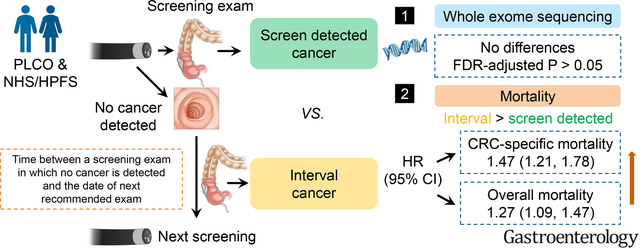
Abstract
Background & Aims:
Interval colorectal cancers (CRC), cancers diagnosed after a screening/surveillance exam in which no cancer is detected, and before the date of next recommended exam, reflect an unprecedented challenge in CRC detection and prevention. To better understand this poorly characterized CRC variant, we examined the clinical and mutational characteristics of interval CRCs in comparison to screen detected CRCs.
Methods:
We included 1175 CRCs documented in the Prostate, Lung, Colorectal, and Ovarian (PLCO) cancer screening trial and 3661 CRCs in the Nurses’ Health Study (NHS) and Health Professionals Follow-Up Study (HPFS). Multivariable Cox models were performed to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) of death risk. Whole-exome sequencing (WES) was conducted in 147 PLCO cases and 796 NHS/HPFS cases.
Results:
A total of 619 deaths (312 CRC-specific) and 2404 deaths (1904 CRC-specific) were confirmed during follow-up of PLCO and NHS/HPFS, respectively. Compared to screen detected CRCs, interval CRCs had a multivariate-adjusted HR (95% CI) of 1.47 (1.21, 1.78) for CRC-specific mortality and 1.27 (1.09, 1.47) for overall mortality (meta-analysis combining all three cohorts). However, we did not observe significant differences in mutational features between interval and screen detected CRCs (FDR adjusted P > .05).
Conclusion:
Interval CRCs had a significantly increased risk of death compared to screen detected CRCs that were not explained by established clinical prognostic factors, including stage at diagnosis. The survival disadvantage of interval CRCs did not appear to be explained by differences in the genomic landscape of tumors characterized by WES.
Keywords: interval colorectal cancer, colonoscopy, screening, whole exome sequencing
Graphical Abstract
INTRODUCTION
Colorectal cancer (CRC) incidence and mortality rates have been declining among adults older than 50 in the US,1 largely attributable to standard endoscopic screening.2 However, the incidence of interval colorectal cancers (CRC), which are defined as “CRCs diagnosed after a screening or surveillance exam in which no cancer is detected, and before the date of next recommended exam” by the World Endoscopy Organization,3 has not decreased 4 or has been rising.5 Interval CRCs are estimated to account for 5–6% of overall CRC incidence,6, 7 and reflect an unprecedented challenge in clinical detection and management of CRC because these cancers are seemingly less preventable by endoscopic screening. Therefore, understanding and preventing this poorly characterized variant of CRC is of great scientific and clinical significance.
Some interval CRCs may arise from lesions overlooked on endoscopy due to poor procedural quality.8, 9 Some interval CRCs may also differ biologically compared with typical CRCs which could lead to differences in their clinical behavior. However, evidence on this topic has been limited and inconsistent.10 Compared to “detected CRCs” in general, interval CRCs have been shown to have better7, worse11, 12 or similar 10, 13–17 survival. Thus far, most efforts to genomically characterize CRC, such as The Cancer Genome Atlas (TCGA), have not taken into account the timing of tumor development relative to endoscopic screening nor do they distinguish tumors identified through routine endoscopic screening from those that become clinically apparent through symptoms.18 A few small-scale investigations on molecular features of interval CRC only included a few molecular markers such as microsatellite instability (MSI)13, DNA mismatch repair deficiency (dMMR),19 and CpG island methylator phenotype (CIMP).20 A more comprehensive annotation of the features that characterize interval CRCs remains lacking.
Therefore, we utilized whole exome sequencing (WES) and clinical data to compare the clinical characteristics and tumor mutational landscape of interval CRCs with screened-detected CRCs within the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer randomized controlled trial (RCT) of flexible sigmoidoscopy (FS) screening. We corroborated these findings in two large prospective cohort studies, the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS).
METHODS
Study Population
PLCO:
We used data from the multicenter, randomized PLCO Cancer Screening Trial of FS, which enrolled 154,900 men and women aged 55 to 74 years between 1993 and 2001.21, 22 Participants were randomized to receive FS at baseline and again at year 3 (for those who were randomized before April 1995) or at year 5 versus usual care. Abnormal findings (e.g., polyp) at FS were followed up with a complete colonoscopy to the cecum. At baseline, demographic information, medical histories, lifestyle habits were collected. The follow-up rate exceeded 95% in all participants. Each participating center’s institutional review board approved the protocol and all study participants provided written informed consent.
NHS/HPFS:
We utilized data from two ongoing prospective US cohorts: the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS). NHS enrolled 121,700 female nurses aged 30–55 years in 1976 and HPFS enrolled 51,529 male health professionals aged 40–75 years at baseline in 1986.23, 24 Participants were mailed questionnaires every two to four years since baseline to collect update data for demographics, lifestyle factors, medical histories, and disease outcomes. Endoscopy information was collected from biennial questionnaires and detailed information of the assessment of lower endoscopy has been described previously.25 Briefly, in NHS, in 1990, the year of first ever and most recent lower endoscopy was queried, including endoscopy status between 1984 and 1988. In both NHS and HPFS, beginning in 1988 and continuing through 2014, participants were asked whether they had undergone either sigmoidoscopy or colonoscopy in the past 2 years and, if so, the reason for the procedure. In 2004, we additionally inquired whether participants’ previously reported endoscopies were sigmoidoscopies or colonoscopies. In every cycle thereafter, responses were recorded separately for sigmoidoscopy and colonoscopy. The response rates in each cohort have been over 90% in most follow-up questionnaire cycles. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required.
Ascertainment of Colorectal Cancer (CRC) Cases
In the PLCO trial, reports of CRCs were collected between 1993 and 2014 through various means including, but not limited to, self-reports through annually mailed study questionnaires and death certificates.21, 22 All incident cancers are confirmed based on medical records, and pathology reports. Details on tumor characteristics, including anatomic site, stage, and tumor histology were abstracted.
In NHS/HPFS, CRC cases were reported on biennial follow-up questionnaires between 1984 and 2014. Additional cases were identified through reporting from family members, postal authorities, or through cross-referencing of the National Death Index. All cases were further confirmed by physicians blinded to demographic and other characteristics of the patients through reviewing medical records, pathology reports, and/or cancer registry data. Information on anatomic site, histology, and stage at presentation were also ascertained during the physicians’ review.25
In all cohorts, proximal cancers were defined as cancers located in the cecum through transverse colon and distal cancers were defined as cancers located in the splenic flexure through rectum.
Identification of Screen detected and Interval CRCs
In PLCO, we conducted a detailed analysis of the circumstances that contributed to CRCs diagnosed within the screening arm. A screening exam was considered positive if a polyp or mass was detected during the FS examination. Screen detected CRCs were defined as CRCs diagnosed on follow-up (within 12 months) of a positive FS screening. Interval CRCs were cancers appearing within 48 months in a segment of the colon that had been previously endoscopically screened.26 To ensure that our proximal interval CRCs would be similar to proximal interval CRCs that would have evaded endoscopic detection through a colonoscopy-only screening approach, we restricted our definition of proximal interval CRCs in PLCO to tumors that arose among individuals within 13–48 months of an endoscopy that covered the territory in which the tumor arose (e.g., a proximal CRC detected after colonoscopy performed for follow-up of abnormal distal findings on FS). Therefore, the definition of interval CRCs encompassed distal CRCs (within the reach of a FS) diagnosed either between 0–48 months after a negative FS or between 13–48 months after a positive FS, as well as proximal CRCs diagnosed between 13–48 months after colonoscopy following a positive FS screening. CRCs diagnosed in the PLCO “usual-care” (control) arm as well as CRCs diagnosed >48 months after the initial screening were classified as “Other CRCs”.
In NHS/HPFS, screen detected CRCs were defined as CRCs diagnosed within the same questionnaire that the participant reported a screening endoscopy. Interval CRCs were defined as distal and proximal CRCs diagnosed 1–5 years after a colonoscopy and distal CRCs diagnosed after a negative FS. Patients confirmed after 5 years since last screening and patients who never underwent an endoscopy were grouped as “Other CRCs” in NHS/HPFS.25
Ascertainment of Death
In PLCO, deaths were ascertained via annual follow-ups and periodic linkage to the National Death Index (NDI). Death certificates were the primary source of identifying date of death and the underlying causes of death. To confirm the cause of the death, an independent Death Review Committee further reviewed deaths among those diagnosed with a confirmed cancer, those whose death certificate stated they died of a cancer, and those whose death certificate was ambiguous as to whether the cause of death was a cancer.21, 22 If the primary cause of a death is due to CRC, it was considered as a CRC-specific death.
In NHS/HPFS, we performed systematic searches of the vital records of states and of the National Death Index (NDI). This search was supplemented by reports from family members and/or by notifications from the postal authorities when a questionnaire or newsletter mailed to a cohort member was returned. Overall, these methods were able to capture more than 98% of the deaths in NHS/HPFS.27 A physician blinded to cohort questionnaire data reviewed death certificates, medical records, tumor registries data, and all other available health information to classify the cause of death according to the International Classification of Diseases (ICD).
Statistical Analyses
Descriptive analyses of patient and tumor characteristics were performed according to cancer type in PLCO and NHS/HPFS (screen detected, interval, others). Continuous variables were shown in means (standard deviations, SDs) and categorical variables were presented as numbers (percentages). Logistic regressions and Fisher’s exact tests were further conducted to compare clinical and mutational characteristics between interval and screen detected cancers, respectively. Person-year accrued from the date of CRC diagnosis until the date of death from any cause or the end of follow-up, whichever came first. Hazards ratios (HRs) and 95% confidence intervals (CIs) of CRC-specific and all-cause mortality of interval CRC were estimated in comparison to screen detected CRC through Cox proportional hazards regression models. Both crude and multivariable adjusted HR (95% CI) were computed. Covariates in multivariable model included age at and year of CRC diagnosis, sex, family history of CRC, tumor location, grade, and stage; missing values were adjusted as an individual level. Analyses were performed separately in PLCO (R v4.1.1, R Studio v1.0.153) and NHS/HPFS (SAS, Unix 9.4) and combined through random-effects meta-analysis (STATA, v15).28 Between-study heterogeneity was assessed by Cochran’s Q statistic.29 All P values were two-sided and P < .05 was considered as statistically significant.
Methods for WES and Copy Number Alteration Analyses
We conducted WES on 147 cases from PLCO and 796 cases from NHS/HPFS with formalin-fixed paraffin embedded (FFPE) tumor and matched adjacent normal tissue pairs.30 In brief, in PLCO, during follow-up, tumor and adjacent normal tissues resected from CRC patients were subsequently fixed in formalin and embedded in paraffin. To obtain tumor-enriched DNA from tissue sections of the FFPE blocks, tumor areas were selected by guide hematoxylin and eosin-stained slides. Genomic DNA was extracted using the QIAGEN QIAamp DNA FFPE Tissue Kit. Normal DNA was extracted from resection margins or other non-tumor areas. Quality of DNA was assessed by the Quant-iT Pico Green dsDNAassay Kit (Invitrogen). DNA specimens then underwent solution-phase hybrid capture with SureSelect v.2 Exome bait (Agilent Technologies), followed by multiplexing of the samples and sequencing on Illumina HiSeq 2000 instruments at the Broad Institute of MIT and Harvard. In NHS/HPFS, WES of tumor DNA extracted from FFPE blocks was carried out as previously described30, 31. The average coverage in tumors and adjacent normal tissue was 85X. For the three cohorts, variant calling/ filtering/annotation was carried out using the Cancer Genome Analysis WES characterization pipeline as previously described.30 Significant chromosomal aberrations were detected using GISTIC (Genomic Identification of Significant Targets in Cancer).32 Data was analyzed with maftools 33 to test for a mutation enrichment (including point mutations, indels and copy number alterations) among interval cancers using a two-sided pairwise and groupwise fisher exact test for mutations present in at least 10 patients.
RESULTS
Baseline Characteristics of Interval CRCs
Our study included a total of 1,175 CRCs (246 screen detected, 182 interval, and 747 others) documented in PLCO (1993–2014) and 3,661 CRCs (494 screen detected, 514 interval, and 2653 others) documented in NHS/HPFS (1984–2014). Baseline characteristics are presented in Table 1 and are relatively consistent in PLCO and NHS/HPFS. Compared to screen detected CRCs, patients with interval CRCs were older and more likely to present at a more advanced stage (Table 1 and Supplementary Table 1). Of note, in our PLCO sample, we restricted our definition of proximal interval CRCs to cancers detected after colonoscopy performed for follow-up of abnormal FS (i.e., after an endoscopy that covered the territory in which the cancer arose). Therefore, the percentage of proximal CRCs defined as interval CRC was comparatively low, reflecting the design of PLCO as a RCT of screening FS rather than colonoscopy.
Table1.
Clinical characteristics of CRC patients according to cancer type in PLCO and NHS/HPFS
| PLCO | NHS/HPFS | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Characteristics | Screen detected (n=246) | Interval (n=182) | Others (n=747) | Screen detected (n=494) | Interval (n=514) | Others (n=2653) |
|
| ||||||
| Age at diagnosis | 65.4 ± 5.6 | 71.2 ± 6.7 | 72.5 ± 6.5 | 70.4 ± 8.4 | 73.0 ± 8.3 | 69.6 ± 9.5 |
| Female | 76 (30.9) | 83 (45.6) | 361 (48.3) | 283 (57.3) | 286 (55.6) | 1770 (66.7) |
| Family history of CRC | 34 (14.8) | 26 (15.1) | 80 (11.5) | 130 (26.3) | 118 (23.0) | 432 (16.3) |
| Tumor location | ||||||
| Cecum | 16 (6.5) | 5 (2.7) | 230 (31.0) | 88 (19.0) | 85 (18.4) | 394 (16.1) |
| Ascending to transverse colon | 25 (10.2) | 13 (7.1) | 344 (46.4) | 158 (34.2) | 150 (32.4) | 707 (28.9) |
| Splenic flexure to sigmoid | 120 (48.8) | 92 (50.5) | 82 (11.1) | 133 (28.8) | 128 (27.6) | 755 (30.8) |
| Rectosigmoid and rectum | 85 (34.6) | 72 (39.6) | 85 (11.5) | 83 (18.0) | 100 (21.6) | 593 (24.2) |
| Stage | ||||||
| Stage I | 148 (65.8) | 53 (30.1) | 188 (25.6) | 157 (39.3) | 114 (28.6) | 454 (21.6) |
| Stage II | 39 (17.3) | 47 (26.7) | 212 (28.9) | 124 (31.1) | 115 (28.9) | 562 (26.7) |
| Stage III | 30 (13.3) | 47 (26.7) | 213 (29.1) | 89 (22.3) | 118 (29.6) | 583 (27.7) |
| Stage IV | 8 (3.6) | 29 (16.5) | 120 (16.4) | 29 (7.3) | 51 (12.8) | 507 (24.1) |
| Tumor differentiation | ||||||
| Well or moderately differentiated | 187 (89.0) | 133 (79.2) | 537 (75.6) | 224 (92.6) | 178 (89.9) | 948 (89.9) |
| Poorly differentiated or undifferentiated | 23 (11.0) | 35 (21.8) | 173 (24.4) | 18 (7.4) | 20 (10.1) | 107 (10.1) |
| Year of diagnosis | ||||||
| ≤ 2000 | 190 (77.2) | 37 (20.3) | 126 (16.9) | 249 (50.4) | 177 (34.4) | 1537 (57.9) |
| > 2000 | 56 (22.8) | 145 (79.7) | 621 (83.1) | 245 (49.6) | 337 (65.6) | 1116 (42.1) |
Notes: Abbreviation: CRC, colorectal cancer; PLCO, Prostate, Lung, Colorectal and Ovarian cancer screening trial; NHS, Nurses’ Health Study; HPFS, Health Professionals Follow-up Study. Continuous variables were presented as mean (standard deviation) and categorical variables were presented as number (percentage). Percentages were calculated among non-missing data.
Survival of Interval CRCs
A total of 619 overall deaths and 312 CRC-specific deaths were documented among 1,175 PLCO cases during a median 6.6 years of follow-up. In 3,661 NHS/HPFS cases, 2,404 overall deaths and 1,940 CRC-specific deaths were documented during a median follow-up of 6.7 years.
In a multivariable model adjusted for age at and year of diagnosis, sex, family history of CRC, tumor location, grade, and stage (Table 2), compared to screen detected CRCs, participants with interval CRCs had an increased risk of CRC-specific mortality [HR (95% CI) = 1.57 (0.98, 2.51) in PLCO, 1.45 (1.18, 1.79) in NHS/HPFS, and 1.47 (1.21, 1.78) when combined through meta-analysis]. Overall mortality was also increased with interval cancers [HR (95% CI) = 1.38 (1.02, 1.86) in PLCO, 1.24 (1.04, 1.47) in NHS/HPFS, 1.27 (1.09, 1.47) when combined]. Through meta-analysis of PLCO and NHS/HPFS, we further explored subgroup associations according to the stage of cancer and we observed poorer survival of stage IV interval cancers than stage IV screen detected cancers after adjusting for all other covariates [HR (95% CI) = 2.06 (1.33, 3.18) for CRC-specific mortality, 2.08 (1.36, 3.19) for overall mortality] (Supplementary Table 2). Those categorized as “Other CRCs,” which included never-screened patients, patients from PLCO control arm (usual care group), and those diagnosed > 48 months (PLCO)/or >60 months (NHS/HPFS) after last endoscopic screening, also had higher mortality compared with screen detected CRCs. No significant heterogeneity was detected between PLCO and NHS/HPFS (All P for heterogeneity > .05) (Table 2).
Table 2.
CRC-specific and overall mortality of interval cancer in PLCO and NHS/HPFS
| CRC-specific mortality | Overall mortality | ||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| No. of cases | No. of events | Crude HR (95% CI) | Multivariable adjusted HR (95% CI) | No. of events | Crude HR (95% CI) | Multivariable adjusted HR (95% CI) | |
|
| |||||||
| PLCO | |||||||
| Screen detected | 246 | 39 | reference | reference | 127 | reference | reference |
| Interval | 182 | 45 | 2.03 (1.32, 3.12) | 1.57 (0.98, 2.51) | 93 | 1.72 (1.31, 2.26) | 1.38 (1.02, 1.86) |
| Others | 747 | 228 | 2.74 (1.94, 3.86) | 1.95 (1.27, 3.00) | 399 | 2.13 (1.73, 2.62) | 1.51 (1.14, 2.01) |
| NHS/HPFSE | |||||||
| Screen detected | 494 | 156 | reference | reference | 251 | reference | reference |
| Interval | 514 | 204 | 1.56 (1.27, 1.92) | 1.45 (1.18, 1.79) | 274 | 1.34 (1.13, 1.59) | 1.24 (1.04, 1.47) |
| Others | 2653 | 1580 | 2.41 (2.05, 2.84) | 2.00 (1.70, 2.37) | 1879 | 1.80 (1.58, 2.06) | 1.68 (1.47, 1.92) |
| PLCO + NHS/HPFS | |||||||
| Screen detected | 740 | 195 | reference | reference | 378 | reference | reference |
| Interval | 696 | 249 | 1.66 (1.33, 2.07) | 1.47 (1.21, 1.78) | 367 | 1.48 (1.16, 1.89) | 1.27 (1.09, 1.47) |
| Others | 3400 | 1808 | 2.47 (2.13, 2.86) | 2.00 (1.71, 2.33) | 2278 | 1.92 (1.64, 2.25) | 1.65 (1.46, 1.86) |
Notes: Abbreviations: CI, confidence interval; HR, hazard ratio; CRC, colorectal cancer; PLCO, Prostate, Lung, Colorectal and Ovarian cancer screening trial; NHS, Nurses’ Health Study; HPFS, Health Professionals Follow-up Study. Multivariable-adjusted HRs (95% CIs) were adjusted for age at diagnosis, year of diagnosis, sex, family history of CRC, tumor location, grade, and stage. Results of NHS/HPFS and PLCO were pooled using random-effects meta-analysis (All P for heterogeneity > .05).
Because PLCO was an RCT of FS, there was relative enrichment of interval CRCs in the distal colorectum within the reach of the FS compared to proximal CRCs. We conducted a secondary analysis restricted to individuals with distal CRCs and our results did not substantively change. Compared with distal screen detected CRCs, the multivariable-adjusted HRs (95% CI) for PLCO and NHS/HPFS combined for distal interval CRC was 1.65 (1.25, 2.16) for CRC-specific mortality, and 1.26 (1.02, 1.56) for overall mortality. In another sensitivity analysis among participants older than 50 including all PLCO cases and 98% of NHS/HPFS cases, we did not observe material changes of estimates [interval CRC vs. screen detected CRC, multivariable-adjusted HRs (95% CI) = 1.46 (1.20, 1.77) for CRC-specific mortality, 1.26 (1.09, 1.47) for overall mortality] compared to main results.
Mutational Landscape of Interval CRCs
We investigated whether specific genetic alterations within interval cancer might be associated with its increased mortality by performing WES on 147 PLCO matched tumor normal pairs and leveraging WES data from a previously published set of 796 matched tumor normal pairs in NHS/HPFS. An overview of our WES dataset in PLCO and NHS/HPFS was shown in Figure 1A and clinical characteristics of these cases were computed and shown in Supplementary Table 3. The characteristic pattern among WES cases was relatively consistent with that of all cases.
Figure 1: Genomic landscape of interval cancers in PLCO and NHS/HPFS.
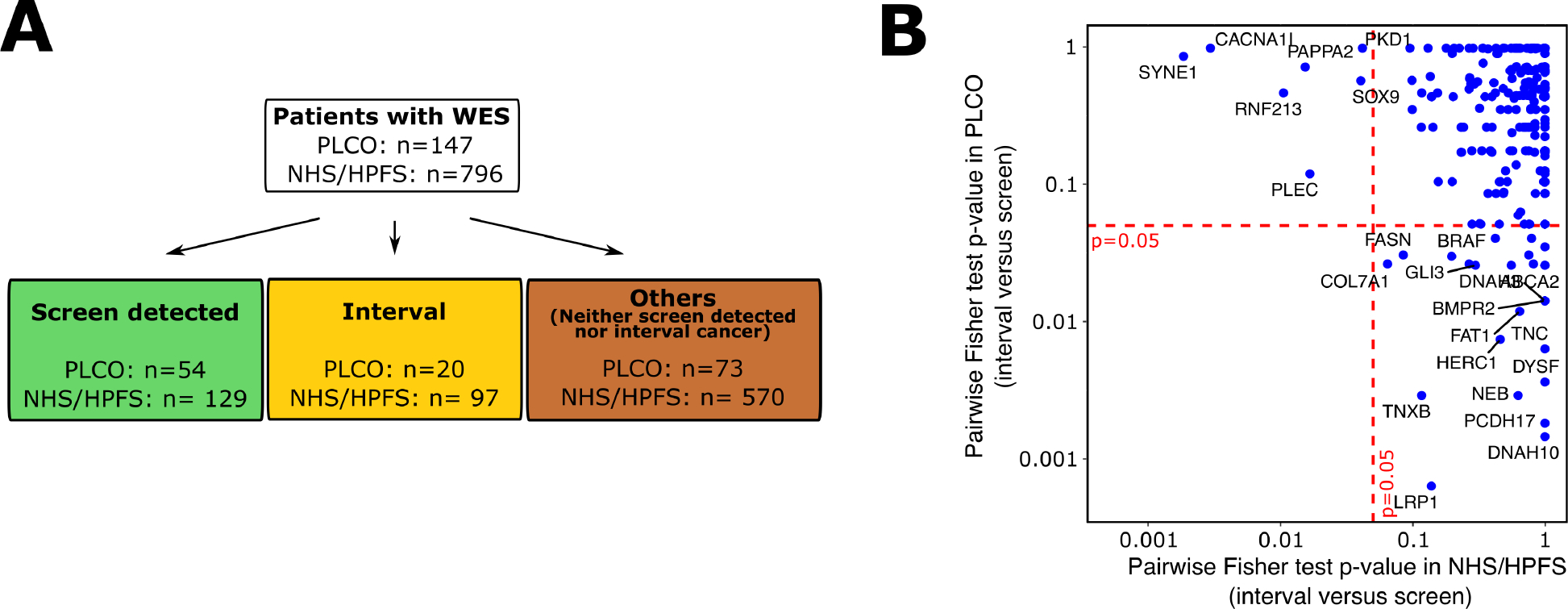
A. Overview of the NHS/HPFS and PLCO cases with WES data. B. Mutation enrichment analysis for interval cancers. For each gene (blue dots), we calculated the Fisher’s test P-value for interval versus screen detected cancers. This analysis was performed for both NHS-HPFS (x axis) and PLCO (y axis). No gene consistently reached statistical significance level (i.e., FDR adjusted P < .05) in both cohorts (i.e., no genes in the lower left quadrant)
In the analysis of WES data, we first identified genomic segments significantly targeted by somatic copy number variations (CNV) in all cohorts using GISTIC (Figure S1A). Ten most frequently mutated genes in PLCO and NHS/HPFS were also identified (Figure S1B). We did not observe significant differences of these variants (i.e., copy number alteration, insertion-deletion mutation, and single based mutations) between interval and screen detected cancers [All False Discovery Rate (FDR) adjusted P > .05] (Supplementary Table 4).
We then performed a mutation enrichment analysis by conducting pairwise Fisher test comparisons for each gene mutated (including point mutations, indels and significant CNVs) in at least 10 patients in both PLCO and NHS/HPFS (Figure 1B). We did not observe any mutation significantly and consistently enriched among interval cancers when compared to screen detected cancers (FDR adjusted P > .05) in both PLCO and NHS/HPFS (i.e., no genes in the lower left quadrant of Figure 1B). Subgroup analyses by cancer stage were further performed and we did not observe significant mutational difference between screen detected and interval cancers within the same cancer stage group (Supplementary Table 5). Similar results were observed when comparing mutational features of interval cancers to all other cancers (i.e., screen detected cancers and those who were neither screen detected nor interval cancers) (Figure S2).
DISCUSSION
We found significantly increased CRC and overall mortality in interval compared to screen-detected cancers. However, we did not observe significant differences in mutational characteristics between interval and screen detected CRCs. Our results were consistent across three prospective cohorts, including a RCT of FS screening.
The increased mortality risk in interval CRCs we observed is consistent with results from a prior population-based retrospective cohort in Canada in which patients with post-colonoscopy CRCs had worse overall survival than those with CRCs detected by any colonoscopy (i.e., colonoscopy of any indication) [HR (95% CI) = 1.25 (1.17, 1.32)].12 A similar pattern was observed in a cohort in Asia which found a higher CRC-specific mortality in post-colonoscopy CRCs compared to cancers detected by any colonoscopy [HR (95% CI) = 1.32 (1.18, 1.49)].11 However, several other studies showed no significant differences in survival between post-colonoscopy CRCs and symptomatic CRCs or CRCs detected by any colonoscopy10, 13–16. On the contrary, a retrospective cohort analysis among Utah residents reported that post-colonoscopy CRCs had better survival compared to CRCs detected by any colonoscopy [age/sex adjusted HR (95% CI) = 0.63 (0.49, 0.81)].7 The differences in these results may be due to variation in the definition of interval CRCs as opposed to post-colonoscopy CRCs as well as in the choice of comparison group. In our study, we identified interval CRCs by time since screening and used screen-detected CRCs as the comparison group. In contrast, most previous studies compared the survival risk of “CRCs diagnosed post colonoscopy of any indication” with “CRCs detected by colonoscopy of any indication” without differentiating between CRCs detected during a screening colonoscopy (i.e., screen detected) versus a colonoscopy following the onset of symptoms. Some previous studies reported that post-colonoscopy CRCs were more likely to be diagnosed at an earlier stage 5, 7, 13, 15, but included symptomatic CRCs in the comparison. We confined our analysis to comparing interval cancers to screen detected cancers. This may explain the survival benefit of post-colonoscopy CRCs observed in one prior study.7
The etiology of interval CRCs is multifactorial. Interval cancers may be a result of cancers that were missed or incompletely excised on initial endoscopy because they are morphologically inconspicuous or due to suboptimal procedural quality. Although one recent study reported no significant relationship between endoscopist quality measures (e.g., specialty, procedure volume) and post-colonoscopy CRC risk,17 multiple previous studies have shown that a lower adenoma detection rate is associated with higher risk of post-colonoscopy CRC.34, 35–37 Specific histologic and morphological polyp subtypes, such as flat adenomas, may be more endoscopically inconspicuous.8, 9, 35, 36 Indeed, emerging data has shown that the some interval CRCs may arise from sessile serrated adenoma (SSA), a morphological polyp subtype that is more difficult to detect with colonoscopy.38
Interval cancers may also develop as new incident lesions with a more rapid and aggressive biological behavior that leads to invasive cancer during the interval between colonoscopies.25, 39 Our data showed higher percentage of interval CRCs than screen detected CRCs (16.5% vs. 3.6% in PLCO, 12.8% vs. 7.3% in NHS/HPFS) were diagnosed with stage IV disease, highlighting the potential aggressive biology of these tumors. Previous studies have examined molecular differences between interval CRCs and non-interval CRCs based on limited panels of known molecular markers. For example, compared to non-interval CRCs, interval CRCs were found more likely to harbor more DNA mismatch repair deficiency (dMMR) and microsatellite instability (MSI)13, 19, 39, 40, as well as CpG island methylator phenotype (CIMP).20, 40 To extend these results, we comprehensively contrasted the genomic landscape of interval cancers with screen detected through WES in PLCO and NHS/HPFS. Our study did not reveal any significant differences in terms of mutational signature or copy number alteration between interval CRCs and screen detected CRCs. These results were consistent with findings from a recent case-control study in which targeted sequencing of a panel of 48 genes that are commonly mutated in CRC failed to identify any significant mutational differences between post-colonoscopy (n=93) and CRCs without previous colonoscopy or with a colonoscopy over 10 years ago (n=79).40
Our study had notable strengths. First, we leveraged data from the PLCO, the only U.S. RCT of endoscopic CRC screening. The screening protocol in PLCO permits rigorous identification of both screen detected and interval CRCs.26 Similarly, in NHS/HPFS, participants provided detailed, biennially updated information on CRC screening and the indications for their exam.25 In contrast, most prior studies lack information on the indication for colonoscopy.7, 11, 12, 10, 13–17 Second, we had detailed information on established predictors for CRC and overall mortality, including stage at diagnosis which allowed us to better assess the specific contribution of interval vs. screen detected status on survival. Third, we conducted our analysis in three separate prospective cohorts and observed similar results, enhancing rigor and reproducibility. Finally, to our knowledge, our study is the first of its kind to use a comprehensive, discovery-based approach through WES technology to pursue genomic characterization of interval CRCs.
We also acknowledge several limitations. First, although our study is the largest to date to examine molecular differences between interval and screen detected CRCs using WES, we still had a relatively limited sample size which may have missed less frequent molecular differences. Also, in our study, we observed higher BRAF mutations among interval (26%) than screen detected (14%) cancers (Figure S1B, Supplementary Table 4); however, the difference did not reach statistical significance level after FDR adjustment (FDR adjusted P = .23). As PLCO is a RCT of screening FS and proximal interval CRCs may be underrepresented in our data, future studies could further explore the molecular characteristics of this CRC variant in data that is enriched with more proximal cancers. We also acknowledge that WES could not capture potential epigenetic changes as well as differences in intronic regions between these CRC subtypes. Besides, we cannot fully exclude residual confounding despite having detailed information on potential confounders, including family history of CRC and tumor characteristics (location, grade, stage). The randomized design of PLCO minimized potential confounding introduced by factors associated with differential screening behavior.
In conclusion, in a RCT of FS and two prospective cohort studies, we observed that interval CRCs had significantly increased risk of CRC-specific and overall mortality compared to screen detected CRCs that were not explained by established clinical prognostic factors, including stage at diagnosis. Intriguingly, the survival disadvantage of interval cancer did not appear to be explained by differences in the genomic landscape of tumors characterized by WES. Future studies are needed to characterize the mechanisms by which interval cancers may exhibit a more aggressive biological profile to better tailor colonoscopic screening and aggressive biological profile to better tailor colonoscopic screening and surveillance.
Supplementary Material
WHAT YOU NEED TO KNOW.
Background and Context:
Interval colorectal cancers diagnosed after colonoscopy screening/surveillance are seemingly less preventable by endoscopic screening and reflect an unprecedented challenge in colorectal cancer detection and prevention. The prognosis and genomic landscape of these cancers in comparison to screened detected cancers remain unclear.
New Findings:
Individuals with interval cancers had a significantly increased risk of death compared to those with screen detected cancers. The genomic landscape of interval cancers was not significantly different from that of screen detected cancers.
Limitations:
Although our study is the largest to date to examine molecular differences between interval and screen detected cancers using whole exome sequencing, our sample size may have been unable to detect less frequent molecular differences between these subtypes.
Impact:
The survival disadvantage associated with interval cancers supports the development of better tailored colonoscopic screening/surveillance strategies and more studies to characterize the mechanisms by which interval cancers may exhibit a more aggressive biological profile.
Lay summary:
Colorectal cancers diagnosed between screening exams (interval cancers) had significantly increased risk of death than screen detected cancers although no significant molecular differences were observed between them at a whole exome sequencing level.
Acknowledgement:
The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries (NPCR) and/or the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program. Central registries may also be supported by state agencies, universities, and cancer centers. Participating central cancer registries include the following: Alabama, Alaska, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Hawaii, Idaho, Indiana, Iowa, Kentucky, Louisiana, Massachusetts, Maine, Maryland, Michigan, Mississippi, Montana, Nebraska, Nevada, New Hampshire, New Jersey, New Mexico, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Puerto Rico, Rhode Island, Seattle SEER Registry, South Carolina, Tennessee, Texas, Utah, Virginia, West Virginia, Wyoming.
Grant support:
This work is supported by NIH grants UM1 CA186107, P01 CA87969, R01 CA49449, U01 CA167552, U01CA182367, R35 CA197735, R35CA253185, and by the Project P Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr. Chan is the Stuart and Suzanne Steele MGH Research Scholar.
Abbreviation:
- CRC
colorectal cancer
- WES
whole exome sequencing
- PLCO
Prostate, Lung, Colorectal and Ovarian
- RCT
randomized controlled trial
- FS
flexible sigmoidoscopy
- NHS
Nurses’ Health Study
- HPFS
Health Professionals Follow-up Study
- HR
hazard ratio
- CI
confidence interval.
Footnotes
Disclosures of conflict of interest: Dr. Giannakis received research funding from Servier and Janssen unrelated to this study. Dr. Schoen receives research support from Freenome, Exact Sciences, and Immunovia. Dr. Chan received research funding from Zoe Ltd and Freenome, research and consulting funds from Pfizer Inc, consulting funds from Bohreinger Ingelheim, and consulting funds from Bayer Pharma AG – all for studies unrelated to the present manuscript. All other authors disclose no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability statement:
The data of this study are available upon reasonable request. Further information including the procedures to obtain and access data from the Nurses’ Health Studies and Health Professionals Follow-up Study is described at https://www.nurseshealthstudy.org/researchers (contact email: nhsaccess@channing.harvard.edu) and https://sites.sph.harvard.edu/hpfs/for-collaborators/.
REFERENCES
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145–164. [DOI] [PubMed] [Google Scholar]
- 2.Edwards BK, Noone AM, Mariotto AB, et al. Annual Report to the Nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer 2014;120:1290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanduleanu S, le Clercq CM, Dekker E, et al. Definition and taxonomy of interval colorectal cancers: a proposal for standardising nomenclature. Gut 2015;64:1257–67. [DOI] [PubMed] [Google Scholar]
- 4.Pullens HJ, Leenders M, Schipper ME, et al. No decrease in the rate of early or missed colorectal cancers after colonoscopy with polypectomy over a 10-year period: a population-based analysis. Clin Gastroenterol Hepatol 2015;13:140–7. [DOI] [PubMed] [Google Scholar]
- 5.Uche-Anya EN, DeCuir N, Lebwohl B. Temporal Trends and Risk Factors for Postcolonoscopy Colorectal Cancer. J Clin Gastroenterol 2019;53:e334–e340. [DOI] [PubMed] [Google Scholar]
- 6.Ertem FU, Ladabaum U, Mehrotra A, et al. Incidence of interval colorectal cancer attributable to an endoscopist in clinical practice. Gastrointest Endosc 2018;88:705–711.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samadder NJ, Curtin K, Tuohy TM, et al. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology 2014;146:950–60. [DOI] [PubMed] [Google Scholar]
- 8.Robertson DJ, Lieberman DA, Winawer SJ, et al. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut 2014;63:949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.le Clercq CM, Bouwens MW, Rondagh EJ, et al. Postcolonoscopy colorectal cancers are preventable: a population-based study. Gut 2014;63:957–63. [DOI] [PubMed] [Google Scholar]
- 10.Singh S, Singh PP, Murad MH, et al. Prevalence, risk factors, and outcomes of interval colorectal cancers: a systematic review and meta-analysis. Am J Gastroenterol 2014;109:1375–89. [DOI] [PubMed] [Google Scholar]
- 11.Cheung KS, Chen L, Seto WK, et al. Epidemiology, characteristics, and survival of post-colonoscopy colorectal cancer in Asia: A population-based study. J Gastroenterol Hepatol 2019;34:1545–1553. [DOI] [PubMed] [Google Scholar]
- 12.Govindarajan A, Rabeneck L, Yun L, et al. Population-based assessment of the outcomes in patients with postcolonoscopy colorectal cancers. Gut 2016;65:971–6. [DOI] [PubMed] [Google Scholar]
- 13.Samadder NJ, Neklason D, Snow A, et al. Clinical and Molecular Features of Post-Colonoscopy Colorectal Cancers. Clin Gastroenterol Hepatol 2019;17:2731–2739.e2. [DOI] [PubMed] [Google Scholar]
- 14.Jodal HC, Løberg M, Holme Ø, et al. Mortality From Postscreening (Interval) Colorectal Cancers Is Comparable to That From Cancer in Unscreened Patients-A Randomized Sigmoidoscopy Trial. Gastroenterology 2018;155:1787–1794.e3. [DOI] [PubMed] [Google Scholar]
- 15.Erichsen R, Baron JA, Stoffel EM, et al. Characteristics and survival of interval and sporadic colorectal cancer patients: a nationwide population-based cohort study. Am J Gastroenterol 2013;108:1332–40. [DOI] [PubMed] [Google Scholar]
- 16.Cha JM, Kim HS, Kwak MS, et al. Features of Postcolonoscopy Colorectal Cancer and Survival Times of Patients in Korea. Clin Gastroenterol Hepatol 2019;17:786–788. [DOI] [PubMed] [Google Scholar]
- 17.Dossa F, Sutradhar R, Saskin R, et al. Clinical and endoscopist factors associated with post-colonoscopy colorectal cancer in a population-based sample. Colorectal Dis 2021;23:635–645. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoffel EM, Erichsen R, Frøslev T, et al. Clinical and Molecular Characteristics of Post-Colonoscopy Colorectal Cancer: A Population-based Study. Gastroenterology 2016;151:870–878.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arain MA, Sawhney M, Sheikh S, et al. CIMP status of interval colon cancers: another piece to the puzzle. Am J Gastroenterol 2010;105:1189–95. [DOI] [PubMed] [Google Scholar]
- 21.Schoen RE, Pinsky PF, Weissfeld JL, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med 2012;366:2345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller AB, Yurgalevitch S, Weissfeld JL. Death review process in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials 2000;21:400s–406s. [DOI] [PubMed] [Google Scholar]
- 23.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med 2012;367:1596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao Y, Bertoia ML, Lenart EB, et al. Origin, Methods, and Evolution of the Three Nurses’ Health Studies. Am J Public Health 2016;106:1573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishihara R, Wu K, Lochhead P, Morikawa T, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013;369:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoen RE, Pinsky PF, Weissfeld JL, et al. Colorectal cancers not detected by screening flexible sigmoidoscopy in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Gastrointest Endosc 2012;75:612–20. [DOI] [PubMed] [Google Scholar]
- 27.Rich-Edwards JW, Corsano KA, Stampfer MJ. Test of the National Death Index and Equifax Nationwide Death Search. Am J Epidemiol 1994;140:1016–9. [DOI] [PubMed] [Google Scholar]
- 28.Smith-Warner SA, Spiegelman D, Ritz J, et al. Methods for pooling results of epidemiologic studies: the Pooling Project of Prospective Studies of Diet and Cancer. Am J Epidemiol 2006;163:1053–64. [DOI] [PubMed] [Google Scholar]
- 29.Pereira TV, Patsopoulos NA, Salanti G, et al. Critical interpretation of Cochran’s Q test depends on power and prior assumptions about heterogeneity. Res Synth Methods 2010;1:149–61. [DOI] [PubMed] [Google Scholar]
- 30.Gurjao C, Zhong R, Haruki K, et al. Discovery and Features of an Alkylating Signature in Colorectal Cancer. Cancer Discov 2021;11:2446–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 2016;17:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gingold-Belfer R, Boltin D, Sneh-Arbib O, et al. Association Between Polyp Detection Rate and Post-Colonoscopy Cancer Among Patients Undergoing Diagnostic Colonoscopy. Clin Gastroenterol Hepatol 2021;19:202–204. [DOI] [PubMed] [Google Scholar]
- 35.Kaminski MF, Regula J, Kraszewska E, et al. Quality indicators for colonoscopy and the risk of interval cancer. The New England journal of medicine 2010;362:1795–803. [DOI] [PubMed] [Google Scholar]
- 36.Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:1298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schottinger JE, Jensen CD, Ghai NR, et al. Association of Physician Adenoma Detection Rates With Postcolonoscopy Colorectal Cancer. Jama 2022;327:2114–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burnett-Hartman AN, Newcomb PA, Potter JD, et al. Genomic aberrations occurring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer research 2013;73:2863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawhney MS, Farrar WD, Gudiseva S, et al. Microsatellite instability in interval colon cancers. Gastroenterology 2006;131:1700–5. [DOI] [PubMed] [Google Scholar]
- 40.Bogie RMM, le Clercq CMC, Voorham QJM, et al. Molecular pathways in post-colonoscopy versus detected colorectal cancers: results from a nested case-control study. Br J Cancer 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this study are available upon reasonable request. Further information including the procedures to obtain and access data from the Nurses’ Health Studies and Health Professionals Follow-up Study is described at https://www.nurseshealthstudy.org/researchers (contact email: nhsaccess@channing.harvard.edu) and https://sites.sph.harvard.edu/hpfs/for-collaborators/.


