Abstract
We have identified and characterized a secreted protein, designated Pic, which is encoded on the chromosomes of enteroaggregative Escherichia coli (EAEC) 042 and Shigella flexneri 2457T. The product of the pic gene is synthesized as a 146.5-kDa precursor molecule which is processed at the N and C termini during secretion, allowing the release of a mature protein (109.8 kDa) into the culture supernatant. The deduced amino acid sequence of Pic shows high homology to autotransporter proteins, particularly a subgroup termed the SPATEs (serine protease autotransporters of the Enterobacteriaceae). Present in all members of this subgroup is a motif similar to the active sites of certain serine proteases. Pic catalyzes gelatin degradation, which can be abolished by disruption of the predicted proteolytic active site. Functional analysis of the Pic protein implicates this factor in mucinase activity, serum resistance, and hemagglutination. Our data suggest that Pic may be a multifunctional protein involved in enteric pathogenesis.
Diarrhea is a major cause of mortality and morbidity worldwide, particularly among children. For all diarrheal pathogens there is a sequence of mucosal interactions requiring mucus attachment and penetration followed by interaction with the tissue and elicitation of host damage (76). Virulence determinants are produced by pathogens to allow them to breach the barriers to infection and to execute this sequence. Adherence is a critical first step in mucosal interactions. It has been suggested that the mucus layer covering the epithelial surface may protect against colonization of the intestinal tract by enteric pathogens by inhibiting their access to enterocytes (20, 84).
Two of the most important enteric bacterial pathogens are Shigella flexneri and diarrheagenic Escherichia coli. Within the species E. coli, there are commensal strains and a variety of pathogenic strains including enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enterohemorrhagic E. coli (EHEC), and enteroaggregative E. coli (EAEC). EAEC has been implicated as an emerging cause of pediatric diarrhea in the developing world (51, 56, 71) and as the causative organism in a number of outbreaks in the developed world (13, 37, 49, 77). EAEC-mediated diarrhea is characterized by the formation of a thick mucus gel on the intestinal mucosa and by mucosal damage. EAEC diarrhea is predominantly secretory in nature; stools contain mucus and often blood but generally no polymorphonuclear leukocytes (17, 50). Although enterotoxins have been described in EAEC (23, 74), the full picture of EAEC pathogenesis has not yet been described.
S. flexneri is closely related genetically to E. coli, and it has been suggested by a number of groups that S. flexneri represents a subspecies of E. coli (40, 64, 69). Nevertheless, S. flexneri elicits a distinctive and complex disease, bacillary dysentery, caused by invasion of the colonic epithelial cells and characterized by an intense inflammatory response (57). Notably, however, many cases of shigellosis are manifested as watery diarrhea, which may be mediated by one or more enterotoxins.
Research on S. flexneri pathogenicity has focused mainly on the plasmid-encoded genes necessary for penetration and intercellular dissemination (48, 72). Likewise, factors associated with EAEC-mediated diarrhea have been localized to a 65-MDa plasmid, which is required for expression of aggregative adherence fimbriae (18) and several putative toxins (23, 73). Nevertheless, evidence exists for chromosomal virulence factors in both S. flexneri (54, 65) and EAEC (19). In this article we report the cloning, nucleotide sequence analysis, and expression of the gene encoding a 116-kDa secreted protein described by Eslava et al. (24), which is located on the chromosome of both EAEC and Shigella strains. We have termed this gene pic and the gene product Pic (for “protein involved in intestinal colonization”). This protein is an extracellular serine protease which displays in vitro mucinolytic activity, serum resistance, and hemagglutination. The protease is synthesized as a large precursor, which is processed during secretion by the autotransporter secretion mechanism.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli 042, a known diarrheal pathogen, was isolated from a child with diarrhea during the course of an epidemiological study in Peru (50). E. coli HB101 was used for genetic manipulations. Strains were passed routinely on Luria-Bertani broth (L-broth) or agar with the following antibiotic supplements where appropriate: ampicillin (100 μg/ml), kanamycin (50 μg/ml), nalidixic acid (50 μg/ml), and tetracycline (10 μg/ml). All strains were stored at −70°C in Trypticase soy broth with 15% glycerol.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| E. coli | ||
| 042 | Wild-type EAEC strain from Peru | 50 |
| HB101 | K-12/B hybrid | 7 |
| KS474 | F− ΔlacX74 galE galK thi rpsL (strA) ΔphoA (PvuII) degP41 (ΔPstI-Kanr) | 4 |
| UT5600 | F−ara-14 leuB6 azi-6 lacY1 tsx67 proC leu-6 trpE38 rfbD1 rpsL109 xyl-5 mtl-1 thi-1 entA403 Δ(ompT-fecC266) ΔompP | 41 |
| JCB517 | araD139 Δ(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi phoR zih12::Tn10 dsbA::Kan-1 | 81 |
| S. flexneri | ||
| 2457T | Wild-type S. flexneri 2a | 21 |
| Plasmids | ||
| pACYC184 | Small low-copy-number cloning vector (Camr, Tetr) | 12 |
| pPic1 | 5.8-kb EcoRI-EcoRV chromosomal fragment of EAEC 042 cloned into pACYC184 (Tetr) | This study |
| pPicS258I | pPic1 (Tetr) S258 to I258 | This study |
Protein preparation and analysis.
Bacteria were harvested in the late logarithmic phase of growth by centrifugation at 16,000 × g for 10 min at 4°C. Envelopes were prepared by a modification of the method outlined by Caffrey and Owen (10). Briefly, envelopes isolated following French pressure lysis of bacterial cells were sedimented by centrifugation (48,000 × g for 60 min at 4°C) and washed twice in 30 ml of 10 mM Tris-HCl (pH 7.2) and once in 3 ml of the same buffer. The standard conditions for sedimentation of envelope fractions were 48,000 × g for 60 min at 4°C. The envelopes were finally resuspended in 1 ml of the same buffer and aliquoted for storage at −70°C for further manipulations.
To prepare culture supernatant fractions, strains were grown overnight at 37°C in 100 ml of L-broth. After centrifugation at 12,000 × g for 10 min, supernatants were concentrated and size fractionated with Ultrafilters (Millipore) with a 100-kDa cutoff.
One-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (43) was performed with 12.5% acrylamide separating gels and 4.5% acrylamide stacking gels. Samples were routinely heated for 5 min at 100°C in Laemmli (43) sample buffer before being loaded. Proteins were detected by staining with Coomassie brilliant blue R250. Western immunoblotting was performed essentially as described by Caffrey et al. (9). Dried skim milk (5%) was used as a blocking reagent. Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) was used as the localizing reagent, and reacting antigens were visualized with 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium as described elsewhere (3).
To determine N-terminal amino acid sequences, the proteins were separated by SDS-PAGE as described by Laemmli (43) and transferred to Immobilon polyvinylidene difluoride membranes (Millipore) prior to amino-terminal sequencing. Amino-terminal sequencing was performed by automated Edman degradation at the Protein and Nucleic Acid Facility, Stanford University, Palo Alto, Calif.
Preparation of antisera.
Rabbit antiserum specific for Pic was raised by subcutaneous injection of Pic preparations in Freund's adjuvant as described previously (33). Preparations of immunoglobulins were adsorbed successively by incubation at 4°C for 24 h with an E. coli protein-agarose suspension (Sigma).
PCR procedures.
Amplifications were performed with 500 ng of purified chromosomal DNA as templates and 0.2 mM each primer in a 100-μl reaction mixture containing 2 U of Taq DNA polymerase, 50 mM each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 10 μl of the manufacturer's buffer (GIBCO BRL). Forty cycles of 1 min of denaturation at 94°C, 1 min of primer annealing at 60°C, and 1 min of extension by Taq polymerase at 72°C were carried out. PCR was performed with the following primers pairs: the forward and reverse primers 5′-GGTACCGGGNATHGTNCGNTCNGAYAT-3′ and 5′-GAGCTCNGTRTCNGCYTGYTGRTT-3′ (where N, H, Y, and R are standard nucleotide abbreviations), which corresponded to the amino acid sequences GIVRSD and NQQADT, respectively, and the forward and reverse primers 5′-CTGAATTCCTCCCCCTTACCGAAGACC-3′ and 5′-TGCCATGTGGCAGCCTGAGTTCACAG-3′, which are located, respectively, 183 bp upstream and 96 bp downstream of the pic gene.
Plasmid and strain construction.
To create a minimal clone expressing Pic (pPic1), a 5.8-kb EcoRI-ScaI chromosomal fragment from E. coli 042 was cloned into the EcoRI-ScaI sites of pACYC184, giving rise to a clone conferring tetracycline resistance. This construction contains the complete Pic coding region, native promoter, and downstream termination motif with an additional 1,000 bp upstream of the ATG initiation codon and 750 bp downstream of the termination codon.
Site-directed mutagenesis was performed with the Quikchange mutagenesis kit (Stratagene) as specified by the manufacturer. The template used for construction of the site-directed mutant (pPicS258I) was pPic1. The oligonucleotides used for this purpose were 5′-GGAGCCCCTGGGGATATTGGTTCTCCTTTGTTTGCTTATG-3′ and 5′-CATAAGCAAACAAAGGAGAACCAATATCCCCAGGGGCTCC-3′. Following mutagenesis, the S258I mutation was confirmed by double-strand sequencing of the area encompassing the serine protease active site.
DNA sequence determination and analysis.
DNA sequence determination was performed on an Applied Biosystems 373A automated sequencer by dye terminator cycle sequencing with Taq polymerase (Perkin-Elmer Corp.) as specified by the manufacturer; sequencing was performed in the Biopolymer Laboratory, Department of Microbiology and Immunology, University of Maryland School of Medicine. The nucleotide sequence was analyzed with the sequence analysis tools on the EXPASY molecular biology server (24a) and the Wisconsin GCG sequence analysis package available through the Center of Marine Biotechnology, University of Maryland. The predicted amino acid sequence of each open reading frame (ORF) was compared with proteins listed in EMBL/GenBank by using the GCG TFASTA program and the BLAST algorithm (National Center for Biotechnology Information). Secondary-structure predictions were performed by the Jähnig (39), Emini et al. (22), or Kyte and Doolittle (42) algorithms, which are available in the HUSAR program package of the Deutsches Krebsforschungszentrum (Heidelberg, Germany). DNA analysis and manipulations were performed by standard methods (3). Plasmid DNA was extracted with the Plasmid Midi Kit (Qiagen Inc.). Purification of DNA fragments and extraction from agarose gel slices were performed with the PCR Wizard Kit (Promega). Plasmid DNA was introduced into appropriate strains by transformation of competent cells by the method of Hanahan (32).
Hemagglutination.
Erythrocytes (RBCs) were obtained as lyophilized preparations from Sigma. They were washed twice in phosphate-buffered saline (PBS) and resuspended to a final concentration of 3% (vol/vol) with 1% mannose (wt/vol). Preparations of Pic were serially diluted in 96-well plates to give a final volume of 100 μl. Subsequently, 100 μl of an RBC suspension was added; after gentle mixing, the reaction mixture was incubated for 20 min at room temperature. Only visible clumping of RBCs was considered to represent a positive result. Inhibition of hemagglutination was performed by the addition of anti-Pic antibodies (diluted 1:50 in PBS) to the serially diluted Pic protein and incubating at room temperature for 30 min prior to the addition of the erythrocytes. The reaction was continued as above.
Protease activity.
Gelatinase and caesinase zymogram analyses were performed by electrophoresing concentrated supernatants of HB101(pPic1) in a precast zymogram gel (Novex). After electrophoresis, the gel was incubated for 30 min at room temperature in zymogram renaturing buffer (2.5% Triton X-100), equilibrated for 30 min with zymogram developing buffer (1.21 g of Tris base, 6.3 g of Tris-HCl, 11.7 g of NaCl, and 0.74 g of CaCl2 per liter), incubated at 37°C for 4 h with fresh developing buffer, and stained with Coomassie blue R-250 for 30 min. Proteolytic activity was easily identified as the presence of clear bands against a dark blue background of nonhydrolyzed proteins.
Mucinase determinations, with ovomucin as the substrate, were performed as described previously (28) with 100 mM Tris-buffered saline (pH 8.0) as the diluent and 1% aqueous cetyltrimethylammonium bromide (CTAB) to precipitate the undigested mucin. The ovomucin was prepared by adding 12 egg whites to 4 liters of ice-cold distilled water. The supernatant was decanted and lightly centrifuged, and the precipitate was dissolved in a minimal amount of 5% NaCl. The mucin was titrated to determine the lowest concentration which in 1 ml would form a clot on addition of 50 μl of the CTAB solution. In the ovomucinase assay, 500-μl preparations of the test substances were added to 500 μl of mucin, which was prepared so that when the mucin was diluted by 50% it would still form a clot. These test mixtures were incubated at 37°C for 30 min, and CTAB was added. The tubes were swirled to determine clot formation. A substance was determined to have mucinase activity if formation of a fibrous clot was absent upon addition of CTAB.
Mucinolytic activity for hog gastric mucin (Sigma), bovine submaxillary mucin (Sigma), and crude mouse large-intestine mucin was detected and quantified by several methods. In the first method, Pic protein preparations were incubated for 24 h at 37°C on a medium containing 1.5% agarose, 1.0% glucose, and 0.5% mucin in L-broth. The plates were subsequently stained with 0.1% amido black in 3.5 M acetic acid. Zones of mucin lysis are observed as discolored halos around colonies or holes bored into the medium (15). In the second method, treated (37°C for 12 h) and untreated mucin specimens were electrophoresed on an SDS–7% polyacrylamide gel as previously described (75). The gel was then incubated in 0.2% periodic acid for 1 h and then in Schiff reagent (PAS) until a color change was evident. The reaction was stopped by the addition of 7.5% acetic acid. In the third method, crude mouse mucus preparations (see below) were treated with Pic protein preparations at 37°C for 12 h before being applied to a gel filtration column containing Sephacryl S400. The column was calibrated with blue dextran, and the position of the excluded void volume is indicated. Fractions (0.8 ml) were collected during chromatography. PAS staining was performed on 300-μl aliquots and measured at 555 nm as previously described (45), and the profiles were compared with those obtained for crude mucus.
Isolation of mouse intestinal mucus glycoprotein.
Mouse mucus glycoprotein was purified essentially as described previously (46). The large intestines of freshly sacrificed mice were dissected out and flushed with sterile saline, and the mucosal surface was exposed by longitudinal dissection. The intestines were placed in 0.2 M NaCl, and the tissue was homogenized for 30 s with a Tissue Tearor (Biospec Products Inc.). Soluble mucus was separated from tissue debris by centrifugation at 6,000 × g for 30 min. This preparation was then layered on a gel filtration column containing Sephacryl S400. Mucin is the predominant glycoprotein eluting in the void volume and was quantitated by a spectrophotometric PAS assay as described previously (45).
Complement inactivation assay.
Normal adult human serum and C9-deficient sera were obtained from Sigma. Complement was inactivated by heating the normal serum at 56°C for 30 min. Samples (20 μl) of normal, C9-deficient, and heat-inactivated serum were incubated with 160 μl of PBS or 160 μl of a Pic preparation (200 μg ml−1) for 30 min at 37°C. After the initial incubation, 20 μl of a DH5α culture, grown to mid-logarithmic phase (optical density at 600 nm = 0.5) in L-broth, was added to the normal serum, the heat-inactivated serum, and the serum pretreated with Pic. Incubation was continued at 37°C. At 0, 15, and 30 min, 10-μl aliquots were removed, spread onto prewarmed agar plates, and incubated overnight at 37°C. In some experiments, organisms were exposed to normal serum containing 10 mM EGTA and MgCl2 to inactivate the alternative pathway (26).
RESULTS
Cloning and sequence analysis of the EAEC pic gene.
We and others have found that many EAEC strains secrete a high-molecular-mass protein of ca. 116 kDa (23, 24, 52). We have previously determined the N-terminal sequence of this protein [GI(V/P)RSDI] and found that it was 100% identical to the predicted product of a S. flexneri gene in the database (accession no. U35656) (19). Of note is that this 4.1-kb gene was predicted to encode a 146-kDa protein on the coding strand and the enterotoxin ShET1 on the antisense strand (see below). We sought to clone and sequence the EAEC gene and to characterize its product. The N-terminal amino acid sequences of two internal peptides, generated by digestion of the 116-kDa EAEC supernatant protein with the endopeptidase Lys-C, were determined and identified as TGDGIVVLNQQADTAGNIQ and XLFVXXAX.
Mixtures of oligonucleotides, corresponding to the amino acid sequences GIVRSD and NQQADT derived from peptide sequencing, were used as primers in PCRs performed with EAEC 042 chromosomal DNA as the template. A 1,198-bp PCR product was obtained and sequenced by primer walking. DNA sequence analysis revealed that this stretch of DNA was 99.9% identical to an internal portion of the S. flexneri gene. In light of this observation, primers which corresponded to sequences positioned 183 bp upstream and 96 bp downstream of the 4.1-kb Shigella gene were designed. As expected, a PCR product of 4,395 bp was amplified. Rather than using PCR, a minimal clone of the gene (pPic1) was constructed by cloning a 5.8-kb native chromosomal segment (EcoRI-ScaI) from E. coli 042 into pACYC184.
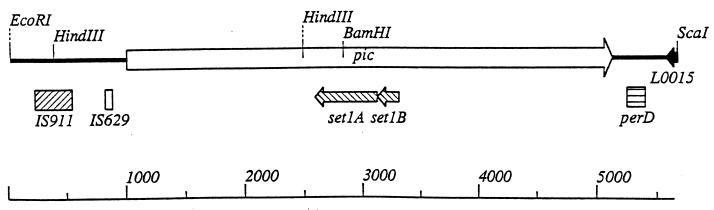
The sequence of the 5.8-kb EAEC chromosomal fragment was determined in its entirety (Fig. 1). The sequence contained a large ORF, designated the pic gene, of 4,116 nucleotides (accession no. AF097644). The insert of pPic1 spanned from 1,000 bp upstream of the pic gene to 750 bp downstream of the termination codon and included a predicted promoter and rho-independent termination motif. The predicted protein (Pic) contained the empirically determined N-terminal amino acid sequences. The overall G+C content of pic is 49.54%. The promoter region contained a −10 sequence (TGTAAA) which was positioned 19 bp upstream of the predicted ATG start codon and a −35 region (TTTACT) which was separated by a further 17 bp. A predicted Shine-Dalgarno site, with the sequence GGAG, was identified 9 bp upstream of the proposed start codon. A sequence similar to a rho-independent transcriptional terminator was present beginning 11 nucleotides beyond the stop codon and contains interrupted inverted repeats with the potential for forming a hairpin structure containing a stem of 10 bases and a loop of 4 bases.
FIG. 1.
Map of the chromosomal fragment cloned into pACYC184. At the 5′ end of the insert are the IS-homologous sequences IS911 and IS629, followed by the pic gene and subsequently by the perD IS-like element. The positions within the pic gene of the set1A and set1B genes which encode the two subunits of the ShET1 enterotoxin are also indicated.
Upstream from the EAEC pic gene are two insertion (IS)-like elements (Fig. 1). A stretch of sequence which is 97% identical to an IS629 element described for Shigella sonnei (47) is located 121 bp upstream of the first ATG codon of pic. This element is 95% identical to an IS1203 element described in pathogenic E. coli O111:H− (58). Preceding this sequence is a stretch of sequence identical to IS911 from S. dysenteriae (60). In addition, 118 bp downstream of pic lies a stretch of sequence homologous to the IS-like ORF perD (30) (Fig. 1). Another 168 bp downstream is a sequence corresponding to an ORF from the cryptic prophage 933L, which delimits the left-hand side of the EHEC O157:H7 locus of enterocyte effacement (59). Although the equivalent flanking sequence in S. flexneri 2457T has not been determined, there is limited sequence upstream of the she gene corresponding to an IS629 element. Rajakumar et al. (65) determined the flanking sequence of the she gene in S. flexneri SBA1336 (a derivative of strain YSH6000T) and found a complete copy of the IS629 element upstream of she. However, no sequence resembling an IS911 element or sequence similar to perD was identified.
Assuming that the first ATG is the start codon, the pic gene encodes a primary translation product of 1,372 amino acids with a molecular mass of 146,450 Da and a calculated isoelectric point of 6.24. In addition to the almost complete identity (99.7%) displayed between the gene from E. coli 042 and that from S. flexneri 2457T, the Pic polyprotein displayed significant homology to a group of autotransporter proteins which we have termed the SPATE subfamily (serine protease autotransporters of the Enterobacteriaceae) and which includes Pet (an enterotoxin of EAEC [23]), EspP (a cytotoxin of EHEC [8]), EspC of EPEC (79), SepA (a putative cytotoxin of S. flexneri [5]), and Tsh (a hemagglutinin-haemoglobin protease from an avian-pathogenic strain of E. coli [55, 63]). Notably, the homologies displayed are not uniformly distributed over the sequences; the N-terminal passenger domain (the secreted protein) displays 49.6, 46.7, 34.6, 31.4, and 31.2% identity to the SepA, Tsh, EspC, EspP, and Pet α-domains, respectively, whereas the C-terminal β-domain (the C-terminal β-barrel) exhibits 76.9, 62.5, 79.4, 80.1, and 79.8% identity, respectively.
The Pic polyprotein possesses several features characteristic of the SPATE subfamily of the autotransporters. First, a consensus serine protease motif (GDSGSP) has been reported for all members of the SPATE subfamily, in addition to the IgA1 proteases and the Hap protein of Haemophilus influenzae (35). At the corresponding site in the polyprotein, the sequence was determined to be GDSGSP. In the IgA1 protease, this site acts as the catalytic site (31); however, a definitive function has not been determined for this motif in any member of the SPATE subfamily. Second, the predicted N terminus possesses the characteristics of an extended signal sequence, with an N-domain possessing six positively charged amino acids (R24RVIKKTCRR), a hydrophobic domain spanning 16 neutral amino acids, and a C-domain which features a sequence (SQA55) compatible with the consensus for a signal peptidase recognition site (38). Indeed, computer-aided analysis of the signal sequence predicts a cleavage site which agrees exactly with the position indicated by N-terminal amino acid sequencing of the secreted protein. Thus, assuming a correct prediction of the start methionine, this signal sequence would be unusually long for E. coli, but it is similar to those predicted for all other members of the SPATE subfamily and a limited number of other autotransporters (35).
Presence of the ShET1 toxin genes.
Contained within the EAEC pic gene, on the complementary noncoding strand, are two consecutive ORFs (Fig. 1) of 186 and 534 nucleotides. These consecutive ORFs are positioned from nucleotides 2486 to 2301 and from 2297 to 1764 with respect to the first nucleotide of the large pic gene. Assuming that the first ATG of each of the consecutive ORFs is the start codon, these genes encode primary translation products of 61 and 177 amino acids, with molecular masses of 6,850 and 19,783 Da, respectively. Comparison of the nucleotide and deduced amino acid sequences with those listed in GenBank databases revealed 100% overall identity to the S. flexneri ShET1 enterotoxin subunits, Set1A and Set1B (25).
Processing of the secreted protein.
Processing of the Pic protein in Shigella has not been described. By N-terminal sequencing, the signal peptidase-processed EAEC Pic protein begins at G56, is composed of 1,317 amino acids, and has a calculated molecular mass of 140,403 Da. In view of the homology to other autotransporters and since the secreted protein has an observed molecular mass of 116 kDa, it seems likely that a further posttranslational cleavage step is required for secretion of the mature protein. Members of the autotransporter family of proteins are exported through the outer membrane of the bacterium via a characteristic C-terminal amphipathic region (β-domain) comprising an even number of antiparallel β sheets; this region of the protein forms a β-barrel structure in the outer membrane, through which the passenger domain of the protein passes (35). The high identity between the β-domains of Pic and other members of the SPATE subfamily suggests that the β-domain functions as an outer membrane translocator. In view of the highly conserved cleavage site (EVN-NLN) between the passenger and β-domains of the SPATE subfamily, the cleavage site was predicted to be between residues N1095 and N1096. Such a cleavage event would result in a secreted protein of 109.8 kDa, a mass which agrees well with the mass of 116 kDa predicted for the secreted protein by SDS-PAGE analysis.
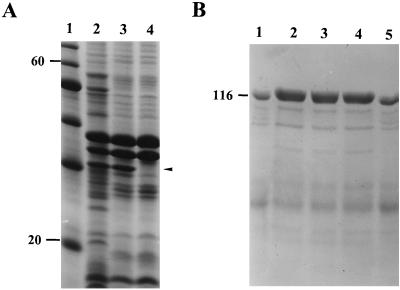
Previous studies have shown that the passenger domains of autotransporters are surface expressed or secreted into the external milieu, while the β-domain remains as an integral membrane-bound protein with features characteristic of outer membrane proteins (35). To determine if this was the case for our protein, the outer membrane β-domain was visualized by SDS-PAGE analysis of envelopes from HB101(pPic1) extracted with Triton X-100 and compared with similar extracts of HB101(pACYC184). These analyses revealed a ca. 30-kDa species in the fractions obtained from HB101(pPic1) that was absent from similar fractions of the control (Fig. 2A).
FIG. 2.
SDS-PAGE analysis of Pic processing. (A) Analysis of Triton X-100-insoluble fractions from HB101(pPic1) (lane 2), HB101(pPicS258I) (lane 3), and HB101(pACYC184) (lane 4). Lane 1 contains molecular mass markers (in kilodaltons). The position of the β-domain is indicated. (B) Analysis of concentrated culture supernatants from HB101(pPic1) (lane 1), JCB517(pPic1) (lane 2), UT5600(pPic1) (lane 3), KS474(pPic1) (lane 4), and HB101(pPicS258I) (lane 5). The position of a molecular mass marker is indicated (in kilodaltons).
To elucidate the organization of the β-domain from the polyprotein, secondary-structure analyses were performed from the putative N1096 cleavage site to the terminal phenylalanine residue. Kyte and Doolittle (42) hydrophobicity analyses did not reveal any linear stretches of hydrophobic amino acids of the type associated with α-helical transmembrane segments. However, predictions according to the algorithm Hβ(i) = [h(i ± 4) + h(i ± 2) + h(i)]/5 of Jähnig (39) indicate with good probability that the β-domain consists of at least 10 membrane-spanning amphipathic β-strands. Four additional β-strands are predicted with lower probability. These β-sheets are interrupted by large external loops and generally short periplasmic loops spanning amino acids 1096 to 1372 of the precursor. Furthermore, regions of high surface probability, predicted by Emini et al. (22), are in good agreement with the positions of the β-strands, since such regions are always located between the β-strands.
The possible role of several membrane-associated enzymes in the processing and export of the secreted protein was investigated. The pPic1 clone was transformed into E. coli JCB517 (dsbA), UT5600 (ompP ompT), and KS474 (degP). The resulting constructions were screened for the correct processing of the polyprotein by SDS-PAGE analysis of concentrated culture supernatants. Each strain yielded a 109.4-kDa species, suggesting that processing of the precursor occurs without the interaction of the OmpP, OmpT, or DegP proteases and in the absence of the DsbA isomerase (Fig. 2B).
We investigated the role of the serine protease motif in proteolytic processing of the polyprotein. By using site-directed mutagenesis, the putative active-site serine residue at position 258 (S258) was changed to isoleucine, generating the construct pPicS258I. Analysis of the proteins in culture supernatants revealed similar levels of protein to those derived from the wild-type clone, pPic1 (Fig. 2B). No difference in the molecular weights of the secreted proteins was observed. Comparison of envelope preparations from HB101(pPic1) and HB101(pPicS258I) by SDS-PAGE revealed the presence of β-domains of similar size (Fig. 2A).
Regulation of Pic expression in EAEC.
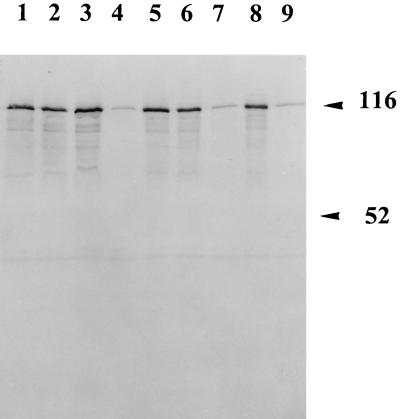
Variations in environmental parameters often affect the level of expression of certain virulence determinants. To determine the parameters involved in Pic expression, strain 042 was grown in L-broth under different conditions to an optical density at 600 nm of 0.5 and concentrated culture supernatants were analyzed for the Pic protein by Western immunoblotting. As previously mentioned, on the antisense strand of pic is encoded the ShET1 enterotoxin, which is iron regulated (25). To determine if expression from the pic gene is iron regulated as well, strain 042 was grown in L-broth supplemented with either 25 μg of ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDA) ml−1 (low iron) or 0.5 mM FeSO4 (high iron). Although 25 μg of EDDA ml−1 induced growth retardation, no significant difference was observed in the levels of protein apparent in preparations from high- or low-iron-containing medium (Fig. 3, lanes 1 and 2).
FIG. 3.
Regulation of Pic expression. An SDS-PAGE analysis of concentrated L-broth culture supernatants of E. coli 042 is shown. Culture conditions: 25 μg of EDDA ml−1 (lane 1), 0.5 mM FeSO4 (lane 2), pH 9.0 (lane 3), pH 5.0 (lane 4), no NaCl (lane 5), 0.4 M NaCl (lane 6), 24°C (lane 7), 37°C and pH 7.0 (lane 8), and 42°C (lane 9). The positions of some molecular mass markers are indicated (in kilodaltons).
Expression of the EHEC EspP protein, a Pic homologue, is regulated by temperature and pH (8). To determine if pH played a role in the regulation of expression, we analyzed the expression of the Pic protein under different pH conditions (pH 5 to 9). In supernatants of cultures grown at pH 5.0 (at 37°C [Fig. 3, lane 4]), less protein was found than in supernatants from cultures grown at pH 9.0 (lanes 3). To exclude the possibility that the protein was degraded under these acidic conditions, the native protein was incubated at pH 5.0 for a comparable length of time and analyzed by SDS-PAGE. No degradation products were visible compared to controls (data not shown). In addition, we analyzed the expression of the Pic protein from E. coli 042 under conditions of different osmolarities. The level of expression of the protein in L-broth containing no NaCl (low osmolarity [Fig. 3, lane 5]) was comparable to that in normal L-broth at 37°C (lane 8) and to that for bacteria grown in L-broth under conditions of high osmolarity (0.4 M NaCl) (lane 6).
Since the expression of many bacterial virulence factors is thermoregulated, it was of interest to determine if the expression of the pic gene was also regulated by temperature. Production of the secreted protein was clearly temperature dependent, since expression was significantly increased when bacteria were grown at 37°C compared to 24 and 42°C (Fig. 3, lanes 7 to 9). In the supernatant from bacteria grown at all temperatures in L-broth, many minor bands were visible after Coomassie blue staining, indicating that even during exponential growth a certain percentage of bacteria were lysed (Fig. 3).
Protease activity.
The observed sequence similarities between the secreted Pic protein and other proteases, in addition to the fact that the polyprotein features a serine protease site, suggests that Pic has proteolytic activity. Consequently, the ability of Pic to act as a protease was tested by separating concentrated supernatants through casein or gelatin zymogram gels. Concentrated supernatants from strains HB101(pPic1) yielded zones of clearing on gelatin gels whereas HB101(pPicS258I) and HB101(pACYC184) did not exhibit proteolytic activity (data not shown). No activity was observed in the casein zymograms. In view of the similarity to the IgA1 proteases, Pic preparations were incubated with human IgA, IgM, and IgG preparations; however, no evidence of immunoglobulin degradation was observed.
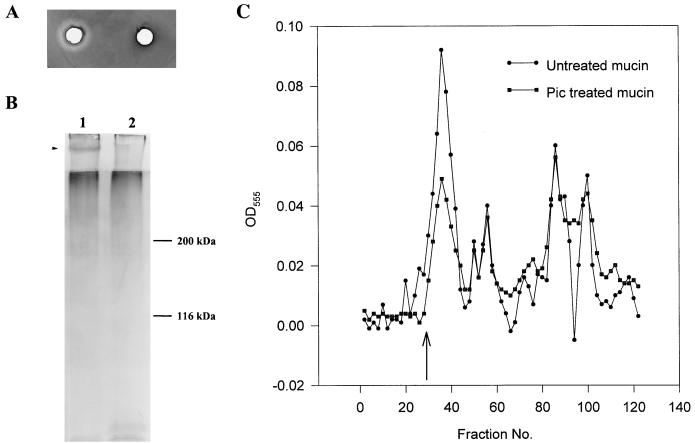
Preliminary experiments had suggested that the 116-kDa protein from S. flexneri was able to cleave ovomucin (53). We observed ovomucinase activity with preparations of the Pic protein from EAEC (data not shown); this activity was inhibited by rabbit antiserum against the Pic protein. To determine whether the Pic protein cleaved mammalian mucin species, several experiments were performed with hog gastric mucin, bovine submaxillary mucin, and mouse crude mucin. First, E. coli 042 and S. flexneri 2457T were grown on agarose plates containing glucose and one of the mucin species listed above (see Materials and Methods). However, no clear zones of mucin lysis were apparent around any of the colonies. In contrast, zones of clearing were apparent when Pic protein was added to wells bored in the agar containing the bovine and murine mucin species (Fig. 4A). Such diffusible mucinolytic activity was absent on plates containing hog gastric mucin. Furthermore, the mutant Pic protein from the serine protease mutant (S258I) failed to produce zones of clearing on mucin species.
FIG. 4.
Mucinolytic activity of the 116-kDa secreted protein. (A) Zones of mucin lysis on bovine submaxillary mucin are visible after treatment with Pic protein and staining with amido black (left). No lysis was visible in the section treated with similar preparations of the Pic protein containing a mutation in the serine protease active site (S258I) (right). (B) SDS-PAGE analysis of untreated crude mouse mucus (lane 1) and mucus treated with Pic protein (lane 2). The gel is PAS stained, and the band which is cleaved is indicated, as are the positions of some molecular mass markers. (C) Sephacryl S-400 column chromatography of untreated crude mouse mucus and mucus treated with the concentrated secreted protein. The column was calibrated with dextran blue, and the position of the excluded void volume is indicated. Fractions (0.8 ml) were collected during chromatography. PAS staining was performed on 300-μl aliquots and measured at 555 nm, and the absorbance has been graphed. OD, optical density.
Samples of crude murine mucus were incubated overnight at 37°C with and without the Pic protein. These samples were analyzed by two methods. First, equal amounts were applied to an SDS–7% polyacrylamide gel, and after electrophoresis the gel was stained with PAS stain for glycoprotein. In the untreated sample, a high-molecular-mass PAS-staining band (>200 kDa) was evident, whereas in the treated sample, this band was absent (Fig. 4B). The large PAS-staining band remained evident in SDS-PAGE profiles of crude mucus after treatment with Pic protein from the (S258I) strain. Second, the treated and untreated samples were applied to a Sephacryl S-400 column, and the eluted fractions were monitored for PAS positivity in a spectrophotometer at 555 nm. On gel filtration, all mucus samples contained both excluded glycoprotein (indicative of native mucin) and lower-molecular-weight glycoproteins. Similar profiles were obtained when the mucus was treated with supernatants from HB101(pACYC184). However, mucus treated with Pic protein preparations produced a gel filtration profile different from that of crude mucus. This was manifested as a reduced height (by ∼50%) of the predominant high-molecular-weight PAS-staining species excluded at the void volume. In addition, the treated mucus yielded more lower-molecular-weight glycoprotein than the normal mucus did (Fig. 4C).
Effect of Pic on serum resistance.
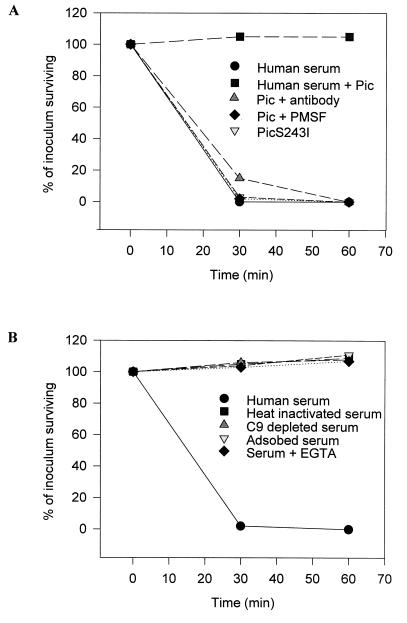
Secreted proteases may exhibit functions other than mucinase activity that promote the intestinal survival of enteric pathogens. Other members of the autotransporter family have been shown to confer serum resistance (1, 26), adhesion (6, 80), and hemagglutination (63). To examine the effect of Pic on serum resistance, E. coli DH5α was tested in a serum bactericidal assay with Pic protein preparations. As expected, DH5α was readily killed by normal human serum (Fig. 5). However, when the normal serum was pretreated with Pic for 30 min at 37°C prior to the addition of DH5α, the bacteria were able to grow normally. In contrast, pretreatment of the Pic protein with anti-Pic antibodies prior to incubation with normal serum abolished the ability of the protein to protect the DH5α bacteria from serum bactericidal activity. To assess whether the serine protease domain of Pic was involved in the serum resistance activity of Pic, the protein was treated with 2 mM phenylmethylsulfonyl fluoride, and washed in 100 volumes of PBS before being incubated with normal serum. This treatment abolished the activity of Pic in the assay. Furthermore, pretreatment of serum with the Pic site-directed mutant S258I did not abolish the antibacterial activity of normal human serum (Fig. 5A).
FIG. 5.
Susceptibility of DH5α to killing by normal human serum. (A) Killing of DH5α is inhibited in the presence of Pic protein preparations but not in the presence of protein from the serine protease mutant (S258I), phenylmethylsulfonyl fluoride (PMSF), or antibodies to Pic. (B) Serum killing of DH5α occurs via the classical pathway of complement. Organisms were exposed to normal serum, antibody-depleted serum, C9-deficient serum, and serum with added EGTA and MgCl2. Data are presented as the percentage of the original inoculum remaining at each time point.
Heat inactivation of the serum completely abolished its antibacterial effect, suggesting that the serum component mediating the killing of DH5α is complement (Fig. 5B). Furthermore, the use of C9-deficient serum in place of normal serum also resulted in loss of the killing effect. To test which of the complement pathways was involved, DH5α was exposed to serum containing 10 mM EGTA and MgCl2. This treatment inactivates the classical pathway by chelating Ca2+ (67). As indicated in Fig. 5, unlike for normal serum, the killing effect was abrogated in Ca2+-depleted serum. Similarly, depletion of antibodies in the serum by adsorption (for 18 h at 4°C) with heat-inactivated DH5α (heated at 100°C for 5 min) eliminated the ability of this serum to kill the bacteria (Fig. 5B). These data suggest that Pic acts on the classical pathway or on a component essential for the early steps of the complement cascade.
Hemagglutination.
Previous experiments by other investigators have shown that Tsh, a close homologue of Pic, is a hemagglutinin of chicken RBCs (63). Therefore, we tested whether Pic was also a hemagglutinin. Protein preparations derived from HB101(pPic1) were incubated with RBCs from different species (as described in Materials and Methods). Preparations of the proteins weakly agglutinated RBCs from rats, pigs, rabbits, horses, and sheep; however, they did not agglutinate human or chicken RBCs (Table 2). Interestingly, the protein preparation agglutinated the rat RBCs at a higher titer than any for other species. Rabbit antiserum against Pic, adsorbed with whole cells, failed to agglutinate RBCs. Preincubation of the protein preparations with anti-Pic antibodies inhibited the hemagglutination of RBCs from all species.
TABLE 2.
Relative hemagglutinating activity of Pic
| Species of origin | Activitya |
|---|---|
| Rat | ++ |
| Chicken | − |
| Human | − |
| Pig | ± |
| Horse | + |
| Sheep | + |
| Rabbit | ± |
Agglutination with 1:8 (++), 1:4 (+), and undiluted (±) Pic protein preparations.
DISCUSSION
In this study, we have characterized the gene encoding a 109.8-kDa extracellular protein which is secreted by two enteric pathogens, S. flexneri 2a and EAEC. Analysis of nucleotide sequence data provides the basis for this distribution. A number of IS-like elements were found flanking the pic gene; of note is the presence of a fragment of an IS629 element and an IS911 element which have been found flanking other members of the SPATE family (35). The flanking sequence in EAEC differs from that of the identical pic gene in Shigella, suggesting that this gene has been acquired by horizontal transfer. The presence of the gene on a pathogenicity island in Shigella (65) suggests that the pic gene may also be located on an as yet uncharacterized pathogenicity island in E. coli 042. This hypothesis is supported by the presence of an ORF from the cryptic prophage 933L, which delimits the left-hand side of the EHEC O157:H7 locus of enterocyte effacement island. Additional evidence for differences in the organization of the chromosomal regions encoding Pic arises from the fact that EAEC 042 lacks the sigA gene, which is present on the Shigella pathogenicity island and encodes another autotransporter (19, 65).
Our analyses of the gene encoding the Pic protein show that Pic has a high level of identity to proteins of the autotransporter family, a rapidly growing group of virulence determinants from gram-negative bacteria (35). The autotransporter family takes its name from its unique secretion mechanism, for which it is hypothesized that the N-terminal amino acid signal sequence directs secretion across the inner membrane (presumably via a Sec-dependent mechanism) and subsequently the C-terminal portion of the precursor forms a β-barrel structure in the outer membrane. The N-terminal portion of the molecule (the so-called passenger domain) is believed to be transported through the β-barrel to the cell surface. Analysis of the C-terminal 277 amino acid residues of Pic suggested the presence of 14 amphipathic β-strands, typical for members of the autotransporter family. An even number of β-strands would place the first and last segments in opposite orientations and would allow closing of the β-barrel, a feature also observed for the trimeric porins such as OmpF (16). In agreement with the postulate that the β-domain forms a β-barrel structure in the outer membrane, the amino acids at the extreme C terminus of Pic (YMF) fit the consensus profile previously derived for other outer membrane proteins (35, 82).
Another feature common among members of the autotransporter family is the presence of an unusually long N-terminal signal peptide (35). For Pic, N-terminal sequencing of the secreted protein indicated cleavage of the precursor molecule at residue 55 (assuming a methionine start site), which agrees with the prediction of a signal peptidase cleavage site between residues 55 and 56 (SQA-GIV). The significance of this extended signal sequence has not been determined.
Based upon sequence comparison with other members of the autotransporter family, the release of mature Pic apparently occurs by proteolysis from the β-domain between residues N1095 and N1096. To further characterize the processing step involved in the maturation of Pic, secretion of the passenger domain was investigated in various E. coli strains possessing pic constructs. Expression of the pic gene in E. coli strains lacking the OmpT and OmpP outer membrane proteases, DsbA (the disulfide bond isomerase) and DegP (the periplasmic protease) indicated that formation of the β-domain and the secreted mature protein was independent of these four enzymes and implied either that an unidentified protease is involved or that autoprocessing may occur. For Hap from Haemophilus influenzae and the IgA1 proteases from Neisseria, this processing step seemed to be the result of autoproteolysis mediated by the serine protease site (36). The presence in Pic of a putative serine protease active site suggested that a similar step could also occur for Pic. However, unlike Hap and the IgA1 protease, site-directed mutagenesis of the active-site serine residue (S258) in the putative serine protease motif did not abolish processing and secretion of the protein.
Pic belongs to a subfamily of autotransporters that feature a serine protease motif in the N-terminal one-third of the passenger domain, including a growing number of SPATEs, the IgA proteases, and Hap. Although the precise role of these proteins in pathogenesis has not been determined, our data support the possibility that Pic and perhaps other secreted proteases of the SPATE class may mediate one or more steps in enteric pathogenesis. Indeed, there is ample evidence for roles (often multiple) of secreted proteases in pathogenesis, including in group A streptococci (44), Entamoeba histolytica (78), Porphyromonas gingivalis (83), and Vibrio cholerae (28).
Pic may play similar or different roles in EAEC and Shigella diarrhea. Clinical observations suggest that EAEC diarrhea is associated with mucosal damage, apparently via elaboration of a cytotoxin, and formation of a thick mucus gel on the intestinal mucosa. Eslava et al. (24) have described two EAEC proteins of 104 kDa and ca. 116 kDa that were isolated from outbreak strains and that, when injected into ileal loops, induced cytotoxic effects on the mucosa. Recent evidence suggests that the 104-kDa protein (Pet) is a plasmid-encoded enterotoxin of EAEC (52). Using molecular methods, we have found that the 116-kDa protein is Pic. Our in vitro studies suggest possible roles for Pic that may be responsible for its effects in both EAEC and Shigella. In vitro, Pic appears to possess more than one relevant phenotype. These functions include the degradation of mucin, serum resistance, and hemagglutination, which could be representative of mucosal binding. However, our data do not support a role for Pic as a cytotoxin (34, 52).
The ability to resist complement-mediated killing is not generally thought to be relevant to intestinal pathogens such as EAEC or S. flexneri. However, both of these organisms are associated with bloody diarrhea, often, in Shigella infections, together with mucosal ulceration. This phenomenon could expose the organisms to increased transudation of complement proteins, which could exert potent antibacterial and inflammatory effects. Even in the absence of mucosal ulceration, complement proteins have been identified in the intestinal tract (2), suggesting that complement resistance may be relevant to intestinal colonization. It is not yet clear how Pic mediates complement resistance, yet our data suggest that this effect is dependent on the protease activity of the protein. We must note, however, that the wild-type strains of our test organisms were not themselves highly susceptible to complement-mediated killing and that we have yet to demonstrate a role for this phenotype in vivo.
A more important role for Pic in pathogenesis may lie in its mucinolytic activity. The pathogenesis of both E. coli and S. flexneri infections requires contact with the mucosal cell surfaces. However, the mucus layer overlying the mucosal surface is considered to be a protective barrier against enteric infections (66). As a result, some enteric pathogens have developed various strategies for penetrating this gel-like layer, in some cases employing heightened flagellar motion and in others elaborating enzymes which degrade the mucus (14, 29). However, little is known about the mechanism by which Shigella and EAEC penetrate the mucus layer. Previous studies have noted that mucus is depleted on the colonic surface during S. flexneri infection (70) and that S. flexneri produces glycosidases or other enzymes which degrade either the peptide or oligosaccharide moieties of the mucin molecules (61, 62). It has also been suggested that these mucinolysins not only promote colonization but also promote Shigella penetration into the intestinal mucosa.
The phenotypes we have identified for Pic suggest that it is involved in the early stages of pathogenesis and most probably promotes intestinal colonization. Indeed, preliminary animal studies with both EAEC and Shigella pic mutants support this hypothesis (34). For this reason, we have adopted the designation Pic rather than the previously proposed she and ShMu; in addition, the designation she has been used previously in E. coli (68) and the she nomenclature suggests the presence of the gene in Shigella only.
S. flexneri 2a is the most common serotype causing bacillary dysentery worldwide (11, 27). In view of the distribution of pic and its putative role in S. flexneri pathogenicity, it is tempting to speculate that Pic contributes to the epidemiologic prevalence of serotype 2a by enhancing the clinical manifestations of the disease. The precise contribution of Pic to Shigella and EAEC pathogenesis and epidemiology requires further investigation.
ACKNOWLEDGMENT
This work was supported by U.S. Public Health Service grant AI33096 to J.P.N.
REFERENCES
- 1.Aebi C, Lafontaine E R, Cope L D, Latimer J L, Lumbley S L, McCracken G H, Jr, Hansen E J. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect Immun. 1998;66:3113–3119. doi: 10.1128/iai.66.7.3113-3119.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andoh A, Fujiyama Y, Sakumoto H, Uchihara H, Kimura T, Koyama S, Bamba T. Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinomas. Clin Exp Immunol. 1998;111:477–483. doi: 10.1046/j.1365-2249.1998.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1989. [Google Scholar]
- 4.Bardwell J C, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 5.Benjelloun-Touimi Z, Sansonetti P J, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17:123–135. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 6.Benz I, Schmidt M A. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect Immun. 1989;57:1506–1511. doi: 10.1128/iai.57.5.1506-1511.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 9.Caffrey P, McVeigh T, Owen P. Western immunoblotting. In: Owen P, Foster T J, editors. Immunochemical and molecular genetic analysis of bacterial pathogens. Amsterdam, The Netherlands: Elsevier Science Publishing; 1988. pp. 255–266. [Google Scholar]
- 10.Caffrey P, Owen P. Purification and N-terminal sequence of the alpha subunit of antigen 43, a unique protein complex associated with the outer membrane of Escherichia coli. J Bacteriol. 1989;171:3634–3640. doi: 10.1128/jb.171.7.3634-3640.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casalino M, Yusuf M W, Nicoletti M, Bazzicalupo P, Coppo A, Colonna B, Cappelli C, Bianchini C, Falbo V, Ahmed H J, et al. A two-year study of enteric infections associated with diarrhoeal diseases in children in urban Somalia. Trans R Soc Trop Med Hyg. 1988;82:637–641. doi: 10.1016/0035-9203(88)90542-1. [DOI] [PubMed] [Google Scholar]
- 12.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobeljic M, Miljkovic-Selimovic B, Paunovic-Todosijevic D, Velickovic Z, Lepsanovic Z, Zec N, Savic D, Ilic R, Konstantinovic S, Jovanovic B, Kostic V. Enteroaggregative Escherichia coli associated with an outbreak of diarrhoea in a neonatal nursery ward. Epidemiol Infect. 1996;117:11–16. doi: 10.1017/s0950268800001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen P S, Laux D C. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 1995;253:309–314. doi: 10.1016/s0076-6879(95)53026-6. [DOI] [PubMed] [Google Scholar]
- 15.Colina A R, Aumont F, Deslauriers N, Belhumeur P, de Repentigny L. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 1996;64:4514–4519. doi: 10.1128/iai.64.11.4514-4519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 17.Cravioto A, Tello A, Navarro A, Ruiz J, Villafan H, Uribe F, Eslava C. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet. 1991;337:262–264. doi: 10.1016/0140-6736(91)90868-p. [DOI] [PubMed] [Google Scholar]
- 18.Czeczulin J R, Balepur S, Hicks S, Phillips A, Hall R, Kothary M H, Navarro-Garcia F, Nataro J P. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun. 1997;65:4135–4145. doi: 10.1128/iai.65.10.4135-4145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czeczulin J R, Whittam T S, Henderson I R, Nataro J P. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–2699. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drumm B, Roberton A M, Sherman P M. Inhibition of attachment of Escherichia coli RDEC-1 to intestinal microvillus membranes by rabbit ileal mucus and mucin in vitro. Infect Immun. 1988;56:2437–2442. doi: 10.1128/iai.56.9.2437-2442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DuPont H L, Hornick R B, Dawkins A T, Snyder M J, Formal S B. The response of man to virulent Shigella flexneri 2a. J Infect Dis. 1969;119:296–299. doi: 10.1093/infdis/119.3.296. [DOI] [PubMed] [Google Scholar]
- 22.Emini E A, Hughes J V, Perlow D S, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eslava C, Navarro-Garcia F, Czeczulin J R, Henderson I R, Cravioto A, Nataro J P. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66:3155–3163. doi: 10.1128/iai.66.7.3155-3163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eslava C, Villaseca J, Morales R, Navarro A, Cravioto A. Abstracts of the 93rd General Meeting of the American Society for Microbiology 1993. Washington, D.C: American Society for Microbiology; 1993. Identification of a protein with toxigenic activity produced by enteroaggregative Escherichia coli, abstr. B105; p. 44. [Google Scholar]
- 24a.EXPASY Molecular Biology Server.6 September 1999, revision date. [Online.] Tools and software packages. http://www.expasy.ch. [8 September 1999, last date accessed.]
- 25.Fasano A, Noriega F R, Liao F M, Wang W, Levine M M. Effect of shigella enterotoxin 1 (ShET1) on rabbit intestine in vitro and in vivo. Gut. 1997;40:505–511. doi: 10.1136/gut.40.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez R C, Weiss A A. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun. 1994;62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreccio C, Prado V, Ojeda A, Cayyazo M, Abrego P, Guers L, Levine M M. Epidemiologic patterns of acute diarrhea and endemic Shigella infections in children in a poor periurban setting in Santiago, Chile. Am J Epidemiol. 1991;134:614–627. doi: 10.1093/oxfordjournals.aje.a116134. [DOI] [PubMed] [Google Scholar]
- 28.Finkelstein R A, Boesman-Finkelstein M, Holt P. Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F. M. Burnet revisited. Proc Natl Acad Sci USA. 1983;80:1092–1095. doi: 10.1073/pnas.80.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freter R, Allweiss B, O'Brien P C, Halstead S A, Macsai M S. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infect Immun. 1981;34:241–249. doi: 10.1128/iai.34.1.241-249.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundy F J, Plaut A G, Wright A. Localization of the cleavage site specificity determinant of Haemophilus influenzae immunoglobulin A1 protease genes. Infect Immun. 1990;58:320–331. doi: 10.1128/iai.58.2.320-331.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 34.Henderson, I. R., and J. P. Nataro. Unpublished data.
- 35.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:337–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 36.Hendrixson D R, de la Morena M L, Stathopoulos C, St. Geme J W., 3rd Structural determinants of processing and secretion of the Haemophilus influenzae hap protein. Mol Microbiol. 1997;26:505–518. doi: 10.1046/j.1365-2958.1997.5921965.x. [DOI] [PubMed] [Google Scholar]
- 37.Itoh Y, Nagano I, Kunishima M, Ezaki T. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J Clin Microbiol. 1997;35:2546–2550. doi: 10.1128/jcm.35.10.2546-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izard J W, Kendall D A. Signal peptides: exquisitely designed transport promoters. Mol Microbiol. 1994;13:765–773. doi: 10.1111/j.1365-2958.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 39.Jähnig F. Structure predictions of membrane proteins are not that bad. Trends Biochem Sci. 1990;15:93–95. doi: 10.1016/0968-0004(90)90188-h. [DOI] [PubMed] [Google Scholar]
- 40.Karaolis D K, Lan R, Reeves P R. Sequence variation in Shigella sonnei (Sonnei), a pathogenic clone of Escherichia coli, over four continents and 41 years. J Clin Microbiol. 1994;32:796–802. doi: 10.1128/jcm.32.3.796-802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufmann A, Stierhof Y D, Henning U. New outer membrane-associated protease of Escherichia coli K-12. J Bacteriol. 1994;176:359–367. doi: 10.1128/jb.176.2.359-367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 43.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Lukomski S, Montgomery C A, Rurangirwa J, Geske R S, Barrish J P, Adams G J, Musser J M. Extracellular cysteine protease produced by streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67:1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantle M, Allen A. A colorimetric assay for glycoproteins based on the periodic acid/Schiff stain. Biochem Soc Trans. 1978;6:607–609. doi: 10.1042/bst0060607. [DOI] [PubMed] [Google Scholar]
- 46.Mantle M, Allen A. Isolation and characterization of the native glycoprotein from pig small-intestinal mucus. Biochem J. 1981;195:267–275. doi: 10.1042/bj1950267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsutani S, Ohtsubo E. Complete sequence of IS629. Nucleic Acids Res. 1990;18:1899. doi: 10.1093/nar/18.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menard R, Dehio C, Sansonetti P J. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 49.Morabito S, Karch H, Mariani-Kurkdjian P, Schmidt H, Minelli F, Bingen E, Caprioli A. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J Clin Microbiol. 1998;36:840–842. doi: 10.1128/jcm.36.3.840-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nataro J P, Deng Y, Cookson S, Cravioto A, Savarino S J, Guers L D, Levine M M, Tacket C O. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis. 1995;171:465–468. doi: 10.1093/infdis/171.2.465. [DOI] [PubMed] [Google Scholar]
- 51.Nataro J P, Steiner T, Guerrant R L. Enteroaggregative Escherichia coli. Emerg Infect Dis. 1998;4:251–261. doi: 10.3201/eid0402.980212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navarro-Garcia F, Eslava C, Villaseca J M, Lopez-Revilla R, Czeczulin J R, Srinivas S, Nataro J P, Cravioto A. In vitro effects of a high-molecular-weight heat-labile enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66:3149–3154. doi: 10.1128/iai.66.7.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noriega, F., and S. Formal. Unpublished data.
- 54.Noriega F R, Liao F M, Formal S B, Fasano A, Levine M M. Prevalence of Shigella enterotoxin 1 among Shigella clinical isolates of diverse serotypes. J Infect Dis. 1995;172:1408–1410. doi: 10.1093/infdis/172.5.1408. [DOI] [PubMed] [Google Scholar]
- 55.Otto B R, van Dooren S J M, Nuijens J H, Luirink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pai M, Kang G, Ramakrishna B S, Venkataraman A, Muliyil J. An epidemic of diarrhoea in south India caused by enteroaggregative Escherichia coli. Indian J Med Res. 1997;106:7–12. [PubMed] [Google Scholar]
- 57.Parsot C, Sansonetti P J. Invasion and the pathogenesis of Shigella infections. Curr Top Microbiol Immunol. 1996;209:25–42. doi: 10.1007/978-3-642-85216-9_2. [DOI] [PubMed] [Google Scholar]
- 58.Paton A W, Paton J C. Characterization of IS1203, an insertion sequence in Escherichia coli O111:H. Gene. 1994;150:67–70. doi: 10.1016/0378-1119(94)90859-1. [DOI] [PubMed] [Google Scholar]
- 59.Perna N T, Mayhew G F, Posfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prere M F, Chandler M, Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prizont R. Degradation of intestinal glycoproteins by pathogenic Shigella flexneri. Infect Immun. 1982;36:615–620. doi: 10.1128/iai.36.2.615-620.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prizont R, Reed W P. Differences in blood group B-specific mucinase activity between virulent and avirulent Shigella flexneri 2a strains. Microb Pathog. 1991;11:129–135. doi: 10.1016/0882-4010(91)90006-v. [DOI] [PubMed] [Google Scholar]
- 63.Provence D L, Curtiss R., III Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect Immun. 1994;62:1369–1380. doi: 10.1128/iai.62.4.1369-1380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pupo G M, Karaolis D K, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island, which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajkumar R, Devaraj H, Niranjali S. Binding of Shigella to rat and human intestinal mucin. Mol Cell Biochem. 1998;178:261–268. doi: 10.1023/a:1006844125976. [DOI] [PubMed] [Google Scholar]
- 67.Rambow A A, Fernandez R C, Weiss A A. Characterization of BrkA expression in Bordetella bronchiseptica. Infect Immun. 1998;66:3978–3980. doi: 10.1128/iai.66.8.3978-3980.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reingold J, Starr N, Maurer J, Lee M D. Identification of a new Escherichia coli She haemolysin homolog in avian E. coli. Vet Microbiol. 1999;66:125–134. doi: 10.1016/s0378-1135(98)00310-1. [DOI] [PubMed] [Google Scholar]
- 69.Rolland K, Lambert-Zechovsky N, Picard B, Denamur E. Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of E. coli. Microbiology. 1998;144:2667–2672. doi: 10.1099/00221287-144-9-2667. [DOI] [PubMed] [Google Scholar]
- 70.Sachdev H P, Chadha V, Malhotra V, Verghese A, Puri R K. Rectal histopathology in endemic Shigella and Salmonella diarrhea. J Pediatr Gastroenterol Nutr. 1993;16:33–38. doi: 10.1097/00005176-199301000-00007. [DOI] [PubMed] [Google Scholar]
- 71.Sang W K, Oundo J O, Mwituria J K, Waiyaki P G, Yoh M, Iida T, Honda T. Multidrug-resistant enteroaggregative Escherichia coli associated with persistent diarrhea in Kenyan children. Emerg Infect Dis. 1997;3:373–374. doi: 10.3201/eid0303.970317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sansonetti P J. Molecular and cellular mechanisms of invasion of the intestinal barrier by enteric pathogens. The paradigm of Shigella. Folia Microbiol. 1998;43:239–246. doi: 10.1007/BF02818608. [DOI] [PubMed] [Google Scholar]
- 73.Savarino S J. Diarrhoeal disease: current concepts and future challenges. Enteroadherent Escherichia coli: a heterogeneous group of E. coli implicated as diarrhoeal pathogens. Trans R Soc Trop Med Hyg. 1993;87(Suppl. 3):49–53. doi: 10.1016/0035-9203(93)90539-3. [DOI] [PubMed] [Google Scholar]
- 74.Savarino S J, Fasano A, Watson J, Martin B M, Levine M M, Guandalini S, Guerry P. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci USA. 1993;90:3093–3097. doi: 10.1073/pnas.90.7.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shuter J, Hatcher V B, Lowy F D. Staphylococcus aureus binding to human nasal mucin. Infect Immun. 1996;64:310–318. doi: 10.1128/iai.64.1.310-318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith H. What happens to bacterial pathogens in vivo? Trends Microbiol. 1998;6:239–243. doi: 10.1016/s0966-842x(98)01250-5. [DOI] [PubMed] [Google Scholar]
- 77.Smith H R, Cheasty T, Rowe B. Enteroaggregative Escherichia coli and outbreaks of gastroenteritis in UK. Lancet. 1997;350:814–815. doi: 10.1016/s0140-6736(05)62611-6. [DOI] [PubMed] [Google Scholar]
- 78.Spinella S, Petek F, Gayral P, Rigothier M C. A novel cysteine protease in Entamoeba histolytica. Arch Med Res. 1997;28:180–181. [PubMed] [Google Scholar]
- 79.Stein M, Kenny B, Stein M A, Finlay B B. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178:6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.St. Geme J W, III, de la Morena M L, Falkow S. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol Microbiol. 1994;14:217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 81.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 83.Tokuda M, Chen W, Karunakaran T, Kuramitsu H K. Regulation of protease expression in Porphyromonas gingivalis. Infect Immun. 1998;66:5232–5237. doi: 10.1128/iai.66.11.5232-5237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wanke C A, Cronan S, Goss C, Chadee K, Guerrant R L. Characterization of binding of Escherichia coli strains which are enteropathogens to small-bowel mucin. Infect Immun. 1990;58:794–800. doi: 10.1128/iai.58.3.794-800.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]