Abstract
Background and Objective. The morbidity and mortality rates of non-small cell lung cancer (NSCLC) remain high. Zhenqi Fuzheng (ZQFZ) granule, which consists of Astragali Radix and Ligustri Lucidi Fructus, is commonly used to improve the immunity of cancer patients. However, the mechanism of ZQFZ granule against NSCLC is still unclear. In this study, the network pharmacology and molecular docking approaches were used to investigate the potential mechanism of ZQFZ granule on NSCLC. Methods. The ingredients in the ZQFZ granule were considered in one study based on UPLC, and the potential targets were predicted in the SwissTargetPrediction database. NSCLC targets were gathered from GeneCards, OMIM, and TTD databases. The ingredient-target-NSCLC network was drawn by Cytoscape. The protein–protein interaction was obtained from the STRING database, and the gene function and biological pathways were analyzed by Metascape. AutoDock Vina was used to verify the molecular docking between the key compounds and core targets, and PyMol visualized the results. Results. 244 targets were related to 13 candidate compounds and 1904 targets were related to NSCLC, of which a total of 106 anti-NSCLC targets were predicted. The compound-target-NSCLC network indicated that sinapinic acid, ferulic acid, asiatic acid, pratensein, and glycitein might be the key components for treating NSCLC. The 41 vital targets (out of 106 targets) above the median calculated by PPI degree were selected for bioinformatics analysis. The top 10 targets out of 41 ranked by MCC were IL-6, SRC, CTNNB1, STAT3, CASP3, TNF, EGFR, MAPK8, HSP90AA1, and PTGS2. ZQFZ granule treatment for NSCLC involved many pathways through KEGG analyses, which included pathways in cancer (hsa05200), proteoglycans in cancer (hsa05205), endocrine resistance (hsa01522), microRNAs in cancer (hsa05206), PI3K-Akt signaling pathway (hsa04151), and IL-17 signaling pathway (hsa04657). Molecular docking studies revealed that sinapinic acid, ferulic acid, asiatic acid, pratensein, and glycitein had good infinity with most core targets. Conclusions. This study indicated that ZQFZ granule with multicompounds could treat NSCLC through multitargets and multipathways.
1. Introduction
GLOBOCAN 2020 showed that there were an estimated 19 million new cancer cases and 10 million deaths worldwide in 2020. The cancer morbidity and mortality rates remained high, with breast cancer ranking first among women and lung cancer being the leading cause of cancer death [1]. Surgery, radiotherapy, chemotherapy, targeted therapy, immunotherapy, and adjuvant therapy with Chinese medicine are the common means of cancer treatment. It is well known that Chinese medicine compounding has a long history with multicomponent and multitarget characteristics. In complementary treatment, Chinese medicine compounding has a great effect on increasing chemotherapy drugs' chemosensitivity, reducing adverse drug reactions and toxicity, relieving patients' pain, and improving life quality particularly [2].
As a traditional Chinese medicine (TCM), ZQFZ granule has the ability to improve immunity and protect bone marrow and adrenal cortex functions [3]. Combined with surgery, chemotherapy, and radiotherapy, ZQFZ granule could effectively reduce adverse effects caused by chemotherapy and promote recovery of immune function, subsequently improving the patient's quality of life and ultimately decreasing the recurrence rate. A prospective, open-label, randomized controlled trial confirmed that TCM treatment could promote NSCLC patients' quality of life, relieve symptoms, and reduce adverse events. The TCM was composed of the immuno-boosting drug called Kangai injection, herbal decoction, and ZQFZ capsule [4].
ZQFZ granule consists of Astragali Radix (AsR, dried root of Astragalus membranaceus, known as HuangQi in Chinese) and Ligustri Lucidi Fructus (LLF, dried ripe fruit of Ligustrum lucidum, known as NvZhenZi in Chinese). Triterpenoids and phenylethanoid glycosides are the two major types of constituents present in AsR [5]. As one of the most commonly used herbal remedies for cancer in China, AsR can activate immune regulation, inhibit cell proliferation, and attenuate adverse effects caused by cytotoxic therapy [6]. Similarly, the chemical constituents of LLF mainly include saponins, flavonoids, and nitrogen-containing chemicals [7]. Studies have shown that LLF can enhance the sensitivity of adriamycin-induced apoptosis, improve immunity, and promote apoptosis, so as to inhibit tumor growth [8]. The classical formulation of ZQFZ is a (2 : 1, w/w) mixture of AsR and LLF. The mixture was decocted twice with 10–30 times water, and the filtrate was concentrated into a thick paste, mixed evenly with sucrose, and dried into granules [9].
However, due to the complexity of the components of Chinese herbal compounds, the mechanisms and targets of reaction are difficult to elucidate. The mechanisms are being studied by examining the impacts of monomeric components, which are not exhaustive. Based on the whole perspective, network pharmacology combines network biology and multipharmacology to compensate for the shortage of multicomponent system research of TCM. In this study, we focused primarily on the active components from a recent study based on ultra-performance liquid chromatography (UPLC) [10] and explored the molecular mechanism of ZQFZ granule for the treatment of NSCLC. The flowchart is shown in Figure 1.
Figure 1.
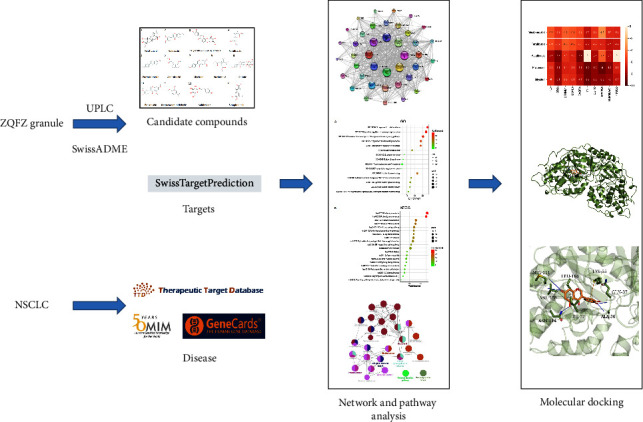
The flowchart.
2. Materials and Methods
2.1. Collection of the Main Ingredients from ZQFZ Granule and ADME Evaluation
The ingredients in ZQFZ granule were considered from one study based on UPLC [10]. The main active compounds were selected according to Lipinski's rule of five (RO5) [11]. The criteria of Lipinski's RO5 were as follows: a molecule weight (MW) should be less than 500, the number of hydrogen bond donors (Hdon) should be less than 5, the number of hydrogen bond acceptors (Hacc) should be less than 10, lipid-water partition coefficient (LogP) no more than 5, and the number of rotatable bonds (Rbon) no more than 10. The chemical structures and canonical SMILES were searched in PubChem (https://pubchem.ncbi.nlm.nih.gov/) and the ADME of compounds were uploaded into SwissADME (https://www.swissadme.ch/) to evaluate the druggability.
2.2. Screening of Targets of the Vital Ingredients from ZQFZ Granule in NSCLC
The SwissTargetPrediction (https://www.swisstargetprediction.ch/) was used to predict the potential targets of vital ingredients from the ZQFZ granule. The targets with a probability greater than zero were included. NSCLC targets were gathered from GeneCards (https://www.genecards.org/), OMIM (https://omim.org/), and TTD (https://db.idrblab.net/ttd/) by the keywords “non-small cell lung cancer” and “non-small cell lung carcinoma.” The common genes, probably the targets for ZQFZ granule to treat NSCLC, were assessed by Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/). An ingredient-disease-target network was visualized by Cytoscape 3.7.2 software.
2.3. Protein–Protein Interaction (PPI) Analysis and Vital Target Screening
The PPI of common genes was analyzed in STRING (https://string-db.org/). The organism was set to Homo sapiens and the confidence of 0.4 was selected as significant. The active interaction sources for PPI analysis and median calculation included known, predicted, and other interactions. The PPI network was analyzed and visualized using the Cytoscape software. The node size represents the degree value and the edge represents the connection between proteins. In this study, the vital genes, which were above the median calculated by degree, were then used for biological function analysis. Moreover, the top 10 proteins ranked by maximal clique centrality (MCC) using the cytoHubba plug-in were selected as core targets.
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways Enrichment Analyses
GO analysis of the vital genes, including the biological process (BP), cellular component (CC), and molecular function (MF), was enriched and analyzed by the Metascape database (https://metascape.org/gp). The screening thresholds were a minimum of 3 genes, p < 0.01 and an enrichment factor greater than 1.5. The same method and settings were used for KEGG pathway enrichment. Finally, the results were plotted as bubble charts using the online bioinformatics tool (https://bioinformatics.com.cn/). Furthermore, the Cytoscape plug-in ClueGo was used for analyzing the relevant KEGG pathway.
2.5. Molecular Docking
To verify the binding of the vital ingredients of ZQFZ granule with predicted core genes, compounds in mol2 format were downloaded from TCMSP (https://tcmsp-e.com/) and the core protein structures were acquired from RCSB PDB (https://www.rcsb.org/). The compounds and proteins were imported to AutoDockTools 1.5.6 software for dehydration, hydrogenation, and other pretreatments. Then, molecular docking for analyzing the binding activity was performed using AutoDock Vina 1.1.2 [12] software, and some great results were visualized using PyMol 4.6.0 software.
3. Results
3.1. The Chemical Structure and ADME Properties of the Vital Ingredients from ZQFZ Granule
A recent study has identified 95 chemical components from ZQFZ granule including 15 batches from 3 producers based on UPLC [10]. The chemical structures of the compounds were searched in the PubChem database, and ADME properties were evaluated in SwissADME. A total of 15 compounds met Lipinski's RO5, indicating that these components had good drug-like properties. Moreover, potential target genes were predicted based on the chemical structure via the SwissTargetPrediction database, and two of them had no targets. Therefore, we selected 13 compounds (Figure 2 and Table 1) as candidate compounds.
Figure 2.
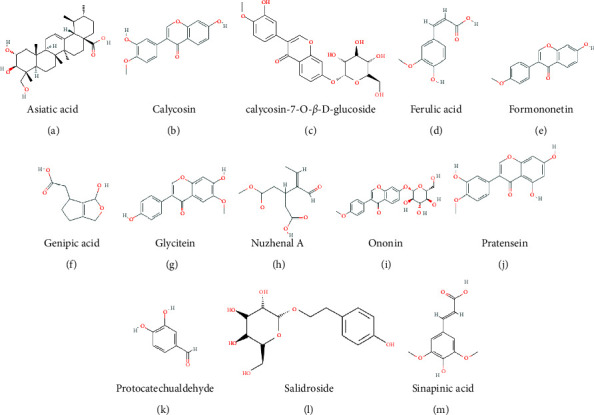
Structures of the 13 candidate compounds extracted from ZQFZ granule.
Table 1.
Pharmacological and molecular properties of the main compounds in ZQFZ granule.
| Name | Formula | MW (g/mol) | Rbon | Hacc | Hdon | TPSA (Å) | LogP | LogS | Log Kp (cm/s) | Type | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Asiatic acid | C30H48O5 | 488.7 | 2 | 5 | 4 | 97.99 | 3.2 | −6.33 | −5.23 | Triterpenoid | AsR |
| Calycosin | C16H12O5 | 284.26 | 2 | 5 | 2 | 79.9 | 2.4 | −3.57 | −6.3 | Flavonoid | AsR |
| Calycosin-7-O-β-D-glucoside | C22H22O10 | 446.4 | 5 | 10 | 5 | 159.05 | 2.65 | −3.05 | −8.57 | Flavonoid | AsR |
| Ferulic acid | C10H10O4 | 194.18 | 3 | 4 | 2 | 66.76 | 1.62 | −2.11 | −6.41 | Organic acid | LLF |
| Formononetin | C16H12O4 | 268.26 | 2 | 4 | 1 | 59.67 | 2.49 | −3.73 | −5.95 | Flavonoid | AsR |
| Genipic acid | C9H12O4 | 184.19 | 2 | 4 | 2 | 66.76 | 1.37 | −0.21 | −8.15 | Organic acid | LLF |
| Glycitein | C16H12O5 | 284.26 | 2 | 5 | 2 | 79.9 | 2.36 | −3.57 | −6.3 | Flavonoid | AsR |
| Nuzhenal A | C10H14O5 | 214.21 | 7 | 5 | 1 | 80.67 | 1.37 | −0.66 | −7.66 | Others | LLF |
| Ononin | C22H22O9 | 430.4 | 5 | 9 | 4 | 138.82 | 2.65 | −3.18 | −8.22 | Flavonoid | AsR |
| Pratensein | C16H12O6 | 300.26 | 2 | 6 | 3 | 100.13 | 2.38 | −4.06 | −5.93 | Flavonoid | — |
| Protocatechualdehyde | C7H6O3 | 138.12 | 1 | 3 | 2 | 57.53 | 0.79 | −1.76 | −6.37 | Others | LLF |
| Salidroside | C14H20O7 | 300.3 | 5 | 7 | 5 | 119.61 | 1.04 | −0.92 | −8.88 | Phenylethanoid | LLF |
| Sinapinic acid | C11H12O5 | 224.21 | 4 | 5 | 2 | 75.99 | 1.63 | −2.16 | −6.63 | Organic acid | LLF |
LLF, Ligustri Lucidi Fructus; AsR, Astragali Radix; —, not detected in LLF or AsR but detected in ZQFZ granule.
3.2. Acquisition of Targets of the Candidate Ingredients from ZQFZ Granule in NSCLC
Based on the SwissTargetPrediction database, a total of 244 targets for the 13 candidate compounds were acquired. In addition, results from GeneCards (https://www.genecards.org/), Online Mendelian Inheritance in Man (OMIM, https://omim.org/), and Therapeutic Target Database (TTD, https://db.idrblab.net/ttd/) for non-small cell lung cancer identified 1904 targets relevant to NSCLC. Furthermore, the Venn diagram reflected the common 106 target genes of the candidate compounds and NSCLC for further analysis (Figure 3(a)). The 106 common targets of ZQFZ granule against NSCLC are listed in Supplementary Table S1. Moreover, these 106 target proteins were grouped into seven different classes according to cellular function via PANTHER (https://www.pantherdb.org/), with protein-modifying enzyme (PC00260, 31.7%) being the most enriched class (Figure 3(b)). These protein-modifying enzymes include protease (PC00190), which covers ACE, CASP3, ECE1, F2, MME, MMP1, MMP12, MMP13, MMP2, MMP9, PLAT, PLAU, and PREP, protein phosphatase (PC00195) that covers ACP1, CDC25A, CDC25C, DUSP1, PTPN1, PTPN11, PTPN6, and PTPRC, and nonreceptor serine/threonine protein kinase (PC00167) including CDK4, ILK, MAPK14, MAPK8, and RAF1 (Figure 3(c)). The results above implied that the candidate ingredients from the ZQFZ granule exerted anti-NSCLC effects through a variety of targets and biological functions.
Figure 3.
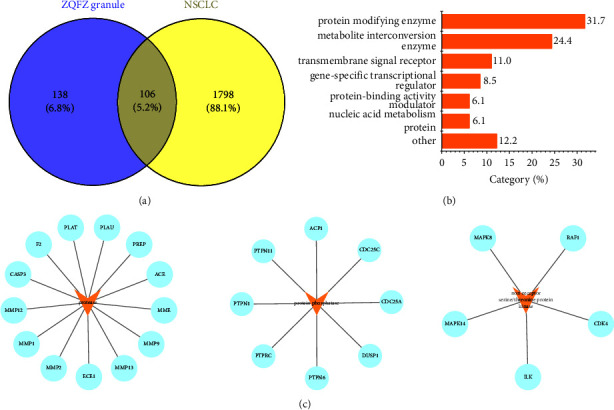
Targets of the candidate ingredients from ZQFZ granule in NSCLC. (a) A Venn diagram applied to acquire the 106 common targets between the 13 candidate compounds from the ZQFZ granule and NSCLC. (b) Panther classification of the 106 common targets. (c) Targeted proteins involved in protein-modifying enzymes (PC00260).
3.3. Construction of a Candidate Ingredients from the ZQFZ Granule Target NSCLC Network
To study the mechanism of ZQFZ granule in the treatment of NSCLC, 106 common targets and 13 candidate compounds were used for constructing the potential ZQFZ granule target NSCLC network. As shown in Figure 4, all compounds were associated with multiple targets, generating a total of 344 component-target connections between the 13 compounds and 106 targets. The network revealed sinapinic acid (degree = 40) had the most targets, followed by ferulic acid (degree = 33), asiatic acid (degree = 30), pratensein (degree = 24), and glycitein (degree = 21), indicating that these compounds from ZQFZ granule were highly likely to become key components for treating NSCLC.
Figure 4.
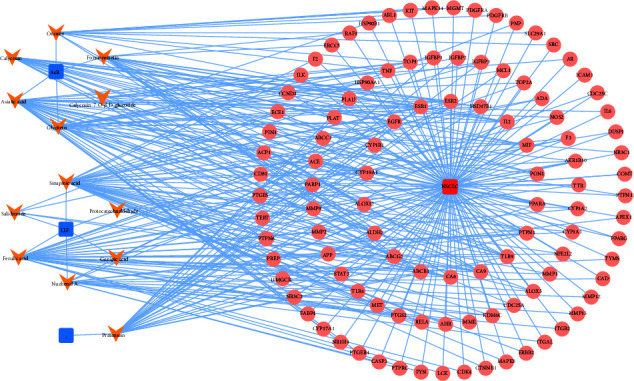
Potential compounds from the ZQFZ granule target NSCLC network. Blue nodes represent the herbs derived from the ZQFZ granule, orange nodes show the vital ingredients from herbs, and pink nodes show shared targets between potential targets of the vital compounds from the ZQFZ granule and NSCLC targets. ZQFZ granule, Zhenqi Fuzheng granule; AsR, Astragali Radix; LLF, Ligustri Lucidi Fructus; —, not detected in LLF or AsR but detected in ZQFZ; NSCLC, non-small cell lung cancer.
3.4. Protein–Protein Interaction (PPI) Analysis
The 106 common targets of the predicted ZQFZ granule and NSCLC were explored in the STRING database and protein interaction relationships were gained (Figure 5(a)). To further study the relationship between these targets, 41 vital genes above the median calculated by degree were selected and the protein interaction relationships were analyzed via Cytoscape software. Table 2 shows the details of the 41 vital targets. The PPI network generated 41 nodes, which represent proteins, and 552 edges which represent the connections between proteins. The larger node means the greater degree (Figure 5(b)). Furthermore, the top 10 targets ranked by MCC using the cytoHubba plug-in were IL-6, SRC, CTNNB1, STAT3, CASP3, TNF, EGFR, MAPK8, HSP90AA1, and PTGS2 (Figure 5(c)) and selected as core targets for interaction with NSCLC.
Figure 5.
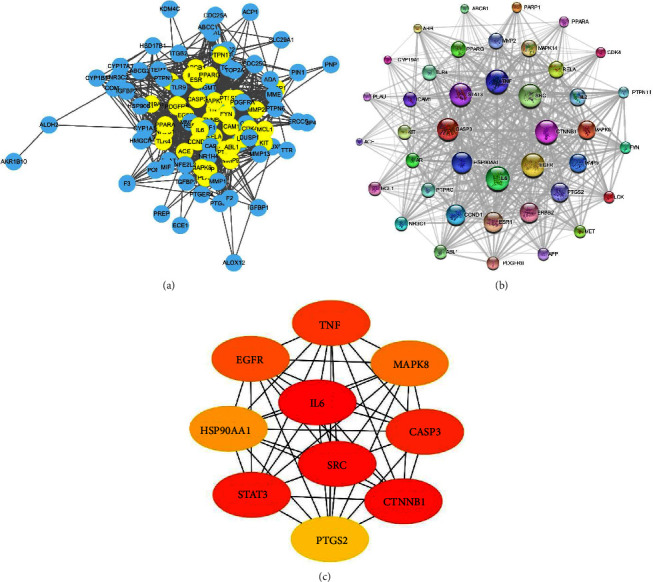
Protein–protein interaction (PPI) network: (a) PPI network of the 106 common targets between the 13 candidate compounds from ZQFZ granule and NSCLC. The PPI was analyzed in the STRING database and visualized by Cytoscape software. Nodes represent target proteins and edges represent interactions among targets. Yellow nodes represent the 41 vital targets above the median calculated by degree. (b) PPI network of the 41 vital targets. The larger the node is, the greater the degree is. (c) The top 10 targets in the PPI network ranked by maximal clique centrality (MCC) using the cytoHubba plug-in.
Table 2.
Information on 41 vital targets of ZQFZ granule against NSCLC.
| Number | Uniprot ID | Gene symbol | Description | Degree |
|---|---|---|---|---|
| 1 | P05231 | IL-6 | Interleukin-6 | 40 |
| 2 | P00533 | EGFR | Epidermal growth factor receptor | 39 |
| 3 | P35222 | CTNNB1 | Catenin beta-1 | 39 |
| 4 | P12931 | SRC | Proto-oncogenetyrosine-protein kinase Src | 39 |
| 5 | P01375 | TNF | Tumor necrosis factor | 39 |
| 6 | P40763 | STAT3 | Signal transducer and activator of transcription 3 | 38 |
| 7 | P42574 | CASP3 | Caspase-3 | 37 |
| 8 | P07900 | HSP90AA1 | Heat shock protein HSP 90-alpha | 35 |
| 9 | P24385 | CCND1 | G1/S-specific cyclin-D1 | 34 |
| 10 | P03372 | ESR1 | Estrogen receptor alpha | 34 |
| 11 | P04626 | ERBB2 | Receptor protein-tyrosine kinase ERBB-2 | 33 |
| 12 | P35354 | PTGS2 | Prostaglandin G/H synthase 2 | 32 |
| 13 | P45983 | MAPK8 | Mitogen-activated protein kinase 8 | 31 |
| 14 | P14780 | MMP9 | Matrix metalloproteinase 9 | 31 |
| 15 | P60568 | IL2 | Interleukin-2 | 29 |
| 16 | Q04206 | RELA | Nuclear factor NF-kappa B p65 subunit | 28 |
| 17 | Q16539 | MAPK14 | Mitogen-activated protein kinase 14 | 28 |
| 18 | P08253 | MMP2 | Matrix metalloproteinase 2 | 27 |
| 19 | P37231 | PPARG | Peroxisome proliferator-activated receptor gamma | 27 |
| 20 | O00206 | TLR4 | Toll-like receptor 4 | 26 |
| 21 | P10721 | KIT | Mast/stem cell growth factor receptor kit | 25 |
| 22 | P10275 | AR | Androgen receptor | 25 |
| 23 | P08575 | PTPRC | Receptor-typetyrosine-protein phosphatase C | 25 |
| 24 | P05362 | ICAM1 | Intercellular adhesion molecule 1 | 25 |
| 25 | P04150 | NR3C1 | Glucocorticoid receptor | 24 |
| 26 | P00519 | ABL1 | Tyrosine-protein kinase ABL1 | 24 |
| 27 | Q07820 | MCL1 | Induced myeloid leukemia cell differentiation protein Mcl-1 | 24 |
| 28 | P09619 | PDGFRB | Platelet-derived growth factor receptor beta | 23 |
| 29 | P05067 | APP | Amyloid-beta precursor protein | 22 |
| 30 | P08581 | MET | Hepatocyte growth factor receptor | 22 |
| 31 | P06239 | LCK | Tyrosine-protein kinase LCK | 21 |
| 32 | P06241 | FYN | Tyrosine-protein kinase FYN | 21 |
| 33 | Q06124 | PTPN11 | Protein-tyrosine phosphatase 2C | 20 |
| 34 | P11802 | CDK4 | Cyclin-dependent kinase 4 | 19 |
| 35 | P09874 | PARP1 | Poly [ADP-ribose]polymerase-1 | 19 |
| 36 | Q07869 | PPARA | Peroxisome proliferator-activated receptor alpha | 19 |
| 37 | P08183 | ABCB1 | ATP-dependent translocase ABCB1 | 17 |
| 38 | P35869 | AHR | Aryl hydrocarbon receptor | 17 |
| 39 | P11511 | CYP19A1 | Cytochrome P450 19A1 | 16 |
| 40 | P00749 | PLAU | Urokinase-type plasminogen activator | 16 |
| 41 | P12821 | ACE | Angiotensin-converting enzyme | 14 |
3.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways Enrichment Analyses
The 41 vital targets between ZQFZ granule and NSCLC were further analyzed by GO enrichment analysis and KEGG pathway analysis by Metascape. The GO results showed 421 entries for BP, 49 entries for CC, and 82 entries for MF. The top 5 BPs, CCs, and MFs are shown in bubble charts (Figure 6(a)). The top 5 biological processes were related to oxidative stress (GO: 0006979), protein phosphorylation (GO: 0001934), and transmembrane receptor protein-tyrosine kinase signaling pathway (GO: 0007169). Cellular components analysis revealed including membrane raft (GO: 0045121), receptor complex (GO: 0043235), and side of the membrane (GO: 0098552). For molecular functions, the targets were enriched in protein kinase binding (GO: 0019901), transmembrane receptor protein-tyrosine kinase activity (GO: 0004714), and protein domain-specific binding (GO: 0019904). The complex list of the top 5 BP, CC, and MF pathway terms enriched in ZQFZ granule against NSCLC is shown in Supplementary Table S2.
Figure 6.
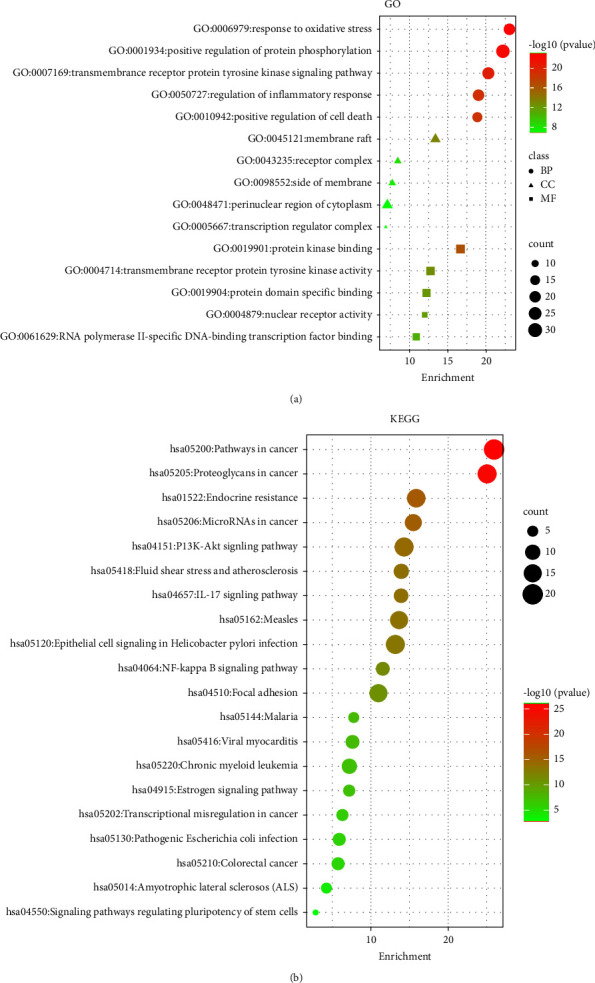
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses. (a) Bubble chart of the top 5 biological processes (BP), cellular component (CC), and molecular function (MF) terms identified by GO enrichment analysis. (b) Bubble chart of the top 20 pathway terms identified by KEGG pathway enrichment analysis. The size of the dots represents gene numbers. The larger is the dot, the higher is the number of genes.
In addition, the KEGG pathway revealed that 206 pathways were involved, and the bubble chart (Figure 6(b)) reflects the top 20 terms. Table 3 displays detailed information about the top 20 pathways related to pathways in cancer (hsa05200), proteoglycans in cancer (hsa05205), endocrine resistance (hsa01522), microRNAs in cancer (hsa05206), PI3K-Akt signaling pathway (hsa04151), and IL-17 signaling pathway (hsa04657). In particular, 8 out of 10 core targets (IL-6, CTNNB1, STAT3, CASP3, EGFR, MAPK8, HSP90AA1, and PTGS2) involved in the first-term pathways in cancer (hsa05200), and 7 core targets (IL-6, SRC, CTNNB1, STAT3, CASP3, TNF, and EGFR) involved in the second-term proteoglycans in cancer (hsa05205). A compound-target-pathway network (Figure 7) shows the connection between the top 20 pathways, targets, and compounds. The predictive diagram of the mechanism of the proteoglycans pathway is displayed in Figure 8.
Table 3.
Top 20 KEGG pathway terms enriched in ZQFZ granule against NSCLC.
| Term | Pathway | P value | Count | Symbols |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 1.105E − 26 | 20 | ABL1/AR/CCND1/CASP3/CDK4/CTNNB1/EGFR/ERBB2/HSP90AA1/IL-6/KIT/MET/MMP2/MMP9/PDGFRB/PPARG/MAPK8/PTGS2/RELA/STAT3 |
| hsa05205 | Proteoglycans in cancer | 8.793E − 26 | 17 | CCND1/CASP3/MAPK14/CTNNB1/EGFR/ERBB2/ESR1/IL-6/MET/MMP2/MMP9/PLAU/PTPN11/SRC/STAT3/TLR4/TNF |
| hsa01522 | Endocrine resistance | 1.345E − 16 | 16 | CCND1/CDK4/MAPK14/EGFR/ERBB2/ESR1/MMP2/MMP9/MAPK8/SRC/RELA/STAT3/CTNNB1/KIT/ABL1/PTGS2 |
| hsa05206 | MicroRNAs in cancer | 3.195E − 16 | 13 | ABL1/CCND1/CASP3/EGFR/ERBB2/MCL1/MET/MMP9/PDGFRB/ABCB1/PLAU/PTGS2/STAT3 |
| hsa04151 | PI3K-Akt signaling pathway | 5.023E − 15 | 17 | CCND1/CDK4/EGFR/ERBB2/HSP90AA1/IL2/IL-6/KIT/MCL1/MET/PDGFRB/RELA/TLR4/ABL1/MAPK8/PTPN11/TNF |
| hsa05418 | Fluid shear stress and atherosclerosis | 1.152E − 14 | 10 | MAPK14/CTNNB1/HSP90AA1/ICAM1/MMP2/MMP9/MAPK8/RELA/SRC/TNF |
| hsa04657 | IL-17 signaling pathway | 1.23E − 14 | 9 | CASP3/MAPK14/HSP90AA1/IL-6/MMP9/MAPK8/PTGS2/RELA/TNF |
| hsa05162 | Measles | 2.238E − 14 | 15 | CCND1/CASP3/CDK4/FYN/IL2/IL-6/MAPK8/RELA/STAT3/TLR4/EGFR/ERBB2/MAPK14/MCL1/PTPN11 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 6.772E − 14 | 17 | CASP3/MAPK14/EGFR/MET/MAPK8/PTPN11/RELA/SRC/CCND1/ESR1/STAT3/ABL1/MMP2/ICAM1/PTGS2/HSP90AA1/FYN |
| hsa04064 | NF-kappa B signaling pathway | 2.867E − 12 | 8 | PARP1/ICAM1/LCK/PLAU/PTGS2/RELA/TLR4/TNF |
| hsa04510 | Focal adhesion | 1.056E − 11 | 15 | CCND1/CTNNB1/EGFR/ERBB2/FYN/MET/PDGFRB/MAPK8/SRC/IL-6/STAT3/MAPK14/KIT/CDK4/PTPN11 |
| hsa05144 | Malaria | 1.676E − 08 | 5 | ICAM1/IL-6/MET/TLR4/TNF |
| hsa05416 | Viral myocarditis | 2.401E − 08 | 8 | ABL1/CCND1/CASP3/FYN/ICAM1/LCK/PTPN11/TNF |
| hsa05220 | Chronic myeloid leukemia | 6.165E − 08 | 10 | ABL1/CCND1/CDK4/PTPN11/RELA/CASP3/SRC/STAT3/KIT/PTGS2 |
| hsa04915 | Estrogen signaling pathway | 6.51E − 08 | 6 | EGFR/ESR1/HSP90AA1/MMP2/MMP9/SRC |
| hsa05202 | Transcriptional misregulation in cancer | 4.888E − 07 | 6 | IL-6/MET/MMP9/PLAU/PPARG/RELA |
| hsa05130 | Pathogenic Escherichia coli infection | 1.25E − 06 | 7 | ABL1/CTNNB1/FYN/TLR4/MET/PTPN11/SRC |
| hsa05210 | Colorectal cancer | 1.777E − 06 | 7 | CCND1/CASP3/CTNNB1/MAPK8/PPARG/ESR1/SRC |
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | 5.683E − 05 | 5 | CASP3/MAPK14/TNF/APP/PTGS2 |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 0.0014409 | 3 | MAPK14/CTNNB1/STAT3 |
Figure 7.
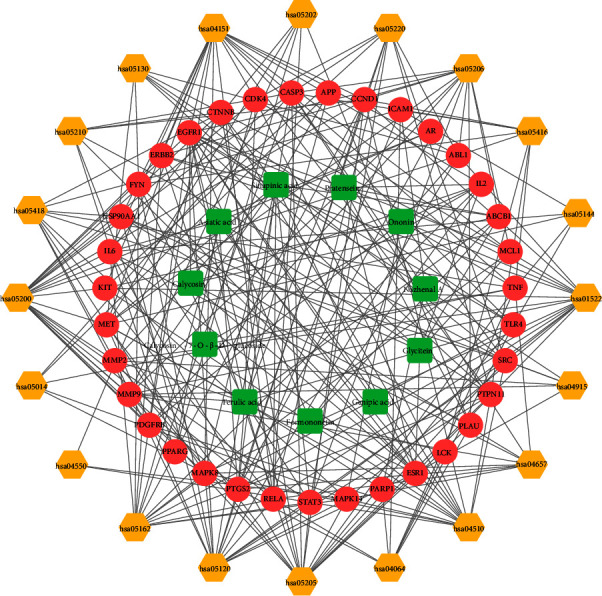
A compound-target-pathway network. The green round rectangle represents the chemical compounds, the pink circles represent the targets, and the yellow hexagon represents the signal pathways.
Figure 8.
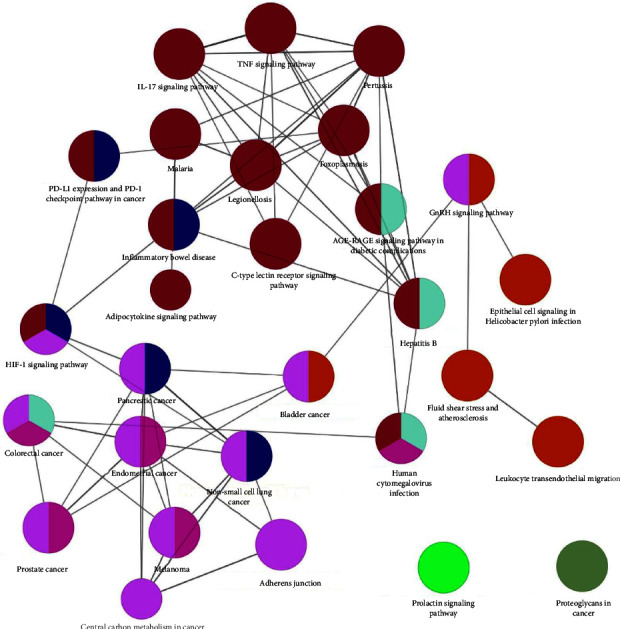
KEGG analysis of the 41 vital targets of ZQFZ granule against NSCLC using the ClueGO plug-in. Each node is a representative enrichment pathway. The nodes imply gene numbers shared between the pathways; the color indicates the enrichment classification of the node.
3.6. Molecular Docking Simulation
According to the potential compounds from the ZQFZ granule target NSCLC network (Figure 4), sinapinic acid, ferulic acid, asiatic acid, pratensein, and glycitein had a high number of targets against NSCLC and were the top 5 compounds ranked by degree. The PDBIDs and resolutions of the core target proteins are listed in Supplementary Table S3. Thus, molecular docking predicted the binding of the five compounds to core targets related to NSCLC. The affinity is usually regarded as the binding effect of the compound to the target; the lower the absolute value of affinity is, the more stable the binding is. As shown in Figure 9, sinapinic acid showed a strong binding to TNF, with an affinity value of −7.1 kcal/mol; ferulic acid showed a strong binding to TNF and HSP90AA1, with affinity values of −7.2 and −7.1 kcal/mol; asiatic acid showed the strong binding to IL-6, SRC, CASP3, EGFR, and HSP90AA1 with affinity values of −7.6, −8.7, −8.7, −7.5, and −7.3 kcal/mol; pratensein showed the strong binding to SRC, CASP3, TNF, EGFR, MAPK8, and HSP90AA1 with affinity values of −8.1, −7.8, −8.9, −8.2, −9.0, and −8.3 kcal/mol; and glycitein showed a strong binding to SRC, CASP3, TNF, EGFR, MAPK8, HSP90AA1, and PTGS2, with affinity values of −8.1, −7.4, −8.9, −8.2, −8.3, −8.3, and −7.1 kcal/mol. In general, an affinity value of less than −4.25 kcal/mol means some binding capacity, the affinity value of less than −5.0 kcal/mol means good binding capacity, and the affinity value of less than −7.0 kcal/mol means strong binding capacity. Table 4 shows some representative docking results with the core targets.
Figure 9.
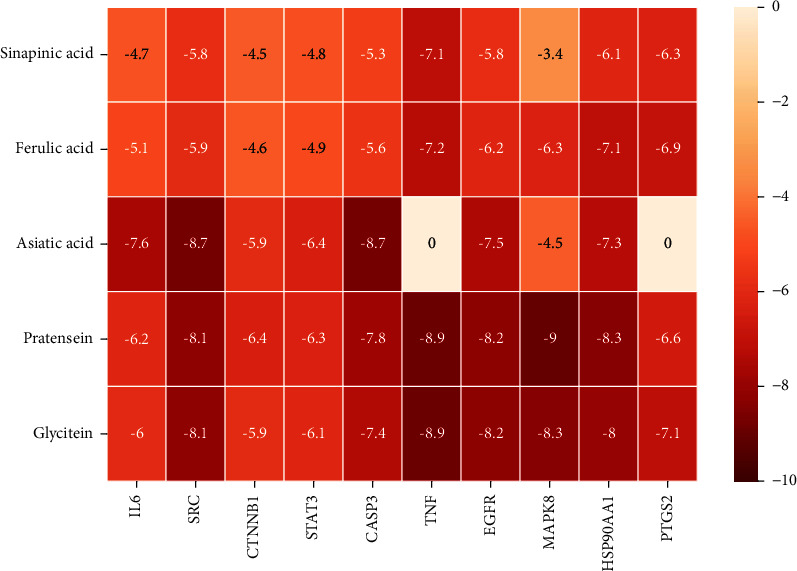
Heat map of the affinity values via molecular docking. Molecular docking of core targets with the top 5 compounds ranked by degree. The darker the box is, the stronger the bond is.
Table 4.
Visualization of the representative molecular docking results.
| Target | Compound | Affinity (kcal/mol) | Zoomed complex | Complex |
|---|---|---|---|---|
| IL-6 | Asiatic acid | −7.6 |
|
|
| SRC | Asiatic acid | −8.7 |
|
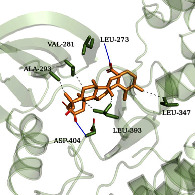
|
| CTNNB1 | Pratensein | −6.4 |
|
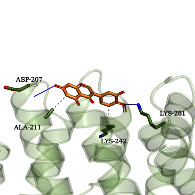
|
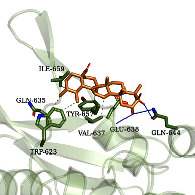
| STAT3 | Asiatic acid | −6.4 |
|
|
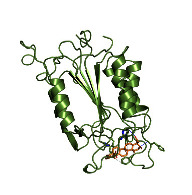
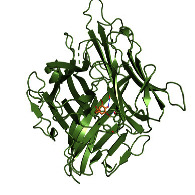
| CASP3 | Asiatic acid | −8.7 |
|
|
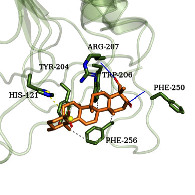
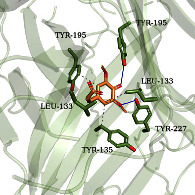
| TNF | Sinapinic acid | −7.1 |
|
|
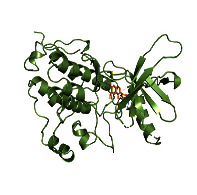
| EGFR | Glycitein | −8.2 |
|
|
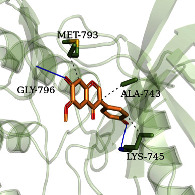
| MAPK8 | Pratensein | −9.0 |
|
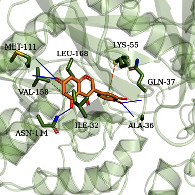
|
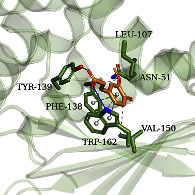
| HSP90AA1 | Ferulic acid | −7.1 |
|
|
| PTGS2 | Glycitein | −7.1 |
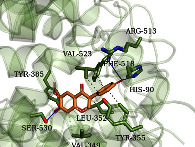
|
|
As shown in Table 4, the structure of asiatic acid could form one hydrogen bond, two hydrogen bonds, and two hydrogen bonds with Tyr103, Tyr32, and Asp56, respectively, in IL-6. Respectively, asiatic acid could interact with Leu273 and Asp404 via one hydrogen bond in SRC. The structure of pratensein could interact with Asp207 and Lys281 through one hydrogen bond in CTNNB1. The structure of asiatic acid could form one hydrogen bond and two hydrogen bonds with Glu638, and Gln644, respectively, in STAT3. The structure of asiatic acid could form one hydrogen bond with Arg207, Phe250, and Phe256, respectively, in CASP3. Sinapinic acid could form one hydrogen bond with Tyr195 and Tyr227 in TNF. Glycitein could interact with Lys745 and Gly796 through one hydrogen bond in EGFR. The structure of pratensein could interact with Ala36, Gln37, and Asn114 through one hydrogen bond, respectively, and Met111 via two hydrogen bonds in MAPK8. The structure of ferulic acid was linked to Asn51, Tyr139, and Trp162 through one hydrogen bond, respectively, in HSP90AA1. One hydrogen bond could combine pratensein with Tyr139 in HSP90AA1. Glycitein could interact with His90, Tyr385, and Arg513 via one hydrogen bond, respectively, and Ser530 via two hydrogen bonds in PTGS2. Furthermore, some known inhibitors for the core targets were used to validate the molecular docking. It is shown in Table 5. The result indicated the affinities of the inhibitors were comparable to the respective compounds of ZQFZ granule. The structures of the known inhibitors for the core targets are listed in Supplementary Table S4.
Table 5.
Molecular docking of the known inhibitors with core targets.
| Inhibitor | CAS ID | Target | Affinity (kcal/mol) |
|---|---|---|---|
| LMT-28 | 1239600-18-0 | IL-6 | −5.7 |
| Src inhibitor 1 | 179248-59-0 | SRC | −8.2 |
| PNU 74654 | 113906-27-7 | CTNNB1 | −5.8 |
| Static | 19983-44-9 | STAT3 | −5.4 |
| TNF-α-IN-1 | 444287-49-4 | TNF | −9.4 |
| EGFR inhibitor | 879127-07-8 | EGFR | −8.6 |
| VER-50589 | 747413-08-7 | HSP90AA1 | −8.4 |
| BUR1 | 23000-46-6 | PTGS2 | −6.2 |
4. Discussion
Lung cancer remains the leading cause of cancer death, accounting for 18% of all cancer deaths [1]. To explore the active ingredients and potential mechanisms of ZQFZ granule for the treatment of NSCLC, this study comprehensively investigated the therapeutic effect of ZQFZ granule in NSCLC.
Generally, compounds for network pharmacology studies are obtained from public databases, while the effect of the content may be ignored, resulting in inaccurate predictions. Especially, active compounds were derived from the study that analyzed 15 batches from 3 producers, which significantly increased the credibility of data [10]. Based on Lipinski's RO5 and SwissTargetPrediction databases, 13 compounds were selected as candidate compounds and 106 potential targets were screened against NSCLC. Moreover, a potential compound from the ZQFZ granule target NSCLC network implied that sinapinic acid, ferulic acid, asiatic acid, pratensein, and glycitein were probably the critical components for NSCLC treatment. Sinapinic acid, one of the most common hydroxycinnamic acids, has an anticancer effect on prostate cancer cells. Treatment with sinapinic acid (1 mM for 72 h) could eliminate half the number of PC-3 and LNCaP cells [13]. Ferulic acid, as a 4-hydroxy-3-methoxycinnamic acid, can inhibit phorbol-12-myristate-13-acetate (PMA)-stimulated invasion of A549 cells at a concentration of ≥100 μM [14]. Asiatic acid, as a natural triterpene, has been proven to enhance the sensitivity of multidrug-resistant A549 cells to cisplatin by downregulating P-glycoprotein (MDR1) and its targets [15]. Glycitein had significant cytotoxic effects on gastric cancer cells by inducing cell apoptosis and G0/G1 phase cell cycle arrest via the ROS-related MAPK/STAT3/NF-κB signaling pathway [16]. Therefore, these compounds play an important role in the treatment of tumors. Notably, there are few studies on the treatment of NSCLC with sinapinic acid, pratensein, and glycitein.
The 106 targets in common between ZQFZ granule and NSCLC were considered potential targets. The top 10 core targets shown in Figure 5(c) included IL-6, SRC, CTNNB1, STAT3, CASP3, TNF, EGFR, MAPK8, HSP90AA1, and PTGS2. These target genes are associated with the regulation of tumorigenesis. IL-6, STAT3, and TNF are involved in immunoactivation and play critical roles in the development of NSCLC. CTNNB1 and EGFR are highly expressed in carcinomas such as lung and breast cancers. SRC is related to endocrine regulation and CASP3 is commonly regarded as the predominant terminal shear enzyme in cell apoptosis. Furthermore, the relationship between the potential compounds and the core targets could be validated by the literature. For instance, sinapinic acid significantly repressed TNF levels and activated CASP3 in acute doxorubicin-induced cardiotoxicity rats [17]. Ferulic acid inhibited UVB-induced TNF and IL-6 protein expression in mice skin tissue, which might be related to the activation of MAPK and NF-kappa B signaling pathways [18]. Asiatic acid induced HT-29 cell apoptosis via CASP3 activation and inhibited the growth and metastasis of breast cancer in mice by downregulating SRC protein expression [19, 20].
The core targets of ZQFZ granule for the treatment of NSCLC were applied to obtain an enrichment map of GO and KEGG pathway analyses via the Metascape database. The results showed BP was involved in oxidative stress, protein phosphorylation, protein-tyrosine kinase signaling pathway, and inflammatory response. CC was primarily concerned with various cell bodies such as membrane, complexes, and cytoplasm. Additionally, MF was mainly associated with protein kinase binding. Furthermore, the main signaling pathways were endocrine resistance, PI3K/Akt, IL-17, and NF-kappa B signaling pathways, while the diseases involved were cancers, atherosclerosis, and measles.
IL-6 is a pleiotropic cytokine with various biological functions in immunity, tissue regeneration, and metabolism. It is highly expressed in lung cancer and negatively correlated with survival [21]. Blocking IL-6 expression can inhibit lung cancer promotion, cell proliferation, angiogenesis markers, and tumor cell-intrinsic STAT3 activation [22]. STAT3 is mainly expressed in naive CD4+ T cells and activated p-STAT3 can modulate the transcriptional activity of target genes associated with tumor cell migration and invasion [23]. IL-6/STAT3 signaling is activated in lung tumorigenesis and metastasis. IL-6 can activate STAT3 signaling and escape host immunity by upregulating and recruiting granulocyte-likemyeloid-derived suppressor cells and type II macrophage polarization [24]. EGFR, a member of the receptor tyrosine kinase ERBB family, is an important oncogene in NSCLC progression. EGFR-TKIs are used for treating NSCLC by modulating the immune microenvironment. However, EGFR-TKI resistance is widely present in NSCLC and promotes immune escape via increased PD-L1 expression and, subsequently, activates the PI3K/AKT/mTOR pathway aberrantly [25, 26]. The PI3K/Akt signaling pathway, a classical mediator that regulates cell growth and inflammation, was found to be activated in this study. There are 17 targets identified in this pathway, and IL-6, TNF, EGFR, MAPK8, and HSP90AA1 are core targets. Tumor necrosis factor (TNF), a cytokine, can combine with extracellular death receptors to activate the apoptosis process and regulate the inflammatory microenvironment via the PI3K/Akt pathway in NSCLC [27, 28]. Upregulation of HSP90AA1 is related to poor overall survival in cancer patients [29]. This could be due to the fact that HSP90AA1 overexpression reduces immune surveillance and resists foreign substances [30].
This is a novel way to study the mechanism of ZQFZ granule in the treatment of NSCLC via a network pharmacologic method. The selected compounds of ZQFZ granule were derived from a recent study based on UPLC. The results may be useful to better understand the multipathway and multitarget regulation of ZQFZ granule against NSCLC.
In total, 13 compounds in the ZQFZ granule and 106 common targets against NSCLC were screened by network pharmacology analysis. GO and KEGG enrichment analyses implied the 13 compounds in the ZQFZ granule could treat NSCLC via multiple NSCLC pathological processes. However, the theoretical observations still need to be validated by clinical studies in the future. This study provided a theoretical basis for the clinical application of ZQFZ granule in NSCLC.
5. Conclusion
This study revealed the pharmacological mechanism of ZQFZ granule in NSCLC based on network pharmacology and molecular docking. GO and KEGG enrichment analyses implied that the 13 compounds in the ZQFZ granule played an important role in the treatment of NSCLC. This study provided a theoretical basis for further research on the clinical application of ZQFZ granule in NSCLC.
Acknowledgments
This study was financially supported by the Research Fund of Medical Specialties Construction Project in the Field of Healthcare in Baoshan District, Shanghai (BSZK-2019-C03) and the Research Fund of Key Specialities Construction Project in Shanghai Baoshan Luodian Hospital, Shanghai (TSZK-2020-03).
Abbreviations
- NSCLC:
Non-small cell lung cancer
- ZQFZ:
Zhenqi Fuzheng
- TCM:
Traditional Chinese medicine
- AsR:
Astragali Radix
- LLF:
Ligustri Lucidi Fructus
- UPLC:
Ultra-performance liquid chromatography
- RO5:
Rule of five
- Hdon:
Hydrogen bond donors
- Hacc:
Hydrogen bond acceptors
- LogP:
Lipid-water partition coefficient
- Rbon:
Rotatable bonds
- PPI:
Protein–protein interaction
- MCC:
Maximal clique centrality
- GO:
Gene ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- BP:
Biological process
- CC:
Cellular component
- MF:
Molecular function
- LUAD:
Lung adenocarcinoma
- LUSD:
Lung squamous cell carcinoma
- OMIM:
Online Mendelian Inheritance in Man
- TTD:
Therapeutic target database.
Contributor Information
Yue Yue Li, Email: liyue429@163.com.
Liang Zhao, Email: zhaoliangphar@163.com.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Yanqing Zhou and Chenxi Wu contributed equally to this work.
Supplementary Materials
Table S1. The 106 common targets of ZQFZ granule against NSCLC. Table S2. Top 5 BP, CC, and MF pathway terms enriched in ZQFZ granule against NSCLC. Table S3. Molecular docking of core targets with top 5 compounds ranked by degree. Table S4. Structures of the known inhibitors for the core targets.
References
- 1.Sung H., Ferlay J., Siegel R. L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians . 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Su X. L., Wang J. W., Che H., et al. Clinical application and mechanism of traditional Chinese medicine in treatment of lung cancer. Chinese Medical Journal . 2020;133(24):2987–2997. doi: 10.1097/CM9.0000000000001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao M., Cao L., Bai M., Chen G. Effect of Zhen Qi Fu Zheng granules on the bone marrow depression model induced by Zidorf. Saudi Journal of Biological Sciences . 2018;25(2):220–225. doi: 10.1016/j.sjbs.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao Z., Chen Z., Han R., et al. Comprehensive TCM treatments combined with chemotherapy for advanced non-small cell lung cancer: a randomized, controlled trial. Medicine (Baltimore) . 2021;100(18) doi: 10.1097/md.0000000000025690.e25690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C. H., Yang X., Wei J. R., et al. Ethnopharmacology, phytochemistry, pharmacology, toxicology and clinical applications of radix astragali. Chinese Journal of Integrative Medicine . 2021;27(3):229–240. doi: 10.1007/s11655-019-3032-8. [DOI] [PubMed] [Google Scholar]
- 6.Jung Y., Jerng U., Lee S. A systematic review of anticancer effects of radix astragali. Chinese Journal of Integrative Medicine . 2016;22(3):225–236. doi: 10.1007/s11655-015-2324-x. [DOI] [PubMed] [Google Scholar]
- 7.Chen B., Wang L., Li L., et al. Fructus ligustri lucidi in osteoporosis: a review of its pharmacology, phytochemistry, pharmacokinetics and safety. Molecules . 2017;22(9):p. 1469. doi: 10.3390/molecules22091469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabiani R., Rosignoli P., De Bartolomeo A., et al. Oxidative DNA damage is prevented by extracts of olive oil, hydroxytyrosol, and other olive phenolic compounds in human blood mononuclear cells and HL60 cells. Journal of Nutrition . 2008;138(8):1411–1416. doi: 10.1093/jn/138.8.1411. [DOI] [PubMed] [Google Scholar]
- 9.Lu M. The preparation method of Zhenqi Fuzheng granule . 2016. [Google Scholar]
- 10.Xue Z., Xu L., Shang Z., Shi X., Ye M., Qiao X. Discovery of minor quality evaluation marker compounds for Chinese patent medicine products using a two-leveled metabolomics strategy. Journal of Chromatography A . 2021;1652 doi: 10.1016/j.chroma.2021.462354.462354 [DOI] [PubMed] [Google Scholar]
- 11.Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies . 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Trott O., Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry . 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eroglu C., Avci E., Vural H., Kurar E. Anticancer mechanism of sinapic acid in PC-3 and LNCaP human prostate cancer cell lines. Gene . 2018;671:127–134. doi: 10.1016/j.gene.2018.05.049. [DOI] [PubMed] [Google Scholar]
- 14.Tsai C. M., Yen G. C., Sun F. M., Yang S. F., Weng C. J. Assessment of the anti-invasion potential and mechanism of select cinnamic acid derivatives on human lung adenocarcinoma cells. Molecular Pharmaceutics . 2013;10(5):1890–1900. doi: 10.1021/mp3006648. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Q., Liao M., Hu H., Li H., Wu L. Asiatic acid (AA) sensitizes multidrug-resistant human lung adenocarcinoma A549/DDP cells to cisplatin (DDP) via downregulation of P-glycoprotein (MDR1) and its targets. Cellular Physiology and Biochemistry . 2018;47(1):279–292. doi: 10.1159/000489806. [DOI] [PubMed] [Google Scholar]
- 16.Zang Y. Q., Feng Y. Y., Luo Y. H., et al. Glycitein induces reactive oxygen species-dependent apoptosis and G0/G1 cell cycle arrest through the MAPK/STAT3/NF-κB pathway in human gastric cancer cells. Drug Development Research . 2019;80(5):573–584. doi: 10.1002/ddr.21534. [DOI] [PubMed] [Google Scholar]
- 17.Bin Jardan Y. A., Ansari M. A., Raish M., et al. Sinapic acid ameliorates oxidative stress, inflammation, and apoptosis in acute doxorubicin-induced cardiotoxicity via the NF-κB-mediated pathway. BioMed Research International . 2020;2020:10. doi: 10.1155/2020/3921796.3921796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambothi K., Prasad N. R., Balupillai A. Ferulic acid inhibits UVB-radiation induced photocarcinogenesis through modulating inflammatory and apoptotic signaling in Swiss albino mice. Food and Chemical Toxicology . 2015;82:72–78. doi: 10.1016/j.fct.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Bunpo P., Kataoka K., Arimochi H., et al. Inhibitory effects of asiatic acid and CPT-11 on growth of HT-29 cells. Journal of Medical Investigation . 2005;52(1-2):65–73. doi: 10.2152/jmi.52.65. [DOI] [PubMed] [Google Scholar]
- 20.Tian M., Chen K., Huang J., et al. Asiatic acid inhibits angiogenesis and vascular permeability through the VEGF/VEGFR2 signaling pathway to inhibit the growth and metastasis of breast cancer in mice. Phytotherapy Research . 2021;35(11):6389–6400. doi: 10.1002/ptr.7292. [DOI] [PubMed] [Google Scholar]
- 21.Barrera L., Montes-Servin E., Barrera A., et al. Cytokine profile determined by data-mining analysis set into clusters of non-small-cell lung cancer patients according to prognosis. Annals of Oncology . 2015;26(2):428–435. doi: 10.1093/annonc/mdu549. [DOI] [PubMed] [Google Scholar]
- 22.Caetano M. S., Zhang H., Cumpian A. M., et al. IL6 blockade reprograms the lung tumor microenvironment to limit the development and progression of K-ras-Mutant lung cancer. Cancer Research . 2016;76(11):3189–3199. doi: 10.1158/0008-5472.can-15-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh J. E., Kreimer S., Walker S. R., et al. Granulin, a novel STAT3-interacting protein, enhances STAT3 transcriptional function and correlates with poorer prognosis in breast cancer. Genes Cancer . 2015;6(3-4):153–168. doi: 10.18632/genesandcancer.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jing B., Wang T., Sun B., et al. IL6/STAT3 signaling orchestrates premetastatic niche formation and immunosuppressive traits in lung. Cancer Research . 2020;80(4):784–797. doi: 10.1158/0008-5472.can-19-2013. [DOI] [PubMed] [Google Scholar]
- 25.Peng S., Wang R., Zhang X., et al. EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Molecular Cancer . 2019;18(1):p. 165. doi: 10.1186/s12943-019-1073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fumarola C., Bonelli M. A., Petronini P. G., Alfieri R. R. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochemical Pharmacology . 2014;90(3):197–207. doi: 10.1016/j.bcp.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R., Dong Y., Sun M., et al. Tumor-associated inflammatory microenvironment in non-small cell lung cancer: correlation with FGFR1 and TLR4 expression via PI3K/Akt pathway. Journal of Cancer . 2019;10(4):1004–1012. doi: 10.7150/jca.26277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balkwill F. TNF-alpha in promotion and progression of cancer. Cancer and Metastasis Reviews . 2006;25(3):409–416. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 29.Fan G., Tu Y., Wu N., Xiao H. The expression profiles and prognostic values of HSPs family members in head and neck cancer. Cancer Cell International . 2020;20(1):p. 220. doi: 10.1186/s12935-020-01296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang X., You X. M., Li L. Q. Expression of HSP90AA1/HSPA8 in hepatocellular carcinoma patients with depression. OncoTargets and Therapy . 2018;11:3013–3023. doi: 10.2147/ott.s159432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The 106 common targets of ZQFZ granule against NSCLC. Table S2. Top 5 BP, CC, and MF pathway terms enriched in ZQFZ granule against NSCLC. Table S3. Molecular docking of core targets with top 5 compounds ranked by degree. Table S4. Structures of the known inhibitors for the core targets.
Data Availability Statement
The data used to support the findings of this study are included within the article.


