Abstract
Genotyping of Coxiella burnetii using multispacer sequence typing (MST) and multiple locus variable number tandem repeat analysis (MLVA) was conducted from infected animals for the first time in the Republic of Korea. C. burnetii was detected by real-time PCR, and followed by MST and MLVA genotyping. The result showed that detected C. burnetii all had the same MLVA genotype, 6-13-2-7-9-10 for markers MS23-MS24-MS27-MS28-MS33-MS34, respectively, and genotype group 61 for MST. The same genotypes were previously identified in Poland. Importantly, this MLVA type was detected in humans in France, suggesting that the Korean strain can also potentially cause Q fever in humans. MST and MLVA were very useful tools for analyzing the molecular epidemiology of C. burnetii and helpful for interpreting the epidemiological relationship between isolates from domestic and international resources.
Keywords: Coxiella burnetii, multiple locus variable number tandem repeat analysis, multispacer sequence typing, Q fever
1. Introduction
C. burnetii is an obligate intracellular bacterium, a causative agent of Query (Q) fever in human and animals [1]. C. burnetii-infected people might have clinical illness after one to three weeks, with symptoms of flu, fever, headache, diarrhea, night sweats, and abdominal or chest pains [2]. The zoonotic disease has been dispersed worldwide and has become an emerging threat to public health [3]. Ruminants are the main natural reservoirs of C. burnetii, which can be found in urine, faeces, milk, or birth products of infected animals [1,4,5]. Human infection mainly occurs via inhalation of contaminated aerosols. However, consumption of raw animal product can also result in the infection of C. burnetii [6]. Identification of the sources of C. burnetii and molecular characterization of the bacterial strain is necessary to identify the link between epidemiological sources and human infection [7,8].
Several techniques have been developed for genotyping and characterization of C. burnetii. Earlier developed methods were based on the analysis of PCR-restriction fragment length polymorphism [9,10], restriction-endonuclease-digested DNA separated by SDS-PAGE [11], and sequence analysis of the com1 and mucZ genes [12]. However, these methods have a limitation of inter- and intra-laboratory reproducibility [13]. Afterwards, the high-resolution multispacer sequence typing (MST) [14] and multiple locus variable number tandem repeat analysis (MLVA) methods [15,16] were developed and used extensively to date for epidemiological tracking and for identifying the phylogenetic relationship of C. burnetii isolates from different regions [17,18].
In the Republic of Korea (ROK), C. burnetii has been widely detected in cattle [19], and in ticks infesting cattle [20]. C. burnetii infection in a human who had contact with cattle was reported in 2020 [21]. The patient worked as a dairy cattle raiser and was diagnosed with C. burnetii infection after one month of fever. Therefore, the pathogen in infected cattle could be a potential source of human infection. However, a molecular method to identify the circulation of the bacterium in infected animals and humans has not been introduced in the country, and therefore it is necessary to determine the sources of human infection and the relationship between C. burnetii isolates from domestic and international resources. Therefore, this study was conducted to characterize the genotype of C. burnetii detected in vaginal swabs of cattle in ROK using MST and MLVA.
2. Materials and Methods
2.1. Samples
A total of eight vaginal swab samples from cattle with C. burnetii infection were used for genotyping analysis. The samples were collected from cattle in the Jeonnam (n = 1) and Chungnam (n = 7) provinces in ROK in 2021.
2.2. DNA Extraction and Detection of C. burnetii
DNA extraction from swab samples was performed using QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany). The procedure of DNA extraction was performed according to the instructions from the kit. Detection of C. burnetii in the samples was performed by ultra-rapid real-time PCR targeting on IS1111 insertion element using primer Cox-F (5′-GTCTTAAGGTGGGCTGCGTG-3′), and Cox-R (5′-CCCCGAATCTCATTGATCAGC-3′); and probe Cox-TM (FAM-AGCGAACCATTGGTATCGGACGTT-TAMRA) [20,22].
2.3. Multiple Locus Variable Number Tandem Repeat Analysis (MLVA)
MLVA was performed by analyzing six microsatellite markers (MS23, MS24, MS27, MS28, MS33, and MS34) [15]. Primers used for amplification of the markers are shown in Table 1. PCR was performed using AccuPower® ProFi Taq PCR PreMix (Bioneer, Daejeon, Korea), and the 20 µL reaction mix consisted of 1 µL (10 pmol) of each primer, 3 µL DNA template, and 15 µL of ddH2O. PCR conditions were 95 °C (3 min), 37 cycles of 95 °C (30 s)–57 °C (30 s)–72 °C (30 s), and 72 °C (5 min). The PCR product of each marker from different samples was loaded in 4% (w/v) agarose gel for electrophoresis to identify the different amplicon size of each marker among the samples. The band in agarose gel was extracted for sequencing analysis. The MS23, MS24, MS28, and MS34 amplicons were directly sequenced using the specific primers; meanwhile, the amplicons of MS27 and MS33 were inserted into plasmid pGEM-T using the pGEM-T® easy vector system (Promega, Madison, WI, USA). The recombinant plasmids carrying MS27 and MS33 were introduced into Escherichia coli DH5α (Enzynomics, Daejeon, Korea) and grown in Luria-Bertani medium. The recombinant plasmids were extracted using an AccuPrep® plasmid mini extraction kit (Bioneer, Daejeon, Korea) and were sequenced using primer M13F (5′-GTA AAA CGA CGG CCA GTG-3′) and M13R (5′-CAG GAA ACA GCT ATG AC-3′) due to the small size of the two markers. Sanger sequencing was performed by Macrogen Inc. (Seoul, Korea). The generated sequence data was aligned with reference sequences of the Nine Mile strain using Clustal X 2.0 [23] to identify the number of tandem repeats. The genotype of the Nine Mile strain is 9-27-4-6-9-5 for markers MS23-MS24-MS27-MS28-MS33-MS34. The genotype was identified by comparing to the MLVA database, available at https://microbesgenotyping.i2bc.paris-saclay.fr/databases (accessed on 20 August 2022).
Table 1.
Primers used for multiple locus variable-number tandem-repeat analysis (MLVA).
| Locus | Primer | Sequence (5′–3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| Ms23 | Ms33-F | GGACAAAAATCAATAGCCCGTA | 157 | [8,15] |
| Ms33-R | GAAAACAGAGTTGTGTGGCTTC | |||
| Ms24 | Ms24-F | ATGAAGAAAGGATGGAGGGACT | 344 | |
| Ms24-R | GATAGCCTGGACAGAGGACAGT | |||
| Ms27 | Ms27-F | TCTTTATTTCAGGCCGGAGT | 89 | |
| Ms27-R | GAACGACTCATTGAACACACG | |||
| Ms28 | Ms28-F | TAGCAAAGAAATGTGAGGATCG | 276 | |
| Ms28-R | ATTGAGCGAGAGAATCCGAATA | |||
| Ms33 | Ms33-F | TCGCGTAGCGACACAACC | 104 | |
| Ms33-R | GTAGCCCGTATGACGCGAAC | |||
| Ms34 | Ms34-F | TGACTATCAGCGACTCGAAGAA | 210 | |
| Ms34-R | TCGTGCGTTAGTGTGCTTATCT |
2.4. Multispacer Sequence Typing (MST)
Ten spacers used for analyzing of MST were previously described by Glazunova et al. [14]. The spacers include Cox2, Cox5, Cox18, Cox20, Cox22, Cox37, Cox51, Cox56, Cox57, and Cox61. Primers used for amplification and sequencing of the ten spacers are shown in Table 2. PCR was performed using AccuPower® ProFi Taq PCR PreMix (Bioneer, Daejeon, Korea), and the 20 µL reaction mix was composed of 1 µL (10 pmol) of each primer, 3 µL DNA template, and 15 µL of ddH2O. PCR conditions were 95 °C (3 min), 37 cycles of 95 °C (30 s)–55 °C (30 s)–72 °C (1 min), and 72 °C (5 min). PCR products were sequenced by Macrogen Inc. (Seoul, Korea). The genotype was identified by comparing to the MST database available at https://ifr48.timone.univ-mrs.fr/mst/coxiella_burnetii/blast.html (accessed on 20 August 2022). Phylogenetic tree showing the relationship among the genotypes was created using MEGA7 software [24].
Table 2.
Primers for multispacer sequence typing (MST).
| Spacer Name | Primer | Sequence (5′–3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| Cox2 | Cox20766 | CAACCCTGAATACCCAAGGA | 397 | [14] |
| Cox21004 | GAAGCTTCTGATAGGCGGGA | |||
| Cox5 | Cox77554 | CAGGAGCAAGCTTGAATGCG | 395 | |
| Cox77808 | TGGTATGACAACCCGTCATG | |||
| Cox18 | Cox283060 | CGCAGACGAATTAGCCAATC | 557 | |
| Cox283490 | TTCGATGATCCGATGGCCTT | |||
| Cox20 | Cox365301 | GATATTTATCAGCGTCAAAGCAA | 631 | |
| Cox365803 | TCTATTATTGCAATGCAAGTGG | |||
| Cox22 | Cox378718 | GGGAATAAGAGAGTTAGCTCA | 383 | |
| Cox378965 | CGCAAATTTCGGCACAGACC | |||
| Cox37 | Cox657471 | GGCTTGTCTGGTGTAACTGT | 463 | |
| Cox657794 | ATTCCGGGACCTTCGTTAAC | |||
| Cox51 | Cox824598 | TAACGCCCGAGAGCTCAGAA | 674 | |
| Cox825124 | GCGAGAACCGAATTGCTATC | |||
| Cox56 | Cox886418 | CCAAGCTCTCTGTGCCCAAT | 479 | |
| Cox886784 | ATGCGCCAGAAACGCATAGG | |||
| Cox57 | Cox892828 | TGGAAATGGAAGGCGGATTC | 617 | |
| Cox893316 | GGTTGGAAGGCGTAAGCCTTT | |||
| Cox61 | Cox956825 | GAAGATAGAGCGGCAAGGAT | 611 | |
| Cox957249 | GGGATTTCAACTTCCGATAGA |
3. Results
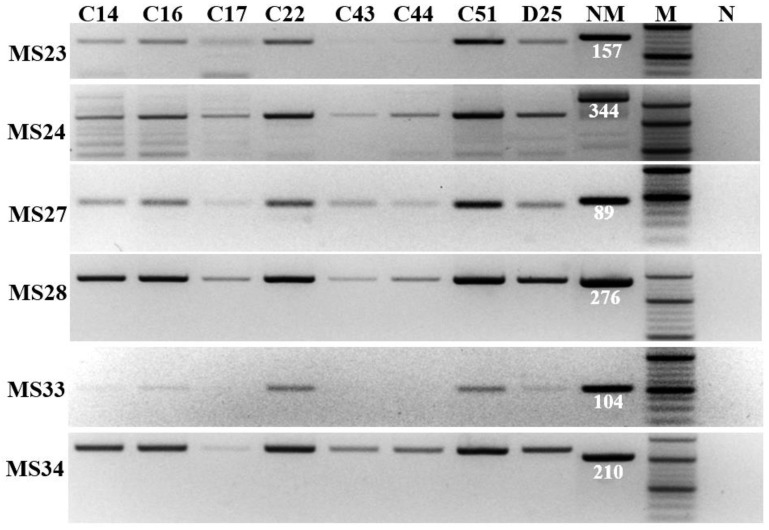
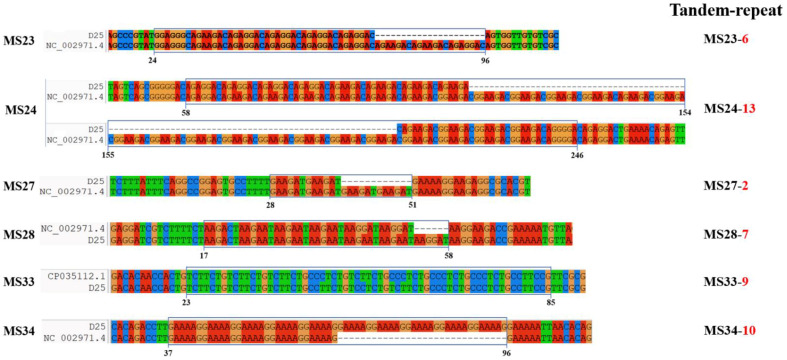
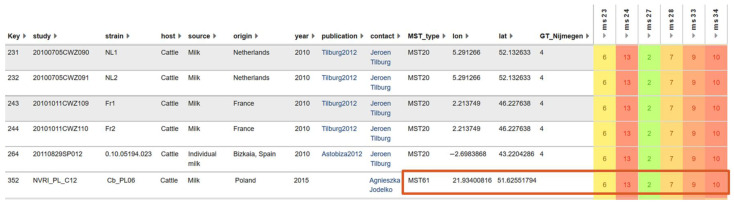
The result of electrophoresis showed that C. burnetii in all eight samples collected from the Jeonnam and Chungnam provinces had the same MLVA type with the same amplicon size of six markers (Figure 1). Sequencing analysis of the six markers from a representative sample, D25 (Figure 2), showed that the genotype of Korean C. burnetii strains is 6-13-2-7-9-10 for markers MS23-MS24-MS27-MS28-MS33-MS34, respectively. This genotype was also reported in various countries in Europe such as the Netherlands, France, Spain, and Poland (Figure 3).
Figure 1.
MLVA typing of C. burnetii. PCR products of six markers (MS23, MS24, MS27, MS28, MS33, and MS34) were amplified from eight vaginal swab samples: C14, C16, C17, C22, C43, C44, C51, and D25. The reference Nine Mile strain (NM) with amplicon size (in base pair long) of each marker is indicated, “M” is 20 bp DNA marker, and “N” is negative control without DNA template.
Figure 2.
Identification of tandem repeat of C. burnetii. Alignment of sequences of six markers (MS23, MS24, MS27, MS28, MS33, and MS34) from the Nile Mile strain (NCBI accession No.: NC_002971 and CP035112) and the detected strain D25 is shown. The region with repeat units in each marker is indicated, and the numbers indicating the positions in the analyzed sequences are presented. Number of tandem repeats of each marker is shown.
Figure 3.
Identification of MLVA type of C. burnetii detected in South Korea. Six markers (MS23, MS24, MS27, MS28, MS33, and MS34) of MLVA of C. burnetii identified in South Korea were compared with the MLVA database. Strains identified in the Netherlands, France, Spain, and Poland showed the same MVLA type, of which the strain Cb_PL06 had the same MST (MST61) and MLVA type with the strain identified in South Korea (marked in the red box).
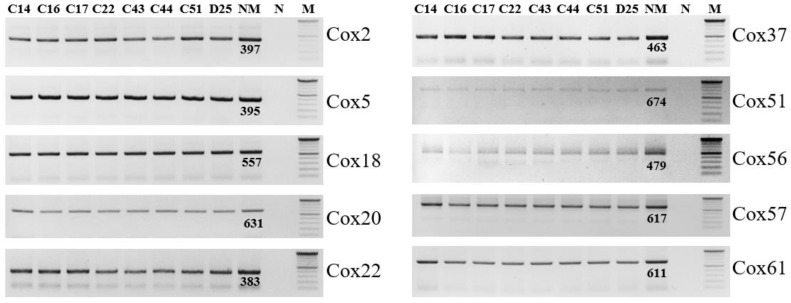
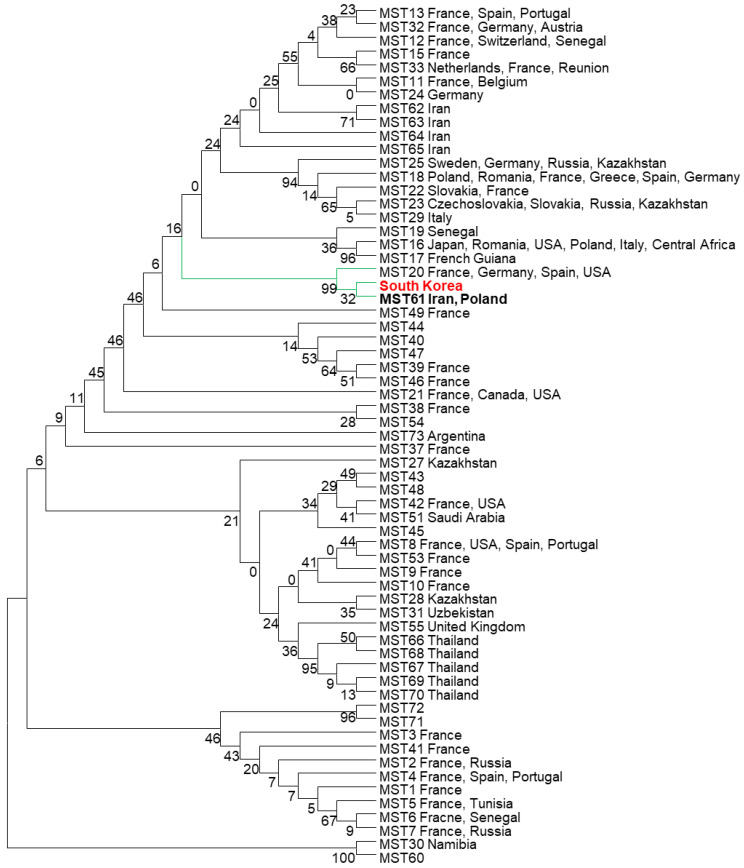
The 10 spacers of MST were amplified and confirmed in agarose gel electrophoresis (Figure 4). Sequencing analysis of the spacers from eight samples showed that C. burnetii in all samples had the same genotype, 3-2-6-1-5-10-4-10-6-5, for the spacers Cox2-Cox5-Cox18-Cox20-Cox22-Cox37-Cox51-Cox56-Cox57-Cox61, respectively. The genotype of the detected C. burnetii belongs to MST group 61, which was reported in cattle milk from Poland and Iran. The genotype has a close relationship with the genotype of MST group 20, which originated from France, Germany, USA, and Spain (Figure 5). The combination of MST and MLVA showed that the strain of C. burnetii identified in South Korea had a close relationship with the strain Cb_PL06 identified in Poland in 2015 with the same MST (MST61) and MLVA type (Figure 3).
Figure 4.
Amplification of 10 spacers for MST analysis. The 10 spacers, including Cox2, Cox5, Cox18, Cox20, Cox22, Cox37, Cox51, Cox56, Cox57, and Cox61, were amplified from eight vaginal swab samples: C14, C16, C17, C22, C43, C44, C51, and D25. “NM” is the Nine Mile strain with amplicon size (in base pair long) of each marker is shown, “M” is 100 bp DNA marker, and “N” is negative control without DNA template.
Figure 5.
Phylogenetic tree showing relationships among the different groups of MST. Maximum-likelihood phylogenetic trees were created from sequences of C. burnetii detected from vaginal swab samples of cattle in South Korea and other MST groups using the Kimura 2-parameter model, γ distribution, and bootstrapping 500 times with MEGA7 software.
4. Discussion
MST and MLVA genotyping of C. burnetii was performed in this study for the first time in South Korea. Only one genotype of C. burnetii was seen from samples collected in two provinces, Jeonnam and Chungnam. The genotype of C. burnetii detected in this study had the same type of MST and MLVA as that detected in cattle milk in Poland [25]. The result suggests that the pathogen in ROK could have originated from Poland.
MST has been established for identification of the geographical relationship of C. burnetii. Meanwhile, MLVA is useful for epidemiological purposes [8,26]. The MLVA genotype identified in this study was detected in humans in France in 2000 (https://microbesgenotyping.i2bc.paris-saclay.fr/databases/view/43 (accessed on 20 August 2022)). The result suggests that the detected C. burnetii strain in cattle in the two provinces could be an important source of causative agents of Q fever in humans in ROK. In addition, C. burnetii was detected and isolated from a patient who worked as a dairy cattle raiser in ROK [21,27]. However, the evidence of direct transmission of C. burnetii from cattle has not been provided. MST and MLVA typing could be useful to determine the relationship between C. burnetii in its vectors, natural reservoirs, and in patients. Therefore, the result of this study initially provides important information for further study on the epidemiological source of Q fever in ROK.
MST and MLVA genotyping of C. burnetii in ROK provide information on geographical distributions of the C. burnetii genotypes, and it is important to establish the global database on the phylogenetic relationship among the geographical strains and epidemiological study on Q fever. Furthermore, it could be useful information for determining the relationship between human infection and the natural reservoirs of the pathogens, by which a high potential source of disease could be suggested.
5. Conclusions
The genotype of C. burnetii based on MST and MLVA was analyzed in ROK for the first time. The group MST61 was identified with the code 3-2-6-1-5-10-4-10-6-5 for spacers Cox2-Cox5-Cox18-Cox20-Cox22-Cox37-Cox51-Cox56-Cox57-Cox61, respectively. In addition, MLVA typing was identified with the tandem repeat 6-13-2-7-9-10 for markers MS23-MS24-MS27-MS28-MS33-MS34, respectively. The same MST and MLVA type of C. burnetii from cattle in Poland was seen. The result demonstrated a close relationship of C. burnetii strains between the two countries. Furthermore, a close relationship between MST61 and MST20, which includes strains that have infected humans, from the same type of MLVA, suggests that the strain identified in ROK could potentially transmit to humans. This study provides the initial result of MST and MLVA typing of C. burnetii that is helpful for analyzing molecular epidemiology of C. burnetii in the country.
Acknowledgments
We would like to thank Bo-Ram Yun for her technical assistance and precious help.
Author Contributions
Conceptualization, Y.S.C. and A.-T.T.; methodology, Y.S.C. and A.-T.T.; software, A.-T.T.; validation, A.-T.T., S.Y.Y., M.-S.Y., S.-S.Y. and Y.S.C.; formal analysis, A.-T.T.; investigation, Y.S.C. and A.-T.T.; data curation, A.-T.T., S.Y.Y., M.-S.Y. and J.-Y.L.; writing—original draft preparation, A.-T.T.; writing—review and editing, A.-T.T., S.Y.Y., M.-S.Y., S.-S.Y. and Y.S.C.; visualization, A.-T.T.; supervision, Y.S.C.; project administration, Y.S.C. and S.-S.Y.; funding acquisition, Y.S.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported by the Animal and Plant Quarantine Agency (Project No. B-1543081-2020-22-03).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shaw E.I., Voth D.E. Coxiella burnetii: A Pathogenic Intracellular Acidophile. Microbiology. 2019;165:1–3. doi: 10.1099/mic.0.000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colville J., Berryhill D.L. Q fever. In: Colville J., Berryhill D.L., editors. Handbook of Zoonoses: Identification and Prevention. Mosby Elsevier; St. Louis, MO, USA: 2007. [Google Scholar]
- 3.Bronner M.B., Haagsma J.A., Dontje M.L., Barmentloo L., Kouwenberg R., Olde Loohuis A., de Groot A., Erasmus V., Polinder S. Long-term impact of a Q-fever outbreak: An evaluation of health symptoms, health-related quality of life, participation and health care satisfaction after ten years. J. Psychosom. Res. 2020;139:110258. doi: 10.1016/j.jpsychores.2020.110258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agerholm J. Coxiella burnetii associated reproductive disorders in domestic animals-a critical review. Acta Vet. Scand. 2013;55:13. doi: 10.1186/1751-0147-55-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurin M., Raoult D. Q fever. Clin. Microbiol. Rev. 1999;12:518–553. doi: 10.1128/CMR.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tissot-Dupont H., Raoult D. Q Fever. Infect. Dis. Clin. N. Am. 2008;22:505–514. doi: 10.1016/j.idc.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Astobiza I., Tilburg J.J., Piñero A., Hurtado A., García-Pérez A.L., Nabuurs-Franssen M.H., Klaassen C.H. Genotyping of Coxiella burnetii from domestic ruminants in northern Spain. BMC Vet. Res. 2012;8:241. doi: 10.1186/1746-6148-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Domenico M., Curini V., Di Lollo V., Massimini M., Di Gialleonardo L., Franco A., Caprioli A., Battisti A., Cammà C. Genetic diversity of Coxiella burnetii in domestic ruminants in central Italy. BMC Vet. Res. 2018;14:171. doi: 10.1186/s12917-018-1499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jager C., Willems H., Thiele D., Baljer G. Molecular characterization of Coxiella burnetii isolates. Epidemiol. Infect. 1998;120:157–164. doi: 10.1017/S0950268897008510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen S.V., Hirai K. Differentiation of Coxiella burnetii isolates by sequence determination and PCR-restriction fragment length polymorphism analysis of isocitrate dehydrogenase gene. FEMS Microbiol. Lett. 1999;180:249–254. doi: 10.1111/j.1574-6968.1999.tb08803.x. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix L.R., Samuel J.E., Mallavia L.P. Differentiation of Coxiella burnetii isolates by analysis of restriction-endonuclease-digested DNA separated by SDS-PAGE. J. Gen. Microbiol. 1991;137:269–276. doi: 10.1099/00221287-137-2-269. [DOI] [PubMed] [Google Scholar]
- 12.Sekeyova Z., Roux V., Raoult D. Intraspecies diversity of Coxiella burnetii as revealed by com1 and mucZ sequence comparison. FEMS Microbiol. Lett. 1999;180:61–67. doi: 10.1111/j.1574-6968.1999.tb08778.x. [DOI] [PubMed] [Google Scholar]
- 13.Massung R.F., Cutler S.J., Frangoulidis D. Molecular typing of Coxiella burnetii (Q fever) Adv. Exp. Med. Biol. 2012;984:381–396. doi: 10.1007/978-94-007-4315-1_19. [DOI] [PubMed] [Google Scholar]
- 14.Glazunova O., Roux V., Freylikman O., Sekeyova Z., Fournous G., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D. Coxiella burnetii genotyping. Emerg. Infect. Dis. 2005;11:1211–1217. doi: 10.3201/eid1108.041354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arricau-Bouvery N., Hauck Y., Bejaoui A., Frangoulidis D., Bodier C.C., Souriau A., Meyer H., Neubauer H., Rodolakis A., Vergnaud G. Molecular characterization of Coxiella burnetii isolates by infrequent restriction site-PCR and MLVA typing. BMC Microbiol. 2006;6:38. doi: 10.1186/1471-2180-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svraka S., Toman R., Skultety L., Slaba K., Homan W.L. Establishment of a genotyping scheme for Coxiella burnetii. FEMS Microbiol. Lett. 2006;254:268–274. doi: 10.1111/j.1574-6968.2005.00036.x. [DOI] [PubMed] [Google Scholar]
- 17.Hornstra H.M., Priestley R.A., Georgia S.M., Kachur S., Birdsell D.N., Hilsabeck R., Gates L.T., Samuel J.E., Heinzen R.A., Kersh G.J., et al. Rapid typing of Coxiella burnetii. PLoS ONE. 2011;6:e26201. doi: 10.1371/journal.pone.0026201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA) FEMS Immunol. Med. Microbiol. 2007;49:22–27. doi: 10.1111/j.1574-695X.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 19.Seo M.G., Ouh I.O., Lee S.H., Kim J.W., Rhee M.H., Kwon O.D., Kim T.H., Kwak D. Prevalence of Coxiella burnetii in cattle at South Korean national breeding stock farms. PLoS ONE. 2017;12:e0177478. doi: 10.1371/journal.pone.0177478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truong A.-T., Yun B.R., Lim J., Min S., Yoo M.S., Yoon S.S., Yun Y.M., Kim J.T., Cho Y.S. Real-time PCR biochip for on-site detection of Coxiella burnetii in ticks. Parasit. Vectors. 2021;14:239. doi: 10.1186/s13071-021-04744-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S.H., Lee J.H., Park S., Lee H.K., Do Hwang S., Jeong H.W., Heo J.Y., Lee Y.S. Isolation of Coxiella burnetii in patients with nonspecific febrile illness in South Korea. BMC Infect. Dis. 2020;20:421. doi: 10.1186/s12879-020-05130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klee S.R., Tyczka J., Ellerbrok H., Franz T., Linke S., Baljer G., Appel B. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. 2006;6:2. doi: 10.1186/1471-2180-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szymańska-Czerwińska M., Jodełko A., Zaręba-Marchewka K., Niemczuk K. Shedding and genetic diversity of Coxiella burnetii in Polish dairy cattle. PLoS ONE. 2019;14:e0210244. doi: 10.1371/journal.pone.0210244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eldin C., Mélenotte C., Mediannikov O., Ghigo E., Million M., Edouard S., Mege J.L., Maurin M., Raoult D. From Q fever to Coxiella burnetii infection: A paradigm change. Clin. Microbiol. Rev. 2017;30:115–190. doi: 10.1128/CMR.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S.H., Heo J.Y., Lee H.K., Lee Y.S., Jeong H.W., Hwang S.D. Clinical and Genetic Features of Coxiella burnetii in a Patient with an Acute Febrile Illness in Korea. J. Korean Med. Sci. 2017;32:1038–1041. doi: 10.3346/jkms.2017.32.6.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.