Abstract
Context
A hypothesis-free genetic association analysis has not been reported for patients with primary hyperparathyroidism (PHPT).
Objective
We aimed to investigate genetic associations with PHPT using both genome-wide association study (GWAS) and candidate gene approaches.
Methods
A cross-sectional study was conducted among patients of European White ethnicity recruited in Tayside (Scotland, UK). Electronic medical records were used to identify PHPT cases and controls, and linked to genetic biobank data. Genetic associations were performed by logistic regression models and odds ratios (ORs). The combined effect of the genotypes was researched by genetic risk score (GRS) analysis.
Results
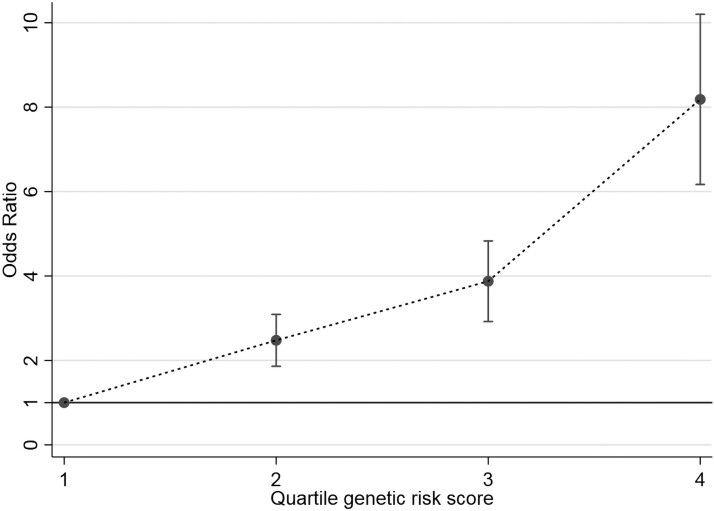
We identified 15 622 individuals for the GWAS that yielded 34 top single-nucleotide variations (formerly single-nucleotide polymorphisms), and LPAR3-rs147672681 reached genome-wide statistical significance (P = 1.2e-08). Using a more restricted PHPT definition, 8722 individuals with data on the GWAS-identified loci were found. Age- and sex-adjusted ORs for the effect alleles of SOX9-rs11656269, SLITRK5-rs185436526, and BCDIN3D-AS1-rs2045094 showed statistically significant increased risks (P < 1.5e-03). GRS analysis of 5482 individuals showed an OR of 2.51 (P = 1.6e-04), 3.78 (P = 4.0e-08), and 7.71 (P = 5.3e-17) for the second, third, and fourth quartiles, respectively, compared to the first, and there was a statistically significant linear trend across quartiles (P < 1.0e-04). Results were similar when stratifying by sex.
Conclusion
Using genetic loci discovered in a GWAS of PHPT carried out in a Scottish population, this study suggests new evidence for the involvement of genetic variants at SOX9, SLITRK5, LPAR3, and BCDIN3D-AS1. It also suggests that male and female carriers of greater numbers of PHPT-risk alleles both have a statistically significant increased risk of PHPT.
Keywords: primary hyperparathyroidism, genetics, genome-wide association study, polygenic risk score
Primary hyperparathyroidism (PHPT) is a commonly encountered endocrine disorder with an estimated prevalence of approximately 0.5% to 1%, which increases with age (1–3). PHPT typically manifests with a raised serum calcium concentration in association with an inappropriate elevation in serum parathyroid hormone (PTH) and usually results from benign parathyroid adenomas (85% of cases) or parathyroid gland hyperplasia (10%-15% of cases), and rarely from parathyroid carcinoma (< 1% of cases). In many instances PHPT is mild and patients are frequently asymptomatic, so many cases can go undiagnosed and untreated, or revealed only incidentally on routine biochemical testing. However, PHPT may be associated with significant morbidity, which includes osteoporosis, renal stones, and nephrocalcinosis (3). Furthermore, previous epidemiological studies of patients with PHPT have indicated there may be an increased all-cause and cardiovascular mortality (3–5).
While the majority of patients presenting with PHPT have a nonfamilial (ie, sporadic) etiology, up to 10% of cases occur as part of a hereditary disorder, either as part of a wider endocrine syndrome or as an isolated endocrinopathy (6). In contrast, familial isolated hyperparathyroidism describes the occurrence of hereditary PHPT as a sole endocrinopathy and 15% to 20% of such kindreds harbor activating mutations in the GCM2 gene but the majority are genetically undefined (7, 8). Another disorder of calcium metabolism, namely familial hypocalciuric hypercalcemia (FHH), is associated with inactivating mutations of the calcium-sensing receptor (CASR) in approximately 65% of cases and with variants in either the GNA11 or AP2S1 genes accounting for the majority of remaining cases (9).
Somatic inactivation and/or loss of heterozygosity of the MEN1 locus is observed in 20% to 40% of sporadic tumors, while whole-exome sequencing studies of parathyroid adenomas reveal recurrent somatic MEN1 mutations in 20% to 30% of tumors (10, 11). Loss of function in CDC73 and CDKN have been implicated in sporadic tumors, while other candidate driver genes have also been implicated in parathyroid tumorigenesis such as the EZH2 and ZFX genes, although variants in such genes are observed at low frequency (12).
Despite progress in understanding the hereditary basis of familial PHPT and knowledge of some of the somatic alterations present in parathyroid tumors, there is a paucity of larger studies investigating whether additional germline genetic variations may contribute to the risk of sporadic PHPT at a population level. Prior GWAS studies evaluating determinants of serum PTH concentration have been reported and identified several potential associated loci, which include those in the vicinity of the CYP24A1, CASR RGS14, CLDN14, RTDR1, RASGEF1, and DPP10 genes (13, 14). Notably, of these genes, CYP24A1, a member of the cytochrome P450 family responsible for inactivation of vitamin D, and CASR, are directly implicated in calcium/PTH homeostasis. However, previous candidate association studies evaluating variants in genes relevant to calcium homeostasis (eg, vitamin D metabolism, CASR), have demonstrated no consistent genetic contribution to PHPT disease risk. To our knowledge a hypothesis-free genome-wide analysis has not been reported for a population PHPT cohort. Thus, in this study, we aimed to investigate genetic associations with PHPT using both GWAS and candidate gene approaches.
Materials and Methods
Discovery Cohort
A cross-sectional study was conducted among individuals older than 18 years from the Genetics of Scottish Health Research register (GoSHARE) and the Genetics of Diabetes and Audit Research Tayside Study (GoDARTS) recruited in Tayside, Scotland (UK). All individuals in these populations are of White ethnicity and have been described previously (15, 16). Electronic medical records (biochemistry, prescribing, hospital admissions, and demographics) were used to ascertain people with PHPT, and were anonymously linked to genetic biobank data by the Health Informatics Centre of the University of Dundee (http://www.dundee.ac.uk/hic). The same data linkage procedure and phenotype definition criteria were applied to these datasets.
Phenotype Definition
Patients with at least 2 recordings of serum-corrected calcium (CCA) greater than 2.55 mmol/L at least 1 year apart, who had a record of PHPT from the International Classification of Diseases (ICD) 10th revision codes (ICD10: E21.0, E21.2, E21.3), and/or who had parathyroid surgery from the Operations and Procedures Codes 4 (OPCS-4: B14, B16, Z13.5) during the study period (1994-2018) were defined as being PHPT case patients. Patients with FHH from ICD codes (ICD10: E83.5) and/or tertiary hyperparathyroidism identified from having an estimated glomerular filtration rate less than or equal to 30 mL/min 36 months before or within 6 months after first raised serum-CCA were excluded. The median serum CCA recorded throughout the study period for each patient was estimated, and control individuals had an average serum CCA concentration of 2.1 to 2.55 mmol/L and never had a record of low or high CCA. A locally determined formula for serum CCA was used: CCA mmol/L = total calcium mmol/L + (0.012 × [39.9 – albumin g/L]), where 39.9 was the Tayside mean serum albumin.
Genetic Data
Genotype data was available from 4 platforms: Illumina HumanOmni Express -12VI platform (Illumina), Affymetrix 6.0 platform (Affymetrix), Illumina Infinium custom GWAS chip (Illumina), and the Global Screening Arrays (GSA) version 2 (Illumina). Imputation of nongenotyped single-nucleotide variations (SNVs, formerly known as single-nucleotide polymorphisms [SNP]) was performed using IMPUTE2 and MINIMAC4 against the haplotype reference consortium (17); calls made with an imputation score of less than 95% were discarded.
Genome-wide Association Study
Standard pre–genome-wide association study (GWAS) quality control was performed (18). Filters were applied to include variants with minor allele frequency greater than 1%. Linkage disequilibrium pruning and concordance between clinically recorded and genetically determined sex was checked. A Hardy-Weinberg equilibrium threshold of P greater than 10e-10 was used for cases and P greater than 10e-6 for controls. GWAS were performed using Plink 2.0. Binary logit models of the outcome of PHPT were compared to population/unselected controls. The model used was PHPT (case vs control) ∼ SNVs + age + age2 + sex + principal components (1–10). GWAS were run on samples of the platforms described previously, and a meta-GWAS across the platforms was also performed using Plink 2.0.
Genetic Association Study on Selected Single-Nucleotide Variations
This study was carried out using the top single SNVs at P less than 5.0e-06 identified from the GWAS (ie, candidate genes). This analysis was performed using a more restricted phenotype definition by which patients had 2 raised CCAs at least 1 year apart with a maximum serum PTH greater than 3 pmol/L and/or a 24-hour urinary excretion greater than 7 mmol/day to be considered as cases by biochemistry. Genetic tests of association were performed by logistic regression models. The combined effect of the genotypes was researched by genetic risk score (GRS) analysis using an unweighted sum of PHPT increasing alleles across the selected SNVs available in our cohort. Participants missing more than 2 of these SNVs were excluded from the analyses. Odds ratios (ORs) from logistic models were also adjusted for age at first CCA recording and sex. Linking correlated alleles were annotated using the LDlink database with the default search options (19). All statistical analyses were conducted using STATA/MP, version 15.1 software (StataCorp).
Results
We identified 88 752 individuals as being eligible for the GWAS after excluding those younger than 18 years and those with FHH or tertiary hyperparathyroidism, of whom 15 622 (778 case patients and 14 844 controls) had available genetic data in the GoDARTS/GoSHARE biobank. PHPT cases were more likely to be female (64.4 vs 45.8%), older (77.1 vs 72.1 years), with diabetes mellitus (80.1 vs 55.8%), and had a higher average serum CCA (2.6 vs 2.3 mmol/L) concentrations than controls (P < 1.0e-03). The GWAS included age, sex, and 2 principal components as covariates to adjust for possible population stratification, which provided an acceptable control; the genomic inflation factor lambda (λ) was 1.03. It yielded 34 top single SNVs at P less than 5.0e-06, but only LPAR3-rs147672681 at chromosome 1 reached genome-wide statistical significance (P < 5.0e-08) (Supplementary Table 1) (20). Supplementary Fig. 1 shows a Manhattan plot of the tested SNVs, and the quantile-quantile plot is shown in Supplementary Fig. 2 (20). All SNVs were in Hardy-Weinberg equilibrium (P < 10e-04).
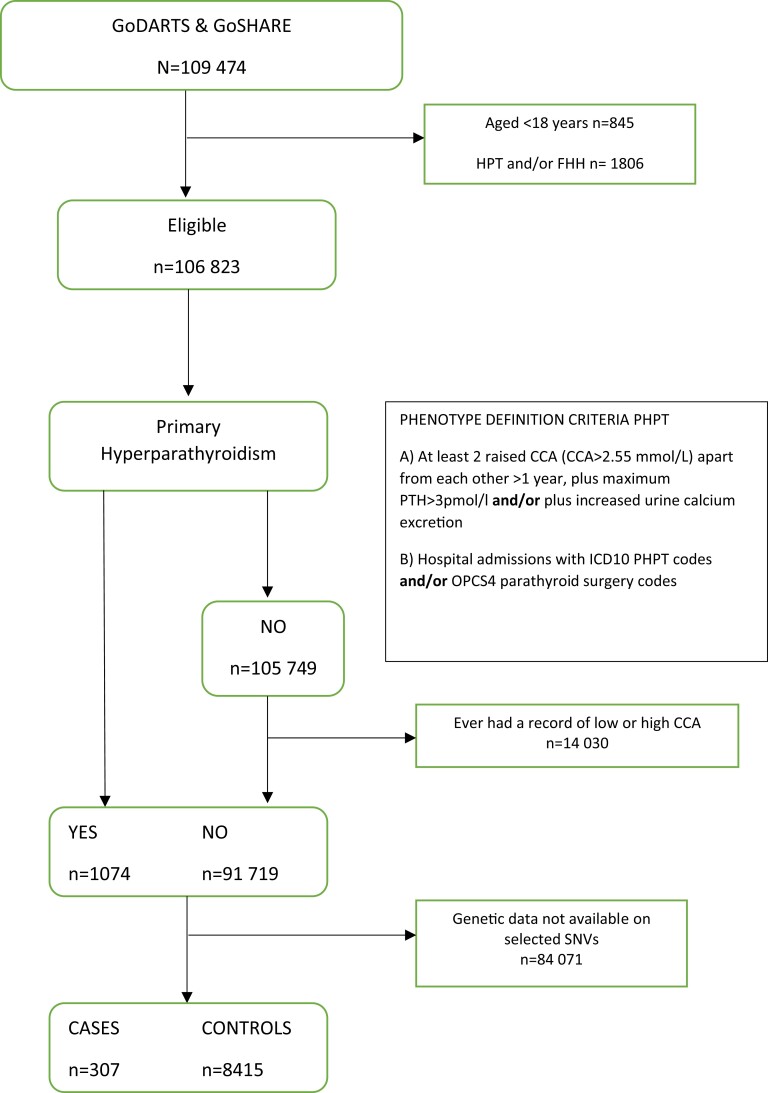
The use of a more restricted phenotype definition identified a study cohort of 8722 individuals (307 case patients and 8415 controls) with available genetic data on the selected SNVs from the GWAS (Fig. 1). PHPT case patients were more likely to be female (65.2 vs 44.2%; P < 1.0e-03), had a higher average serum PTH (9.4 vs 6.5 pmol/L; P < 1.0e-03) and CCA (2.6 vs 2.3 mmol/L; P < 1.0e-03) concentrations than controls (Table 1).
Figure 1.
Flowchart describing the study cohort's generation process and the patients included in the genetic association study of the selected genome-wide association study–identified loci. CCA, serum corrected calcium; FHH, familial hypocalciuric hypercalcemia; HPT, tertiary hyperparathyroidism; ICD10, International Classification of Diseases, 10th revision; OPCS4, Classification of Surgical Operations and Procedures version 4; PHPT, primary hyperparathyroidism; PTH, parathyroid hormone.
Table 1.
Description of genotyped patients with primary hyperparathyroidism (cases) and their comparison controls (n = 8722)
| Cases | Controls | ||
|---|---|---|---|
| Characteristic | (n = 307) | (n = 8415) | P |
| n (%) | |||
| Female sex | 200 (65.2) | 3722 (44.2) | < .001 |
| Diabetes mellitus | 252 (82.1) | 4960 (58.9) | < .001 |
| Mean (SD) | |||
| Age, y | 77.5 (10.1) | 74.3 (10.7) | < .001 |
| Serum-corrected calcium, mmol/La | 2.6 (2.5-2.8) | 2.3 (2.2-2.4) | < .001 |
| Serum PTH, pmol/La,b | 9.4 (5.6-15.8) | 6.5 (3.7-11.6) | < .001 |
Abbreviation: PTH, parathyroid hormone.
Median (interquartile range) of measurements recorded throughout the study period.
Sample size for serum PTH (n = 873; 299 case patients and 574 comparison cohort).
Table 2 shows the results of the genetic association study of the selected 34 genetic variants. An association was found for 19 noncoding SNVs, with 4 reaching the Bonferroni statistical significance threshold at 1.5e-03 (ie, 0.05/34). The nearest genes to these 4 loci were SOX9 (SRY-box transcription factor 9), SLITRK5 (SLIT and NTRK-like family member 5), LPAR3 (lysophosphatidic acid receptor 3), and BCDIN3D-AS1 (BCDIN3D antisense RNA 1). Further adjustment of logistic regression models for age and sex did not change the size and direction of the effect estimates. Age- and sex-adjusted ORs for the effect alleles of SOX9-rs11656269 (OR = 2.12; 95% CI, 1.56-4.04; P = 8.7e-05), SLITRK5-rs185436526 (OR = 3.09; 95% CI, 1.73-5.53; P = 1.4e-04), LPAR3-rs147672681 (OR = 2.05; 95% CI, 1.35-3.11; P = 7.0e-04), and BCDIN3D-AS1-rs2045094 (OR = 1.51; 95% CI, 1.18-1.94; P = 8.5e-04) showed increased risks for PHPT.
Table 2.
Association of single-nucleotide variations with primary hyperparathyroidism at a statistical significance level of P less than 5.0e-02
| SNV | CHR | Position | Gene | A1/A | MAF | Cases | Controls | OR (95% CI)a | P | OR (95% CI)b | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11656269 | 17 | 70125348 | SOX9 | C/T | 0.0371 | 221 | 4814 | 2.13 (1.47-3.10) | 7.2e-05 c | 2.12 (1.46-3.09) | 8.7e-05 c |
| rs185436526 | 13 | 88808800 | SLITRK5 | C/T | 0.0107 | 244 | 5283 | 3.15 (1.77-5.60) | 8.9e-05 c | 3.09 (1.73-5.53) | 1.4e-04 c |
| rs147672681 | 1 | 85353665 | LPAR3 | G/A | 0.0311 | 244 | 5300 | 1.92 (1.27-2.89) | 1.9e-03 | 2.05 (1.35-3.11) | 7.0e-04 c |
| rs2045094 | 12 | 50229509 | BCDIN3D-AS1 | G/T | 0.1298 | 238 | 5201 | 1.52 (1.19-1.94) | 7.4e-04 c | 1.51 (1.18-1.94) | 8.5e-04 c |
| rs35928058 | 6 | 124221344 | NKAIN2 | T/C | 0.0137 | 237 | 5202 | 2.28 (1.29-4.03) | 4.2e-03 | 2.49 (1.41-4.27) | 1.7e-03 |
| rs145504087 | 2 | 197905320 | ANKRD44 | G/C | 0.0258 | 236 | 5182 | 2.02 (1.29-3.17) | 2.0e-03 | 2.00 (1.27-3.15) | 2.6e-03 |
| rs62282716 | 3 | 150633140 | CLRN1-AS1 | A/G | 0.0126 | 231 | 5082 | 2.41 (1.36-4.28) | 2.6e-03 | 2.43 (1.36-4.34) | 2.7e-03 |
| rs138472039 | 4 | 168206250 | LINC02174 | C/G | 0.0145 | 244 | 5305 | 2.38 (1.37-4.12) | 1.9e-03 | 2.29 (1.32-3.99) | 3.2e-03 |
| rs140216208 | 5 | 75841080 | IQGAP2 | T/G | 0.0149 | 237 | 5185 | 2.13 (1.22-3.73) | 7.7e-03 | 2.33 (1.32-4.11) | 3.3e-03 |
| rs45618533 | 1 | 74847747 | FPGT-TNNI3K | A/G | 0.0711 | 213 | 4648 | 1.60 (1.16-2.19) | 3.5e-03 | 1.57 (1.15-2.16) | 4.7e-03 |
| rs345322 | 1 | 108412324 | VAV3 | A/G | 0.0293 | 242 | 5220 | 1.61 (1.23-2.95) | 3.6e-03 | 1.85 (1.19-2.88) | 5.7e-03 |
| rs113075488 | 2 | 4501263 | NPM1P48 | T/C | 0.0252 | 240 | 5257 | 1.77 (1.12-2.81) | 1.4e-02 | 1.82 (1.15-2.90) | 1.1e-02 |
| rs183670739 | 8 | 112729353 | RP11-58O3.2 | G/T | 0.0114 | 242 | 5326 | 2.13 (1.14-3.98) | 1.8e-02 | 2.24 (1.19-4.21) | 1.2e-02 |
| rs11168417 | 12 | 48515720 | PFKM | T/C | 0.1463 | 243 | 5353 | 1.33 (1.05-1.69) | 1.8e-02 | 1.34 (1.06-1.71) | 1.5e-02 |
| rs182479997 | 6 | 147801672 | RP11-15G8.1 | G/T | 0.0138 | 241 | 5286 | 2.05 (1.15-3.65) | 1.4e-02 | 2.04 (1.14-3.66) | 1.6e-02 |
| rs113622331 | 8 | 141520126 | CHRAC1 | A/T | 0.0188 | 234 | 5041 | 1.92 (1.12-3.29) | 1.8e-02 | 1.85 (1.07-3.20) | 2.6e-02 |
| rs79264750 | 7 | 46959622 | AC004901.1 | A/T | 0.0159 | 243 | 5226 | 1.93 (1.08-3.47) | 2.6e-02 | 1.88 (1.05-3.38) | 3.5e-02 |
| rs148035035 | 6 | 21705164 | CASC15 | A/G | 0.0165 | 240 | 5260 | 1.86 (1.06-3.27) | 2.9e-02 | 1.82 (1.03-3.21) | 3.7e-02 |
| rs77959286 | 10 | 55934767 | PCDH15 | C/T | 0.0227 | 246 | 5353 | 1.59 (0.97-2.59) | 6.4e-02 | 1.64 (1.01-2.70) | 4.9e-02 |
Abbreviations: A1, coded allele; CHR, chromosome; MAF, minor allele frequency; OR, odds ratio; SNV, single-nucleotide variation.
Univariate logistic regression models.
Age- and sex-adjusted logistic regression models.
SNVs reaching the Bonferroni statistical significance threshold at α = 0.05/34 = 1.5e-03.
When these 19 SNVs were combined into a GRS using an unweighted sum of PHPT-risk alleles across the selected SNVs available (ie, PHPT-GRS), there was a statistically significant association with the risk of PHPT in 5482 individuals (238 case patients and 5244 controls) after excluding those individuals missing more than 2 of these SNVs. An OR of 2.51 (95% CI, 1.56-4.04; P = 1.6e-04), 3.78 (95% CI, 2.35-6.07; P = 4.0e-08), and 7.71 (95% CI, 4.78-12.43; P = 5.3e-17) for the second, third, and fourth quartiles, respectively, compared to the first quartile of PHPT-GRS scores were found. There was a highly significant linear trend across quartiles of PHPT-GRS scores (chi-square for linear trend extended Mantel-Haenszel test = 98.9; P < 1.0e-04) as shown in Fig. 2. Stratifying by sex, these results were similar for men (n = 3089) and women (n = 2393) (see Supplementary Table 2) (20). Carriers of greater numbers of PHPT-risk alleles were associated with a statistically significant increased risk of PHPT.
Figure 2.
Association of primary hyperparathyroidism (PHPT)-based genetic risk score (GRS) with the risk of developing PHPT (n = 5482). Odds ratios (± SE) for quartiles of the GRS. Chi-square for linear trend (extended Mantel-Haenszel test) = 98.92 (P < 5.0e-04).
Discussion
This record-linkage study used electronic databases to investigate genetic associations with PHPT using genetic loci discovered in a GWAS within a Scottish population. We identified 4 loci located in chromosomes 1, 12, 13, and 17 (mapped genes LPAR3, BCDIN3D-AS1, SLITRK5, and SOX9, respectively), and by using a GRS this study showed that carriers of greater numbers of PHPT-risk alleles were associated with a statistically significant increased risk of PHPT.
GoDARTS/GoSHARE databases are longitudinal cohorts and thus more than one biochemical measurement (ie, serum CCA, PTH, 24-hour urinary excretion) was available for the majority of participants. This allowed the use of biochemistry records to identify patients with at least 2 raised serum CCAs apart from each other for more than 1 year but exclude those having an estimated glomerular filtration rate less than or equal to 30 mL/min within 36 months before or 6 months after the first raised serum CCA (ie, tertiary hyperparathyroidism). The application of these criteria confirmed the identification of cases. Unfortunately, data on serum vitamin D were not available at the time of this study.
Population stratification is always a threat to the validity of a GWAS. The best way to avoid this bias is ensuring the study cohort is derived from a genetically homogeneous population. A genomic inflation factor value close to unity (ie, λ = 1.03) reflects no evidence of population stratification in our GWAS (21). GWAS is an approach used to investigate genetic variants spanning the entire genome and has several limitations such as the small effect sizes of most associations (22). By further limiting the analysis to a few numbers of selected SNVs (ie, candidate genes), it is possible to increase the statistical power to detect statistically significant effects. Thus, after running the GWAS, we performed a genetic association study with selected top single SNVs from the GWAS and a more restricted PHPT definition by considering serum PTH concentration and urine calcium excretion in addition to serum CCA to identify cases. This analysis contributed also to assessing the robustness of our GWAS findings by examining the extent to which results are affected by changes in phenotype definition. This approach confirmed the association of LPAR3 and revealed 3 additional loci that had been suggestive in the GWAS (ie, BCDIN3D-AS1, SLITRK5, and SOX9).
Complications of PHPT are represented by skeletal, kidney, and/or gastrointestinal involvement (23). The present study identified 4 loci mainly related to the skeleton (ie, calcium mobilization, osteoblasts, chondrocytes, and cranial vault traits), and thus relevant to calcium metabolism. SOX9 encodes a transcription factor that plays key roles during embryonic development both in skeletal development and sex determination. In particular, SOX9 is required for chondrocyte differentiation and cartilage formation. It negatively regulates maturation and calcification of chondrocytes through upregulation of PTH-related protein (24, 25). More recently, SOX9 has been demonstrated to be required postnatally to prevent growth-plate closure (26). A GWAS meta-analysis on vault measures in a sample of 4419 healthy individuals of European ancestry found a significant association at SOX9 (27). Thus, SOX9 is implicated in many functions related to bone and cartilage physiology, although how this might influence serum calcium and/or parathyroid function remains to be determined. Little is known about the rs2045094 variant that occurs in the antisense transcript of BCDIN3D, a highly conserved member of the Bin3 methyltransferase family involved in methylation of cytoplasmic transfer RNA (28). The physiological role of this protein is not well defined although overexpression of BCDIN3D is reported in breast cancer, where it is associated with a tumorigenic phenotype and poor prognosis. The further 2 variants occur in proximity to SLITRK5 and LPAR3, respectively. SLITRK5 encodes a transmembrane protein selectively expressed in osteoblasts and acts as a negative regulator of hedgehog signaling that may act to inhibit osteoblast differentiation (29). The LPAR3 gene encodes a G protein–coupled receptor that functions as a receptor for lysophosphatidic acid and mediates lysophosphatidic acid-evoked calcium mobilization, although any relationship to parathyroid biology is unknown. However, LPAR3 is also reported to harbor potential tumorigenic properties (30).
There is a longstanding controversy as to whether serum measurements of total calcium should be adjusted for albumin concentration, and if so which formula is the most appropriate. Locally determined formulas (ie, like ours) usually perform better than formulas taken from the literature. Pekar et al (31) reported that when serum total calcium and CCA were compared to ionized calcium results, the outcomes were very similar for those with normal albumin levels, which relates to the vast majority of our patient cohort. The authors also indicated that, compared to total calcium, CCA tended to overestimate the calcium state for patients with hypoalbuminemia. If so, it is possible that CCA misclassified some patients with low albumin levels, but there is no reason to believe that the probability of patients being misclassified occurred differently across case patients and controls in this study (ie, nondifferential misclassification). Nondifferential misclassification will generally bias an effect estimate toward the null. In other words, our genetic effects of association with PHPT might be closer to the null than it would be were there no misclassification at all. In addition, because our algorithm for patient identification made use of longitudinal data (ie, > one biochemical measurement for each patient over time), the effect of any potential misclassification of the outcome due to the calcium state would be much lower than using cross-sectional data (ie, just one measurement for each patient). Although this is unlikely to affect the findings of this study as most patients had normal serum albumin, we recognize that ionized calcium is the best test to performed, but that in most clinical settings it is not practical yet.
We acknowledge that an independent cohort was not included in this study for replication because longitudinal biochemistry data (ie, > one biochemical measurement for individuals over time) from a different population was not available to apply our criteria for identification of case patients and controls. As expected, a phenome-wide association study on hyperparathyroidism and other disorders of the parathyroid gland (ICD10: E21.0-E21.5) using UK Biobank cross-sectional data (GeneATLAS database, http://geneatlas.roslin.ed.ac.uk) did not replicate any of our top signals (32). Important differences in study design (phenome-wide association study vs GWAS), type of data (cross-sectional vs longitudinal data) and phenotype definition (any hyperparathyroidism and other disorders of parathyroid gland vs PHPT) may have accounted for that (33). Failure to replicate an independent genetic effect may also provide important information about the complexity of the underlying genetic architecture (34).
We also acknowledge that a weighted GRS, defined as weighted sums of risk alleles of SNVs, is a more powerful alternative to a single SNV approach or the use of unweighted GRS for detection of genetic associations. However, appropriate external weights from an independent study must be available. Thus, we made use of an unweighted GRS because no external weights were available because of the novelty of this study. This novel GRS was statistically significantly associated with the risk of PHPT, and there was a highly significant linear trend across quartiles of PHPT-GRS scores that was similar for men and women.
In conclusion, this record-linkage study used electronic databases to investigate hypothesis-free genetic associations with PHPT, using genetic loci discovered in a GWAS carried out in a Scottish population. We used a new approach to define PHPT cases, based on routine biochemistry data and hospital admissions, to achieve a reasonably representative phenotype of this disease. This study suggests new evidence for the involvement of genetic variants in the vicinity of SOX9, SLITRK5, LPAR3, and BCDIN3D-AS1. It also suggests that carriers of greater numbers of PHPT-risk alleles are associated with a statistically significant increased risk of PHPT in men and women. Our results need to be verified in an independent cohort and at the molecular level to confirm hypothesized pathways of PHPT mechanisms.
Acknowledgments
We acknowledge the support of the Health Data Research UK (HDRUK) and the Health Informatics Centre (HIC) at the University of Dundee (Scotland, UK) for managing and supplying the anonymized data. We thank Dr Cyrielle Maroteau (Centre for Pharmacogenetics and Pharmacogenomics, University of Dundee) for her support in performing the GWAS. We also thank Dr Philip Appleby (Population Health & Genomics, University of Dundee) for his contribution on arranging the genetic data.
All analyses were performed on anonymized data sets. This study was approved by the East of Scotland Research Ethics Service–EoSRES (HIC data sets V2, REC ref. 18/ES/0126, IRAS ID 143637), and informed consent was obtained for all participants.
Abbreviations
- CASR
calcium-sensing receptor
- CCA
corrected calcium
- FHH
familial hypocalciuric hypercalcemia
- GoDARTS
Genetics of Diabetes and Audit Research Tayside Study
- GoSHARE
Genetics of Scottish Health Research register
- GRS
genetic risk score
- GWAS
genome-wide association study
- ICD
International Classification of Diseases
- OR
odds ratio
- PHPT
primary hyperparathyroidism
- PTH
parathyroid hormone
- SNV
single-nucleotide variation
Contributor Information
Enrique Soto-Pedre, Division of Population Health & Genomics, School of Medicine, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK.
Paul J Newey, Division of Population Health & Genomics, School of Medicine, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK; Department of Endocrinology and Diabetes, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK.
Sundararajan Srinivasan, Division of Population Health & Genomics, School of Medicine, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK.
Moneeza K Siddiqui, Division of Population Health & Genomics, School of Medicine, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK.
Colin N A Palmer, Division of Population Health & Genomics, School of Medicine, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK; Centre for Pharmacogenetics and Pharmacogenomics, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK.
Graham P Leese, Division of Population Health & Genomics, School of Medicine, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK; Department of Endocrinology and Diabetes, Ninewells Hospital & Medical School, University of Dundee, Dundee DD1 9SY, UK.
Financial Support
This work was supported by the NHS Tayside Research Endowments. We also acknowledge the National Institute for Health Research using Official Development Assistance (ODA) funding (INSPIRED 16/136/102) for access to databases; M.K.S. and C.N.P. are funded through the same grant.
Author Contributions
E.S.P. researched/analyzed data and wrote the manuscript; P.J.N. planned the study, researched data, and wrote the manuscript; S.S. analyzed data and wrote the manuscript; M.K.S. researched data and wrote the manuscript; C.N.P. contributed to data analysis and to discussion; and G.P.L planned the study, researched data, contributed to the discussion, and reviewed/edited the manuscript.
Disclosures
The authors have nothing to disclose.
Data Availability
These are consented data and because of their sensitive nature are stored in secure computing environments. Data can be shared based on specific requests but as such are not publicly available.
References
- 1. Yu N, Donnan PT, Murphy MJ, Leese GP. Epidemiology of primary hyperparathyroidism in Tayside, Scotland, UK. Clin Endocrinol (Oxf). 2009;71(4):485–493. [DOI] [PubMed] [Google Scholar]
- 2. Yeh MW, Ituarte PH, Zhou HC, et al. Incidence and prevalence of primary hyperparathyroidism in a racially mixed population. J Clin Endocrinol Metab. 2013;98(3):1122–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bilezikian JP. Primary hyperparathyroidism. J Clin Endocrinol Metab. 2018;103(11):3993–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu N, Donnan PT, Flynn RWV, et al. Increased mortality and morbidity in mild primary hyperparathyroid patients. The Parathyroid Epidemiology and Audit Research Study (PEARS). Clin Endocrinol (Oxf). 2010;73(1):30–34. [DOI] [PubMed] [Google Scholar]
- 5. Yu N, Donnan PT, Leese GP. A record linkage study of outcomes in patients with mild primary hyperparathyroidism: the Parathyroid Epidemiology and Audit Research Study (PEARS). Clin Endocrinol (Oxf). 2011;75(2):169–176. [DOI] [PubMed] [Google Scholar]
- 6. Newey PJ, Hannan FM, Wilson A, Thakker RV. Genetics of monogenic disorders of calcium and bone metabolism. Clin Endocrinol (Oxf). Published online December 21, 2021. doi:10.1111/cen.14644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding C, Buckingham B, Levine MA. Familial isolated hypoparathyroidism caused by a mutation in the gene for the transcription factor GCMB. J Clin Invest. 2001;108(8):1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guan B, Welch JM, Sapp JC, et al. GCM2-activating mutations in familial isolated hyperparathyroidism. Am J Hum Genet. 2016;99(5):1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorvin CM, Frost M, Malinauskas T, et al. Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias. Hum Mol Genet. 2018;27(21):3720–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cromer MK, Starker LF, Choi M, et al. Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J Clin Endocrinol Metab. 2012;97(9):E1774–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newey PJ, Nesbit MA, Rimmer AJ, et al. Whole-exome sequencing studies of nonhereditary (sporadic) parathyroid adenomas. J Clin Endocrinol Metab. 2012;97(10):E1995–E2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simonds WF. Genetics of hyperparathyroidism, including parathyroid cancer. Endocrinol Metab Clin North Am. 2017;46(2):405–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson-Cohen C, Lutsey PL, Kleber ME, et al. Genetic variants associated with circulating parathyroid hormone. J Am Soc Nephrol. 2017;28(5):1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matana A, Brdar D, Torlak V, et al. Genome-wide meta-analysis identifies novel loci associated with parathyroid hormone level. Mol Med. 2018;24(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKinstry B, Sullivan FM, Vasishta S, et al. Cohort profile: the Scottish research register SHARE. A register of people interested in research participation linked to NHS data sets. BMJ Open. 2017;7(2):e013351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hébert HL, Shepherd B, Milburn K, et al. Cohort profile: Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS). Int J Epidemiol. 2018;47(2):380–381j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCarthy S, Das S, Kretzschmar W, et al. Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soto E, Newey P, Srinivasan S, Siddiqui M, Palmer C, Leese G. Supplementary data for “Identification of 4 new loci associated with primary hyperparathyroidism (PHPT) and a polygenic risk score for PHPT.” Mendeley Data V2. Deposited August 30, 2022. 10.17632/3ckp3w9vpb.2 [DOI] [PMC free article] [PubMed]
- 21. Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. [DOI] [PubMed] [Google Scholar]
- 22. Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20(8):467–484. [DOI] [PubMed] [Google Scholar]
- 23. Silverberg SJ, Clarke BL, Peacock M, et al. Current issues in the presentation of asymptomatic primary hyperparathyroidism: proceedings of the Fourth International Workshop. J Clin Endocrinol Metab. 2014;99(10):3580–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang W, Chung UI, Kronenberg HM, de Crombrugghe B. The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc Natl Acad Sci U S A. 2001;98(1):160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amano K, Hata K, Sugita A, et al. Sox9 family members negatively regulate maturation and calcification of chondrocytes through up-regulation of parathyroid hormone-related protein. Mol Biol Cell. 2009;20(21):4541–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haseeb A, Kc R, Angelozzi M, et al. SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation. Proc Natl Acad Sci U S A. 2021;118(8):e2019152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roosenboom J, Lee MK, Hecht JT, et al. Mapping genetic variants for cranial vault shape in humans. PLoS One. 2018;13(4):e0196148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomita K, Liu Y. Human BCDIN3D is a cytoplasmic tRNAHis-specific 5-monophosphate methyltransferase. Front Genet. 2018;9:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun J, Shin DY, Eiseman M, et al. SLITRK5 Is a negative regulator of hedgehog signaling in osteoblasts. Nat Commun. 2021;12(1):4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ihara T, Hosokawa Y, Kumazawa K, et al. An in vivo screening system to identify tumorigenic genes. Oncogene. 2017;36(14):2023–2029. [DOI] [PubMed] [Google Scholar]
- 31. Pekar JD, Grzych G, Durand G, et al. Calcium state estimation by total calcium: the evidence to end the never-ending story. Clin Chem Lab Med. 2020;58(2):222–231. [DOI] [PubMed] [Google Scholar]
- 32. Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50(11):1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4(6):e5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
These are consented data and because of their sensitive nature are stored in secure computing environments. Data can be shared based on specific requests but as such are not publicly available.