Abstract
Met encodes a receptor tyrosine kinase (MET) that is expressed during development and regulates cortical synapse maturation. Conditional deletion of Met in the nervous system during embryonic development leads to deficits in adult contextual fear learning, a medial prefrontal cortex (mPFC)‐dependent cognitive task. MET also regulates the timing of critical period plasticity for ocular dominance in primary visual cortex (V1). However, the underlying circuitry responsible remains unknown. Therefore, this study determines the broad expression patterns of MET throughout postnatal development in mPFC and V1 projection neurons (PNs), providing insight into similarities and differences in the neuronal subtypes and temporal patterns of MET expression between cortical areas. Using a transgenic mouse line that expresses green fluorescent protein (GFP) in Met+ neurons, immunofluorescence and confocal microscopy were performed to visualize MET‐GFP+ cell bodies and PN subclass‐specific protein markers. Analyses reveal that the MET expression is highly enriched in infragranular layers of mPFC, but in supragranular layers of V1. Interestingly, temporal regulation of the percentage of MET+ neurons across development not only differs between cortical regions but also is distinct between lamina within a cortical region. Further, MET is expressed predominantly in the subcerebral PN subclass in mPFC, but the intratelencephalic PN subclass in V1. The data suggest that MET signaling influences the development of distinct circuits in mPFC and V1 that underlie subcerebral and intracortical functional deficits following Met deletion, respectively.
Keywords: cortico‐cortical, MET receptor tyrosine kinase, molecular heterogeneity, neuronal subclasses, projection neurons, subcerebral
Percentage of MET‐expressing neurons in medial prefrontal and primary visual (V1) cortices varies between layer and age in mice. The predominant subclass of MET+ projection neurons in mPFC is subcortical, including corticothalamic, whereas it is intratelencephalic in V1, demonstrating stark heterogeneity between cortical regions.
1. INTRODUCTION
The cerebral cortex comprises diverse cell types and circuits required to subserve higher‐order brain processes. This diversity arises during development, when molecular expression of cortical neurons becomes heterogeneous and changes over time to meet developmental demands, including those related to experience‐dependent maturation of circuits (Bruno et al., 2009; Dantzker & Callaway, 1998; Trevino et al., 2021; Zhong et al., 2018). This occurs before entering a more stable state in adulthood. Despite its complexity, great strides have been made in understanding the development of cortical heterogeneity. For cortical projection neurons (PNs), heterogeneity has been studied broadly, in the context of areal patterning (Bhaduri et al., 2021; C. H. Chen et al., 2011; Levitt et al., 1997) and lamina identity within a cortical area (R. Luo et al., 2011; Molinard‐Chenu et al., 2020; Qian et al., 2020), as well as more specifically, at the level of a cell subclass—defined by broad neuronal projection differences within a cortical layer (Gerfen et al., 2018; Hatanaka et al., 2016; Kassai et al., 2008; Kast & Levitt, 2019; Molyneaux et al., 2007; Tsyporin et al., 2021)—and, most recently, at the level of a cell type, which further refines subclass identity based on RNA composition (Z. Zhang et al., 2021; E. J. Kim et al., 2020; C. Luo et al., 2017; Zeisel et al., 2015). These efforts have led to a greater understanding of the molecular heterogeneity of mature PNs. Currently, however, there is a more limited understanding of the generalizability of neuronal cell types identified in one cortical region at one stage of development to other cortical regions and developmental timepoints. Identification of molecules exhibiting discrete temporal and spatial patterns in the developing cortex will narrow this knowledge gap.
The c‐MET receptor tyrosine kinase (MET) is expressed transiently in cortical PNs during the peak period of synaptogenesis, with greatly reduced expression during adolescence (K. Chen et al., 2021; Eagleson et al., 2016; Judson et al., 2009). Selective deletion of Met in cells that arise from the dorsal pallium results in precocious electrophysiological and molecular maturation of excitatory synapses. In contrast, extending cortical Met expression past its normal temporal decline results in synapses remaining in a more immature state (K. Chen et al., 2021; Ma et al., 2022). Thus, the timing of the downregulation of MET expression modulates the timing of synapse maturation and stabilization, with functional consequences at the circuit and behavioral level. For example, the critical period for ocular dominance in the primary visual cortex (V1) is closed prematurely or opened later by deleting or extending Met expression, respectively (K. Chen et al., 2021). Disruption of the temporal regulation of MET also impacts medial prefrontal cortex (mPFC)‐mediated functions, including social cognition (Ma et al., 2022) and contextual fear memory (Heun‐Johnson & Levitt, 2017; Thompson & Levitt, 2015; Xia et al., 2021). Notably, while deficits in contextual fear memory are apparent in adults when Met expression is reduced or eliminated developmentally from all neural cells (Heun‐Johnson & Levitt, 2017; Thompson & Levitt, 2015) or from cells arising from the dorsal pallium (Xia et al., 2021), there is no effect on the onset of expression of contextual fear memory in weanling mice (unpublished data). Therefore, while MET expression is involved in appropriate emergence of visual circuit patterning, the receptor appears to be necessary for long‐term cognitive capabilities in adults, perhaps reflecting differences in the specific circuits expressing MET in V1 and mPFC.
The identity of PNs contributing to MET‐expressing circuits in V1 and mPFC is not known, although recent studies in mice provide insight. For example, in granular V1, visual experience drives cell‐type differentiation of PNs in layers 2/3, but not infragranular layer 5 or 6 (Cheng et al., 2022), suggesting experience‐driven critical periods may involve supragranular plasticity, while neurons in infragranular layers are stable at the molecular level before eye opening. In contrast, social deficits in autism mouse models are driven by abnormalities in layer 5 subcortical (SC) PNs in granular mPFC (Brumback et al., 2018). Similarly, cognitive flexibility is driven by activity of infragranular PNs but not supragranular intratelencephalic (IT) PNs (Nakayama et al., 2018; Spellman et al., 2021). These data indicate that, although MET is expressed in all cerebral cortical areas (Judson et al., 2009), there may be regional differences in the specific populations that express the receptor. Thus far, detailed analyses have focused on primary somatosensory cortex (S1), in which MET is enriched in ITPNs. In this region, MET is also expressed in a smaller subset of SCPNs but is excluded from nearly all granule neurons and corticothalamic (CT) PNs (Kast et al., 2019). Whether the expression patterns of MET in S1 are recapitulated in V1 and mPFC remain unknown. The developmental contributions of MET signaling, as well as the different roles of ITPNs and SCPNs in V1 and mPFC, however, raise questions regarding the specificity of MET‐expressing PN phenotypes across cortical regions. Here, experiments were designed to determine the developmental expression patterns of MET in mPFC and V1 PNs, providing greater insight into (1) the timing and changes over time of MET expression by cortical PNs during development and (2) the similarities and differences in temporal and spatial MET expression in mPFC and V1.
2. MATERIALS AND METHODS
2.1. Animals
A MetEGFP BAC transgenic mouse line was rederived from the Met‐EGFP Bacterial Artificial Chromosome (BX139), obtained from the GENSAT repository at Rockefeller University (RRID:SCR_002721), on an FVB background, as previously described in Kamitakahara et al. (2017) and Kast et al. (2019). Multiplex in situ hybridization in the brainstem raphe and neocortex has validated that the expression of green fluorescent protein (GFP) recapitulates endogenous Met transcript expression in founder lines (Kamitakahara et al., 2017; Kast et al., 2017; Kast et al., 2019). Founder mice were then backcrossed with C57BL/6J mice (The Jackson Laboratory, RRID:IMSR_JAX:000664) for at least 10 generations. Backcrossed female and male mice homozygous for the Met‐EGFP transgene were bred in our facility to produce the homozygous MetGFP pups used in this study. MetGFP mice express EGFP under the control of the Met promoter, which permits visualization of cell bodies that express MET. Mice were housed on ventilated racks with a 13:11 h light:dark cycle (lights on at 06:00 h, lights off at 19:00 h) at 22°C with ad libitum access to water and a standard chow diet (PicoLab Rodent Diet 20, #5053, St. Louis, MO). Animal care and procedures were in accordance with the guidelines set forth by the Children's Hospital Los Angeles Institutional Animal Care and Use Committee.
2.2. Immunofluorescence staining
Brain tissue for immunofluorescence staining was collected at several ages between postnatal day (P) 2 and P35 from male and female MetGFP mice. Mice used at P2 were anesthetized by cold exposure followed by acute decapitation. Brains were then dissected at room temperature in 0.1 M phosphate‐buffered saline (PBS) and immersed in fixative (4% paraformaldehyde [Sigma, St. Louis, MO] in PBS, pH 7.4) at 4°C for 12–18 h. Mice used on or after P4 were deeply anesthetized by intraperitoneal injection of ketamine:xylazine (100 mg/kg:10 mg/kg, Henry Schein, Melville, NY). Animals were then transcardially perfused with fixative, followed by immediate dissection and immersion of the brain in fixative at 4°C for 2 h. Following post‐fixation, brains of all ages were cryoprotected by sequential incubation in 10%, 20%, and 30% sucrose in PBS, embedded in Tissue‐Tek® Optimal Cutting Temperature Compound (VWR, Radnor, PA), frozen over liquid nitrogen vapors, and stored at −80°C until cryosectioning. Coronal sections (20 µm) were collected at −20°C and mounted onto superfrost plus microscope slides (VWR, Radnor, PA) in a series of five. Slides were stored at −80°C until immunofluorescence staining. For immunostaining, slides were defrosted at room temperature for 10 min, dried at 60°C for 15 min in a hybridization oven, and washed in PBS at room temperature for 10 min. Sections were blocked and permeabilized at room temperature for 1 h in PBS containing 5% Normal Donkey Serum (Jackson ImmunoResearch, West Grove, PA) and 0.3% Triton X‐100 (Sigma, St. Louis, MO), then incubated overnight at room temperature in one or more primary antibody diluted in 0.1% Triton X‐100 in PBS. Primary antibodies (characterizations below) used were chicken antigreen fluorescent protein (GFP; Abcam Cat# ab13970, RRID:AB_300798), rat anti‐CTIP2 (Abcam Cat# ab18465, RRID:AB_2064130), rabbit anti‐NeuN (Millipore Cat# ABN78, RRID:AB_10807945), mouse anti‐SATB2 (Abcam Cat # ab51502, RRID:AB_882455), rabbit anti‐DARPP‐32 (Cell Signaling Technology Cat# 2306; RRID:AB_823479), and rabbit anti‐PCP4 (J Morgan Laboratory, St. Jude's). Sections were washed five times for 5 min each at room temperature with 0.2% Tween‐20 (Sigma, St. Louis, MO) in PBS, then incubated at room temperature for 1 h in diluted Alexa Fluor ® F(ab′)2 conjugated secondary antibodies (1:500; Abcam) in 0.1% Triton X‐100 in PBS and protected from light, hereon. Sections were washed three times for 5 min each with 0.2% Tween‐20 in PBS. Sections were then incubated in DAPI (1:15,000; ThermoFisher Scientific Cat# D1306) diluted in PBS for 8 min, followed by two washes in PBS for 5 min each. Sections were embedded with a coverslip using ProLong Gold antifade mountant (ThermoFisher Scientific Cat# P36930), and the mounting media cured for at least 24 h before acquiring images using confocal microscopy.
2.3. Antibody characterization
The chicken anti‐GFP polyclonal antibody (Abcam, Cat# ab13970, used at 1:500) immunogen is a recombinant full‐length protein, corresponding to GFP. Samples from transgenic mice expressing GFP analyzed by Western blot using this antibody exhibit a single 25 kDa band (manufacturer's datasheet). Immunofluorescence staining with the antibody recapitulates endogenous Met transcript in the MetGFP line (Kamitakahara et al., 2017; Kast et al., 2017; Kast et al., 2019).
The rat anti‐CTIP2 [25B6] monoclonal antibody (Abcam, Cat# ab18465, used at 1:500) immunogen is a recombinant fragment corresponding to human CTIP2 with a GTS fusion. The antibody detects two bands representing CTIP2 at about 120 kDa and is highly expressed in brain and in malignant T‐cell lines derived from patients with adult T‐cell leukemia/lymphoma (manufacturer's datasheet). The antibody stained cell nuclei in infragranular layers of mouse cortex in a pattern that is identical with previous reports of CTIP2 expression (Arlotta et al., 2005).
The rabbit anti‐NeuN polyclonal antibody (Millipore, Cat# ABN78, used at 1:500) immunogen is a GST‐tagged recombinant fragment corresponding to first 97 amino acids from the N‐terminal region of murine NeuN. The antibody detects bands ∼48/42 kDa. Uncharacterized bands may be observed in some lysates (manufacturer's datasheet). NeuN is a neuronal marker (Dent et al., 2010).
The mouse anti‐SATB2 [SATBA4B10]–C‐terminal monoclonal antibody (Abcam, Cat # ab51502, used at 1:100) immunogen is a recombinant fragment corresponding to human SATB2 (C terminal). The antibody detects a single band at 82 kDa (manufacturer's datasheet).
The rabbit anti‐DARPP‐32 monoclonal antibody (Cell Signaling Technology, Cat# 2306, used at 1:250) is produced by immunizing animals with a synthetic peptide corresponding to residues surrounding Glu160 of human DARPP‐32 (manufacturer's datasheet).
The antiserum to PCP4 (PEP‐19) (J Morgan Laboratory, St. Jude's, used at 1:3000) immunogen is a 13 a.a. peptide of PCP4 VAIQSQFRKFQKK (Ziai et al., 1988). The specificity of detection for these antibodies has also been confirmed by colocalization of ISH signals (Watakabe et al., 2012).
2.4. Imaging and analyses
Two cortical regions, mPFC (corresponding to areas 24a, 25, and 32 in Paxinos & Franklin, 2019) and V1 (corresponding to V1 in Paxinos & Franklin, 2019), were analyzed. For qualitative analyses, the images of mPFC or V1 were acquired on a Zeiss LSM 700 inverted confocal microscope using a 10×/0.45 Plan‐APOCHROMAT or a Leica STELLARIS 5: 10×/0.40 HC PL APO CS2, respectively. For quantitative analyses, the images of mPFC were acquired on a Zeiss LSM 700 inverted confocal microscope using a 20×/0.8NA Plan‐APOCHROMAT objective lens, using refractive index correction, and images of V1 were acquired on a Leica STELLARIS 5 inverted confocal microscope using a 20×/.75 air lens. Images were collected at 2 µm z‐stacks through the entire thickness of the section at 1AU (Zeiss: 0.313 × 0.313 × 2 µm; Leica: 0.757 × 0.757 × 2 µm). Three brain sections, at least 100 µm apart and corresponding to rostral, middle, and caudal mPFC or V1, were imaged and counted per animal. Some brains were used for mPFC and V1 analyses, while other brains were used for only one brain region. For each analysis, no more than two mice of the same sex from a single litter were used. Further, at least three litters and approximately equal numbers of males and females are represented in each analysis to minimize potential litter and sex effects. Criteria for an immunolabeled cell, for which a positive profile included the fluorescent signal representing the marker of interest and a DAPI+ nucleus, were established by two researchers and validated through independent counts to confirm inter‐rater reliability. Manual counts of immunolabeled cell bodies in discrete cortical layers were then performed using the “cell counter” plugin in FIJI software version 2.3.0 (https://fiji.sc/, RRID:SCR_002285). First, images were cropped to the layer of interest, based on DAPI and CTIP2 immunostaining. The thickness of each cortical layer crop varied based on the depth of the layer to capture the full thickness, while the width of the cortical crop was held consistent in each brain region (mPFC: 321 µm; V1: 861 µm) The number of cells immunolabeled with MET‐GFP or with a specific marker, as well as the number of cells immunolabeled with MET‐GFP and the marker, were counted for each section. The percentage of colabeled cells was determined for each section, and percentages for the three sections were averaged to obtain one value per animal (per brain region).
2.5. Statistical analysis
Data were statistically analyzed and graphed using GraphPad PRISM software version 9.1.2 (http://www.graphpad.com/, RRID:SCR_002798) and then figures of the graphs were created using Microsoft PowerPoint version 16.63, with the exception of the graphical abstract, which was created with BioRender (http://biorender.com, RRID:SCR_018361). Mean ± standard error of the mean is reported in the text, with individual values and sample sizes reported in the figure legends. For each analysis, sample size was determined to obtain at least 80% power with α = .05 (SPH Analytics, statistical power calculator using average values). D'Agostino and Pearson normality test, when n ≥ 8, or Shapiro Wilk normality test, when n < 8, was used to determine normal distribution for each data set. For data with normal distributions, two‐tailed unpaired t‐test (test statistic: t), ordinary one‐way ANOVA (equal standard deviation; test statistic: F), or Welch's ANOVA (not equal standard deviation; test statistic: W) were used to determine statistically significant differences in percentage of colocalization between ages. For the ANOVAs, if the omnibus test detected a significant difference, a post hoc Tukey's multiple comparisons test (ordinary one‐way ANOVA) or Dunnett's T3 multiple comparisons test (Welch's ANOVA) was performed to determine the ages at which differences occurred. For data without normal distributions, the Kruskal‐Wallis test (test statistic: H) was performed to determine statistically significant differences in percentage of colocalization between ages. If the omnibus test detected a significant difference, a post hoc Dunn's multiple comparisons test was performed to determine at which ages the differences occurred.
3. RESULTS
The MetGFP transgenic mouse line was used to the profile GFP‐expressing (MET‐GFP+) neurons in the medial prefrontal cortex (mPFC) and primary visual cortex (V1) across early postnatal development. There are two patterns of MET‐GFP+ expression that are described: spatial, related to enrichment differences in laminar location, and temporal, related to changes in the percentage of neurons that express MET‐GFP over developmental time. In mPFC, MET‐GFP+ neurons are largely restricted to layers 5 and 6 (Figure 1a). Quantitative analysis reveals no significant effect of age on the percentage of layer 5 MET‐GFP+ neurons, defined by coexpression with NeuN, with approximately one‐fifth double‐labeled across the first 5 postnatal weeks (F = 1.8263; p = .1457; P2: 13.84 ± 0.82%; P7: 20.87 ± 1.25%; P12: 19.23 ± 1.62%; P15: 18.29 ± 2.90%; P21: 18.50 ± 4.15%; P35: 17.20 ± 1.32%; Figure 1b). This contrasts with layer 6, in which there is a significant effect of age on the percentage of neurons expressing MET‐GFP over the same time period (F = 52.7200; p < .0001; Figure 1c). Post hoc analyses demonstrate a significant reduction in the percentage of neurons expressing MET‐GFP after the first postnatal week. Specifically, the mean percentage at P2 (57.40 ± 2.59%) and P7 (49.83 ± 2.07%) is significantly different from those at P12 (26.57 ± 1.91%), P15 (28.49 ± 3.68%), P21 (21.88 ± 1.10%), and P35 (17.12 ± 2.14%), as are those between P15 and P35; all other comparisons are not significantly different (Table 1). In comparison to mPFC, the laminar distribution of MET‐GFP+ neurons is substantially different in V1. Most notably, in addition to the infragranular layers, MET‐GFP+ neurons are abundant in layers 2/3 (Figure 1d), similar to previous findings in primary somatosensory cortex (Kast et al., 2019). The high packing density of neurons in relatively immature layers 2/3 at P4 precluded quantitative analysis at that age, but analyses at later ages revealed no significant difference in the percentage of MET‐GFP+ neurons at P7 and P21 (t = 1.8817; p = .0843; P7: 18.10 ± 2.51%; P21: 25.50 ± 3.02%; Figure 1e). Further, the temporal pattern of changes in the percentage of MET‐GFP+ neurons in layers 5 and 6 are distinct from that in mPFC. Specifically, in V1 layer 5, there is a significant effect of age on the percentage of neurons that express MET‐GFP (H = 8.1109; p = .0112; Figure 1f), driven by a decrease over the first postnatal week (p = .0209; P4: 15.95 ± 8.61%; P7: 7.25 ± 1.02%); there is no significant difference between P4 or P7 and P21 (P21: 7.77 ± 1.24%; Table 2). There also is a significant effect of age in layer 6 (W = 7.7554; p = .0148; Figure 1g), driven by a decrease between P4 (13.33 ± 0.85%) and P21 (9.96 ± 0.29%); P7 (12.91 ± 1.59%) does not differ significantly from either P4 or P21 (Table 3). Collectively, the data suggest there are divergent laminar patterns of MET expression in mPFC and V1 neurons, as well as distinct differences in the stability of expression over time.
FIGURE 1.
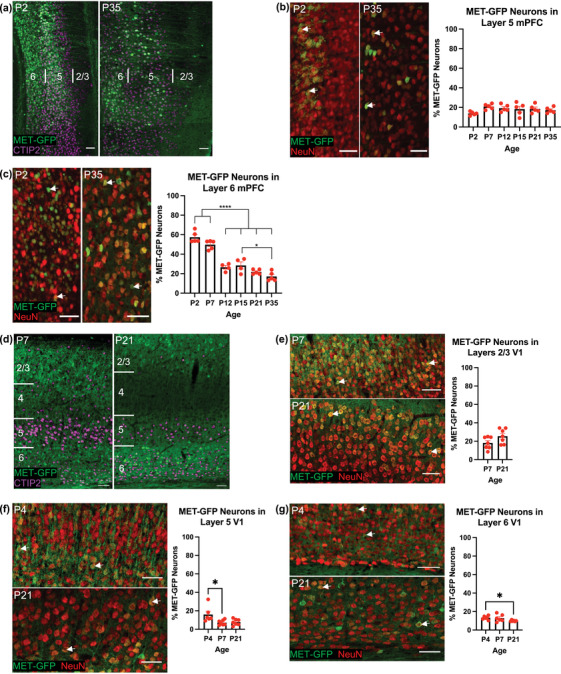
The spatial and temporal expression of MET‐GFP in mPFC and V1 neurons across developmental ages. (a) Representative images at P2 (left panel) and P35 (right panel) of MET‐GFP (green) expression in mPFC, with CTIP2 (magenta) as marker for infragranular layers 5 and 6. Boundaries of layers 2/3, 5, and 6 are denoted. (b) Representative images at P2 (left panel) and P35 (middle panel) of MET‐GFP (green) and NeuN (red) overlayed expressions in layer 5 mPFC. Arrows denote examples of colocalization between and MET‐GFP and NeuN. Quantification of the percentage of layer 5 mPFC neurons that express MET‐GFP at developmental ages between P2 and P35 (right panel) (n = 5 for each age). There is no significant effect of age, analyzed by ordinary one‐way ANOVA. (c) Representative images at P2 (left panel) and P35 (middle panel) of MET‐GFP (green) and NeuN (red) overlayed expression in layer 6 mPFC. Arrows denote examples of colocalization between NeuN and MET‐GFP. Quantification of the percentage of layer 6 mPFC neurons that express MET‐GFP at developmental ages between P2 and P35 (right panel). n = 5 for P2, P7, P21, and P35, n = 4 for P12 and P15. “*” indicates p ≤ .05, ‘****’ indicates p < .0001, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (d) Representative images at P7 (left panel) and P21 (right panel) of MET‐GFP (green) expression in V1, with CTIP2 (magenta) as marker for infragranular layers 5 and 6. Boundaries of layers 2/3, 5, and 6 are denoted. (e) Representative images at P7 (top left panel) and P21 (bottom left panel) of MET‐GFP (green) and NeuN (red) overlayed expressions in layers 2/3 V1. Arrows denote examples of colocalization between and MET‐GFP and NeuN. Quantification of the percentage of layer 2/3 V1 neurons that express MET‐GFP at developmental ages between P7 and P21 (right panel). n = 7 for each age. There is no significant difference between ages, analyzed by unpaired t test. (f) Representative images at P7 (top left panel) and P21 (bottom left panel) of MET‐GFP (green) and NeuN (red) overlayed expressions in layer 5 V1. Arrows denote examples of colocalization between and MET‐GFP and NeuN. Quantification of the percentage of layer 5 V1 neurons that express MET‐GFP at developmental ages between P4 and P21 (right panel). n = 6 for P4, n = 7 for P7 and P21. “*” indicates p ≤ .05, analyzed by Kruskal‐Wallis test followed by Dunn's multiple comparisons test. (g) Representative images at P7 (top left panel) and P21 (bottom left panel) of MET‐GFP (green) and NeuN (red) overlayed expressions in layer 6 V1. Arrows denote examples of colocalization between and MET‐GFP and NeuN. Quantification of the percentage of layer 6 V1 neurons that express MET‐GFP at developmental ages between P4 and P21 (right panel). n = 6 for each age. “*” indicates p ≤ .05, analyzed by Welch's ANOVA test followed by Dunnett's T3 multiple comparisons test. All scale bars = 50 µm. The brightness and contrast of each channel were increased globally in images for visualization purposes
TABLE 1.
Post hoc analyses of MET‐GFP+ neurons in layer 5 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P2 vs. P7 | .1953 |
| P2 vs. P12 | <.0001 |
| P2 vs. P15 | <.0001 |
| P2 vs. P21 | <.0001 |
| P2 vs. P35 | <.0001 |
| P7 vs. P12 | <.0001 |
| P7 vs. P15 | <.0001 |
| P7 vs. P21 | <.0001 |
| P7 vs. P35 | <.0001 |
| P12 vs. P15 | .9934 |
| P12 vs. P21 | .7223 |
| P12 vs. P35 | .0881 |
| P15 vs. P21 | .3821 |
| P15 vs. P35 | .0262 |
| P21 vs. P35 | .6572 |
TABLE 2.
Post hoc analyses of MET‐GFP+ neurons in layer 5 V1
| Dunn's multiple comparisons test | Adjusted p value |
|---|---|
| P4 vs. P7 | .0209 |
| P4 vs. P21 | .0791 |
| P7 vs. P21 | >.9999 |
TABLE 3.
Post hoc analyses of MET‐GFP+ neurons in layer 6 V1
| Dunnett's T3 multiple comparisons test | Adjusted p value |
|---|---|
| P4 vs. P7 | .9928 |
| P4 vs. P21 | .0248 |
| P7 vs. P21 | .2999 |
In the cerebral cortex, the MET expression is enriched in excitatory projection neurons (PNs; Eagleson et al., 2011; Judson et al., 2009). In layer 5, two broad subclasses of PNs, long‐range subcerebral PNs (SCPNs) and intratelencephalic PNs (ITPNs), can be identified using molecular markers (Woodworth et al., 2012). The SCPN subclass broadly includes PNs that project subcortically to target regions including midbrain, brain stem, and spinal cord, while the ITPN subclass broadly includes PNs that project to cortical targets (Harris & Shepherd, 2015). We next determined if the divergent patterns of MET expression in infragranular layers of mPFC and V1 are reflected by differential coexpression of MET with CTIP2, which is involved in the development of SCPNs (Arlotta et al., 2005), or with SATB2, which enables neurons to project cortically (ITPNs) through repression of Ctip2 (Alcamo et al., 2008). First, we determined the proportion of CTIP2+ (Figure 2a,b) or SATB2+ (Figure 2c,d) layer 5 neurons that express MET‐GFP in each region. In mPFC, there is no significant effect of age on the percentage of CTIP2+ neurons that express MET‐GFP (F = 0.5279; p = .5982; Figure 2e). Specifically, approximately 20% of CTIP2+ neurons express MET‐GFP across the three ages examined (P7: 19.97 ± 1.31%; P15: 22.43 ± 2.48%; P21: 19.91 ± 1.63%). In V1, however, MET‐GFP is almost completely absent from the CTIP2+ population at P7 (1.28 ± 0.32%). Further, there is a significant effect of age on the percentage of CTIP2+ neurons that express MET‐GFP (F = 4.4144; p = .0192; Figure 2f), with post hoc analyses demonstrating a significant increase at P15 (5.82 ± 2.60%) and P21 (6.79 ± 1.09%) compared to P7; the percentage of V1 CTIP2 neurons that express MET‐GFP at P12 (4.68 ± 0.70%) is not statistically different from any other age analyzed (Table 4). In mPFC, similar to the CTIP2+ population, there is no significant effect of age on the percentage of SATB2+ neurons that express MET‐GFP (F = 0.5994; p = .5592; Figure 2g), with approximately 10% of SATB2+ neurons expressing MET‐GFP at the three ages examined (P7: 12.60 ± 0.86%; P15: 11.51 ± 2.06%; P21: 9.87 ± 0.87%). A similar percentage of SATB2+ neurons express MET‐GFP in V1, again with no significant effect of age (F = 0.4538; p = .7182; P7: 10.57 ± 1.54%; P12: 9.96 ± 0.96%; P15: 9.56 ± 1.73%; P21: 8.40 ± 1.17%; Figure 2h). Together, these data suggest that less than one quarter of the total layer 5 CTIP2+ and SATB2+ population of neurons express MET in mPFC and V1, consistent with MET expression in less than one quarter of the total neurons in layer 5 of these regions (Figure 1b,f).
FIGURE 2.
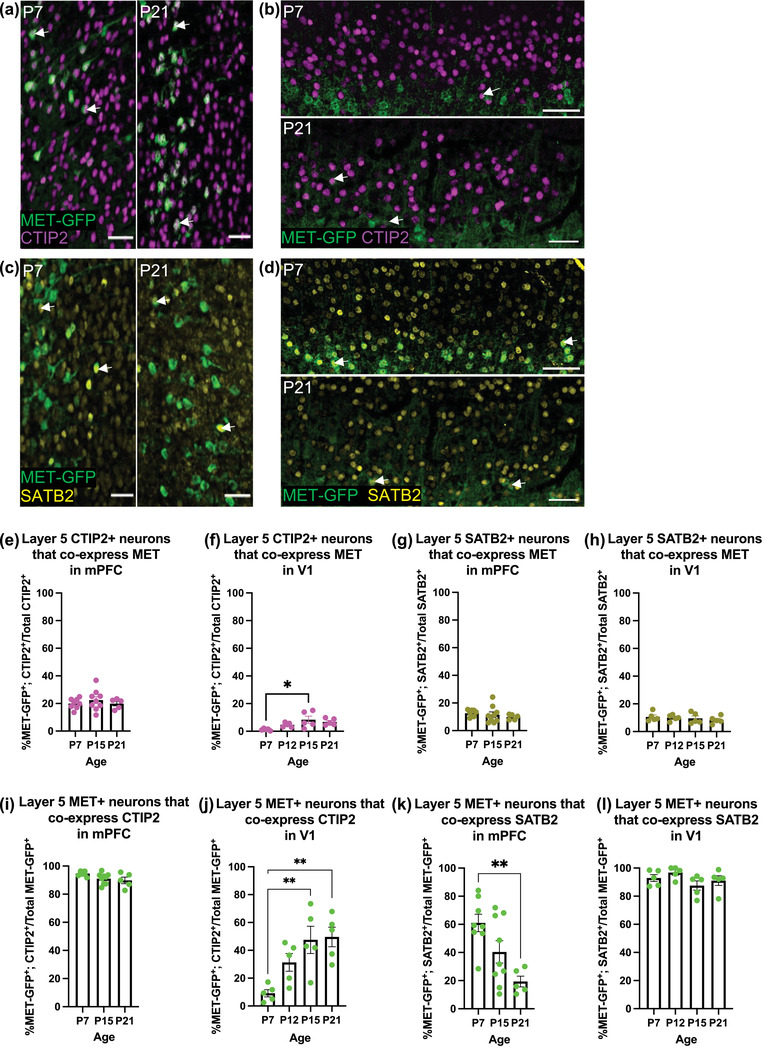
Colocalization analyses of MET‐GFP with CTIP2 and SATB2 in layer 5 mPFC and V1 across developmental ages. (a) Representative images at P7 (left) and P21 (right) of MET‐GFP (green) and CTIP2 (magenta) overlayed expressions in layer 5 mPFC. Arrows denote examples of colocalization between MET‐GFP and CTIP2. (b) Representative images at P7 (top) and P21 (bottom) of MET‐GFP (green) and CTIP2 (magenta) overlayed expressions in layer 5 V1. Arrows denote examples of colocalization between MET‐GFP and CTIP2. (c) Representative images at P7 (left) and P21 (right) of MET‐GFP (green) and SATB2 (yellow) overlayed expressions in layer 5 mPFC. Arrows denote examples of colocalization between MET‐GFP and SATB2. (d) Representative images at P7 (top) and P21 (bottom) of MET‐GFP (green) and SATB2 (yellow) overlayed expressions in layer 5 V1. Arrows denote examples of colocalization between MET‐GFP and SATB2. (e) Quantification of the percentage of layer 5 mPFC CTIP2+ neurons that co‐express MET‐GFP at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. There is no significant effect of age, analyzed by an ordinary one‐way ANOVA. (f) Quantification of the percentage of layer 5 V1 CTIP2+ neurons that co‐express MET‐GFP at P7, P12, P15, and P21. n = 5 for each age. “*” indicates p ≤ .05, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (g) Quantification of the percentage of layer 5 mPFC SATB2+ neurons that co‐express MET‐GFP at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. There is no significant effect of age, analyzed by an ordinary one‐way ANOVA. (h) Quantification of the percentage of layer 5 V1 SATB2+ neurons that co‐express MET‐GFP at P7, P12, P15, and P21. n = 5 for each age. There is no significant effect of age, analyzed by an ordinary one‐way ANOVA. (i) Quantification of the percentage of layer 5 mPFC MET‐GFP+ neurons that co‐express CTIP2 at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. There is no significant difference between ages, analyzed by an ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (j) Quantification of the percentage of layer 5 V1 MET‐GFP+ neurons that co‐express CTIP2 at P7, P12, P15, and P21. n = 5 for each age. “**” indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (k) Quantification of the percentage of layer 5 mPFC MET‐GFP+ neurons that co‐express SATB2 at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. “**” indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (l) Quantification of the percentage of layer 5 mPFC MET‐GFP+ neurons that co‐express SATB2 at P7, P12, P15, and P21. n = 5 for each age. There is no significant effect of age, analyzed by an ordinary one‐way ANOVA. All scale bars = 50 µm. The brightness and contrast of each channel were increased globally in images for visualization purposes
TABLE 4.
Post hoc analyses of CTIP2+ neurons that co‐express MET‐GFP in layer 5 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .3818 |
| P7 vs. P15 | .0161 |
| P7 vs. P21 | .0719 |
| P12 vs. P15 | .3138 |
| P12 vs. P21 | .7396 |
| P15 vs. P21 | .8675 |
Next, we determined the percentage of the total population of layer 5 MET‐GFP+ neurons that express CTIP2 or SATB2. In mPFC, there is a significant effect of age on the percentage of MET‐GFP+ neurons expressing CTIP2 (F = 3.5450; p = .0492; Figure 2i). Post hoc analyses reveal no significant difference between individual ages, with approximately 90% of MET‐GFP+ neurons expressing CTIP2 over time (P7: 94.72 ± 0.57%; P15: 91.06 ± 1.28%; P21: 89.81 ± 2.32%; Table 5). In V1, there is also a significant effect of age on the percentage of MET‐GFP+ neurons that express CTIP2 in layer 5 (F = 7.3029; p = .0027; Figure 2j). In contrast to mPFC, however, few MET‐GFP+ neurons express CTIP2 at P7 (9.13 ± 2.58%). Post hoc analyses demonstrate a significant increase in MET‐GFP+ neurons that express CTIP2 at P15 (47.51 ± 9.76%) and P21 (49.61 ± 7.04%) compared to P7, such that approximately half of MET‐GFP+ neurons also express CTIP2 by the end of the second postnatal week; there is no significant difference between P12 (31.33 ± 14.22%) and any other age analyzed (Table 6). In mPFC, there is a significant age effect on the percentage of MET+ neurons that express SATB2 (F = 7.4194; p = .0042; Figure 2k), with post hoc analyses demonstrating a significant decrease between P7 (61.01 ± 6.18%) and P21 (19.38 ± 3.83%); P15 (40.40 ± 7.94%) is not significantly different than P7 or P21 (Table 7). In contrast, in V1 MET‐GFP+ neurons expressing SATB2 are abundant in layer 5, with approximately 90% of the MET‐GFP+ neurons expressing SATB2, and there is no significant effect of age (F = 1.8300; p = .1823; P7: 92.92 ± 2.46%; P12: 96.82 ± 1.86%; P15: 87.58 ± 3.31%; P21: 91.07 ± 3.45%; Figure 2l). Together, these data suggest that the majority of MET‐expressing layer 5 neurons are molecularly distinct between cortical regions, expressing CTIP2 in mPFC but SATB2 in V1.
TABLE 5.
Post hoc analyses of MET‐GFP+ neurons that co‐express CTIP2 in layer 5 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P15 | .1155 |
| P7 vs. P21 | .0654 |
| P15 vs. P21 | .8086 |
TABLE 6.
Post hoc analyses of MET‐GFP+ neurons that co‐express CTIP2 in layer 5 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .1480 |
| P7 vs. P15 | .0061 |
| P7 vs. P21 | .0039 |
| P12 vs. P15 | .3798 |
| P12 vs. P21 | .2804 |
| P15 vs. P21 | .9963 |
TABLE 7.
Post hoc analyses of MET‐GFP+ neurons that co‐express SATB2 in layer 5 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P15 | .0944 |
| P7 vs. P21 | .0032 |
| P15 vs. P21 | .1476 |
In the previous analyses, over 100% of the MET‐GFP+ population were accounted for at each age when considering MET‐GFP co‐expression with either CTIP2 or SATB2, alone, indicating there is a population of MET‐GFP neurons that co‐expresses both CTIP2 and SATB2. In this context, while initial reports indicated that CTIP2 and SATB2 are expressed in largely nonoverlapping populations of SCPNs and ITPNs, respectively, via repression mechanisms (Alcamo et al., 2008; Baranek et al., 2012; Britanova et al., 2008; B. Chen et al., 2008), more recent studies have demonstrated coexpression of SATB2 and CTIP2 in subpopulations of excitatory PNs (Harb et al., 2016; Leone et al., 2015; Lickiss et al., 2012; Paolino et al., 2020). These studies further showed that a double‐labeled cell represents either a SCPN or an ITPN, rather than a PN that sends collaterals to both targets. We therefore performed a more nuanced analysis of the molecular phenotype of layer 5 MET‐GFP+ neurons, in which a CTIP2+; SATB2+ cell represents either an SCPN or ITPN, CTIP2+(SATB2−) cells represent SCPNs, and SATB2+(CTIP2−) cells represent ITPNs, in mPFC (Figure 3a) and V1 (Figure 3b). In mPFC at P7, approximately 55% of MET‐GFP+ neurons are CTIP2+; SATB2+, 40% are CTIP2+(SATB2−), and 5% are SATB2+(CTIP2−). There are significant effects of age for the percentage of MET‐GFP+ neurons that are CTIP2+; SATB2 + (F = 8.5190; p = .0023; Figure 3c) or CTIP2+(SATB2−) (F = 7.0515; p = .0051; Figure 3d). Post hoc analyses demonstrate a significant decrease in CTIP2+; SATB2+ MET‐GFP+ neurons between P7 (56.62 ± 6.00%) and P21 (13.06 ± 2.75%; Table 8) that is paralleled by a significant increase in the percentage of CTIP2+(SATB2−) MET‐GFP+ neurons between P7 (38.10 ± 6.07%) and P21 (76.76 ± 3.76%; Table 9). For both populations, the percentage of MET‐GFP+ neurons at P15 (CTIP2+; SATB2+: 33.58 ± 7.98%; CTIP2+(SATB2−): 57.48 ± 7.45%) is not significantly different from P7 or P21 (Tables 8 and 9). There is no significant effect of age on the percentage of MET‐GFP+ neurons expressing SATB2+(CTIP2−) in mPFC (F = 1.4951; p = .2494; P7: 4.39 ± 0.71%; P15: 6.82 ± 1.05%; P21: 6.32 ± 1.77%; Figure 3e). The percentage of MET‐GFP+ neurons in mPFC is stable across the second and third postnatal week (Figure 1b). These findings indicate that over this time period, at least 75% of the layer 5 MET population in mPFC represent the SCPN subclass, with the majority initially coexpressing CTIP2 and SATB2, but subsequently downregulating SATB2. In contrast, less than 10% of the MET‐GFP+ population is the ITPN subclass, with the remaining approximately 15% that continue to coexpress both CTIP2 and SATB2 representing either class. Notably, age‐dependent coexpression of SATB2 and CTIP2 in the MET‐GFP+ subset of neurons follows a near identical pattern to all layer 5 mPFC neurons. Specifically, there is a significant effect of age on the percentage of CTIP2+ neurons that express SATB2, independent of MET‐GFP, (F = 9.2545; p = .0016; Figure 3f), with post hoc analyses demonstrating a significant difference between P7 (59.77 ± 4.62%) and P21 (23.84 ± 3.64%), and between P15 (49.02 ± 6.09%) and P21; all other comparisons are not significantly different (Table 10). Together, these findings indicate that the downregulation of SATB2 in the MET+ SCPN subpopulation is similar to that in the whole SCPN population. The pattern of CTIP2+; SATB2+ in V1 layer 5 is strikingly different from that in mPFC. At P7, less than 10% of MET‐GFP+ neurons are CTIP2+; SATB2+, few are CTIP2+(SATB2−), and almost 90% are SATB2+(CTIP2−). There is a significant effect of age on the percentage of MET‐GFP+ neurons expressing CTIP2 and SATB2 (F = 5.7723; p = .0071; Figure 3g). Post hoc analyses demonstrate a significant increase between P7 (6.18 ± 2.22%) and P15 (38.40 ± 10.05%), and P7 and P21 (43.86 ± 6.97%), with all other comparisons not significantly different (Table 11). There is no significant effect of age on the percentage of MET‐GFP+ neurons that are CTIP2+(SATB2−) (H = 3.5917; p = .3091; P7: 2.95 ± 1.21%; P12: 1.35 ± 0.97%; P15: 9.11 ± 4.54%; P21: 5.76 ± 2.48%; Figure 3h). There also is an effect of age on the percentage of SATB2+(CTIP2−) MET‐GFP+ neurons (F = 6.5618; p = .0042; Figure 3i). Post hoc analyses reveal that, paralleling the increase in the percentage of CTIP2+; SATB2+ MET‐GFP+ layer 5 V1 neurons (Figure 3g), there is a significant decrease in the percentage of SATB2+(CTIP2−) MET‐GFP+ neurons between P7 (86.74 ± 2.78%) and P15 (38.40 ± 10.05%), and P7 and P21 (47.21 ± 7.51%); all other comparisons are not significantly different (P12: 66.84 ± 7.09%; Table 12). As in mPFC, the percentage of MET‐GFP+ neurons is stable across the second and third postnatal week in V1 (Figure 1f). The data indicate, however, that, in contrast to mPFC, over 85% of the layer 5 MET‐GFP+ population in V1 is the ITPN subclass. This ITPN population initially expresses SATB2, but not CTIP2, and then upregulates CTIP2 across the second and third postnatal weeks. In contrast, less than 10% of the MET‐GFP+ population in this layer is the SCPN subclass, with the remaining approximately 5% that expresses both subclass markers at P7 representing either ITPNs or SCPNs. We note that, independent of MET‐GFP, there is no significant effect of age on the percentage of the V1 SATB2 layer 5 population that also express CTIP2 at these ages (F = 0.4250; p = .7377; P7: 31.79 ± 2.03%; P12: 34.76 ± 3.54%; P15: 37.18% ± 11.95%; P21: 32.46% ± 3.34%; Figure 3j), suggesting that there is a bias towards the suppression of CTIP2 in the MET‐GFP+ subpopulation of ITPNs early postnatally.
FIGURE 3.
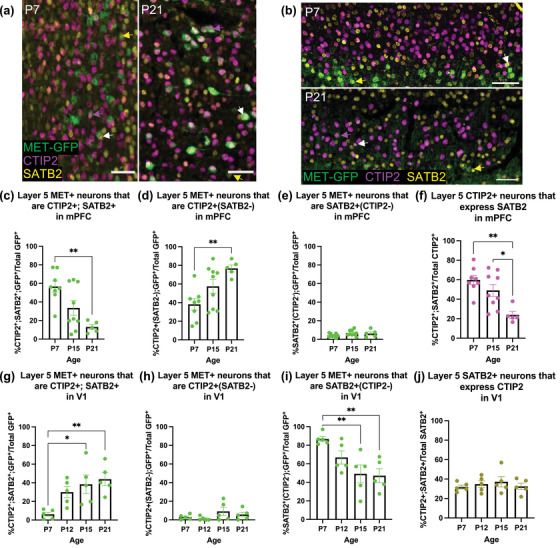
Colocalization analyses of MET‐GFP+ neurons that co‐express CTIP2 and/or SATB2 in layer 5 mPFC and V1 across developmental ages. (a) Representative images at P7 (left panel) and P21 (right panel) of MET‐GFP (green), CTIP2 (magenta), and SATB2 (yellow) overlayed expressions in layer 5 mPFC. White arrows denote examples of colocalization between MET‐GFP, CTIP2, and SATB2. Magenta arrows denote examples of MET‐GFP+; CTIP2+(SATB2−) neurons. Yellow arrows denote examples of MET‐GFP+; SATB2+(CTIP2−) neurons. (b) Representative images at P7 (top panel) and P21 (bottom panel) of MET‐GFP (green), CTIP2 (magenta), and SATB2 (yellow) overlayed expressions in layer 5 V1. White arrows denote examples of colocalization between MET‐GFP, CTIP2, and SATB2. Magenta arrows denote examples of MET‐GFP+; CTIP2+(SATB2−) neurons. Yellow arrows denote examples of MET‐GFP+; SATB2+(CTIP2−) neurons. (c) Quantification of the percentage of layer 5 mPFC MET‐GFP+ neurons that co‐express both CTIP2 and SATB2 at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. “**” indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (d) Quantification of the percentage of layer 5 mPFC MET‐GFP+ neurons that are CTIP2+(SATB2−) at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. ‘**’ indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (e) Quantification of the percentage of layer 5 mPFC MET‐GFP+ neurons that are SATB2+(CTIP2−) at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. There is no significant effect of age, analyzed by ordinary one‐way ANOVA. (f) Quantification of the percentage of layer 5 mPFC CTIP2+ neurons that express SATB2 at P7, P15, and P21. n = 8 for P7, n = 9 for P15, n = 5 for P21. “*” indicates p ≤ .05, “**” indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (g) Quantification of the percentage of layer 5 V1 MET‐GFP+ neurons that co‐express both CTIP2 and SATB2 at P7, P12, P15, and P21. n = 5 for each age. “*” indicates p ≤ .05, “**” indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (h) Quantification of the percentage of layer 5 V1 MET‐GFP+ neurons that are CTIP2+(SATB2−) at P7, P12, P15, and P21. n = 5 for each age. There is no significant effect of age, analyzed by Kruskal‐Wallis test. (i) Quantification of the percentage of layer 5 V1 MET‐GFP+ neurons that are SATB2+(CTIP2−) at P7, P12, P15, and P21. n = 5 for each age. “**” indicates p < .01, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (j) Quantification of the percentage of layer 5 V1 SATB2+ neurons that express CTIP2 at P7, P12, P15, and P21. n = 5 for each age. There is no significant effect of age, analyzed by ordinary one‐way ANOVA. All scale bars = 50 µm. The brightness and contrast of each channel were increased globally in images for visualization purposes
TABLE 8.
Post hoc analyses of MET‐GFP+ neurons that are CTIP2+; SATB2+ in layer 5 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P15 | .0525 |
| P7 vs. P21 | .0019 |
| P15 vs. P21 | .1515 |
TABLE 9.
Post hoc analyses of MET‐GFP+ neurons that are CTIP2+(SATB2−) in layer 5 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P15 | .0999 |
| P7 vs. P21 | .0040 |
| P15 vs. P21 | .1683 |
TABLE 10.
Post hoc analyses of CTIP2+ neurons that co‐express SATB2 in layer 5 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .1108 |
| P7 vs. P15 | .0215 |
| P7 vs. P21 | .0070 |
| P12 vs. P15 | .8250 |
| P12 vs. P21 | .5068 |
| P15 vs. P21 | .9431 |
TABLE 11.
Post hoc analyses of MET‐GFP+ neurons that are CTIP2+; SATB2+ in layer 5 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .2443 |
| P7 vs. P15 | .0095 |
| P7 vs. P21 | .0064 |
| P12 vs. P15 | .3383 |
| P12 vs. P21 | .2550 |
| P15 vs. P21 | .9973 |
TABLE 12.
Post hoc analyses of MET‐GFP+ neurons that are SATB2+(CTIP2−) in layer 5 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P2 vs. P9 | <.0001 |
| P2 vs. P15 | <.0001 |
| P9 vs. P15 | .9977 |
We next focused on determining the phenotype of MET‐GFP+ neurons in layer 6, which contains two major PN subclasses—corticothalamic (CT) and IT (Woodworth et al., 2012). In mPFC, DARPP‐32 serves as a marker of CTPNs (Ouimet, 1991). There is a significant age effect on the percentage of MET‐GFP+ neurons that express DARPP‐32 (F = 25.0550; p < .0001; Figure 4a,b). Post hoc analyses demonstrate a significant increase in this percentage between P7 (29.30 ± 1.85%) and P9 (P9: 57.33 ± 2.45%), and P7 and P15 (57.62 ± 4.25%); there is no significant difference between P9 and P15 (Table 13). This increase is paralleled by a decrease in the percentage of MET‐GFP+ neurons in layer 6 mPFC over the same period (Figure 1c), suggesting that as the percentage of MET‐GFP+ neurons decrease, those that continue expressing MET are most likely in the CTPN subclass. There also is an age effect on the percentage of DARPP‐32 neurons that co‐express MET‐GFP in layer 6 mPFC (F = 26.2137; p < .0001; Figure 4c). Post hoc comparisons reveal at P2, over 60% of DARPP‐32 neurons co‐express MET (64.76 ± 2.99), which decreases significantly to approximately 40% by P9 (43.23 ± 2.76); all other comparisons are not significant (P15: 35.90 ± 2.97; Table 14). These results, together with the finding that the percentage of MET‐GFP neurons decreases after P7 (Figure 1c), suggest that MET is expressed in a large population of CTPNs in the first postnatal week, and as the percentage of MET‐GFP neurons decreases, the total population of CTPNs that co‐express MET also decreases.
FIGURE 4.
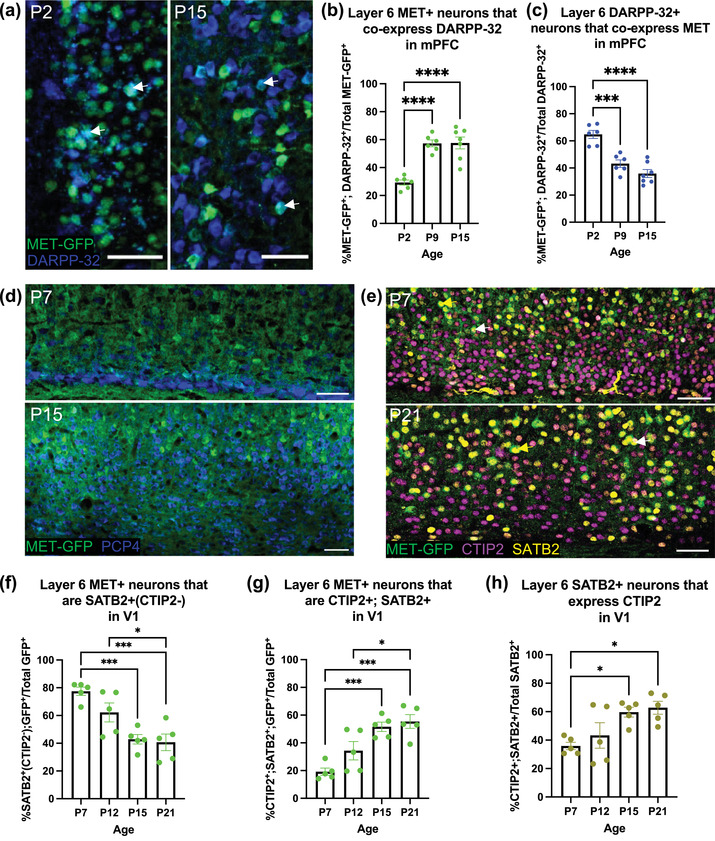
Colocalization analysis of MET‐GFP with layer 6 projection neuron markers, in mPFC and V1 across developmental ages. (a) Representative images at P2 (left panel) and P15 (right panel) of MET‐GFP (green) and DARPP‐32 (blue) overlayed expressions in layer 6 mPFC. Arrows denote examples of colocalization between MET‐GFP and DARPP‐32. (b) Quantification of the percentage of layer 6 mPFC MET‐GFP+ neurons that co‐express DARPP‐32 at P2, P9, and P15. n = 6 for P2 and P9, n = 7 for P15. “****” indicates p < .0001, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (c) Quantification of the percentage of layer 6 mPFC DARPP‐32+ neurons that co‐express MET‐GFP at P2, P9, and P15. n = 6 for P2 and P9, n = 7 for P15. “***” indicates p < .001, “****” indicates p < .0001, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (d) Representative images at P7 (top panel) and P15 (bottom panel) of MET‐GFP (green) and PCP4 (blue) overlayed expressions in layer 6 V1. (e) Representative images at P7 (top panel) and P21 (bottom panel) of MET‐GFP (green), CTIP2 (magenta), and SATB2 (yellow) overlayed expressions in layer 6 V1. White arrows denote examples of colocalization between MET‐GFP, CTIP2, and SATB2. Magenta arrows denote examples of MET‐GFP+; CTIP2+(SATB2−) neurons. (f) Quantification of the percentage of layer 6 V1 MET‐GFP+ neurons that are SATB2+(CTIP2−) at P7, P12, P15, and P21. n = 5 for each age. “*” indicates p ≤ .05, “***” indicates p < .001, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (g) Quantification of the percentage of layer 6 V1 MET‐GFP+ neurons that co‐express both CTIP2 and SATB2 at P7, P12, P15, and P21. n = 5 for each age. “*” indicates p ≤ .05, “***” indicates p < .001, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. (h) Quantification of the percentage of layer 6 V1 SATB2+ neurons that express CTIP2 at P7, P12, P15, and P21. n = 5 for each age. “*” indicates p ≤ .05, analyzed by ordinary one‐way ANOVA followed by Tukey's multiple comparisons test. All scale bars = 50 µm. The brightness and contrast of each channel were increased globally in images for visualization purposes.
TABLE 13.
Post hoc analyses of MET‐GFP+ neurons that co‐express DARPP‐32 in layer 6 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P2 vs. P9 | .0003 |
| P2 vs. P15 | <.0001 |
| P9 vs. P15 | .2045 |
TABLE 14.
Post hoc analyses of DARPP‐32+ neurons that co‐express MET‐GFP in layer 6 mPFC
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .1876 |
| P7 vs. P15 | .0010 |
| P7 vs. P21 | .0005 |
| P12 vs. P15 | .0674 |
| P12 vs. P21 | .0377 |
| P15 vs. P21 | .9900 |
DARPP‐32 is not a validated marker of CTPNs in V1 and its developmental expression patterns have not been mapped in this region. We observed low expression of DARPP‐32 at P7 and widespread expression across all cortical layers at P21 (data not shown), indicating that in V1, DARPP‐32 is not a selective marker of CTPNs. Therefore, PCP4, a previously validated marker of CTPNs in primary sensory areas (Watakabe et al., 2012), was used to identify this population, as reported previously in S1 cortex (Kast et al., 2019). In V1, there is little overlap of MET‐GFP and PCP4 at any age examined, demonstrating that MET is largely excluded from CTPNs (Figure 4d). Rather, at P7 the majority of MET‐GFP+ layer 6 neurons in V1 are ITPNs, expressing SATB2+(CTIP2−) (77.46 ± 3.02%; Figure 4e,f). There is, however, a significant age effect on the percentage of MET‐GFP+ neurons expressing SATB2 alone (F = 11.7631; p = .0003; Figure 4f), with post hoc analyses demonstrating a significant decrease between P7 and P15 (P15: 42.85 ± 3.42%), P7 and P21 (P21: 40.67 ± 5.93%), and P12 (62.23 ± 6.85%) and P21; all other comparisons are not significantly different (Table 15). Given the stable expression of MET‐GFP+ neurons in layer 6 V1 across these ages (Figure 1g), we next determined whether the decline in MET‐GFP+ neurons expressing SATB2 alone reflected an increase in those co‐expressing CTIP2 and SATB2. Indeed, there is a significant effect of age on percentage of MET‐GFP+ neurons that are CTIP2+; SATB2+ (F = 12.9124; p = .0002; Figure 4g), with post hoc analyses demonstrating a significant increase in the mean percentage between P7 (19.18 ± 2.58%) and P15 (51.60 ± 3.39%), P7 and P21 (55.42 ± 4.98%), and P12 (34.34 ± 6.60%) and P21; all other comparisons were not significantly different (Table 16). A similar pattern is observed when considering the entire layer 6 V1 SATB2 population, for which there is a significant effect of age on the percentage of SATB2 neurons that co‐express CTIP2 (F = 5.5736; p = .0082; Figure 4h). Post hoc analyses demonstrate a significant increase in this measure between P7 (35.88 ± 2.60%) and P15 (59.67 ± 3.48%), and P7 and P21 (62.75 ± 4.56%), with all other comparisons not statistically significant (P12: 43.27 ± 8.94%; Table 17). Together, these data indicate that the MET neurons in layer 6 of V1 are predominantly ITPNs and that the increasing co‐expression of CTIP2 in the MET population is consistent for the entire ITPN population.
TABLE 15.
Post hoc analyses of MET‐GFP+ neurons that are SATB2+(CTIP2−) in layer 6 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .1384 |
| P7 vs. P15 | .0008 |
| P7 vs. P21 | .0003 |
| P12 vs. P15 | .0780 |
| P12 vs. P21 | .0255 |
| P15 vs. P21 | .9365 |
TABLE 16.
Post hoc analyses of MET‐GFP+ neurons that are CTIP2+; SATB2− in layer 6 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P15 | .3127 |
| P7 vs. P21 | .0011 |
| P15 vs. P21 | .0168 |
TABLE 17.
Post hoc analyses of SATB2+ neurons that co‐express CTIP2 in layer 6 V1
| Tukey's multiple comparisons test | Adjusted p value |
|---|---|
| P7 vs. P12 | .7764 |
| P7 vs. P15 | .0330 |
| P7 vs. P21 | .0149 |
| P12 vs. P15 | .1887 |
| P12 vs. P21 | .0946 |
| P15 vs. P21 | .9777 |
4. DISCUSSION
The present study determined specific temporal and spatial patterns of MET expression—a protein that is present transiently during development and modulates the timing of cerebral cortical synapse development and maturation by cortical PNs. Here, we examined during postnatal development two architecturally distinct cortical regions, agranular, association mPFC, and granular primary sensory V1. The analyses revealed discrete temporal, laminar, and PN subclass‐specific patterns in mPFC and V1. First, in mPFC, MET is expressed in layers 5 and 6, with very limited expression in layer 2/3 neurons. In contrast, in V1, similar to findings reported in S1 (Kast et al., 2019), MET is enriched in layers 2/3, expressed in different PN subclasses in layers 5 and 6, and nearly absent from layer 4 neurons. Second, in mPFC, the percentage of MET+ neurons is stable across the first 5 postnatal weeks in layer 5, but declines after the first week in layer 6. In V1, MET expression is stable in layers 2/3 between P7 and P21, declines modestly in the first postnatal week in layer 5, and declines by P21 in layer 6. The data reveal differences in the temporal regulation of MET between the two cortical areas, and at a more discrete level, layer‐specific temporal regulation within a given cortical area. Finally, there is an enrichment of MET expression in ITPNs in V1, similar to that reported previously in S1 (Kast et al., 2019). Both primary sensory neocortical areas contrast with mPFC, in which MET is expressed primarily in SCPNs, including CTPNs. Given the role of MET signaling as a modulator of synapse development and maturation, the data indicate that the receptor operates within discrete circuits for each cortical region.
Previous studies using Western blot analyses indicated that MET protein levels in mouse cortex peak in the second postnatal week (Eagleson et al., 2016; Judson et al., 2009). These analyses included whole cortex and as such do not reflect potential region or layer‐specific differences in expression across time. Further, the detection of the MET protein is a combination of the receptor synthesized locally by PNs in the cerebral cortex as well as the receptor expressed in projections from different sources (e.g., hippocampus), in which MET protein is trafficked down the axon to presynaptic terminals. Finally, Western blot analyses do not discriminate between a general reduction in quantity of protein expressed across all MET‐expressing neurons and a reduction in the number of MET‐expressing neurons over time. As such, the current analyses add key complementary spatial information, demonstrating region‐ and layer‐specific trajectories of MET+ neurons in the cortex during postnatal development. Given that downregulation of MET acts as a modulator for the timing of synapse maturation and stabilization, the present findings pose interesting questions. Future studies will need to address whether sustained MET expression in layer 5 mPFC and layers 2/3 V1 are indicative of a more prolonged maturation process for the synapses of these PNs compared to those in which MET is turned off earlier in postnatal development. It also will be important to identify comparable proteins that regulate synapse maturation in PNs that do not express MET during development. The demonstration of substantial differences in spatial, specifically interlaminar and between cortical regions, and temporal expression of MET suggests that there are distinct molecular regulatory mechanisms upstream of Met.
Among the most striking findings of the current study is the difference in MET‐expressing cortical PN subclasses between mPFC and V1. This may reflect differences in function and circuit organization between the two regions. mPFC, an agranular association cortex involved in social communication and executive functions, can be considered a hub that communicates with many other cortical and subcortical regions, issuing top‐down control of behaviors and learning (Q. Anastasiades & Carter, 2021; Reinert et al., 2021; Zarr & Brown, 2016; Zhang et al., 2021). Neurons in infragranular, but not supragranular, layers of mPFC have been implicated in cognitive flexibility (Nakayama et al., 2018). SCPNs in mPFC are involved in active behavioral states (Warden et al., 2012), decision‐making behavior for goals that involve both reward and punishment (C. K. Kim et al., 2017), and social‐spatial learning (Murugan et al., 2017). Additionally, there are abnormalities specific to SCPNs in animal models of autism, and these neurons affect social behavior (Brumback et al., 2018). In contrast, V1 is a granular sensory cortex, with a major input from the lateral geniculate nucleus (LGN) in the thalamus. The LGN relays visual information directly from the retina to layer 4 of V1, in which visual information, such as static versus moving objects, orientation, and pattern recognition, are processed and further relayed to other brain regions (Glickfeld et al., 2013; Mazade & Alonso, 2017; Resulaj, 2021). Projections from V1 to subcortical structures are involved in oculomotor reflex‐driven functions, such as optokinetic nystagmus (Liu et al., 2016), and visually‐evoked innate behaviors (Liang et al., 2015). Projections from V1 to cortical structures are involved in relaying information for further visual processing, such as motion direction discrimination (Marques et al., 2018), contour detection and discrimination (van Kerkoerle et a1., 2018), and spatial attention (Tiesinga & Buia, 2009). ITPNs in V1 have also been shown to process coincidental multisensory events (Knöpfel et al., 2019) and enhance neural selectivity to learn visual discrimination tasks (Poort et al., 2015). As aforementioned, MET is predominantly expressed in infragranular layers of mPFC and, more specifically, in SCPNs, including CTPNs, rather than ITPNs, reflecting a role in mPFC circuits that connect outside of the cortex. In V1, MET expression in supragranular ITPNs and in predominantly infragranular ITPNs, rather than in SCPNs or CTPNs, indicate that in primary sensory cortices, MET functions mainly in neurons that connect to other cortical regions. Based on the known roles of SCPNs in mPFC and ITPNs in V1, this suggests that MET expression is enriched in developing circuits that are involved in higher‐order functions. We note that in both mPFC and V1, there was a minor population of layer 5 neurons that remained dual‐labeled with CTIP2 and SATB2, so their PN subclass remained ambiguous; however, this did not change the overall conclusions. Future detailed tracing studies will determine specific connectivity maps of MET+ mPFC and V1 neurons.
Maturation and refinement of V1 are affected by and result in numerous developmental processes that occur postnatally in mice: retinotopy occurs by P8 (Cang et al., 2005), eye‐opening around P14, orientation selection around the period of eye‐opening, independent of visual stimulus (Rochefort et al., 2011), and the critical period for ocular dominance between P19 and P32 (Gordon & Stryker, 1996). With the abundant expression of MET in supragranular layers 2/3 remaining unchanged from P7 to P21, it can first be inferred that eye‐opening, and subsequent visual information reaching V1, does not influence the percentage of neurons expressing MET. Further, MET modulates the timing of the critical period for ocular dominance (K. Chen et al., 2021). Thus, the sustained MET expression until P21 in layers 2/3 may suggest that MET+ neurons in these layers modulate the timing of the critical period, rather than the infragranular MET+ neurons, in which downregulation occurs earlier postnatally. This is supported by a recent report demonstrating visual experience‐driven maturation in layers 2/3, but not layer 5 or 6 (Cheng et al., 2022). Conversely, the temporal regulation of infragranular MET expression may play a role in developmental processes that occur prior to the critical period. Nevertheless, follow‐up studies are needed to test the layer‐specific contributions of MET in V1 developmental processes.
It is notable that MET is predominantly expressed in a particular PN subclass in each region, but not in all neurons of that PN subclass. Expression of MET within only a subpopulation of the PN subclass may allow for asynchrony of synapse maturation, even within neurons projecting to the same brain region. While differences in timing of maturation of circuits subserving different functions has been reported (Hensch, 2004; Huttenlocher & Dabholkar, 1997; Moyer & Zuo, 2018), at a finer level, differences in timing of maturation of synapses within circuits involved in the same function may also be advantageous. For example, critical period plasticity represents a time when circuits have maximum opportunity to undergo experience‐based modification, but this is at the expense of the brain being in a more vulnerable state (Hensch, 2003; Nelson & Gabard‐Durnam, 2020). Closure of critical periods allow the brain to function in a more stable, resilient state. We speculate that if the timing of closure of a critical period of plasticity was completely uniform for all circuits that underlie a specific function, this could lead to greater vulnerability, for which an insult may affect each circuit equally. Mechanisms that allow for differential timing of maturation of circuits for a given function would provide opportunities for select circuits to stabilize, while others remain in a more plastic state, leading to further optimization. Therefore, the expression of MET within subpopulations of PN subclasses may contribute to differential timing of maturation.
While this study focused on one synapse‐enriched receptor, it underscores the dynamic process of development, during which many molecular and structural changes are occurring. The temporal features of our study emphasize the need to model neurodevelopmental disorders that include developmental phenotypes to capture biological mechanisms associated with transient processes of maturation that may be disrupted during development. Even developmental studies using single‐cell sequencing may not capture the spatial and temporal dynamics of genes and proteins of interest due to limitations in spatial resolution and the practical limitations of sampling a large number of developmental timepoints. Lastly, while the cortex, as a whole, has properties and functions that are unique compared to subcortical brain structures, cortical regions are heterogeneous, such that findings in one region do not necessarily translate to other regions. Continuing to compare and contrast the development of different cortical regions will provide greater understanding of the emergence and maturation of specific functions and their underlying molecular contributions.
AUTHOR CONTRIBUTIONS
Alexandra L. Lanjewar, Kathie L. Eagleson, and Pat Levitt conceived and designed the research. Alexandra L. Lanjewar, Sonum Jagetia, and Zuhayr M. Khan performed the experiments. Alexandra L. Lanjewar, Kathie L. Eagleson, and Pat Levitt interpreted the results of the experiments. Alexandra L. Lanjewar, Sonum Jagetia, and Zuhayr M. Khan prepared figures. Alexandra L. Lanjewar drafted the manuscript. All authors edited and revised the manuscript. All authors approved of the final version of the manuscript.
CONFLICT OF INTERESTS
None declared.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/cne.25418.
ACKNOWLEDGMENTS
This work was supported by National Institute of Mental Health R01 MH067842. We thank Dr. James Morgan for providing anti‐PCP4 antibodies. We thank Amanda Whipple for assistance with animal husbandry and Dr. Esteban Fernandez of the Children's Hospital Los Angeles Cellular Imaging Core for assistance with image acquisition and analysis. We thank Dr. Hsiao‐Huei Wu for assistance in conceptualizing and creating the graphical summary.
Lanjewar, A. L. , Jagetia, S. , Khan, Z. M. , Eagleson, K. L. , & Levitt, P. (2023). Subclass‐specific expression patterns of MET receptor tyrosine kinase during development in medial prefrontal and visual cortices. Journal of Comparative Neurology, 531, 132–148. 10.1002/cne.25418
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Alcamo, E. A. , Chirivella, L. , Dautzenberg, M. , Dobreva, G. , Fariñas, I. , Grosschedl, R. , & McConnell, S. K. (2008). Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron, 57(3), 364–377. 10.1016/j.neuron.2007.12.012 [DOI] [PubMed] [Google Scholar]
- Anastasiades, P. G. , & Carter, A. G. (2021). Circuit organization of the rodent medial prefrontal cortex. Trends in Neurosciences, 44(7), 550–563. 10.1016/j.tins.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta, P. , Molyneaux, B. J. , Chen, J. , Inoue, J. , Kominami, R. , & Macklis, J. D. (2005). Neuronal subtype‐specific genes that control corticospinal motor neuron development in vivo. Neuron, 45(2), 207–221. 10.1016/j.neuron.2004.12.036 [DOI] [PubMed] [Google Scholar]
- Baranek, C. , Dittrich, M. , Parthasarathy, S. , Bonnon, C. G. , Britanova, O. , Lanshakov, D. , Boukhtouche, F. , Sommer, J. E. , Colmenares, C. , Tarabykin, V. , & Atanasoski, S. (2012). Protooncogene Ski cooperates with the chromatin‐remodeling factor Satb2 in specifying callosal neurons. Proceedings of the National Academy of Sciences of the United States of America, 109(9), 3546–3551. 10.1073/pnas.1108718109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri, A. , Sandoval‐Espinosa, C. , Otero‐Garcia, M. , Oh, I. , Yin, R. , Eze, U. C. , Nowakowski, T. J. , & Kriegstein, A. R. (2021). An atlas of cortical arealization identifies dynamic molecular signatures. Nature, 598(7879), 200–204. 10.1038/s41586-021-03910-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova, O. , de Juan Romero, C. , Cheung, A. , Kwan, K. Y. , Schwark, M. , Gyorgy, A. , Vogel, T. , Akopov, S. , Mitkovski, M. , Agoston, D. , Sestan, N. , Molnár, Z. , & Tarabykin, V. (2008). Satb2 is a postmitotic determinant for upper‐layer neuron specification in the neocortex. Neuron, 57(3), 378–392. 10.1016/j.neuron.2007.12.028 [DOI] [PubMed] [Google Scholar]
- Brumback, A. C. , Ellwood, I. T. , Kjaerby, C. , Iafrati, J. , Robinson, S. , Lee, A. T. , Patel, T. , Nagaraj, S. , Davatolhagh, F. , & Sohal, V. S. (2018). Identifying specific prefrontal neurons that contribute to autism‐associated abnormalities in physiology and social behavior. Molecular Psychiatry, 23(10), 2078–2089. 10.1038/mp.2017.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, R. M. , Hahn, T. T. , Wallace, D. J. , de Kock, C. P. , & Sakmann, B. (2009). Sensory experience alters specific branches of individual corticocortical axons during development. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 29(10), 3172–3181. 10.1523/JNEUROSCI.5911-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang, J. , Rentería, R. C. , Kaneko, M. , Liu, X. , Copenhagen, D. R. , & Stryker, M. P. (2005). Development of precise maps in visual cortex requires patterned spontaneous activity in the retina. Neuron, 48(5), 797–809. 10.1016/j.neuron.2005.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. , Wang, S. S. , Hattox, A. M. , Rayburn, H. , Nelson, S. B. , & McConnell, S. K. (2008). The Fezf2‐Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America, 105(32), 11382–11387. 10.1073/pnas.0804918105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. H. , Panizzon, M. S. , Eyler, L. T. , Jernigan, T. L. , Thompson, W. , Fennema‐Notestine, C. , Jak, A. J. , Neale, M. C. , Franz, C. E. , Hamza, S. , Lyons, M. J. , Grant, M. D. , Fischl, B. , Seidman, L. J. , Tsuang, M. T. , Kremen, W. S. , & Dale, A. M. (2011). Genetic influences on cortical regionalization in the human brain. Neuron, 72(4), 537–544. 10.1016/j.neuron.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Ma, X. , Nehme, A. , Wei, J. , Cui, Y. , Cui, Y. , Yao, D. , Wu, J. , Anderson, T. , Ferguson, D. , Levitt, P. , & Qiu, S. (2021). Time‐delimited signaling of MET receptor tyrosine kinase regulates cortical circuit development and critical period plasticity. Molecular Psychiatry, 26(8), 3723–3736. 10.1038/s41380-019-0635-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S. , Butrus, S. , Tan, L. , Xu, R. , Sagireddy, S. , Trachtenberg, J. T. , Shekhar, K. , & Zipursky, S. L. (2022). Vision‐dependent specification of cell types and function in the developing cortex. Cell, 185(2), 311–327.e24. 10.1016/j.cell.2021.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzker, J. L. , & Callaway, E. M. (1998). The development of local, layer‐specific visual cortical axons in the absence of extrinsic influences and intrinsic activity. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 18(11), 4145–4154. 10.1523/JNEUROSCI.18-11-04145.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, M. A. , Segura‐Anaya, E. , Alva‐Medina, J. , & Aranda‐Anzaldo, A. (2010). NeuN/Fox‐3 is an intrinsic component of the neuronal nuclear matrix. FEBS Letters, 584(13), 2767–2771. 10.1016/j.febslet.2010.04.073 [DOI] [PubMed] [Google Scholar]
- Eagleson, K. L. , Campbell, D. B. , Thompson, B. L. , Bergman, M. Y. , & Levitt, P. (2011). The autism risk genes MET and PLAUR differentially impact cortical development. Autism Research: Official Journal of the International Society for Autism Research, 4(1), 68–83. 10.1002/aur.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson, K. L. , Lane, C. J. , McFadyen‐Ketchum, L. , Solak, S. , Wu, H. H. , & Levitt, P. (2016). Distinct intracellular signaling mediates C‐MET regulation of dendritic growth and synaptogenesis. Developmental Neurobiology, 76(10), 1160–1181. 10.1002/dneu.22382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen, C. R. , Economo, M. N. , & Chandrashekar, J. (2018). Long distance projections of cortical pyramidal neurons. Journal of Neuroscience Research, 96(9), 1467–1475. 10.1002/jnr.23978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickfeld, L. L. , Histed, M. H. , & Maunsell, J. H. (2013). Mouse primary visual cortex is used to detect both orientation and contrast changes. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(50), 19416–19422. 10.1523/JNEUROSCI.3560-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, J. A. , & Stryker, M. P. (1996). Experience‐dependent plasticity of binocular responses in the primary visual cortex of the mouse. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 16(10), 3274–3286. 10.1523/JNEUROSCI.16-10-03274.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb, K. , Magrinelli, E. , Nicolas, C. S. , Lukianets, N. , Frangeul, L. , Pietri, M. , Sun, T. , Sandoz, G. , Grammont, F. , Jabaudon, D. , Studer, M. , & Alfano, C. (2016). Area‐specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. eLife, 5, e09531. 10.7554/eLife.09531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K. , & Shepherd, G. (2015). The neocortical circuit: themes and variations. Nature Neuroscience, 18, 170–181. 10.1038/nn.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka, Y. , Namikawa, T. , Yamauchi, K. , & Kawaguchi, Y. (2016). Cortical divergent projections in mice originate from two sequentially generated, distinct populations of excitatory cortical neurons with different initial axonal outgrowth characteristics. Cerebral Cortex (New York, N.Y.: 1991), 26(5), 2257–2270. 10.1093/cercor/bhv077 [DOI] [PubMed] [Google Scholar]
- Hensch, T. K. (2003). Controlling the critical period. Neuroscience Research, 47(1), 17–22. 10.1016/s0168-0102(03)00164-0 [DOI] [PubMed] [Google Scholar]
- Hensch, T. K. (2004). Critical period regulation. Annual Review of Neuroscience, 27, 549–579. 10.1146/annurev.neuro.27.070203.144327 [DOI] [PubMed] [Google Scholar]
- Heun‐Johnson, H. , & Levitt, P. (2017). Differential impact of Met receptor gene interaction with early‐life stress on neuronal morphology and behavior in mice. Neurobiology of Stress, 8, 10–20. 10.1016/j.ynstr.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher, P. R. , & Dabholkar, A. S. (1997). Regional differences in synaptogenesis in human cerebral cortex. The Journal of Comparative Neurology, 387(2), 167–178. [DOI] [PubMed] [Google Scholar]
- Judson, M. C. , Bergman, M. Y. , Campbell, D. B. , Eagleson, K. L. , & Levitt, P. (2009). Dynamic gene and protein expression patterns of the autism‐associated met receptor tyrosine kinase in the developing mouse forebrain. The Journal of Comparative Neurology, 513(5), 511–531. 10.1002/cne.21969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitakahara, A. , Wu, H. H. , & Levitt, P. (2017). Distinct projection targets define subpopulations of mouse brainstem vagal neurons that express the autism‐associated MET receptor tyrosine kinase. The Journal of Comparative Neurology, 525(18), 3787–3808. 10.1002/cne.24294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassai, H. , Terashima, T. , Fukaya, M. , Nakao, K. , Sakahara, M. , Watanabe, M. , & Aiba, A. (2008). Rac1 in cortical projection neurons is selectively required for midline crossing of commissural axonal formation. The European Journal of Neuroscience, 28(2), 257–267. 10.1111/j.1460-9568.2008.06343.x [DOI] [PubMed] [Google Scholar]
- Kast, R. J. , & Levitt, P. (2019). Precision in the development of neocortical architecture: From progenitors to cortical networks. Progress in Neurobiology, 175, 77–95. 10.1016/j.pneurobio.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast, R. J. , Wu, H. H. , & Levitt, P. (2019). Developmental connectivity and molecular phenotypes of unique cortical projection neurons that express a synapse‐associated receptor tyrosine Kinase. Cerebral Cortex (New York, N.Y.: 1991), 29(1), 189–201. 10.1093/cercor/bhx318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast, R. J. , Wu, H. H. , Williams, P. , Gaspar, P. , & Levitt, P. (2017). Specific connectivity and unique molecular identity of MET receptor tyrosine kinase expressing serotonergic neurons in the caudal dorsal raphe nuclei. ACS Chemical Neuroscience, 8(5), 1053–1064. 10.1021/acschemneuro.7b00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. K. , Ye, L. , Jennings, J. H. , Pichamoorthy, N. , Tang, D. D. , Yoo, A. W. , Ramakrishnan, C. , & Deisseroth, K. (2017). Molecular and circuit‐dynamical identification of top‐down neural mechanisms for restraint of reward seeking. Cell, 170(5), 1013–1027.e14. 10.1016/j.cell.2017.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. J. , Zhang, Z. , Huang, L. , Ito‐Cole, T. , Jacobs, M. W. , Juavinett, A. L. , Senturk, G. , Hu, M. , Ku, M. , Ecker, J. R. , & Callaway, E. M. (2020). Extraction of distinct neuronal cell types from within a genetically continuous population. Neuron, 107(2), 274–282.e6. 10.1016/j.neuron.2020.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöpfel, T. , Sweeney, Y. , Radulescu, C. I. , Zabouri, N. , Doostdar, N. , Clopath, C. , & Barnes, S. J. (2019). Audio‐visual experience strengthens multisensory assemblies in adult mouse visual cortex. Nature Communications, 10(1), 5684. 10.1038/s41467-019-13607-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone, D. P. , Heavner, W. E. , Ferenczi, E. A. , Dobreva, G. , Huguenard, J. R. , Grosschedl, R. , & McConnell, S. K. (2015). Satb2 regulates the differentiation of both callosal and subcerebral projection neurons in the developing cerebral cortex. Cerebral Cortex (New York, N.Y.: 1991), 25(10), 3406–3419. 10.1093/cercor/bhu156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, P. , Barbe, M. F. , & Eagleson, K. L. (1997). Patterning and specification of the cerebral cortex. Annual Review of Neuroscience, 20, 1–24. 10.1146/annurev.neuro.20.1.1 [DOI] [PubMed] [Google Scholar]
- Lickiss, T. , Cheung, A. F. , Hutchinson, C. E. , Taylor, J. S. , & Molnár, Z. (2012). Examining the relationship between early axon growth and transcription factor expression in the developing cerebral cortex. Journal of Anatomy, 220(3), 201–211. 10.1111/j.1469-7580.2011.01466.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F. , Xiong, X. R. , Zingg, B. , Ji, X. Y. , Zhang, L. I. , & Tao, H. W. (2015). Sensory cortical control of a visually induced arrest behavior via corticotectal projections. Neuron, 86(3), 755–767. 10.1016/j.neuron.2015.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. H. , Huberman, A. D. , & Scanziani, M. (2016). Cortico‐fugal output from visual cortex promotes plasticity of innate motor behaviour. Nature, 538(7625), 383–387. 10.1038/nature19818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C. , Keown, C. L. , Kurihara, L. , Zhou, J. , He, Y. , Li, J. , Castanon, R. , Lucero, J. , Nery, J. R. , Sandoval, J. P. , Bui, B. , Sejnowski, T. J. , Harkins, T. T. , Mukamel, E. A. , Behrens, M. M. , & Ecker, J. R. (2017). Single‐cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science (New York, N.Y.), 357(6351), 600–604. 10.1126/science.aan3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, R. , Jeong, S. J. , Jin, Z. , Strokes, N. , Li, S. , & Piao, X. (2011). G protein‐coupled receptor 56 and collagen III, a receptor‐ligand pair, regulates cortical development and lamination. Proceedings of the National Academy of Sciences of the United States of America, 108(31), 12925–12930. 10.1073/pnas.1104821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Wei, J. , Cui, Y. , Xia, B. , Zhang, L. , Nehme, A. , Zuo, Y. , Ferguson, D. , Levitt, P. , & Qiu, S. (2022). Disrupted timing of MET signaling derails the developmental maturation of cortical circuits and leads to altered behavior in mice. Cerebral cortex (New York, N.Y.: 1991), 32(8), 1769–1786. 10.1093/cercor/bhab323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, T. , Summers, M. T. , Fioreze, G. , Fridman, M. , Dias, R. F. , Feller, M. B. , & Petreanu, L. (2018). A role for mouse primary visual cortex in motion perception. Current Biology: CB, 28(11), 1703–1713.e6. 10.1016/j.cub.2018.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazade, R. , & Alonso, J. M. (2017). Thalamocortical processing in vision. Visual Neuroscience, 34, E007. 10.1017/S0952523817000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinard‐Chenu, A. , Fluss, J. , Laurent, S. , Laurent, M. , Guipponi, M. , & Dayer, A. G. (2020). MCF2 is linked to a complex perisylvian syndrome and affects cortical lamination. Annals of Clinical and Translational Neurology, 7(1), 121–125. 10.1002/acn3.50949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux, B. J. , Arlotta, P. , Menezes, J. R. , & Macklis, J. D. (2007). Neuronal subtype specification in the cerebral cortex. Nature Reviews. Neuroscience, 8(6), 427–437. 10.1038/nrn2151 [DOI] [PubMed] [Google Scholar]
- Moyer, C. E. , & Zuo, Y. (2018). Cortical dendritic spine development and plasticity: Insights from in vivo imaging. Current Opinion in Neurobiology, 53, 76–82. 10.1016/j.conb.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Murugan, M. , Jang, H. J. , Park, M. , Miller, E. M. , Cox, J. , Taliaferro, J. P. , Parker, N. F. , Bhave, V. , Hur, H. , Liang, Y. , Nectow, A. R. , Pillow, J. W. , & Witten, I. B. (2017). Combined social and spatial coding in a descending projection from the prefrontal cortex. Cell, 171(7), 1663–1677.e16. 10.1016/j.cell.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, H. , Ibañez‐Tallon, I. , & Heintz, N. (2018). Cell‐Type‐specific contributions of medial prefrontal neurons to flexible behaviors. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 38(19), 4490–4504. 10.1523/JNEUROSCI.3537-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, 3rd, C. A. , & Gabard‐Durnam, L. J. (2020). Early adversity and critical periods: Neurodevelopmental consequences of violating the expectable environment. Trends in Neurosciences, 43(3), 133–143. 10.1016/j.tins.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet, C. C. (1991). DARPP‐32, a dopamine and cyclic AMP‐regulated phosphoprotein, is present in corticothalamic neurons of the rat cingulate cortex. Brain Research, 562(1), 85–92. 10.1016/0006-8993(91)91190-c [DOI] [PubMed] [Google Scholar]
- Paolino, A. , Fenlon, L. R. , Kozulin, P. , Haines, E. , Lim, J. , Richards, L. J. , & Suárez, R. (2020). Differential timing of a conserved transcriptional network underlies divergent cortical projection routes across mammalian brain evolution. Proceedings of the National Academy of Sciences of the United States of America, 117(19), 10554–10564. 10.1073/pnas.1922422117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos, G. , & Franklin, K. B. (2019). Paxinos and Franklin's the mouse brain in stereotaxic coordinates. Academic Press. [Google Scholar]
- Poort, J. , Khan, A. G. , Pachitariu, M. , Nemri, A. , Orsolic, I. , Krupic, J. , Bauza, M. , Sahani, M. , Keller, G. B. , Mrsic‐Flogel, T. D. , & Hofer, S. B. (2015). Learning enhances sensory and multiple non‐sensory representations in primary visual cortex. Neuron, 86(6), 1478–1490. 10.1016/j.neuron.2015.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, X. , Su, Y. , Adam, C. D. , Deutschmann, A. U. , Pather, S. R. , Goldberg, E. M. , Su, K. , Li, S. , Lu, L. , Jacob, F. , Nguyen, P. , Huh, S. , Hoke, A. , Swinford‐Jackson, S. E. , Wen, Z. , Gu, X. , Pierce, R. C. , Wu, H. , Briand, L. A. , … Ming, G. L. (2020). Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell, 26(5), 766–781.e9. 10.1016/j.stem.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resulaj, A. (2021). Projections of the mouse primary visual cortex. Frontiers in Neural Circuits, 15, 751331. 10.3389/fncir.2021.751331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert, S. , Hübener, M. , Bonhoeffer, T. , & Goltstein, P. M. (2021). Mouse prefrontal cortex represents learned rules for categorization. Nature, 593(7859), 411–417. 10.1038/s41586-021-03452-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochefort, N. L. , Narushima, M. , Grienberger, C. , Marandi, N. , Hill, D. N. , & Konnerth, A. (2011). Development of direction selectivity in mouse cortical neurons. Neuron, 71(3), 425–432. 10.1016/j.neuron.2011.06.013 [DOI] [PubMed] [Google Scholar]
- Spellman, T. , Svei, M. , Kaminsky, J. , Manzano‐Nieves, G. , & Liston, C. (2021). Prefrontal deep projection neurons enable cognitive flexibility via persistent feedback monitoring. Cell, 184(10), 2750–2766.e17. 10.1016/j.cell.2021.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, B. L. , & Levitt, P. (2015). Complete or partial reduction of the Met receptor tyrosine kinase in distinct circuits differentially impacts mouse behavior. Journal of Neurodevelopmental Disorders, 7, 35. 10.1186/s11689-015-9131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiesinga, P. H. , & Buia, C. I. (2009). Spatial attention in area V4 is mediated by circuits in primary visual cortex. Neural Networks: The Official Journal of the International Neural Network Society, 22(8), 1039–1054. 10.1016/j.neunet.2009.07.010 [DOI] [PubMed] [Google Scholar]
- Trevino, A. E. , Müller, F. , Andersen, J. , Sundaram, L. , Kathiria, A. , Shcherbina, A. , Farh, K. , Chang, H. Y. , Pașca, A. M. , Kundaje, A. , Pașca, S. P. , & Greenleaf, W. J. (2021). Chromatin and gene‐regulatory dynamics of the developing human cerebral cortex at single‐cell resolution. Cell, 184(19), 5053–5069.e23. 10.1016/j.cell.2021.07.039 [DOI] [PubMed] [Google Scholar]
- Tsyporin, J. , Tastad, D. , Ma, X. , Nehme, A. , Finn, T. , Huebner, L. , Liu, G. , Gallardo, D. , Makhamreh, A. , Roberts, J. M. , Katzman, S. , Sestan, N. , McConnell, S. K. , Yang, Z. , Qiu, S. , & Chen, B. (2021). Transcriptional repression by FEZF2 restricts alternative identities of cortical projection neurons. Cell Reports, 35(12), 109269. 10.1016/j.celrep.2021.109269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkoerle, T. , Marik, S. A. , Meyer Zum Alten Borgloh, S. , & Gilbert, C. D. (2018). Axonal plasticity associated with perceptual learning in adult macaque primary visual cortex. Proceedings of the National Academy of Sciences of the United States of America, 115(41), 10464–10469. 10.1073/pnas.1812932115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden, M. R. , Selimbeyoglu, A. , Mirzabekov, J. J. , Lo, M. , Thompson, K. R. , Kim, S. Y. , Adhikari, A. , Tye, K. M. , Frank, L. M. , & Deisseroth, K. (2012). A prefrontal cortex‐brainstem neuronal projection that controls response to behavioural challenge. Nature, 492(7429), 428–432. 10.1038/nature11617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe, A. , Hirokawa, J. , Ichinohe, N. , Ohsawa, S. , Kaneko, T. , Rockland, K. S. , & Yamamori, T. (2012). Area‐specific substratification of deep layer neurons in the rat cortex. The Journal of Comparative Neurology, 520(16), 3553–3573. 10.1002/cne.23160 [DOI] [PubMed] [Google Scholar]
- Woodworth, M. B. , Greig, L. C. , Kriegstein, A. R. , & Macklis, J. D. (2012). SnapShot: Cortical development. Cell, 151(4), 918–918.e1. 10.1016/j.cell.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, B. , Wei, J. , Ma, X. , Nehme, A. , Liong, K. , Cui, Y. , Chen, C. , Gallitano, A. , Ferguson, D. , & Qiu, S. (2021). Conditional knockout of MET receptor tyrosine kinase in cortical excitatory neurons leads to enhanced learning and memory in young adult mice but early cognitive decline in older adult mice. Neurobiology of Learning and Memory, 179, 107397. 10.1016/j.nlm.2021.107397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarr, N. , & Brown, J. W. (2016). Hierarchical error representation in medial prefrontal cortex. NeuroImage, 124(Pt A), 238–247. 10.1016/j.neuroimage.2015.08.063 [DOI] [PubMed] [Google Scholar]
- Zeisel, A. , Muñoz‐Manchado, A. B. , Codeluppi, S. , Lönnerberg, P. , La Manno, G. , Juréus, A. , Marques, S. , Munguba, H. , He, L. , Betsholtz, C. , Rolny, C. , Castelo‐Branco, G. , Hjerling‐Leffler, J. , & Linnarsson, S. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single‐cell RNA‐seq. Science (New York, N.Y.), 347(6226), 1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Weber, M. A. , & Narayanan, N. S. (2021). Medial prefrontal cortex and the temporal control of action. International Review of Neurobiology, 158, 421–441. 10.1016/bs.irn.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Zhou, J. , Tan, P. , Pang, Y. , Rivkin, A. C. , Kirchgessner, M. A. , Williams, E. , Lee, C. T. , Liu, H. , Franklin, A. D. , Miyazaki, P. A. , Bartlett, A. , Aldridge, A. I. , Vu, M. , Boggeman, L. , Fitzpatrick, C. , Nery, J. R. , Castanon, R. G. , Rashid, M. , … Callaway, E. M. (2021). Epigenomic diversity of cortical projection neurons in the mouse brain. Nature, 598(7879), 167–173. 10.1038/s41586-021-03223-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S. , Zhang, S. , Fan, X. , Wu, Q. , Yan, L. , Dong, J. , Zhang, H. , Li, L. , Sun, L. , Pan, N. , Xu, X. , Tang, F. , Zhang, J. , Qiao, J. , & Wang, X. (2018). A single‐cell RNA‐seq survey of the developmental landscape of the human prefrontal cortex. Nature, 555(7697), 524–528. 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- Ziai, M. R. , Sangameswaran, L. , Hempstead, J. L. , Danho, W. , & Morgan, J. I. (1988). An immunochemical analysis of the distribution of a brain‐specific polypeptide, PEP‐19. Journal of Neurochemistry, 51(6), 1771–1776. 10.1111/j.1471-4159.1988.tb01158.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


