Abstract
Anubias Schott (Araceae) have high ornamental properties as aquarium plants. However, the genus has difficulties in species identification, and the mechanism of its adaptation to the aquatic environment is unknown. To better identify species and understand the evolutionary history of Anubias, the plastomes of Anubias barteri Schott, A. barteri var. nana (Engl.) Crusio, and A. hastifolia Engl., were sequenced. The sizes of the plastomes of Anubias ranged from 169,841 bp to 170,037 bp. These plastomes were composed of conserved quadripartite circular structures and comprised 112 unique genes, including 78 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes. The comparative analysis of genome structure, repeat sequences, codon usage and RNA editing sites revealed high similarities among the Anubias plastomes, indicating the conservation of plastomes of Anubias. Three spacer regions with relatively high nucleotide diversity, trnL-CAA-ndhB, ycf1-ndhF, and rps15-ycf1, were found within the plastomes of Anubias. Phylogenetic analysis, based on 75 protein-coding genes, showed that Anubias was sister to Montrichardia arborescens (L.) Schott (BS = 99). In addition, four genes (ccsA, matK, ndhF, and ycf4) that contain sites undergoing positive selection were identified within the Anubias plastomes. These genes may play an important role in the adaptation of Anubias to the aquatic environment. The present study provides a valuable resource for further studies on species identification and the evolutionary history of Anubias.
Keywords: Anubias, aquarium plants, plastomes, positive selection
1. Introduction
The genus Anubias Schott (Araceae, Alismatales) consists of eight perennially herbaceous species endemic to the western and central tropical Africa [1]. Anubias plants tend to grow on the banks of small streams, rough rocks, or driftwoods in tropical humid forests and are sometimes completely submerged [1,2]. They can be specified as aquatic plants because, although they are not physiologically bound to water, they are able to tolerate longer periods of submergence [2,3]. They are widely cultivated as aquarium plants, owing to their aquatic life form (helophytes or rheophytes), exotic appearance and easy maintenance [2,4]. Plants of this genus have been commercialized around the world as highly demanded aquatic ornamental plants [5]. To date, numerous polymorphic cultivars have been developed, such as A. barteri ‘marble’ and A. barteri var. nana ‘petite’ [6]. Although Anubias species can be identified by using mostly characteristics of the inflorescence [1], the inflorescence is often not available, due to its slow growth rate [7]. Leaf blades within the genus Anubias are highly plastic in their morphology [1,2]. Moreover, there are still many taxonomic problems, such as incomplete species names and synonyms among the Anubias species, making classification inconsistent both on the markets and in aquariums [5,8]. Considering their high economic value, there is a need for further improvement of species identification within the genus.
Accurate species identification of plants is fundamental for their utility, breeding new cultivars, and implementation of conservation priorities and biosecurity monitoring. Historically, identification of the majority of plant species has been based on the analysis of morphological variation, which has resulted in low reliability of species identification due to phenotypic plasticity [9]. The analysis of DNA sequence variation can provide useful information for species identification and for phylogenetic analysis [10]. The chloroplast (cp) genomes of plants are relatively conserved in size, organization, gene content and order, giving them unique values in comparative genomics and phylogenetics, compared to the nuclear and mitochondrial genomes [11]. To date, only one cp genome (A. heterophylla Engl.) in the genus Anubias has been reported [12]. Therefore, it is necessary to acquire more cp genomes within the genus Anubias.
Understanding the mechanism of plant adaptations to the aquatic environment is an important topic in evolutionary biology. Aquatic plants occupy stressful water habitats characterized by low light levels, reduced carbon and oxygen availability, and mechanical damage through wave exposure [13]. To survive in the aquatic environment, aquatic plants have undergone a series of morphologic and physiologic adaptive changes. For example, the aerenchyma in roots, stems, and leaves enhances the capture and transportation of oxygen [14,15]. At the metabolic level, aquatic plants might change the level of glycolytic fluxes and ethanolic fermentation [16]. Although all of the Anubias species have adapted to the aquatic environment, the related mechanism has rarely been reported.
In this study, we sequenced and assembled the plastomes of A. barteri Schott, A. barteri var. nana (Engl.) Crusio, and A. hastifolia Engl; then, we compared them with the published plastome sequences of A. heterophylla and other genera in the family Araceae. Furthermore, the phylogenetic relationships among Anubias and other genera in the Arum family were reconstructed based on consensus protein-coding genes. Our main objectives were to (1) explore the size range and structure of the Anubias plastomes; (2) detect highly variable regions as the bases of developing molecular markers for species identification; (3) construct a phylogenetic tree for investigating the interspecific relationships within Anubias, as well as the relationships among Anubias and other genera in Araceae; (4) identify the protein-coding genes under positive selection within the four plastomes of Anubias.
2. Materials and Methods
2.1. Plant Material, Plastome Sequencing, Assembly and Annotation
The samples of cultivated plants, including A. barteri (voucher number: JYH085), A. barteri var. nana (voucher number: JYH087), and A. hastifolia (voucher number: JYH086), were collected by Yunheng Ji on August 27, 2018, from Kunming World Horticultural Expo Garden, Yunnan, China. The voucher specimens were deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN, Yunheng Ji, jiyh@mail.kib.ac.cn). Total genomic DNA of three Anubias plants was extracted from 20 mg silica gel-dried leaf tissues using the CTAB method [17]. Then, genomic DNA was fragmented into 500 bp fragments to construct a paired-end library according to the manufacturer’s protocol (Illumina, San Diego, CA, USA) and finally sequenced on the Illumina HiSeq 2000 system.
Raw reads were assembled using the software GetOrganelle v1.7.1 [18], with the default parameters, and the plastome sequence of A. heterophylla (GenBank accession number: MN046884) was set as a reference. The assembled genomes were annotated using the PGA (Plastid Genome Annotator) software [19] and the online software, Geseq [20]. The preliminary annotated sequences were manually corrected for start and stop codons and intron/exon boundaries in Geneious prime 2019.2.1 [21]. All the tRNA genes were further verified using the online software tRNAscan-SE, version 2.0 [22], with the default parameters. Furthermore, circular genome maps of Anubias were visualized using the online program OrganellarGenomeDRAW version 1.3.1 [23]. Finally, newly sequenced plastomes of A. barteri, A. barteri var. nana, and A. hastifolia were deposited into GenBank with the accession numbers OP279443, OP279444, and MW984413, respectively.
2.2. Comparative Plastome Analysis
To better understand the interspecific variation of the plastomes in the genus Anubias, one published and three newly sequenced plastomes of Anubias were compared. Moreover, 27 published plastomes (Table S1) in the family Araceae were added to determine the intergeneric variation among Anubias and other genera. Genome rearrangements within the family Araceae, including Anubias and other genera, were identified using the Mauve alignment [24], after removing all the IRa regions. Interspecific variations among the complete plastomes in the genus Anubias were performed in the online program mVISTA [25] with the shuffle-LAGAN model, using A. heterophylla as a reference. The boundaries of the LSC (large single-copy), SSC (small single-copy), and IRs (inverted repeats) of the plastomes of Anubias and other genera were visualized with IRscope [26].
2.3. Repeats, Nucletide Diversity, Codon Usage and RNA Editing Sites
The REPuter program [27] was used to analyze the numbers of forward, palindromic, reverse and complement repeats of four Anubias plastomes with the following parameters: Hamming distance was 3, and minimal repeat size was 30 bp. The Simple Sequence Repeats (SSRs) of four Anubias plastomes were detected using the MIcroSAtellite identifcation tool (MISA) [28], including mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotides, with a minimum number of 8, 4, 4, 3, 3, and 3, respectively.
Four Anubias plastomes were aligned using MAFFT v7.450 [29]. Nucleotide diversity analysis was performed using DnaSP v.6.12.01 software [30] with the parameters of 600 bp in window length and 200 bp in step size.
To identify the frequency of synonymous codon and codon biases within the genus Anubias, the relative synonymous codon usage (RSCU) [31] of the protein-coding genes was analyzed in CondoW v1.4.2 software [32], after removing the sequences less than 300 bp [33]. The RNA editing sites in the plastomes of Anubias were analyzed by a predictive RNA editor for plants (PREP-cp) with the default settings [34].
2.4. Phylogenetic Analysis
To examine the phylogenetic positions of Anubias, 31 aroid taxa, including 3 Anubias taxa, were sequenced in this study and 28 taxa in the family Araceae downloaded from the GenBank were selected in this study (Table S1). The species Zamioculcas zamiifolia (Lodd.) Engl. was set as the outgroup. A maximum likelihood (ML) tree was constructed based on 75 protein-coding genes (Table S2) shared by the 31 plastomes. Each selected protein-coding gene sequence alignment was performed with MAFFT v.7.450 and concatenated to a supermatrix. The ML tree was constructed using the RAxML-HPC2 program [35] on the XSEDE resource in the CIPRES Science Gateway [36] with the GTRGAMMA model. Bootstrap support values were obtained with 1000 bootstrap replicates.
2.5. Positive Selection Analysis
To identify positively selected protein-coding genes in the genus Anubias, each protein-coding gene sequence matrix was aligned using MAFFT v.7.450. The stop codons were again manually deleted in each aligned sequence. The phylogenetic tree for each protein-coding gene was constructed using the FastTree 2.1.11 plugin [37] of Geneious prime 2019.2.1. The ratio of nonsynonymous (dN) and synonymous substitution (dS) (ω = dN/dS) was calculated using the site-specific model (M0, M1a, M2a, M3, M7, M8, and M8a) based on likelihood ratio tests (LRTs) with statistically significant p values (<0.05) performed in CODEML algorithms [38] implemented in EasyCondelML v1.4 software [39].
3. Results
3.1. Plastome Features
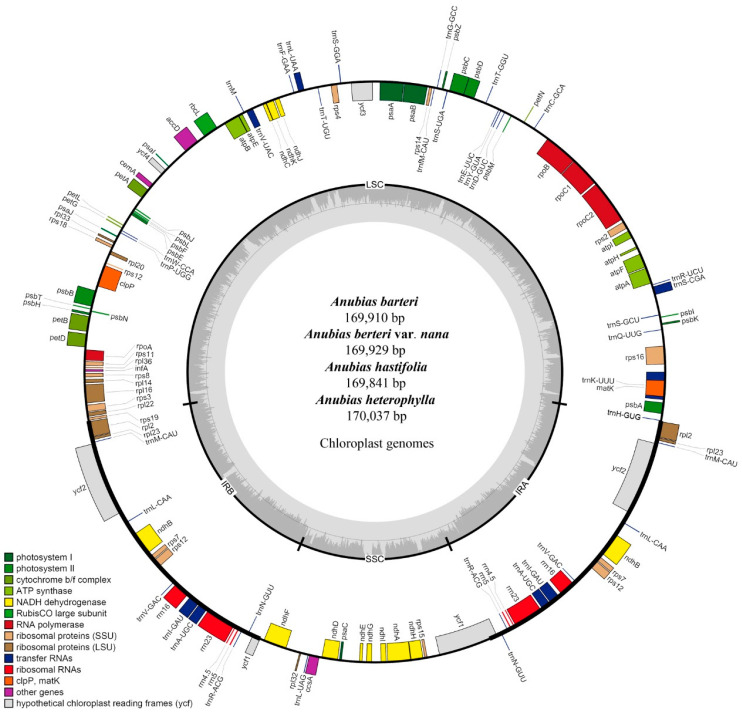
Complete plastomes of A. barteri, A. barteri var. nana, and A. hastifolia were assembled with genome sizes of 169,910 bp, 169,929 bp, and 169,841 bp in length, respectively (Figure 1). The sizes within the plastomes of Araceae in this study ranged between 158,177 bp (Anchomanes hookeri Schott) and 175,906 bp (Zantedeschia elliottiana Engl.). Four Anubias plastomes were composed of a circular conserved quadripartite structure, with similar gene content and genome organization. Each sample within the Anubias plastomes comprised a total of 112 unique genes, including 78 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes (Table 1).
Figure 1.
Circular map of chloroplast genomes of four Anubias plants. Genes shown inside of the inside layer circle are transcribed clockwise, whereas those genes outside of this circle are transcribed counterclockwise. The colored bars indicate the known protein-coding genes, tRNA, and rRNA. The darker gray area of the inner circle denotes the GC content, while the lighter gray area indicates the AT content of the genome. LSC, large single-copy; SSC, small single-copy; IR, inverted repeat.
Table 1.
List of genes identified in four Anubias plastomes.
| Category of Genes | Group of Gene | Name of Gene |
|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4.5 ×2, rrn5 ×2, rrn16 ×2, rrn23 ×2 |
| Transfer RNA genes | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU *, trnL-UAA *, trnL-UAG, trnM, trnM-CAU ×2, trnP-UGG, trnQ-UUG, trnR-UCU, trnS-CGA, trnS-GCU, trnS-GGA, trnS-UGA, trnT-UGU, trnT-GGU, trnV-UAC *, trnY-GUA, trnW-CCA, trnfM-CAU, trnA-UGC *,×2, trnI-GAU *,×2, trnL-CAA ×2, trnN-GUU ×2, trnR-ACG ×2, trnV-GAC ×2 | |
| Ribosomal protein (small subunit) | rps2, rps3, rps4, rps7×2, rps8, rps11, rps12 **,×2, rps14, rps15, rps16 *, rps18, rps19 | |
| Ribosomal protein (large subunit) | rpl2 *,×2, rpl14, rpl16 *, rpl20, rpl22, rpl23 ×2, rpl32, rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Translational initiation factor | infA | |
| Genes for photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3 **, ycf4 |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of cytochrome | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of Rubisco | rbcL | |
| Subunits of NADH dehydrogenase | ndhA *, ndhB *,×2, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Other genes | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA | accD | |
| Synthesis gene | ccsA | |
| ATP-dependent protease | clpP ** | |
| Component of TIC complex | ycf1 ×2 | |
| Genes of unknown function | Conserved open reading frames | ycf2 ×2 |
Note: ×2: Two gene copies in IR regions; *: With one intron; **: With two introns.
The overall GC content percentages of A. barteri (35.2%), A. barteri var. nana (35.2%), A. hastifolia (35.1%), and A. heterophylla (35.1%) were similar, and this number ranged between 34.7% (Schismatoglottis calyptrata (Roxb.) Zoll. & Moritzi) and 37.0% (Anchomanes hookeri) within Araceae (Table S1). The LSC length ranged between 75,594 bp (Anchomanes hookeri) and 94,702 bp (Arisaema franchetianum Engl.), with an average length of 91,000 bp; the SSC length ranged between 8432 bp (Zantedeschia elliottiana) and 24,871 bp (Pinellia peltata C.Pei), with an average length of 20,216 bp; and the IR length ranged between 24,982 bp (Pinellia peltata) and 39,445 bp (Zantedeschia elliottiana), with an average length of 27,553 bp (Table S1). The Mauve alignment revealed that no genome rearrangements existed within the plastomes of Anubias, but did exist among other 6 genera (such as Anchomanes Schott, Arisarum Mill., Zantedeschia Spreng.) in the family Araceae (Figure S1).
3.2. Contraction and Expansion of INVERTED repeats
Comprehensive comparative analysis of the junction was performed among the 31 taxa for the contraction and expansion in JLB (LSC/IRb), JSB (IRb/SSC), JSA (SSC/IRa), and JLA (IRa/LSC). Four Anubias plastomes shared similar junction structures. JLB junctions were located between the rps19 gene and the rpl2 gene in four Anubias plastomes and other studied Araceae plastomes, with the exception of Zantedeschia elliottiana, Zomicarpella amazonica Bogner, and Anchomanes hookeri, which presented the JLB within the rpl22, rpl2, rpl23 genes, respectively. JSB and JSA junctions were relatively conserved in four Anubias plastomes but highly variable in other analyzed Araceae plastomes. In four Anubias plastomes, the JSB junctions were characterized by the presence of a truncated copy of the ycf1 gene. This phenomenon was also found in seven other plastomes (Philodendron lanceolatum Schott, Homalomana occulta, Aglaonema costatum N.E.Br., Syngonium angustatum Schott, Pistia stratiotes L., Xanthosoma helleborifolium (Schott) Schott, and Zomicarpella amazonica) in this study. JSA junctions were completely included in the ycf1 gene in four Anubias plastomes and in the other seven plastomes mentioned above in the JSB junctions. JLA junctions were highly conserved in four Anubias plastomes and other plastomes with the presence of rpl2 and trnH genes, except in Zantedeschia elliottiana (with the presence of rps19 and psbA genes) and Anchomanes hookeri (with the presence of trnQ and psbK genes) (Figure S2).
3.3. Repeats, Nucleotide Diversity, Codon Usage and RNA Editing Sites
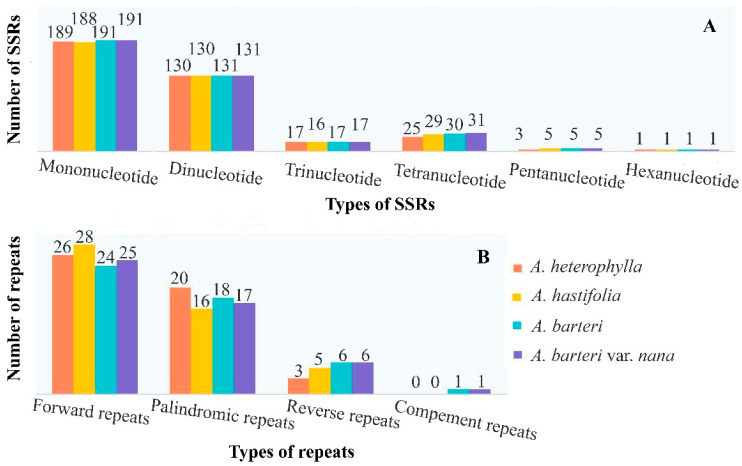
The number of SSRs observed in four Anubias plastomes ranged between 365 (A. heterophylla) and 376 (A. barteri var. nana). Mononucleotide A/T repeats were the most abundant types of repeats in four Anubias plastomes (Figure 2A, Table S3). Analysis of the long repeats (forward, reverse, palindromic, and complement repeats) showed high consistency in repeat number among four Anubias plastomes. Forward repeats were the most abundant types of repeats in four Anubias plastomes. Complement repeats were the most unusual types of repeats in A. barteri and A. barteri var. nana, while reverse repeats were the most unusual types of repeats in A. heterophylla and A. hastifolia (Figure 2B, Table S4).
Figure 2.
Four types of long repeats in four Anubias plastomes. (A) Number of SSRs; (B) Number of four types of long repeats.
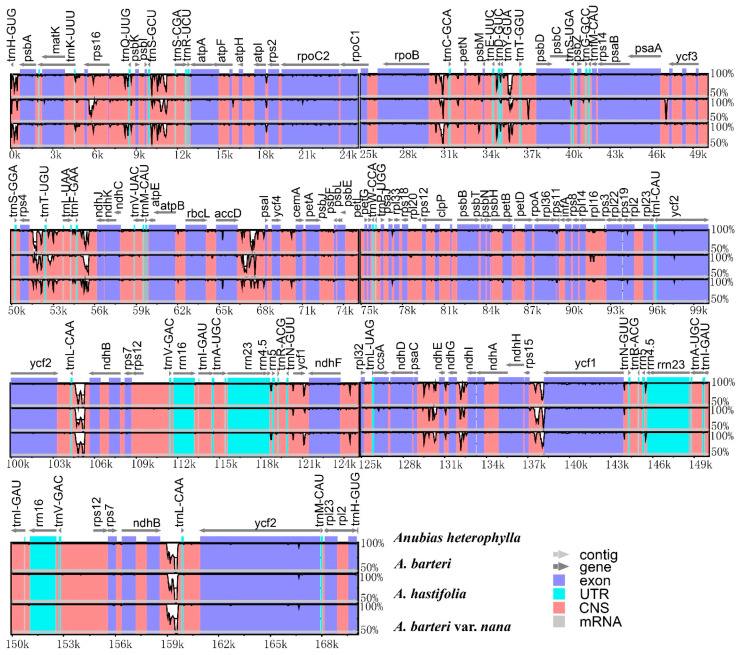
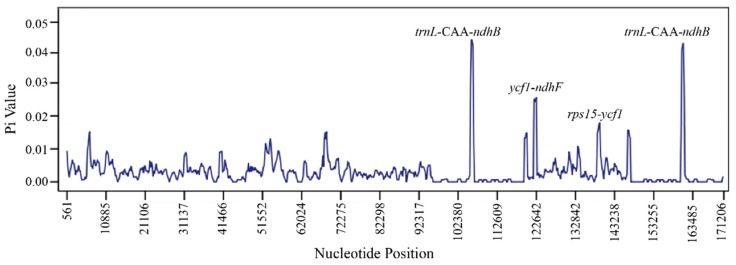
Interspecific variation among four plastomes of Anubias conducted in the online program mVISTA showed that the coding region was more conserved than the noncoding region (Figure 3). Furthermore, the nucleotide diversity (Pi) of four Anubias plastomes was analyzed. Sliding window analysis detects some regions with high Pi values, e.g., trnL-CAA-ndhB, ycf1-ndhF, and rps15-ycf1 spacer regions with Pi values of 0.042, 0.025, and 0.017, respectively (Figure 4).
Figure 3.
Alignment visualization of four Anubias plastomes with the mVISTA program.
Figure 4.
Nucleotide diversity and hotspot regions of four Anubias plastomes.
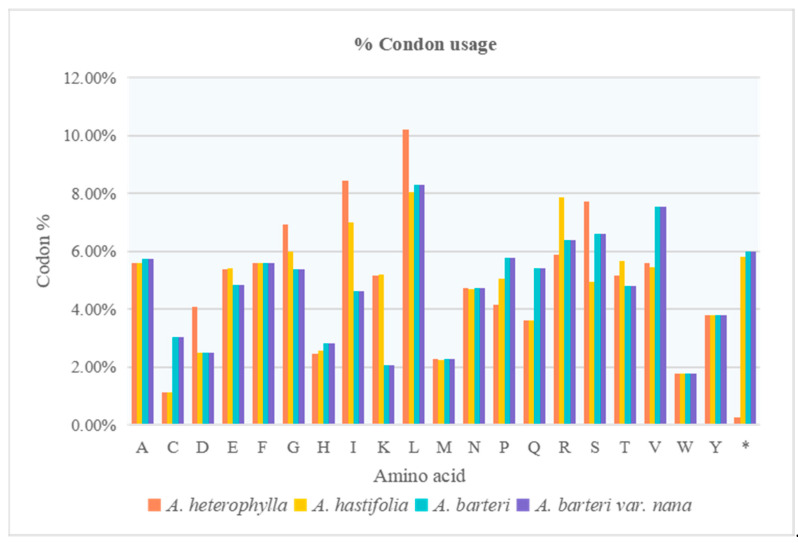
Codon usage of four Anubias plastomes were compared. The plastomes of A. heterophylla, A. hastifolia, A. barteri, and A. barteri var. nana exhibited 21,145; 21,358; 21,361; and 21,361 codons, respectively. Leucine (L) was the most frequently coded amino acid in all of the compared plastomes. Cysteine (C) was the least coded amino acid in the plastomes of A. heterophylla and A. hastifolia, while tryptophan (W) was the least coded amino acid in the plastomes of A. barteri and A. barteri var. nana (Figure 5). The highest RSCU values were 1.94, 1.93, 1.93, and 1.93, while the lowest values were all 0.29 in A. heterophylla, A. hastifolia, A. barteri, and A. barteri var. nana (Table S5). Codon usage was biased toward adenine (A) and thymine (T) in all of the compared Anubias plastomes. RNA editing analysis showed similarities with respect to genes and the position of editing sites in the coding genes. We predicted RNA editing sites in 23 to 24 genes among four Anubias plastomes (Table S6). The matK gene contained one RNA editing site in the plastome of A. hastifolia with the conversion of TCA to TTA. The rpl2 genes contained ACG as a start codon instead of ATG in four Anubias plastomes.
Figure 5.
Codon Usage in four Anubias plastomes. * stop condon.
3.4. Phylogenetic Analysis
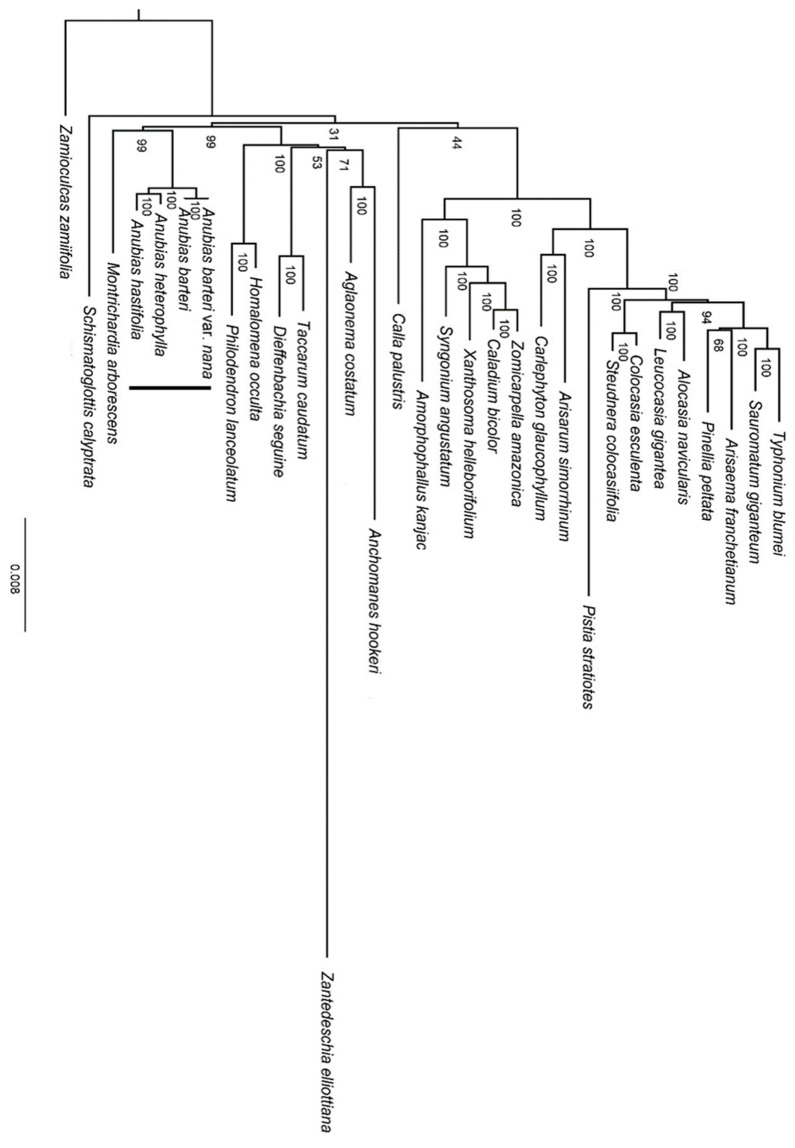
In the phylogenetic analysis (Figure 6), the four representatives of Anubias formed a monophyletic group with strong support (BS = 100), in which A. heterophylla was sister to A. hastifolia (BS = 100), with A. barteri and A. barteri var. nana being sister to the clade of A. heterophylla and A. hastifolia (BS = 100). The genus Anubias was sister to Montrichardia arborescens (L.) Schott with robust support (BS = 99).
Figure 6.
ML phylogenetic tree reconstruction of Anubias and other genera in Araceae based on 75 consensus CDSs.
3.5. Selective Pressure Analysis
A total of 78 consensus protein-coding genes of four Anubias plastomes were selected to estimate the selective pressure. Four genes, ccsA, matK, ndhF, and ycf4, were identified to have undergone positive selection with ω values of 20.827, 21.224, 58.206, and 137.628, respectively (Table 2, Table S7).
Table 2.
Positive selection sites in four Anubias plastomes.
| Gene Name | Positive Sites |
|---|---|
| ccsA | 90 Y 0.935, 91 F 0.843, 92 R 0.636, 198 H 0.937, 211 Y 0.598, 216 L 0.625 |
| matK | 37 L 0.630, 46 E 0.618, 94 F 0.607, 95 D 0.933, 214 R 0.660, 393 P 0.615, 459 P 0.936 |
| ndhF | 49 N 0.579, 53 V 0.566, 291 M 0.568, 463 Q 0.575, 571 D 0.568, 644 G 0.871 |
| ycf4 | 129 G 0.886 |
4. Discussion
4.1. Chloroplast Genome Features and Comparisons
Three complete plastomes of Anubias were obtained in this study. The comparative analysis of the plastomes within the genus was first reported. The plastomes of A. hastifolia, A. barteri, and A. barteri var. nana were extremely similar to the published A. heterophylla in terms of genome size, genome structure, and gene content, indicating that the plastomes of Anubias were relatively conserved. Similar genome structures and gene contents were also reported in other plastomes of Araceae [12,40,41]. In all analyzed plastomes, the regions with higher GC content are located in IR regions or coding sequences, and the same findings are found in previous studies [12,40,41].
The contraction and expansion of the IR borders are common evolutionary events in most angiosperms (e.g., monocots) [42]. In this study, a comparative analysis of IR borders revealed that the functional ycf1 gene existed in the JSA junction and that the other pseudogene ycf1 copy was located at the JSB junction among four Anubias plastomes. The same phenomenon was also found in seven analyzed plastomes within the family Araceae in this study and other angiosperm plastomes in previous studies [12,41].
SSRs, also known as microsatellites, have been widely applied as molecular markers for population genetics and species delimitation in aquatic plants [43]. SSR analysis in this study revealed that mononucleotide A/T repeats were the most abundant type of repeats in four Anubias plastomes, which was similar to the previous studies in other species [41,44]. SSRs of Anubias reported for the first time in this study can act as potential markers for genetic diversity studies of the genus. Our study also revealed the numbers of four types of oligonucleotide repeats, in which forward repeats were the most abundant types of repeats. These repeats may play an important role in the generation of substitutions and InDels, as previous studies of nuclear and cp genomes have revealed [45,46].
Codon usage bias patterns in genomes can be used to reveal phylogenetic relationships between organisms, horizontal gene transfers, and the molecular evolution of genes and identify selective forces [47]. RSCU values, as an important parameter, have been used for evaluating the codon usage bias degree [48]. A higher RSCU value (RSCU > 1) denotes the more frequently used codon in a gene, and a lower RSCU value (RSCU < 1) denotes the less frequently used codon [31]. High RSCU values of the codons are probably related to amino acid functions that avoid transcriptional errors in cp genomes [49]. Our study revealed that the highest abundance of the amino acid was leucine in four Anubias plastomes, which is similar to previous studies [41].
RNA editing is described as a posttranscriptional change in RNA nucleotides that alters a cytosine (C) to uridine (U) at a specific codon [50]. This is the key mechanism by which RNA maturation avoids incorrect mutations and enriches genetic information [51]. Our analysis revealed that the ndhB gene contained the most RNA editing sites, within 12 potential RNA editing sites, which is similar to previous reports in other species [44]. Additionally, we also observed that the rpl2 gene contained ACG as a start codon instead of ATG in four Anubias plastomes, and this phenomenon was first reported in the maize plastome [52]. RNA editing mostly occurs in the first and second bases of codons, resulting in the conversion of hydrophilic amino acids to hydrophobic amino acids [53]. Research on RNA editing can improve our understanding of the gene expression and molecular evolution mechanisms of Anubias.
Saad et al. [4] revealed that the matK gene had shown high potential as DNA barcoding for selected Anubias species in their research. Our results also supported that the matK gene had a high variability in the coding region. However, the coding region was more conserved than the noncoding region, which is consistent with previous studies in other species [44]. Hence, three intergenic spacer regions (trnL-CAA-ndhB, ycf1-ndhF, and rps15-ycf1) with higher level of variation were selected in this study, which could be used for further development in applications such as DNA barcoding and phylogenetic reconstruction.
4.2. Phylogenetic Inference
Although the phylogenetic position of the genus among the monocots was previously known, the accuracy of the phylogenetic relationship needs to be confirmed with more evidence because of the insufficient sample size (21–23/75 genera in the subfamily Aroideae) [12,40]. In our study, we extended the taxon sampling to 27 genera within the subfamily Aroideae. Although the sampling size is still limited, our result was consistent with previous studies with robust support (BS = 99) [12,40]. The result was also supported by the same basic number of chromosomes (x = 24) shared by Anubias and Montrichardia [54]. In addition, the uncertain position of Calla L. and Schismatoglottis Zoll. & Moritzi in the phylogenies of Araceae reported in a previous study [12] was still unresolved in our study with low support values. We propose that future phylogenetic studies of Araceae should be focused on wider taxa sampling and nuclear genomes.
4.3. Adaptations to the Aquatic Environment
All of the Anubias species have adapted to the aquatic environment, indicating that this species might have evolved mechanisms to respond to different abiotic stresses that occur underwater, such as low light level, reduced carbon and oxygen availability, and mechanical damage through wave exposure. For example, A. barteri may adapt to low light level by increasing leaf area and chlorophyll content and to low O2 and CO2 concentration underwater through forming adventitious roots [55]. Considering the characteristics of their slow growth rate [7] and unusual morphological anatomy (e.g., no aerenchyma tissues differentiated in the adventitious root of A. barteri) [55], we speculate that Anubias possibly adopts the low-oxygen quiescence strategy (LOQS, an energy-saving mode) [56] to adapt to the aquatic environment. However, this needs to be confirmed by future studies.
With the exception of morphological, physiological and anatomical adaptations, some molecular adaptations (e.g., gene loss, gene positive selection) of aquatic plants could play an important role in the aquatic environment [57,58]. In this study, there was no gene loss other than four genes (ccsA, matK, ndhF, and ycf4) under positive selection with significant selective sites in Anubias plastomes. Previous studies have revealed the ccsA gene with positive selection in some aquatic plants such as Oryza L. [59], Zosteraceae [58], and some species of Lythraceae [60]. The ccsA gene is required for the biogenesis of c-type cytochromes at the step of heme attachment, and its function is linked to electron transfer in respiration and photosynthesis [61,62]. The matK gene has been reported to be under positive selection in some aquatic or hygrophilous plants (e.g., Oryza, Lupinus L.) [59,63]. This gene encodes an intron maturase that is involved in the splicing of group II RNA transcriptional introns, and its function is linked to plastid translation and photosynthesis [64,65,66]. Currently the ndhF gene under positive selection has not yet been reported in aquatic plants, but has been reported in some land plants, such as Debregeasia Gaudich. (Urticaceae) [67], Rheum L. (Polygonaceae) [68], and Limonium Mill. (Plumbaginaceae) [69]. The ndhF gene is a subunit of NADH-dehydrogenase, and its functions are linked to cyclic electron flow around photosystem I, essential for photosynthesis [70,71]. Some studies have reported the ycf4 gene with positive selection in several plants, such as some species of Zingiberaceae [72], Lythraceae [60], and Orchidaceae [73]. The ycf4 gene encodes a protein as a nonessential assembly factor for photosystem I, and it may have additional functions in chloroplasts deficient in photosystem II repair [74,75]. Further studies of Anubias plants chloroplast genomes should determine whether the positive selection of these genes is related to the adaptations to the aquatic environment.
5. Conclusions
In this study, three complete plastomes of Anubias were newly sequenced, and the structures of four plastomes within the genus were compared. Through comparative analyses, some important genetic features (such as IR contraction and expansion, SSRs, codon usage and RNA editing sites) were obtained. Three spacer regions (trnL-CAA-ndhB, ycf1-ndhF, and rps15-ycf1) with a high potential to be developed for DNA barcoding were found in the Anubias plastomes. Phylogenetic analysis showed that Anubias was sister to Montrichardia with robust support. Four genes (ccsA, matK, ndhF, and ycf4) were identified to have undergone positive selection. These results could provide valuable information for further studies on species identification and the evolutionary history of Anubias, especially its molecular adaptations to the aquatic environment. However, more species sampling of Anubias and other genera in the family Araceae is needed to facilitate our understanding of the phylogeny and evolutionary history within Anubias in the future.
Acknowledgments
We also thank Yunheng Ji (Kunming Institute of Botany, CAS) for providing materials used for this work and Linbo Jia (Kunming Institute of Botany, CAS) for providing valuable ideas for the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13112043/s1, Figure S1: Gene rearrangement analyses among Anubias and other genera in Araceae by Mauve alignment; Figure S2: Contraction and expansion of inverted repeats of Anubias and other genera in Araceae; Table S1: Plastome characteristics in Anubias and other genera in Araceae; Table S2: 75 protein-coding genes used to construct a ML phylogenetic tree; Table S3: Number of SSRs in four Anubias plastomes; Table S4: Number of long repeats in four Anubias plastomes; Table S5: Codon Usage in four Anubias plastomes; Table S6: RNA editing sites in the four Anubias plastomes; Table S7: Positive selection sites in four Anubias plastomes.
Author Contributions
Conceptualization, L.L. and C.L.; Data curation, L.L. and K.H.; Formal analysis, L.L. and C.L.; Funding acquisition, W.L.; Methodology, L.L. and C.L.; Project administration, W.L.; Software, C.L. and K.H.; Supervision, W.L.; Validation, L.L., C.L. and K.H.; Visualization, K.H.; Writing—original draft, L.L. and C.L.; Writing—review and editing, L.L., C.L. and W.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be made available on request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Key Research and Development Program of Shanxi Province of China (Grant/Award Number: 2019SF-305) and the Fourth National Survey of Traditional Chinese Medicine Resources (Grant/Award Number: 2019-6).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Crusio W.E. A revision of Anubias Schott (Araceae) Meded. Landbouwhogesch. 1979;79:1–48. [Google Scholar]
- 2.Mayo S., Bogner J., Boyce P. The Genera of Araceae. Royal Botanical Gardens, Kew; London, UK: 1997. pp. 180–182. [Google Scholar]
- 3.Cook C. The number and kinds of embryo-bearing plants which have become aquatic: A survey. Perspect. Plant Ecol. Evol. Syst. 1999;2:79–102. doi: 10.1078/1433-8319-00066. [DOI] [Google Scholar]
- 4.Saad M., Sahidin N., Bhassu S. DNA Barcoding of Selected Aquarium Plants, Anubias Species Using Chloroplast Marker; Proceedings of the 18th Scientific Meeting Malaysian Society for Molecular Biology and Biotechnology (MSMBB); Kuala Lumpur, Malaysia. 18–20 August 2009; [DOI] [Google Scholar]
- 5.Brunel S. Pathway analysis: Aquatic plants imported in 10 EPPO Countries. EPPO Bull. 2009;39:201–213. doi: 10.1111/j.1365-2338.2009.02291.x. [DOI] [Google Scholar]
- 6.Oyedeji A., Abowei J. The classification, distribution, control and economic importance of aquatic plants. Int. J. Fish. Aquat. Sci. 2012;1:118–128. [Google Scholar]
- 7.Sholichah L., Yamin M., Ginanjar R., Meilisza N. Anubias (Anubias sp.) propagation trough hydroponic culture technique. J. Phys. Conf. Ser. 2020;1422:012024. doi: 10.1088/1742-6596/1422/1/012024. [DOI] [Google Scholar]
- 8.Han S.Q. Master’s Thesis. Zhejing University; Hangzhou, China: 2005. Study on Ornamental Water Grass Species and Cultivation Technique. [Google Scholar]
- 9.Duminil J., Di Michele M. Plant species delimitation: A comparison of morphological and molecular markers. Plant Biosyst. 2009;143:528–542. doi: 10.1080/11263500902722964. [DOI] [Google Scholar]
- 10.Hollingsworth P., Li D.Z., van der Bank M., Twyford A. Telling plant species apart with DNA: From barcodes to genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150338. doi: 10.1098/rstb.2015.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouin G., Daoud H., Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008;49:827–831. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Henriquez C., Abdullah, Ahmed I., Carlsen M., Zuluaga A., Croat T., Mckain M. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae) Genomics. 2020;112:2349–2360. doi: 10.1016/j.ygeno.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Chen L.Y., Lu B., Morales-Briones D.F., Moody M.L., Liu F., Hu G.W., Huang C.H., Chen J.M., Wang Q.F. Phylogenomic Analyses of Alismatales Shed Light into Adaptations to Aquatic Environments. Mol. Biol. Evol. 2022;39:msac079. doi: 10.1093/molbev/msac079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung J., Lee S.C., Choi H.-K. Anatomical patterns of aerenchyma in aquatic and wetland plants. J. Plant Biol. 2008;51:428–439. doi: 10.1007/BF03036065. [DOI] [Google Scholar]
- 15.Nowak J.S., Ono J., Cronk Q.C. Anatomical study of an aquatic mustard: Subularia aquatica (Brassicaceae) Aquat. Bot. 2010;93:55–58. doi: 10.1016/j.aquabot.2010.02.004. [DOI] [Google Scholar]
- 16.Billet K., Genitoni J., Bozec M., Renault D., Barloy D. Aquatic and terrestrial morphotypes of the aquatic invasive plant, Ludwigia grandiflora, show distinct morphological and metabolomic responses. Ecol Evol. 2018;8:2568–2579. doi: 10.1002/ece3.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle J., Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 18.Jin J.J., Yu W.B., Yang J.B., Song Y., de Pamphilis C., Yi T.S., Li D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu X.J., Moore M., Li D.Z., Yi T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:50. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E., Fischer A., Bock R., Greiner S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan P., Lowe T. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling A., Mau B., Blattner F., Perna N. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frazer K., Pachter L., Poliakov A., Rubin E., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amiryousefi A., Hyvönen J., Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 27.Kurtz S., Choudhuri J., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-Web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K., Standley D. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J., Guirao-Rico S., Librado P., Ramos-Onsins S., Sánchez-Gracia A. DnaSP6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 31.Sharp P., Li W.H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peden J. Ph.D. Thesis. University of Nottingham; Nottingham, UK: 1999. Analysis of Codon Usage. [Google Scholar]
- 33.Wright F. The Effective Number of Codons Used in A Gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 34.Mower J. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller M., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees; Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; [DOI] [Google Scholar]
- 37.Price M., Dehal P., Arkin A. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z.H., Nielsen R. Synonymous and Nonsynonymous Rate Variation in Nuclear Genes of Mammals. J. Mol. Evol. 1998;46:409–418. doi: 10.1007/PL00006320. [DOI] [PubMed] [Google Scholar]
- 39.Gao F.L., Chen C.J., Arab D., Du Z.G., He Y.H., Ho S. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henriquez C., Arias T., Pires J., Croat T., Schaal B. Phylogenomics of the plant family Araceae. Mol. Phylogenet. Evol. 2014;75:91–102. doi: 10.1016/j.ympev.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Abdullah, Henriquez C., Mehmood F., Hayat A., Sammad A., Waseem S., Waheed M., Matthews P., Croat T., Poczai P., et al. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae) Genomics. 2021;113:183–192. doi: 10.1016/j.ygeno.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 42.Wang R.J., Cheng C.L., Chang C.C., Wu C.L., Su T.M., Chaw S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu S.Q., Li G.J., Yang J.J., Hou H.W. Aquatic Plant Genomics: Advances, Applications, and Prospects. Int. J. Genom. 2017;2017:6347874. doi: 10.1155/2017/6347874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong F., Lin Z.C., Lin J., Ming R., Zhang W.P. Chloroplast Genome of Rambutan and Comparative Analyses in Sapindaceae. Plants. 2021;10:283. doi: 10.3390/plants10020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mcdonald M., Wang W.C., Huang H.D., Leu J.Y. Clusters of Nucleotide Substitutions and Insertion/Deletion Mutations Are Associated with Repeat Sequences. PLoS Biol. 2011;9:e1000622. doi: 10.1371/journal.pbio.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed I., Biggs P., Matthews P., Collins L., Hendy M., Lockhart P. Mutational Dynamics of Aroid Chloroplast Genomes. Genome Biol. Evol. 2012;4:1316–1323. doi: 10.1093/gbe/evs110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parvathy S., Udayasuriyan V., Bhadana V. Codon usage bias. Mol. Biol. Rep. 2022;49:539–565. doi: 10.1007/s11033-021-06749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaBella A., Opulente D., Steenwyk J., Hittinger C., Rokas A. Variation and selection on codon usage bias across an entire subphylum. PLoS Genet. 2019;15:e1008304. doi: 10.1371/journal.pgen.1008304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raman G., Park S., Lee E., Park S. Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Sci. Rep. 2019;9:5028. doi: 10.1038/s41598-019-41377-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maier R., Zeltz P., Kössel H., Bonnard G., Gualberto J., Grienenberger J. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 51.Wang M.X., Liu H., Ge L.Q., Xing G.W., Wang M., Song W.N., Nie X.J. Identification and Analysis of RNA Editing Sites in the Chloroplast Transcripts of Aegilops tauschii L. Genes. 2016;8:13. doi: 10.3390/genes8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoch B., Maier R., Appel K., Igloi G., Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991;353:178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- 53.Shikanai T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell. Mol. Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen G. Cytology and systematics of Araceae. Nord. J. Bot. 1989;9:119–166. doi: 10.1111/j.1756-1051.1989.tb02111.x. [DOI] [Google Scholar]
- 55.Săndulescu E.B., Stavrescu-Bedivan M.M., Vasile Scăețeanu G., Șchiopu T. Morpho-Anatomic features and chemical compounds in some aquatic plant species—Preliminary data; Proceedings of the Agro for Life. Life for Agriculture. Scientific Papers. Series A. Agronomy; Bucharest, Romania. 5–7 June 2014; pp. 441–447. [DOI] [Google Scholar]
- 56.Schulze E.-D., Beck E., Buchmann N., Clemens S., Müller-Hohenstein K., Scherer-Lorenzen M. Plant Ecology. 2nd ed. Springer; Berlin, Germany: 2019. pp. 143–162. [DOI] [Google Scholar]
- 57.Peredo E.L., King U.M., Les D.H. The Plastid Genome of Najas Flexilis: Adaptation to Submersed Environments Is Accompanied by the Complete Loss of the NDH Complex in an Aquatic Angiosperm. PLoS ONE. 2013;8:e68591. doi: 10.1371/journal.pone.0068591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J., Zang Y., Shang S., Liang S., Zhu M., Wang Y., Tang X. Comparative Chloroplast Genomes of Zosteraceae Species Provide Adaptive Evolution Insights into Seagrass. Front. Plant Sci. 2021;12:741152. doi: 10.3389/fpls.2021.741152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang B., Gao L., Li J., Zhou Y., Su Y.J., Wang T. Adaptive evolution of the chloroplast genome in AA-genome Oryza species. Chin. Sci. Bull. 2014;59:1975–1983. doi: 10.1360/N972014-00127. [DOI] [Google Scholar]
- 60.Gu C.H., Ma L., Wu Z.Q., Chen K., Wang Y.X. Comparative analyses of chloroplast genomes from 22 Lythraceae species: Inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 2019;19:281. doi: 10.1186/s12870-019-1870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie Z.Y., Merchant S. The Plastid-encoded ccsA Gene Is Required for Heme Attachment to Chloroplast c-type Cytochromes. J. Biol. Chem. 1996;271:4632–4639. doi: 10.1074/jbc.271.9.4632. [DOI] [PubMed] [Google Scholar]
- 62.Mavridou D., Ferguson S., Stevens J. Cytochrome c assembly. IUBMB Life. 2013;65:209–216. doi: 10.1002/iub.1123. [DOI] [PubMed] [Google Scholar]
- 63.Tang Y.W., Zhao Y.L., Li C.Y., Yang G.Y., Peng J., Xu Z.G. New insights into the evolutionary characteristic between the New World and Old World Lupinus species using complete chloroplast genomes. All Life. 2021;14:414–427. doi: 10.1080/26895293.2021.1926341. [DOI] [Google Scholar]
- 64.Barthet M., Hilu K. Expression of matK: Functional and evolutionary implications. Am. J. Bot. 2007;94:1402–1412. doi: 10.3732/ajb.94.8.1402. [DOI] [PubMed] [Google Scholar]
- 65.Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. An Organellar Maturase Associates with Multiple Group II Introns. PNAS. 2010;107:3245–3250. doi: 10.1073/pnas.0909400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hertel S., Zoschke R., Neumann L., Qu Y.J., Axmann I., Schmitz-Linneweber C. Multiple Checkpoints for the Expression of the Chloroplast-Encoded Splicing Factor MatK. Plant Physiol. 2013;163:1686–1698. doi: 10.1104/pp.113.227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang R.N., Milne R., Du X.Y., Liu J., Wu Z.Y. Characteristics and Mutational Hotspots of Plastomes in Debregeasia (Urticaceae) Front. Genet. 2020;11:729. doi: 10.3389/fgene.2020.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J.J., Liu H.X., Mao S.Z., Zhao B., Huang S.X. Adaptive evolution of the ndhF gene in the genus Rheum (Polygonaceae) Guihaia. 2016;36:101–106. [Google Scholar]
- 69.Darshetkar A.M., Maurya S., Lee C., Bazarragchaa B., Batdelger G., Janchiv A., Jeong E.J., Choi S., Choudhary R.K., Kim S.-Y. Plastome analysis unveils Inverted Repeat (IR) expansion and positive selection in Sea Lavenders (Limonium, Plumbaginaceae, Limonioideae, Limonieae) PhytoKeys. 2021;175:121–139. doi: 10.3897/phytokeys.175.61054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munekage Y., Hashimoto M., Miyake C., Tomizawa K.-I., Endo T., Tasaka M., Shikanai T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature. 2004;429:579–582. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 71.Rumeau D., Becuwe-Linka N., Beyly A., Louwagie M., Garin J., Peltier G. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell. 2005;17:219–232. doi: 10.1105/tpc.104.028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li D.M., Li J., Wang D.R., Xu Y.C., Zhu G.F. Molecular evolution of chloroplast genomes in subfamily Zingiberoideae (Zingiberaceae) BMC Plant Biol. 2021;21:558. doi: 10.1186/s12870-021-03315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong W.L., Wang R.N., Zhang N.Y., Fan W.B., Fang M.F., Li Z.H. Molecular evolution of chloroplast genomes of orchid species: Insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018;19:716. doi: 10.3390/ijms19030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krech K., Ruf S., Masduki F., Thiele W., Bednarczyk D., Albus C., Tiller N., Hasse C., Schöttler M., Bock R. The Plastid Genome-Encoded Ycf4 Protein Functions as a Nonessential Assembly Factor for Photosystem I in Higher Plants. Plant Physiol. 2012;159:579–591. doi: 10.1104/pp.112.196642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dogra V., Duan J., Lee K., Kim C. Impaired PSII proteostasis triggers an UPR-like response in the var2 mutant of Arabidopsis thaliana. J. Exp. Bot. 2019;70:3075–3088. doi: 10.1093/jxb/erz151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.