU/ml with bacteria washed in phosphate-buffered saline, and the bacteria were tested for reactivity with MAbs (1 μg/ml) as described before (4). In indicated cases, titrations were done with MAbs diluted in serial twofold steps.
TABLE 1.
MAbs used in this study
| MAb | Specificity | Isotypea | Sourceb | Reference |
|---|---|---|---|---|
| 4D2 | H type 1 | IgM | R. Negrini | 2, 23, 24, 29; |
| 6H3 | Lex (mono or trimeric) | IgM | R. Negrini | 2, 23, 24, 29; |
| 54.1F6A | Lex (tri or polymeric) | IgM | G. J. van Dam | 33 |
| Hp151 | Ley | IgG | R. Negrini | 2, 23, 24, 29; |
| NAM61-1A2 | i antigen | IgM | D. Blanchard | 8 |
| 3C10 | H type 2 | IgM | D. Blanchard | 20 |
Ig, immunoglobulin.
R. Negrini, Biotechnology Laboratory, General Hospital, Brescia, Italy; G. J. van Dam, Department of Parasitology, University of Leiden, The Netherlands; D. Blanchard, Regional Blood Transfusion Service, Nantes, France.
Fucosyltransferase assays.
α3-Fucosyltransferase activity was determined as follows (18, 26). Bacterial cell extract (12.5 μl) was incubated with 20 μM GDP-fucose (Sigma), 100,000 cpm of GDP-[3H]fucose (Amersham), 5 mM N-acetyllactosamine (Sigma), 5 mM MnCl2, 1 mM ATP, buffered to pH 7.2 with 50 mM HEPES-NaOH in a total volume of 50 μl. Reaction mixtures were incubated for 1 h at 37°C, and the reactions were stopped by the addition of 1 ml of mixed-bed resin slurry AG1-X8 (Cl− form; Bio-Rad) at 1:4 (wt/vol) in water. The mixtures were then vortexed briefly and centrifuged for 5 min at 20,000 × g at room temperature. The radioactivity in 600 μl of supernatant was measured by scintillation counting. Allowance was made for nonspecific breakdown of labeled nucleotide sugar and transfer to endogenous acceptors by performing control reactions in the absence of acceptor.
DNA sequencing.
Poly(C) tracts and terminal repeats of the α3-fucosyltransferase genes futA (HP0379 orthologs) and futB (HP0651 orthologs) were sequenced with several primers in both strands. The following primers were used: HPFT-3 (TGGCAAACCCTCTTTTCAAAG), HPFT-4 (GTGTAATGCTGACTTAAAAT), HPFT-5 (TAGCCCTAATCAAGCCTTTG), HPFT-12 (TGTGCTGAGTTTGGATCCATATGTTCCAACCCCTATTA), HPFT-13 (TTCTAAAGTGGATTCTGAAAT), HPFT-14 (GAGTGGGCGAAAGAGAGATTG), HPFT-15 (CCTAAATTAGCTTAAAGGATAACC), HPFT-16 (GCGATGATAGCGCAAGGGGTTTGA), HPFT-17 (AAGGCATTCTCAAATAACGATC), HPFT-18 (GAATTTTTTAACCCATCTCCC), HPFT-19 (AGAGGACATGCTCAAAAACCC), Kanr-F (CTATGAAGCGCCATATTTAA), and Kanr-R (TTTAGACATCTAAATCTAGG). Sequencing was carried out on α3-fucosyltransferase gene fragments amplified by PCR. DNA fragments containing futA were amplified from H. pylori genomic DNA with primers HPFT-15 and HPFT-16 and sequenced with primers HPFT-3, HPFT-4, HPFT-12, HPFT-15, and HPFT-17 [poly(C) region] and HPFT-9, HPFT-11, HPFT-16, and HPFT-18 (terminal zipper-like repeat region). futB was amplified with HPFT-5 and HPFT-3 or HPFT-5 and HPFT-19 and sequenced with HPFT-3, HPFT-4, HPFT-17, HPFT-5, and HPFT-12 [poly(C) region] and HPFT-9, HPFT-11, and HPFT-19 (terminal zipper-like repeat region). DNA sequencing reactions were performed with AmpliTaq FS with dye terminators (Perkin-Elmer Cetus) and analyzed on an Applied Biosystems 373 automated sequencer. As the sequencing of polynucleotide C tracts is prone to errors (15), the relevant region of α3-fucosyltransferase genes from each strain was sequenced several times with template DNA from separate PCR amplifications. The sequence data were compiled with the Lasergene software package (DNASTAR). The final assessment of C-tract length was done by one of us (S.L.M.), unaware of serological information.
Construction of α3-fucosyltransferase knockout mutants. (i) Mutagenesis of cloned H. pylori α3-fucosyltransferase genes.
The source of the futB gene was clone p15M19, previously isolated from an H. pylori plasmid gene (18). The plasmid was linearized at the unique BssHII site within the futB gene, blunt ended with Klenow polymerase, and dephosphorylated with shrimp alkaline phosphatase (Amersham-Pharmacia Biotech). A Campylobacter coli chloramphenicol (Cm) resistance marker cassette (36), excised from a clone in pUC20 with HincII, was ligated to the linearized p15M19, and the resulting plasmid was used to transform Escherichia coli XL1-Blue (Stratagene) to chloramphenicol resistance. The futA gene was amplified from H. pylori 26695 genomic DNA by PCR with primers positioned approximately 1 kb from each end of futA, i.e., HP0379 in the published sequence (5′-TTCTAAAGTGGATTCTGAAAT-3′ and 5′-GAGTGGGCGAAAGAGAGATTG-3′). The fragment was cloned into pGEM T-easy (Promega). The resulting plasmid, designated pHP0379, was linearized with AccB7I at the unique site within the futA gene, blunt-ended with T4 polymerase plus all four deoxynucleoside triphosphates, and dephosphorylated with shrimp alkaline phosphatase. A C. coli kanamycin (Km) resistance marker (32) was obtained as a 1.4-kb EcoRI fragment from a clone containing the amplified cassette in pGEM. The fragment was blunt ended with Klenow polymerase plus all four deoxynucleoside triphosphates and ligated to the linearized pHP0379, and the resulting plasmid was used to transform E. coli XL1-Blue to kanamycin resistance. Correct insertion of the Cmr marker into pHP0651 and of Kmr into pHP0379 was confirmed by restriction mapping and nucleotide sequencing; the resulting plasmids were designated pHP0651::Cmr and pHP0379::Kmr, respectively.
(ii) Homologous recombination in H. pylori.
futA and futB were inactivated by homologous recombination with the above-mentioned plasmids, which contain disrupted copies of the respective genes, flanked on either side by approximately 1 kb of homologous sequence. DNA was introduced by electroporation. For selection of the transformants, suspensions were spread onto Columbia chocolate agar plates containing 20 μg of chloramphenicol/ml (in the case of futB disruptants) or 20 μg of kanamycin/ml (for futA disruptants). Single colonies were streaked on fresh antibiotic-containing plates, and transformants were grown. A double-knockout mutant (4187E-KO379/651) was produced by inactivating futB in an established futA-disrupted (Kanr) strain. Genomic DNA from all transformants was analyzed by PCR and Southern hybridization to confirm that the recombination had occurred at the intended location.
Structural analysis of LPS.
Methylation linkage analysis and fast atom bombardment-mass spectrometry (FAB-MS) of purified LPS of strain 4187E and its α3-fucosyltransferase knockout mutants was performed. Methylation linkage analysis was carried out by the NaOH-dimethyl sulfoxide-CH3I procedure (9) and with characterization of permethylated alditol acetate derivatives by gas-liquid chromatography-MS in the electron impact mode. Methylated material was used for positive-ion FAB-MS, which was performed on a Jeol JMS-AX505H mass spectrometer with thioglycerol as the matrix. A 6-kV Xenon beam was used to produce pseudomolecular ions, which were then accelerated to 3 kV, and their mass was analyzed. Product ion scan was performed on metastable ions created in the first free field with a source pressure of 5 × 10−5 torr. The interpretation of positive-ion mass spectra of the permethylated LPS derivatives was done as previously described (10).
RESULTS
Molecular mechanisms of reversible phase variation from Lex to i antigen and back to Lex.
The molecular genetic mechanisms underlying the phase variation of NCTC 11637 to variant K4.1 were investigated; K4.1 switches back to K5.1, which has a serotype similar to those of NCTC 11637 and phase variant 2b (Table 2). NCTC 11637 expresses polymeric Lex, as measured by the strong reactivity with MAb 54.1F6A. MAb 6H3, specific for monomeric Lex, did not react; strain NCTC 11637 also weakly expresses Ley and strongly expresses H type 1. In addition, it reacts weakly with 3C10, a MAb specific for H type 2; no nonfucosylated polylactosamines (i.e., i antigen) could be detected. In contrast, phase variant K4.1 mainly expresses i antigen and H type 1 but does not express Lex or Ley. DNA sequence analysis revealed that in both the parent, NCTC 11637, and the variant K4.1, the poly(C) repeat in futB contains nine C residues (C9) (Table 2). However, the futA genes differ in repeat length: the phase variant has an additional C residue (C11) relative to NCTC 11637 (C10). Predicted translations of both the futA and futB genes reveal that full coding integrity is maintained with a C10 repeat while expansion or reduction by one residue introduces a frameshift which truncates the coding sequence. Thus, NCTC 11637 has one intact α3-fucosyltransferase (futA) and expresses Lex and Ley while in the phase variant K4.1 both genes are truncated and α3-fucosylated epitopes cannot be produced. The connection between polynucleotide repeat length and Lex/y serotype is further illustrated by a second variant, K5.1, derived from K4.1 itself. The poly(C) repeat lengths of futB and futA in K5.1 were found to be C9 and C10, respectively, i.e., K5.1 represents a reversion to the NCTC 11637 genotype. One would therefore expect K5.1 to show an Lex/y serotype similar to that of NCTC 11637, as indeed was found to be the case (Table 2). We conclude from these results that phase-variable expression of Lex/y epitopes in NCTC 11637 and variants K4.1 and K5.1 is the result of changes in poly(C) repeat length in futA. It is interesting that switching of the α3-fucosyltransferase genes appears to have some effect on α2-fucosylation. Loss of Lex and Ley expression in variant K4.1 relative to NCTC 11637 is not accompanied by an increase in expression of the α2-fucosylated core structure of Ley, H type 2. Since in vitro evidence suggests that the H. pylori α3-fucosyltransferases have no detectable α2-fucosyltransferase activity (13, 18), this observation suggests that α3-fucosylation precedes α2-fucosylation in H. pylori Ley biosynthesis. This was in sharp contrast with the strong expression of H type 1 in both NCTC 11637 and K4.1, and evidently the α2-fucosyltransferase is able to fucosylate Galβ1-3GlcNAc. HP093-HP094 codes for an α2-fucosyltransferase enzyme that is involved in Ley synthesis (35); we propose the name futC to designate H. pylori α2-fucosyltransferase genes (HP093-HP094 and orthologs). Evidently, for H type 1 biosynthesis also, α2-fucosyltransferase enzyme activity is required; we have evidence that the same gene (futC) is involved in Ley and H type 1 biosyntheses (see below). In NCTC 11637 and the variants K4.1 and K5.1, the number of leucine zipper-like 7-amino-acid repeats near the termini of the α3-fucosyltransferase genes (18) was invariant (eight repeats in futB and two in futA), suggesting that this region is not involved in LPS phase variation.
TABLE 2.
Reactivitya of MAbs with H. pylori strains in relation to α3-fucosyltransferase gene C-tract length
| Strainb | Specificity of MAb used
|
Fucosyltransferase gened
|
||||||
|---|---|---|---|---|---|---|---|---|
| Mono-Lex (6H3)c | Poly-Lex (54.1F6A)c | i antigen | H type 1 | H type 2 | Ley | futB | futA | |
| NCTC 11637 | 0.2 | 2.5 | 0.2 | 2.5 | 0.8 | 1.1 | C9 (off) R8 | C10 (on) R2 |
| K5.1 | 0.2 | 1.6 | 0.1 | 1.6 | NDe | 0.5 | C9 (off) R8 | C10 (on) R2 |
| 2b | 0.4 | 2.5 | 0.2 | 2.3 | ND | 0.9 | C9 (off) R8 | C10 (on) R2 |
| K4.1 | 0.2 | 0.2 | 2.5 | 2.5 | 0.4 | 0.4 | C9 (off) R8 | C11 (off) R2 |
| 1c | 0.4 | 2.5 | 0.1 | 0.9 | 0.7 | 2.5 | C10 (on) R8 | C10 (on) R2 |
| 4187E | 2.5 | 2.5f | 0.1 | 1.3 | 0.1 | 2.5g | C10 (on) R7 | C10 (on) R2 |
| 4187E-KO651 | 0.5 | 2.5f | 0.3 | 2.5 | 0.1 | 1.1 | ND | ND |
| 4187E-KO379 | 2.5 | 2.5f | 1.0 | 1.0 | 0.1 | 2.5g | ND | ND |
| 4187E-KO379/651 | 0.1 | 0.1 | 2.5 | 2.5 | 0.1 | 0.5 | ND | ND |
Optical density at 492 nm in ELISA.
Strains K5.1, 2b, K4.1, and 1c are LPS-phase variants of strain NCTC 11637.
MAbs 6H3 and 54.1F6A recognize monomeric and polymeric Lex, respectively.
futB and futA designate the orthologs of H. pylori 26695 α3-fucosyltransferase genes HP0651 and HP0379, respectively. C, C-tract length; R, number of terminal repeats.
ND, not done.
In titration, this MAb reacted 128-fold more with 4187E-KO651 than with strain 4187E and 64-fold less with 4187E-KO379 than with the 4187E parent.
In titration, MAb Hp 151 (anti-Ley) reacted equally well with strains 4187E and 4187E-KO379.
Molecular mechanisms of phase variation from Lex to Lex plus Ley.
We next investigated phase variation from NCTC 11637 to its variant 1c, described before (4). Most strikingly, compared to NCTC 11637, variant 1c has a strongly enhanced Ley reactivity and markedly reduced H type 1 expression (Table 2). In addition, strain 1c reacted with MAb 6H3, specific for monomeric Lex; the reactivities of variant 1c and strain NCTC 11637 with MAb 3C10 were similar. Sequencing data show that 1c has a C10 repeat in futB while NCTC 11637 has a C9 tract in that gene, implying that both α3-fucosyltransferase genes are intact (on) in 1c, whereas in NCTC 11637, only futA is functional. Once again, changes in the α3-fucosyltransferase gene status appear to influence the expression of an α2-fucosylated epitope; it may be that having both genes switched on in variant 1c increases the availability of terminal Lex, a precursor of Ley (see below).
In order to confirm the link between α3-fucosyltransferase gene status and serotype and to establish whether futA and futB play completely interchangeable roles in LPS biosynthesis, we wished to compare the serotype of NCTC 11637 (futA on; futB off) with that of its “mirror” variant (futA off; futB on), but no such variant was found. We therefore constructed mutant strains in which one or both genes were permanently inactivated.
α3-Fucosyltransferase knockout mutants.
The role of futA and futB in LPS biosynthesis was studied in greater detail by insertional mutagenesis. Since our interests lay in the potential role of α3-fucosylation in H. pylori infection, this work was conducted with a strain (4187E) previously validated in a mouse model of H. pylori colonization (19). 4187E has a C10 repeat in both α3-fucosyltransferase genes (Table 2). Sequence analysis confirmed that, accordingly, both reading frames are intact (data not shown); both genes are on. Isogenic α3-fucosyltransferase mutant strains were constructed by introducing kanamycin or chloramphenicol resistance markers into futA or futB as described in Materials and Methods. A double mutant with both α3-fucosyltransferase genes disrupted was also constructed. Correct insertion of the resistance cassette into the intended target gene was confirmed by Southern hybridization and PCR analysis. Since the two α3-fucosyltransferase genes have a high degree of sequence similarity, primers specific to flanking genes were used in conjunction with resistance cassette and α3-fucosyltransferase primers to ensure that only the intended gene had been disrupted.
In ELISA, strain 4187E behaved almost identically to strain 1c, which also has both α3-fucosyltransferase genes on. It showed strong monomeric and polymeric Lex expression, and it also strongly expressed Ley; no reaction with the H type 2 MAb was observed in 4187E or in its knockout mutants; this is striking, since H type 2 equals Ley minus α3-fucose. A mutant strain in which futB had been disrupted (4187E-KO651) showed an altered serological phenotype with greatly reduced Ley expression and increased reactivity to H type 1. Reactivity to the monomeric (terminal) Lex antibody 6H3 is reduced in 4187E-KO651, but titrations with MAb 54.1F6A revealed a 128-fold increase in polymeric Lex expression. As can be seen from Table 2, the overall ELISA profile of 4187E-KO651 is very similar to that of NCTC 11637, in which futB is switched off (see above).
Compared to the disruption of gene futB, inactivation of gene futA had different effects: no change in Ley or H type 1 expression was observed in the knockout (4187E-KO379), while reactivity with 54.1F6A strongly decreased and a modest reactivity with the anti-i MAb was detected. We infer that the two α3-fucosyltransferase enzymes have different fine specificities, which justifies the use of distinct gene names (futA and futB).
Inactivation of both α3-fucosyltransferase genes (strain 4187E-KO379/651) completely abolished Lex and Ley, while a strong expression of the i antigen and H type 1 was observed; this serotype was similar to that of NCTC 11637 variant K4.1, which has both α3-fucosyltransferase genes switched off. The increased i-antigen expression is easily understood as an unmasking of the Lex/y lactosamine scaffold in the absence of α3-fucosylation.
The increase in H type 1 expression seen in K4.1 and 4187E-KO379/651 is more difficult to explain but may reflect increased α2-fucosylation of type 1 (Galβ1-3GlcNAc) structures in the absence of competing Lex-type acceptors.
α3-Fucosyltransferase gene C-tract measurements and Lewis antigen expression in other strains.
Chemical structural analysis of strain J-223 has revealed that it carries H type 1 and i-antigen structures in its LPS (21). When tested in our ELISA system, J-223 reacted only with MAbs specific for H type 1 and i antigen, towards which a strong reaction was observed. The serotype of J-223 is thus identical to that of K4.1 and 4187E-KO379/651. We anticipated that J-223 was therefore likely to have both futA and futB switched off; this was confirmed by sequence analysis (not shown). Structural, serological, and sequence data for J-223 thus support the hypothesis that C repeat length in the α3-fucosyltransferase genes futA and futB determines Lex/y expression.
The role of an active futB gene product in determining a strong Ley expression was further investigated in strains N6, SS-1, 26695, P466, and J99. These strains all strongly expressed Ley together with Lex, as shown in ELISA; sequence analysis of futB demonstrated that it was on in all strains (not shown).
α3-Fucosyltransferase enzyme activity in 4187E α3-fucosyltransferase double-knockout mutant strain.
We used N-acetyllactosamine, a good acceptor for H. pylori α3-fucosyltransferase enzyme activity, to measure this activity in sonicates of strain 4187E-KO379/651. No such activity could be detected.
Structural information about LPS of strain 4187E and its knockouts.
The expression of Lewis blood group antigens in the LPS of strain 4187E and its α3-fucosyltransferase knockout mutants was also investigated by chemical methods. The structure of the O-chain region was investigated by FAB-MS analysis of the methylated intact LPS (21). The FAB-MS spectrum of strain 4187E methylated LPS revealed the presence of H type 1 (m/z 638→228), Lex (m/z 638→432), Ley (m/z 812→402), Galβ1-4GlcNac (LacNac) (m/z 464→432), traces of Lea (m/z 638→402), and LacNac-Lex (m/z 1087→881). Mutant 4187E-KO651 expressed H type 1, Lex, and Ley. Methylation linkage analysis of this mutant showed a significant decrease in 2-substituted galactose and terminal fucose and an increase in terminal galactose, implying a decrease in the formation of Ley and/or H type 1 expression in this LPS. The FAB-MS spectrum of mutant 4187E-KO379 showed the presence of H type 1, Lex, and Ley, with small amounts of terminal galactose being observed in the linkage analysis compared with amounts of the 2-substituted galactose. Thus, in 4187E-KO379, Lex expression seems to be weaker than H type 1 and/or Ley, both of which contain 2-substituted Gal. FAB-MS of the LPS of the double knockout clearly showed that Lex and Ley were no longer present and that this LPS expressed only H type 1, the i antigen (LacNAc-LacNac) [m/z 464→432, 913→881], and an H type 1-LacNAc-LacNAc sequence [m/z 1087→1055, 1536, and 1985]. No 3,4-substituted GlcNAc was observed in the linkage analysis, which confirmed the absence of Lex in this LPS. No Lea was observed in any of the 4187E knockouts. The simultaneous expression of H type 1 (type 1 chain), Lex, Ley, and i antigen (type 2 chains) by strain 4187E places this strain in the LPS category of the glycotype F family (21).
DISCUSSION
In this paper we provide evidence that phase variation in the LPS of H. pylori takes place through changes in the length of poly(C) repeats of the α3-fucosyltransferase genes futA and futB. Our data suggest that the LPS serotypes of phase variants and clinical isolates are determined, at least in part, by the on or off status of α3-fucosyltransferase genes. Other genes play a role in LPS biosynthesis: previously we have shown that GlcNAcT activity determines the serotypes of several phase variants (4), and for expression of Ley (35) or H type 1 antigens, α2-fucosyltransferase enzyme activity is required. The data also show that the genes futA and futB code for α3-fucosyltransferase enzymes with different specificities. The phenotypes and genotypes of LPS phase variants and α3-fucosyltransferase knockouts (Table 2) reveal that only strains with an intact futB reading frame contain terminal mono- and oligomeric Lex. We infer from this that the futB gene product efficiently fucosylates lactosamine at the residues at the nonreducing terminus of the O-antigen chain and fucosylates internal units less efficiently. The presence of terminal Lex is required for α2-fucosylation in H. pylori, and hence strains that have an active futB gene also strongly express Ley. Titration with MAb 54.1F6A, which reacts with polymeric Lex, revealed that disruption of futA caused a significant decrease (64-fold) in polymeric Lex expression. It would therefore appear that the futA gene product has an acceptor preference which is complementary to that of the futB gene product: it preferentially fucosylates internal lactosamines. The resulting nonterminal polymeric Lex structures do not provide a precursor for Ley synthesis; this is consistent with the low expression of Ley by strains which have only the futA gene intact. Similar fine specificities have been described for mammalian enzymes (11, 14). The differences between the two H. pylori α3-fucosyltransferase enzymes may be related to the different numbers of terminal leucine zipper-like repeats in these genes (Table 2). Experiments with chimeric α3-fucosyltransferase gene constructs could be used to explore this.
Strikingly, inactivation of the futB gene product, either by insertional mutagenesis or by phase variation, leads to a strongly increased H type 1 expression (Table 2). This may reflect competition between different α2-fucosyltransferase receptors. We infer that the Lex terminus, synthesized by the futB gene product, is such a good acceptor for the α2-fucosyltransferase enzyme that it effectively competes with the Galβ1→3GlcNAc acceptor termini, hampering H type 1 synthesis. This competition disappears on inactivation of the futB gene product, when Ley is formed inefficiently and more H type 1 can be formed. These data suggest that the same α2-fucosyltransferase links fucose to both type 2 (Lex) and type 1 (Galβ1→3GlcNAc) acceptors. futC (HP093/94 and orthologs) codes for this α2-fucosyltransferase enzyme (35).
The lack of H type 2 structures in the 4187E α3-fucosyltransferase double-knockout strain 4187E-KO379/651 and in strains J223 and K4.1 demonstrates that α2-fucosylation of the i chain does not take place efficiently in H. pylori. We infer that the final step of Ley biosynthesis in H. pylori is the α2-fucosylation of an Lex terminus. This is consistent with the reported negligible activity of the futB gene product with Fucα1→2Galβ1→4Glc, a good acceptor for human α3-fucosyltransferase (12, 18). For the synthesis of Ley in mammals, the prescribed order of fucosylation is the opposite of what is observed in H. pylori, i.e., first α2-fucosylation, which is a prerequisite for addition of α3-fucose (37). From the presence of the H type 1 epitope in NCTC 11637, K4.1, and J-223, we conclude that the futC gene product is able to link fucose to C-2 of β1→3-linked Gal.
The serological data on LPS of strain 4187E are confirmed by the chemical-structural information. The small amounts of 2-substituted Gal and terminal fucose, detected chemically in LPS of strain 4187E-KO651, are in agreement with the weak Ley expression detected by serology. The small amount of terminal Gal compared to the amount of 2-substituted Gal detected by structural means in LPS of strain 4187E-KO379 is in agreement with the decreased Lex expression detected by serology. Both by serology and by means of structural analysis, H type 1 and i antigen, but no Lex or Ley, were found in the double knockout mutant 4187E-KO370/651; the same applies to strain J-223, in which both futA and futB are also off.
The biological role of H. pylori LPS phase variation is still unsettled. Data from experimental-infection studies of monkeys clearly suggest an adaptive role (39). Four monkeys were colonized with the same strain. Bacteria of that strain isolated from animals that expressed Ley in their gastric mucosa also strongly expressed Ley in their LPS; bacteria isolated from monkeys that expressed Lex also strongly expressed Lex. These data suggest that the Lewis phenotype of the pathogen can vary and can adapt to that of the host. Whether this is also the case in humans is controversial: one study reported a relationship between the Lewis phenotype of the host and that of the colonizing strain of H. pylori (38); this was not confirmed in another study (30). A related question is whether the expression of Lewis antigens by H. pylori per se has any relevance to infection and disease. We are currently studying whether the Lex/y-deficient mutant strain 4187E-KO379/651 is as able to colonize mice as its parent strain.
ACKNOWLEDGMENTS
We thank R. Negrini, D. Blanchard, and G. van Dam for providing MAbs. We thank R. Alm, T. Boren, S. Krakowka, A. Labigne, A. Lee, and H. P. Wirth for strains. We thank R. Pot and A. Bart for wordprocessing.
REFERENCES
- 1.Alm R A, Ling L L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Jiang Q, Taylor D E, Voviv G F, Trust T J. Comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Appelmelk B J, Simoons-Smit I M, Negrini R, Moran A P, Aspinall G O, Forte J G, De Vries T, Quan H, Verboom T, Maaskant J J, Ghiara P, Kuipers E J, Bloemena E, Tadema T M, Townsend R R, Tyagarajan K, Crothers J M, Jr, Monteiro M A, Savio A, de Graaff J. Potential role of molecular mimicry between Helicobacter pylori lipopolysaccharide and host Lewis blood group antigens in autoimmunity. Infect Immun. 1996;64:2031–2040. doi: 10.1128/iai.64.6.2031-2040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appelmelk B J, Negrini R, Moran A P, Kuipers E J. Molecular mimicry between Helicobacter pylori and the host. Trends Microbiol. 1997;5:70–73. doi: 10.1016/S0966-842X(96)10084-6. [DOI] [PubMed] [Google Scholar]
- 4.Appelmelk B J, Shiberu B, Trinks C, Tapsi N, Zheng P Y, Verboom T, Maaskant J, Hokke C H, Schiphorst W E C M, Blanchard D, Simoons-Smit I M, van den Einden D H, Vandenbroucke-Grauls C M J E. Phase variation in Helicobacter pylori lipopolysaccharide. Infect Immun. 1998;66:70–76. doi: 10.1128/iai.66.1.70-76.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aspinall G O, Monteiro M A, Pang H, Walsh E J, Moran A P. Lipopolysaccharide of the Helicobacter pylori type strain NCTC 11637 (ATCC 43504): structure of the O antigen and core oligosaccharide regions. Biochemistry. 1996;35:2489–2497. doi: 10.1021/bi951852s. [DOI] [PubMed] [Google Scholar]
- 6.Aspinall G O, Monteiro M A. Lipopolysaccharides of Helicobacter pylori strains P466 and MO19: structures of the O antigen and core oligosaccharide strains. Biochemistry. 1996;35:2498–2504. doi: 10.1021/bi951853k. [DOI] [PubMed] [Google Scholar]
- 7.Berg D E, Hoffman P, Appelmelk B J, Kusters J G. The Helicobacter pylori genome sequence: genetic factors for long life in the gastric mucosa. Trends Microbiol. 1997;5:468–474. doi: 10.1016/s0966-842x(97)01164-5. [DOI] [PubMed] [Google Scholar]
- 8.Blanchard D, Bernard D, Loirat M J, Frioux Y, Guimbretiere J, Guimbretiere L. Characterization of murine monoclonal antibodies directed to fetal erythrocytes. Rev Fr Transfus Hemobiol. 1992;35:239–254. doi: 10.1016/s1140-4639(05)80102-9. [DOI] [PubMed] [Google Scholar]
- 8a.Breton C, Oriol R, Imberty A. Conserved structural features in eukaryotic and prokaryotic fucosyltransferases. Glycobiology. 1998;8:87–94. doi: 10.1093/glycob/8.1.87. [DOI] [PubMed] [Google Scholar]
- 9.Ciucanu I, Kerek F. A simple method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 10.Dell A, Azadi P, Tiller P, Thomas-Oates J, Jennings H J, Beurret M, Michon F. Analysis of oligosaccharide epitopes of meningococcal lipopolysaccharides by fast-atom bombardment mass spectrometry. Carbohydr Res. 1990;200:59–76. doi: 10.1016/0008-6215(90)84182-t. [DOI] [PubMed] [Google Scholar]
- 11.De Vries T, Norberg T, Lonn H, van den Eijnden D H. The use of human milk fucosyltransferase in the synthesis of tumor-associated trimeric X determinants. Eur J Biochem. 1993;216:769–777. doi: 10.1111/j.1432-1033.1993.tb18197.x. [DOI] [PubMed] [Google Scholar]
- 12.De Vries T, Palcic M P, Schoenmakers P S, Van den Eijnden D H, Joziasse D H. Acceptor specificity of GDP-Fuc:Galβ1→4GlcNAc-R α3-fucosyltransferase VI (FUCT VI) expressed in insect cells as soluble, secreted enzyme. Glycobiology. 1997;7:921–927. doi: 10.1093/glycob/7.7.921. [DOI] [PubMed] [Google Scholar]
- 13.Ge Z, Chan N W C, Palcic M, Taylor D E. Cloning and heterologous expression of an α1,3-fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J Biol Chem. 1997;272:21357–21363. doi: 10.1074/jbc.272.34.21357. [DOI] [PubMed] [Google Scholar]
- 14.Howard D R, Fukuda M, Fukuda M N, Stanley P. The GDP-fucose:N-acetylglucosaminide 3-α-L-fucosyltransferases of LEC11 and LEC12 Chinese hamster ovary mutants exhibit novel specificities for glycolipid substrates. J Biol Chem. 1987;262:16830–16837. [PubMed] [Google Scholar]
- 15.Jennings M P, Hood D W, Peak I R A, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase variable expression of the lacto-N-neotetraose terminal LPS structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 16.Kuipers E J. Helicobacter pylori and the risk and management of associated diseases: gastritis, ulcer disease, atrophic gastritis and gastric cancer. Aliment Pharmacol Ther. 1997;11(Suppl. 1):71–88. doi: 10.1046/j.1365-2036.11.s1.5.x. [DOI] [PubMed] [Google Scholar]
- 17.Logan P H, Berg D E. Genetic diversity of Helicobacter pylori. Lancet. 1996;348:1462–1463. doi: 10.1016/s0140-6736(05)65885-0. [DOI] [PubMed] [Google Scholar]
- 18.Martin S L, Edbroke M R, Hodgman T C, van den Eijnden D H, Bird M I. Lewis x biosynthesis in Helicobacter pylori: molecular cloning of an α-(1,3)-fucosyltransferase gene. J Biol Chem. 1997;272:21349–21356. doi: 10.1074/jbc.272.34.21349. [DOI] [PubMed] [Google Scholar]
- 19.McColm A. Nonprimate animal models of H. pylori infection. In: Clayton C A, Mobley H L T, editors. Helicobacter pylori protocols. Totowa, N.J: Humana Press; 1997. pp. 235–252. [Google Scholar]
- 20.Mollicone R, Cailleau A, Imberty A, Gane P, Perez S, Oriol R. Recognition of the blood group H type 2 trisaccharide epitope by 28 monoclonal antibodies and three lectins. Glycoconj J. 1996;13:263–271. doi: 10.1007/BF00731501. [DOI] [PubMed] [Google Scholar]
- 21.Monteiro M A, Chan K H, Rasko D A, Taylor D E, Zheng P Y, Appelmelk B J, Wirth H P, Yang M, Blaser M J, Hynes S O, Moran A P, Perry M B. Simultaneous expression of type 1 and type 2 Lewis blood group antigens by Helicobacter pylori lipopolysaccharides. Molecular mimicry between H. pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. J Biol Chem. 1998;273:11533–11543. doi: 10.1074/jbc.273.19.11533. [DOI] [PubMed] [Google Scholar]
- 22.Monteiro M A, Zheng P Y, Appelmelk B J, Perry M B. The lipopolysaccharide of H. mustelae type strain ATCC 43772 expresses the monofucosyl A type 1 histo-blood group epitope. FEMS Microbiol Lett. 1997;154:103–109. doi: 10.1111/j.1574-6968.1997.tb12630.x. [DOI] [PubMed] [Google Scholar]
- 23.Negrini R, Lisato L, Zanella I, Cavazzini S, Gullini S, Villanacci V, Poiesi C, Albertini A, Ghielmi S. Helicobacter pylori infection induces antibodies cross-reacting with human gastric mucosa. Gastroenterology. 1991;101:437–445. doi: 10.1016/0016-5085(91)90023-e. [DOI] [PubMed] [Google Scholar]
- 24.Negrini R, Savio A, Poiesi C, Appelmelk B J, Buffoli F, Paterlini A, Cesari P, Graffeo M, Vaira D, Franzin G. Antigenic mimicry between Helicobacter pylori and gastric mucosa in the pathogenesis of body atrophic gastritis. Gastroenterology. 1996;111:655–665. doi: 10.1053/gast.1996.v111.pm8780570. [DOI] [PubMed] [Google Scholar]
- 25.O’Croinin T O, Clyne M, Drumm B. Molecular mimicry of ferret gastric epithelial blood group antigen by Helicobacter mustelae. Gastroenterology. 1998;114:690–696. doi: 10.1016/s0016-5085(98)70582-7. [DOI] [PubMed] [Google Scholar]
- 26.Prieels J P, Monnom D, Dolmans M, Beyer T A, Hill R L. Co-purification of the Lewis blood group N-acetylglucosaminide alpha-1,4-fucosyltransferase and an N-acetylglucosaminide alpha-1,3-fucosyltransferase from human milk. J Biol Chem. 1981;256:10456–10463. [PubMed] [Google Scholar]
- 27.Roche R J, Moxon E R. Phenotypic variation of carbohydrate surface antigens and the pathogenesis of Haemophilus influenzae infections. Trends Microbiol. 1995;3:304–309. doi: 10.1016/s0966-842x(00)88959-3. [DOI] [PubMed] [Google Scholar]
- 28.Saunders N J, Peden J F, Hood D W, Moxon E R. Simple sequence repeats in the H. pylori genome. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 29.Simoons-Smit I M, Appelmelk B J, Verboom T, Negrini R, Penner J L, Aspinall G O, Moran A P, Fei S F, Shi B S, Rudnica W, de Graaff J. Typing of Helicobacter pylori with monoclonal antibodies against Lewis antigens in lipopolysaccharide. J Clin Microbiol. 1996;34:2196–2200. doi: 10.1128/jcm.34.9.2196-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor D E, Rasko D A, Sherburne R, Ho C, Jewell L D. Lack of correlation between Lewis antigen expression by Helicobacter pylori and gastric epithelial cells in infected patients. Gastroenterology. 1998;115:1113–1122. doi: 10.1016/s0016-5085(98)70082-4. [DOI] [PubMed] [Google Scholar]
- 31.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischman R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Bordovsky M, Karp P D, Smith H O, Fraser C M, Venter C J. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Dam G J, Bergwerff A A, Thomas-Oates J E, Rotmans J P, Kamerling J P, Vliegenthart J F, Deelder A M. The immunologically reactive O-linked polysaccharide chains derived from circulating cathodic antigen isolated from human blood fluke Schistosoma mansoni have Lewis x as repeating unit. Eur J Biochem. 1994;225:467–482. doi: 10.1111/j.1432-1033.1994.00467.x. [DOI] [PubMed] [Google Scholar]
- 34.Van Putten J P M, Robertson B D. Molecular mechanisms and implications for infection of lipopolysaccharide variation in Neisseria. Mol Microbiol. 1995;16:847–853. doi: 10.1111/j.1365-2958.1995.tb02312.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang G, Rosko D A, Sherburne R, Taylor D E. Molecular genetic basis for the variable expression of Lewis y antigen in Helicobacter pylori: analysis of the α(1,2)fucosyltransferase gene. Mol Microbiol. 1999;31:1265–1274. doi: 10.1046/j.1365-2958.1999.01268.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 37.Watkins W M, Greenwell P, Yates A D, Johnson P H. Regulation of expression of carbohydrate blood group antigens. Biochimie. 1988;70:1597–1611. doi: 10.1016/0300-9084(88)90295-7. [DOI] [PubMed] [Google Scholar]
- 38.Wirth H P, Yang M, Peek R M, Jr, Tham K T, Blaser M J. H. pylori Lewis expression is related to the host phenotype. Gastroenterology. 1997;113:1091–1098. doi: 10.1053/gast.1997.v113.pm9322503. [DOI] [PubMed] [Google Scholar]
- 39.Wirth H P, Yang M, Dubois A, Berg D E, Blaser M J. Host Lewis phenotype-dependent selection of H. pylori Lewis expression in Rhesus monkeys. Gut. 1998;43:A26. doi: 10.1096/fj.05-5529fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q L, Gotschlich E C. Variations of gonococcal lipopolysaccharide structure is due to alterations in poly-G tracts in lgt genes encoding glycosyltransferases. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]