Abstract
Protein kinases play a key role in signal transduction pathways in both eukaryotic and prokaryotic cells. Using in vivo expression technology, we have identified several promoters in Pseudomonas aeruginosa which are preferentially activated during infection of neutropenic mice. One of these promoters directs the transcription of a gene encoding a putative protein kinase similar to the enzymes found in eukaryotic cells. The full characterization of this protein, termed PpkA, is presented in this communication. The ppkA gene encodes a 1,032-amino-acid polypeptide with an N-terminal catalytic domain showing all of the conserved residues of protein kinases with the substrate phosphorylation specificities for serine and threonine residues. The catalytic domain is linked to the rest of the protein by a short proline-rich segment. The enzymes showed anomalous migration behavior when analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, which could be attributed to autophosphorylation activity. The full-length enzyme was expressed as an oligohistidine fusion protein and was shown to phosphorylate several artificial protein substrates. Both autophosphorylation and phosphorylation of added substrates were strongly reduced by a single-amino-acid substitution in the catalytic domain of PpkA. Although PpkA appears to be differentially phosphorylated by autocatalysis, the levels of phosphorylation have minimal effect on its overall enzymatic activity. Our results, therefore, indicate the operation of a novel protein phosphorylation mechanism during transduction of signals in P. aeruginosa, and this pathway may be important in regulating the expression of virulence factors by this pathogen during certain phases of infection.
In eukaryotic cells, regulation of signal transduction pathways through enzymatic protein phosphorylation at serine, threonine, or tyrosine residues is a widely distributed mechanism, with hundreds of protein kinases described to date (9). In prokaryotic cells, protein phosphorylation also plays a role in signal transduction involving almost exclusively phosphorylation mechanisms that are carried out by the two-component regulatory proteins, which are responsible for transcriptional activation of genes and for chemotaxis. The sensors of the two-component regulatory family are histidine autokinases that activate transcription by phosphorylation of specific aspartic acid residues of their cognate response regulators. Only recently, protein kinases similar to eukaryotic enzymes have been described in bacteria, including enzymes found in Myxococcus xanthus, Yersinia pseudotuberculosis, Streptomyces coelicolor, and Anabaena sp. strain PCC7 (17, 29). Sequences having significant similarities to the catalytic domains of Ser/Thr protein kinases have also been identified in the genomes of several archaea (21) and by the inspection of the current databases for the mycobacterial genome-sequencing projects. Although the best-characterized protein kinases are those with specificity for serines or threonines, tyrosine phosphorylation has recently been demonstrated in Escherichia coli (7).
There is good evidence that most of the prokaryotic Ser/Thr kinases carry out phosphorylation of their target proteins within the bacterial-cell cytoplasm. The Streptomyces Ser/Thr protein kinase AfsK phosphorylates the global regulator AfsR, which controls the production of certain antibiotics (15). Several distinct Ser/Thr protein kinases described in M. xanthus are involved in fruiting body formation and spore production (10, 30). It is very likely that all of these proteins play some role in signal transduction. In Bacillus subtilis, a pair of serine-specific protein kinases (RsbT and RsbW) participate in a pathway responsible for the transduction of environmental stress signals to the transcription factor ςB (11, 28). A bacterial protein kinase with a markedly different function is the Y. pseudotuberculosis product YpkA, which is translocated from the bacteria into eukaryotic cells, where it functions in altering the host cellular functions by phosphorylating target proteins in the mammalian cytoplasm (8).
Selective expression of genes of pathogenic bacteria in response to environmental signals also depends on efficient signal transduction systems for transmitting environmental information to the transcriptional machinery. Signal transduction with two-component regulatory systems is used by a number of different bacteria to control the expression of virulence factors (5). In Pseudomonas aeruginosa, these systems regulate the expression of genes specifying the alginate biosynthetic enzymes (4), as well as formation of adhesins and flagella (20). Although the molecular details of signal transduction have been reasonably well characterized, the environmental stimuli that activate the autophosphorylation of the sensory components are largely unknown.
We have previously described a genetic system for the identification of genes in P. aeruginosa that are active during infection. This technique is a direct adaptation of the in vivo expression technology (IVET) initially described by Mahan et al. (14). We have shown that a number of genes that are preferentially expressed during infection of neutropenic mice (23) or during bacterial growth in human respiratory mucus (24) can be identified by IVET. Sequencing of one such chromosomal fragment revealed a gene with similarity to the catalytic domains present in Ser/Thr protein kinases of eukaryotic and prokaryotic origin. Activation of this gene in vivo could imply a role for phosphorylation of serines and threonines in target proteins that are important for P. aeruginosa virulence. Here we describe the complete molecular characterization of this P. aeruginosa protein kinase (called PpkA) and provide additional information on its potential role in P. aeruginosa virulence.
MATERIALS AND METHODS
Bacterial culture conditions.
Cultures of E. coli and P. aeruginosa were routinely propagated in Luria broth, tryptone-yeast extract or low-phosphorus medium as indicated. Antibiotics were added at the following concentrations (in micrograms per milliliter): ampicillin, 100; kanamycin, 50 (for E. coli); and carbenicillin, 150 (for P. aeruginosa).
Plasmid construction.
One of the P. aeruginosa genes identified previously as induced in neutropenic mice (np6) was used as a probe to screen a library of P. aeruginosa DNA fragments cloned in vector pVK102 (23) by a colony hybridization method. Several cosmid clones were identified which contained sequences hybridizing with the np6 probe, and one such cosmid clone, pVNP3, was used for further subcloning of the np6-containing region. A 4.0-kb SalI fragment of pVNP3 was cloned into pUC18 to create pCSN5, and a 6.5-kb EcoRI fragment was cloned, in both orientations, in pUC19 to create pNR8 and pNR12.
The insert of pCSN5 was sequenced and found to contain the 5′ end of an open reading frame with areas of high homology to the conserved domains of eukaryotic protein kinases. The DNA fragments containing the sequence of the 3′ end of the ppkA gene were identified by Southern blot analysis of digests of pVNP3, and a 2.9-kb XhoI fragment from this region was cloned into pZero2.0 (Invitrogen) to create pZX10. This fragment was also sequenced to give the remainder of the ppkA gene sequence.
To construct plasmids expressing PpkA with an oligohistidine tag, an NdeI site at the ATG start codon of ppkA was created which allowed its cloning into plasmid pET15BVP. The NdeI site was created by PCR mutagenesis with the oligonucleotides BglII/NdeI (5′CGGCCCTGGTGGTCAGATCTTTCTGAGACGCATATGGACATAGAACTTCC3′) and lat3′ (5′CACGTAGAGCACCGAGAA3′) and the plasmid pCSN5 as the template.
This mutagenesis also introduced a BglII site 19 bases to the 5′ side of the ATG initiation codon. The PCR yielded a 900-bp fragment which was digested with BglII and XhoI, and the resulting fragment was cloned into pZero2.0 digested with BamHI and XhoI. This created plasmid pZNX8, which has the 5′ end of the ppkA gene with an NdeI site at the ATG start. pZNX8, carrying the NdeI site at the ATG start of ppkA through the ppka internal XhoI site, was digested with XhoI and XbaI and combined with the 1.8-kb XhoI-XbaI fragment of pNR8 to create pZXX2. Plasmid pZXX2 was digested with NdeI and BamHI, and the resultant 2.8-kb fragment was cloned into pET15BVP, which had been digested with NdeI and BamHI, to create pHPKA2.1. The DNA segment containing the 3′ sequence of ppkA was restored after digestion of pHPKA2.1 with EcoRI and treatment with Klenow fragment of DNA polymerase followed by digestion with NotI. The large vector fragment was combined with the DNA fragment produced by digesting pZX10 with XhoI, treatment with Klenow polymerase, and then digestion with NotI. This produced pHFAX7-1.
To create a substitution of arginine for lysine at codon 37 of ppkA, PCR mutagenesis was used. Oligonucleotides lat3′ and lys/arg-c (5′CGCTGCAGCGCAAGGTGGCGCTGCGCGTGATGGCCGCGGCGCTGGCCGC3′) were used with pCSN5 as the template. The 1.3-kb PCR product was digested with PstI and XhoI, and the 800-bp fragment was cloned into pZero2.0, creating pZPX-4. The same PCR product was cloned into the EcoRV site of pZero2.0, creating pZV3. The 1.1-kb PstI-NotI fragment of pZV3 was combined with the fragment generated by digesting pZXX2 with EcoRI, treating it with Klenow polymerase, and then digesting it with PstI and the fragment generated by digesting pZX10 with XhoI, treating it with Klenow polymerase, and then digesting it with NotI. The product of this three-fragment ligation was pZXA-5. The 4.0-kb fragment produced by digesting pZXA-5 with PstI and XmnI was combined with the 800-bp fragment produced by digesting pZPX4 with PstI-XhoI and the 1.6-kb XmnI-XhoI fragment of pZX10 to create pXX453.
The DNA fragment carrying the mutagenized codon was excised from pXX453 as a 3.1-kb NdeI-BamHI fragment and was ligated into pET15BVP digested with NdeI-BamHI to create pETXX-11. This plasmid expresses the full-length ppkA gene, with a product (PpkA-K37R) which contains an N-terminal oligohistidine extension and a substitution of an arginine for lysine at position 37.
DNA sequencing.
The inserts in the plasmids pCSN5 and pZX10 were sequenced with the use of universal oligonucleotides of the vectors and synthetic oligonucleotides based on the generated sequence. The DNA was sequenced in both directions by the chain termination method. The Big Dye Terminator reactions (Perkin Elmer Applied Biosystems) were analyzed on the ABI 377 DNA sequencer.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Whole bacterial cells were dissolved in 2% sodium dodecyl sulfate (SDS)–10% β mercaptoethanol–25 mM Tris HCl (pH 7.5)–5% glycerol (SB). The samples were denatured by boiling them for 10 min and separated on 7% SDS-polyacrylamide gels (12), and the proteins were electrophoretically transferred to nitrocellulose (22).
For detecting 32P-labeled proteins, the filters were briefly air dried, wrapped in Saran Wrap, and subjected to autoradiography. The labeled protein bands were quantified with the Molecular Dynamics PhosphorImager.
To determine the relative levels of PpkA in the samples, the filters were blocked with 5% nonfat dry milk in phosphate-buffered saline and 0.1% Tween 20 for 1 h and then probed with anti-His thrombin monoclonal antibody (a gift from M. Katze) (1:1,000) for 3 h and counterprobed with alkaline phosphatase-labeled goat anti-mouse antibody (Kirkegaard and Perry Laboratories), and the signal was detected with Nitro Blue Tetrazolium–5-bromo-4-chloro-3-indolyl phosphate (BCIP) mix (Bethesda Research Laboratories).
For separating the products of the in vitro kinase assays, the reactions were stopped by adding 4× SDS sample buffer (SB) and by SDS–15% PAGE. The gels were dried and subjected to autoradiography. Signal intensities were quantitated with a Molecular Dynamics PhosphorImager.
Purification of PpkA from P. aeruginosa.
The plasmids pHFAX7-1 and pETXX-11 were introduced into P. aeruginosa PAO ADD 1976, which carries the chromosomally incorporated gene for T7 RNA polymerase under the control of the lac repressor. The transformed strains were grown in 2× yeast extract-typtone medium at 37°C until an optical density at 600 nm of 1.0 was reached, at which time isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM final concentration) was added. The cultures were incubated for an additional 4 h, after which the bacteria were harvested and resuspended in lysis buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA, 10% sucrose, 100 μg of DNase I/ml, 50 μg of RNase A/ml). The cells were frozen, thawed, and passed through a French pressure cell at 20,000 lb/in2. The lysate was cleared of cell debris by centrifugation at 5,000 × g for 5 min, and the supernatant was centrifuged at 40,000 × g for 60 min. The pellet, which contained the majority of PpkA, was resuspended in a buffer of 10 mM NaHPO4 (pH 7.2) and solubilized after the addition of 4 volumes of 6 M guanidine-HCl, 500 mM NaCl, and 20 mM NaHPO4, pH 7.5. This material was bound to Probond resin (Invitrogen) preequilibrated in 8 M urea, 500 mM NaCl, and 20 mM NaHPO4 (pH 7.5) and was washed in batch mode twice with 4 volumes of 8 M urea, 500 mM NaCl, and 20 mM NaHPO4 (pH 7.5), followed by two more washes with 4 volumes of 8 M urea, 500 mM NaCl, and 20 mM NaHPO4 (pH 6.0). The resin was then packed into a column, and fractions were eluted with 8 M urea, 500 mM NaCl, and 20 mM NaHPO4 (pH 4.0). After the pH was adjusted to 7.2 with NaOH, the protein composition of each fraction was determined by SDS-PAGE. The fractions containing PpkA were combined and loaded on a column of Macro-Prep ceramic hydroxyapatite type I (80 μm) (CHAP; Bio-Rad) preequilibrated with 4 M urea, 250 mM NaCl, and 10 mM NaHPO4 (pH 7.2). The column was washed with 4 M urea, 250 mM NaCl, and 10 mM NaHPO4 (pH 7.2) followed by extensive washing with 4 M urea and 10 mM NaHPO4 (pH 7.2). The elution of bound protein was accomplished with an NaHPO4 gradient (10 to 200 mM) in 4 M urea at pH 7.2. The fractions containing PpkA were pooled and concentrated with Millipore Ultra 4 concentration units.
Labeling of cells with [32P] orthophosphate.
P. aeruginosa PAO ADD 1976 carrying either pET15BVP, pHFAX7-1, or pETXX-11 was grown in 2× YT medium until a density corresponding to an optical density at 600 nm of 1.0 was reached. The cells were pelleted and resuspended in low-phosphate medium [50 mM MOPS (morpholinepropanesulfonic acid; pH 7.3), 50 mM KCl, 0.8 mM MgSO4, 0.8 mM CaCl2, 0.3 mM KH2PO4, 0.5 g of sodium citrate/liter, 1 g of (NH4)2SO4/liter, 0.4% glucose, 0.2 μg of vitamin B1/ml, and each amino acid at 25 μg/ml] (16) with 1 mM IPTG and carbenicillin at 75 μg/ml. The cultures were grown with shaking at 37°C for 1 h. Rifampin was added to 150 μg/ml, and the culture was incubated with shaking for an additional 30 min. ortho-32P (NEN) was added to 10 μCi/ml of culture for 1 h, and the cells were pelleted. The cell pellets were resuspended in 1× SB.
In vitro protein kinase assays.
The kinase reactions routinely contained 750 ng of the enzyme in kinase buffer (25 mM HEPES [pH 7.4], 25 mM glycerolphosphate, 25 mM MgCl2, 2 mM dithiothreitol, 2 mM NaF) with 25 μg of each substrate and 10 μCi (3,000 Ci/mmol) of [γ-32P]ATP. Typically, after a 20-min incubation at 20°C the reactions were stopped by the addition of 4× SB. The labeled products were separated by SDS-PAGE and visualized by autoradiography. The incorporation of phosphate into proteins was quantified with a Molecular Dynamics PhosphorImager.
Purification of PpkA from cultures induced with IPTG and uninduced.
To compare the activity of PpkA from induced and uninduced cultures, PAO ADD 1976 pHFAX7-1 was grown as previously described in 2× YT medium at 37°C. No IPTG was added when the cultures reached log phase, however. The cultures were allowed to grow for an additional 4 h, and the cells were harvested. The PpkA was purified as described previously with Probond resin. Peak fractions were identified by SDS-PAGE. Those fractions were combined, and the pH was adjusted to pH 7.2 with NaOH. The protein was concentrated, and the buffer was changed to 4 M urea and 10 mM NaHPO4 (pH 7.2) with Millipore Ultra 4 concentration units. The protein concentration was estimated from SDS-PAGE gels loaded with known amounts of a standard protein stained with Coomassie brilliant blue.
Comparison of kinase activity of PpkA from uninduced and induced cultures of PAO ADD 1976 pHFAX7-1.
The kinase reactions contained 100 ng of PpkA (from uninduced or induced cultures) in kinase buffer with 50 μg of myelin basic protein (MBP) and 20 μCi (3,000 Ci/mmol) of [γ-32P]ATP in a 100-μl total reaction volume. Aliquots (15 μl) were taken at various time points. The reactions were stopped by adding 5 μl of 4× SB. The labeled products were separated by SDS-PAGE, visualized by autoradiography, and quantitated with a Molecular Dynamics PhosphorImager.
Nucleotide sequence accession number.
The ppkA nucleotide sequence data was deposited in the GenBank database under accession no. AF160512.
RESULTS
An in vivo-expressed gene of P. aeruginosa encodes a Ser/Thr protein kinase.
IVET was previously used to identify the promoters of a number of genes that are activated when P. aeruginosa infects neutropenic mice (23). One such promoter was found to transcribe a gene with a partial sequence identity to the catalytic domains of eukaryotic protein kinases (23). The chromosomal DNA fragment which was located adjacent to the reporter gene (purEK) of the IVET selection vector was used as a probe to screen a bank of P. aeruginosa DNA in a cosmid vector, pVK102, by colony hybridization. Subclones were generated from one of the probe-reactive cosmids which contained the full-length gene, and these clones were used for determination of the complete nucleotide sequence of the gene encoding the putative protein kinase, which will be referred to as PpkA (Pseudomonas protein kinase A). The purEK reporter gene of the IVET selection vector was fused 477 bp into the 5′ coding region of the ppkA gene, as shown in Fig. 1A.
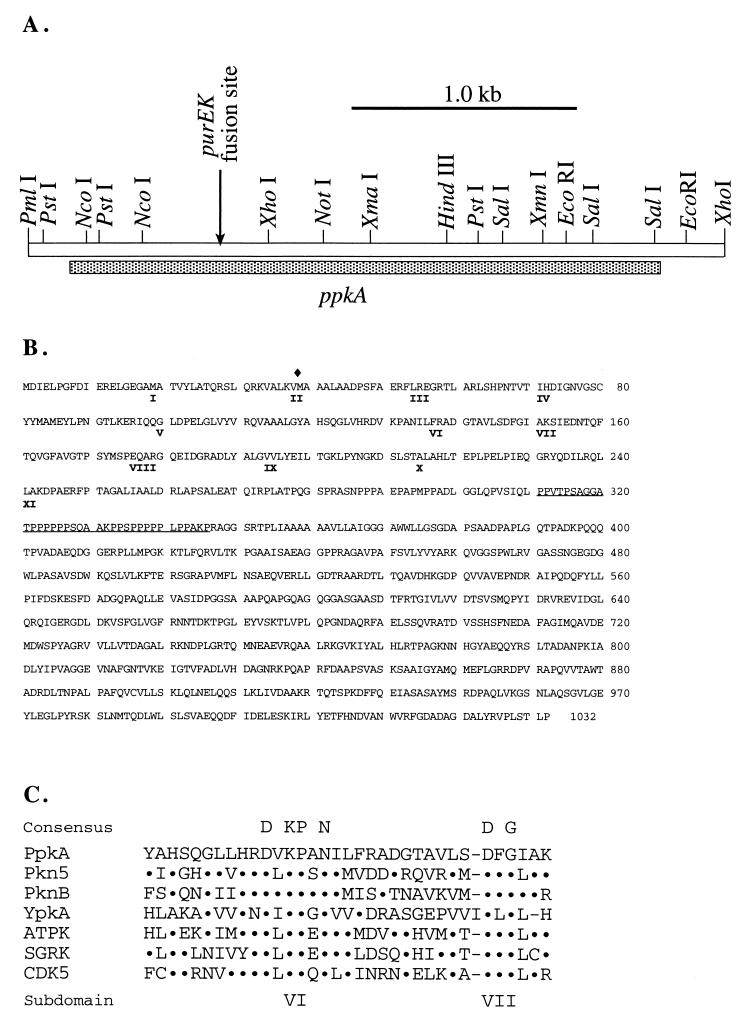
FIG. 1.
Restriction map of the ppkA locus and sequence analysis of the ppkA gene. (A) Restriction map of the PmlI-XhoI chromosomal segment containing ppkA. Shown is the location of the open reading frame encoding PpkA (shaded bar). The site at which the promoter-containing fragment of ppkA was fused to the purEK gene in the IVET selection system is indicated by an arrow. (B) Deduced amino acid sequence of the ppkA gene product. The roman numerals denote the positions of conserved subdomains present in the eukaryotic and prokaryotic Ser/Thr protein kinases. The diamond indicates the site of the K37R mutation. The underlined amino acids indicate the unusually proline-rich (53%) region of the protein. (C) Comparison of the amino acid sequences of the most conserved subdomains (VI and VII) of the catalytic sites of Ppka with the sequences of the other protein kinases. Pkn5, M. xanthus P54737; PknB, Mycobacterium leprae P54744; YpkA, Yersinia enterocolitica 1401295; ATPK1, ribosomal protein S6 kinase of Arabidopsis thaliana P42818; SGRK, serum- and glucocorticoid-regulated rat protein kinase A48094; CDK5, human cell division protein kinase Q00535.
The ppkA gene encodes a protein of 1,032 amino acids with several notable features (Fig. 1B). The subdomains of PpkA, which are present in all protein kinases, are located in the N-terminal 250 amino acids (9). This region of PpkA has significant homology with other members of the protein kinase family. The remaining portion of the protein has no significant similarity to any other protein sequence in the data banks. The similarities among the various protein kinases are strongest in the conserved subdomains VI and VII (Fig. 1C). Subdomain VI contains the consensus sequence DVKPAN, which indicates that the enzyme is a Ser/Thr-specific protein kinase (9). Computer-assisted comparison (BLAST and National Center for Biotechnology Information) (1) of the first 250 amino acids of PpkA with other protein kinases showed comparably high scores for both eukaryotic and prokaryotic Ser/Thr kinases, indicating that this sequence similarity reflects the conservation of the domains related to the enzymatic mechanism of phosphorylation.
The most striking feature of the region outside of the catalytic domain is the unusual clustering of proline residues between residues 280 and 355, in a segment immediately adjacent to the catalytic domain. This 75-amino-acid region contains 29 prolines, with a very high concentration in two segments at positions 287 to 297 and at 322 to 346. This portion of the protein may play a role in the binding of PpkA to other proteins, such as substrates, regulators, or cofactors (26). No other motifs could be detected in PpkA with the computer-assisted analysis programs BLOCKS (Fred Hutchinson Cancer Research Center, Seattle, Wash.) and MOTIF (GenomeNet, Kyoto University, Kyoto, Japan).
Expression of PpkA in P. aeruginosa.
In order to characterize the enzymatic properties of PpkA, an expression system was used to allow overproduction and purification of PpkA. Site-directed mutagenesis was used to create a unique NdeI site at the ATG codon in ppkA, which allowed cloning of ppkA into the broad-host-range expression vector pET15BVP (2). This construct (pHFAX7-1) directs expression of PpkA from a T7 promoter and specifies a full-length PpkA protein with an additional N-terminal 20-amino-acid extension, which includes a polyhistidine region (six residues). This N-terminal extension not only facilitates affinity purification of PpkA but also provides a means of detection for PpkA, using a monoclonal antibody directed towards this 20-amino-acid peptide addition.
Plasmid pHFAX7-1 was introduced into P. aeruginosa PAO ADD 1976, which carries a chromosomally integrated T7 RNA polymerase gene under the control of the lac repressor. Induction of T7 RNA polymerase synthesis with IPTG was followed by SDS-PAGE analysis and immunodetection of PpkA with an antibody directed toward the N-terminal oligohistidine-containing extension. When extracts were analyzed by SDS-PAGE and Western immunoblotting, a broad and diffuse band with a mobility of ca. 130 kDa was detected in extracts expressing PpkA and was absent in cells carrying the vector plasmid pET15BVP.
In order to characterize the protein kinase activity of PpkA, a mutant enzyme was also engineered by site-directed mutagenesis of a codon for an amino acid residue in the catalytic site. A highly conserved lysine in subdomain II (corresponding to K37 in PpkA), which has been shown to be essential for the binding of ATP by other protein kinases (13), was changed to arginine. When the plasmid pETXX-11 (pET15BVP based), which contains the gene encoding this mutant protein (PpkA-K37R), was introduced into P. aeruginosa PAO ADD 1976, the product was detected as a single sharp band migrating with the mobility of a 120-kDa protein (Fig. 2). This is in contrast to the broad band for the wild-type protein, which migrated slightly more slowly. Furthermore, in contrast to overexpression of PpkA, the mutant protein was usually recovered in much higher yields than its wild-type counterpart. The differences in migration of the two enzymes could be the consequence of alterations in the relative electrophoretic mobilities of the proteins, resulting from the Arg-for-Lys substitution. Alternatively, the mobility during electrophoresis in SDS may reflect different posttranslational modifications that either the wild-type or mutant protein undergoes in P. aeruginosa during its synthesis. This possibility was examined in detail (see below).
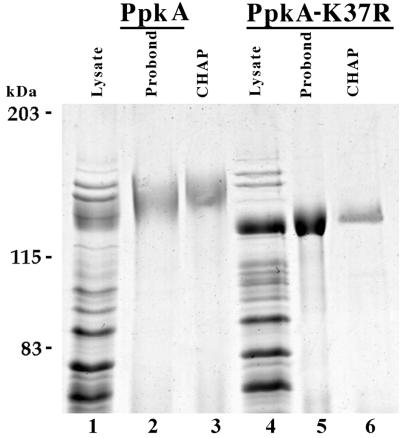
FIG. 2.
Overexpression and purification of wild-type PpkA and its active-site mutant PpkA-K37R. Plasmids expressing wild-type PpkA (pHFAX7-1) and mutant PpkA-K37R (as oligohistidine-tagged fusions) were introduced into P. aeruginosa PAO ADD 1976. The T7 polymerase gene was induced with IPTG, and the protein was purified from cell lysates by sequential nickel affinity (Probond) and ceramic (CHAP) chromatography steps, as described in Materials and Methods. Representative samples from each of the purification steps were analyzed by SDS-PAGE and Coomassie blue staining. Molecular mass markers are indicated in kilodaltons. Lanes 1 to 3, P. aeruginosa PAO ADD 1976 (pHFAX7-1); lanes 4 to 6, P. aeruginosa PAO ADD 1976 (pETXX-11); lanes 1 and 4, total cell lysate; lanes 2 and 5, Probond column fraction; lanes 3 and 6, CHAP column fraction.
Protein kinase activity of PpkA.
To purify PpkA and PpkA-K37R, cell lysates were separated into soluble and insoluble fractions by differential centrifugation. Both the wild-type and mutant forms of PpkA were present in the insoluble fraction, presumably forming inclusion bodies (data not shown). The insoluble material was collected by low-speed centrifugation and dissolved in guanidinium HCl. Purification of PpkA and of PpkA-K37R was accomplished by nickel affinity chromatography in the presence of 4 M urea followed by additional chromatography on CHAP columns. SDS-PAGE analysis of samples of pooled fractions from each step that contained the highest concentrations of proteins that reacted with the antibody to the 20-amino-acid tag is shown in Fig. 2. Greater than 90% purity, as estimated by SDS-PAGE, was obtained for each protein preparation. Interestingly, a slight increase in the mobilities of these proteins was observed during the course of the purification; however, Western blotting with the monoclonal antibody directed toward the tag showed that the purified preparation corresponded to the proteins detected in Coomassie blue-stained gels.
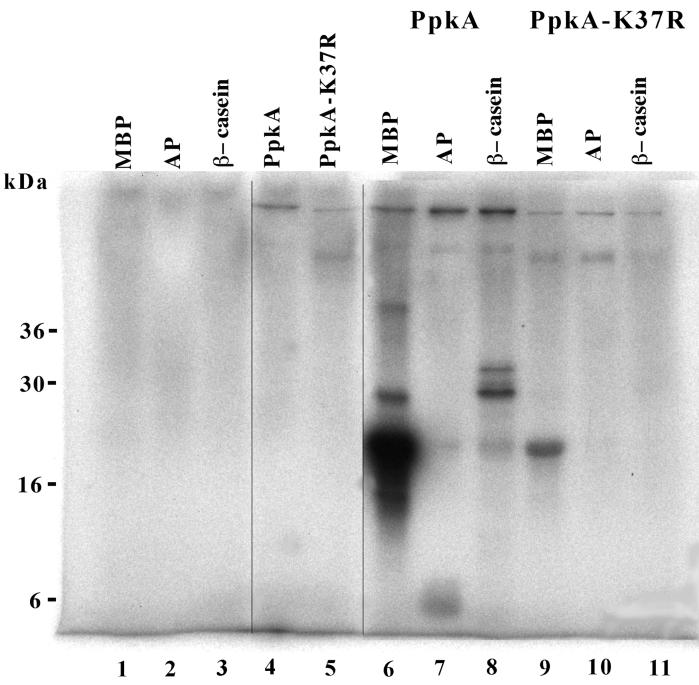
The ability of PpkA to phosphorylate a variety of exogenous substrates was examined. Purified PpkA or PpkA-K37R was added to reaction mixtures containing [γ-32P]ATP and three different protein substrates: MBP, β-casein, and aprotinin. The reaction products were separated by SDS-PAGE, and the labeled proteins were identified by autoradiography of the dried gels. As shown in Fig. 3, PpkA phosphorylated all three substrates to different extents, with the most extensive incorporation of 32P into MBP. In contrast, the mutant PpkA-K37R only slightly phosphorylated MBP and did not phosphorylate β-casein or aprotinin. The relative levels of MBP phosphorylation catalyzed by the wild-type and the mutant forms of PpkA were determined by densitometric analysis of the PhosphorImager-scanned autoradiograms. When corrected for the amount of enzyme present, the conservative amino acid substitution in PpkA-K37R drastically affected the ability of the enzyme to phosphorylate MBP, with only 2.5% of the residual activity of the wild-type PpkA remaining in the mutant. Furthermore, almost no phosphorylation of β-casein or aprotinin by the PpkA-K37R mutant was observed, and this prevented a meaningful comparison between the two enzymes with additional substrates. Low-level enzymatic activity of similar mutations in the ATP binding domains of other protein kinases has also been reported (13).
FIG. 3.
Protein kinase activity of PpkA and PpkA-K37R. Purified PpkA and PpkA-K37R were incubated with [32P]ATP and exogenous substrates as described in Materials and Methods. The proteins were separated by SDS–15% PAGE and visualized by autoradiography. Lane 1, MBP; lane 2, aprotinin (AP); lane 3, β-casein; lane 4, PpkA; lane 5, PpkA-K37R; lane 6, PpkA with MBP; lane 7, purified PpkA with aprotinin; lane 8, purified PpkA with β-casein; lane 9, PpkA-K37R with MBP; lane 10, PpkA-K37R with aprotinin; lane 11, PpkA-K37R with β-casein.
Phosphorylation of PpkA in P. aeruginosa.
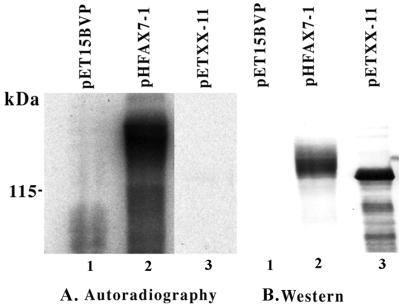
In order to determine whether PpkA itself is a substrate of another P. aeruginosa protein kinase, in vivo labeling with radioactive phosphorus was employed. P. aeruginosa PAO ADD 1976 carrying pHFAX7-1 or pETXX-11 was grown in low-phosphate medium (16) in the presence of IPTG. Host cell protein expression was inhibited by rifampin treatment, and the cells were incubated with ortho-[32P]phosphate followed by the analysis of extracts by SDS-PAGE. Figure 4 shows a comparison of the extent of incorporation of 32P into PpkA and the active-site mutant PpkA-K37R following blotting of SDS-PAGE-resolved proteins onto nitrocellulose. The same blot was also subjected to Western analysis with antibody directed toward the N-terminal oligohistidine-containing extension, thus allowing for the approximation of the quantity of PpkA and PpkA-K37R present in the extracts. When the levels of phosphorylated enzyme were corrected for differences in the protein present, the incorporation of 32P into wild-type PpkA was significantly higher than the radioactive phosphorus detected in the enzyme with the active-site mutation. The extent of labeling of PpkA-K37R was ca. 3% of that of the wild type. This value approximately corresponds to the relative phosphorylation activity of PpkA-K37R with MBP as a substrate (Fig. 4). The close correlation between the protein kinase activities of the wild type and the active-site mutant of PpkA, along with the ability of these proteins to serve as phosphorylation substrates, suggests that the observed in vivo labeling of PpkA is due to the autophosphorylation activity of this enzyme. Furthermore, the diffuse bands of lower mobility during SDS-PAGE observed for PpkA, when compared with PpkA-K37R, are very likely the consequence of differentially phosphorylated forms of the wild-type enzyme that have different electrophoretic mobilities.
FIG. 4.
In vivo phosphorylation of PpkA and PpkA-K37R. P. aeruginosa PAO ADD 1976 carrying pET15BVP (empty vector), pHFAX7-1 (expressing wild-type PpkA), or pETXX-11 (expressing PpkA-K37R) was grown in phosphate-limiting conditions and labeled with ortho-32P as described in Materials and Methods. Total cell lysates were separated by SDS-PAGE and blotted to nitrocellulose membranes. The labeled proteins were visualized by autoradiography (A), and the same blot was probed with antibody to the polyhistidine tag (B). (A) Lane 1, PAO ADD 1976 pET15BVP; lane 2, PAO ADD 1976 pHFAX7-1; lane 3, PAO ADD 1976 pETXX-11. (B) Lane 1, PAO ADD 1976 pET15BVP; lane 2, PAO ADD 1976 pHFAX7-1; lane 3, PAO ADD 1976 pETXX-11.
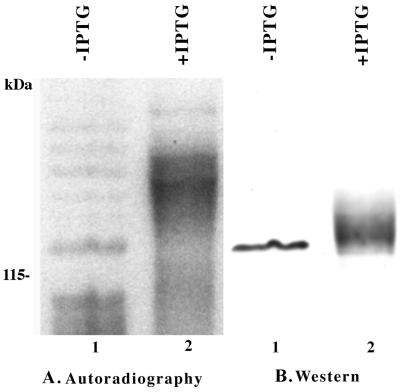
To directly address the possibility that PpkA is autophosphorylated and that this modification alters its protein kinase activity, the relatively underphosphorylated protein was purified from P. aeruginosa PAO ADD 1976 without prior induction with IPTG. We took advantage of the observation that in the absence of inducer this strain, carrying plasmid pHFAX7-1, expresses a low level of a relatively homogeneous and apparently underphosphorylated form of PpkA. Figure 5A is the autoradiograph of an extract of P. aeruginosa PAO ADD 1976 pHFAX7-1 grown in phosphate-limiting medium with or without the addition of IPTG and labeled with ortho-32P as previously described. Figure 5B shows the relative amounts of PpkA in the same membrane probed with the antibody to the oligohistidine-containing extension. Based on the comparison of the autoradiograph and Western blot, we concluded that in the absence of IPTG a relatively small amount of the enzyme is made and it is underphosphorylated or phosphorylated at a single site while the overexpressed form appears to be multiply phosphorylated.
FIG. 5.
In vivo phosphorylation of PpkA. Duplicate cultures of P. aeruginosa PAO ADD 1976 carrying pHFAX7-1 (PpkA) were grown to mid-exponential phase in phosphate-limiting medium followed by induction of one of the cultures with IPTG and rifampin treatment. The cells were labeled with ortho-32P for 1 h. Total cell lysates were separated by SDS-PAGE and blotted to nitrocellulose. The labeled proteins were visualized by autoradiography. The same membranes were then probed with the anti-tag antibody. (A) Lane 1, P. aeruginosa PAO ADD 1976 pHFAX7-1 (uninduced); lane 2, P. aeruginosa PAO ADD 1976 pHFAX7-1 (induced). (B) Lane 1, P. aeruginosa PAO ADD 1976 pHFAX7-1 (uninduced); lane 2, P. aeruginosa PAO ADD 1976 pHFAX7-1 (induced).
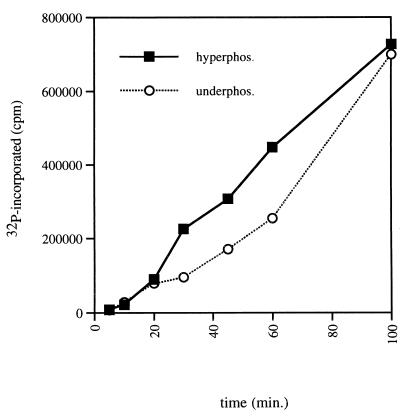
We purified this underphosphorylated form of the protein from extracts and compared its activity to that of the higher-molecular-weight hyperphosphorylated form. The two forms of PpkA were compared in an in vitro kinase assay with MBP and [γ-32P]ATP. Comparison of the kinase activities of PpkA purified from induced (hyperphosphorylated) and uninduced (underphosphorylated) extracts is shown in Fig. 6 and reveals only minimal differences in the activities of the two forms of the protein. Autophosphorylation at multiple sites, therefore, does not appear to play a major role in mediating the kinase activity of PpkA and may modulate another activity of the enzyme.
FIG. 6.
Phosphorylation of MBP by purified PpkA. Two forms of PpkA were purified from cultures of P. aeruginosa PAO ADD 1976 pHFAX7-1 with and without induction with IPTG, respectively. Normalized amounts (100 ng) of the purified underphosphorylated and hyperphosphorylated proteins were incubated with [γ-32P]ATP and MBP. Aliquots were removed at various time points, and the proteins were separated by SDS–15% PAGE. The amount of 32P incorporation into MBP was quantitated following phosphorimage analysis of the gels.
DISCUSSION
Modification of proteins by phosphorylation at a limited number of amino acid residues is a general method of controlling activities of proteins in both eukaryotic and prokaryotic cells. The existence of hundreds of different serine/threonine or tyrosine kinases in eukaryotic cells suggests a central role of protein phosphorylation in regulating virtually every function in a living cell, ranging from environmental response to development. The less ubiquitous histidine kinases, which are members of the bacterial two-component signal-transducing systems, are most likely present in all bacterial species, as suggested by microbial genomic analysis. The number of distinct histidine kinases varies within the bacterial species and may reflect the range of niches that a particular bacterium can occupy, with a concomitant need to process a large number of different environmental signals.
This view of distinct prokaryotic and eukaryotic modes of signal transduction by specific protein phosphorylation mechanisms has recently been altered by discovery of two-component signal transducers in plants and a variety of lower eukaryotes (27). Similarly, protein kinases with specificities for serines, threonines, and tyrosines have been described in bacteria. This further underscores the importance of protein phosphorylation as a key means of signal transduction in all living cells, where this mechanism allows not only efficient unidirectional signal transduction but also an efficient means of rapid amplification or deamplification of the transduced signal by interplay of phosphorylation and dephosphorylation steps catalyzed by protein kinases and phosphatases, respectively.
Here we report the identification of a gene, ppkA, which encodes a protein with a sequence and functional characteristics that resemble those of eukaryotic serine/threonine kinases. The ppkA gene was identified with the IVET system to study genes in P. aeruginosa that are preferentially expressed following infection of neutropenic mice. The deduced sequence of the ppkA gene product revealed complete conservation of all of the subdomains that make up the enzymatic portion of Ser/Thr protein kinases. We have demonstrated that ppkA encodes a functional enzyme capable of phosphorylating several exogenous substrates in vitro. Moreover, PpkA is itself phosphorylated by an autocatalytic mechanism, as shown by incorporation of added phosphorus into PpkA expressed in P. aeruginosa and lack of phosphorylation of the enzyme by a mutant with a substitution of an amino acid in its active site.
The analysis of the polypeptide sequence of PpkA revealed some resemblance to a modular organization. The amino-terminal quarter of the 1,032-amino-acid polypeptide contains the catalytic domain of the enzyme. Immediately adjacent to the catalytic domain is a proline-rich sequence. The C-terminal two-thirds of the protein lacks any distinguishing features or motifs. The presence of a high concentration of prolines in a 35-amino-acid region immediately adjacent to the catalytic domain suggests a specialized function for this segment that may be related to the enzymatic activity of PpkA. Alternatively, the proline-rich segment may simply function as a “linker” sequence between the amino-terminal catalytic domain and the carboxy-terminal domain of unknown function. Finally, we cannot exclude the possibility that this proline-rich region is a binding domain for the natural substrate(s) of PpkA or for any other P. aeruginosa protein which modulates the function of PpkA in bacteria.
The purification of PpkA as an oligohistidine-tagged fusion protein revealed an unusual migration pattern when the protein was analyzed by SDS-PAGE. The enzyme migrated as a diffuse band with an Mr of approximately 130,000, suggesting a size which is significantly larger than the size predicted by the sequence. This anomalous migration may be due to autophosphorylation as well as the high proline content of PpkA, since an active-site mutation yields a species that migrates somewhat faster but is still significantly larger than the Mr of 112,000 predicted from the sequence. Anomalous migration of proline-rich proteins has been observed previously for a number of proteins, including cytoadherence proteins of Mycoplasma pneumoniae (18) and the tracheal colonization factor produced by Bordetella pertussis (6). Moreover, the diffuse anomalous migration of the enzyme very likely reflects the different phosphorylated species, similar to that observed for Pkn9, a Ser/Thr kinase recently described in M. xanthus (10), and Pkn6, another Ser/Thr kinase described in M. xanthus (29). In vivo phosphorylation of PpkA further revealed that the most significant incorporation of [32P]phosphate is seen in slower-migrating species relative to the protein detected by Western immunoblotting. This difference in mobility of the hyperphosphorylated protein relative to the total enzyme is in agreement with a similar difference seen between wild-type PpkA and the active-site mutant PpkA-K37R, where the wild-type, highly phosphorylated PpkA migrates more slowly than the underphosphorylated active-site mutant of PpkA. A similar pattern of migration during SDS-PAGE was observed for the Ser/Thr kinase Pkn6 of M. xanthus (29). We interpret these results as evidence for continuous phosphorylation of PpkA following its expression, resulting in a heterogeneous mixture of molecules, with the most hyperphosphorylated and slower-migrating species present as a relatively minor fraction of the total population of PpkA molecules.
We compared the specific activities of the underphosphorylated and hyperphosphorylated forms of the enzyme. With MBP as a substrate, no significant differences were found between the underphosphorylated species, purified from the lysates of P. aeruginosa expressing low levels of PpkA as a result of the leaky tac promoter in the cloning vector pMMB67, and the putative phosphorylated species found in fully induced cultures where ppkA is transcribed from a promoter controlled by bacteriophage T7 RNA polymerase. This result suggests that although PpkA is capable of undergoing autophosphorylation, this modification does not alter its general catalytic activity. Therefore, unlike many eukaryotic protein kinases, autophosphorylation serves a function other than modifying the activity of the enzyme. However, autophosphorylation may still play a role in controlling the enzymatic activity of PpkA on its native substrates, perhaps stabilizing the enzyme-substrate complex or altering interaction of PpkA with an unknown regulatory protein that escaped detection under conditions in which PpkA is overproduced.
PpkA lacks any recognizable segments with similarities to known export signals, and we were unable to detect this protein in an extracellular form. Moreover, we were unable to identify any fraction of the enzyme in the extracellular medium by using recombinant constructs of PpkA (data not shown). Therefore, PpkA is most likely retained in the P. aeruginosa cytoplasm. PpkA, therefore, differs from YpkA, the extracellular enzyme of Y. pseudotuberculosis, which functions after being targeted to mammalian cells. The cytoplasmic localization of PpkA suggests that it functions within the P. aeruginosa cell and is most likely part of a signal transduction pathway. At this time, the nature of the substrate(s) of PpkA is not known. Given the high-level expression of PpkA during P. aeruginosa infection of neutropenic mice, compared to that in bacteria grown in standard laboratory media (23), we hypothesize that this enzyme plays a role in transmitting an environmental signal, either directly or through a transcriptional activator, to genes that may function in virulence or allow bacterial adaptation to a mammalian environment during the initial phase of infection. We constructed a deletion derivative of ppkA, and this mutant was tested in several animal models of infection. No difference in virulence compared to that of the wild type was detected in the agar bead model (2a), in a murine corneal-scratch model (6a), or in a mouse gut colonization model (19a). Interestingly, this mutant showed no effect on virulence in a neutropenic-mouse model (10a), even though the ppkA gene is overexpressed when P. aeruginosa infects immunosuppressed mice. Collectively, these observations suggest that ppkA may respond to a general signal present in the mammalian host but the virulence genes that it controls may not be essential for survival of the bacteria in the particular infection model. The substrates of PpkA, however, may still be essential for more relevant infections, such as the chronic respiratory infection of humans with cystic fibrosis. The elucidation of the signaling pathway involving PpkA, including the identification of all of the substrates of this enzyme, will require the utilization of techniques that detect subtle phosphorylation differences in proteins when PpkA is present at physiological levels. Overexpression of the wild-type enzyme in P. aeruginosa results in a severe growth defect, as observed in low cell yields in cultures where ppkA was induced, presumably due to artificial phosphorylation of one or several essential proteins. Nevertheless, the involvement of Ser/Thr kinases in the pathogenic process and the existence of these enzymes in other bacterial pathogens (19) suggest that they may play a broad role in sensing the various environments encountered by the bacteria and provide distinct response mechanisms which are different from those mediated by classical two-component systems. It may also be interesting to determine whether a particular signaling pathway simultaneously utilizes the two-component phosphorelay mechanism and phosphorylation at Ser/Thr residues of target proteins. Such interplay between the two mechanisms of protein phosphorylation has been suggested for regulation of the yeast Hog1 kinase through a pathway involving two-component protein kinases and protein kinases of the MAPK family (26).
After the manuscript was completed, we became aware of a paper by Wang et al. (25) reporting characterization of a gene which is identical to the ppkA gene reported here. The major discrepancy between these independent reports is the demonstration of a virulence defect in a P. aeruginosa strain, constructed by Wang and coworkers, which carries a mutation virtually identical to the one constructed by us. As indicated, we were unable to show an effect on virulence in several animal models, including the neutropenic-mouse model. The basis of the differences in the virulence properties of the two very similar mutants in unclear.
ACKNOWLEDGMENTS
This work was supported by grant AI-41479 from the NIH.
We thank Gita Bangera, Lisa Stevens, Phil Bergman, and Jeffrey Ichikawa for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Boyd, J. Personal communication.
- 3.Carrera A C, Alexandrov K, Roberts T M. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci USA. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deretic V, Mohr C D, Martin D W. Mucoid Pseudomonas aeruginosa in cystic fibrosis: signal transduction and histone-like elements in the regulation of bacterial virulence. Mol Microbiol. 1991;5:1577–1583. doi: 10.1111/j.1365-2958.1991.tb01903.x. [DOI] [PubMed] [Google Scholar]
- 5.Dzieman M, Mekalanos J J. Two-component signal transduction and its role in the expression of bacterial virulence. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 305–317. [Google Scholar]
- 6.Finn T M, Stevens L A. Tracheal colonization factor: a Bordetella pertussis secreted virulence determinant. Mol Microbiol. 1995;16:625–634. doi: 10.1111/j.1365-2958.1995.tb02425.x. [DOI] [PubMed] [Google Scholar]
- 6a.Fleiszig, S. Personal communication.
- 7.Freestone P, Grant S, Toth I, Norris V. Identification of phosphoproteins in Escherichia coli. Mol Microbiol. 1995;15:573–580. doi: 10.1111/j.1365-2958.1995.tb02270.x. [DOI] [PubMed] [Google Scholar]
- 8.Hakansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 9.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 10.Hanlon W A, Inouye M, Inouye S. Pkn9, a Ser/Thr protein kinase involved in the development of Myxococcus xanthus. Mol Microbiol. 1997;23:459–471. doi: 10.1046/j.1365-2958.1997.d01-1871.x. [DOI] [PubMed] [Google Scholar]
- 10a.Jin, S. Personal communication.
- 11.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Yu J C, Shin D Y, Pierce J H. Characterization of a protein kinase C-Δ (PKC-Δ) ATP binding mutant. An inactive enzyme that competitively inhibits wild type PKC-Δ enzymatic activity. J Biol Chem. 1995;270:8311–8318. doi: 10.1074/jbc.270.14.8311. [DOI] [PubMed] [Google Scholar]
- 14.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto A, Hong S, Ishizuka H, Horinouchi S, Beppu T. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene. 1994;146:47–56. doi: 10.1016/0378-1119(94)90832-x. [DOI] [PubMed] [Google Scholar]
- 16.Munoz-Dorado J, Inouye S, Inouye M A. Gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a Gram-negative bacterium. Cell. 1991;67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 17.Ogawara H, Urabe H. Cloning, sequencing and expression of serine/threonine kinase-encoding genes from Streptomyces coelicolor A3(2) Gene. 1995;153:99–104. doi: 10.1016/0378-1119(94)00789-u. [DOI] [PubMed] [Google Scholar]
- 18.Ogle K F, Lee K K, Krause D C. Nucleotide sequence analysis reveals novel features of the phase-variable cytoadherence accessory protein HMW3 of Mycoplasma pneumoniae. Infect Immun. 1992;60:1633–1641. doi: 10.1128/iai.60.4.1633-1641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peirs P, DeWit L, Braibant M, Huygen K, Content J A. Serine/threonine protein kinase from Mycobacterium tuberculosis. Eur J Biochem. 1997;244:604–612. doi: 10.1111/j.1432-1033.1997.00604.x. [DOI] [PubMed] [Google Scholar]
- 19a.Ramphal, R. Personal communication.
- 20.Simpson D A, Ramphal R, Lory S. Characterization of Pseudomonas aeruginosa fliO, a gene involved in flagellar biosynthesis and adherence. Infect Immun. 1995;63:2950–2957. doi: 10.1128/iai.63.8.2950-2957.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Mushegian A, Lory S, Jin S. Large scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Li C, Yang H, Mushegian A, Jin S. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J Bacteriol. 1998;180:6764–6768. doi: 10.1128/jb.180.24.6764-6768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wurgler-Murphy S M, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C. Bacterial signalling involving eukaryotic-type protein kinases. Mol Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Inouye M, Inouye S. Reciprocal regulation of the differentiation of Myxococcus xanthus by Pkn5 and Pkn6, eukaryotic-like Ser/Thr protein kinases. Mol Microbiol. 1996;20:435–447. doi: 10.1111/j.1365-2958.1996.tb02630.x. [DOI] [PubMed] [Google Scholar]