SUMMARY
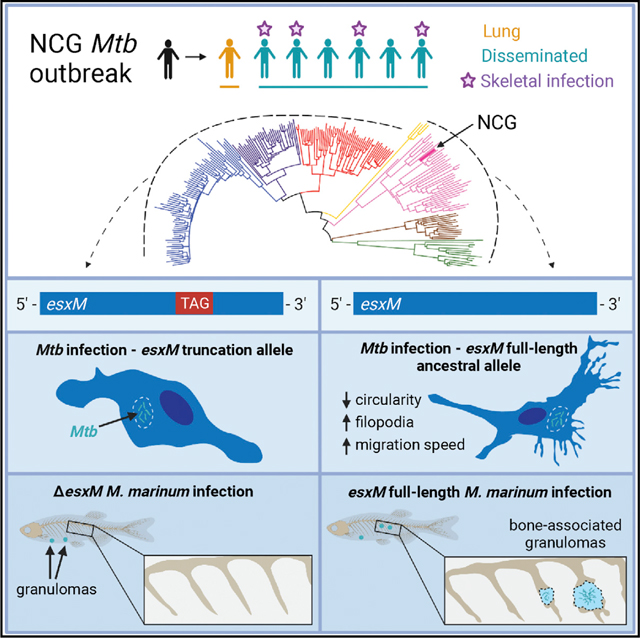
The human pathogen Mycobacterium tuberculosis typically causes lung disease but can also disseminate to other tissues. We identified a M. tuberculosis (Mtb) outbreak presenting with unusually high rates of extrapulmonary dissemination and bone disease. We found that the causal strain carried an ancestral full-length version of the type VII-secreted effector EsxM rather than the truncated version present in other modern Mtb lineages. The ancestral EsxM variant exacerbated dissemination through enhancement of macrophage motility, increased egress of macrophages from established granulomas, and alterations in macrophage actin dynamics. Reconstitution of the ancestral version of EsxM in an attenuated modern strain of Mtb altered the migratory mode of infected macrophages, enhancing their motility. In a zebrafish model, full-length EsxM promoted bone disease. The presence of a derived nonsense variant in EsxM throughout the major Mtb lineages 2, 3, and 4 is consistent with a role for EsxM in regulating the extent of dissemination.
Graphical Abstract
In brief
Tuberculosis outbreak with a unique clinical manifestation unveils previously unappreciated insights into pathogen evolution and host-pathogen interactions.
INTRODUCTION
Mycobacterium tuberculosis (Mtb), the etiologic agent of the disease tuberculosis (TB), is a pathogen of critical public health importance.1 TB is predominantly a pulmonary disease, but 15%–20% of cases present with extrapulmonary manifestations.2 Extrapulmonary infections can be difficult to diagnose and treat. Skeletal TB, in turn, is a relatively uncommon manifestation of extrapulmonary TB, affecting only 148 of 7,174 (~2%) reported TB cases in the US in 2020.2 Although skeletal TB, or Pott’s Disease, has long been recognized, with characteristic TB-induced spinal deformities described in humans from as far back as ancient Egypt,3 the specific bacterial factors that influence dissemination, tissue tropism, and bone disease are not well characterized.
Upon its establishment as a distinct species, Mtb evolved largely clonally.4–7 Although horizontal gene transfer likely contributed to its initial evolution as a human pathogen,8 Mtb thereafter seems to have undergone relatively few horizontal gene transfer events, with some structural variants arising through genomic deletions.7,9,10 Mtb has classically been grouped into at least seven discrete lineages. So-called “modern” lineages, which include lineages 2, 3, and 4, (L2, L3, and L4) are broadly distributed throughout the world.9 Other lineages (L1, L5, L6, and L7) are more geographically constrained.11 L1 strains, although geographically limited, still account for many overall cases and have been previously reported to induce distinct inflammatory phenotypes and differentially modulate innate immune signaling in the human host.12–15
Pathogenic mycobacteria achieve dissemination through a remarkable ability to spread within their hosts. They hijack host macrophages as both a major replicative niche and for delivery to distal locations within and between tissues.16 Macrophage function and motility influence dissemination of mycobacterial disease through a number of mechanisms, including macrophage survival and cell death, and efflux from initial nidi to new sites.17–20 The dynamics of granuloma formation, dissolution, and resolution also influence the trajectory of infection and dissemination in zebrafish and macaque models.21–24
Macrophage and granuloma behavior is heavily influenced by dedicated bacterial effectors secreted through type VII secretion systems. The ESX-1 system, notably absent in the attenuated BCG vaccine strain,25–28 plays multiple roles in virulence, most prominently in the permeabilization of the phagosomal membrane in infected macrophages.29–34 Mutants defective for the small secreted effectors specific to the ESX-1 secretion system—EsxA and EsxB—display altered virulence.35 Similarly, the paralogous ESX-3 system regulates important pathogenhost interactions, including iron acquisition,36,37 and, through interactions of the small secreted effectors EsxG and EsxH with the ESCRT complex, host membrane trafficking, and damage response.38–40
ESX-5, the most recently evolved of the paralogous type VII secretion systems, is found only in the slow-growing pathogenic mycobacteria, including Mtb and Mycobacterium marinum.41,42 ESX-5 has been implicated in secretion of Mtb’s abundant PE and PPE family proteins43,44 and the CpnT toxin.45 However, a biological role of any of the small secreted effectors specific to ESX-5, including EsxM and EsxN, has not fully been examined.
Here, we describe an unusual outbreak of Mtb with high rates of extrapulmonary dissemination and bone disease. We uncover a functional variant in the ESX-5 secreted effector EsxM that precisely coincides with a transition from the ancestral allele present in Mtb lineages 1, 5, 6, and 7 to a derived truncation allele in lineages L2–L4. The ancestral version of EsxM present in the outbreak strain, and generally in L1 strains, leads to alterations in the modality of infected host macrophage migration and the rate of egress of infected macrophages from granulomas. Mtb generally requires airborne lung-to-lung transmission, so we therefore propose that limiting the degree of dissemination to tissues outside the lung may be advantageous with respect to the likelihood of transmission. We infer that the stop codon in EsxM was introduced in the most recent common ancestor of the L2–L4 Mtb strains, leading to decreased rates of dissemination and skeletal disease for strains carrying this variant.
RESULTS
A North Carolina outbreak with high rates of skeletal disease
We investigated a TB outbreak with extremely high rates of disseminated and skeletal disease (Figures 1A and 1B). The index case, a man originally from Vietnam, was diagnosed with pulmonary TB after over a year of symptoms, and a contact investigation was carried out.46 Seven secondary cases of active TB were identified (Figures 1A and 1B), and six of the seven (86%) presented with extrapulmonary disease. Remarkably, though the reported frequency of skeletal disease is 2% of all US cases,2 four of the extrapulmonary cases (57% of cases in the outbreak) had skeletal disease. The binomial probability of observing four or more bone cases among seven TB patients if the probability of each case having bone TB is equal to the population proportion is approximately 5×10−6. Two of the bone TB cases had a single site of disease in the spine, and the other two had diffuse bony disease involving the ribs, scapula, iliac crest, spine, and sternum (Figures 1C–1E). One of the patients with diffuse bone disease had concurrent pulmonary involvement but no other site, and the other had no site of disease outside the bones. All secondary case patients were US-born (one White, the rest Black), HIV-negative, genetically unrelated to the index case and each other, and seemingly otherwise immunocompetent. The strains isolated from each patient had identical genotypes by mycobacterial interspersed repetitive units-variable number tandem repeats (MIRU-VNTR) typing and spoligotyping, suggesting a single transmitting strain, which we refer to as NCG (Figure S1).
Figure 1. An outbreak in North Carolina with high rates of extrapulmonary and bone disease.
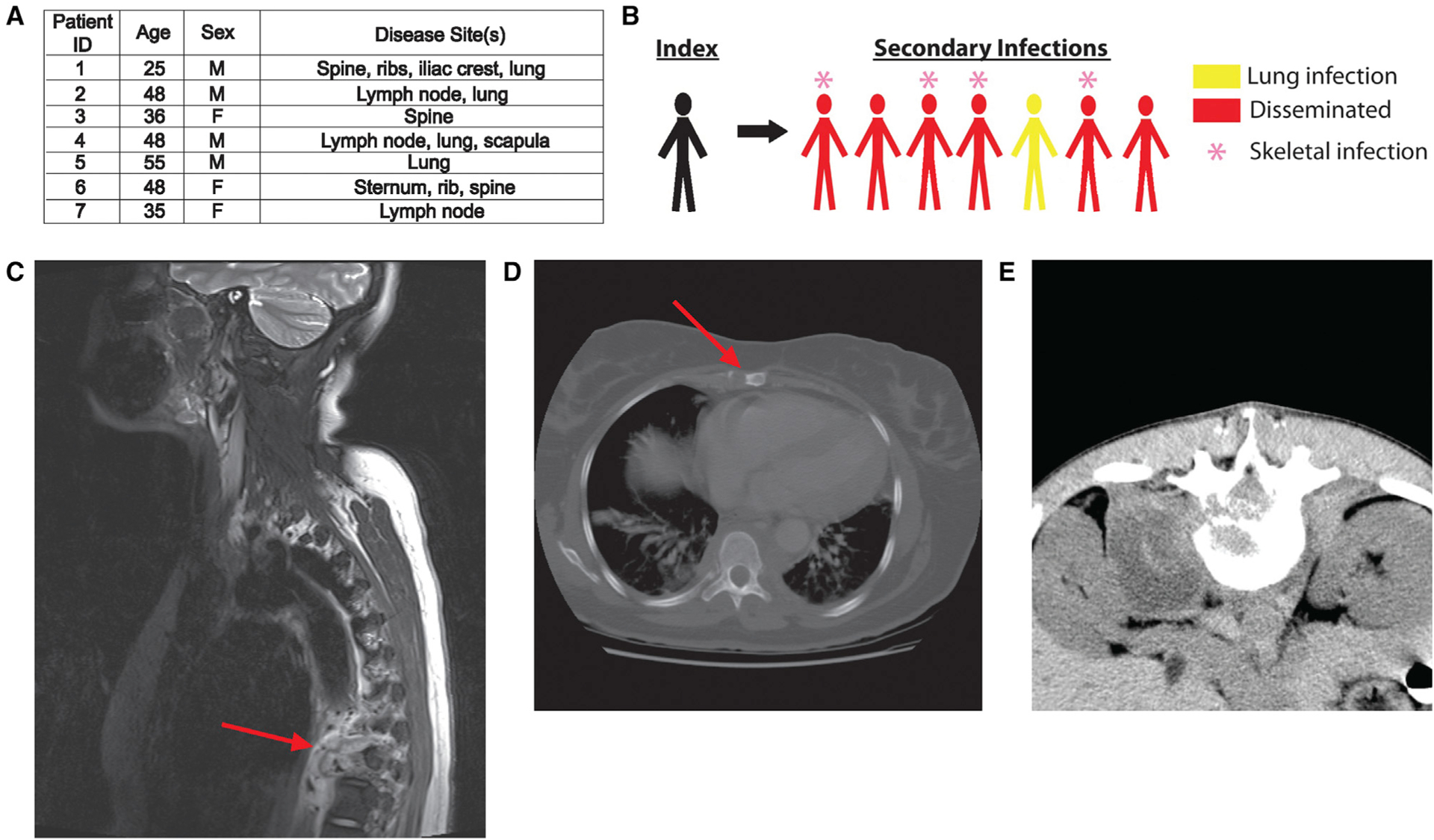
(A) Characterization of all secondary cases of the NCG outbreak strain. All patients were HIV negative and had no known immunodeficiencies.
(B) Schematic depicting index and secondary cases in the NCG outbreak.
(C) Destructive lesion of T-spine vertebra 9 with extensive paraspinous abscess (red arrow) caused by Mtb NCG infection.
(D) Lytic lesion of the sternum (red arrow) caused by Mtb NCG infection.
(E) CT scan of paraspinous abscess caused by Mtb NCG infection.
The NCG outbreak strain is a Lineage 1 strain
Extant Mtb strains are generally classified by lineage, with so-called “ancestral” strains being from L1, and L5–L7 and so-called “modern” strains being from L2–L4.4,47,48 The “modern” lineages are the most prevalent lineages in the Americas, Europe, and parts of Africa and Asia.9 Though L1 is responsible for substantial disease burden worldwide, it is comparatively restricted geographically to areas bordering the Indian Ocean; L5–L7 cause the least worldwide burden and are extremely restricted geographically.11 Initial genotyping placed NCG evolutionarily in the Manila clade of L1 (Figure S1A).
After whole genome sequencing, we compared NCG to 37 other L1 strains and identified all NCG-specific variants (Table S1). We did not identify any obvious functional null or gain-of-function variants (structural variants or early stop co-dons) unique to NCG that would easily explain the clinical phenotype, although it is possible that other variant types contribute to the high rate of bone disease. Given anecdotal reports of higher extrapulmonary dissemination rates overall in “ancestral” line-age strains,15,49–52 we considered whether variants shared among multiple, or all, L1 strains might contribute to clinical course.
We first interrogated 225 strains, including NCG, representing multiple strains from each Mtb lineage.6,53 We called variants against the L4 H37Rv reference genome and identified 35,787 SNPs across the 225 strains. After removal of genes associated with drug-resistance and those found in repetitive regions of the Mtb genome (including PE/PPE and PE-PGRS genes), we used the remaining 31,839 SNPs to construct neighbor-joining and maximum-likelihood phylogenies that placed NCG among L1 strains, confirming the initial genotyping results (Figure 2A).
Figure 2. Identification of the NCG outbreak strain as a Lineage 1 strain.
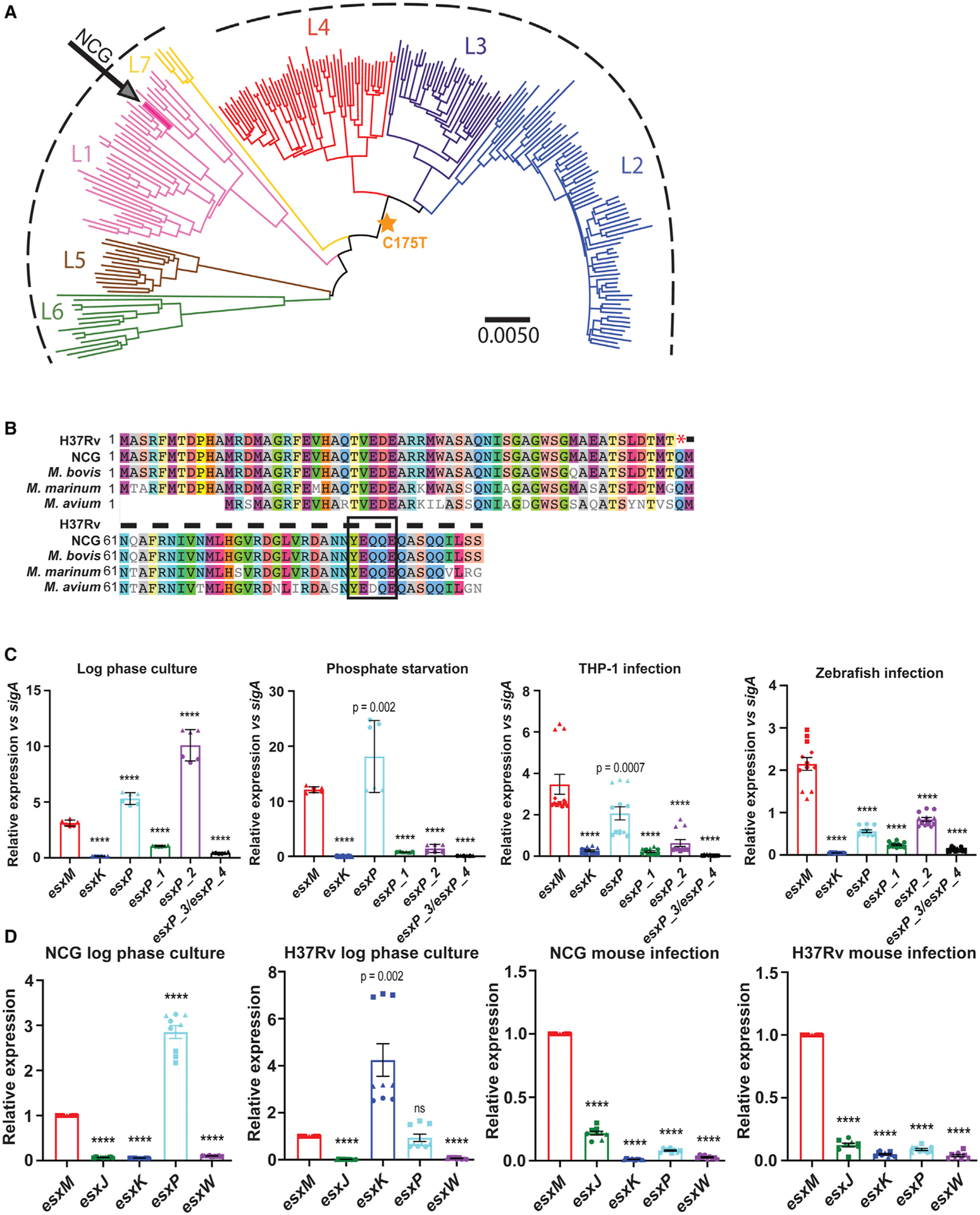
(A) Whole genome phylogenetic analysis based on reads from Comas et al.,6 identifying NCG as a member of the Manila clade of the Lineage 1 (L1) strains. Arrow denotes NCG. Gold star indicates emergence of shared C175T variant common to all L2, L3, and L4 strains. Scale bar indicates substitutions per variant as described in Saelens et al..54
(B) Amino acid alignment of EsxM for Mtb and other mycobacterial species, noting the early stop codon in modern strains predicted to truncate the protein and eliminate C-terminal secretion sequences (boxed area).
(C) Quantitative RT-PCR analysis showing mean transcript levels of esxM and paralogs relative to sigA in WT M. marinum cultured in vitro and upon THP-1 infections (2 dpi) and in vivo larval zebrafish (4 dpi) infections. Log phase culture and phosphate starvation–error bars represent SD, data pooled from two independent experiments with three technical replicates for each. THP-1 and in vivo infection error bars represent SEM, data pooled from three independent experiments with three technical replicates. One-way ANOVA with Dunnett’s multiple comparisons test.
(D) Mean transcript levels of esxM paralogs relative to esxM in Mtb NCG and H37Rv strains cultured in vitro in 7H9 media and upon mouse infection. Bacterial RNA isolated from C57BL/6J mouse lungs at 6 wpi after i.v. infection with Mtb NCG or H37Rv strains. Error bars represent SEM, three independent experiments with three technical replicates each. Statistical comparison by one sample t test. ****p < 0.0001.
See also Figures S1 and S2 and Tables S1, S2, and S3.
We next comprehensively assessed lineage- and strain-specific variants across the 225 strains (Table S1). When we compared variants appearing in extant strains from the ancestral lineages (L1, and L5–L7) and modern lineages (L2, L3, and L4), we discovered an ancestral variant present in all L1, and L5–L7 strains (including NCG) in the gene encoding the putative secreted effector EsxM that had not been previously noted as specific to these lineages (Figure 2A and Table S1).
The full-length esxM variant is an ancestral variant specific to a subset of Mtb lineages
Like other type VII secretion loci, the ESX-5 locus contains two small Esx proteins encoded in tandem.35 esxM, a member of the ~100 amino acid WXG-100 gene family, is located within the ESX-5 locus. It has previously been characterized as a pseudogene in the H37Rv reference genome due to an early stop codon that truncates ~40% of the protein. We found that the “ancestral lineage” Mtb strains (L1 and L5–L7), in contrast, contain the complete open reading frame and do not carry the early stop mutation (Figure 2A). The characteristic EsxM truncation identified in “modern” strains (L2–L4) occurs at codon position 59, upstream of the C-terminal regions implicated in secretion.55,56
To replicate these findings in other collections containing L1, we analyzed sequencing data from 3,236 diverse previously published strains from different continents.57,58 We found the same association of full-length EsxM with the L1 branch and the truncated variant with the discrete L2–L4 branch (Figures S1B and S1C).
The full-length EsxM variant had previously been reported in one set of clinical strains,59 but had not been explicitly associated with the L1 and L5–L7 lineages historically referred to as ancestral. We therefore asked whether the stop-loss variant in EsxM is the ancestral or the derived allele. Other members of the Mtb complex such as Mycobacterium bovis as well as closely related mycobacterial species, including Mycobacterium avium and M. marinum, share the stop-loss variant (glutamine residue at amino acid 59) with the ancestral lineage strains of Mtb. (Figure 2B). Therefore, we conclude that the full-length EsxM variant is ancestral, and the early-stop variant found in L2–L4 strains of Mtb is more recently derived.
Analysis of additional L1 strains reveals an epidemiological association with osteomyelitis
Although not formally tested in large case-control studies, several epidemiological studies, while not definitive, have suggested that L1 strains carrying the ancestral allele present with higher rates of extrapulmonary disease.15,49–52 To assess more systematically and in a larger dataset whether there was an association of L1 strains (full-length EsxM) with bone disease, we identified one of the few datasets with whole genome information that includes systematic reporting on osteomyelitis.58 The ten-year UK study included over 1,600 patients, with representation of the four most common Mtb lineages. Using this dataset, we found that infection with L1 strains (full-length EsxM) was associated with bone disease (p = 0.0001, OR 2.5, 95% CI 1.6–4.0) compared to the lineages with the derived esxM allele (L2–L4) (Table S2). Thus, we decided to examine the role of the full-length EsxM variant in dissemination and bone disease, as well as the regulation and function of EsxM in both Mtb and the closely related pathogen M. marinum.
Conserved organization and infection-dependent regulation of the ESX-5 locus in M. marinum and M. tuberculosis
Though the EsxM variant is a strong candidate for a functional variant due to the early stop codon, we considered whether close ESX-5 paralogs might compensate, leaving the organism functionally unchanged. To test this hypothesis computationally, we calculated if any of the close EsxM paralogs showed a change in their rate of evolution in the modern lineages, which would be consistent with subfunctionalization resulting in little or no overall functional change; however, we did not detect a significant change in the rate of evolution for any of these paralogs (Table S3). Further arguing against this hypothesis of complementation between paralogs is that these paralogous loci are differentially regulated at the transcriptional level,60 and mutations result in specific, non-redundant phenotypes in M. marinum.61
M. marinum, a close relative of the Mtb complex, encodes full-length EsxM, and, apart from ESX-2, shares orthologous ESX loci with Mtb. The esxM gene is found within the M. marinum ESX-5 locus, and its encoded amino acid sequence is 87% identical to EsxM in the L1 Mtb strains.62The principal ESX-5 locus is conserved in both M. marinum and Mtb and contains the small effectors EsxM and EsxN within a core secretion apparatus.41,63 Both M. marinum and Mtb contain four additional loci encoding small EsxM/N-related proteins that are secreted by the ESX-5 machinery. Consistent with previous analyses,41,61 our investigation of synteny and sequence conservation revealed matching homologous regions between M. marinum and Mtb and closely corresponding EsxM/EsxN-like proteins (Figure S2A).
Because EsxM/EsxN proteins and their paralogs have high levels of sequence homology, we sought to investigate the regulation of these loci in varying contexts, including conditions previously reported to alter ESX-5 transcription in Mtb.60 We developed a qRT-PCR-based assay targeting divergent sequences at the 5′ end of each esxM- and esxM-like paralog transcript for M. marinum and Mtb. In M. marinum, we found that although esxM was substantially expressed during growth in liquid cultures, two other esxM-like transcripts were more abundant (Figure 2C). Upon phosphate starvation, the relative expression of esxM increased and was exceeded by only one paralog (Figure 2C). However, EsxM became the clear predominant paralog during infection, both in cell culture and during infection of zebrafish larvae, a natural host (Figure 2C).
We applied the same strategy to bacterial RNA from Mtb to investigate whether infection-dependent upregulation of esxM is conserved. We analyzed two Mtb strains: H37Rv, a L4 strain containing the early stop codon in EsxM, and the L1 NCG outbreak strain. Paralleling results from M. marinum, esxM transcript was expressed substantially in broth cultures but was not the most abundant paralog for either strain (Figure 2D). RNA-seq-based analysis of both Mtb strains grown in broth validated our qRT-PCR assay, with concordant results in transcript levels between the two assays (Figure S2B).
To test whether esxM was differentially regulated during Mtb infection, we infected C57Bl/6J mice intravenously (i.v.) with each strain and extracted bacterial RNA from mouse lungs at 6 weeks post-infection. We found that Mtb esxM became the predominantly expressed paralog during in vivo infections, with a dramatic upregulation of the esxM transcript (Figure 2D) for both the L1 NCG outbreak strain and the L4 H37Rv strain. These results are also consistent with previously reported cell culture infection experiments using Mtb.57 Thus, in cell culture, zebrafish, and mouse infections, esxM remained the predominant paralog, and hence our top candidate for contributing to the clinical dissemination phenotype.
Characterization of a M. marinum esxM mutant
In humans, M. marinum infections are temperature limited but can often result in tenosynovitis and osteomyelitis.64 In zebrafish and other teleosts, M. marinum infections can disseminate widely, often with bone involvement.65 To test the hypothesis that esxM promotes dissemination, we genetically disrupted esxM in M. marinum to study dissemination, particularly to bone, using the natural zebrafish host.
To first test the impact of EsxM on protein secretion, we grew the WT (full-length EsxM), ΔesxM, and the complemented strain (extra-chromosomal expression of esxM from the hsp60 promoter) in Sauton’s media in vitro. We performed mass spectrometry-based proteomic analysis of the secreted protein fractions. Importantly, the deletion and restoration of EsxM did not have widespread impacts on the M. marinum secretome. EsxM itself was only detected in culture filtrates from the complemented (and overexpressed) strain, likely due to low levels of expression and/or regulation of secretion under standard conditions, consistent with the lower levels of transcription we identified in broth-grown cultures (Table S4). However, we were able to detect EsxN, the putative secreted partner of EsxM, from wild-type WT M. marinum culture filtrates, and found that deletion of the EsxM partner resulted in the absence of secreted EsxN, a phenotype that was fully complemented upon restoration of EsxM (Figure S2C). In contrast, other ESX-5 substrates were secreted at similar or increased levels in the esxM mutants, suggesting that any phenotype observed was specific to EsxM and not an indirect consequence of a general ESX-5 secretion defect (Figure S2C and Table S4).
As additional confirmation that EsxM could be secreted, we examined the expression of a tagged, complementing version of EsxM introduced into WT or esxM mutants under both standard and low-phosphate conditions. For both WT and mutant bacterial strains, we observed robust secretion of EsxM, detectable in the culture filtrate under low phosphate conditions (Figures S2D and S2E), an in vitro condition under which ESX-5 components are also transcriptionally induced (Figure 2C).60
Mutations in esxM result in decreased dissemination independent of burden
Early events of mycobacterial infection largely rely on macrophages infected with intracellular bacteria. We first sought to identify dissemination of WT M. marinum possessing an intact copy of esxM via the egress of infected macrophages from early granulomas, a mechanism by which mycobacteria disseminate within their host.23 We infected larval zebrafish at 2 days post-fertilization (dpf). Due to the site of infection, most initial granulomas form ventrally. Intravital time-lapse experiments with longitudinal imaging over 15 h between 4 and 5 dpi captured trafficking of infected macrophages from nascent granulomas to sites above the midline, where dorsal granulomas can be established (Figure 3A and Video S1).
Figure 3. Dissemination in vivo is dependent on esxM.
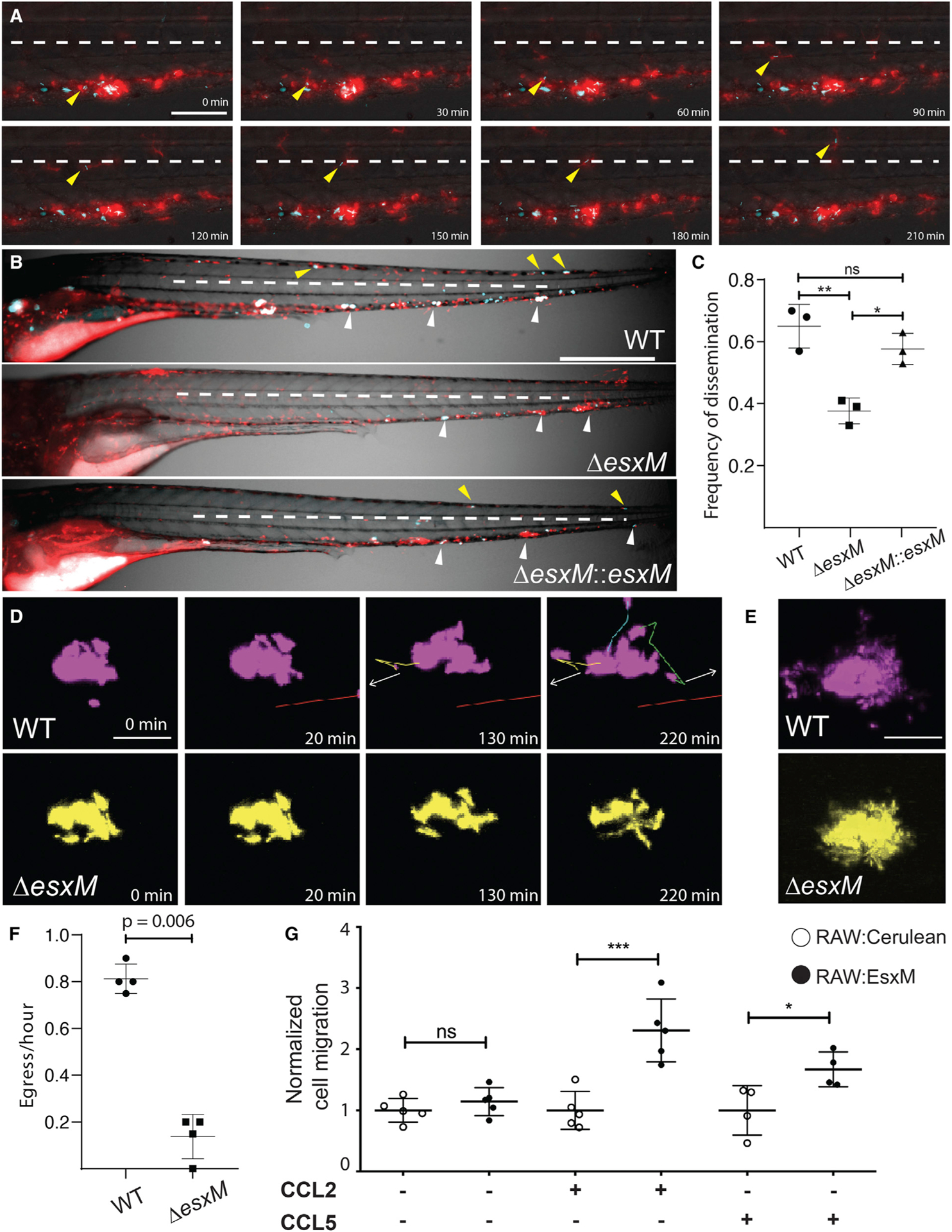
(A) A macrophage infected with WT M. marinum departs from an early granuloma and migrates above the midline (dashed white line). Scale bar, 50 μm.
(B) Representative images from WT M. marinum, ΔesxM, and ΔesxM:esxM infections in larval zebrafish. 2 dpf zebrafish with red fluorescent macrophages are infected ventrally in the caudal vein with cerulean-expressing M. marinum. Scale bar, 500 μm.
(C) At 5 dpi, ΔesxM demonstrates a reduced frequency in dissemination above the midline (white dashed line) to the dorsal side of the animal adjusting for burden across all three groups. One-way ANOVA with Tukey’s post-test, data from three biological replicates, mean ± SD shown. n > 35/experiment.
(D) Co-infection with WT and ΔesxM M. marinum results in larval granulomas populated by both strains. Time-lapse imaging reveals WT M. marinum egressing from the granuloma (tracking lines), while ΔesxM remains confined within. White arrows indicate direction of egress. Scale bar, 50 μm.
(E) Projection of time-lapse imaging in (D) showing complete range of movement for macrophages infected with each strain over 220 min; Scale bar 50 μm.
(F) Quantification of rate of egress for WT and ΔesxM M. marinum from co-populated granulomas. Long-term monitoring of 4 mixed granulomas from 4 independent animals. Student’s paired t test, mean ± SD shown.
(G) Migration of RAW 264.7 cells expressing fluorescent cerulean control or EsxM through transwell membranes in the presence and absence of chemokine. One-way ANOVA with Bonferonni’s post-test. *p < 0.05, **p < 0.01, ***p < 0.001.
To quantitate dissemination to distal points, we measured the rate at which infection spread above the midline (Figure 3A). We found that WT M. marinum (EsxM full-length) disseminated to the dorsal side of the larval zebrafish midline in ~70% of samples by 5 days post-infection (dpi) (Figures 3B and 3C). In contrast, ΔesxM mutants displayed a 2-fold reduction in dorsal dissemination (Figures 3B and 3C). The mutant could be complemented through constitutive extra-chromosomal expression of esxM (Figures 3B and 3C).
We observed no significant difference in bacterial burden in ΔesxM at the 5 dpi time point (Figure S3A), indicating that the initial dorsal dissemination phenotype is not a downstream effect of decreased overall burden. We also asked, using a slightly lower starting dose, whether overexpression of esxM in a WT M. marinum strain could enhance rates of dissemination; we observed a trend toward increased dissemination, although this was not statistically significant (Figure S3B). Overall, these experiments suggest that modulation of bacterial EsxM levels is associated with dissemination of infection to new sites and tissues.
Cell-autonomous effects of EsxM on infected macrophages in granulomas in vivo
Although we observed macrophages infected with EsxM-expressing M. marinum disseminating at a higher rate, this could arise through non-cell-autonomous effects of EsxM expression—for example, changing the overall inflammatory environment within the granuloma. To address this possibility, we co-infected larval zebrafish with differentially labeled WT and ΔesxM M. marinum and assessed dissemination from granulomas composed of mixed infected macrophages in equal proportions. Importantly, individual macrophages within the granuloma contained only one strain or the other, enabling testing of cell-autonomous effects. In mixed granulomas, granuloma macrophages infected with ΔesxM bacteria egressed at a dramatically lower rate than adjacent macrophages infected with WT M. marinum (Figures 3D–3F and Video S2). Thus, the esxM macrophage dissemination phenotype appears to be cell autonomous, suggesting local intracellular modulation of the infected host cell.
To assess whether secreted EsxM might directly modulate macrophage-mediated dissemination, we examined whether EsxM expression was sufficient to alter macrophage migration behavior and motility in the absence of any other bacterial proteins or lipids. In cultured mammalian macrophages, we found that cells heterologously expressing full-length EsxM from Mtb migrated through transwell membranes across a gradient at a higher rate than control macrophages in the presence of chemokines (Figure 3G).
To investigate macrophage behavior in response to a defined inflammatory stimulus, we performed zebrafish larval tail fin transections and measured recruitment of macrophages to the wound site. To understand the role of full-length EsxM in macrophage migration, we created a set of transgenic zebrafish lines that expressed bacterial EsxM (M. marinum EsxM, codon optimized for expression in zebrafish) from a macrophage-specific promoter (Tg(mfap4:esxM-p2a-tdTomatoxt49)) (Figures 4A and 4B). Macrophages producing EsxM arrived at the wound after tail transection much faster and in greater numbers than control macrophages from a matched macrophage transgenic line expressing only the fluorophore (Figures 4C–4E and Video S3). We also found that esxM-expressing macrophages demonstrated an elevated maximum migration velocity compared to control macrophages (Figure 4F). As an additional control, we generated a transgenic line that drove macrophage-specific expression of the truncated EsxM found in modern lineages of Mtb (Tg(mfap4:esxM_Q59X-p2a-tdTomatoxt50)). These macrophages behaved similarly to controls (Figures 4C–4F and Video S3). As a control for the function of other ESX-secreted proteins, we created a transgenic line in which the paralogous protein EsxB from the ESX-1 locus was driven in macrophages (Tg(mfap4:esxB-p2a-mNeonGreenxt51)). EsxB-expressing macrophages were indistinguishable from WT macrophages in these assays (Figures S3C and S3D).
Figure 4. Macrophage-specific expression of ancestral full-length EsxM, but not its paralogs or modern, truncated EsxM, alters macrophage motility.
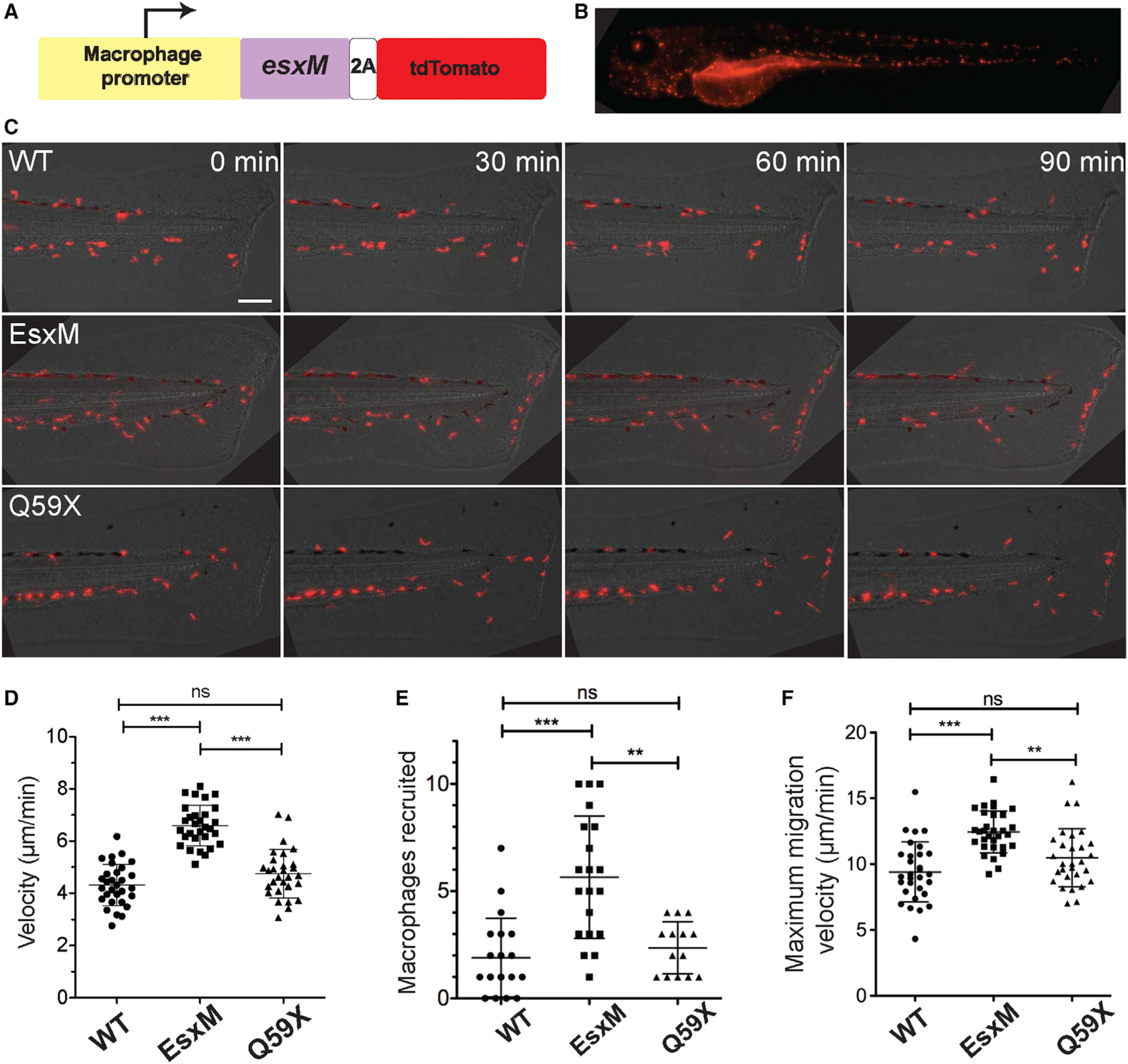
(A) Transgene design (not to scale) for macrophage-specific expression of EsxM. The macrophage-specific zebrafish promoter mfap4 drives expression of the mycobacterial protein within zebrafish macrophages. 2A peptide uncouples EsxM protein (98 AA) from tdTomato.
(B) Representative larval zebrafish expressing the mfap4:esxM-p2A-tdTomato transgene. Scale bar, 500 μm.
(C) Increased macrophage motility in the absence of infection for EsxM-expressing transgenic zebrafish lines. Scale bar, 100 μm.
(D) Migration and velocity measured in response to tail fin transection. One-way ANOVA, Dunn’s multiple comparison test. Data representative of three biological replicates.
(E) Number of macrophages recruited to tail wound 90 min post-transection. Macrophages express fluorescent TdTomato, EsxM, or truncated EsxM (Q59X) via a macrophage-specific promoter (mfap4). One-way ANOVA with Dunn’s multiple comparison test. Data representative of three biological replicates.
(F) Maximum velocity of macrophages migrating to tail wound. One-way ANOVA with Dunn’s multiple comparison test. Data representative of three biological replicates. Mean ± SD shown on all graphs. **p < 0.01, ***p < 0.001.
We returned to our mixed granuloma model to assess the effects of EsxM on macrophage motility in a natural infection. We assayed the velocity of zebrafish macrophages infected with WT or ΔesxM M. marinum as they departed granulomas. ΔesxM-infected macrophages departed established granulomas at a much lower rate overall, and the subset of ΔesxM-infected macrophages that did depart migrated at a lower velocity (Figures 3D and S3E). Overall, a strikingly low number of ΔesxM-infected macrophages departed established granulomas in the mixed infection experiments (Figure 3D).
To gain further insight into the observed cell-autonomous effect of EsxM on migration velocity, we measured the migration velocity of infected macrophages not in granulomas during the mixed infection. Here, we also observed a significant decrease in the migrating velocity of ΔesxM-infected macrophages compared to those infected with WT M. marinum, despite an overall higher rate of migration velocity relative to macrophages within granulomas (Figure S3F). Together, these results suggest that full-length EsxM is both required and sufficient for characteristic alterations in macrophage behavior, including egress from the initial granulomas that form during infection.
Reconstitution of ancestral EsxM in an attenuated modern Mtb strain enhances macrophage motility
We next determined whether EsxM could enhance the migratory behavior of its primary host cell type in the context of infection with Mtb. We reconstituted the ancestral protein in an attenuated modern L4 Mtb strain. Using mc26020, a double deletion (ΔlysAΔpanCD) auxotrophic mutant from the H37Rv background66 (i.e., a strain of Mtb that harbors the derived Q59X allele in esxM), we investigated whether expression of the ancestral, full-length esxM in Mtb enhances the migratory capacity of infected macrophages. We found that RAW 264.7 macrophages infected with full-length esxM-expressing mc26020 migrated directionally in a transwell assay at a higher rate than cells infected with control mc26020, both in a serum gradient and in the presence of chemokines (Figure 5A). These results indicate that full-length, ancestral esxM expressed in a L4 Mtb strain promotes mammalian host cell migration during Mtb infection. During these experiments, we observed that the ancestral EsxM-expressing strain induced a distinct morphology of the infected macrophages compared to control infections, with the emergence of increased numbers of membrane spikes resembling filopodia (Figure S4A).
Figure 5. Reintroduction of ancestral EsxM into modern Mtb reprograms macrophage motility through cytoskeletal alterations.
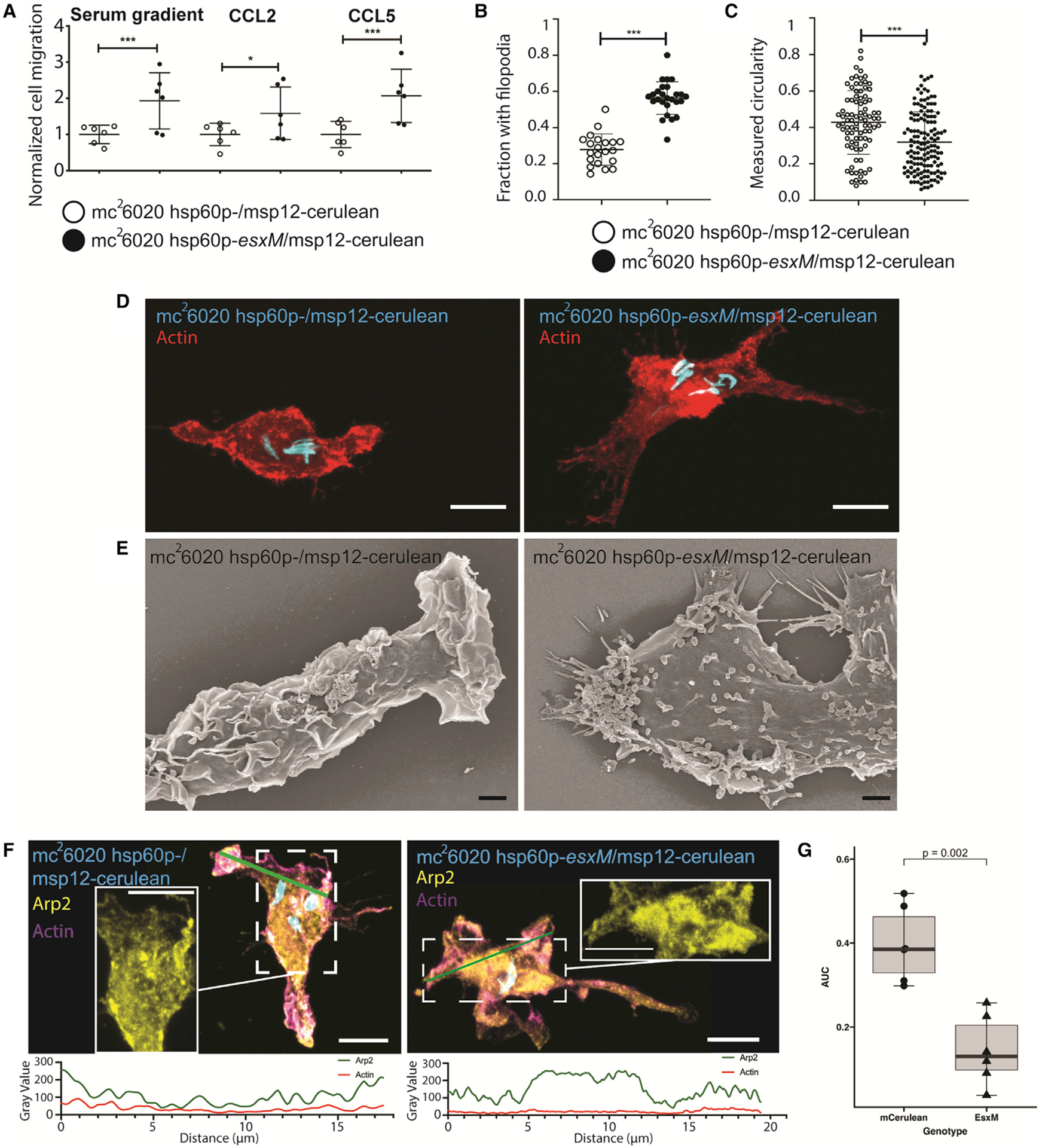
(A) RAW264.7 cell migration during infection with Mtb strains mc26020 (Mtb6020 msp12:cerulean) or mc26020 constitutively expressing esxM (Mtb6020 hsp60p:esxM/msp12:cerulean) from an episomal plasmid across transwell membranes in the presence and absence of chemokine. Mean ± SD shown. One-way ANOVA with Bonferroni’s correction.
(B) Quantitation of the presence of filopodia on BLaER1 cells infected with Mtb6020 msp12:cerulean or Mtb6020 hsp60p:esxM/msp12:cerulean. Two-tailed paired Student’s t test, mean ± SD shown.
(C) Measured circularity of BLaER1 cells infected with Mtb6020 msp12:cerulean or Mtb6020 hsp60p:esxM/msp12:cerulean.
(D) BLaER1 cells infected with Mtb6020 hsp60p∷esxM/msp12:cerulean demonstrate increased filopodia and reduced circularity compared to BLaER1 cells infected with Mtb6020 msp12∷cerulean. Bacteria in cyan and actin in red. Scale bar, 8 μm.
(E) Electron micrographs of BLaER1 cells infected with Mtb6020 hsp60p∷esxM/msp12∷cerulean demonstrate increased filopodia compared to BLaER1 cells infected with Mtb6020 msp12∷cerulean. Scale bar, 2 μm.
(F) BLaER1 cells infected with Mtb6020 msp12∷cerulean demonstrate peripheral localization of Arp2, compared to interior localization of Arp2 in cells infected with Mtb6020 hsp60p∷esxM/msp12∷cerulean. Insets display fluorescence intensity of Arp2 and actin along the green line, with interior localization in Mtb6020 hsp60p∷esxM/msp12∷cerulean infections.
(G) Quantification of Arp2 localization from (F). Plot of the percentage of the area under the curve (AUC) for fluorescence in the Arp2 channel for the interval 0% to 25% of the normalized distance across the longest cell axis normalized to AUC from the middle 50% (25%–75%) of the total cell length. Each data point represents a single cell. Statistics from Student’s t test. Scale bar, 8 μm. *p < 0.05, **p < 0.01, ***p < 0.001.
All images are representative of at least two biologically independent experiments.
See also Figure S4.
To explore this phenotype further in a human cell line, we used BLaER1 cells that, after transdifferentiation into macrophage-like cells, recapitulate critical aspects of macrophage biology.67,68 We infected transdifferentiated BLaER1 cells with Mtb strain mc26020 with a plasmid expressing cerulean fluorescent protein or an identical plasmid expressing cerulean fluorescent protein and ancestral esxM. Similar to the result in RAW 264.7 cells, infection of the human macrophage cell line with Mtb expressing ancestral EsxM resulted in increased filopodial protrusions (Figures 5B and 5D). In addition, transdifferentiated BLaER1 cells infected with Mtb expressing ancestral EsxM took on a more extended morphology relative to those infected with the control Mtb strain (Figures 5C and 5D).
To characterize EsxM-dependent macrophage alterations more fully, we performed electron microscopy on infected macrophages from the Mtb mc26020 strain with the control plasmid or the mc26020 strain expressing ancestral, full-length EsxM. We found dramatic differences at the macrophage surfaces. Macrophages infected with the control strain had easily recognizable lamellipodia and prominent membrane ruffling, whereas those infected with Mtb expressing ancestral EsxM instead displayed prominent filopodia and extensive membrane blebs at the edge of the infected macrophage (Figure 5E).
These alterations closely resembled those described for a macrophage-specific knockout of Arpc2 in mouse macrophages.69 In addition, Arpc2−/− macrophages, under some conditions, display increased random migration speeds as well as increased velocities during migration to chemoattractants.69 We therefore asked whether EsxM expression may disrupt this pathway, leading to the observed Arpc2-like phenotype in terms of increased migration and altered morphology (Figures 5A and 5F). Though Arp2 localizes to the periphery of macrophages infected with the modern Mtb strain, there is consistent disruption of Arp2 localization in macrophages infected with Mtb expressing full-length EsxM, with Arp2 confined to the interior (Figures 5F, 5G, S4B and S4C). In addition, treatment of zebrafish larvae with Arp2/3 inhibitor CK66670,71 pharmacologically rescued the decreased migration velocity of ΔesxM-infected macrophages, indicating that EsxM may regulate the host cytoskeletal axis in vivo (Figure S5 and Video S4).
To examine how the differences we observed influence migration, we performed live imaging of transdifferentiated BLaER1 macrophages infected with the two Mtb strains that differ only in the expression of full-length EsxM. The leading edge of macrophages infected with the control Mtb strain displayed classical lamellipodia and ruffles that predicted the direction of movement (Figure 6A). In contrast, prominent filopodial projections marked the leading edge of the cells infected with Mtb expressing ancestral EsxM (Figure 6A). Thus, ancestral EsxM is required and sufficient in a variety of contexts to alter macrophage migratory dynamics and subsequent mycobacterial dissemination.
Figure 6. Altered macrophage motility and morphology during M. tuberculosis infection with full-length EsxM variants.
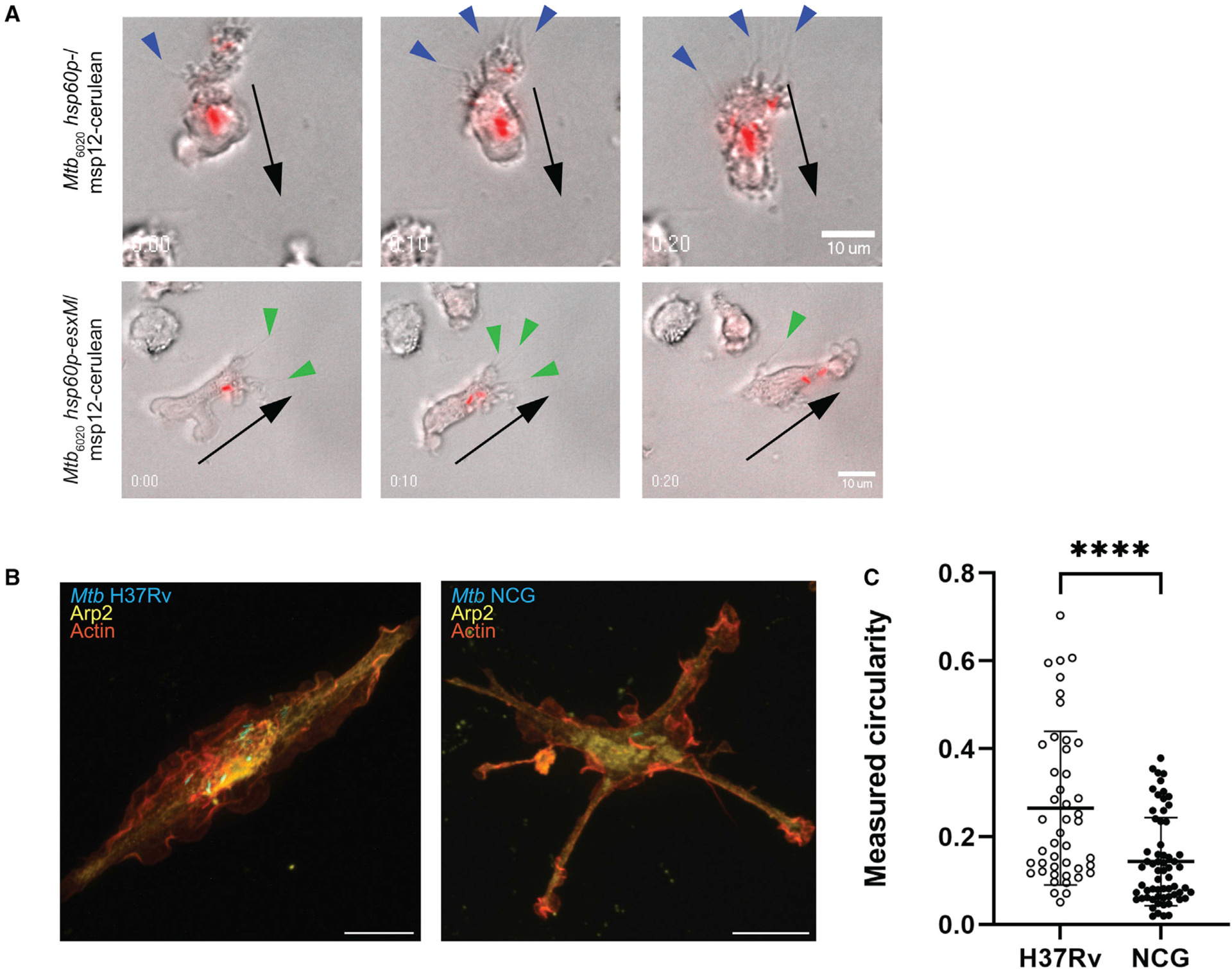
(A) Time-lapse imaging of BLaER1 cells infected with Mtb6020 msp12∷cerulean or Mtb6020 hsp60p∷esxM/msp12∷cerulean. Bacteria false-colored red. Spiky projections, likely retraction fibers, (blue arrowheads) localize to the lagging edge in BLaER1 cells infected with Mtb6020 msp12∷cerulean. Filopodial projections (green arrowheads) localize to the leading edge in BLaER1 cells infected with Mtb6020 hsp60p∷esxM/msp12∷cerulean. Black arrows indicate the direction of cell migration.
(B and C) Infection of BLaER1 cells with Lineage 4 (L4) Mtb H37Rv results in infected cells with membrane ruffles. Infection with the Lineage 1 (L1) Mtb NCG outbreak strain results in an increase in spiky projections and (C) a decrease in circularity, two-tailed paired Student’s t test, mean ± SD shown. ****p < 0.0001. All images representative from at least two biologically independent experiments.
See also Figures S4 and S5 and Video S4.
To confirm that the reconstitution experiments reflect the biology of the outbreak strain itself, we performed BLaER1 macrophage infections with cerulean-labeled versions of the L4 strain H37Rv and the NCG outbreak strain. Although we were not able to perform macrophage motility assays under BSL3 conditions, we found that cells infected with the NCG strain displayed similar alterations in morphology, with more extended branches than H37Rv-infected cells (Figures 6B, 6C, and S4D).
Ancestral esxM promotes dissemination to bone in vivo
We next explored in vivo whether this altered macrophage motility might contribute to the outbreak’s clinical phenotype using a zebrafish infection model.72,73 Like human bone, zebrafish bone consists of osteocytes, bone-lining cells, osteoblasts, and mono- and multinucleate osteoclasts.74 Key signaling molecules that regulate bone-remodeling cells are conserved between humans and zebrafish.72,75 Human genetic variants affecting bone physiology and development have translated into similar alterations in zebrafish bone.76–80
In cases of TB skeletal disease in humans, the spine is the most common site of infection.81 Likewise, spinal deformities in zebrafish have long been known to be a sign of mycobacterial infection in aquaculture.65 We infected the osteoblast reporter line Tg(Ola.Sp7:mCherry-Eco.NfsB)pd46 and examined M. marinum-induced pathology in zebrafish bone. Periostitis and lytic lesions are a well-documented consequence of skeletal TB in humans.82 We found that infection with M. marinum led to an increased signal from osteoblasts near sites of M. marinum infection and erosion of bone in contact with mycobacterial infection (Figure 7A–7B′).
Figure 7. Ancestral esxM promotes spinal dissemination.
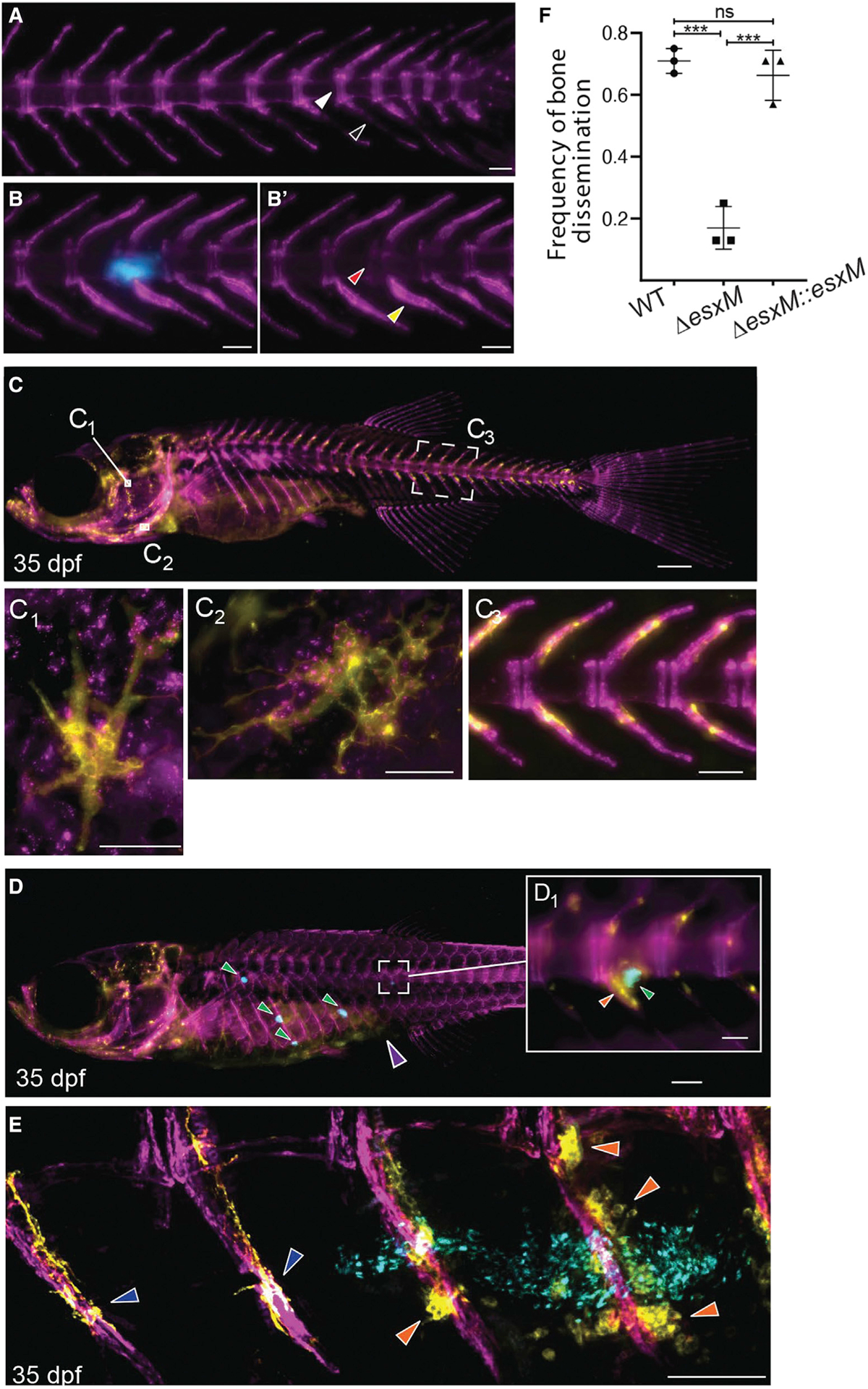
(A) Representative image of sham-injected osteoblast line 3 weeks post-injection. Intervertebral spacing is tightly delineated (white arrowhead), and osteoblast signal at hemal arches is typical of healthy zebrafish (black arrowhead). Scale bar, 500 μm.
(B) Representative image of cerulean M. marinum-infected osteoblast line, three weeks post-infection (wpi) showing bone-associated infection. Scale bar, 100 μm.
(B′) Same animal as in (B), showing only osteoblasts. M. marinum has invaded into the intervertebral space, giving rise to a vertebral lesion (red arrowhead) and proliferation of osteoblasts proximal to infection (yellow arrowhead). Scale bar, 100 μm.
(C) Dual transgenic line at 35 dpf labeling osteoblasts in purple from the Sp7/osterix promoter and osteoclasts from the acp5a/TRAP promoter in yellow. Scale bar, 500 μm.
(C1-C3) Insets show tight association of osteoblasts and osteoclasts along the spine (C3). Yellow multinucleate osteoclasts are also visible in the head (C1 and C2). C1 inset scale bar, 500 μm C2 inset scale bar, 50 μ. C3 inset scale bar, 100 μm.
(D) Intraperitoneal infection of juvenile animals with cerulean-fluorescent M. marinum results in frequent dissemination to spine (approximately 70% of animals). Scale bar, 500 μm.
(D1) High magnification view of boxed area of (D) showing association of granuloma with spine and alterations in normal osteoclast distribution. Scale bar, 100 μm.
(E) PACT-based clearing techniques in adults and juveniles enable high-resolution imaging of sites of bone disease. Confocal image of dorsal region of spine showing unaffected vertebrae with typical osteoclast behavior (dark blue arrowheads) and altered osteoclast behavior (orange arrowheads) in segments associated with granulomas (bacteria in cyan). Scale bar, 100 μm.
(F) Quantitation of dissemination to spine at 14 dpi in intraperitoneal infections. n ≥ 7 per group for 3 biological replicates. One-way ANOVA with Tukey’s post-test, data from three biological replicates, mean ± SD shown. ***p < 0.001.
We next developed an osteoclast-specific transgenic line using upstream regulatory sequences of the zebrafish tartrate-resistant acid phosphatase gene (Tg(acp5a:mNeonGreen-CAAX)xt52), which labeled osteoclasts specifically (Figure 7C). Using a dual osteoblast-osteoclast transgenic line derived from crossing the two lines, we infected zebrafish at 3 weeks post-fertilization (wpf) and traced the dissemination of M. marinum to skeletal sites two weeks later (Figures 7D and 7E). We found consistent and substantial dissemination of infection to bone at this time point (Figures 7D–7F). At sites of spinal infections in juvenile zebrafish, we noted a strong response by proximal osteoclasts and osteoblasts alike. Osteoclasts near M. marinum appeared in greater number and altered morphology (Figure 7E). Despite the close developmental relationship between macrophages and osteoclasts, we did not find any evidence of osteoclasts infected with M. marinum.
Finally, we infected juvenile zebrafish intraperitoneally with WT, ΔesxM, and the complemented mutant at 3 wpf and assessed the presence of spinal infections at 2 weeks post-infection (wpi). Although juvenile zebrafish infected with WT M. marinum developed spinal infections in ~70% of samples by 2 wpi, this proportion was reduced to ~20% in ΔesxM infections (Figures 7D–7F). The mutant could be complemented through constitutive extra-chromosomal expression of esxM (Figures 7D–7F).
DISCUSSION
Investigation of outbreak strains with extreme phenotypes can lead to mechanistic understanding of critical host-pathogen interactions that promote or restrain virulence and transmission. Here, we link an unusual Mtb outbreak with high rates of dissemination and skeletal disease to a specific Mtb-secreted effector that is present in the so-called “ancestral” lineages but truncated in major Mtb lineages L2–L4, the most broadly distributed lineages geographically.
Full-length EsxM is both necessary and sufficient to drive in vivo changes in the motility of infected macrophages, rates of granuloma egress and dissemination, and ultimately interaction with bone. Notably, reintroduction of the full-length EsxM into a modern Mtb strain resulted in a change in the migration modality of infected macrophages. Generally, lamellipodia at the leading edge are associated with macrophage migration within interstitial tissues.83 In cells infected with an Mtb strain expressing reconstituted ancestral EsxM, a dramatic reprogramming of macrophage motility occurred, from lamellipodia-led migration to motility marked by filopodia at the leading edge and membrane blebbing. We hypothesize that ancestral and generalist mycobacterial pathogens benefit from dissemination via enhanced macrophage motility, and that the derived truncation of EsxM in L2–L4 curtails overall dissemination to some degree.
Our finding that the L2–L4 Mtb strains contain a derived, truncated version of EsxM suggests its functional change would have significantly altered their properties during human infection. Infection of non-transmissible tissue sites such as bone would not promote transmission.84 Inversely, mutations that increase the likelihood of spreading to new hosts would be selected for, provided there is a sufficiently large host population and therefore may influence the transmission dynamics of these strains.85 The derived EsxM variant may shape the properties of Mtb strains from L2–L4, perhaps by promoting residence in the lungs, the site of disease most effective for transmission, and reducing the frequency of unproductive dissemination. Nevertheless, L1 strains remain prevalent in areas bordering the Indian Ocean and may indeed be well adapted to their particular niche or host population,11 and it is notable that the outbreak with high rates of bone disease occurred among a host population where L1 strains are rare. Host genetic variants may also influence the rates of extrapulmonary and skeletal disease.86,87
We identified two large-scale studies with mixed strain populations consisting of L1–L4; one in Vietnam and one in Birmingham, UK.57,58 The Vietnamese dataset did not include any information about rates of osteomyelitis, but both studies suggested a substantial transmission advantage for the lineages that had the derived, truncated EsxM allele over the full-length allele,57,58 at least in these settings. Overall prevalence of the different lineages as well as the emergence and expansion of specific sub-lineages may be driven by genetic lineage and strain-specific factors, but may also be influenced by human migration patterns, changes in ecological niche, or epidemic dynamics over time.11,88–90
The Birmingham, UK study, one of the few large-scale published studies to include detailed information about TB bone disease combined with whole genome sequencing, revealed an additional association of L1 with higher rates of bone disease.58 Infection with the modern L2–L4 strains can certainly still result in bone disease,91,92 but in the Birmingham study, osteomyelitis occurs at substantially lower rates for the L2–L4 strains carrying the derived EsxM truncation than for the Lineage 1 strains with the ancestral allele.
Some additional properties of L2–L4 strains relative to L1 strains are likely attributable to a characteristic deletion present in L2–L4 strains associated with resistance to oxidative stress and hypoxia.10 Our data reveal that the emergence of Mtb L2–L4, the lineages that are most broadly distributed globally, was also marked by an inactivating mutation in esxM. We find that the ancestral version of this protein present in L1, and L5–L7 strains, as well as almost all pathogenic mycobacterial species, enhances macrophage motility—reprogramming macrophage migration in infected cells—and promotes dissemination. Results analyzing intrahost strain evolution from human autopsy studies have suggested similar patterns of intra-lung spread and inter-organ spread, indicating that there may be shared dissemination mechanisms, including macrophage trafficking.93,94 Although numerous sequence variants separate the Mtb lineages, acquisition of the derived esxM variant may have contributed to the specialization of L2–L4 to their current human niches and to the clinical properties and progression of infections with these strains.
Limitations of the study
Though we identified a strong association between the full-length ancestral esxM allele and bone disease and identified a functional role for this effector using animal models, there are likely other bacterial variants or host factors that contribute to the extremely high rates of bone disease in the North Carolina outbreak. In addition, the in vivo regulation of the ESX-5 small, secreted proteins is complex. esxM becomes the predominant transcribed paralog in cell culture and animal models of infection, both in M. marinum and Mtb, but our evolutionary analysis also revealed strong conservation of other esxM paralogs (Table S3) compared to published whole genome analysis of the overall rate of Mtb evolution.48,95 The roles of these paralogs during mycobacterial growth and pathogenesis remain to be defined. Additionally, although the zebrafish model recapitulates many important aspects of bone disease and dissemination, there are limited mammalian models in which bone dissemination has been established and can be studied.96 Finally, the distinction between dissemination and bone tropism remains an open question, as well as the precise nature of the in vivo initiation event for the bone-associated granulomas we observe.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, David Tobin (david.tobin@duke.edu).
Materials availability
All materials and lines generated in this study are available from the lead contact.
Data and code availability
Genomic sequencing and RNA-seq data are available at NCBI (BioProjects PRJNA540867 and PRJNA872173)
Raw and processed mass spectrometry files are archived and available at MassIVE and PRIDE repositories under accession numbers MSV000090143 and PDX036131.
Original code is deposited and publicly available on GitHub and Zenodo. DOIs are listed in the Key Resource Table.
All other data are available in the main text or supplementary figures.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Arp2 | Abcam | Cat#ab49674; RRID:AB_867730 |
| goat anti-mouse Alexa Fluor 647 | Thermo Fisher | Cat#A21236; RRID:AB_2535805 |
| rabbit anti-HA-Tag | Cell Signaling Technology | Cat#C29F4; RRID:AB_1549585 |
| mouse anti-RNAP | BioLegend | Cat#8RB13; RRID:AB_2566583 |
| Bacterial and virus strains | ||
| Mycobacterium marinum M strain/pMSP12:cerulean | Oehlers et al.97 | N/A |
| Mycobacterium marinum M strain/pMSP12:tomato | Cambier et al.98 | N/A |
| Mycobacterium marinum ΔesxM | This paper | N/A |
| Mycobacterium marinum ΔesxM + hsp60:esxM | This paper | N/A |
| Mycobacterium tuberculosis 6020 auxotroph | Sambandamurthy et al.66 | N/A |
| Mycobacterium tuberculosis 6020 + msp 12::cerulean | This paper | N/A |
| Mycobacterium tuberculosis 6020 + msp 12::cerulean-hsp60:esxM | This paper | N/A |
| Mycobacterium tuberculosis H37Rv | BEI Resources, NIAID | NR-13648 |
| Mycobacterium tuberculosis NCG | This paper | N/A |
| Mycobacterium tuberculosis H37Rv/msp12:cerulean | This paper | N/A |
| Mycobacterium tuberculosis NCG/msp12:cerulean | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 7H10 | Difco | Cat#262710 |
| Trizol | Ambion | Cat#15596026 |
| Sodium chloride | Fisher Scientific | Cat#S271 |
| Potassium chloride | VWR | Cat#BDH9258 |
| Calcium chloride | VWR | Cat#BDH9224 |
| Magnesium chloride | Ward’s Scientific | Cat#470301 |
| 1-phenyl-2-thiourea | Sigma-Aldrich | Cat#P7629 |
| Tricaine-S (MS-222) | Syndel | ANADA#200–226 |
| Low melting point agarose | Fisher Scientific | Cat#BP165–25 |
| CK-666 Arp2/3 inhibitor | Sigma-Aldrich | Cat#182515 |
| CK-689 inactive control | Sigma-Aldrich | Cat#182517 |
| Dimethyl sulfoxide (DMSO) | Fisher Scientific | Cat#BP231 |
| BP Clonase II | Thermo Fisher | Cat#11789020 |
| HyClone HyQTase Cell Detachment Reagent | Fisher Scientific | Cat#SV3003001 |
| DAPI Fluoromount-G | SouthernBiotech | Cat#0100–20 |
| RPMI-1640 | Sigma-Aldrich | Cat#R8758 |
| GlutaMAX Supplement | Thermo Fisher | Cat#35050061 |
| Phorbol-12-myristate-13-acetate (PMA) | Sigma-Aldrich #R8758 | Cat#P148 |
| Sodium pyruvate | Gibco | Cat#11360 |
| Recombinant human IL-3 | Peprotech | Cat#200–03 |
| Recombinant human M-CSF | Peprotech | Cat#300–25 |
| β-Estradiol | Sigma-Aldrich | Cat#E2758 |
| Alexa Fluor 555 phalloidin stain | Thermo Fisher | Cat#A34055 |
| Hygromycin B solution | Invitrogen | Cat#10687010 |
| 7H9 | Difco | Cat#271310 |
| OADC | Sigma-Aldrich | Cat#M0678 |
| Neonate-80 | Fisher Scientific | Cat#BP337 |
| TRIzol Reagent | Invitrogen | Cat#15596026 |
| RNase-free DNase I | New England BioLabs | Cat#M0303S |
| Luna universal qPCR master mix | New England BioLabs | Cat#M3003X |
| Tyloxapol | Sigma-Aldrich | Cat#T8761 |
| cOmplete EDTA-free protease inhibitor cocktail | Millapore Sigma | Cat#11836170001 |
| Super-Signal West Pico PLUS | Thermo Fisher | Cat#34580 |
| Super-Signal West Femto Maximum Sensitivity | Thermo Fisher | Cat#34095 |
| Prolong Gold Anti-fade mounting solution | Invitrogen | Cat#P36934 |
| Critical commercial assays | ||
| mMessage mMachine T7 Kit | Thermo Fisher | Cat#AM1344 |
| RNeasy Plus Mini Kit | Qiagen | Cat#74134 |
| Zymo Direct-zol RNA Miniprep Kit | Thomas Scientific | Cat#1159U94 |
| DC Protein Assay Kit | Bio-Rad | Cat#5000111 |
| Pierce Micro BCA Protein Assay Kit | Thermo Fisher | Cat#23235 |
| Deposited data | ||
| Mtb genomic sequencing | NCBI BioProject | PRJNA540867 |
| Mtb H37Rv and NCG RNA sequencing | NCBI BioProject | PRJNA872173 |
| Raw mass spectrometry files | MassIVE repository | MSV000090143 |
| Processed mass spectrometry files | PRIDE repository | PDX036131 |
| Experimental models: Cell lines | ||
| BLaER1 | Rappino et al.67 | N/A |
| RAW 264.7 | ATCC | ATCC product TIB-71 |
| THP-1 | ATCC | ATCC product TIB-202 |
| Experimental models: Organisms/strains | ||
| Zebrafish (Danio rerio), *AB wildtype strain | ZIRC | ZDB-GENO-960809–7 |
| Zebrafish, Tg(mfap4:tomato-caax)xt6 | Walton et al.99 | N/A |
| Zebrafish, Tg(mfap4:esxM-p2a-tdTomato) xt49 | This paper | N/A |
| Zebrafish, Tg(mfap4:esxM_Q59X-p2a-tdTomato) xt50 | This paper | N/A |
| Zebrafish, Tg(mfap4:esxB-p2a- mNeonGreen) xt51 | This paper | N/A |
| Zebrafish, Tg(Ola.Sp7:mCherry-Eco.NfsBp46 | Singh et al.100 | N/A |
| Zebrafish, Tg(acp5a:mNeonGreen-CAAX)xt52 | This paper | N/A |
| Mouse, C57Bl/6J | Jackson Laboratories | RRID:IMSR_JAX:000,664 |
| Recombinant DNA | ||
| Oligonucleotides | This paper | Available in Table S5 |
| Software and algorithms | ||
| RNA seq analysis original code | This paper | https://doi.org/10.5281/zenodo.6981721 |
| Phylogenetic analysis original code | This paper | https://github.com/vertgenlab/gonomics |
| BWA | Li and Durbin101 | N/A |
| ClustalW2 | Larkin et al.102 | N/A |
| FIJI/ImageJ2, 2.5.0 | Rueden et al.103; Schindelin et al.104 | N/A |
| iTol | Letunic and Bork105 | N/A |
| Kallisto v0.48 | Bray et al.106 | N/A |
| MtrackJ, 1.5.1 | Meijering et al.107 | N/A |
| RStudio | RStudio Team (2022). RStudio: Integrated Development Environment for R. | N/A |
| RAxML | Stamatakis108 | N/A |
| SAMtools | Li et al.109 | N/A |
| Sleuth v0.30.0 | Pimentel et al.110 | N/A |
| VarScan | Koboldt et al.111 | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics statement
All zebrafish husbandry and experiments were approved by the Duke University Animal Care and Use Committee (protocol A091-20-04). Mouse studies were performed in strict accordance using the recommendations from the Guide for the Care and Use of Laboratory Animals of the National Institute of Health and the Office of Laboratory Animal Welfare. Mouse studies were performed at Duke University using protocols approved by the Duke Institutional Animal Care and Use Committee (IACUC) (protocol A221-20-11) in a manner designed to minimize pain and suffering in M. tuberculosis-infected animals. Any animal that exhibited severe disease signs was immediately euthanized in accordance with IACUC approved endpoints.
Human subjects
Approval was obtained from the Duke University Medical Center Institutional Review Board to obtain the NCG isolate and to use de-identified clinical data for preparation of the manuscript. The index case was male; the outbreak comprised of three female patients and four male patients, with ages ranging from 25 to 55 years old. Detailed sex and age information is contained in Figure 1A.
Zebrafish husbandry
Zebrafish were maintained on a 14 h light/10 h dark cycle. Water conditions within the system were maintained at 28°C between pH7.0–7.3 and conductivity 600–700 μS. Zebrafish were fed twice per day—once per day with dry food and once per day with Artemia.
Zebrafish lines
All zebrafish strains used were in the *AB wildtype background. Zebrafish experiments were performed with the approval of the Duke University Animal Care and Use Committee (protocol A091-20-04). Larvae were raised in E3 medium with methylene blue for the first 24 h postfertilization (hpf). Thereafter, larvae were raised in E3 medium supplemented with 45 μg/mL 1-phenyl-2-thiourea (PTU) to halt pigmentation. Tg(mfap4:tomato-caax)xt6, Tg(Ola.Sp7:mCherry-Eco.NfsB)pd46 (a kind gift from Dr. Kenneth Poss’s laboratory) have been previously described.97,99,100
Mice
Male C57BL/6J were purchased from the Jackson Laboratory (#0664). All mice were housed in a specific pathogen-free facility under standard conditions (12h light/dark, food and water ad libitum). Mice were infected with Mtb between 8 and 12 weeks of age. All mice were male.
METHOD DETAILS
Mouse infections
For in vivo experiments, 1×106 CFU of prepared H37Rv or NCG strains were delivered via i.v. tail vein injection, resulting in an infectious dose (Day 1 CFU) of 105 in the spleen and 104 in the lung. Groups of 3–4 mice per bacterial strain were infected. At 6 weeks post-infection, mice were euthanized, and organs were harvested then homogenized in a FastPrep-24 (MP Biomedicals). Bacterial burden was quantified by dilution plating half the organ on 7H10 agar and counting colony forming units (CFU) after three weeks of growth. The other half of the lung was placed in trizol, homogenized and frozen until RNA extraction.
Larval and juvenile zebrafish infections
2 dpf larval zebrafish were anesthetized with tricaine (MS-222, final concentration 0.016%). Approximately 200 fluorescent bacteria (FB) of M. marinum were injected into the caudal vein of each larval zebrafish with a borosilicate needle. The infected zebrafish were subsequently recovered in E3 medium containing PTU. Any embryos with infection initially seeded above the midline or damaged in the process of injection were removed from the experiment. Fluorescent bacteria were quantified as described in.112 Mixed infection experiments were set up as described above, except that both WT M. marinum msp12∷tdtomato and ΔesxM M. marinum msp12::mCerulean were loaded into the same needle.
Juvenile zebrafish (21 dpf) were anesthetized with tricaine (final concentration 0.016%). Approximately 200 FB were injected into the peritoneum of each juvenile zebrafish using a borosilicate needle. Infected fish were recovered in clean zebrafish system water and maintained in 1 L beakers in an incubator set to 28.5°C with a 14 h light/dark cycle. Animals were fed and observed daily, removing waste and changing over approximately half of the water for fresh fish system water. Fish were monitored for signs of distress and euthanized if they appeared moribund.
PACT clearing of juvenile zebrafish
Juvenile zebrafish were infected as described above. At 2 wpi, zebrafish were euthanized by tricaine overdose and PACT-mediated clearing was performed as previously described.113
Wound recruitment assays
Larval zebrafish at 3 dpf were anesthetized with tricaine (final concentration 0.016%) and the tail fin was amputated with a sterile no. 11 Miltex razor. For imaging, zebrafish were immobilized in 0.8% low-melt agarose (Fisher BP165) and imaged on an inverted Zeiss axio observer Z1 (20× objective, 0.645 μm/pixel) immediately for 90 min post-wounding.
Zebrafish drug treatment
Larval zebrafish were infected with M. marinum at 2 dpf as described above. At 3dpi, infected zebrafish were treated with 50 μM CK666 (Millipore Sigma) or 50 μM CK689 (Millipore Sigma) for 30 min in media supplemented with 0.5% DMSO. The treated zebrafish were then anesthetized with tricaine (final concentration 0.016%) and mounted in 0.8% low-melt agarose (Fisher BP165) supplemented with the appropriate drug. Zebrafish were imaged for up to 12 h post treatment with an X-Light V2 spinning disk confocal imaging system (Biovision). Migrating macrophages were tracked in ImageJ using the MTrackJ plugin.
Generation of transgenic constructs
To visualize zebrafish osteoclasts, primers (see methods table, acp5a -F, acp5a -R) containing XhoI (5′) and KpnI (3′) restriction digest sites were designed to target the 4047 base pair sequence immediately upstream of the tartrate-resistant acid phosphatase gene (acp5a) contained on the CH211–276A17 BAC clone obtained from the Children’s Hospital Oakland Research Institute (CHORI). The amplicon was cloned into p5E-MCS of the Gateway Cloning system using XhoI and KpnI restriction digest sites. The acp5a:m-NeonGreen-CAAX transgene was subsequently constructed by recombining p5E acp5a, pME mNeonGreen-CAAX and p3E polyA into pDestTol2pA2 to generate pDestTol2; acp5a:mNeonGreen-CAAX.
To generate entry clones for expression of mycobacterial proteins (EsxM, truncated EsxM [Q59X]) in zebrafish macrophages, primers targeting the gene sequences were flanked with attB1 and attB2 sites and recombined with BP clonase into the pDONR 221 entry clone (see methods table). The EsxB entry clone was derived from the Mtb ORFeome (BEI Resources). Macrophage expression constructs were subsequently constructed by recombining: p5E mfap4, pME EsxM, and p3E p2a-tdTomato into pDest-Tol2pA2 to generate pDestTol2; mfap4:esxM-p2a-tdTomato; p5E mfap4, pME EsxM (Q59X) and p3E p2a-tdTomato into pDest-Tol2pA2 to generate pDestTol2; mfap4:esxM(Q59X)-p2a-tdTomato; and p5E mfap4, pME EsxB, and p3E p2a-mNeonGreen into pDestTol2pA2 to generate pDestTol2; mfap4:esxB-p2a-mNeonGreen.
Generation of transgenic zebrafish lines
Transgenic zebrafish were generated via Tol2 transgenesis.114 Tol2 mRNA was generated from T3TS-Tol2115 using an mMessage mMachine kit (Life Technologies). Transgenes were assembled using Gateway Multisite cloning (Invitrogen) according to.116 Single-cell embryos were collected immediately after fertilization and injected with 1 nL of a transgenesis mixture of 25 ng/μL of Tol2 mRNA and 50 ng/μL transgenesis construct. Injected embryos were screened for fluorescence and raised to adulthood. Founders were identified and outcrossed to *AB zebrafish to establish transgenic lines.
RAW 264.7 transwell migration assays
RAW 264.7 macrophages were seeded in 60mm petri dishes to ~80% confluency. Cell concentration was determined, and cells split to seed two 60mm petri dishes with 106 cells/mL and incubated with fresh DMEM +10% FBS at 37°C with 5% CO2. After 18 h, one plate was infected with the M. tuberculosis double deletion auxotrophic strain mc26020 (ΔlysAΔpanCD)66 carrying the kanamycin-resistant msp12:mCerulean plasmid, and the other was infected with mc26020 containing the same plasmid with the addition of the hsp60 promoter driving expression of esxM. Each plate was infected at an MOI of 5 and media was supplemented with 24 μg/mL panthothenate and 80 μg/mL L-lysine. Infection occurred over 3 h at 37°C with 5% CO2. Media was aspirated off, and cells were washed with PBS. Fresh media containing pantothenate, L-lysine and 200 μg/mL gentamicin was added, and cells were incubated at 37°C with 5% CO2 for 1 h. Cells were starved in DMEM +1% FBS with 20 μg/mL gentamicin, pantothenate, and L-lysine and incubated at 37°C with 5% CO2 for 18 h. Cells were washed and dislodged with HyQTase (HyClone SV30030–01), and 1 × 106 cells were resuspended in serum-free DMEM with pantothenate and L-lysine into the upper chambers of 24-well transwell plates, with 600 μL of DMEM +10% FBS in the lower chambers. Cells were incubated at 37°C with 5% CO2. After 20 h of incubation, insert membranes containing migrated cells were fixed with 3.6% PFA and DAPI stained. Fixed membranes were imaged at 2.5× magnification and quantified for cell number using the Zen analysis suite.
RAW 264.7 transduction
The lentiviral vectors for stable macrophage expression of esxM-3X FLAG and cerulean-3X FLAG were generated through Gateway recombination. The primers targeting the gene sequences were flanked with attB1 and attB2 sites and the PCR amplified products were recombined with pDONR 211. Macrophage expression constructs were subsequently generated by recombining pDONR 221: esxM-3X FLAG/pDONR 221: cerulean-3X FLAG with pLX301. High titer lentivirus packed with the expression vectors were prepared and used to infect RAW 264.7 cells as described.117 RAW 264.7 cells transduced with pLX301: esxM-3X FLAG/pLX301:cerulean-3X FLAG were selected using 5 μg/mL puromycin.
THP-1 macrophage infection
THP-1 monocytes were cultured at 37°C in 5% CO2 in RPMI 1640 medium supplemented with 10% (v/v) Fetal bovine serum and 2 mM GlutaMAX (Gibco). For each experiment, four 10 cm culture dishes treated with 25ug/mL poly-D-Lysine were seeded with the THP-1 cells at a density of 2 × 107 cells per dish, differentiated with 50 ng/mL of phorbol-12-myristate13-acetate (PMA) for two days, and infected 24 h later. The differentiated cells were infected with the single cell suspension of WT M. marinum:msp12∷cerulean at a MOI of 1:4. After incubation with the bacteria for 4 h at 33°C in 5% CO2 in antibiotic-free complete RPMI media, cells were washed with 1X PBS and treated with 200 μg/mL gentamicin for 1 h to kill any remaining extracellular bacilli. The cells were then washed in 1X PBS and incubated with complete RPMI containing 20 μg/mL gentamicin at 33°C in 5% CO2. The cells were harvested at 2 dpi for total RNA isolation.
BLaER1 cell transdifferentiation and infection
BLaER1 cells, a kind gift from T. Graf (Barcelona), were cultured in complete RPMI-1640 medium (supplemented with 2mM L-Glutamine, 1mM Sodium Pyruvate, 100 U/mL Penicillin, 100 μg/mL Streptomycin, and 10% (v/v) heat-inactivated FCS) at 37°C/5%CO2. They were transdifferentiated in 48-well plates at a concentration of 2.5 × 105 cells/well by using the transdifferentiation media (10 ng/mL hrIL-3 (Peprotech 200–03), 10 ng/mL hr-M-CSF (Peprotech 300–25), 100 nM β-Estradiol (Sigma E2758) in complete RPMI medium) and incubating at 37°C/5%CO2 for 8 days. The transdifferentiated cells were infected with Mtb 6020 cerulean and Mtb 6020 esxM at MOI 5 and incubated at 37°C/5%CO2 for 18–20 h. The infected BLaER1 cells were detached from the 48-well plate using HyClone HyQTase (GE Healthcare Life Sciences SV30030.01), spun down, resuspended in complete RPMI and placed on collagen-coated coverslips in 24-well plates. The immunofluorescence assay was performed using Anti-Arp2 antibody (Abcam, ab49674) as primary antibody and goat Anti-mouse Alexa Fluor 647 (Thermo Fisher, A21236) as a secondary antibody plus phalloidin stain (Alexa Fluor 555, Thermo Fisher A34055). Coverslips were mounted on slides using Mowiol and images were taken with a Zeiss 880 AiryScan or with a Biovision spinning disk confocal microscope. Analysis of the images was performed using FIJI/ImageJ.
Scanning electron microscopy
BLaER1 cells were transdifferentiated in response to 17β-estradiol in the presence of a cytokine mix (hrIL-3 and hr-M-CSF-1) in 48-well plates for 8 days at 37°C/5%CO2. Then the cells were infected with either Mtb 6020 msp12∷cerulean or Mtb 6020 msp12∷cerulean/hsp60∷esxM at an MOI 5, at 37°C/5%CO2 for 24h. After that the cells were detached with HyQtase, washed and seeded on 12mm round collagen coated coverslips in a 24-well plate and incubated for 37°C/5%CO2 for an additional 24h. Cells were fixed with 2.5% glutaraldehyde/0.15M sodium phosphate buffer (pH7.4) at room temperature then processed as described in.69
BSL3 infections and analysis using M. tuberculosis NCG and H37Rv strains
Wild type BLaER1 cells were cultured in complete RPMI-1640 medium at 37°C/5%CO2. RPMI 1640 medium supplemented with 2mM L-Glutamine, 1mM Sodium Pyruvate, and 10% (v/v) heat-inactivated FCS. They were transdifferentiated in Poly-D-Lysine (25ug/ml) treated Tissue Culture Slides (Mat Tek CCS-4) at a concentration of 2.5 × 105 cells/chamber by using the transdifferentiation media (10 ng/ml hrIL-3 (Peprotech 200–03), 10 ng/ml hr-M-CSF (Peprotech 300–25), 100 nM β-Estradiol (Sigma E2758) in complete RPMI medium) and incubating at 37°C/5%CO2/7 days. The transdifferentiated cells were infected with Mtb NCG msp12:cerulean and H37Rv msp12:cerulean at MOI 10, incubated at 37°C/5%CO2/72h and fixed under BSL3 conditions and then stained with Phalloidin (Alexa Fluor Plus 555 A30106). Coverslips were mounted on slides by using Prolong Gold Anti-fade mounting solution (Invitrogen P36934) and images were taken with spinning disk confocal microscopy. Analysis of the images was performed using Fiji.
Bacterial strains
M. marinum strain containing msp12:tdTomato98 was a kind gift from Lalita Ramakrishnan (University of Cambridge). The msp12∷cerulean M. marinum strain has been previously published.97 All strains were grown in either Middlebrook 7H9 media or Middlebrook 7H10 plates supplemented with OADC (10%) and 50 μg/mL hygromycin and 25 μg/mL kanamycin. The recombinant M. tuberculosis double deletion auxotrophic strains (ΔlysAΔpanCD) Mtb6020 msp12∷cerulean and Mtb6020 hsp60p:esxM/msp12∷cerulean were grown in Middlebrook 7H9 media or Middlebrook 7H10 plates supplemented with OADC (10%), 50 μg/mL hygromycin, 25 μg/mL kanamycin, 24 μg/mL panthothenate and 80 μg/mL L-lysine as described.66 For consistency, single use, frozen aliquots for both the M. marinum and Mtb strains were prepared for infection as described in.112 All Mtb strains used for mouse infections (H37Rv or NCG background) were grown in Middlebrook 7H9 medium containing oleic acid-albumin-dextrose-catalase (OADC), 0.2% glycerol, and 0.05% Tween 80 to log-phase with shaking (200 rpm) at 37°C. Prior to all in vivo infections, cultures were washed, resuspended in PBS (PBS) containing 0.05% Tween 80, and sonicated before diluting to desired concentration (see below).
Construction of plasmids
A kanamycin-resistant plasmid containing the msp12 promoter sequence driving the Cerulean fluorescent protein was constructed for transformation into the hygromycin-resistant ΔesxM M. marinum mutant. The hygromycin-resistance gene was excised from the msp12∷cerulean plasmid that has been previously published (Oehlers et al. 2015) and replaced with the aph gene for kanamycin resistance. The kanamycin-resistant msp12∷cerulean plasmid was amplified using the msp12:cerulean_KanR-F and msp12:cerulean_KanR-R primers listed in the methods table to complement the ΔesxM M. marinum mutant via In-Fusion Cloning (Clontech). Primers were designed to amplify the hsp60 promoter sequence immediately upstream of esxM with a C-terminal HA-tag. Primer sequences used are displayed in Table S5. The two PCR products were joined by In-Fusion cloning.
Generating the esxM deletion in M. marinum
An esxM gene deletion was generated in an M. marinum M strain by using the ORBIT system.118 In this system an esxM gene targeting oligonucleotide and a payload plasmid were cotransformed into a RecT- and Int-expressing M. marinum strain (Mm:pkM444, Addgene #108319) via homologous recombination. A culture of M. marinum containing the pKM444 Kanr plasmid was started overnight by adding 100–150ul of fresh saturated stock culture in 30mL 7H9 + 10% OADC +0.05% Tween-80 + 0.2% glycerol +25ug/mL Kanamycin in a 125mL flask at 33°C/130rpm/24–48h. At an OD600 ~ 0.5, anhydrotetracycline (ATc) was added at a final concentration of 500 ng/mL and incubated again for 8h, then 3mL of 2M Glycine was added to the culture and incubated overnight (19.5h in total after ATc induction). Bacterial cells were spun down in 50mL conical tubes at 4000 rpm/10min/RT and the supernatant was discarded. The pellet was gently resuspended in 2mL of 10% glycerol and then 10% glycerol was added up to 30 mL. Cells were mixed by inverting the tube and spun at 4000 rpm/10min/RT. After the 10% glycerol wash and spin steps were repeated, cells were resus-pended in 2 mL 10% glycerol and aliquoted in sterile Eppendorf tubes.
Mm:pkM444 electrocompetent aliquots (380ul) were combined with 1mg of the attP-containing oligonucleotide targeting esxM gene and 200ng of an attB-containing plasmid (pK464 payload plasmid, Addgene #108322). The cells and DNA were mixed by pipetting and transferred to ice-cooled electroporation cuvettes (0.2cm path length). The cells were electroporated with a gene Pulser Xcell set for 25msec (2.5kV, 650Ω and 50uF). After electroporation, the cells were suspended in 900ul of 7H9 without antibiotics at room temperature and up to 2mL in T25 flasks and incubated overnight at 33°C/130rpm. The next day, two 0.5 mL aliquots of the culture were spread on 7H10 plates with 50ug/mL Hygromycin and incubated at 33°C/14 days.
The recombinant candidate colonies were picked and streaked on fresh 7H10 plates with 50ug/mL Hygromycin. The deletion was confirmed by PCR analysis and sequencing. An aliquot of Mm:pKM444 electrocompetent cells with 200ng of the payload plasmid pKM464 but with no oligomer was used as a negative control. The positive control consisted in Mm:pKM444 electrocompetent cells with 200ng of the pKM464 plasmid and the rpsL 70mer used to confirm RecT expression and conferring streptomycin resistance.
Protein secretion assays
Protein secretion assays were performed as described previously.119 Briefly, M. marinum strains were grown in 7H9 (Middlebrook) broth supplemented with 0.1% Tween 80 and 20 μg/mL Kanamycin to retain the mCerulean plasmid during in vitro growth. Cultures were grown to saturation, and sub-cultured to an OD600 of 0.8 into 50mL of Sauton’s broth supplemented with Kanamycin and 0.01% Tween 80. After 48 h, the M. marinum cells were collected by centrifugation (pellet), and the spent culture media (supernatant) was collected via filtration. The culture supernatant was filtered through 0.2μm Nalgene Stericup with polyethersulfone (PES) filters and 500μL of phenylmethylsulfonyl fluoride (PMSF) at a concentration of 174.1 μg/mL. Supernatants were concentrated 50- to 100-fold using a 3kDa molecular weight cut-off Amicon filter (Millipore). The M. marinum cells were resuspended in 500μL of PBS with PMSF at a concentration of 174.2 μg/mL and lysed using a bead beater (BioSpec). The resulting lysate was clarified by centrifugation. The protein concentrations of the resulting pellet and supernatant fractions were measured using a Micro BCA assay (Pierce).
Proteomics
Short term culture filtrates (described above) of WT:msp12:cerulean, ΔEsxM:msp12:cerulean, and ΔEsxM:msp12:cerulean:H-sp60:esxM were digested with trypsin for proteomics analysis as in.119 Briefly, 100 μg of protein was denatured with 5% SDS, alkylated with iodoacetamide and digested with trypsin using S-Trap reactors (Protifi, NY) according to manufacturer’s instructions. Following digestion, peptides were desalted and dried down prior to nano LC-MS/MS analysis as described.
Samples were resuspended in 0.1% formic acid and water, to 1 mg/mL concentration. 100ng of each sample was injected in triplicate into a Bruker nanoElute and timsTOF Pro LC-MS system. 90-min 600 nL/min gradients were used on a 75 μm × 100 mm PepSep column with C18 ReproSil AQ stationary phase at 1.9μm particle size, 120Å pore size. nano-ESI was used as the method of ionization, with a spray voltage of 1700V. MS was set to Parallel accumulation, serial fragmentation Data Dependent Mode (PASEF-DDA) with a mass range of 100–1700 m/z, ion mobility range of 0.6–1.6 v*s/cm2, and ramp and accumulation times of 100ms. Each precursor consisted of 10 PASEF ramps for a cycle time of 1.17 s. Precursors were filtered to contain only charges from 2 to 5. MS/MS collision energy settings were set to ramp from 20 eV at 0.6 ion mobility to 70 eV at 1.6 ion mobility. Instrument tune parameters were set to default for proteomic studies with the following differences: quadrupole low mass set to 20 m/z, focus pre-TOF pre-pulse storage set to 5μs.120
Raw ‘.d’ files were subjected to peptide spectral mass matching using MaxQuant and quantification was performed using Label Free Quantification within MaxQuant as in.121 The most current M. marinum FASTA was downloaded from Uniprot and data were filtered to a false discovery rate of 0.01 (1%). Data were normalized to mCerulean and MPT64 to control for cell lysis and sample preparation variances as in.121 Data were performed in analytical triplicate and biological replicate.
RNA isolation and quantitative RT-PCR analysis
WT M. marinum grown in Middlebrook 7H9 media or Pi low Sauton’s media (described elsewhere) was harvested at an OD600 of 1 and the pellets were resuspended in RLT plus buffer supplied with the RNeasy Plus kit (QIAGEN). The suspended bacterial pellets were then lysed using 0.7 mm zirconia beads (BioSpec Products) in a BeadBug homogenizer (Benchmark Scientific) at 4000 RPM for 35 s and this process was repeated three more times. Bacterial RNA was then isolated using the RNeasy Plus kit by following the manufacturer’s protocol. A similar protocol was followed for the lysis and isolation of total RNA from the WT M. marinum-infected THP-1 Monolayers and zebrafish larvae at 2 dpi & 4 dpi respectively. Total RNA was isolated from 50 fish for each experiment. Each fish was infected with a starting dose of ~250 bacilli. Mtb H37Rv and NCG grown in Middlebrook 7H9 media were harvested at an OD600 of 1 and the pellets were resuspended in TRIzol reagent (Invitrogen). The suspended bacterial pellets were then lysed using 0.7 mm zirconia beads as described previously. Subsequently, bacterial RNA was isolated using the Direct-zol RNA Miniprep kit (Zymo Research) and treated with RNAse-free DNAse I (NEB). Mtb H37Rv and NCG infected mouse lung tissues were harvested at 6 wpi in TRIzol reagent (described elsewhere), and the tissues were initially disrupted in 2 mm zirconia beads followed by the bacterial lysis using 0.7 mm zirconia beads. Total RNA was then isolated using the Direct-zol RNA Miniprep kit as described above.
For in vitro grown cultures, 1 μg of RNA was converted into cDNA using the LunaScript RT Super-Mix Kit (NEB). Appropriate no reverse transcriptase controls were included to check for potential DNA contamination. 1 μL of 1:10 diluted cDNA was used as a template for each reaction of quantitative RT-PCR using the Luna Universal qPCR Master Mix (NEB). For in vivo infections, total RNA was converted into cDNA as described above and 4 μL of undiluted cDNA was used as a template for each reaction of quantitative RT-PCR. Primers were designed to generate 150 bp – 190 bp amplicons. Due to the very close homology between the esxM paralogs of M. marinum and Mtb, forward primers were designed to anneal to the divergent 5′ UTR of their transcripts (Table S5) to differentiate among these paralogs.57,122 Transcript levels were normalized to sigA for M. marinum and esxM for Mtb.
Protein secretion assay and western blotting
Cultures were initially grown in Middlebrook 7H9 media to log phase and then washed and grown in complete Sauton’s media (30 mM DL-asparagine, 7 mM sodium citrate, 3mM potassium phosphate dibasic (Pi), 4 mM magnesium sulfate, 0.2 mM ferric ammonium citrate, 5.7 μM zinc sulfate, and 4.8% glycerol adjusted to a pH of 7.4) with 0.05% tyloxapol. For experiments performed in Pi-normal condition, bacteria were washed and resuspended in 200 mL of detergent-free complete Sauton’s media at an initial OD600 of 0.5 and incubated with shaking at 130 RPM for five days at 33°C to an OD600 of 0.8–1.0.123 For Pi-low conditions, bacteria were initially cultured in modified Sauton’s media containing 250 μM potassium phosphate dibasic with 0.05% tyloxapol and then washed and grown in 200 mL of detergent-free Sauton’s media containing 2.5 μM potassium phosphate dibasic for 7 days60 Cultures were then centrifuged (3000 × g for 20 min) and culture supernatants and pellets were collected. Culture supernatants were double filtered with Steriflip-GP 0.22 μm filters (EMD Millipore) and concentrated ~100–fold using Centricon Plus – 70, 3 kDa molecular weight cut-off centrifugal filters (Millipore). Proteins from the culture supernatants were then precipitated using 10% trichloroacetic acid overnight at 4°C and the pellets were washed twice with ice-cold acetone at maximum RPM. The pellets were then resuspended in 250 μL of extraction buffer (50 mM Tris-Cl – pH 7.5, 6% SDS, and 5 mM EDTA) with cOmplete EDTA-free protease inhibitor cocktail (Sigma-Aldrich). For cell lysate preparation, bacterial pellets were dissolved in 2 mL of extraction buffer with the protease inhibitor cocktail and lysed using 0.7 mm zirconia beads (BioSpec Products). The lysate was then separated from the cellular debris by centrifugation at maximum RPM. The culture filtrate and cell lysate protein concentrations were quantified using the DC protein assay kit (Bio-Rad).
For immunoblot analysis, 30 μg of culture filtrates and cell lysates were subjected to SDS-PAGE and transferred to the PVDF membrane (Bio-Rad). The membranes were probed with rabbit α-HA-Tag (C29F4, Cell Signaling Technology, 1:1000 dilution) and mouse α-RNAP (8RB13, BioLegend, 1:5000 dilution) overnight at 4°C followed by 1 h incubation at room temperature with appropriate secondary antibodies (goat-anti-rabbit (1:5000 dilution), goat-anti-mouse (1:2500 dilution) conjugated with HRP, Thermo Scientific). After brief incubation with Super-Signal West Pico PLUS (Thermo Scientific) or Super-Signal West Femto maximum sensitivity Substrate (1:3 dilution, Thermo Scientific), the blots were exposed to Carestream Kodak Biomax MR film (Sigma-Aldrich) for signal detection.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA sequencing of M. tuberculosis bacterial transcripts
Mtb H37Rv and NCG grown in Middlebrook 7H9 media were harvested at an OD600 of 1 and the pellets were resuspended in TRIzol reagent (Invitrogen). The suspended bacterial pellets were then lysed using 0.7 mm zirconia beads as described previously. Subsequently, bacterial RNA was isolated using the Direct-zol RNA Miniprep kit (Zymo Research) and treated with RNAse-free DNAse I (NEB). Library prep using Illumina Stranded Total RNA Prep Ligation with Ribo-Zero Plus (Illumina) was performed by the Duke Center for Genomic and Computational Biology (GCB). The prepared libraries were subsequently sequencing using the NextSeq 500 High-Output platform to generate 75 bp single-end reads.
Transcript abundances were quantified using Kallisto v0.48113 in single-end mode, with an estimated fragment length of 250 and a SD of 35, and sequence based bias correction. H37Rv cDNA was used as the index for pseudoalignment. Visualization of the data was done in RStudio using Sleuth v0.30.0.110 Raw reads deposited at NCBI in Bioproject PRJNA872173.
Genome assembly and variant calling
The NCG isolate was sequenced on Illumina HiSeq to >500X coverage. Reads are available to download under BioProject ID PRJNA540867. 50 base pair paired-end fastq sequence reads were aligned against the H37Rv reference genome using BWA.101 Variants were called with SAMtools and filtered with VarScan for a minimum read depth of 10, a consensus quality score of 20, and a minimum variant frequency of 0.75.111,109 SNPs adjacent to indels were discarded. Drug resistance-associated mutations and genes in repetitive regions (PE/PPE and PGRS) of the genome were discarded, resulting in 31,839 SNPs used for phylogenetic analysis. For lineage analysis, we included SNPs from these regions (35,787 total) since a portion likely represent true variation among global strains.
The reads aligned to esxM were inspected manually for every strain included in the initial analysis. Furthermore, the 27 base pair sequence that includes the invariant sequence 10 bp upstream and 14 bp downstream of the TAG stop codon in esxM was searched in the fastq files of every isolate included in the initial analysis shown in Figure 2, where the sequence was only found in isolates from the so-called “modern” lineages.
Phylogenetic methods
All trees were based on genome-wide SNPs derived according to the parameters specified above. A superset of SNPs was constructed for each strain with reference alleles occupying sites for which no variants were detected using custom Perl scripts. These SNPs informed neighbor-joining and maximum-likelihood methods of phylogeny construction. Neighbor-joining methodology was implemented with ClustalW2 using pairwise similarity scores of SNP supersets as a measure of genetic distance.124 A maximum-likelihood phylogeny was generated with RAxML using a GTR model of nucleotide substitution.108 For each method, 1000 bootstrap replicates provided support for nodes on the tree. Trees were visualized with FigTree (tree.bio.ed.ac.uk/software/figtree).
When replicating the phylogenetic findings on larger datasets and estimating the selective pressure acting on these genes, we utilized two previously published datasets from Vietnam and Birmingham, UK.57,58 We mapped the sequencing reads to the H37Rv reference genome with BWA.101 Similar to the original analysis, we then used SAMtools109 and VarScan111 to call substitutions. We removed libraries where there was support for more than one allele to be called present at the early stop codon in esxM, or if there was insufficient support to call either allele as present. We also removed five libraries that did not have both reads available, that could not be confidently assigned to a lineage, or that had an excess of sites that could not be confidently called (SAMN26038068, SAMN26038018, ERR9117709, ERR9117721, SAMN07658239). These filters removed 52 libraries from the UK dataset and none from the Vietnam set. We then used an alignment of only variant sites to construct an approximately maximum likelihood tree.125 We then used the RELAX hypothesis testing framework126 to calculate the dN/dS rate ratios for both ancestral and modern lineages, as well as assess if there had been a relaxation, or intensification, of the selective pressure in “modern” lineage strains compared to “ancestral” lineage strains. During this hypothesis testing we followed the guidance of the software to remove identical sequences for computational efficiency. We visualized the phylogenies with iTOL.127
In silico analyses
Multiple Sequence Alignments were performed using Clustal Omega128 and the aligned sequences were visualized using Jalview v2.11.2.0.129
Microscopy
Conventional and time-lapse fluorescent microscopy was carried out on a Zeiss Axio Observer Z1 inverted microscope with an Xcite 120Q light source (Lumen Dynamics) and an MRm camera (Zeiss). Imaging of PACT-cleared juvenile zebrafish was performed on an Andor XD spinning disk confocal microscope. Time-lapse and other confocal spinning disk microscopy was performed on a Zeiss Axio Observer Z1 inverted microscope with an X-Light v2 (Crest Optics) imaging system. Images were processed with ImageJ.
Fiji/ImageJ
Infection burden was measured as the number of pixels above background using a constant threshold in ImageJ as described in.112 Dissemination above the midline was defined as colocalization of M. marinum and macrophages above the midline at 5 dpi. Following PACT treatment of juvenile zebrafish,113 animals were scored for dissemination by scanning the spine for fluorescent M. marinum. Macrophage migration following partial tail transection was tracked in ImageJ using the MTrackJ plugin. Quantitation of cell culture BLaER1 results was performed using automated plugin “Analyze Particles …” for Circularity as well as blinded analysis of filopodia frequency.
For Arpc2 cellular distribution analysis, cells were analyzed in ImageJ using in-house Jython scripts. Briefly, macrophage differentiated BLaER1 cells were infected with M. tuberculosis mc26020 expressing either full-length EsxM or the mCerulean fluorescent protein. These were imaged singly at 100×, maximum intensity projected, and then a cellular outline is calculated from a channel that is well-distributed across the cell body (in this case, β-actin). This outline is then used to mask the cell and clear any external background. These cells are then visualized in the 3D Surface Plot plugin in FIJI/ImageJ, where the longest X axis is identified and the cells arranged in the XZ orientation along that axis. Smoothing is applied (value = 8.5 in this analysis) and all axes are removed and the background is changed to black before exporting to a static 2D image, which is then used for quantitation by scanning for >0 pixels on a binarized image and writing those coordinates to a.csv, which can then be further analyzed in R. The analysis in R consists of identifying the maximum Y coordinate at each pixel and using that to calculate the area under the curve for each cell across a defined interval of the cell (in percent X distance). Here, our analysis compared the edge 25% of the AUC for each condition, normalized to the AUC for that cell in the middle 50% (25–75%). This allows comparison of the distal/central fluorescence intensity ratio for each cell, which is then plotted in ggplot2. The script allows for arbitrary numbers of cells to be analyzed and compared at once, provided each cell is given a discrete identifier.
Cells were maximum-intensity projected and then the isolated using the “Analyze Particles …” function. The area outside of the cell was cleared and then the cells were visualized individually using “3D Surface Plot.” These plots were used to identify the longest axis along the cell and then exported to black background RGB images that were then binarized in 8-bit and each data-containing pixel was captured to .csv files. These files were then processed in R to identify the maximum outline of the curves and then the areas under the curve were calculated for each individual cell for a given interval, which were then compared using a t test.
Mass spectrometry statistical analysis
Fold-change data were determined by the ratio of the average of triplicate measurements from the LFQ proteomics data (Table S4). % Coefficient of variation were determined. The propagated average CV for the entire dataset was 8.04% and 8.02% for ΔesxM/WT and Complemented/WT respectively. In cases where protein abundance was significantly determined in one sample (eg WT) but missing in another we limited these values to 2′8 fold-change, the approximate dynamic range of the experiment. Significance was determined using Welsh’s modified T test; multiple testing was performed using the Benjamini-Hochberg with an alpha of 0.05. Corrected critical vales were p = 0.0164, and 0.0200 for ΔesxM/WT and Complemented/WT respectively. Samples were performed in biological duplicate.
Statistics
Statistical tests utilized for each experiment are indicated in the figure legends. All statistical analyses were performed using Prism (GraphPad Software) or using R with specified packages when specifically noted.
| Software | |
|---|---|
| Name, Version | Citation |
| BWA | Li and Durbin101 |
| ClustalW2 | Larkin et al.102 |
| FIJI/ImageJ2, 2.5.0 | Schindelin et al.104Rueden et al.103 |
| iTol | Letunic and Bork105 |
| Kallisto v0.48 | Bray et al.106 |
| MtrackJ, 1.5.1 | Meijering et al.107 |
| R studio | RStudio Team (2022). RStudio: Integrated Development Environment for R. |
| RAxML | Stamatakis108 |
| SAMtools | Li et al.109 |
| Sleuth v0.30.0 | Pimentel et al.110 |
| VarScan | Koboldt et al.111 |
Supplementary Material
Highlights.
High rates of disseminated and skeletal disease during a tuberculosis outbreak
Lineage 1 strains of Mycobacterium tuberculosis carry the ancestral version of EsxM
Ancestral, full-length EsxM alters the macrophage cytoskeleton and enhances motility
Ancestral EsxM promotes granuloma efflux, dissemination, and bone disease
ACKNOWLEDGMENTS
We thank R. Asrican, R. Beerman, M. Braunstein, L. Cameron, L. Dolat, Y. Gao, S. Harris, E. Hunt, R. Meade, S. Miller, K. Murphy, C. Pyle, R. Vancini, and C. Xander for experimental advice and assistance; E. Hunt, I. Padmanaban, and A. Yu for zebrafish care; T. Graf for BLaER1 cells; M. Bagnat, J. Bear, T. Ioerger, D. Pickup, M. Welch, and members of the Tobin laboratory for helpful discussions; and M. Cronan and L. Dolat for comments on the manuscript. Graphical abstract created in part with BioRender. This work was funded by National Institutes of Health grants AI125517 (D.M.T., J.E.S., and S.L.), AI130236, AI127115 (D.M.T), AI142127, AI149147, AI106872 (P.A.C.), White-head Scholar Awards (C.B.L. and C.M.S), an NIH Director’s New Innovator Award 1DP2-GM146458-01 (C.M.S.), and a Vallee Scholar Award (D.M.T.). Biocontainment work performed in the Duke Regional Biocontainment Laboratory received partial support for construction from NIAID (UC6-AI058607).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.10.019.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.World Health Organization (2021). Global Tuberculosis Report 2021 (World Health Organization). https://www.who.int/publications/i/item/9789240037021.
- 2.Centers for Disease Control and Prevention (CDC) (2021). Reported Tuberculosis in the United States, 2020 (CDC: US Department of Health and Human Services; ). [Google Scholar]
- 3.Crubezy E, Ludes B, Poveda JD, Clayton J, Crouau-Roy B, and Montagnon D (1998). Identification of Mycobacterium DNA in an Egyptian Pott’s disease of 5, 400 years old. Comptes rendus de l’Academie des sciences. Serie III, Sciences de la vie 321, 941–951. 10.1016/s0764-4469(99)80009-2. [DOI] [PubMed] [Google Scholar]
- 4.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, et al. (2002). A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99, 3684–3689. 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orgeur M, and Brosch R (2018). Evolution of virulence in the Mycobacterium tuberculosis complex. Curr. Opin. Microbiol 41, 68–75. 10.1016/j.mib.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 6.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, et al. (2013). Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet 45, 1176–1182. 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepperell CS (2022). Evolution of Tuberculosis Pathogenesis. Annu. Rev. Microbiol 76, 661–680. 10.1146/annurev-micro-121321-093031. [DOI] [PubMed] [Google Scholar]
- 8.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, et al. (2013). Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet 45, 172–179. 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagneux S, and Small PM (2007). Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis 7, 328–337. 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 10.Bottai D, Frigui W, Sayes F, Di Luca M, Spadoni D, Pawlik A, Zoppo M, Orgeur M, Khanna V, Hardy D, et al. (2020). TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat. Commun 11, 684. 10.1038/s41467-020-14508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill MB, Shockey A, Zarley A, Aylward W, Eldholm V, Kitchen A, and Pepperell CS (2019). Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia. Mol. Ecol 28, 3241–3256. 10.1111/mec.15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnan N, Malaga W, Constant P, Caws M, Thi Hoang Chau T, Salmons J, Thi Ngoc Lan N, Bang ND, Daffe M, Young DB, et al. (2011). Mycobacterium tuberculosis lineage influences innate immune response and virulence and is associated with distinct cell envelope lipid profiles. PLoS One 6, e23870. 10.1371/journal.pone.0023870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portevin D, Gagneux S, Comas I, and Young D (2011). Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 7, e1001307. 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiens KE, and Ernst JD (2016). The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent. PLoS Pathog. 12, e1005809. 10.1371/journal.ppat.1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saelens JW, Viswanathan G, and Tobin DM (2019). Mycobacterial Evolution Intersects With Host Tolerance. Front. Immunol 10, 528. 10.3389/fimmu.2019.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst JD (2012). The immunological life cycle of tuberculosis. Nat. Rev. Immunol 12, 581–591. 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 17.Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, Sherman DR, Gerner MY, and Urdahl KB (2018). Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe 24, 439–446.e4. e434. 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Divangahi M, Behar SM, and Remold H (2013). Dying to live: how the death modality of the infected macrophage affects immunity to tuberculosis. Adv. Exp. Med. Biol 783, 103–120. 10.1007/978-1-4614-6111-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amaral EP, Costa DL, Namasivayam S, Riteau N, Kamenyeva O, Mittereder L, Mayer-Barber KD, Andrade BB, and Sher A (2019). A major role for ferroptosis in Mycobacterium tuberculosis-induced cell death and tissue necrosis. J. Exp. Med 216, 556–570. 10.1084/jem.20181776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, and Ramakrishnan L (2007). Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe 2, 29–39. 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronan M, Beerman R, Rosenberg A, Saelens J, Johnson M, Oehlers S, Sisk D, Jurcic Smith K, Medvitz N, Miller S, et al. (2016). Macrophage Epithelial Reprogramming Underlies Mycobacterial Granuloma Formation and Promotes Infection. Immunity 45, 861–876. 10.1016/j.immuni.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cronan MR, Hughes EJ, Brewer WJ, Viswanathan G, Hunt EG, Singh B, Mehra S, Oehlers SH, Gregory SG, Kaushal D, and Tobin DM (2021). A non-canonical type 2 immune response coordinates tuberculous granuloma formation and epithelialization. Cell 184, 1757–1774.e14. e1714. 10.1016/j.cell.2021.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis JM, and Ramakrishnan L (2009). The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136, 37–49. 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin PL, Ford CB, Coleman MT, Myers AJ, Gawande R, Ioerger T, Sacchettini J, Fortune SM, and Flynn JL (2014). Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med 20, 75–79. 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, and Small PM (1999). Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284, 1520–1523. 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 26.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et al. (2003). The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 100, 12420–12425. 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, and Stover CK (1996). Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol 178, 1274–1282. 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pym AS, Brodin P, Brosch R, Huerre M, and Cole ST (2002). Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol 46, 709–717. 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 29.Lewis K, Liao R, Guinn K, Hickey M, Smith S, Behr M, and Sherman D (2003). Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis 187, 117–123. 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley SA, Raghavan S, Hwang WW, and Cox JS (2003). Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA 100, 13001–13006. 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augenstreich J, Arbues A, Simeone R, Haanappel E, Wegener A, Sayes F, Le Chevalier F, Chalut C, Malaga W, Guilhot C, et al. (2017). ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 19, e12726. 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- 32.Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, Hopkinson-Woolley D, Brosch R, and Ramakrishnan L (2017). Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. USA 114, 1371–1376. 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manzanillo P, Shiloh M, Portnoy D, and Cox J (2012). Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11, 469–480. 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, and Enninga J (2012). Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8, e1002507. 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groschel MI, Sayes F, Simeone R, Majlessi L, and Brosch R (2016). ESX secretion systems: mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol 14, 677–691. 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 36.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, and Rubin EJ (2009). Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. USA 106, 18792–18797. 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serafini A, Boldrin F, Palu G, and Manganelli R (2009). Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J. Bacteriol 191, 6340–6344. 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittal E, Skowyra ML, Uwase G, Tinaztepe E, Mehra A, Koster S, et al. (2018). Mycobacterium tuberculosis Type VII Secretion System Effectors Differentially Impact the ESCRT Endomembrane Damage Response. mBio 9, e01765–18. 10.1128/mBio.01765-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portal-Celhay C, Tufariello JM, Srivastava S, Zahra A, Klevorn T, Grace PS, Mehra A, Park HS, Ernst JD, Jacobs WR Jr., and Philips JA (2016). Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4(+) T-cell activation. Nat Microbiol 2, 16232. 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehra A, Zahra A, Thompson V, Sirisaengtaksin N, Wells A, Porto M, Koster S, Penberthy K, Kubota Y, Dricot A, et al. (2013). Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog. 9, e1003734. 10.1371/journal.ppat.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah S, and Briken V (2016). Modular Organization of the ESX-5 Secretion System in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol 6, 49. 10.3389/fcimb.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, and Beyers AD (2001). The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome biology 2. RESEARCH0044. 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ates LS, Ummels R, Commandeur S, van der Weerd R, Sparrius M, Weerdenburg E, Alber M, Kalscheuer R, Piersma SR, Abdallah AM, et al. (2015). Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria. PLoS Genet. 11, e1005190. 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jimenez C, Parra M, Cadieux N, Brennan MJ, Appelmelk BJ, and Bitter W (2009). PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol. Microbiol 73, 329–340. 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 45.Izquierdo Lafuente B, Ummels R, Kuijl C, Bitter W, and Speer A (2021). Mycobacterium tuberculosis Toxin CpnT Is an ESX-5 Substrate and Requires Three Type VII Secretion Systems for Intracellular Secretion. mBio 12. 10.1128/mBio.02983-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Person AK, Goswami ND, Bissette DJ, Turner DS, Baker AV, Gadkowski LB, Naggie S, Erlandson K, Chen L, Lalani T, et al. (2010). Pairing QuantiFERON gold in-tube with opt-out HIV testing in a tuberculosis contact investigation in the Southeastern United States. AIDS patient care and STDs 24, 539–543. 10.1089/apc.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, and Gagneux S (2008). High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6, e311. 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, and Gagneux S (2010). Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet 42, 498–503. 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caws M, Thwaites G, Dunstan S, Hawn TR, Thi Ngoc Lan N, Thuong NTT, Stepniewska K, Huyen MNT, Bang ND, Huu Loc T, et al. (2008). The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 4, e1000034. 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Click ES, Moonan PK, Winston CA, Cowan LS, and Oeltmann JE (2012). Relationship between Mycobacterium tuberculosis Phylogenetic Lineage and Clinical Site of Tuberculosis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 54, 211–219. 10.1093/cid/cir788. [DOI] [PubMed] [Google Scholar]
- 51.Pareek M, Evans J, Innes J, Smith G, Hingley-Wilson S, Lougheed KE, Sridhar S, Dedicoat M, Hawkey P, and Lalvani A (2013). Ethnicity and mycobacterial lineage as determinants of tuberculosis disease phenotype. Thorax 68, 221–229. 10.1136/thoraxjnl-2012-201824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seraphin MN, Doggett R, Johnston L, Zabala J, Gerace AM, and Lauzardo M (2017). Association between Mycobacterium tuberculosis lineage and site of disease in Florida, 2009–2015. journal of molecular epidemiology and evolutionary genetics in infectious diseases 55, 366–371. 10.1016/j.meegid.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Comas I, Hailu E, Kiros T, Bekele S, Mekonnen W, Gumi B, Tschopp R, Ameni G, Hewinson RG, Robertson BD, et al. (2015). Population Genomics of Mycobacterium tuberculosis in Ethiopia Contradicts the Virgin Soil Hypothesis for Human Tuberculosis in Sub-Saharan Africa. Curr. Biol. : CB 25, 3260–3266. 10.1016/j.cub.2015.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saelens JW, Lau-Bonilla D, Moller A, Medina N, Guzman B, Calderon M, Herrera R, Sisk DM, Xet-Mull AM, Stout JE, et al. (2015). Whole genome sequencing identifies circulating Beijing-lineage Mycobacterium tuberculosis strains in Guatemala and an associated urban outbreak. Tuberculosis 95, 810–816. 10.1016/j.tube.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CMJE, Luirink J, and Bitter W (2012). General secretion signal for the mycobacterial type VII secretion pathway. Proc. Natl. Acad. Sci. USA 109, 11342–11347. 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Champion PAD, Stanley SA, Champion MM, Brown EJ, and Cox JS (2006). C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313, 1632–1636. 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- 57.Holt KE, McAdam P, Thai PVK, Thuong NTT, Ha DTM, Lan NN, Lan NH, Nhu NTQ, Hai HT, Ha VTN, et al. (2018). Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat. Genet 50, 849–856. 10.1038/s41588-018-0117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker TM, Choisy M, Dedicoat M, Drennan PG, Wyllie D, Yang-Turner F, Crook DW, Robinson ER, Walker AS, Smith EG, and Peto TE (2022). Mycobacterium tuberculosis transmission in Birmingham, UK, 2009–19: An observational study. Lancet Reg Health Eur 17, 100361. 10.1016/j.lanepe.2022.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uplekar S, Heym B, Friocourt V, Rougemont J, and Cole ST (2011). Comparative genomics of Esx genes from clinical isolates of Mycobacterium tuberculosis provides evidence for gene conversion and epitope variation. Infect. Immun 79, 4042–4049. 10.1128/IAI.05344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elliott SR, and Tischler AD (2016). Phosphate starvation: a novel signal that triggers ESX-5 secretion in Mycobacterium tuberculosis. Mol. Microbiol 100, 510–526. 10.1111/mmi.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah S, Cannon JR, Fenselau C, and Briken V (2015). A Duplicated ESAT-6 Region of ESX-5 Is Involved in Protein Export and Virulence of Mycobacteria. Infect. Immun 83, 4349–4361. 10.1128/IAI.00827-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, et al. (2008). Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18, 729–741. 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ates LS, van der Woude AD, Bestebroer J, van Stempvoort G, Musters RJP, Garcia-Vallejo JJ, Picavet DI, Weerd R.v. d., Maletta M, Kuijl CP, et al. (2016). The ESX-5 System of Pathogenic Mycobacteria Is Involved In Capsule Integrity and Virulence through Its Substrate PPE10. PLoS Pathog. 12, e1005696. 10.1371/journal.ppat.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson MG, and Stout JE (2015). Twenty-eight cases of Mycobacterium marinum infection: retrospective case series and literature review. Infection 43, 655–662. 10.1007/s15010-015-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matthews JL (2004). Common diseases of laboratory zebrafish. Methods Cell Biol. 77, 617–643. 10.1016/s0091-679x(04)77033-8. [DOI] [PubMed] [Google Scholar]
- 66.Sambandamurthy VK, Derrick SC, Jalapathy KV, Chen B, Russell RG, Morris SL, and Jacobs WR Jr. (2005). Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect. Immun 73, 1196–1203. 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rapino F, Robles E, Richter-Larrea J, Kallin E, Martinez-Climent J, and Graf T (2013). C/EBPa Induces Highly Efficient Macrophage Trans-differentiation of B Lymphoma and Leukemia Cell Lines and Impairs Their Tumorigenicity. Cell Rep. 3, 1153–1163. 10.1016/j.celrep.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Gaidt MM, Morrow A, Fairgrieve MR, Karr JP, Yosef N, and Vance RE (2021). Self-guarding of MORC3 enables virulence factor-triggered immunity. Nature 600, 138–142. 10.1038/s41586-021-04054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rotty JD, Brighton HE, Craig SL, Asokan SB, Cheng N, Ting JP, and Bear JE (2017). Arp2/3 Complex Is Required for Macrophage Integrin Functions but Is Dispensable for FcR Phagocytosis and In Vivo Motility. Dev. Cell 42, 498–513.e6. e496. 10.1016/j.dev-cel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hetrick B, Han M, Helgeson L, and Nolen B (2013). Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol 20, 701–712. 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, and Pollard TD (2009). Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034. 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mackay EW, Apschner A, and Schulte-Merker S (2013). A bone to pick with zebrafish. BoneKEy Rep. 2, 445. 10.1038/bone-key.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knight RD, and Schilling TF (2006). Cranial neural crest and development of the head skeleton. Adv. Exp. Med. Biol 589, 120–133. 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- 74.Witten PE, and Huysseune A (2009). A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol. Rev. Camb. Phil. Soc 84, 315–346. 10.1111/j.1469-185X.2009.00077.x. [DOI] [PubMed] [Google Scholar]
- 75.To TT, Witten PE, Renn J, Bhattacharya D, Huysseune A, and Winkler C (2012). Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development 139, 141–150. 10.1242/dev.071035. [DOI] [PubMed] [Google Scholar]
- 76.Asharani PV, Keupp K, Semler O, Wang W, Li Y, Thiele H, Yigit G, Pohl E, Becker J, Frommolt P, et al. (2012). Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am. J. Hum. Genet 90, 661–674. 10.1016/j.ajhg.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-Glez V, Valencia M, Caparros-Martin JA, Aglan M, Temtamy S, Tenorio J, Pulido V, Lindert U, Rohrbach M, Eyre D, et al. (2012). Identification of a mutation causing deficient BMP1/mTLD proteolytic activity in autosomal recessive osteogenesis imperfecta. Hum. Mutat 33, 343–350. 10.1002/humu.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laue K, Pogoda HM, Daniel P, van Haeringen A, Alanay Y, von Ameln S, Rachwalski M, Morgan T, Gray M, Breuning M, et al. (2011). Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am. J. Hum. Genet 89, 595–606. 10.1016/j.ajhg.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eames BF, Yan YL, Swartz ME, Levic DS, Knapik EW, Post-lethwait JH, and Kimmel CB (2011). Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 7, e1002246. 10.1371/journal.pgen.1002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simpson MA, Hsu R, Keir LS, Hao J, Sivapalan G, Ernst LM, Zackai EH, Al-Gazali LI, Hulskamp G, Kingston HM, et al. (2007). Mutations in FAM20C are associated with lethal osteosclerotic bone dysplasia (Raine syndrome), highlighting a crucial molecule in bone development. Am. J. Hum. Genet 81, 906–912. 10.1086/522240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johansen IS, Nielsen SL, Hove M, Kehrer M, Shakar S, Woyen AV, Andersen PH, Bjerrum S, Wejse C, and Andersen AB (2015). Characteristics and Clinical Outcome of Bone and Joint Tuberculosis from 1994 to 2011: A Retrospective Register-Based Study in Denmark, 61 (Clinical infectious diseases : an official publication of the Infectious Diseases Society of America), pp. 554–562. 10.1093/cid/civ326. [DOI] [PubMed] [Google Scholar]
- 82.Leonard MK, and Blumberg HM (2017). Musculoskeletal Tuberculosis. Musculoskeletal Tuberculosis. Microbiology spectrum 5. 10.1128/microbiolspec.TNMI7-0046-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barros-Becker F, Lam PY, Fisher R, and Huttenlocher A (2017). Live imaging reveals distinct modes of neutrophil and macrophage migration within interstitial tissues. J. Cell Sci 130, 3801–3808. 10.1242/jcs.206128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blaser MJ, and Kirschner D (2007). The equilibria that allow bacterial persistence in human hosts. Nature 449, 843–849. 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- 85.Otchere ID, Coscolla M, Sanchez-Buso L, Asante-Poku A, Brites D, Loiseau C, Meehan C, Osei-Wusu S, Forson A, Laryea C, et al. (2018). Comparative genomics of Mycobacterium africanum Lineage 5 and Lineage 6 from Ghana suggests distinct ecological niches. Sci. Rep 8, 11269. 10.1038/s41598-018-29620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fernando SL, Saunders BM, Sluyter R, Skarratt KK, Goldberg H, Marks GB, Wiley JS, and Britton WJ (2007). A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am.J. Respir. Crit. Care Med 175, 360–366. 10.1164/rccm.200607-970OC. [DOI] [PubMed] [Google Scholar]
- 87.Xie X, Li J, Gu F, Zhang K, Su Z, Wen Q, Sui Z, Zhou P, and Yu T (2021). Genetic Determinants for Bacterial Osteomyelitis: A Focused Systematic Review of Published Literature. Front. Genet 12, 654792. 10.3389/fgene.2021.654792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Q, Ma A, Wei L, Pang Y, Wu B, Luo T, Zhou Y, Zheng HX, Jiang Q, Gan M, et al. (2018). China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nat Ecol Evol 2, 1982–1992. 10.1038/s41559-018-0680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, Fenner L, Rutaihwa L, Borrell S, Luo T, et al. (2016). Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet 48, 1535–1543. 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blower SM, McLean AR, Porco TC, Small PM, Hopewell PC, Sanchez MA, and Moss AR (1995). The intrinsic transmission dynamics of tuberculosis epidemics. Nat Med 1, 815–821. 10.1038/nm0895-815. [DOI] [PubMed] [Google Scholar]
- 91.Vyazovaya A, Mokrousov I, Solovieva N, Mushkin A, Manicheva O, Vishnevsky B, Zhuravlev V, and Narvskaya O (2015). Tuberculous spondylitis in Russia and prominent role of multidrug-resistant clone Mycobacterium tuberculosis Beijing B0/W148. Antimicrob. Agents Chemother 59, 2349–2357. 10.1128/AAC.04221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taylor GM, Young DB, and Mays SA (2005). Genotypic analysis of the earliest known prehistoric case of tuberculosis in Britain. J. Clin. Microbiol 43, 2236–2240. 10.1128/JCM.43.5.2236-2240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krishnan N, Robertson BD, and Thwaites G (2010). The mechanisms and consequences of the extra-pulmonary dissemination of Mycobacterium tuberculosis. Mycobacterium tuberculosis. Tuberculosis (Edinb) 90, 361–366. 10.1016/j.tube.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 94.Lieberman TD, Wilson D, Misra R, Xiong LL, Moodley P, Cohen T, and Kishony R (2016). Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat Med 22, 1470–1474. 10.1038/nm.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pepperell CS, Casto AM, Kitchen A, Granka JM, Cornejo OE, Holmes EC, Birren B, Galagan J, and Feldman MW (2013). The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 9, e1003543. 10.1371/journal.ppat.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kager LM, Runge JH, Nederveen AJ, Roelofs JJ, Stoker J, Maas M, and van der Poll T (2014). A new murine model to study musculoskeletal tuberculosis (short communication). Tuberculosis 94, 306–310. 10.1016/j.tube.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 97.Oehlers SH, Cronan MR, Scott NR, Thomas MI, Okuda KS, Walton EM, Beerman RW, Crosier PS, and Tobin DM (2015). Interception of host angiogenic signalling limits mycobacterial growth. Nature 517, 612–615. 10.1038/nature13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, and Ramakrishnan L (2014). Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222. 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walton EM, Cronan MR, Beerman RW, and Tobin DM (2015). The Macrophage-Specific Promoter mfap4 Allows Live, Long-Term Analysis of Macrophage Behavior during Mycobacterial Infection in Zebrafish. PLoS One 10, e0138949. 10.1371/journal.pone.0138949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh S, Holdway J, and Poss K (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879–886. 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 103.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529. 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Letunic I, and Bork P (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 107.Meijering E, Dzyubachyk O, and Smal I (2012). Methods for cell and particle tracking. Methods Enzymol. 504, 183–200. 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 108.Stamatakis A (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pimentel H, Bray NL, Puente S, Melsted P, and Pachter L (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690. 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 111.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, and Ding L (2009). VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285. 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takaki K, Davis JM, Winglee K, and Ramakrishnan L (2013). Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc 8, 1114–1124. 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cronan MR, Rosenberg AF, Oehlers SH, Saelens JW, Sisk DM, Jurcic Smith KL, Lee S, and Tobin DM (2015). CLARITY and PACT-based imaging of adult zebrafish and mouse for whole-animal analysis of infections. Dis Model Mech 8, 1643–1650. 10.1242/dmm.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, and Mishina M (2004). A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144. 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 115.Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, and Ekker SC (2006). Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169. 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dynam 236, 3088–3099. an official publication of the American Association of Anatomists. 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 117.Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. (2011). A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661. 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murphy KC, Nelson SJ, Nambi S, Papavinasasundaram K, Baer CE, and Sassetti CM (2018). ORBIT: A New Paradigm for Genetic Engineering of Mycobacterial Chromosomes. mBio 9. 10.1128/mBio.01467-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cronin RM, Ferrell MJ, Cahir CW, Champion MM, and Champion PA (2022). Proteo-genetic analysis reveals clear hierarchy of ESX-1 secretion in Mycobacterium marinum. Proc. Natl. Acad. Sci. USA 119, e2123100119. 10.1073/pnas.2123100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meier F, Brunner AD, Koch S, Koch H, Lubeck M, Krause M, Goedecke N, Decker J, Kosinski T, Park MA, et al. (2018). Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteomics 17, 2534–2545. 10.1074/mcp.TIR118.000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bosserman RE, Thompson CR, Nicholson KR, and Champion PA (2018). Esx Paralogs Are Functionally Equivalent to ESX-1 Proteins but Are Dispensable for Virulence in Mycobacterium marinum. J. Bacteriol 200, e00726–17. 10.1128/JB.00726-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang S, Zhou W, Tang W, Zhang Y, Hu Y, and Chen S (2021). Genome-scale analyses of transcriptional start sites in Mycobacterium marinum under normoxic and hypoxic conditions. BMC Genom. 22, 235. 20210406 ed. 10.1186/s12864-021-07572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zulauf KE, Sullivan JT, and Braunstein M (2018). The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation. PLoS Pathog. 14, e1007011. 10.1371/journal.ppat.1007011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thompson JD, Gibson TJ, and Higgins DG (2002). Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics/Editoral Board. 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 125.Price MN, Dehal PS, and Arkin AP (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, and Scheffler K (2015). RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol 32, 820–832. 10.1093/mol-bev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Letunic I, and Bork P (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Waterhouse AM, Procter JB, Martin DMA, Clamp M, and Barton GJ (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequencing and RNA-seq data are available at NCBI (BioProjects PRJNA540867 and PRJNA872173)
Raw and processed mass spectrometry files are archived and available at MassIVE and PRIDE repositories under accession numbers MSV000090143 and PDX036131.
Original code is deposited and publicly available on GitHub and Zenodo. DOIs are listed in the Key Resource Table.
All other data are available in the main text or supplementary figures.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Arp2 | Abcam | Cat#ab49674; RRID:AB_867730 |
| goat anti-mouse Alexa Fluor 647 | Thermo Fisher | Cat#A21236; RRID:AB_2535805 |
| rabbit anti-HA-Tag | Cell Signaling Technology | Cat#C29F4; RRID:AB_1549585 |
| mouse anti-RNAP | BioLegend | Cat#8RB13; RRID:AB_2566583 |
| Bacterial and virus strains | ||
| Mycobacterium marinum M strain/pMSP12:cerulean | Oehlers et al.97 | N/A |
| Mycobacterium marinum M strain/pMSP12:tomato | Cambier et al.98 | N/A |
| Mycobacterium marinum ΔesxM | This paper | N/A |
| Mycobacterium marinum ΔesxM + hsp60:esxM | This paper | N/A |
| Mycobacterium tuberculosis 6020 auxotroph | Sambandamurthy et al.66 | N/A |
| Mycobacterium tuberculosis 6020 + msp 12::cerulean | This paper | N/A |
| Mycobacterium tuberculosis 6020 + msp 12::cerulean-hsp60:esxM | This paper | N/A |
| Mycobacterium tuberculosis H37Rv | BEI Resources, NIAID | NR-13648 |
| Mycobacterium tuberculosis NCG | This paper | N/A |
| Mycobacterium tuberculosis H37Rv/msp12:cerulean | This paper | N/A |
| Mycobacterium tuberculosis NCG/msp12:cerulean | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 7H10 | Difco | Cat#262710 |
| Trizol | Ambion | Cat#15596026 |
| Sodium chloride | Fisher Scientific | Cat#S271 |
| Potassium chloride | VWR | Cat#BDH9258 |
| Calcium chloride | VWR | Cat#BDH9224 |
| Magnesium chloride | Ward’s Scientific | Cat#470301 |
| 1-phenyl-2-thiourea | Sigma-Aldrich | Cat#P7629 |
| Tricaine-S (MS-222) | Syndel | ANADA#200–226 |
| Low melting point agarose | Fisher Scientific | Cat#BP165–25 |
| CK-666 Arp2/3 inhibitor | Sigma-Aldrich | Cat#182515 |
| CK-689 inactive control | Sigma-Aldrich | Cat#182517 |
| Dimethyl sulfoxide (DMSO) | Fisher Scientific | Cat#BP231 |
| BP Clonase II | Thermo Fisher | Cat#11789020 |
| HyClone HyQTase Cell Detachment Reagent | Fisher Scientific | Cat#SV3003001 |
| DAPI Fluoromount-G | SouthernBiotech | Cat#0100–20 |
| RPMI-1640 | Sigma-Aldrich | Cat#R8758 |
| GlutaMAX Supplement | Thermo Fisher | Cat#35050061 |
| Phorbol-12-myristate-13-acetate (PMA) | Sigma-Aldrich #R8758 | Cat#P148 |
| Sodium pyruvate | Gibco | Cat#11360 |
| Recombinant human IL-3 | Peprotech | Cat#200–03 |
| Recombinant human M-CSF | Peprotech | Cat#300–25 |
| β-Estradiol | Sigma-Aldrich | Cat#E2758 |
| Alexa Fluor 555 phalloidin stain | Thermo Fisher | Cat#A34055 |
| Hygromycin B solution | Invitrogen | Cat#10687010 |
| 7H9 | Difco | Cat#271310 |
| OADC | Sigma-Aldrich | Cat#M0678 |
| Neonate-80 | Fisher Scientific | Cat#BP337 |
| TRIzol Reagent | Invitrogen | Cat#15596026 |
| RNase-free DNase I | New England BioLabs | Cat#M0303S |
| Luna universal qPCR master mix | New England BioLabs | Cat#M3003X |
| Tyloxapol | Sigma-Aldrich | Cat#T8761 |
| cOmplete EDTA-free protease inhibitor cocktail | Millapore Sigma | Cat#11836170001 |
| Super-Signal West Pico PLUS | Thermo Fisher | Cat#34580 |
| Super-Signal West Femto Maximum Sensitivity | Thermo Fisher | Cat#34095 |
| Prolong Gold Anti-fade mounting solution | Invitrogen | Cat#P36934 |
| Critical commercial assays | ||
| mMessage mMachine T7 Kit | Thermo Fisher | Cat#AM1344 |
| RNeasy Plus Mini Kit | Qiagen | Cat#74134 |
| Zymo Direct-zol RNA Miniprep Kit | Thomas Scientific | Cat#1159U94 |
| DC Protein Assay Kit | Bio-Rad | Cat#5000111 |
| Pierce Micro BCA Protein Assay Kit | Thermo Fisher | Cat#23235 |
| Deposited data | ||
| Mtb genomic sequencing | NCBI BioProject | PRJNA540867 |
| Mtb H37Rv and NCG RNA sequencing | NCBI BioProject | PRJNA872173 |
| Raw mass spectrometry files | MassIVE repository | MSV000090143 |
| Processed mass spectrometry files | PRIDE repository | PDX036131 |
| Experimental models: Cell lines | ||
| BLaER1 | Rappino et al.67 | N/A |
| RAW 264.7 | ATCC | ATCC product TIB-71 |
| THP-1 | ATCC | ATCC product TIB-202 |
| Experimental models: Organisms/strains | ||
| Zebrafish (Danio rerio), *AB wildtype strain | ZIRC | ZDB-GENO-960809–7 |
| Zebrafish, Tg(mfap4:tomato-caax)xt6 | Walton et al.99 | N/A |
| Zebrafish, Tg(mfap4:esxM-p2a-tdTomato) xt49 | This paper | N/A |
| Zebrafish, Tg(mfap4:esxM_Q59X-p2a-tdTomato) xt50 | This paper | N/A |
| Zebrafish, Tg(mfap4:esxB-p2a- mNeonGreen) xt51 | This paper | N/A |
| Zebrafish, Tg(Ola.Sp7:mCherry-Eco.NfsBp46 | Singh et al.100 | N/A |
| Zebrafish, Tg(acp5a:mNeonGreen-CAAX)xt52 | This paper | N/A |
| Mouse, C57Bl/6J | Jackson Laboratories | RRID:IMSR_JAX:000,664 |
| Recombinant DNA | ||
| Oligonucleotides | This paper | Available in Table S5 |
| Software and algorithms | ||
| RNA seq analysis original code | This paper | https://doi.org/10.5281/zenodo.6981721 |
| Phylogenetic analysis original code | This paper | https://github.com/vertgenlab/gonomics |
| BWA | Li and Durbin101 | N/A |
| ClustalW2 | Larkin et al.102 | N/A |
| FIJI/ImageJ2, 2.5.0 | Rueden et al.103; Schindelin et al.104 | N/A |
| iTol | Letunic and Bork105 | N/A |
| Kallisto v0.48 | Bray et al.106 | N/A |
| MtrackJ, 1.5.1 | Meijering et al.107 | N/A |
| RStudio | RStudio Team (2022). RStudio: Integrated Development Environment for R. | N/A |
| RAxML | Stamatakis108 | N/A |
| SAMtools | Li et al.109 | N/A |
| Sleuth v0.30.0 | Pimentel et al.110 | N/A |
| VarScan | Koboldt et al.111 | N/A |


