Abstract
During infection, circulating blood monocytes migrate from the vasculature to the extravascular compartments where they mature into tissue macrophages. The maturation process prepares the cell to actively participate in the inflammatory and the immune responses, and many transcription factors have been found to be involved. Here we report on a novel role for nuclear factor κB (NF-κB) in this process. Its accumulation in the cytoplasm of differentiated macrophages is responsible for the enhanced ability of the cell to respond to lipopolysaccharide (LPS) stimulation, as determined by tumor necrosis factor alpha (TNF-α) secretion. Differentiation of the human monocytic cell line THP-1 into macrophage-like cells was induced by exposure of the cells to phorbol myristate acetate. DNA-bindable NF-κB was not detected in the cytoplasm of undifferentiated THP-1 cells but accumulated in the cytoplasm of the cells following differentiation. No TNF-α was detected in the media of resting differentiated and nondifferentiated THP-1 cells. Stimulation with LPS of differentiated cells induced the production of higher levels of TNF-α than stimulation of nondifferentiated cells. This hyperresponsiveness to LPS was found in the mRNA and secreted TNF-α levels. Furthermore, stimulation with LPS induced the translocation of NF-κB from the cytoplasm into the nucleus. This translocation process was more rapid in the differentiated cells than in the nondifferentiated cells, and the resultant accumulated levels of NF-κB in the nucleus were higher. The DNA-bindable NF-κB was identified as a heterodimer of p65 and p50. The results suggest that NF-κB accumulation in the cytoplasm during maturation of monocytes to macrophages primes the cells for enhanced responsiveness to LPS and results in the rapid secretion of inflammatory mediators, such as TNF-α, by mature macrophages following LPS challenge.
Macrophages play a key role in the orchestration and execution of the innate and adaptive arms of the immune response to bacterial infection. During the infective process, circulating blood monocytes migrate from the vasculature into the extravascular compartment under the influence of many different endogenous and exogenous factors. In the tissues they differentiate to macrophages (2). Upon differentiation, the cell loses its ability to replicate and its antibacterial properties are markedly enhanced, allowing it to participate in the inflammatory and immune responses. The differentiation process is a complex one and is controlled by the expression or activation of several transcription factors (30). However, the events during terminal differentiation of the macrophage leading to its enhanced antibacterial activities are poorly understood.
Activated macrophages elicit many of their effects via the secretion of soluble inflammatory mediators. Lipopolysaccharide (LPS) derived from gram-negative bacteria is considered to be the most potent activator of the macrophage secretory response. Tumor necrosis factor alpha (TNF-α) is one of the earliest major proinflammatory mediators secreted by macrophages when stimulated with LPS in vivo and in vitro (17, 18). TNF-α has been implicated in the pathogenesis of several inflammatory diseases, such as septic shock (19), rheumatoid arthritis (15, 16), multiple sclerosis (21), and periodontal disease (25), and its production has been suggested as a possible target for therapy in these diseases.
The intracellular events that mediate LPS-induced TNF-α secretion have been the subjects of intense research. TNF-α is not secreted from intracellular stores but is synthesized de novo in response to an effective stimulus. The stimulus is thought to act via several nuclear factors, and nuclear factor κB (NF-κB) was found to have an important role in the regulation of TNF-α gene transcription (9, 10, 22, 27, 28). NF-κB, a heterodimer of p65 and p50 proteins and the rel family, is an inducible eukaryotic transcription factor which exists in the cytoplasm of most cells (12). Several stimulants, including bacterial LPS, induce the phosphorylation of IκB and the subsequent release and activation of NF-κB. The activated NF-κB translocates from the cytoplasm to the nucleus, where it binds to specific binding sites in the TNF-α promoter region and activates TNF-α gene transcription (5, 28).
Here we report a novel role for NF-κB in the terminal differentiation of monocytes to macrophages, that of enhancing the ability of the differentiated macrophage to respond to LPS stimulation. Using a phorbol myristate acetate (PMA)-induced cell differentiation model of the human monocytic cell line THP-1 (29), we studied the relationship between THP-1 cell maturation and NF-κB levels before and after LPS stimulation. TNF-α production was chosen as the outcome variable for studying the effect of the differentiation process on the functional activity of the macrophage and for correlating it to the levels and compartmentalization of NF-κB.
MATERIALS AND METHODS
Cell culture.
The human monocytic cell line THP-1 (American Type Culture Collection, Manassas, Va.) was maintained in RPMI 1640 media supplemented with 2 mM l-glutamine–100 U of penicillin per ml–100 μg of streptomycin per ml–25 mM HEPES (C-RPMI) and 5% fetal bovine serum (all from Gibco BRL, Gaithersburg, Md.). For the induction of cell differentiation, cells (5 × 105 to 106 per ml) were seeded in macrophage serum-free medium (macrophage-SFM; Gibco BRL) with 2 to 200 nM PMA for 24 h (29). After incubation, nonattached cells were removed by aspiration, and the adherent cells were washed with C-RPMI three times. THP-1 cells in macrophage SFM with no PMA were used as control (undifferentiated) cells.
For cell stimulation, the cells were further incubated with or without LPS for the indicated periods (23) in fresh C-RPMI with 2% heat-inactivated human AB serum (Sigma, St. Louis, Mo.), while undifferentiated cells were used as controls. The LPS was extracted from Porphyromonas gingivalis A7436 by a hot-phenol-water method and further purified by cesium chloride isopyknic density gradient centrifugation as described previously (24).
Detection of secreted TNF-α.
Human TNF-α was quantified by an enzyme-linked immunosorbent assay (ELISA) as previously described (24). Briefly, 96-well ELISA plates (Maxisorp; Nunc, Naperville, Ill.) were coated with an anti-TNF-α monoclonal antibody (R&D Systems, Minneapolis, Minn.) in a coating buffer (carbonate-bicarbonate buffer, pH 9.6), followed by overnight incubation at 4°C. The wells were blocked overnight (4°C) with 2% bovine serum albumin in coating buffer; samples were then added and incubated overnight (4°C). Goat anti-TNF-α polyclonal antibody (R&D Systems) was added, followed by donkey anti-goat horseradish peroxidase conjugate (Sigma). o-Phenylenediamine was used as the substrate. The reaction was stopped by the addition of 4 N sulfuric acid, and optical density was read in a Vmax microplate reader (Molecular Devices, Palo Alto, Calif.) at 490 to 600 nm.
Semiquantification of TNF-α mRNA accumulation.
Total cellular RNA was recovered from the cells by a single-step method (7), and the TNF-α mRNA was assessed by semiquantitative reverse transcription (RT)-PCR as previously described (23, 27), with 6.6 nM [α-32P]dCTP (3,000 Ci/mmol; NEN). Successful isolation of RNA was monitored by detection of β-actin mRNA by RT-PCR performed in parallel. One-tenth of the PCR product was analyzed on 3% NuSieve GTG (FMC BioProducts, Rockland, Maine)–1% agarose (Gibco BRL) gel and stained with 0.5 μg of ethidium bromide per ml. After photographs were taken, the signal bands were cut from the gel and the radioactivity was determined by liquid scintillation spectroscopy. Counts per minute were converted to molecules of mRNA by using a standard curve with double-stranded cDNA (data not shown). In all experiments, the presence of possible contaminants and background radioactivity in the gel were evaluated by control reactions in which RT and amplification were carried out on RNA-free samples.
Identification of newly transcribed TNF-α RNA.
The transcription rate was determined by a nuclear run-on transcription assay (13). Cells were lysed in 1 ml of cell lysis buffer (10 mM Tris-HCl, [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 0.65% Nonidet P-40) by repeated pipetting. Cell lysates were transferred into microcentrifuge tubes, vortexed vigorously for 10 s, and incubated on ice for 5 min. Nuclei were pelleted by centrifugation (5 min at 800 × g, 4°C). The nuclei were then resuspended in 100 μl of buffer (50 mM Tris-HCl [pH 8.5], 5 mM MgCl2, 0.1 mM EDTA, 40% glycerol) and stored at −70°C until assayed. The in vitro transcription assay was performed in 200 μl of transcription buffer (5 mM Tris-HCl [pH 8.0], 2.5 mM MgCl2, 150 mM KCl, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, and 2.5 mM dithiothreitol) with 50 μCi of [α-32P]UTP (3,000 Ci/mmol; NEN) at 30°C for 30 min. RNA in the reaction mixture was isolated by the single-step method (7), washed twice with 75% ethanol, and dissolved in 100 μl of H2O. DNA slot blot membranes (Hybond-N+; Amersham, Arlington Heights, Ill.) containing 5 μg of plasmid DNA with human β-actin cDNA (661 bp) in a vector, 5 μg of plasmid DNA with human TNF-α cDNA (374 bp) in a vector, and 5 μg of plasmid vector pGEM7Z (3,000 bp; Promega, Madison, Wis.) as a control were hybridized with equal numbers of ethanol-precipitable counts (5 × 105 cpm) of 32P-labeled RNA for 16 h at 65°C in a hybridization buffer (8). The membranes were washed finally with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.5% sodium dodecyl sulfate at 65°C for 10 min, and the signals were visualized by autoradiography at −70°C for a week in the presence of an intensifying screen.
Detection of NF-κB in the nucleus and the cytoplasm.
Nuclear and cytoplasmic fractions from THP-1 cells were extracted after cells were washed with cold phosphate-buffered saline (2.7 mM KCl, 1.2 mM KH2PO4, 8.1 mM Na2HPO4, 138 mM NaCl; pH 7.4) (1, 14). Cytoplasmic extraction was obtained in 400 μl of hypotonic buffer (10 mM HEPES [pH 7.9 at 4°C], 1.5 mM MgCl2, 10 mM KCl, 5× proteinase inhibitor mixture [20 μg of 4-amidinophenyl-methanesulfonyl fluoride, 25 μg of antipain, 20 μg of aprotinin, 5 μg of nitobestatin, 20 μg of chymostatin, 25 μg of 3,4-dichloroisocoumarin, 50 μg of E-64, 10 μg of leupeptin, 10 μg of pepstatin A, and 10 μg of phosphoramidon per ml, 50 μM bezamidine, and 50 μM sodium metabisulfite], 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol). Nuclear extracts were sequentially obtained in 150 μl of extraction buffer (0.6 M KCl, 20 mM HEPES [pH 7.9 at 4°C], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1× proteinase inhibitor mixture, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol). Nuclear extracts were dialyzed against a buffer (20 mM HEPES [pH 7.9 at 4°C], 20% glycerol, 100 mM KCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) at 4°C for 1 h. Cell extracts were stored at −70°C until assayed. Protein concentrations in the extracts were determined with a protein assay kit (Bio-Rad, Richmond, Calif.). An electrophoretic mobility shift assay (EMSA) was performed to detect DNA-bindable NF-κB (6). NF-κB consensus double-stranded oligonucleotides (Promega) were end labeled with [γ-32P]ATP (NEN Research Products). Nuclear or cytoplasmic extracts (2 μg) were incubated with a labeled oligonucleotide probe (35 fmol) in binding buffer (4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris-HCl [pH 7.5], 50 ng of poly[dI-dC]) at room temperature for 20 min and then separated by electrophoresis with high-ionic-strength 4% polyacrylamide gel (acrylamide/bisacrylamide at a ratio of 80:1, 50 mM Tris-HCl, 380 mM glycine, 2 mM EDTA, 2.5% glycerol) at 100 V. After the gel was dried, autoradiography was performed with an intensifying screen at −70°C. Identification of the DNA-bindable NF-κB was carried out by supershift EMSA (4), with antibodies against the different NF-κB subunits. Protein samples (2 μg) were mixed with 2 μl of each antiserum for 15 min at room temperature before the EMSA procedure was performed as described above. The antibodies were a generous gift from W. C. Greene, Duke University Medical Center, Durham, N.C.
RESULTS
PMA-induced differentiation of THP-1 cells.
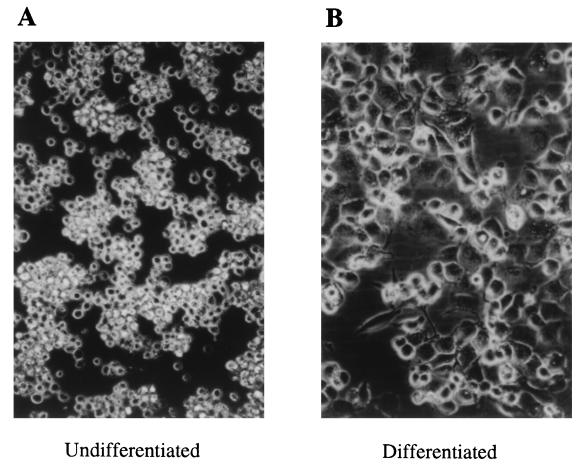
THP-1 cells are premonocytes, committed to the monocytic cell lineage. They grow in suspension and do not adhere to the plastic surfaces of the culture plates (Fig. 1A). For the induction of terminal differentiation to macrophage-like cells, THP-1 cells were cultured in the presence of 2, 20, and 200 nM PMA for 4 days. After 20 h of culture with 200 nM PMA, the cells adhered to the dish bottom and had the morphological characteristics of macrophages (Fig. 1B). Flow cytometry analysis revealed that these cells expressed high levels of CD14, a macrophage-specific differentiation antigen, compared to untreated THP-1 cells. PMA (20 nM) also induced adherence and spreading of the cells after 3 to 4 days of culture. However, a dose of 2 nM PMA was ineffective for the induction of maturation. Based on these results, further experiments using 200 nM PMA for 24 h cell differentiation were conducted.
FIG. 1.
Induction of differentiation in THP-1 cells by PMA. THP-1 cells were incubated for 24 h without (A) or with (B) PMA (200 nM). The cells were photographed at ×200 magnification with a phase-contrast inverted microscope.
TNF-α secretion and mRNA expression in differentiated and undifferentiated THP-1 cells.
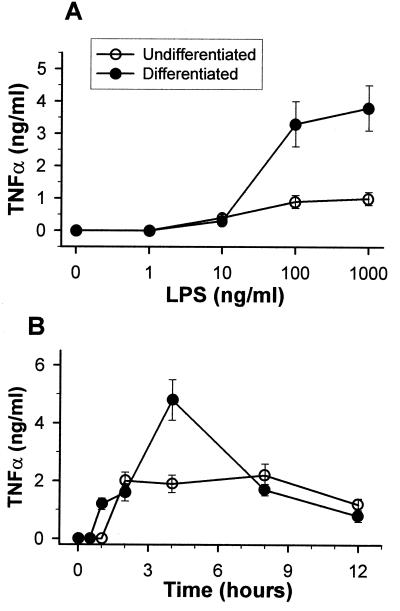
Unstimulated differentiated or undifferentiated THP-1 cells did not secrete any TNF-α into the culture media during the 12-h incubation period (Fig. 2A). Stimulation with LPS (100 ng/ml) of undifferentiated cells induced detectable levels of TNF-α in the media within 2 h (Fig. 2B). These levels remained stable for 8 h and then started to decline. Exposure of the differentiated THP-1 cells to the same LPS concentration stimulated the secretion of TNF-α within 1 h. TNF-α levels peaked within 4 h and then declined. The peak levels of TNF-α in LPS-stimulated differentiated cells were more than 2.5-fold higher than levels secreted by LPS-stimulated undifferentiated cells. Both differentiated and undifferentiated THP-1 cells responded to LPS doses above 10 ng/ml in a dose-dependent manner (Fig. 2A).
FIG. 2.
LPS-induced TNF-α secretion from differentiated and undifferentiated THP-1 cells. Differentiation to macrophages was induced by incubation of the cells with 200 nM PMA for 24 h. (A) Dose response of TNF-α secretion. Cells (106) were washed, and different doses of LPS as indicated in the graph were added to the culture medium. Medium was harvested for TNF-α analysis by ELISA after 6 h of incubation. (B) Kinetics of TNF-α secretion. Cells (106) were washed, and LPS (100 ng/ml) was added to the culture medium. Medium was harvested for TNF-α analysis by ELISA at the intervals indicated in the graph. This is one representative experiment of three. Results are presented as means ± standard errors.
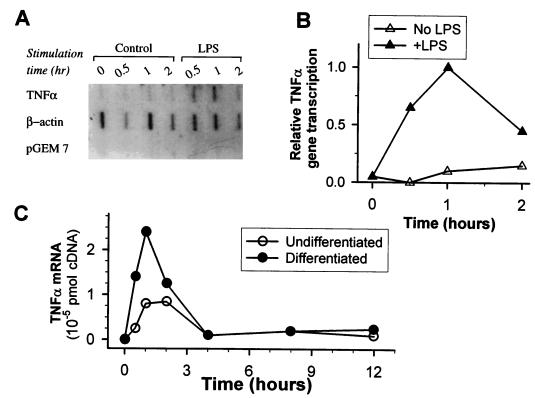
Nuclear run-on analysis revealed that PMA-differentiated THP-1 cells did not express any new TNF-α mRNA prior to LPS stimulation. Active TNF-α gene transcription started within 30 min of LPS challenge, peaked at 1 h, and decreased at 2 h (Fig. 3A and B). In parallel, RT-PCR analysis showed that TNF-α mRNA accumulation also began within 30 min and peaked 1 to 2 h after stimulation (Fig. 3C). The magnitude of the response in the undifferentiated cells was threefold lower than that in the differentiated cells.
FIG. 3.
Analysis of TNF-α gene transcription and mRNA accumulation in THP-1 cells. (A) Nuclear run-on assay for newly transcribed TNF-α mRNA. Differentiation into macrophages was induced by incubation of the cells with 200 nM PMA for 24 h. Nuclei were harvested from LPS-stimulated and nonstimulated cells at different intervals, and in vitro transcription of the TNF-α gene was analyzed with specific probes for TNF-α. β-Actin was used as the internal control, and pGEM 7 vector was used as the negative control. (B) Densitometric analysis of the bands in panel A. (C) Kinetics of TNF-α mRNA accumulation in LPS-stimulated differentiated and undifferentiated THP-1 cells. Cells were stimulated with 100 ng of LPS per ml, and mRNA was extracted in the indicated time intervals. TNF-α mRNA was quantified by RT-PCR with [32P]dCTP in relation to a standard curve.
Expression of DNA-bindable NF-κB in differentiated and undifferentiated macrophages.
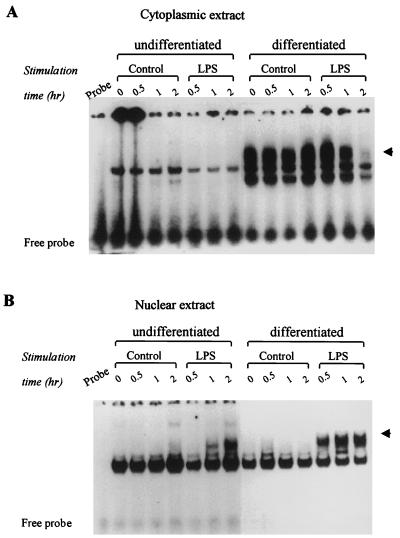
NF-κB was detected in the cytoplasm and nucleus by EMSA with consensus NF-κB-bindable DNA motifs as probes. NF-κB was not detected in the cytoplasm of undifferentiated THP-1 cells. In contrast, NF-κB was found to accumulate in cytoplasm of the differentiated cells (Fig. 4A). A drastic decrease in cytoplasmic NF-κB was observed after LPS challenge, and approximately 70% of the cytoplasmic NF-κB had disappeared from the cytoplasm 2 h after LPS challenge (Fig. 4A). In the nucleus, NF-κB was not detected prior to LPS stimulation. In both undifferentiated and differentiated cells (Fig. 4B) LPS stimulation resulted in detectable levels of NF-κB in the nucleus. In the differentiated cells NF-κB was detectable 0.5 h after LPS stimulation and reached a plateau after 1 h. In the undifferentiated cells NF-κB was only detected in the nucleus 1 h post-LPS stimulation and levels continued to accumulate until the end of the experiment (Fig. 4B). However, the NF-κB levels were always lower than the levels achieved in the differentiated THP-1 cell nuclei.
FIG. 4.
Effect of LPS on the accumulation of DNA-bindable NF-κB in the cytoplasm (A) and in the nuclei (B) of differentiated and undifferentiated THP-1 cells. Differentiation was induced by incubation of THP-1 cells (10 × 106 cells/100-mm-diameter plate) with 200 nM PMA for 24 h. After being washed, adherent cells were incubated with or without LPS (100 ng/ml) for the indicated time intervals. Cytoplasmic and nuclear extracts were obtained and then used for EMSA with NF-κB-specific 32P-labeled DNA probes. Specific bindings are shown by arrowheads. Data are representative of two independent experiments.
Characterization of the DNA-bindable cytoplasmic NF-κB-like protein.
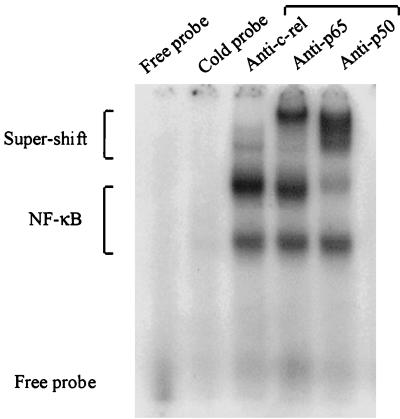
To identify the DNA-bindable NF-κB-like proteins in cytoplasm, antisera specific to the NF-κB-related proteins (c-Rel, p65, and p50) were used to induce supershifts in the EMSA gels by binding to the NF-κB consensus oligonucleotide probe and the target protein (Fig. 5). Anti-p65 serum eliminated part of the LPS-induced band complex and caused this band to migrate slower on the gel. Anti-p50 serum reduced the intensity of the LPS-induced band complex and caused a broad supershifted band. Anti-c-Rel serum had only a slight effect and did not reduce the LPS-induced band complex (Fig. 5). The anti-p50 serum recognized the p50 subunit in both the upper and lower parts of the bands, indicating the presence of the heterodimer p50-p65 and the homodimer p50-p50 in the identified complexes. The same supershift pattern was observed when nuclear extracts were used (data not shown).
FIG. 5.
Identification of the NF-κB subunits. The NF-κB in the differentiated THP-1 cells was detected by supershift EMSA with specific antibodies to the different NF-κB subunits. Cytoplasmic extracts used in Fig. 4 were preincubated with the indicated antiserum before the binding reaction with the NF-κB-specific probe. Data are representative of two independent experiments.
DISCUSSION
The present study investigated the expression of NF-κB in monocyte-like cells, before and after the induction of differentiation into macrophages in vitro. The results demonstrate that in parallel to multiple changes in cell morphology and function during the differentiation process, there is an accumulation of functional proteins from the NF-κB family in the cytoplasm of the cell, as indicated by their ability to bind consensus DNA sequences related to NF-κB binding sites in the mammalian genome. However, due to their location in the cytoplasmic compartment of the cell, far from the cellular DNA, they are unable to induce cellular events. Stimulation by LPS induced the translocation of the NF-κB proteins from the cytoplasm into the nucleus, where they can bind DNA and activate transcription of the target genes (hypothesis summarized in Fig. 6). These findings suggest that the maturation process primes the cell, by the induction of NF-κB in the cytoplasm, for a rapid and enhanced response to an additional stimulus, such as LPS.
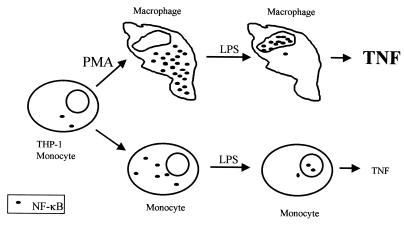
FIG. 6.
Schematic representation of the interrelationship between monocyte differentiation, NF-κB accumulation and translocation, and TNF-α secretion. When exposed to the phorbol ester PMA, THP-1 monocytes differentiate to mature macrophages. During the differentiation process, NF-κB accumulates in the cytoplasm. LPS stimulation induces the translocation of NF-κB into the nucleus, followed by the secretion of TNF-α. In the undifferentiated THP-1 cells, there are only low levels of NF-κB in the cytoplasm, which causes the cells to respond to LPS stimulation in a much slower way and by the secretion of lower levels of TNF-α, compared to the differentiated cells.
NF-κB is thought to be one of the more important nuclear factors in mammalian cells. Several stimulants such as LPS, superantigen, phorbol esters, and cytokines are known to induce the appearance of NF-κB in the cytoplasm and its translocation into the nucleus (12). In addition, many genes, including genes encoding cytokines, are known to include NF-κB binding sites in their promoter regions (3, 22), and one of these genes is the TNF-α gene (26). Earlier studies had suggested that NF-κB may not be necessary for LPS-induced TNF-α gene transcription (11), but more recent studies showed clearly that NF-κB plays an important role in the activation of the TNF-α gene by LPS in human monocytes and macrophages (5, 28). Our results have demonstrated that although THP-1 cells respond to LPS by the secretion of TNF-α, the induction of differentiation enhanced TNF-α transcription and secretion. The enhanced production of TNF-α correlated with the accumulation of NF-κB in the cytoplasm prior to the stimulus and with its rapid translocation into the nucleus after stimulation. These results, together with the fact that NF-κB is an important regulator of TNF-α production, suggest indirectly that there is a connection between these two events. NF-κB in the cytoplasm of differentiated THP-1 cells translocated to the nucleus within 30 min after LPS stimulation, and this translocation precedes the peak in the transcription rate of TNF-α. Furthermore, the levels of NF-κB translocated to the nucleus in differentiated THP-1 cells were higher than those in undifferentiated cells. It is important to note that differentiated THP-1 cells showed low levels of TNF-α gene transcription without LPS stimulation (data not shown) but that no NF-κB was detected in the nuclei of these cells without LPS stimulation. In addition, no TNF-α secretion was detected without LPS stimulation. It is possible that an additional factor(s) other than NF-κB is involved in the activation of transcription of the TNF-α gene and that NF-κB is needed also for induction of cofactors involved in TNF-α secretion (27). This assumption is supported by experiments showing that LPS-induced TNF-α transcription in CD14 “knockout” macrophages occurred without detectable NF-κB binding (20). While NF-κB is involved in the major LPS-induced pathway, which starts from the CD14 receptor, NF-κB might not be essential for an alternative CD14-independent pathway. However, LPS activation of a CD14-independent pathway requires much higher doses than those used in the present study (20, 23). The data suggest that the priming of the cells by the induction of cytoplasmic NF-κB might be limited to the major LPS-induced signaling pathway, which is CD14 dependent.
The cytoplasmic NF-κB-like proteins which bind oligonucleotide probes specific for NF-κB were identified by antibodies against the p65 and p50 subunits of NF-κB. Anti-p50 caused the signal to shift almost completely, while anti-p65 antiserum caused only partial shift. Anti-c-Rel caused minimal shift of the protein-DNA complex. These results suggest that proteins from the NF-κB family in THP-1 cells expressed following exposure to phorbol ester exist as p50-p50 homodimers, p50-p65 heterodimers, or heterodimers of p50 with an unidentified protein. The ability of cytoplasmic NF-κB to bind specific DNA probes indicates that IκB may dissociate from NF-κB in the cytoplasm during the differentiation process. However, the dissociated NF-κB remains in the cytoplasm of the differentiated macrophages and may need some other factor(s) to assist its translocation into the nucleus upon LPS stimulation.
In conclusion, the present study has demonstrated that the maturation of monocytes to macrophages enhanced their responsiveness to LPS. This hyperresponsiveness coincides with the accumulation of NF-κB in the cytoplasm, which rapidly translocated into the nucleus for the induction of TNF-α gene transcription upon stimulation with LPS. These findings support the hypothesis that NF-κB plays an important role in the differentiation of monocytes into macrophages, preparing them to respond rapidly to infection.
ACKNOWLEDGMENTS
We are grateful to Warner C. Greene for providing the antibodies to NF-κB. The excellent technical assistance of Barbara J. Gordon and Martha Warbington is highly appreciated.
This work was supported by a grant from the Chief Scientist of the Ministry of Health of Israel (L.S.), a Grant-in-Aid for Scientific Research (B, 09470425; C, 09671951) by the Ministry of Education, Science, Sports and Culture of Japan (S.T.), NIH grant DE-10709 (S.A.), and USPHS grant DE06436 (T.E.V.D.).
REFERENCES
- 1.Abmayer S M, Workman J L. Preparation of nuclear and cytoplasmic extracts from mammalian cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1990. pp. 12.1.1–12.1.9. [Google Scholar]
- 2.Auger M J, Ross J A. The biology of the macrophage. In: Lewis C E, McGee J O, editors. The macrophage. New York, N.Y: IRL Press; 1992. pp. 3–74. [Google Scholar]
- 3.Baeuerle P A. The inducible transcription activator NF-κB: regulation by distinct protein subunits. Biochim Biophys Acta. 1991;1072:63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- 4.Ballard D W, Dixon E P, Peffer N J, Bogerd H, Doerre S, Stein B, Greene W C. The 65-kDa subunit of human NF-kappa B functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc Natl Acad Sci USA. 1992;89:1875–1879. doi: 10.1073/pnas.89.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondeson J, Browne K A, Brennan F M, Foxwell B M J, Feldmann M. Selective regulation of cytokine induction by adenoviral gene transfer of IκB into human macrophages: lipopolysaccharide-induced, but not zymosan-induced, proinflammatory cytokines are inhibited, but IL-10 is nuclear factor-κB independent. J Immunol. 1999;162:2939–2945. [PubMed] [Google Scholar]
- 6.Chodosh L A. Mobility shift DNA-binding assay using gel electrophoresis. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1988. pp. 12.2.1–12.2.10. [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart M A, Baeuerle P, Vassalli P. Regulation of tumor necrosis factor alpha transcription in macrophages: involvement of four κB motifs and of constitutive and inducible forms of NF-κB. Mol Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drouet C, Shakhov A N, Jongeneel C V. Enhancers and transcription factors controlling the inducibility of tumor necrosis factor alpha promoter in primary macrophages. J Immunol. 1991;147:1694–1700. [PubMed] [Google Scholar]
- 11.Goldfeld A E, Doyle C, Maniatis T. Human tumor necrosis factor alpha gene regulation by virus and lipopolysaccharide. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grilli M, Chiu J J, Lenardo M J. NF-kappa B and Rel: participants in a multiform transcriptional regulatory system. Int Rev Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- 13.Groudine M, Peretz M, Weintraub H. Transcriptional regulation of hemoglobin switching on chicken embryos. Mol Cell Biol. 1989;9:281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K A, Bindereif A, Green M R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 15.Macnaul K L, Hutchinson N I, Parsons J N, Bayne E K, Tocci M J. Analysis of IL-1 and TNF alpha gene expression in human rheumatoid synoviocytes and normal monocytes by in situ hybridization. J Immunol. 1990;145:4154–4166. [PubMed] [Google Scholar]
- 16.McCartney-Francis N, Allen J B, Mizel D E, Albina J E, Xie Q, Nathan C F, Wahl S M. Suppression of arthritis by inhibitor of nitric oxide synthase. J Exp Med. 1993;178:749–754. doi: 10.1084/jem.178.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–3063. [PubMed] [Google Scholar]
- 18.Nussler A K, Billiar T R. Inflammation, immunoregulation and inducible nitric oxide synthase. J Leukoc Biol. 1993;54:171–178. [PubMed] [Google Scholar]
- 19.Parrillo J E. Pathogenetic mechanisms of septic shock. N Engl J Med. 1993;328:1471–1477. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- 20.Perera P Y, Vogel S N, Detore G R, Haziot A, Goyert S M. CD14-dependent and CD14-independent signaling pathways in murine macrophages from normal and CD14 knockout mice stimulated with lipopolysaccharide or taxol. J Immunol. 1997;158:4422–4429. [PubMed] [Google Scholar]
- 21.Selmaj K, Raine C S, Cannella B, Brosnan C F. Identification of lymphotoxin and tumor necrosis factor in multiple sclerosis lesions. J Clin Investig. 1991;87:949–954. doi: 10.1172/JCI115102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakhov A N, Collart M A, Vassalli P, Nedospasov S A, Jongeneel C V. κB-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of tumor necrosis factor alpha gene in primary macrophages. J Exp Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapira L, Takashiba S, Amar S, Van Dyke T E. Porphyromonas gingivalis lipopolysaccharide stimulation of human monocytes: dependence on serum and CD14 receptor. Oral Microbiol Immunol. 1994;9:112–117. doi: 10.1111/j.1399-302x.1994.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 24.Shapira L, Takashiba S, Champagne C, Amar S, Van Dyke T E. Involvement of protein kinase C and protein tyrosine kinase in lipopolysaccharide-induced TNF alpha and IL-1 beta production by human monocytes. J Immunol. 1994;153:1818–1824. [PubMed] [Google Scholar]
- 25.Stashenko P, Jandinski J J, Fujiyioshi P, Rynar J, Socranski S S. Tissue levels of bone resorptive cytokines in periodontal disease. J Periodontol. 1991;62:504–509. doi: 10.1902/jop.1991.62.8.504. [DOI] [PubMed] [Google Scholar]
- 26.Takashiba S, Shapira L, Amar S, Van Dyke T E. Cloning and characterization of human TNF alpha promoter region. Gene. 1993;131:307–308. doi: 10.1016/0378-1119(93)90314-s. [DOI] [PubMed] [Google Scholar]
- 27.Takashiba S, Van Dyke T E, Shapira L, Amar S. Lipopolysaccharide-inducible and salicylate-sensitive nuclear factor(s) on human tumor necrosis factor alpha promoter. Infect Immun. 1995;63:1529–1534. doi: 10.1128/iai.63.4.1529-1534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trede N S, Tsytsykova A V, Chatila T, Goldfeld A E, Geha R S. Transcriptional activation of human TNF-alpha promoter by superantigen in human monocytic cells: role of NF-kappa B. J Immunol. 1995;155:902–908. [PubMed] [Google Scholar]
- 29.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in culture human monocytic leukemia cells by phorbol diester. Cancer Res. 1982;42:1530–1536. [PubMed] [Google Scholar]
- 30.Valledor A F, Borras F E, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998;63:405–417. doi: 10.1002/jlb.63.4.405. [DOI] [PubMed] [Google Scholar]