Abstract
Clostridium perfringens enterotoxin (CPE), a single polypeptide of 319 amino acids, has a unique multistep mechanism of action. In the first step, CPE binds to claudin proteins and/or a 50-kDa eukaryotic membrane protein receptor, forming a small (∼90-kDa) complex. This small complex apparently then associates with a 70-kDa eukaryotic membrane protein, resulting in formation of a large complex that induces the onset of membrane permeability alterations. To better define the boundaries of CPE functional regions and to identify specific amino acid residues involved in various steps of CPE action, in this study we subjected the cloned cpe gene to random mutagenesis in XL-1 Red strains of Escherichia coli. Seven CPE random mutants with reduced cytotoxicity for Vero cells were phenotypically characterized for the ability to complete each step in CPE action. Five of these seven recombinant CPE (rCPE) random mutants (G49D, S59L, R116S, R137G, and S167P) exhibited binding characteristics similar to those of rCPE or native CPE, while the Y310C and W226Stop mutants showed reduced binding and no binding, respectively, to brush border membranes. Interestingly, two completely nontoxic mutants (G49D and S59L) were able to bind and form small complex but they did not mediate any detectable large complex formation. Another strongly attenuated mutant, R116S, formed reduced amounts of an anomalously migrating large complex. Collectively, these results provide further support for large complex formation being an essential step in CPE action and also identify the CPE region ranging from residues ∼45 to 116 as important for large complex formation. Finally, we also report that limited removal of extreme N-terminal CPE sequences, which may occur in vivo during disease, stimulates cytotoxic activity by enhancing large complex formation.
The gram-positive, spore-forming, anaerobe Clostridium perfringens is a notorious toxin-producing pathogen. One biomedically important toxin expressed by some C. perfringens isolates is C. perfringens enterotoxin (CPE), which has been implicated as a virulence factor in several human gastrointestinal illnesses (15, 16). These CPE-associated illnesses include C. perfringens type A food poisoning, which ranks as the second most commonly reported food-borne disease in the United States, as well as non-food-borne diarrheas such as antibiotic-associated diarrhea and sporadic diarrhea.
Recent studies indicate that CPE has a unique multistep mechanism of action that involves, at a minimum, (i) CPE binding to receptors, which may include claudins or a 50-kDa membrane protein (10, 11, 25), (ii) formation of a small (∼90-kDa) CPE-containing complex in plasma membranes (25), (iii) an association between this small complex and additional membrane proteins, forming a large (>160-kDa) CPE-containing complex (10, 20, 25, 26), (iv) the rapid induction of massive small molecule permeability changes in the plasma membranes of sensitive eukaryotic cells (13, 14, 18, 21), which may result from the partial insertion of CPE into membranes (24), and (iv) a collapse of the cellular colloid-osmotic equilibrium (14, 19), which results in cell death (9, 17).
Understanding the structure-function relationship of a bacterial toxin is necessary to fully appreciate how that toxin interacts with mammalian cells. Several previous studies have investigated the structure-function relationship for CPE, a 35-kDa single polypeptide that lacks significant amino acid homology with other known toxins. For example, receptor binding activity has been mapped to the extreme C terminus of the CPE protein (4, 7, 8, 12), and it was shown that C-terminal CPE fragments, such as CPE171-319 (a CPE fragment consisting of residues 171 to 319), are nontoxic (5, 7, 8, 12). Deletion mutagenesis analysis (12) and proteolytic cleavage studies (2, 3, 6) demonstrated that the toxic activity of CPE increases two- to threefold when up to the first 45 N-terminal amino acids are removed from the native toxin protein. This stimulation of toxicity does not result from the activated CPE fragments possessing enhanced binding ability (6). While these activated CPE fragments have been shown to form more large complex in mammalian membranes than does native CPE (12), it remains unclear whether the limited removal of N-terminal CPE sequences directly promotes large complex formation or, instead, promotes the earlier step of small complex formation. Interestingly, deletion mutagenesis studies have also demonstrated that removing additional N-terminal amino acids beyond residue 45 from native CPE causes a complete loss of cytotoxic activity, with these nontoxic CPE deletion fragments failing to form large complex. Collectively, these structure-function data indicate that, like many bacterial toxins, CPE segregates its functional regions responsible for receptor binding and toxic activities, with receptor binding activity mapping to the toxin's extreme C terminus and toxic activity requiring amino acid residues located between positions 45 and 171 of the enterotoxin protein.
Despite the progress described above, knowledge of CPE's structure-function relationship remains rudimentary. The boundaries of the toxicity region present in the N-terminal half of the CPE molecule remain poorly defined, it has yet to be determined whether amino acids residing in this N-terminal toxicity region are required for biologic activity because they directly promote small versus large complex formation, and it remains unclear why limited removal of extreme N-terminal sequences activates CPE biologic activity. By phenotypically characterizing recombinant CPE (CPE) random mutants and a previously generated rCPE45-319 deletion mutant (12), we have now addressed these important questions about the CPE structure-function relationship.
MATERIALS AND METHODS
Materials.
Native CPE was purified from C. perfringens NCTC 8239, and the biological activity of this purified toxin was assayed as described previously (22). Aliquots (2 mg) of purified native CPE were radioiodinated, as described previously (21), using lactoperoxidase-glucose oxidase (Bio-Rad) and 2 mCi of Na125I (17 mCi/mg; ICN Radiochemicals). Rabbit intestinal brush border membranes (BBMs) were prepared from the small intestines of female New Zealand White rabbits by the method of Sigrist et al. (23). All restriction enzymes and other recombinant DNA reagents were purchased from Boehringer Mannheim (Indianapolis, Ind.), unless otherwise noted.
Random mutagenesis of pJKFLt-1.
Construction of pJKFLt-1, which contains the entire cpe open reading frame (ORF) ligated (in frame) into the pTrc-HisA vector, has been described previously (12). Plasmid pJKFLt-1 encodes rCPE, a fusion protein consisting of full-length CPE coupled to a short, vector-encoded N-terminal His6-containing sequence whose presence assists purification using metal affinity chromatography. Previous studies (12) have established that rCPE exhibits biologic activity indistinguishable from that of native CPE produced by C. perfringens.
To obtain random CPE point mutants, pJKFLt-1 was initially transformed into the XL-1 Red Epicurian coli (Stratagene) random-mutator strain of Escherichia coli. Following selection on plates containing Luria agar plus ampicillin (100 μg/m), the XL-1 Red(pJKFLt-1) transformants were inoculated into 5 ml of Luria-Bertani medium containing ampicillin (300 μg/ml). After incubation for 24 h at 37°C with shaking, which allows random mutations to occur in pJKFLt-1, plasmid DNA was prepared from the mutagenized XL-1 Red(pJKFLt-1) cultures, using a Wizard Plus plasmid purification kit. This purified, mutagenized, plasmid DNA was then used to transform XL-1 Blue, which stopped the further accumulation of mutations in pJKFLt-1. XL-1 Blue(pJKFLt-1) transformants were selected on LA+Amp plates and screened for expression of possible rCPE mutants as described below.
Vero cell screening of lysates prepared from E. coli XL-1 Blue(pJKFLt-1) transformants.
Individual XL-1 Blue transformants carrying either the wild-type pJKFLt-1 plasmid, i.e., XL-1 Blue(wild-type pJKFLt-1) transformants, or a pJKFLt-1 plasmid that had been previously subjected to random mutagenesis in XL-1 Red, i.e., XL-1 Blue(randomly mutagenized pJKFLt-1) transformants, were inoculated into 2 ml of SOB broth plus ampicillin (100 μg/ml). These cultures were incubated for 4 h at 37°C with shaking, and expression of rCPE species was then induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. Following overnight incubation of the induced culture at 37°C with shaking, a sonicated culture lysate was prepared by using a W-375 Heat Systems-Ultrasonic Inc. cup sonicator.
Lysates possessing reduced CPE-like cytotoxic activity were identified by a Vero cell morphologic damage assay. Briefly, 96-well plates were seeded with 104 Vero cells per well. After these cultures were grown for 2 days at 37°C, either 5, 2.5, or 1.25 μl of culture lysate prepared from XL-1 Blue(wild-type pJKFLt-1) or XL-1 Blue(randomly mutagenized pJKFLt-1) was added to a Vero cell culture well. After 20 min of incubation at 37°C, the lysate-treated Vero cells were inspected, by light microscopy, for the development of CPE-like morphologic damage (17).
Western blot analysis of rCPE species expression by XL-1 Blue(randomly mutagenized pJKFLt-1) transformants.
When a lysate prepared from a XL-1 Blue(randomly mutagenized pJKFLt-1) transformant showed reduced or no cytotoxic activity on Vero cells, that transformant was inoculated into 100 ml of SOB+Amp (100 μg/ml). The culture was incubated for 3 h at 37°C with shaking before expression of its rCPE species was induced with IPTG (0.5 mM, final concentration). The induced culture was incubated for 3 h at 37°C, with shaking, before the culture was centrifuged and cell pellets were frozen at −20°C. After thawing, a sonicated lysate was prepared from each cell pellet, and the concentration of rCPE species present in the lysate was determined by quantitative Western immunoblotting, performed as described previously (12) except that Supersignal chemiluminescent substrate (Pierce Chemical Company) was used. For each lysate preparation, quantitation was determined twice with three different sample volumes. Densitometric scans of autoradiographs were performed with a ScanJet Plus (Hewlett-Packard), using the DeskScan II 2.3 program, and peak area integrations were determined by using the 1-D Process & Report program (Zeineh Biomedical Instruments).
Nucleotide sequencing.
The cpe ORF present in those XL-1 Blue(randomly mutagenized pJKFLt-1) transformants found to express high levels of a rCPE species with reduced (or no) cytotoxic activity against Vero cells was subjected to automated nucleotide sequencing analysis at the Core Support Facility of the Department of Molecular Genetics and Biochemistry, University of Pittsburgh School of Medicine.
Talon resin affinity enrichment of rCPE random mutants.
XL-1 Blue(randomly mutagenized pJKFLt-1) transformants that (i) carry a cpe ORF with a single, randomly generated CPE point mutation and (ii) express high levels of this rCPE mutant were grown and harvested as described above. The cell pellet was then resuspended in Tris-saline (10 mM Tris, 50 mM saline [pH 8]) and frozen at −20°C until use. After sonication, the lysate supernatants were batch incubated with Talon metal affinity resin (Clontech, Palo Alto, Calif.) for 30 min at room temperature with constant mixing. This lysate-resin mixture was then pelleted by low-speed centrifugation, the unbound lysate supernatant was removed, and the resin was washed twice with Tris-saline. The washed resin was transferred to a plastic column, which was then washed twice with 10 ml of Tris-saline. Bound proteins were eluted in 1-ml fractions of 50 mM imidazole in Tris-saline. Chromatography fractions containing eluted protein were then pooled and dialyzed overnight at 4°C against 50 mM Tris-HCl (pH 8), in order to remove imidazole and NaCl.
The amount of rCPE species present in each Talon affinity-enriched preparation was determined by both quantitative Western immunoblot and quantitative native immunodot blot analyses (12) with purified native CPE as a standard. Except for preparations containing the W226Stop mutant (see below), strong immunoreactivity was always detected with both of these serologic assays. Further, except for preparations containing the W226Stop rCPE mutant, results from both Western immunoblot and native immunodot blot analyses always closely agreed (data not shown) regarding the amount of rCPE species present in each affinity-enriched preparation. Since it has been shown previously (12) that (i) Western immunoblots almost exclusively detect the presence of a very strong linear epitope located in the extreme C terminus of CPE and (ii) native immunodot blots almost exclusively detect the presence of conformational epitopes in CPE, the fact that both Western immunoblots and native immunodot blots produced similar estimates of how much of each rCPE missense mutant was present in affinity-enriched preparations strongly suggests that these six mutants retain most, if not all, of the seroreactivity exhibited by native CPE.
However, the W226Stop mutant was observed (data not shown) to exhibit much stronger immunoreactivity in the native immunodot blot assay than in the Western immunoblot assay, probably because this mutant lacks the very strong linear epitope located in the extreme C terminus of native CPE (12). Consequently, native immunodot blot analysis was used to calculate the amount of W226Stop mutant present in preparations after Talon affinity enrichment.
With respect to purity, all Talon affinity-enriched preparations (except the preparation containing the W226Stop rCPE random mutant [see below]) ran on sodium dodecyl sulfate (SDS)-polyacrylamide gels as a single Coomassie blue-staining band, which always comigrated with the immunoreactive rCPE species present in that affinity-enriched sample (data not shown). Quantitative Western immunoblot analyses indicated that these rCPE species represented ∼60 to 70% of the total protein present in each affinity-enriched preparation.
Talon affinity-enriched preparations containing the W226Stop rCPE random mutant ran on SDS-polyacrylamide gels as two Coomassie blue-staining bands (data not shown). Western immunoblot analysis indicated that the lower band, which migrated at the expected size (32 kDa) of the W226Stop rCPE mutant and was the stronger of the sample's two Coomassie blue-staining bands, comigrated with the immunoreactive rCPE species present in this affinity-enriched preparation. Quantitative native immunodot blot analysis (12) indicated that the W226Stop rCPE mutant represented ∼60% of the total protein present after affinity enrichment.
Binding inhibition by rCPE random mutants.
Binding inhibition experiments were performed as described previously (12). Briefly, BBMs (100 μg) were preincubated in the presence or absence of increasing amounts (1 to 1,000 nM, final concentrations) of either an affinity-enriched rCPE random mutant or affinity-enriched rCPE (prepared as described for the CPE random mutants) for 20 min at room temperature (RT). The binding mixtures were then incubated for an additional 20 min at RT in the presence of 125I-CPE (0.5 μg). After the BBMs were washed several times with phosphate-buffered saline, the BBM-associated radioactivity was determined with a Packard gamma counter.
Total (i.e., 100%) binding shown for each sample corresponds to the amount of 125I-CPE that bound to BBMs in the absence of any rCPE species competitor. Percent total binding values reflect 125I-CPE binding in the presence of various concentrations of rCPE species competitors; these total binding values were calculated from three independent assays, each of which contained triplicate data points. The concentration (nanomolar) of each rCPE species needed to inhibit total 125I-CPE binding by 50% was then extrapolated by graphing percent total binding values versus each concentration of rCPE species competitor tested. Results are presented as mean ± standard error (SE).
Small complex formation by rCPE random mutants.
The ability of each rCPE random mutant to form small complex was assessed by using affinity-enriched 125I-labeled preparations of each rCPE species. Because radioiodination required the preparation of relatively large (milligram quantities) amounts of each rCPE species, larger-scale (5-liter) cultures of XL-1 Blue(pJKFLt-1) transformants were grown for Talon affinity enrichment; however, these cultures were otherwise processed and stored at −20°C as described above. After thawing, sonication, and centrifugation, lysate supernatants were treated with ammonium sulfate (15%, final concentration). Precipitated proteins were then resuspended in Tris-saline and desalted on PD-10 columns (Pharmacia). The rCPE species present in the desalted sample was enriched by using Talon metal affinity resin as described above, and the eluted protein from the Talon column was pooled, dialyzed overnight at 4°C against sterile H2O, and lyophilized.
These preparations, which contained approximately 2 mg of either an affinity-enriched rCPE random mutant or affinity-enriched rCPE, were then radioiodinated by using the same procedure as used for radiolabeling native CPE except that 1 mCi of Na125I (ICN) was used in the iodination reaction. Radioiodination did not affect the cytotoxic activity of any rCPE species (data not shown). The concentration of rCPE species present in each radioiodinated preparation was then assessed by Western blotting or native immunodot blotting, as described above for quantitating unlabeled rCPE species in crude lysates.
To induce small complex formation in the absence of appreciable large complex formation (25), 7 μg (∼900 nM, final concentration) of each radioiodinated rCPE species was incubated, in the presence or absence of a 50-fold excess of unlabeled CPE, with 100 μg of BBMs for 5 min at 4°C. After washing, these BBMs were extracted in 1% Triton X-100 for 30 min at 4°C. The resultant detergent extracts were then electrophoresed on a 6% native polyacrylamide gel containing 0.1% Triton X-100, as described before for detection of small complex (25). These gels were then autoradiographed, and the amount of small complex present in each lane was determined by densitometric evaluation, using a minimum of four gel analyses, as described above.
Large complex formation by rCPE random mutants.
An aliquot (100 or 200 nM, final concentration) of each affinity-enriched rCPE random mutant or affinity-enriched rCPE was incubated at RT with 100 μg of BBMS for 20 min. After washing, these BBMs were resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer without 2-mercaptoethanol and incubated at RT for 20 min to extract membrane proteins. The detergent extracts were then resolved by SDS-PAGE on 6% gels, transferred to nitrocellulose, and Western blotted as described above. Previous studies (20, 25, 26) have established that only specifically-bound CPE species form large complex.
To determine the amount of large complex formed by each rCPE species, a minimum of four Western blots analyses were evaluated by densitometry as described above. These data were then used to compare large complex formation by each rCPE random mutant versus rCPE.
86Rb release assay to quantify the cytotoxic properties of each rCPE random mutant.
The cytotoxic properties of each affinity-enriched rCPE random mutant were assessed by measuring the mutant's ability to induce CPE-like release of 86Rb from radiolabeled Vero cells, as previously described (20, 21). Briefly, 5-day-old, confluent monolayers of Vero cells were labeled with 86RbCl (NEN Life Sciences Products, Inc.), washed twice with warm Hanks' balanced salt solution, and incubated for 15 min at 37°C with Hanks' balanced salt solution containing increasing amounts (0.5 to 25 μg) of one affinity-enriched rCPE random mutant or affinity-enriched rCPE. The supernatant was then removed from the monolayers and counted in a gamma counter.
The percent maximal release induced by each rCPE random mutant or rCPE was calculated as 100 × (86Rb release due to sample −spontaneous 86Rb release)/(maximal 86Rb release −spontaneous 86Rb release). Maximal release represents the total cytoplasmic radioactivity present at the end of radiolabeling, while spontaneous release represents the amount of radiolabel released from monolayers in the absence of any sample challenge.
To ensure that all 86Rb release values detected above spontaneous release were mediated by the rCPE species present in that affinity-enriched sample, an aliquot (0.5 μg) of each rCPE random mutant or rCPE was also preincubated in the presence of a 50-fold excess of either monoclonal antibody (MAb) 3C9 (a CPE-neutralizing MAb) or MAb 10G6 (a nonneutralizing anti-CPE MAb) prior to use in the 86Rb release assay. In all cases, MAb 3C9, but not MAb 10G6, neutralized all 86Rb release above background (data not shown).
Evaluation of possible gross conformational alterations in rCPE random mutants by trypsin digestion.
Each affinity-enriched rCPE random mutant or affinity-enriched rCPE was added to a separate microcentrifuge tube containing phosphate-buffered saline plus trypsin (at a 1:15 molar ratio of trypsin to rCPE species). This mixture was then incubated at room temperature for 0, 5, 15, 30, or 60 min. Aliquots of the mixture were then removed, added to SDS-PAGE sample buffer, and boiled for 5 min. These samples were subjected to SDS-PAGE on 12% gels, electrotransferred to nitrocellulose, and Western blotted as described above.
Phenotypic characterization of an rCPE45-319 deletion fragment.
Construction of plasmid pJKC45t-1 has been described previously (12). This plasmid carries a cpe gene fragment encoding amino acids 45 to 319 of native CPE ligated (in frame) into the pTrc-HisC vector, which allows the expression of a fusion protein consisting of rCPE45-319, i.e., native CPE positions 45 to 319, coupled to a short, vector-encoded N-terminal His6-containing sequence.
The expression, affinity enrichment, and phenotypic characterization of rCPE45-319 were performed as described above for the rCPE random mutants except that 125I-rCPE45-319 was used to characterize both small and large complex formation by this rCPE species.
RESULTS
Cytotoxicity screening of XL-1 Blue(randomly mutagenized pJKFLt-1) transformants.
Culture lysates from 1,000 independent XL-1 Blue(randomly mutagenized pJKFLt-1) transformants were screened for their cytotoxic activity against Vero cells. Lysates from 10 of these transformants appeared to be completely noncytotoxic in this initial screening; that is, addition of even high concentrations of these lysates did not induce any detectable damage to Vero cells. Lysates from 12 lysates were strongly attenuated; that is, CPE-like morphologic damage was detected only at the highest dose of lysate tested. Sixty-four variants were found to be somewhat attenuated; that is, high or medium doses of these lysates damaged Vero cells, but low doses of these lysates had no effect on these cells.
Control experiments demonstrated that lysates prepared from XL-1 Blue transformants containing the pTrcHisA vector were noncytotoxic at all concentrations tested, while XL-1 Blue transformants carrying a wild-type pJKFLt-1 plasmid not subjected to mutagenesis in XL-1 Red were reproducibly cytotoxic at all doses tested.
Western immunoblot analysis of rCPE species expression by XL-1 Blue(randomly mutagenized pJKFLt-1) transformants.
To distinguish pJKFLt-1 random mutations that affect rCPE expression from those affecting rCPE cytotoxicity activity, lysates were prepared from the 86 XL-1 Blue(randomly mutagenized pJKFLt-1) transformants whose lysates showed reduced cytotoxic activities for Vero cells. When these lysates were screened for expression of rCPE species by CPE-specific Western blotting, it was found (data not shown) that 71 of the 86 XL-1 Blue(randomly mutagenized pJKFLt-1) transformants express moderate to high levels of apparently full-size rCPE. One transformant carrying a randomly mutagenized pJKFLt-1 was found to express high levels of a rCPE species that migrates with a significantly lower Mr than does full-length rCPE. The remaining 14 XL-1 Blue(randomly mutagenized pJKFLt-1) transformants expressed rCPE species poorly or not at all.
Combining these Western blot results with the previous cytotoxicity screening data, we concluded that we had obtained 72 XL-1 Blue(randomly mutagenized pJKFLt-1) transformants that may produce an rCPE random mutant with reduced cytotoxic activity for Vero cells.
Nucleotide sequencing analysis of the cpe ORF present in XL-1 Blue(randomly mutagenized pJKFLt-1) transformants showing reduced cytotoxic activity.
For subsequent phenotypic characterization experiments, it was necessary to identify XL-1 Blue(mutagenized pJKFLt-1) transformants that express high levels of an attenuated rCPE random mutant with a single point mutation. Therefore, the cpe ORF was nucleotide sequenced in those transformants expressing high levels of an apparently attenuated rCPE random mutant. This sequencing analysis revealed the presence of a cpe ORF with a single point mutation in seven XL-1 Blue(mutagenized pJKFLt-1) transformants that express high levels of a rCPE mutant with reduced cytotoxicity. These seven XL-1 Blue transformants produce seven CPE variants, G49D, S59L, R116S, R137G, S167P, W226Stop, and Y310C.
These sequencing results were consistent with Western blot analyses indicating that six of these seven transformants express an rCPE fusion protein containing a full-length CPE moiety, but the W226Stop mutant expresses a truncated CPE fragment.
Characterization of binding properties of rCPE random mutants.
After affinity enrichment, the binding properties of the seven rCPE random mutants with a single point mutation were evaluated by using a well-established competitive binding assay (12). In control experiments, the reliability of this system was confirmed when it was shown that (i) preincubation of BBMs with affinity-enriched rCPE decreased the ability of these BBMs to subsequently bind 125I-CPE (Fig. 1) but (ii) similar preincubation of BBMs with even very high concentrations of protein eluted from Talon columns chromatographed with lysates prepared from XL-1 Blue(pTrcHisA) transformants did not affect the subsequent ability of these BBMs to bind 125I-CPE (data not shown).
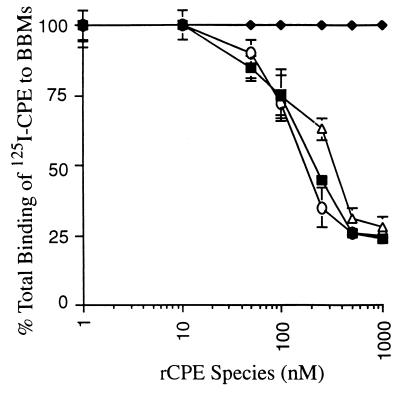
FIG. 1.
Inhibition of 125I-CPE binding to BBMs by rCPE random mutants. BBMs were preincubated with increasing concentrations (10 to 1,000 nM) of each rCPE random mutant (○, G49D; ▵, Y310C; ⧫, W226 Stop random mutant) or rCPE (■) as a binding competitor prior to the addition of 125I-CPE. Results are shown for three representative mutants. Total binding was determined as the binding of 125I-CPE in the absence of competitor. Data shown are mean percentage of total binding (± SE) of three independent experiments in which all points were performed in triplicate. Points without error bars had an SE too small to depict.
When affinity-enriched samples of the seven rCPE random mutants were tested with this assay, preincubation of BBMs with even high concentrations of rCPE W226Stop mutant did not affect the ability of these BBMs to subsequently bind 125I-CPE (Fig. 1), indicating that this rCPE random mutant does not bind to BBM receptors. However, when BBMs were preincubated with enriched samples of rCPE random mutants G49D, S59L, R116S, R137G, S167P, and Y310C, subsequent binding of 125I-CPE to these membranes was significantly reduced (Fig. 1); i.e., these mutants bind to BBMs. Kinetic analyses (Fig. 1 and Table 1) revealed that the competitive binding activities exhibited by rCPE random mutants G49D, S59L, R116S, R137G, and S167P were very similar to that of rCPE or native CPE. However, the rCPE random mutant Y310C reproducibly tested as a slightly (∼2-fold) less efficient binding competitor than rCPE or native CPE.
TABLE 1.
Comparative activities of rCPE species
| Species | Concn (nM) needed for binding | Fold reduction in:
|
||
|---|---|---|---|---|
| Small complex formationb | Large complex formationc | Amt (μg) needed for cytotoxicityd | ||
| rCPE | 200 | 0.6 ± 0.1 | ||
| G49D | 200 | —e | >20 | >25 |
| S59L | 200 | — | >20 | >25 |
| R116S | 200 | — | 3 | >25 |
| R137G | 300 | — | 2 | 1.9 ± 0.2 |
| S167P | 200 | — | 3 | 2.1 ± 0.2 |
| W226Stop | >1,000 | >20 | >20 | >25 |
| Y310C | 400 | 2 | 2 | 2.8 ± 0.6 |
Concentration of rCPE species needed to cause 50% inhibition of 125I-CPE binding. SE was <100 nm for all samples. Only the Y310C and W226Stop mutants had binding ability significantly different (P < 0.05) from that of rCPE.
Relative to small complex formation by rCPE. SE for all random mutants was ±<1-fold reduction except for W226Stop, which did not form any detectable small complex.
Relative to large complex formation by rCPE. SE was ±<1-fold reduction for R116S, R137G, S167P, and Y310C; SE could not be calculated for G49D, S59L, and W226Stop because these mutants failed to form any detectable large complex.
Amount of rCPE species needed to cause 50% 86Rb release from Vero cells. SE values are shown for mutants inducing 50% 86Rb release at doses of <25 μg.
—, no reduction.
Small complex formation by rCPE random mutants.
The next phenotypic property evaluated for the seven rCPE random mutants was the ability to form small complex. We initially attempted to assess this trait by performing immunoblotting with small complex gels. However, this approach was unsuccessful even when rCPE or native CPE was used, probably because the high concentration of Triton X-100 present in small complex gels interferes with protein transfer onto nitrocellulose membranes.
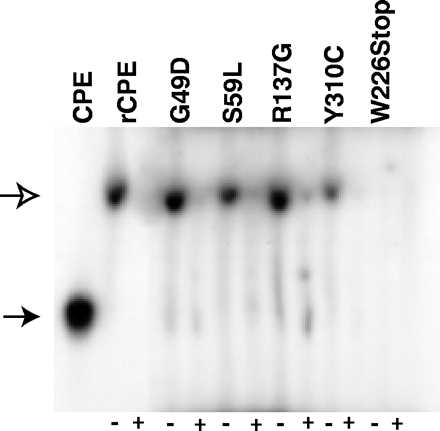
It was possible to assess small complex formation by using 125I-labeled preparations of each affinity-enriched rCPE random mutant, which were incubated with BBMs at 4°C (to inhibit large complex formation [24]), followed by subsequent detection of small complex by gel electrophoresis and autoradiography. These analyses showed (Fig. 2) that the rCPE W226Stop mutant did not form any small complex, as would be expected since this mutant also does not bind to BBMs (Fig. 1 and Table 1). However, the G49D, S59L, R116S, R137G, S167P, and Y310C random rCPE mutants were all able to form small complex (Fig. 2). Densitometric scans of three repetitions of Fig. 2 gels showed that each of these rCPE random mutants makes about the same amount of small complex as does rCPE (Table 1 and Fig. 2) or native CPE (data not shown), except for the Y310C mutant, which appears to be about twofold less efficient at forming small complex (Fig. 2 and Table 1).
FIG. 2.
Small complex formation by 125I-rCPE random mutants. The ability of 125I-rCPE random mutants to form small complex, in the presence (+) or absence (−) of unlabeled CPE, when incubated with BBMs at 4°C was detected by Triton X-100 PAGE and autoradiography (see Materials and Methods). The open arrow indicates the migration of small complex, while the closed arrow indicates the migration of noncomplexed (free) CPE. The gel shown is representative of four repetitions of this experiment.
Large complex formation by rCPE random mutants.
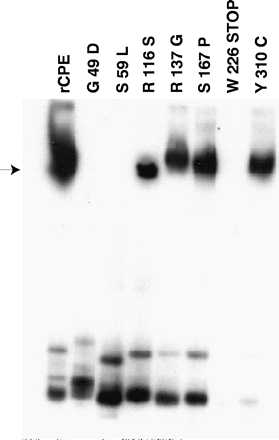
When the ability of the rCPE random mutants to form a high-Mr membrane species resembling large complex was assessed by using a previously described Western immunoblot procedure (12), it was found (Fig. 3) that BBMs incubated for 20 min at RT with the R116S, R137G, S167P, or Y310C mutant formed a high-Mr membrane species that resembles large complex. However, these four rCPE random mutants were slightly (∼2 to 3-fold) less efficient than rCPE at promoting formation of their high-Mr membrane species (Table 1). Furthermore, while the high-Mr membrane species made by the R137G, S167P, and Y310C random rCPE mutants closely comigrates with the large complex made by rCPE (or native CPE [data not shown]), the large complex made by the R116S mutant reproducibly migrated slightly faster than the large complex made by rCPE.
FIG. 3.
Large complex formation by rCPE random mutants. The ability of rCPE random mutants to form large complex after incubation at 24°C with BBMs was demonstrated by Western immunoblot analysis of extracted membrane samples. The arrow on the left indicates the migration of large complex in this gel system. The gel shown is representative of four repetitions of this experiment.
In contrast, several rCPE random mutants did not form any detectable amounts of a high-Mr, large complex-like species in BBMs. Given the inability of the W226Stop mutant to bind or form small complex, the failure of this rCPE random mutant to promote large complex formation (Fig. 3) was expected. Of greater interest, the G49D and S59L mutants, which bind and form small complex about as efficiently as rCPE, also failed to promote any detectable formation of large complex, even when very high concentrations of these mutants were added to BBMs (Fig. 3 and Table 1).
Cytotoxic properties of rCPE random mutants.
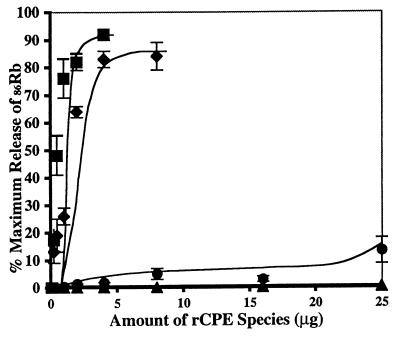
When the seven rCPE random mutants were tested (Fig. 4) for their CPE-like cytotoxic properties by using a 86Rb release assay (12), the R137G, S167P, and Y310C mutants all demonstrated an ∼2- to 3-fold reduction (relative to rCPE) in cytotoxic activity (Fig. 4 and Table 1). The R116S random mutant, which formed an anomalous large complex in the experiments represented in Fig. 3, demonstrated a greatly reduced (but still detectable) cytotoxic activity (Fig. 4 and Table 1).
FIG. 4.
Cytotoxicity of rCPE random mutants for Vero cells. Vero cells were labeled with 86Rb and then treated with increasing amounts of rCPE random mutants (⧫, Y310C; ●, R116S; ▴, G49D) or rCPE (■) for 15 min. Data are expressed as percentage of maximal release after correction for background spontaneous 86Rb release. These representative results are the means of three separate experiments. Error bars represent SE; points without SEs had values too small to depict.
No cytotoxic activity was detected (Fig. 4 and Table 1) for either the G49D or S59L mutant, each of which failed to form any large complex in the experiments represented in Fig. 3. Similarly, no cytotoxic activity was detected for the W226Stop mutant, which did not bind and did not form small or large complex.
Analysis of rCPE random mutants for gross conformational changes.
To assess whether some or all of the rCPE random mutants might have undergone gross conformational changes that could explain their loss of functional activities in previous experiments (Fig. 1 to 4), two approaches were used. First, native immunodotblot analysis (12) revealed that all seven rCPE random mutants exhibited strong immunoreactivity with polyclonal antibodies raised against native CPE (data not shown), which is consistent with these mutants retaining native CPE conformation.
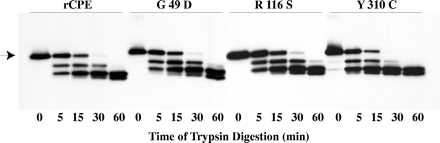
The second approach for assessing conformational integrity of the rCPE random mutants involved comparing their trypsin sensitivity against the trypsin sensitivity of rCPE. Results shown in Fig. 5 indicate that the G49D, R116S, and Y310C random mutants exhibit similar trypsin sensitivity similar to that of rCPE. Since the S59L, R137G, and S167P mutants also showed trypsin digestion patterns similar to that of rCPE (data not shown), these results strongly suggest that none of these six full-length rCPE point mutants has undergone gross conformational changes.
FIG. 5.
Conformational analysis of rCPE species by trypsin digestion. rCPE species were incubated at a 1:15 molar ratio with trypsin at RT. Samples were removed from the reaction tube at the times shown, and trypsin digestion was stopped by boiling for 5 min in SDS-PAGE sample buffer. Samples were resolved by SDS-PAGE on a 12% gel followed by Western immunoblot analysis. The arrow on the left indicates where undigested rCPE species migrate in this gel system. rCPE random mutants S59L, R137G, and S167P also exhibited trypsin digestion patterns similar to that of rCPE (data not shown).
Phenotypic characterization of an rCPE45-319 deletion fragment.
A recent study (12) from our laboratory reported that rCPE45-319, a CPE deletion fragment containing only residues 45 to 319 of native CPE, exhibits cytotoxic activity two- to threefold greater than that of rCPE. That previous study also showed that although this rCPE45-319 fragment binds similarly to rCPE, it forms about twofold more large complex than does rCPE. However, it has not yet been distinguished whether the increased cytotoxic activity noted for the rCPE45-319 fragment specifically results from this fragment possessing enhanced ability to form large complex, or (since small complex formation is a precursor for large complex formation [25]) if this deletion fragment has an enhanced ability to form small complex, which then leads to greater large complex formation.
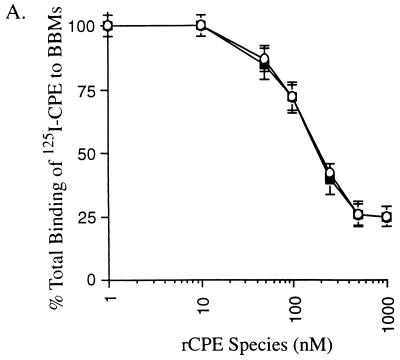
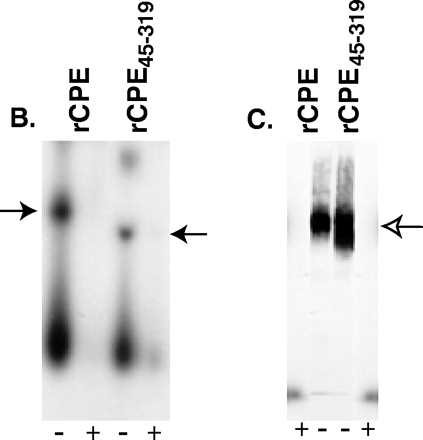
The radioiodination protocol developed in this study for radiolabeling affinity-enriched rCPE random mutants allowed us to distinguish between these two possibilities. As reported previously for unlabeled rCPE45-319, 125I-rCPE45-319 was found to be ∼2-fold more cytotoxic, on a molar basis, than either 125I-rCPE or unlabeled rCPE; i.e., the dose of 125I-rCPE45-319 needed to cause 50% maximum release of cytoplasmic 86Rb label from Vero cells was 0.25 μg, while 0.5 μg of 125I-rCPE or unlabeled rCPE was needed to obtain this same effect. Also consistent with previous reports on unlabeled rCPE45-319, we observed that 125I-rCPE45-319 binds similarly to 125I-CPE (Fig. 6A) but forms about twofold more large complex in BBMs than does either 125I-rCPE (Fig. 6C) or unlabeled rCPE (not shown). However, since we observed that this rCPE45-319 species forms about the same amount of small complex in BBMs as does 125I-rCPE (Fig. 6B), it is now apparent that the enhanced large complex formation and cytotoxicity exhibited by the rCPE fragment cannot be attributed to enhanced small complex formation.
FIG. 6.
Phenotypic characterization of the rCPE45-319 deletion fragment. (A) Abilities of rCPE (■) and the rCPE45-319 deletion fragment (○) to inhibit subsequent binding of 125I-CPE to BBMs. As for Fig. 1, BBMs were incubated with increasing concentrations of rCPE or rCPE45-319 prior to the addition of 125I-CPE. Results shown represent the mean ± SE from three independent experiments. (B) Small complex-forming ability of 125I-rCPE45-319. The closed arrow indicates the migration of small complex formed by 125I-rCPE or 125I-rCPE45-319 (the faster migration of the rCPE45-319 small complex is due to the fact that the rCPE45-319 deletion fragment is smaller than rCPE). The gel shown is representative of three repetitions of this experiment. (C) Large complex-forming ability of 125I-rCPE45-319. The open arrow indicates the migration of large complex in this gel system. The result shown is representative of three repetitions of this experiment.
DISCUSSION
Random mutagenesis often provides valuable insights into the structure-function relationship of a protein, particularly when (as for CPE) detailed structural information is unavailable for that protein. In this study, seven random rCPE mutants with attenuated cytotoxic activity were identified and characterized, in depth, for their phenotypic properties. The missense mutations present in six of these seven attenuated random mutants are distributed throughout the CPE protein, with the notable exception of two regions, (i) the toxin's extreme N terminus and (ii) C-terminal sequences located between residues 167 and 290, the residue which marks the start of the putative receptor binding region located in the extreme C terminus of CPE (4). The failure of our study to identify any attenuated rCPE random mutants with missense mutations in the first 45 CPE amino acid residues is consistent with previous reports (2, 3, 12) indicating that removal of up to the first 45 amino acids of CPE increases, rather than decreases, toxic activity. It remains unclear whether our failure to identify any attenuated random mutants with missense mutations in the CPE region extending from residues ∼170 to 290 indicates that this region is functionally silent or that we simply failed to obtain the correct missense mutant needed to demonstrate phenotypic activity in this CPE region. Further exploration of this issue is ongoing in our laboratory.
The seven rCPE random mutants with attenuated toxicity that were obtained in the current study have provided important new information about the CPE structure-function relationship. For example, only two of these random mutants, W226Stop and Y310C, exhibit impaired binding activity. The inability of the nontoxic W226Stop mutant to exhibit any detectable receptor binding activity is fully consistent with this truncation mutant lacking the C-terminal amino acid 290–319 sequence that contains the putative receptor binding region of CPE. Furthermore, the slight attenuation of binding activity measured for the Y310C random mutant appears sufficient to account for most, it not all, of the slight reductions in small complex formation, large complex formation, and cytotoxicity also noted for this mutant. Therefore, results with both the W226Stop and Y310C random mutants support the importance of the CPE 290–319 region for receptor binding. However, the fact that the Y310C mutant, which has a relatively significant amino acid substitution, exhibits only slightly reduced binding activity suggests that CPE residue 310 is not particularly important for receptor binding. Our laboratory is now performing site-directed mutagenesis studies to further explore this CPE receptor binding region.
Even more significant information has been provided by the G49D and S59L random mutants, which lack cytotoxic activity despite having rCPE-like abilities to bind and form small complex. Significantly, both the G49D and S59L mutants are unable to mediate any formation of large complex. This finding is important for understanding CPE action because the specific inability of these completely nontoxic mutants to form large complex provides strong new evidence supporting the importance of large complex formation for CPE action. The observed failure of our G49D random mutant to form large complex is consistent with results from a previous deletion mutagenesis study (12), which suggested that at least one amino acid residing between residues 45 and 53 of CPE is directly or indirectly important for large complex formation. In that earlier study, a CPE45-319 fragment with enhanced biologic activity was shown to form ∼2-fold more large complex than rCPE, while a nontoxic rCPE53-319 fragment did not form any large complex. That earlier study did not establish whether the failure of the rCPE53-319 fragment to form large complex results from a specific blockage of large complex formation or, instead, results from an impairment in the earlier step of small complex formation. Even using alternative approaches to detect small complex formation, we still could not evaluate small complex formation by the rCPE53-319 fragment in this study (the low expression of soluble rCPE53-319 precluded radioiodination of this species). However, by showing that the G49D random mutant has a specific impairment in its ability to make large (rather than small) complex, we have now conclusively established that the CPE 45–53 region contains some amino acids that are specifically required for large complex formation. Further, our results demonstrating that the completely nontoxic S59L random mutant also has a specific block in large complex formation now extend the boundaries of the large complex-forming region of CPE to encompass at least residues 45 to 59 of native CPE (see below). We are currently exploring this region by site-directed mutagenesis to identify which of CPE residues 45 to 59 are critical for large complex formation.
The inability of the G49D and S59L mutants to mediate large complex formation, even though they perform all known earlier steps in CPE action, also provides a valuable insight for understanding the CPE structure-function relationships by allowing us to identify the first two individual CPE amino acid residues (residues 49 to 59) that are specifically involved in large complex formation. Secondary structure modeling using the Jpred server (1) predicts that in wild-type CPE, residue 49 should be present in a loop region, while residue 59 should be located in a region of β-sheet. All six secondary structure algorithms used by the Jpred server predict that changing residue 49 from glycine to aspartic acid should not significantly affect CPE secondary structure. Jpred analysis is less clear as to whether altering residue 59 from serine to leucine should change CPE secondary structure; i.e., three of the Jpred algorithms predict that a S59L substitution should disrupt the β-sheet structure in the CPE region containing residue 59, but the other three Jpred algorithms predict that this mutation should not affect CPE secondary structure. Our planned site-directed mutagenesis of the CPE49-59 region may help clarify the structural basis by which the G49D and S59L rCPE mutants are unable to form large complex.
Of the remaining three rCPE random mutants (R116S, R137G, and S167P), only the R116S mutant demonstrated more than a twofold attenuation in cytotoxicity. Interestingly, this mutant was found to form about the same amount of a high- Mr, large complex-like membrane species as do the slightly attenuated R137G or S167P random mutants. However, we observed that the high-Mr species made by R116S migrates anomalously relative to the large complex made by R137G, S167P, or rCPE. This observation suggests that amino acids beyond residue 59 may also contribute to large complex formation or stability. Determining the basis for the anomalous migration of the R116S large complex awaits the complete elucidation of the large complex formation process.
The development of a method using 125I-labeled rCPE derivatives to evaluate small complex formation in this study has also allowed us to address the molecular basis by which removal of limited N-terminal sequences from native CPE produces an activated toxin fragment. Consistent with a previous report (12), we demonstrated that relative to rCPE or native rCPE, the two- to threefold increase in cytotoxicity attributed to the rCPE45-319 deletion fragment is not due to increased binding ability. Importantly, we now show that the two- to threefold-greater large complex formation attributed to rCPE45-319 in this and a previous study (12) does not result from this fragment possessing enhanced small complex-forming ability. This finding holds potential pathogenic importance since intestinal proteases have been shown (2, 3, 6) to remove limited N-terminal CPE sequences and activate CPE activity, making it plausible that the enhanced large complex-forming abilities noted for N-terminal deletion fragments such as rCPE45-319 contribute to the pathogenesis of CPE-associated gastrointestinal diseases.
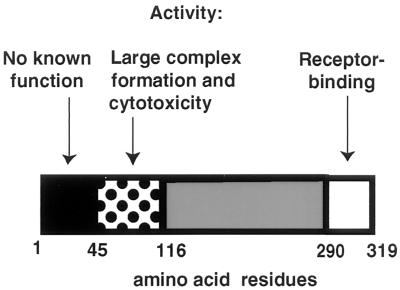
Finally, an updated map of CPE structure-function relationships incorporating the new information from this study is shown in Fig. 7. Strong evidence now implicates the presence of a receptor binding region at the extreme C terminus of the toxin. It remains unclear whether these C-terminal sequences are also responsible for small complex formation; however, no experimental evidence currently distinguishes CPE binding from small complex formation (consistent with this, all of our binding-capable rCPE random mutants also make small complex). In contrast, extreme N-terminal CPE sequences encompassing residues 1 to 45 appear to be biologically silent. The present study now clearly indicates that CPE sequences located between residues 45 and ∼120 are important for large complex formation. We speculate that removal of the biologically silent CPE residues 1 to 45 may increase the large complex-forming and cytotoxic abilities of CPE fragments such as rCPE45-319 because removal of these extreme N-terminal CPE residues improves the accessibility of a eukaryotic protein involved in large complex formation to CPE residues 45 to 116, which are required for large complex formation. Further experimentation is required to refine the map shown in Fig. 7; findings obtained with these genetic approaches must eventually be considered within the three-dimensional context of the enterotoxin, when this information becomes available.
FIG. 7.
Updated map of the CPE structure-function relationship, depicting (dotted box) the importance of CPE residues 45 to 116 for large complex formation and cytotoxicity, as indicated by this study. Also shown (white box) is the previously identified (12) C-terminal region from residues 290 to 319 responsible for receptor binding. Removal of the N-terminal region from residues 1 to 45 (black box) results in increased cytotoxicity and large complex formation (reference 12 and this study).
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI19844 from the National Institute of Allergy and Infectious Diseases.
The first two authors contributed equally to this work.
REFERENCES
- 1.Cuff J A, Barton G J. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins Struct Funct Genet. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 2.Granum P E, Richardson M. Chymotrypsin treatment increases the activity of Clostridium perfringens enterotoxin. Toxicon. 1991;29:445–453. doi: 10.1016/0041-0101(91)90227-i. [DOI] [PubMed] [Google Scholar]
- 3.Granum P E, Whitaker J R, Skjelkvale R. Trypsin activation of enterotoxin from Clostridium perfringens type A. Biochim Biophys Acta. 1981;668:325–332. doi: 10.1016/0005-2795(81)90165-3. [DOI] [PubMed] [Google Scholar]
- 4.Hanna P C, McClane B A. A recombinant C-terminal toxin fragment provides evidence that membrane insertion is important for Clostridium perfringens enterotoxin cytotoxicity. Mol Microbiol. 1991;5:225–230. doi: 10.1111/j.1365-2958.1991.tb01843.x. [DOI] [PubMed] [Google Scholar]
- 5.Hanna P C, Mietzner T A, Schoolnik G K, McClane B A. Localization of the receptor-binding region of Clostridium perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides. The 30 C-terminal amino acids define a functional binding region. J Biol Chem. 1991;266:11037–11043. [PubMed] [Google Scholar]
- 6.Hanna P C, Wieckowski E U, Mietzner T A, McClane B A. Mapping functional regions of Clostridium perfringens type A enterotoxin. Infect Immun. 1992;60:2110–2114. doi: 10.1128/iai.60.5.2110-2114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna P C, Wnek A P, McClane B A. Molecular cloning of the 3′ half of the Clostridium perfringens enterotoxin gene and demonstration that this region encodes receptor-binding activity. J Bacteriol. 1989;171:6815–6820. doi: 10.1128/jb.171.12.6815-6820.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horiguchi Y, Akai T, Sakaguchi G. Isolation and function of a Clostridium perfringens enterotoxin fragment. Infect Immun. 1987;55:2912–2915. doi: 10.1128/iai.55.12.2912-2915.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulkower K I, Wnek A P, McClane B A. Evidence that alterations in small molecule permeability are involved in the Clostridium perfringens type-A enterotoxin-induced inhibition of macromolecular synthesis in Vero cells. J Cell Physiol. 1989;140:498–504. doi: 10.1002/jcp.1041400314. [DOI] [PubMed] [Google Scholar]
- 10.Katahira J, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J Cell Biol. 1997;136:1239–1247. doi: 10.1083/jcb.136.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katahira J, Sugiyama H, Inoue M, Horiguchi Y, Matsuda M, Sugimoto N. Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem. 1997;272:26652–26658. doi: 10.1074/jbc.272.42.26652. [DOI] [PubMed] [Google Scholar]
- 12.Kokai-Kun J F, McClane B A. Deletion analysis of the Clostridium perfringens enterotoxin. Infect Immun. 1997;65:1014–1022. doi: 10.1128/iai.65.3.1014-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda M, Ozutsumi K, Iwashi H, Sugimoto N. Primary action of Clostridium perfringens type A enterotoxin on HeLa and Vero cells in the absence of extracellular calcium: rapid and characteristic changes in membrane permeability. Biochem Biophys Res Commun. 1986;141:704–710. doi: 10.1016/s0006-291x(86)80229-7. [DOI] [PubMed] [Google Scholar]
- 14.McClane B A. Osmotic stabilizers differentially inhibit permeability alterations induced in Vero cells by Clostridium perfringens enterotoxin. Biochim Biophys Acta. 1984;777:99–106. doi: 10.1016/0005-2736(84)90501-7. [DOI] [PubMed] [Google Scholar]
- 15.McClane B A. Clostridium perfringens. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C: ASM Press; 1997. pp. 305–326. [Google Scholar]
- 16.McClane, B. A., D. M. Lyerly, J. S. Moncrief, and T. D. Wilkins. Enterotoxic clostridia: Clostridium perfringens type A and Clostridium difficileASM PressWashington, D.C, in press.
- 17.McClane B A, McDonel J L. The effects of Clostridium perfringens enterotoxin on morphology, viability and macromolecular synthesis. J Cell Physiol. 1979;99:191–200. doi: 10.1002/jcp.1040990205. [DOI] [PubMed] [Google Scholar]
- 18.McClane B A, McDonel J L. Characterization of membrane permeability alterations induced in Vero cells by Clostridium perfringens enterotoxin. Biochim Biophys Acta. 1980;600:974–985. doi: 10.1016/0005-2736(80)90499-x. [DOI] [PubMed] [Google Scholar]
- 19.McClane B A, McDonel J L. Protective effects of osmotic stabilizers on morphological and permeability alterations induced in Vero cells by Clostridium perfringens enterotoxin. Biochim Biophys Acta. 1981;641:401–409. doi: 10.1016/0005-2736(81)90496-x. [DOI] [PubMed] [Google Scholar]
- 20.McClane B A, Wnek A P. Studies of Clostridium perfringens enterotoxin action at different temperatures demonstrate a correlation between complex formation and cytotoxicity. Infect Immun. 1990;58:3109–3115. doi: 10.1128/iai.58.9.3109-3115.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClane B A, Wnek A P, Hulkower K I, Hanna P C. Divalent cation involvement in the action of Clostridium perfringens type A enterotoxin. J Biol Chem. 1988;263:2423–2435. [PubMed] [Google Scholar]
- 22.McDonel J L, McClane B A. Production, purification and assay of Clostridium perfringens enterotoxin. Methods Enzymol. 1988;165:94–103. doi: 10.1016/s0076-6879(88)65018-x. [DOI] [PubMed] [Google Scholar]
- 22a.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Sigrist H, Ronner P, Semenza G. A hydrophobic form of the small-intestinal sucrase-isomaltase complex. Biochim Biophys Acta. 1975;406:433–446. doi: 10.1016/0005-2736(75)90022-x. [DOI] [PubMed] [Google Scholar]
- 24.Wieckowski E, Kokai-Kun J F, McClane B A. Characterization of membrane-associated Clostridium perfringens enterotoxin following pronase treatment. Infect Immun. 1998;66:5897–5905. doi: 10.1128/iai.66.12.5897-5905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieckowski E U, Wnek A P, McClane B A. Evidence that an ∼50kDa mammalian plasma membrane protein with receptor-like properties mediates the amphiphilicity of specifically-bound Clostridium perfringens enterotoxin. J Biol Chem. 1994;269:10838–10848. [PubMed] [Google Scholar]
- 26.Wnek A P, McClane B A. Preliminary evidence that Clostridium perfringens type A enterotoxin is present in a 160,000-Mr complex in mammalian membranes. Infect Immun. 1989;57:574–581. doi: 10.1128/iai.57.2.574-581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]