Abstract
Blood cells carry important information that can be used to represent a person’s current state of health. The identification of different types of blood cells in a timely and precise manner is essential to cutting the infection risks that people face on a daily basis. The BCNet is an artificial intelligence (AI)-based deep learning (DL) framework that was proposed based on the capability of transfer learning with a convolutional neural network to rapidly and automatically identify the blood cells in an eight-class identification scenario: Basophil, Eosinophil, Erythroblast, Immature Granulocytes, Lymphocyte, Monocyte, Neutrophil, and Platelet. For the purpose of establishing the dependability and viability of BCNet, exhaustive experiments consisting of five-fold cross-validation tests are carried out. Using the transfer learning strategy, we conducted in-depth comprehensive experiments on the proposed BCNet’s architecture and test it with three optimizers of ADAM, RMSprop (RMSP), and stochastic gradient descent (SGD). Meanwhile, the performance of the proposed BCNet is directly compared using the same dataset with the state-of-the-art deep learning models of DensNet, ResNet, Inception, and MobileNet. When employing the different optimizers, the BCNet framework demonstrated better classification performance with ADAM and RMSP optimizers. The best evaluation performance was achieved using the RMSP optimizer in terms of 98.51% accuracy and 96.24% F1-score. Compared with the baseline model, the BCNet clearly improved the prediction accuracy performance 1.94%, 3.33%, and 1.65% using the optimizers of ADAM, RMSP, and SGD, respectively. The proposed BCNet model outperformed the AI models of DenseNet, ResNet, Inception, and MobileNet in terms of the testing time of a single blood cell image by 10.98, 4.26, 2.03, and 0.21 msec. In comparison to the most recent deep learning models, the BCNet model could be able to generate encouraging outcomes. It is essential for the advancement of healthcare facilities to have such a recognition rate improving the detection performance of the blood cells.
Keywords: blood cell, new BCNet framework, deep transfer learning, multi-class identification, verification and validation
1. Introduction
Blood cancer, lung cancer, breast cancer, and colon cancer are just a few of the many disorders for which microscopic image analysis plays a crucial role in diagnosis and early detection [1,2]. Leukemia is another term for cancer of the blood [3]. This malignancy begins in the bone marrow and manifests as an abnormal number or shape of white blood cells (leucocytes) in the blood [4,5,6]. In 2020, it was estimated that 19.3 million people have been diagnosed with cancer (18.1 million without nonmelanoma skin cancer) and that nearly 10 million people would lose their lives to the disease [7]. Indeed, cancer is responsible for one out of every six deaths around the world, especially in developing nations, where cancer accounts for a staggering 70% of all fatalities [8]. It is anticipated that there will be 1,918,030 new cancer cases and 609,360 cancer-related fatalities in the United States in the year 2022. Out of these deaths, about 350 each day will be attributable to lung cancer, which is the leading cause of cancer-related mortality. The Cancer Statistics Center predicts that there will be 60,650 new cases of acute leukemia and 24,000 deaths from the disease in 2022, with an incidence rate of 14.2 per 100,000 people and a mortality rate of 6.1 per 100,000 people over that time period [9]. Leukemia is a potentially fatal disease that can be slowed or stopped with early treatment. As a result, the prompt diagnosis and treatment of Leukemia is urgently required [10,11]. Hematological diagnosis relies heavily on microscopic examination and the cell classification of blood [12,13]. Diagnosis of hematological malignancies such as Acute Myeloid Leukemia (AML) begins with a morphological analysis of leukocytes either from peripheral blood or the bone marrow [14] Cytomorphology is especially important in the standard French-American-British (FAB) categorization of acute myeloid leukemias [15]. The cytomorphological analysis of leukocytes is a standard aspect of hematological diagnostic workup that has so far resisted automation and is typically carried out by educated human professionals. Because of the high degree of intra/inter-observer variation that is difficult to account for and the scarcity of appropriately qualified experts, cytomorphological classification is a laborious and time-consuming process [8].
Medical experts rely on the medical imaging modalities such as computed tomography (CT), microscopic blood smear images, Magnetic Resonance Imaging (MRI), X-ray, and ultrasound (US) to diagnose health challenges and assign treatment prescriptions [16,17]. Researchers and developers are able to deliver smart solutions for medical imaging diagnoses thanks to the AI-based potential functionalities of machine learning and deep learning technologies [18,19,20,21,22,23]. Recently, several smart Computer-Aided Diagnosis (CAD) models have been created to handle automatic medical diagnosis purposes. This is to support the medical staff in performing a fast and accurate examination and diagnosis results, especially during the epidemic or pandemic health situation. The haematological examinations are a daily routine in any hospital or medical centre. The role of deep learning CAD system is important in terms of automatically identifying the variety of the patient’s haematological conditions with blood cancer or leukaemia. The microscopic blood smear imagining technique is used for diagnosing leukemia by analyzing and identifying the different blood cells [24]. In the pathology lab assessments, white blood cells’ (WBC) classification and identification subclasses is attained. In addition, the lab examination of leukocytes includes monocytes, lymphocytes, neutrophils, basophils, and eosinophils. To identify the different types of blood cells in a rapid way and fulfil the physician’s requirements is a big challenge. Thus, AI technologies should interact and contribute by providing smart examination solutions without user intervention. The need of AI smart solutions motivates us to develop a novel schema of the CAD system based on the new BCNet deep learning model.
Recently, various investigations on blood cell classification using ML and Deep Learning have been carried out [3,4,5,6,25,26,27,28].There are a number of different optimization algorithms that may be used for training deep neural networks, and the one that is chosen can have an effect on how well the DL model functions. Many hyper-parameters can be adjusted to improve the neural network’s performance. Every one of these factors affects network speed in some way, but not all do so to the same extent. How well the algorithm converges to a solution, or whether it explodes, could depend on the optimizer that has been selected. Among the available optimizers are ADAM, RMSP, and SGDM. However, the great majority of recent image classification efforts using the DL approach have relied on the ADAM optimizer. To the best of our knowledge, no prior research has investigated the impact of competing optimizers on DL’s efficacy. The main contributions of this study are briefly summarized here:
A deep learning schema of the CAD system based on the newly deep learning BCNet is proposed in order to identify multiclass blood cells rapidly and automatically.
The multiple class identification task is conducted in terms of improving the overall classification performance.
A comprehensive evaluation experiment is conducted to investigate the reliability and feasibility of the proposed BCNet using multiple optimizers and different state-of-the-art deep learning models such as DensNet, ResNet, Inception, and MobileNet.
The rest of the paper is organized in the following manner. Section 2 introduces the survey of the latest related works. Section 3 explains in detail the technical schema of the proposed BCNet. The assessment performance results are explained and discussed in Section 4 and Section 5, respectively. The findings of this study are discussed in the conclusion section.
2. Related Works
Several artificial intelligence models have been employed for over two decades to recognize the different types of blood cells. Red blood cells (RBCs), platelets (PRBCs), and white blood cells (WBCs) are the three basic types of blood cells; all three play important roles in the human immune response [29,30,31].
Medical professionals can benefit from the use of ML and DL methods for WBC categorization because it requires less work on their part and yields faster, more accurate results. Variations in the ratio of neutrophils, eosinophils, basophils, monocytes, and lymphocytes between healthy and diseased patients are readily apparent and play a significant role in diagnosis [32]. The combinatory approach of machine learning and the deep learning-based approach for the classification of WBC images were able to achieve 97.57% accuracy [33]. Changhun et al. proposed a W-Net model in a combination of CNN with RNN with DCGANs for image synthesizing later used for WBC classification, and attained an accuracy of 97% for a 5 class dataset [34]. César Cheuque et al. proposed the MLCNN detection of white blood cell Faster RCNN used to extract Region of interest later with Mobilenet based model is used to train the classification framework gained performance accuracy of 98.4% [35]. In continuation Next BCNet [36] to address the blood cell classification for three classes via transfer learning approach with ResNet18 as backbone model for learning and noted 96.78% accuracy. A deep learning based AI framework artificial intelligence-based microscopy image classifier for blood cell classification is proposed with transfer learning methods and realized an accuracy of 98.6% [37]. Furthermore, Acevedo et al. in [32] used the HPBC dataset to build their deep learning framework for classification of blood cells. Classifying different types of blood cells is an important activity that aids hematologists in making intelligent treatment decisions. Using the BCCD datasets, which are four-class datasets, Liang et al. presented a combination of RNN and CNN models to classify blood cells. These models included Inception-v3, ResNet-50, and Xception networks, and were then merged with an LSTM block to achieve the classification [38]. A transfer learning strategy was proposed by Acevedo et al. with the purpose of automating the classification of blood cells. They have utilized the VGG-16 and Inception-V3 as feature extractors in conjunction with the SVM and SoftMax as classifiers for the eight different types of balanced and imbalanced human peripheral blood cells (HPBC). They obtained the data using the CellaVision equipment, and they determined that the classification accuracy was 96.2% [32]. For the purpose of classifying WBC, Almezhghwi et al. have implemented several distinct kinds of Deep CNN models, including VGG, ResNet, and DenseNet, on the datasets provided by LSIC. Neutrophils, eosinophils, lymphocytes, monocytes, and basophils are the several types of white blood cells that make up the WBC. Data augmentation is accomplished by the utilization of transformation strategies, and in addition to that, they have implemented GANs for the purpose of producing datasets. They enhanced their accuracy to 98.8% and attained this result [39]. To categorize WBC [40], it is common practice to first segment the cells, extract defining properties, and then classify them. Such a method relies heavily on proper segmentation and has a poor degree of accuracy. Additionally, the precision of classification models is affected by insufficient datasets or unbalanced classes for related samples when using deep learning for medical diagnoses. To deal with such tasks, the authors in [40] proposed using a Deep Convolutional Generative Adversarial Network (DC-GAN) or a Residual neural network (ResNet). The experimental results show that the model has a high accuracy (91.7%) when it comes to classifying WBC images. Inadequate data samples and the uneven distribution of classes are two problems that this model addresses and corrects. When compared to other network architectures, the proposed technique provides the highest accuracy for classifying WBC images [40,41,42]. Wijesinghe et al. [43] have also presented a new method for the classification of WBC images. K-means cluster analysis using RGB color components and manual thresholding is used to determine the region of interest. The manually cropped WBC images then classified them using a VGG-16-based method. On custom data, they achieved a 97.4% detection rate and a 95.89% classification rate, respectively. Classification, localization, and detection of Leukocytes were proposed by Zhao et al. [44] Databases from Cellavision and Jiashan have been used for detection, while ALL-IDB has been used for classification. Pipelining begins with WBC identification using morphological methods, then moves on to color and granularity features for classification. An SVM Classifier has been used to classify the data as either basophil or eosinophil. On the other hand, a CNN/random forest hybrid model was used to classify the remaining classes, which included neutrophils, lymphocytes, and monocytes. In the end, 92.8% accuracy was achieved. Using DL models including ResNet, AlexNet, GoogLeNet, ZFNet, VGGNet, DenseNet, and Cell3Net, Qin et al. [29] suggested a classification technique for leukocytes with 40 classes. For all 40 courses, the achieved accuracy was 76.84 percent. In continuation, Zheng et al. [45] have proposed a self-supervised approach to blood cell segmentation. To extract the foreground from the background, they used an unsupervised technique based on K-means clustering. After that, they utilized a supervised method to extract features from RGB and HSV colors. Classification was then performed using the SVM on data from CellaVision and the Jiangxi Tecom Science Corporation of China. Their error rates were 3.18 and 0.69 for the CellaVision and Jiangxi Tecom Science Corporation and China datasets, respectively. Sajjad et al. have introduced a mobile-cloud-based model to handle segmentation followed by the classification of leukocytes into their five separate groups, as explained in [46]. Blood cells were distinguished using K-means and morphological procedures. In all, 1030 WBC blood smear samples were given by Hayatabad Medical Center (HMC). Segmented regions are analyzed for texture, geometry, and statistical data, then compared using different classifiers as EMC SVM. The segmentation strategy has a nucleus accuracy of 95.7%, a cytoplasm accuracy of 91.3%, and a classification accuracy of 98.6%.
3. Materials and Methods
In this section, we discuss the technical aspects of our work, including the dataset definition, the proposed BCNet architecture, and the components that make up the BCNet.
3.1. Dataset
A public dataset of human peripheral blood cells (also known as HPBC) is utilized in the process of constructing and assessing various models, including the deep learning BCNet. The blood cell images were created by Anna et al. using data that was acquired from the medical clinic in Barcelona between the years 2015 and 2019. [32]. The data repository consists a total of 17,092 images of blood cells of healthy individuals. The dataset is publicly available via https://data.mendeley.com/datasets/snkd93bnjr/1 (accessed on 21 June 2022).
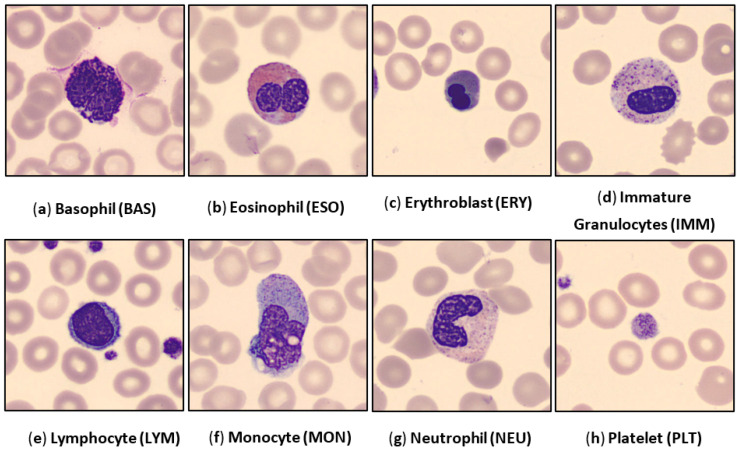
The blood cell images are extracted from the whole microscopic scan slides involving eight subcategories, which are Basophil (BAS), Eosinophil (EOS), Erythroblast (ERY), Immature Granulocytes (IMM), Lymphocyte (LYM), Monocyte (MON), Neutrophil (NEU), and Platelet (PLT). All blood cell images are in the RGB color space in the ‘jpg’ format and with a size of 360 × 360 pixels. Each image was annotated by the pathologists of the hospital clinic of Barcelona. Captured images were obtained from individuals without infections hematologic or oncologic disease and during blood sample collection individuals have not undergone any pharmacologic treatments. Some examples of blood cell images per class are depicted in Figure 1.
Figure 1.
Examples of the human peripheral blood cell (HPBC) images over all eight classes.
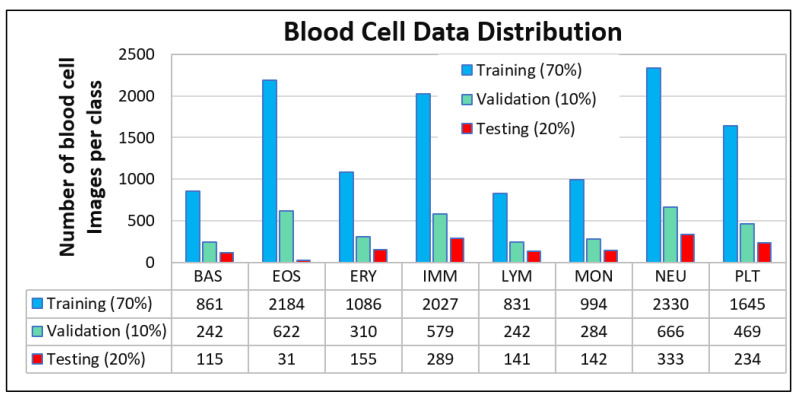
Moreover, the data distribution of all images used in this study is summarized over all classes, as demonstrated in Figure 2.
Figure 2.
HPBC dataset distribution over eight different classes of the human peripheral blood cell (HPBC) images. The images were randomly split per class into 70% (training), 10% (validation), and 20% (testing).
3.2. Pre-Processing
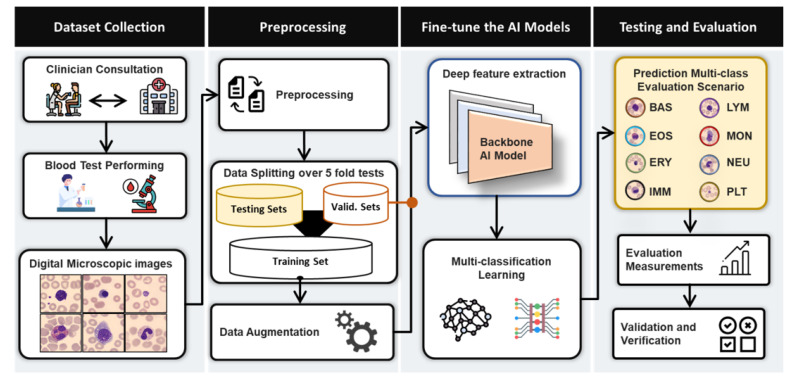
The initial step in the image classification procedure, depicted in Figure 3, involves the pre-processing of blood cell images. It is used to improve the proposed BCNet’s overall performance by cleaning and preparing the images. Bi-cubic interpolation is used to scale all images to a final resolution of 224 × 224 pixels. Meanwhile, normalization of intensity is performed so that all pixel values fall inside the range [0, 255]. The augmentation strategy of the training set is performed to enlarge the dataset size and improve the overall performances, as performed in our previous works [47,48].
Figure 3.
Abstract view paradigm of the proposed BCNet for blood cell multi-class identifications.
3.3. Data Preparation: Training, Validation, and Testing
To fine-tune and assess the proposed BCNet, the images per class are divided into three categories: the training set (70%), validation set (10%), and testing set (20%). Figure 2 depicts the number of blood cell images per category for each class. The trainable parameters such as bias and weights of the proposed CNN model are optimized via the training process by utilizing the training–validation sets. Subsequently, the overall performance of the proposed deep learning model is assessed by an evaluation set. Furthermore, the proposed BCNet model is evaluated through five-fold tests for training, validation, and evaluation sets. To deliver a robust and feasible CAD system for the detection and classification of HPBC data into eight classes, the k-fold cross-validation strategy plays an important role in the case of lower datasets per each class.
3.4. The Proposed Deep Learning Framework
The BCNet has been developed to tackle the blood cell classification problem and to improve the overall identification performance as shown in Figure 3. It can be extended and generalized to be used for different domains such as biomedical imaging with different modalities such as X-ray, MRI, CT, US, microscopic imaging, satellite imaging, etc. In the training process, the deep learning BCNet model has been adopted based on different key factors of the width and depth of the network that can be seen in the previous ones: ResNet [49] and WideResNet [50]. Such factors are helpful for training the models and attaining a higher identification performance. To build and develop the BCNet, we used the EfficientNet model as a base network, which is adopted based on the fundamental blocks of mobile inverted bottleneck convolution (MBConv) from MobileNet [51]. In [38], the EfficientNet model proposed the fundamental relationship, which explains the width and depth parameters’ efficacy for better classification accuracy. The architecture was proposed by updating the base EfficientNetB0 with different optimizers. However, the EfficientNet models were pre-trained on ImageNet datasets and outperformed other state-of-the-art deep learning models in terms of Top-1 accuracy [52]. This means that the EfficientNetB0 could be the best baseline model since it has low computational complexity, scaling of depth and width, and residual blocks with skip connections to improve the overall classification accuracy [51,52].
3.5. BCNet Deep Learning Architecture
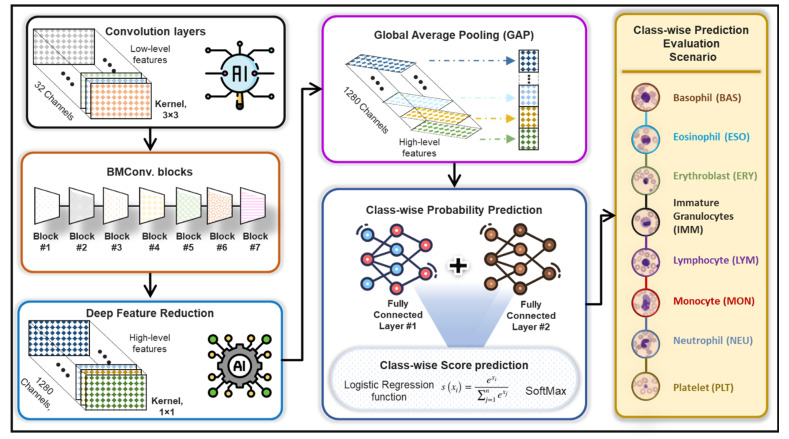
The proposed BCNet framework for the human blood cell classification is depicted in Figure 3, while Figure 4 shows the detailed deep learning structures. The base backbone model for deep feature extraction was built based on the EfficientNet-B0 network using seven MBConv blocks, convolution pooling, and fully connected layers [52]. In the proposed BCNet architecture, we adopted and modified the deep learning architecture by adding different layers to concatenate and figure out the extracted deep features and concise them as an average based on the Global Average Pooling (GAP) layer for reducing the occurrence of overfitting [53]. After that, two fully connected layers were added for better class-wise prediction probabilities of all eight classes. The number of neurons per dense layer were determined experimentally to fit our problem of eight blood cell classes using 1024 and eight nodes, respectively. Lastly, the logistic regression function of SoftMax was added to predict the classification scores for each class. Meanwhile, the convolution layer with kernel size of 1 × 1 and number of channels of 1280 was used mainly to decrease the derived deep feature map dimensionality [54]. The local response normalization (LRN) layers were utilized for all MBConv blocks and convolutional layers leading to better prediction performance of the proposed BCNet. However, it was prone to overfitting and lacked the generalization ability of neural architectures. To overcome such a challenge, a dropout strategy with a rate of 0.5 was assigned and used to handle overfitting and BCNet [55]. The dropout rate was essentially used with all convolutional layers as well as dense layers to drop some neural nodes and minimize the number of trainable parameters, reduce the overfitting, and speed-up the learning process [56]. The technical overview of the BCNet architecture is summarized in detail, as demonstrated in Table 1. For training over 5-fold tests, the same training settings and model parameters were used with training/validation sets. The evaluation was achieved via the testing sets. We experimentally fine-tuned the AI models to achieve the best accuracy based on the trail-based error approach [57,58]. All of our experiments are conducted utilizing the various optimization functions available in ADAM, RMSP, and SGDM.
Figure 4.
Feature extraction methodology of the proposed BCNet framework.
Table 1.
Technical Parameters of the proposed BCNet network.
| Stage | Operatory | Spatial Resolution Hi × Wi | Channel, Ci | Layer, Li |
|---|---|---|---|---|
| 1 | Conv., k3 × 3 | 224 × 224 | 32 | 1 |
| 2 | MBConv1, k3 × 3 | 112 × 112 | 16 | 1 |
| 3 | MBConv6, k3 × 3 | 112 × 112 | 24 | 2 |
| 4 | MBConv6, k3 × 3 | 56 × 56 | 40 | 2 |
| 5 | MBConv6, k3 × 3 | 28 × 28 | 80 | 3 |
| 6 | MBConv6, k3 × 3 | 14 × 14 | 112 | 3 |
| 7 | MBConv6, k3 × 3 | 14 × 14 | 192 | 4 |
| 8 | MBConv6, k3 × 3 | 7 × 7 | 320 | 1 |
| 9 | Conv1 × 1, | 7 × 7 | 1280 | 1 |
| 10 | GAP | |||
| 11 | 2 Dense Layers | 8 nodes | ||
| 12 | SoftMax | 8 nodes: Number of blood cell classes. | ||
Conv: Convolution neural network. MBConv: Mobile Inverted convolution blocks. FC: Fully connected. GAP: Global average pooling.
3.6. Performance Metrics
The evaluation of the proposed BCNet is carried out for every fold test via a weighted objective metrics strategy including overall accuracy (Az.), F1-score, recall or sensitivity (SE), specificity (SP), Matthews correlation coefficient (MCC), positive predictive value (PPV), and negative predictive value (NPV). The weighted-class strategy is adopted to overcome the class imbalance per test set that contains unbalanced images from different classes. A multi-class confusion matrix is used to derive the evaluation parameters. The evaluation results shown in the result section are achieved over a 5-fold cross-validation test to investigate the reliability and feasibility of the proposed BCNet. The definition of the evaluation metrics is summarized in Equations (1)–(7) [20,59,60,61,62,63]. True positive (TP), true negative (TN), false positive (FP), and false negative (FN) are derived via a multi-class confusion matrix for each fold test. Due to the misbalancing testing images, the weighted evaluation strategy is used to derive all of the evaluation metrics [40].
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
4. Experimental Results
Here, we present and discuss the evaluation classification results obtained using a 5-fold test on the proposed BCNet system. Three optimizers (ADAM, RMSP, and SGDM) are used to conduct comparison evaluations, and the results are recorded and analyzed. Table 2 summarizes the results of the 5-fold test conducted with the cross-validation method applied to the HPBC dataset. The proposed model is a deep learning BCNet model built on the same DL framework, with specific training settings applied to improve the model’s reliability and performance. The classification results with multiple optimizers are reported in Table 1 over 5 folds.
Table 2.
Evaluation classification results (%) of the proposed BCNet with different optimizers for blood cell classification over 5-fold tests.
| No. of Fold | Optimizer | SE | SP | Az. | MCC | F1-Score | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Fold 1 | ADAM | 93.89 | 98.14 | 97.53 | 90.93 | 91.66 | 90.44 | 98.55 |
| RMSP | 95.53 | 98.9 | 98.47 | 95.11 | 95.61 | 95.8 | 98.84 | |
| SGD | 93.12 | 98.53 | 97.83 | 92.55 | 93.36 | 93.89 | 98.47 | |
| Fold 2 | ADAM | 97.91 | 99.26 | 99.04 | 97.74 | 97.74 | 98.02 | 99.12 |
| RMSP | 98.3 | 99.26 | 99.13 | 98.15 | 98.3 | 98.33 | 99.22 | |
| SGD | 93.12 | 98.53 | 97.83 | 92.55 | 93.36 | 93.89 | 98.47 | |
| Fold 3 | ADAM | 96.8 | 98.88 | 98.53 | 96.28 | 96.79 | 96.82 | 98.87 |
| RMSP | 95.09 | 98.64 | 98.13 | 94.37 | 95.1 | 95.21 | 98.63 | |
| SGD | 96.6 | 98.85 | 98.49 | 96.05 | 96.59 | 96.61 | 98.84 | |
| Fold 4 | ADAM | 97.16 | 98.99 | 98.7 | 96.73 | 97.15 | 97.15 | 98.86 |
| RMSP | 97.1 | 98.96 | 98.67 | 96.66 | 97.09 | 97.12 | 98.96 | |
| SGD | 96.63 | 98.93 | 98.57 | 96.13 | 96.62 | 96.63 | 98.88 | |
| Fold 5 | ADAM | 96.8 | 98.88 | 98.53 | 96.28 | 96.79 | 96.82 | 98.87 |
| RMSP | 95.09 | 98.64 | 98.13 | 94.37 | 95.1 | 95.21 | 98.63 | |
| SGD | 96.6 | 98.85 | 98.49 | 96.05 | 96.59 | 96.61 | 98.84 | |
| Avg. (%) | ADAM | 96.51 | 98.83 | 98.47 | 95.59 | 96.03 | 95.85 | 98.85 |
| RMSP | 96.22 | 98.88 | 98.51 | 95.73 | 96.24 | 96.33 | 98.86 | |
| SGD | 95.21 | 98.74 | 98.24 | 94.67 | 95.30 | 95.53 | 98.70 |
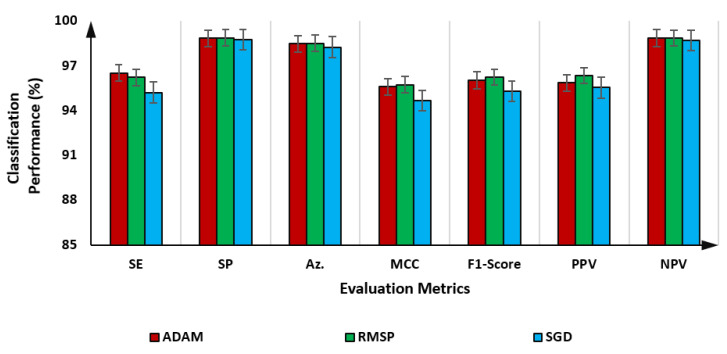
As presented in Table 2, the blood cell classification performance of the proposed BCNet with the RMSP optimizer is the best with almost all fold tests. The average evaluation results over the 5-fold test with all optimizers indicating the error rates are shown in Figure 5. It is shown that the RMSP slightly outperforms other optimizers achieving an overall accuracy of 98.51%, while the ADAM and SGD optimizers achieve 98.47% and 98.24%, respectively. Meanwhile, the BCNet could achieve a promising performance with F1 scores of 96.03%, 96.24%, and 95.30% using ADAM, RMSP, and SGD optimizers, respectively. The evaluation results in terms of the multiclass confusion matrix are depicted for the proposed BCNet with each optimizer, as in the following figures.
Figure 5.
Evaluation classification results of the proposed BCNet as an average over 5-fold tests using ADAM, RMSP, and SGD optimizers.
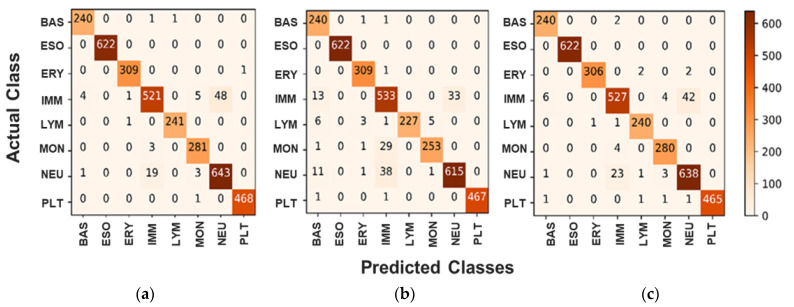
As it is shown in all confusion matrices, the low rate of false rates with different classes is recorded. In the case of class Ig (see Figure 6a–c), the majority of the false rate is recorded with the wrongly 48 classified cases as a Neutrophil class, while five and four cases are classified as Monocytes and Basophil, respectively. In the case of the Neutrophil class, the major classification mistake happened as a cross-similarity with the Ig class with 19 images. Such discussion has proven that both Ig and Neutrophil images have similarities in figure, shape, or size, and these comparable nature features could negatively affect the classification performance of any AI-based models. We can also notice that the performance of BCNet with ADAM and RMSP optimizers is much closer to each other since the TP cases are almost the same, whereas, the classification performance with SGD is slightly lower.
Figure 6.
Confusion Matrix for Multiclass scenario for the proposed BCNet with different optimizers. (a) ADAM optimizer, (b) RMSP optimizer, and (c) SGD optimizer.
5. Discussion
Deep learning based on convolutional networks has recently had excellent and successful results in the analysis of medical images for many applications. In this work, we have proposed an AI-based BCNet framework that aims to automatically, accurately, and rapidly recognize multi-class human blood cells, and helps in decision-making for assisting physicians and other medical staff. At the same time, it could assist in the detection of infections based on various blood cell malignancies such as leukaemia, anaemia, etc. Existing studies provided insights on AI-based deep learning for predicting multiple diseases from a specific medical image modality. In our proposed model, we studied a DL-based model for blood cell classification, which consists of eight classes with a publicly available dataset. The proposed model plays a critical role in our model which gets the best detection performance compared with the latest machine and deep learning models. We have used a transfer learning-based approach with the EffieNetB0 model by modifying this model with three optimizers to get the best achievements. In fact, the BCNet is able to detect and classify eight classes: Basophil, Eosinophil, Erythroblast, Ig, Lymphocyte, Monocyte, Neutrophil, and Platelet. To the best of our knowledge, this is the highest number of classes on which a BCNet-based model has been tested in contrast to the existing literature (they used four or five classes). The results of our proposed BCNet, which are presented in Table 2 and Table 3, achieved a competitive overall classification accuracy of 98.51% with an RMSP optimizer and 98.47% with an ADAM optimizer. This study concluded that BCNet with different optimizers achieves promising identification performance with slightly better achievements using the RMSP optimizer. All evaluation results are summarized in the results section.
Table 3.
Performance comparison of the proposed BCNet against the latest AI models with respect to the number of the learning parameters and the computation costs.
| AI Model | Number of Trainable Parameters (million) | Training Time Per Epoch (sec.) | Testing Time/Image (msec.) |
|---|---|---|---|
| DenseNet 201 | 18.10 | 286 | 18.13 |
| ResNet 50 | 23.55 | 148 | 11.41 |
| Inception V3 | 21.78 | 128 | 9.18 |
| MobileNet V2 | 2.23 | 123 | 7.36 |
| The proposed BCNet | 4.017 | 122 | 7.15 |
5.1. Comparative Results of the BCNet and Other DL Models
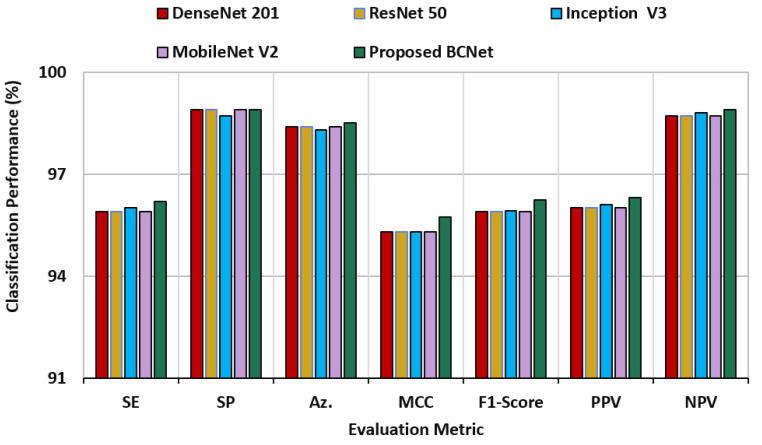
For direct comparison using the same dataset, four deep learning models of DenseNet 201, ResNet50, Inception-V3, and Mobilenet-V2 are adopted and used. These AI models are selected to perform such direct comparison due to their promising classification performance in the research domain [21,37,47,58,59,64,65]. Such comparison is important to investigate the reliability of the proposed model with the trusted ones. The comparison evaluation results among the BCNet and other deep learning models is presented in Figure 7. This result is recorded over 5-fold tests with the same training sets of all models. It is proven that the proposed BCNet could successfully handle and classify all images properly and outperform all other models in terms of all seven metrics.
Figure 7.
Direct comparison results of blood cell classification using the proposed BCNet and other deep learning models of DenseNet 201, ResNet50, Inception-V3, and Mobilenet-V2.
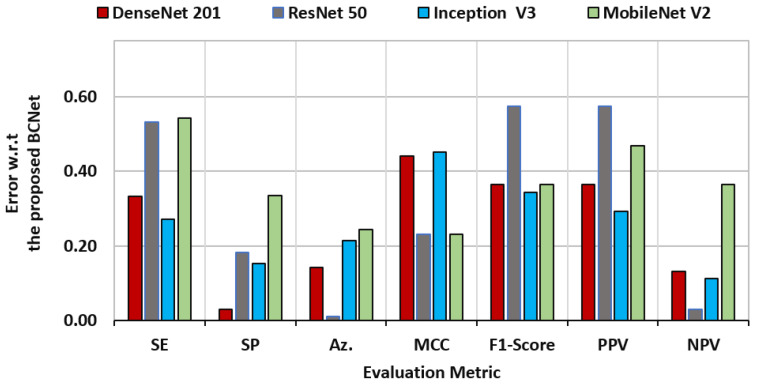
To explain the rate of change among different models, the standard deviation was measured to check the model diversity with respect to the proposed BCNet model. The performance characteristics regarding this study for all AI models (i.e., DenseNet [1], ResNet [5], Inception [9], and MobileNet [20]) based on the BCNet as a reference are shown in Figure 8. It was clearly indicated that the ResNet had the closest accuracy performance compared with our proposed model, since the prediction error was 0.010. On the other hand, the ResNet showed the worst error deviation in terms PPV and F1-score. The closest model with BCNet was the Inception model in terms of sensitivity and F1-score, with error rates of 0.334 and 0.365, respectively. The DesNet model achieved the best match, with the BCNet achieving the lowest specificity error deviation with 0.030.
Figure 8.
Evaluation error performance of the latest deep learning models of DenseNet 201, ResNet 50, Inception V3, and Mobile Net V2 with respect of the proposed BCNet model.
The performance comparison of the proposed BCNet against other AI models (i.e., DenseNet, ResNet, Inception, and MobileNet) was carried out in terms of a number of trainable parameters, costs of training time consumption for each epoch, and testing time for a single blood cell image, as shown in Table 3. The proposed BCNet model outperformed the AI models of DenseNet, ResNet, Inception, and MobileNet in terms of the testing time of a single blood cell image by 10.98, 4.26, 2.03, and 0.21 millisecond (msec), respectively. Based on the comparison in Table 3, the heaviest model was the ResNet model, while the MobileNet was comparatively lightweight compared with BCNet. The proposed BCNet model provided promising evaluation performance as well as outperforming the state-of-the-art AI models without regard to the inferencing and testing time cost.
The comparison of the performance of various models with proposed BCNet is carried out overall in terms of several trainable parameters, testing time for each image, and costs of training time consumption for each epoch with models such as DenseNet, ResNet, Inception, MobileNet and Proposed BCNet. The proposed model, with less training and testing costs, has shown promising results.
5.2. Comparison between Proposed BCNet and Previously Published Models
A comparison between our proposed BCNet against the latest deep learning works for blood cell classification is performed and summarized in Table 4. The proposed BCNet could provide a promising and competitive evaluation result for multiple classes.
Table 4.
Evaluation comparison results for blood cell identification via the proposed BCNet against the latest deep learning works in the literature.
| Reference | Data | Methods | Az. (%) |
|---|---|---|---|
| Zhao et al., 2017 [44] | Cell vision, ALL-IDB, Jiashan | CNN, SVM, and random forest | 92.80 |
| Journal et al., 2021 [66] | Collected, BCCD data set | Two DCNN | 95.17 (Precession) |
| Acevedo et al., 2019 [32] | Private | CNN + Transfer learning | 96 |
| Qin et al., 2018 [29] | Private | CNN | 76.84 |
| Ma et al., 2020 [40] | BCCD | DCGAN + Transfer learning | 91.7 |
| Baydilli and Atila 2020, [67] | LISC | Capsule network | 96.86 |
| Rui Liu et al., 2022 [37] | HPBC | Transfer Learning | 96.83 |
| The proposed BCNet | HPBC images | BCNet | 98.51 |
5.3. Ablation Study
We performed this ablation study to show the proposed model capability in improving the overall classification evaluation performance with/without the additional layers on the base model. To achieve this study, the comparison evaluation results against the baseline model were conducted and the derived evaluation results are reported in Table 5. We retrained and evaluated the proposed model against its base model using three optimizers: Adam, RMSP, and SGD. It was clearly shown that the prediction accuracy performance was improved due to our modifications by 1.94%, 3.33%, and 1.65% in terms of using ADAM, RMSP, and SGD optimizers, respectively. Similarly, the performance was improved in terms of F1-scores by 1.14%, 2.13%, and 0.75% with respect to the ADAM, RMSP, and SGD optimizers, respectively.
Table 5.
The compression evaluation result of the ablation study.
| AI Models | SE | SP | Az. | MCC | F1-Score | PPV | NPV | |
|---|---|---|---|---|---|---|---|---|
| Baseline Model | ADAM | 95.8 | 97.88 | 96.53 | 93.38 | 94.89 | 94.82 | 96.88 |
| RMSP | 94.09 | 95.66 | 95.18 | 94.37 | 94.11 | 94.21 | 95.63 | |
| SGD | 93.50 | 96.65 | 96.59 | 95.05 | 94.55 | 95.61 | 97.84 | |
| The Proposed BCNet | ADAM | 96.51 | 98.83 | 98.47 | 95.59 | 96.03 | 95.85 | 98.85 |
| RMSP | 96.22 | 98.88 | 98.51 | 95.73 | 96.24 | 96.33 | 98.86 | |
| SGD | 95.21 | 98.74 | 98.24 | 94.67 | 95.30 | 95.53 | 98.70 |
5.4. Limitations and Future Work
The most significant limitation of the AI-based medical application is due to the scarcity of the annotated benchmark datasets since the labeling process is expensive and it takes time for the experts to do it. In the future, we plan to check the capability of the BCNet with other medical imaging such as magnetic resonance imaging (MRI), Ultrasound (US), X-ray, and so on. Furthermore, we plan to continue improving the performance behavior and providing more interesting blood cell identification performance by applying the newly impressive AI technologies such as explainable AI, federated learning, etc.
6. Conclusions
One of the most significant issues that the field of medicine is currently facing is the classification of blood cells. This is especially true given the growing number of infection cases and the difficulty in identifying them at an early stage. Within the scope of this work, we have investigated the classification of blood cells by utilizing transfer deep learning, BCNet. An exhaustive experimental investigation is carried out with several different optimizers in order to assess the dependability and practicability of the suggested model. At the same time, we examine the similarities and differences between a variety of transfer learning models and various optimizers. The RMSP optimizer was used to produce the best possible accuracy of 98.51%, which exceeds other deep learning models that are currently available. When applied to the classification of blood cells into eight different classes, our suggested method achieves a high level of accuracy. In order to demonstrate that the transfer learning model with the RMSP activation function is an effective one, we compared this study to a large number of other similar works.
Acknowledgments
This work is supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project Number PNURSP2022R206, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2022-00166402). This research was supported by an Institute for Information & Communications Technology Promotion (IITP) grant funded by the Korean government (MSIT) (No. 2017-0-00655) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT), Korea.
Abbreviations
| AI | Artificial Intelligence |
| BAS | Basophils |
| CAD | Computer-Aided Diagnosis |
| Conv | Convolution |
| CNN | Convolution Neural Network |
| DL | Deep Learning |
| ERY | Erythroblasts |
| EOS | Eosinophils |
| FC | Fully connected |
| GAP | Global Average Pooling |
| HPBC | Human Peripheral Blood Cells |
| LRN | local response normalization |
| LYM | Lymphocytes |
| MBConv | Mobile inverted bottleneck convolution |
| ML | Machine Learning |
| NEU | Neutrophils |
| MRI | Magnetic Resonance Imaging |
| MON | Monocytes |
| PLT | Platelets |
| RBC | Red Blood Cells |
| WBC | White Blood Cells |
Author Contributions
Conceptualization, software, methodology, formal analysis, visualization, and writing, C.C., A.Y.M. and M.A.A.-A.; data curation, resources, C.C., J.V.B.B., K.H. and W.R.N.; validation, resources, M.B.B.H.; funding acquisition, proofreading, editing, N.F.M. and N.A.S.; project administration, supervision, and proofreading, revising, M.A.A.-A., Y.M.K. and T.-S.K. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Not applicable since this study did not involve humans.
Data Availability Statement
The dataset used in this study is publicly available via https://data.mendeley.com/datasets/snkd93bnjr/1 (accessed on 21 June 2022).
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Funding Statement
This work is supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project Number PNURSP2022R206, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li Y., Chen J., Xue P., Tang C., Chang J., Chu C., Ma K., Li Q., Zheng Y., Qiao Y. Computer-Aided Cervical Cancer Diagnosis Using Time-Lapsed Colposcopic Images. IEEE Trans. Med. Imaging. 2020;39:3403–3415. doi: 10.1109/TMI.2020.2994778. [DOI] [PubMed] [Google Scholar]
- 2.Chen R.J., Lu M.Y., Wang J., Williamson D.F.K., Rodig S.J., Lindeman N.I., Mahmood F. Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis. IEEE Trans. Med. Imaging. 2020;41:757–770. doi: 10.1109/TMI.2020.3021387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raphael R.T., Joy K.R. Segmentation and Classification Techniques of Leukemia Using Image Processing: An Overview; Proceedings of the 2019 International Conference on Intelligent Sustainable Systems; Palladam, India. 21–22 February 2019; pp. 378–384. [DOI] [Google Scholar]
- 4.Chin Neoh S., Srisukkham W., Zhang L., Todryk S., Greystoke B., Lim C.P., Hossain M.A., Aslam N. An Intelligent Decision Support System for Leukaemia Diagnosis using Microscopic Blood Images. Sci. Rep. 2015;5:14938. doi: 10.1038/srep14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agaian S., Madhukar M., Chronopoulos A.T. Automated Screening System for Acute Myelogenous Leukemia Detection in Blood Microscopic Images. IEEE Syst. J. 2014;8:995–1004. doi: 10.1109/JSYST.2014.2308452. [DOI] [Google Scholar]
- 6.Amin M.M., Kermani S., Talebi A., Oghli M.G. Recognition of Acute Lymphoblastic Leukemia Cells in Microscopic Images Using K-Means Clustering and Support Vector Machine Classifier. J. Med. Signals Sens. 2015;5:49. doi: 10.4103/2228-7477.150428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M., Gavin A., Visser O., Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 9.Matek C., Schwarz S., Spiekermann K., Marr C. Human-level recognition of blast cells in acute myeloid leukaemia with convolutional neural networks. Nat. Mach. Intell. 2019;1:538–544. doi: 10.1038/s42256-019-0101-9. [DOI] [Google Scholar]
- 10.Shree K.D., Janani B. Classification of Leucocytes for Leukaemia Detection. Res. J. Eng. Technol. 2019;10:59–66. doi: 10.5958/2321-581X.2019.00011.4. [DOI] [Google Scholar]
- 11.Baig R., Rehman A., Almuhaimeed A., Alzahrani A., Rauf H.T. Detecting Malignant Leukemia Cells Using Microscopic Blood Smear Images: A Deep Learning Approach. Appl. Sci. 2022;12:6317. doi: 10.3390/app12136317. [DOI] [Google Scholar]
- 12.Bain B.J. Diagnosis from the blood smear. N. Engl. J. Med. 2005;353:498–507. doi: 10.1056/NEJMra043442. [DOI] [PubMed] [Google Scholar]
- 13.Fey M.F., Buske C. Acute myeloblastic leukaemias in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013;24((Suppl. 6)):vi138–vi143. doi: 10.1093/annonc/mdt320. [DOI] [PubMed] [Google Scholar]
- 14.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett J.M., Catovsky D., Daniel M.T., Flandrin G., Galton D.A., Gralnick H.R., Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann. Intern. Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 16.Mohammed Z.F., Abdulla A.A. An efficient CAD system for ALL cell identification from microscopic blood images. Multimed. Tools Appl. 2021;80:6355–6368. doi: 10.1007/s11042-020-10066-6. [DOI] [Google Scholar]
- 17.Wang M., Lee K.C.M., Chung B.M.F., Bogaraju S.V., Ng H.C., Wong J.S.J., Shum H.C., Tsia K.K., So H.K.H. Low-Latency In Situ Image Analytics With FPGA-Based Quantized Convolutional Neural Network. IEEE Trans. Neural Netw. Learn. Syst. 2021;33:2853–2866. doi: 10.1109/TNNLS.2020.3046452. [DOI] [PubMed] [Google Scholar]
- 18.Rastogi P., Khanna K., Singh V. LeuFeatx: Deep learning–based feature extractor for the diagnosis of acute leukemia from microscopic images of peripheral blood smear. Comput. Biol. Med. 2022;142:105236. doi: 10.1016/j.compbiomed.2022.105236. [DOI] [PubMed] [Google Scholar]
- 19.Atteia G., Alhussan A.A., Samee N.A. BO-ALLCNN: Bayesian-Based Optimized CNN for Acute Lymphoblastic Leukemia Detection in Microscopic Blood Smear Images. Sensors. 2022;22:5520. doi: 10.3390/s22155520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyat M.B., Lai D., Khan F.I., Zhang Y. Sleep Bruxism Detection Using Decision Tree Method by the Combination of C4-P4 and C4-A1 Channels of Scalp EEG. IEEE Access. 2019;7:102542–102553. doi: 10.1109/ACCESS.2019.2928020. [DOI] [Google Scholar]
- 21.Samee N.A., Atteia G., Meshoul S., Al-antari M.A., Kadah Y.M. Deep Learning Cascaded Feature Selection Framework for Breast Cancer Classification: Hybrid CNN with Univariate-Based Approach. Mathematics. 2022;10:3631. doi: 10.3390/math10193631. [DOI] [Google Scholar]
- 22.Samee N.A., Alhussan A.A., Ghoneim V.F., Atteia G., Alkanhel R., Al-Antari M.A., Kadah Y.M. A Hybrid Deep Transfer Learning of CNN-Based LR-PCA for Breast Lesion Diagnosis via Medical Breast Mammograms. Sensors. 2022;22:4938. doi: 10.3390/s22134938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alhussan A.A., Samee N.M.A., Ghoneim V.F., Kadah Y.M. Evaluating Deep and Statistical Machine Learning Models in the Classification of Breast Cancer from Digital Mammograms. [(accessed on 1 June 2022)];Int. J. Adv. Comput. Sci. Appl. 2021 12:1–11. doi: 10.14569/IJACSA.2021.0121033. Available online: https://www.academia.edu/en/68424683/Evaluating_Deep_and_Statistical_Machine_Learning_Models_in_the_Classification_of_Breast_Cancer_from_Digital_Mammograms. [DOI] [Google Scholar]
- 24.Habibzadeh Motlagh M., Jannesari M., Rezaei Z., Totonchi M., Baharvand H. Automatic white blood cell classification using pre-trained deep learning models: ResNet and Inception; Proceedings of the Tenth International Conference on Machine Vision (ICMV 2017); Vienna, Austria. 13–15 November 2017; p. 105. [DOI] [Google Scholar]
- 25.Pan C., Park D.S., Yang Y., Yoo H.M. Leukocyte image segmentation by visual attention and extreme learning machine. Neural Comput. Appl. 2012;21:1217–1227. doi: 10.1007/s00521-011-0522-9. [DOI] [Google Scholar]
- 26.Mohapatra S., Patra D., Satpathi S. Image analysis of blood microscopic images for acute leukemia detection; Proceedings of the 2010 International Conference on Industrial Electronics, Control and Robotics; Rourkela, India. 27–29 December 2010; pp. 215–219. [DOI] [Google Scholar]
- 27.Hegde R.B., Prasad K., Hebbar H., Singh B.M.K., Sandhya I. Automated Decision Support System for Detection of Leukemia from Peripheral Blood Smear Images. J. Digit. Imaging. 2020;33:361–374. doi: 10.1007/s10278-019-00288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goutam D., Sailaja S. Classification of acute myelogenous leukemia in blood microscopic images using supervised classifier; Proceedings of the 2015 IEEE International Conference on Engineering and Technology (ICETECH); Coimbatore, India. 20 March 2015; [DOI] [Google Scholar]
- 29.Qin F., Gao N., Peng Y., Wu Z., Shen S., Grudtsin A. Fine-grained leukocyte classification with deep residual learning for microscopic images. Comput. Methods Programs Biomed. 2018;162:243–252. doi: 10.1016/j.cmpb.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 30.Mundhra D., Cheluvaraju B., Rampure J., Rai Dastidar T. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; Cham, Switzerland: 2017. Analyzing microscopic images of peripheral blood smear using deep learning; pp. 178–185. [DOI] [Google Scholar]
- 31.Çınar A., Arslan S. And neutrophils on white blood cells using hybrid Alexnet—GoogleNet—SVM. SN Appl. Sci. 2021;3:1–11. doi: 10.1007/s42452-021-04485-9. [DOI] [Google Scholar]
- 32.Acevedo A., Alférez S., Merino A., Puigví L., Rodellar J. Recognition of peripheral blood cell images using convolutional neural networks. Comput. Methods Programs Biomed. 2019;180:105020. doi: 10.1016/j.cmpb.2019.105020. [DOI] [PubMed] [Google Scholar]
- 33.Elhassan T.A.M., Rahim M.S.M., Swee T.T., Hashim S.Z.M., Aljurf M. Feature Extraction of White Blood Cells Using CMYK-Moment Localization and Deep Learning in Acute Myeloid Leukemia Blood Smear Microscopic Images. IEEE Access. 2022;10:16577–16591. doi: 10.1109/ACCESS.2022.3149637. [DOI] [Google Scholar]
- 34.Jung C., Abuhamad M., Mohaisen D., Han K., Nyang D.H. WBC image classification and generative models based on convolutional neural network. BMC Med. Imaging. 2022;22:94. doi: 10.1186/s12880-022-00818-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheuque C., Querales M., León R., Salas R., Torres R. An Efficient Multi-Level Convolutional Neural Network Approach for White Blood Cells Classification. Diagnostics. 2022;12:248. doi: 10.3390/diagnostics12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z., Lu S., Wang S.H., Górriz J.M., Zhang Y.D. BCNet: A Novel Network for Blood Cell Classification. Front. Cell Dev. Biol. 2022;9:813996. doi: 10.3389/fcell.2021.813996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Liu R., Dai W., Wu T., Wang M., Wan S., Liu J. AIMIC: Deep Learning for Microscopic Image Classification. Comput. Methods Programs Biomed. 2022;226:107162. doi: 10.1016/j.cmpb.2022.107162. [DOI] [PubMed] [Google Scholar]
- 38.Liang G., Hong H., Xie W., Zheng L. Combining Convolutional Neural Network With Recursive Neural Network for Blood Cell Image Classification. IEEE Access. 2018;6:36188–36197. doi: 10.1109/ACCESS.2018.2846685. [DOI] [Google Scholar]
- 39.Almezhghwi K., Serte S. Improved Classification of White Blood Cells with the Generative Adversarial Network and Deep Convolutional Neural Network. Comput. Intell. Neurosci. 2020;2020:6490479. doi: 10.1155/2020/6490479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L., Shuai R., Ran X., Liu W., Ye C. Combining DC-GAN with ResNet for blood cell image classification. Med. Biol. Eng. Comput. 2020;58:1251–1264. doi: 10.1007/s11517-020-02163-3. [DOI] [PubMed] [Google Scholar]
- 41.Iqbal M.S., Abbasi R., Bin Heyat M.B., Akhtar F., Abdelgeliel A.S., Albogami S., Fayad E., Iqbal M.A. Recognition of mRNA N4 Acetylcytidine (ac4C) by Using Non-Deep vs. Deep Learning. Appl. Sci. 2022;12:1344. doi: 10.3390/app12031344. [DOI] [Google Scholar]
- 42.An Effective and Lightweight Deep Electrocardiography Arrhythmia Recognition Model Using Novel Special and Native Structural Regularization Techniques on Cardiac Signal. [(accessed on 11 October 2022)]. Available online: https://www.hindawi.com/journals/jhe/2022/3408501/ [DOI] [PMC free article] [PubMed] [Retracted]
- 43.Wijesinghe C.B., Wickramarachchi D.N., Kalupahana I.N., De Seram L.R., Silva I.D., Nanayakkara N.D. Fully Automated Detection and Classification of White Blood Cells; Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC); Montreal, QC, Canada. 20–24 July 2020; pp. 1816–1819. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J., Zhang M., Zhou Z., Chu J., Cao F. Automatic detection and classification of leukocytes using convolutional neural networks. Med. Biol. Eng. Comput. 2017;55:1287–1301. doi: 10.1007/s11517-016-1590-x. [DOI] [PubMed] [Google Scholar]
- 45.Zheng X., Wang Y., Wang G., Liu J. Fast and robust segmentation of white blood cell images by self-supervised learning. Micron. 2018;107:55–71. doi: 10.1016/j.micron.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Sajjad M., Khan S., Jan Z., Muhammad K., Moon H., Kwak J.T., Rho S., Baik S.W., Mehmood I. Leukocytes Classification and Segmentation in Microscopic Blood Smear: A Resource-Aware Healthcare Service in Smart Cities. IEEE Access. 2017;5:3475–3489. doi: 10.1109/ACCESS.2016.2636218. [DOI] [Google Scholar]
- 47.Chola C., Benifa J.V.B., Guru D.S., Muaad A.Y., Hanumanthappa J., Al-antari M.A., Alsalman H., Gumaei A.H. Gender Identification and Classification of Drosophila melanogaster Flies Using Machine Learning Techniques. Comput. Math. Methods Med. 2022;2022:4593330. doi: 10.1155/2022/4593330. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Al-masni M.A., Al-antari M.A., Choi M.T., Han S.M., Kim T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018;162:221–231. doi: 10.1016/j.cmpb.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 49.He K. Deep Residual Learning for Image Recognition; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Las Vegas, NV, USA. 27–30 June 2016. [Google Scholar]
- 50.Zagoruyko S., Komodakis N. Wide Residual Networks. arXiv. 20161605.07146 [Google Scholar]
- 51.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Salt Lake City, UT, USA. 18–23 June 2018; pp. 4510–4520. [DOI] [Google Scholar]
- 52.Tan M., Le Q.V. EfficientNet: Rethinking model scaling for convolutional neural networks; Proceedings of the 36th International Conference on Machine Learning ICML; Long Beach, CA, USA. 9–15 June 2019; pp. 10691–10700. [Google Scholar]
- 53.Lin M., Chen Q., Yan S. Network in network; Proceedings of the 2nd International Conference on Learning Representations, ICLR 2014; Banff, AB, Canada. 14–16 April 2014; pp. 1–10. [Google Scholar]
- 54.Al-masni M.A., Al-antari M.A., Park J.M., Gi G., Kim T.Y., Rivera P., Valarezo E., Choi M.T., Han S.M., Kim T.S. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput. Methods Programs Biomed. 2018;157:85–94. doi: 10.1016/j.cmpb.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 55.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- 56.Al-antari M.A., Al-masni M.A., Choi M.T., Han S.M., Kim T.S. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int. J. Med. Inform. 2018;117:44–54. doi: 10.1016/j.ijmedinf.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Chola C., Mallikarjuna P., Muaad A.Y., Bibal Benifa J.V., Hanumanthappa J., Al-antari M.A. A Hybrid Deep Learning Approach for COVID-19 Diagnosis via CT and X-ray Medical Images. Comput. Sci. Math. Forum. 2021;2:13. [Google Scholar]
- 58.Al-antari M.A., Hua C.H., Bang J., Lee S. Fast deep learning computer-aided diagnosis of COVID-19 based on digital chest x-ray images. Appl. Intell. 2021;51:2890–2907. doi: 10.1007/s10489-020-02076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ukwuoma C.C., Qin Z., Belal Bin Heyat M., Akhtar F., Bamisile O., Muad A.Y., Addo D., Al-antari M.A. A Hybrid Explainable Ensemble Transformer Encoder for Pneumonia Identification from Chest X-ray Images. J. Adv. Res. 2022. in press . [DOI] [PMC free article] [PubMed]
- 60.Bin Heyat M.B., Akhtar F., Abbas S.J., Al-Sarem M., Alqarafi A., Stalin A., Abbasi R., Muaad A.Y., Lai D., Wu K. Wearable Flexible Electronics Based Cardiac Electrode for Researcher Mental Stress Detection System Using Machine Learning Models on Single Lead Electrocardiogram Signal. Biosensors. 2022;12:427. doi: 10.3390/bios12060427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heyat M.B., Akhtar F., Khan A., Noor A., Benjdira B., Qamar Y., Abbas S.J., Lai D. A novel hybrid machine learning classification for the detection of bruxism patients using physiological signals. Appl. Sci. 2020;10:7410. doi: 10.3390/app10217410. [DOI] [Google Scholar]
- 62.Sultana A., Rahman K., Heyat M.B., Sumbul, Akhtar F., Muaad A.Y. Role of Inflammation, Oxidative Stress, and Mitochondrial Changes in Premenstrual Psychosomatic Behavioral Symptoms with Anti-Inflammatory, Antioxidant Herbs, and Nutritional Supplements. Oxid. Med. Cell. Longev. 2022;2022:3599246. doi: 10.1155/2022/3599246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sultana A., Begum W., Saeedi R., Rahman K., Bin Heyat M.B., Akhtar F., Son N.T., Ullah H. Experimental and Computational Approaches for the Classification and Correlation of Temperament (Mizaj) and Uterine Dystemperament (Su’-I-Mizaj Al-Rahim) in Abnormal Vaginal Discharge (Sayalan Al-Rahim) Based on Clinical Analysis Using Support Vector Mach. Complexity. 2022;2022:5718501. doi: 10.1155/2022/5718501. [DOI] [Google Scholar]
- 64.Tripathi P., Ansari M.A., Gandhi T.K., Heyat MB B., Akhtar F., Ukwuoma C.C., Muaad A.Y., Kadah Y.M., Al-Antari M.A., Li J.P., et al. Ensemble Computational Intelligent for Insomnia Sleep Stage Detection via the Sleep ECG Signal. IEEE Access. 2022;10:108710–108721. doi: 10.1109/ACCESS.2022.3212120. [DOI] [Google Scholar]
- 65.Mestetskiy L.M., Guru D.S., Benifa J.V.B., Nagendraswamy H.S., Chola C. Gender identification of Drosophila melanogaster based on morphological analysis of microscopic images. Vis. Comput. 2022;2022:1–13. doi: 10.1007/s00371-022-02447-9. [DOI] [Google Scholar]
- 66.Journal A.I., Yao X., Sun K., Bu X., Zhao C., Jin Y. Classification of white blood cells using weighted optimized deformable convolutional neural networks convolutional neural networks. Artif. Cells Nanomed. Biotechnol. 2021;49:147–155. doi: 10.1080/21691401.2021.1879823. [DOI] [PubMed] [Google Scholar]
- 67.Baydilli Y.Y., Atila Ü. Classification of white blood cells using capsule networks. Comput. Med. Imaging Graph. 2020;80:101699. doi: 10.1016/j.compmedimag.2020.101699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset used in this study is publicly available via https://data.mendeley.com/datasets/snkd93bnjr/1 (accessed on 21 June 2022).