Abstract
PknB is a member of the newly discovered eukaryotic-like protein serine/threonine kinase (PSTK) family of proteins. The pknB gene was cloned and expressed in Escherichia coli. The active recombinant protein was purified and shown to be reactive with antiphosphoserine antibodies, as well as with antibodies to the phosphorylated eukaryotic Ser/Thr kinases mitogen-activated protein kinase kinase 3 and 6, P38, and Creb. In vitro kinase assays demonstrated that PknB is a functional kinase that is autophosphorylated on serine/threonine residues and is also able to phosphorylate the peptide substrate myelin basic protein. Analysis of pknB expression in Mycobacterium tuberculosis indicates the presence of pknB mRNA in (i) organisms grown in vitro in bacteriological media, (ii) a murine macrophage in vitro infection model, and (iii) in vivo alveolar macrophages from a patient with tuberculosis.
Tuberculosis is the primary cause of mortality due to an infectious disease in the world today. The causative agent of tuberculosis is the intracellular pathogen Mycobacterium tuberculosis. The ability of M. tuberculosis to enter macrophages and to avoid intracellular killing is believed to be pivotal to its virulence. The host immune response to tuberculosis is complex and involves T cells, mononuclear phagocytes, and cytokines (21). Mycobacteria have been suggested to have the ability to subvert normal host immune response mechanisms in order to enhance their intracellular survival. For example, there is evidence to indicate that mycobacteria prevent macrophage acidification (29, 34), inhibit antigen processing (23), and attenuate gamma interferon action (31) and protein kinase C activation in the macrophage (5). These macrophage deactivation mechanisms are considered to be survival strategies of mycobacteria within the host. Induction of these evasive mechanisms in M. tuberculosis most probably involves the ability of the organism to adapt its responses to external signals.
Protein phosphorylation is a principal mechanism by which extracellular signals are translated into cellular responses. Protein phosphorylation is carried out by specific protein kinases and is coupled to dephosphorylation reactions carried out by protein phosphatases. In bacteria, the molecular system that is responsible for stimulus response coupling involves the so-called two-component system consisting of histidine kinase sensors and their associated response regulators (33). In contrast, protein phosphorylation in eukaryotes occurs mainly on phosphoester (serine, threonine, or tyrosine) residues. The eukaryotic protein kinases and protein phosphatases are the backbone of signal transduction pathways. Phosphoester protein kinases and their coupled phosphatases were previously thought to be unique to eukaryotes. Recently, evidence arising from the accumulation of bacterial genome sequencing data and the use of antiphosphoprotein antibodies has revealed that some prokaryotes also contain phosphoester kinases and phosphatases (7, 28, 40).
In prokaryotes, protein serine/threonine kinases (PSTKs) have been shown to be involved in two different processes, development and pathogenicity. Bacteria capable of differentiation into a new developmental state, including Streptomyces (19, 25, 37) and Anabaena (40–42) spp. and Myxococcus xanthus (13, 35, 36, 43), contain a large number of PSTK genes in their genomes. In these bacteria, kinases are involved in the control of the late stages of development, sporulation, or secondary metabolite production. Alternatively, PSTKs have been shown to be involved in the survival of human pathogens within the host, as exemplified by the Yersinia pseudotuberculosis plasmid-encoded protein kinase yopO (12) or the Pseudomonas aeruginosa PSTK (39). Interestingly, both of these kinases have been shown to be required for the full virulence of these pathogens in mouse models.
Previously, we have shown that M. tuberculosis encodes at least eight eukaryotic-like protein kinases (3). Furthermore, we have demonstrated that six proteins are phosphorylated in vitro (3), suggesting the presence of functional kinases in M. tuberculosis. The completion of the M. tuberculosis genome sequencing project provided a complete list of these eukaryotic-like protein kinases and phosphatases that now form the M. tuberculosis PSTK family (6). As of today, this family is composed of at least 11 protein kinases and four protein phosphatases. The M. tuberculosis PSTK pknD gene has previously been cloned and analyzed and shown to encode a functional serine/threonine kinase (27). This paper describes the cloning, expression, and molecular characterization of PknB, a putative M. tuberculosis protein serine/threonine kinase encoded by an open reading frame that resides near the origin of replication. The data shows that pknB encodes a functional kinase that is constitutively transcribed in M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains, vectors, and culture conditions.
M. tuberculosis H37Rv NCTC 7416 was obtained from the National Collection of Type Cultures, London, United Kingdom. Escherichia coli DH5α (Clontech Laboratories, Inc., Palo Alto, Calif.) and E. coli BL21(DE3) (Novagen R & D) were used for maintenance of plasmids and expression of foreign proteins, respectively. The plasmid pET-22b (Novagen) was used as an expression vector in E. coli BL21(DE3). E. coli strains were cultured on Luria-Bertani (LB) agar or broth with or without selective antibiotics. Mycobacterial strains were cultured in Middlebrook 7H9 broth or 7H10 agar (Difco) supplemented with OADC (Difco), Tween 80, and glycerol.
Amplification and cloning of pknB.
Genomic DNA of M. tuberculosis H37Rv was prepared as described previously (3). The open reading frame Rv 0014c which codes for PknB was amplified from this DNA with the following primers: 1, 5′-AAATACATATGACCACCCCTTCCCA-3′; and 2, 5′-TTAAGCTTCTACTGGCCGAACTCA-3′. Primers 1 and 2 contained NdeI and HindIII restriction sites, respectively. PCR was performed with Taq polymerase obtained from Gibco BRL by using 2 mM MgCl2 and 5% dimethyl sulfoxide. Annealing temperatures were 58 and 63°C. The PCR products were separated on a 1% agarose gel. The appropriate PCR product was ligated into the vector pCR2.1 of the TA cloning kit (Invitrogen) and transformed into E. coli DH5α or INVF′α by standard chemical transformation procedure. Clones containing the vector were selected on LB-plus-ampicillin (100 μg/ml) plates, and plasmid DNA was digested with restriction endonucleases NdeI and HindIII (Fermentas). Restriction enzyme-digested plasmids were isolated with a QIAquick gel extraction kit (Qiagen Ltd.). A corresponding digestion was also applied to plasmid pET-22b, and the two products were ligated together with T4 DNA ligase to obtain the plasmid pYA102 (Fig. 1).
FIG. 1.
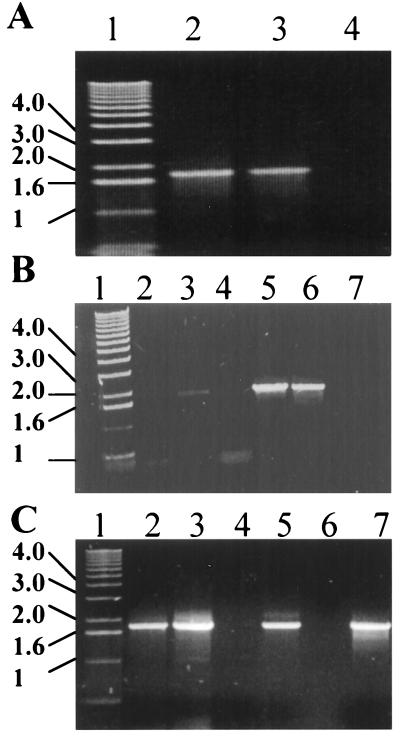
Chromosomal location and cloning of PknB. (A) Schematic map describing the chromosomal location of the pknB open reading frame. (B) PCR amplification of pknB by using gene-specific primers as described in Materials and Methods. Lanes: 1, lambda HindIII molecular size markers; 2, PCR without 5′ primer; 3, PCR without 3′ primer; 4, PCR without template DNA; 5, complete reaction using M. tuberculosis H37Rv DNA as the template. (C) The expression plasmid pYA102.
Expression and purification of PknB.
Competent cells of E. coli BL21(DE3) were prepared according to the CaCl2 method (30) and were transformed by the heat shock method for 2 min at 42°C with 100 ng of pYA102. The transformed E. coli cells were then plated onto LB agar supplemented with ampicillin (100 μg/ml). Single colonies were inoculated into 5 ml of LB broth also containing ampicillin (100 μg/ml). After overnight incubation at 37°C with shaking, the individual cultures were diluted 1:100 in the same medium and incubation was continued at 37°C with shaking. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM when the optical density at 600 nm reached 0.6. Cultures were centrifuged at 5,000 × g for 15 min at room temperature, and pellets were lysed in B-Per (Pierce) bacterial protein extraction reagent. Proteins were separated by centrifugation (15,000 × g, 4°C, 15 min) into soluble and insoluble fractions. PknB inclusion bodies contained in the insoluble fractions were purified from E. coli membrane proteins by washing in a solution of 10% B-Per reagent and centrifugation (45,000 × g, 90 min, 4°C). PknB was separated by sodium dodecyl sulfate–7.5% polyacrylamide electrophoresis (SDS-PAGE) and stained with Coomassie blue or transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The N-terminal amino acid sequence was verified after electrophoresis of samples in SDS-PAGE gels and electroblotting onto PVDF membranes. Edman degradation was performed, and the sequence of the first 10 amino acids from the NH2 terminus was determined at the University of British Columbia Protein Sequencing Laboratory. In order to obtain soluble protein, PknB inclusion bodies were resuspended in 1× phosphate-buffered saline (PBS) (pH 7.4) and slowly added drop-wise to a solution of 16 M urea and 2 M dithiothreitol (DTT) to make a final concentration of 8 M urea and 1 M DTT. Soluble PknB was then dialyzed via a Spectra/Por 8000 cellulose membrane (VWR Scientific) against 200 volumes of 1× Tris-buffered saline (pH 7.4) at 4°C for 16 to 24 h. The sample was then centrifuged for 15 min at 4°C and 15,000 × g (Baxter), and approximately 20 mg of protein was loaded onto a 50-ml Macro-Prep SE agarose size-exclusion column (Bio-Rad), which was used as a desalting column. Proteins were eluted over time at 4°C with a size-exclusion buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM DTT, and 0.01 mM EDTA (18). The purity of PknB was tested by subjecting samples to SDS-PAGE followed by Coomassie blue staining. SDS-PAGE gels were prepared by the method of Laemmli (17). The gels were stained with Coomassie blue R-250 or silver stain. Protein concentrations were determined by the Bradford protein assay reagent (Bio-Rad).
Production of polyclonal antiserum to PknB.
The gel electrophoresis band corresponding to PknB was excised from an SDS–7.5% PAGE gel and homogenized in PBS. Homogenized gels were mixed with Titremax adjuvant (1:1 [vol/vol]) and were injected subcutaneously into 8-week-old BALB/c mice. Two weeks after the immunization, the animals were bled and sera were prepared. Horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) antibodies and enhanced chemiluminescence reagents were used to detect antibodies bound in Western blots.
In vitro kinase assay.
PknB autophosphorylation and myelin basic protein (MBP) phosphorylation were determined by an in vitro kinase assay. Twenty-microgram samples of PknB protein were added to 15 μl of kinase buffer {20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 7.2], 10 mM MgCl2, 10 mM MnCl2} with or without 50 μg of MBP (Sigma), and the reaction was started by addition of 1 μCi of [γ-32P]ATP (Mandel Scientific). Incubation was performed at room temperature. One-third of the incubation mixture was then loaded onto P81 phosphocellulose filter paper (Baxter) for incorporation measurements, and to the remainder 3× Laemmli sample buffer was added to stop the reaction. The latter mixture was boiled for 5 min and resolved by SDS-PAGE. The gels were electroblotted onto nitrocellulose or PVDF membranes (Bio-Rad) and then exposed to Kodak X-Omat/AR film. Radioisotope levels in filter paper assays were determined by scintillation counting (Beckman LS 1800).
Phosphoamino acid analysis, Western blotting, and cross-reactivity with antieukaryotic PSTKs.
Autophosphorylated PknB was excised from PVDF membranes and subjected to acid hydrolysis as described by Kamps and Sefton (15). Samples were spotted onto a cellulose thin-layer chromatography plate (Eastman, Rochester, N.Y.) and subjected to two-dimensional liquid thin-layer chromatography. Control phosphotyrosine, phosphothreonine, and phosphoserine amino acids were stained with ninhydrin, and radiolabelled amino acids were visualized via autoradiography. Phosphoamino acid analysis was also performed by loading PknB on SDS-PAGE gels followed by Western blot analysis with antiphosphoserine and antiphosphothreonine monoclonal antibodies (Sigma) as described in the manufacturer's instructions. Soluble PknB was electrophoresed on an SDS–7.5% PAGE gel and transferred to nitrocellulose by semidry electroblotting (Ancos). Blots were blocked with 4% skim milk (Difco, Detroit, Mich.) in 1× PBS (pH 7.5) overnight at 4°C on a shaker. Blots were washed with 1× PBS–0.1% Tween 20 and incubated for 2 h in 1× PBS–Tween (pH 7.5) with either mouse monoclonal antiphosphoserine (1/500) (Sigma), mouse monoclonal antiphosphothreonine (1/100) (Sigma), rabbit antiphospho Creb Ser-133 (1/10,000) (New England Biolabs), rabbit antiphospho p38 Tyr-182 (1/10,000) (New England Biolabs), rabbit antiphospho mitogen-activated protein kinase kinase 3 and 6 (MKK3/6) Ser 189-202 (1/10,000), or rabbit anti-ERK1 (1/10,000) (Kinetek Pharmaceuticals, Vancouver, British Columbia, Canada). Blots were washed for 45 min in 1× PBS–Tween and incubated with 1/20,000 of either goat anti-rabbit IgG (heavy plus light chain) or goat anti-mouse IgG (heavy plus light chain) HRP-conjugated antibody (Bio-Rad). Blots were incubated in Super Signal reagent (Pierce, Rockford, Ill.) and exposed by using Kodak X-Omat/AR film.
Murine macrophage infection.
The mouse macrophage cell line J774.2 (American Type Culture Collection) was seeded and maintained in 100-mm-diameter tissue culture plates in RPMI 1640 medium (Gibco) containing 10 mM HEPES, 2 mM l-glutamine, and 10% heat-inactivated fetal calf serum (FCS). For reverse transcription (RT)-PCR analysis, approximately 107 cells of M. tuberculosis H37Rv cultured in RPMI 1640 medium supplemented with 5% FCS were added to each J774.2 plate and incubated for 24 h. After 24 h, the cells were washed twice with warm RPMI 1640 medium containing 1% FCS. Total RNA was isolated after 24 and 72 h.
Isolation of human alveolar macrophages.
Alveolar macrophages were obtained from bronchoalveolar lavage fluid from resected human lungs. Bronchoalveolar lavage fluid was filtered through sterile gauze and centrifuged at 450 × g at room temperature for 7 min. Erythrocytes were lysed by treatment with distilled water, and cells were washed twice with sterile 1× PBS. Cells were resuspended in RPMI 1640 culture medium supplemented with 10% heat-inactivated FCS. Alveolar macrophages were plated in 60-mm-diameter tissue culture plates at 2 × 106 cells/ml. Cells were allowed to adhere overnight at 37°C. Nonadherent cells were removed by gently rinsing the plates with warmed RPMI 1640 medium supplemented with 5% FCS. The protocol for using human biological samples was approved by the University of British Columbia Ethics Committee.
cDNA preparation and RT-PCR.
RNA was prepared in a P3-level laboratory as follows. Exponentially growing M. tuberculosis (H37Rv), J774.2 cells infected with M. tuberculosis or adherent alveolar macrophages from BAL fluid, were harvested by centrifugation at 3,000 × g for 10 min. Pellets were resuspended in 2 ml of Tween-saline (0.8% [wt/vol] NaCl, 0.05% [vol/vol] Tween 80) followed by centrifugation at 17,000 × g for 1 min. The mycobacterial pellets were resuspended in 1 ml of guanidinium isothiocyanate buffer (5 M guanidinium isothiocyanate, 25 mM sodium acetate [pH 6.0], 1% N-lauryl sarcosine, 10 mM DTT), and approximately 1 ml of 0.1-mm-diameter zirconium beads was added. Mycobacteria were then subjected to disruption in a bead-beater device for 3 min. Nucleic acid was prepared from the upper aqueous lysate by a series of chloroform and phenol-chloroform-isoamyl alcohol extractions followed by ethanol precipitation. To remove DNA template, RNA was treated with RNase-free DNase (RQ1; Promega) twice at a concentration of 1 U/μg of RNA. DNase was removed by phenol-chloroform-isoamyl alcohol extraction, followed by extraction with chloroform and ethanol precipitation. RNA pellets were resuspended in diethylpyrocarbonate-treated water, and the absence of DNA contamination was confirmed by PCR with specific primers for pknB. RNA concentrations were determined by measuring absorbance at 260 nm. Total RNA was heated at 94°C for 3 min before being cooled on ice for 5 min. RNA was reverse transcribed by adding 1× PCR buffer, 1.5 mM MgCl2, 200 μM each of the four deoxynucleotide triphosphates, 5 nM random hexamers or specific downstream nucleotides, and 50 U of murine leukemia virus reverse transcriptase (Perkin-Elmer). RNA was reverse transcribed at 42°C for 3 h. Murine leukemia virus was inactivated by incubation at 99°C for 3 min. Amplified products were produced routinely with either 20 ng of genomic M. tuberculosis DNA, as a positive control, or 5 to 10 ng of cDNA which was added to 20 μl containing 1× PCR buffer, 200 μM deoxynucleotide triphosphates, 4% dimethyl sulfoxide, 200 pmol of each primer, and 1 U of Taq polymerase (Fermentas). The primers for RT-PCR analysis were the same primers used for the gene cloning. The amplification program consisted of preamplification denaturation at 95°C for 3 min, followed by 35 cycles of 60 s of denaturation at 94°C, 60 s of annealing at 58°C, and a 90-s extension at 72°C. Ten percent of the reaction product was run on a 1% agarose gel and visualized by staining with ethidium bromide.
RESULTS
Cloning and expression of the pknB gene.
Because of the minute quantities of regulatory proteins in bacterial cells and the large volumes of pathogenic M. tuberculosis culture needed for purification of proteins, a recombinant expression approach was taken to characterize PknB at the biochemical level. The M. tuberculosis pknB gene was cloned and expressed under the control of a T7 promoter in the E. coli expression vector pET-22b. By using the primers described in Materials and Methods, the open reading frame encoding PknB was successfully amplified by PCR from M. tuberculosis H37Rv genomic DNA to give a 1,877-bp fragment (Figure 1) and cloned into the T7 expression vector pET-22b. The map of the resulting plasmid described as pYA102 is shown in Fig. 1C.
The pknB gene of M. tuberculosis was expressed from pYA102 following treatment of exponentially growing pYA102-E. coli BL21(DE3) transformed cells with 1 mM IPTG at 37°C for 4 h (Fig. 2). As shown by SDS-PAGE and Coomassie blue staining, IPTG induced a protein approximately 68 kDa in size (Fig. 2A). This expressed band was visible in both whole-cell lysates and postsonication supernatant within 2 h of IPTG induction. Further purification attempts revealed that PknB was present in the form of insoluble inclusion bodies. The inclusion bodies remained as stable insoluble aggregates even following multiple washes with detergent solutions. To verify that the recombinant protein present in the inclusion bodies is identical to the predicted protein encoded by the M. tuberculosis pknB gene, we performed N-terminal amino acid sequencing on the IPTG-inducible protein. The first 10 amino acids of this IPTG-induced band were shown to be identical to the amino acid sequence of the pknB gene product derived from the M. tuberculosis genome sequence database.
FIG. 2.
PknB expression. (A) SDS-PAGE analysis of PknB expression in E. coli. Lanes: 1, molecular size markers; 2, negative control pET-22b in E. coli BL21 after IPTG induction; 3, cell extract of pYA102 expressing PknB after IPTG induction; 4, pellet of cell extract of pYA102 expressing PknB after IPTG induction; 5, PknB inclusion bodies after washes with detergent; 6, PknB after urea-DTT treatment; 7, PknB after size-exclusion chromatography. (B) Western analysis using mouse anti-PknB polyclonal antibodies. Lanes: 1, negative control pET-22B; 2, pellet of cell extract of pYA102 expressing PknB after IPTG induction; 3, PknB inclusion bodies; 4, PknB after urea-DTT treatment; 5, PknB after size-exclusion chromatography.
Production of polyclonal antibodies against PknB.
As described in Materials and Methods, the PknB protein band was excised from an SDS-PAGE gel and used to immunize mice. The polyclonal mouse anti-PknB serum reacted with the PknB inclusion bodies as well as with PknB from all downstream purification steps (Fig. 2B). The polyclonal serum was specific for PknB as it did not react with fractions prepared from E. coli BL21 cells containing the expression vector without the pknB gene. These anti-PknB antibodies also reacted with cell extracts obtained from M. tuberculosis (data not shown) and were able to inhibit PknB activity (Fig. 3C).
FIG. 3.
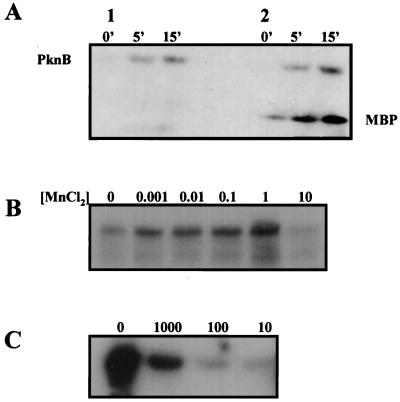
In vitro kinase assay. SDS-PAGE analysis of PknB labelled with [γ-32P]ATP. (A) Time course detection of PknB phosphorylation. Lanes: 1, PknB autophosphorylation; 2, MBP phosphorylation mediated by PknB. (B) MnCl2 concentration effect on PknB autophosphorylation. Units are in millimolar concentrations. (C) Effect of serial dilutions of anti-PknB antibodies on MBP phosphorylation by PknB.
Purification and renaturation of PknB from inclusion bodies.
Soluble, recombinant PknB in E. coli was obtained when bacteria were grown at 28°C. However, the low yield obtained under these conditions and the need for a long purification process prompted us to develop an alternate strategy to obtain active protein from inclusion bodies rather than from the soluble material. A three-step denaturation and purification process was used. As described in Materials and Methods, the approach taken involved solubilization of the inclusion bodies in a highly concentrated solution of urea and DTT followed by dialysis and size-exclusion chromatography (18). The progression of PknB purification and solubilization is shown in Fig. 2A. Dialysis was used to remove urea and DTT, and size-exclusion chromatography was used to further purify refolded PknB in an active form. As shown in Table 1, the purification fold increase for PknB was 58 while the yield of pure active PknB through size-exclusion chromatography was 3.48% of the total cell proteins present prior to purification. The 96.52% loss of the protein may be due to the formation of PknB multimers or incompletely renatured PknB molecules that migrate at a different molecular weight than refolded PknB. These values are similar to those obtained for other recombinant proteins that were recovered from inclusion bodies formed in E. coli (18).
TABLE 1.
Recovery of PknB from inclusion bodiesa
| Recovery step | Total protein (mg) | Sp act (cpm/min μg of protein−1) | Total activity | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 1,207 | 4,220 | 5.10 × 109 | 100 | 1 |
| Inclusion bodies | 593 | 4,420 | 2.63 × 109 | 51.6 | 2.03 |
| Macro Prep SE | 21 | 8,460 | 1.78 × 108 | 3.48 | 58 |
One liter of E. coli BL21 containing the expression plasmid pYA102 produced 6.77 g (wet weight) of cells after induction with IPTG as described in Materials and Methods. Data shown are those for 1 liter of culture and are representative of several experiments. Specific activity measurements are based upon an in vitro kinase assay using MBP as an alternative to the unidentified natural substrate and thus are artificial and are presented solely for the purpose of purification evaluation.
PknB possesses intrinsic kinase activity.
As shown in Fig. 3, in vitro kinase assays revealed that PknB is capable of phosphorylating itself and a conventional in vitro substrate for serine/threonine kinases, MBP from bovine brain. Incorporation of γ-32P from ATP into PknB occurred very rapidly, reaching a maximum in 15 min (Fig. 3A). This kinase activity was enhanced in the presence of up to 1 mM Mn2+. Mn2+ at concentrations of 10 mM and above inhibits PknB autophosphorylation (Fig. 3B) as well as the phosphorylation of MBP. As shown in Fig. 3C, the mouse anti-PknB polyclonal antibodies block MBP phosphorylation by PknB. The enzyme is completely inactivated when it is incubated with 1/100 dilution of 1:1 (vol/vol) of anti-PknB antibodies.
PknB is autophosphorylated on serine and threonine amino acid residues.
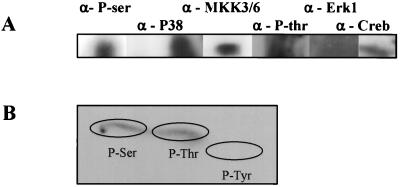
To demonstrate that PknB is a eukaryotic-like serine/threonine protein kinase, specific antibodies against phosphoserine and phosphothreonine were used to detect PknB in Western blots. As shown in Fig. 4, specific antiphosphoserine and antiphosphothreonine antibodies recognized PknB, thus confirming that the refolded PknB is phosphorylated on serine/threonine amino acids as suggested by their homology to other serine/threonine kinases. Most interestingly, refolded PknB is also recognized by antibodies against the phosphorylated eukaryotic signaling elements MKK3 and MKK6, P38, and Creb (Fig. 4A). Phosphoamino acid analysis using acid hydrolysis followed by two-dimensional thin-layer chromatography confirmed that PknB is phosphorylated on both serine and threonine residues and not tyrosine (Fig. 4B).
FIG. 4.
PknB cross-reactivity with eukaryotic phosphoprotein antibodies and phosphoamino acid analysis. (A) Cross activity of PknB with antibodies against phosphoproteins was determined by SDS–7.5% PAGE analysis followed by Western blot analysis. (B) Phosphoamino acid analysis of autophosphorylated PknB was performed by excision of radioactively labelled PknB from a PVDF membrane followed by acid hydrolysis and two-dimensional thin-layer chromatography. The positions of the unlabeled phosphoamino acids standards are encircled.
PknB is constitutively transcribed and is detected in alveolar macrophages from a patient with tuberculosis.
An important aspect of the life cycle of M. tuberculosis is its ability to survive within host macrophages. It was therefore considered pertinent to study whether the expression of M. tuberculosis pknB was affected by the transition from an extracellular to an intracellular environment. Hence, the expression of pknB in vitro and within host macrophages was examined. RT-PCR analysis of RNA extracted from M. tuberculosis growing in culture resulted in the amplification of a fragment of DNA of the expected size (1,600 bp) (Fig. 5A). This indicated that the pknB gene is expressed in the organism growing in in vitro culture. As shown in Fig. 5, RT-PCR analysis of cells from the murine macrophage cell line J774.2 infected with M. tuberculosis resulted in amplification of pknB when total RNA was used as a template. No amplification was observed with cDNA prepared from uninfected mouse macrophages. The intensity of specific pknB expression within murine macrophages appeared to depend upon the duration of infection. Thus, pknB expression was increased between 24 and 72 h of infection (Fig. 5B, lanes 3 and 5). This could suggest either an increase in the expression of pknB as a part of adaptation of M. tuberculosis to the change in the environment or an increase in the number of bacteria associated with J774.2 cells over the additional period of incubation, given that Mehta et al. (22) showed a fourfold increase in J774-associated M. tuberculosis after 3 days of incubation.
FIG. 5.
Transcription analysis of M. tuberculosis pknB. (A) RT-PCR using in vitro-grown M. tuberculosis RNA. Lanes: 1, 1-kb DNA ladder; 2, pknB amplified with cDNA made from M. tuberculosis RNA as a template; 3, pknB amplified from M. tuberculosis genomic DNA preparation; 4, no-DNA-template negative control. (B) Expression upon infection of mouse macrophages. Lanes: 1, 1-kb DNA ladder; 2, cDNA from uninfected control cells after 24 h of incubation; 3, pknB amplified from cDNA from M. tuberculosis infected cells after 24 h of incubation; 4, cDNA from noninfected control cells after 72 h; 5, pknB amplified from cDNA from M. tuberculosis-infected cells after 72 h; 6, pknB amplified from genomic M. tuberculosis DNA; 7, no-DNA template. (C) Expression of pknB in alveolar macrophages from a pulmonary tuberculosis patient. Lanes: 1, 1-kb DNA ladder; 2 to 5, pknB amplified with cDNA derived from alveolar macrophages from a patient suffering from pulmonary tuberculosis; 6, cDNA from noninfected control cells; 7, pknB amplified from genomic M. tuberculosis DNA.
Infection with M. tuberculosis is usually acquired by inhalation, and it is believed that during the initial establishment of infection organisms reside primarily in alveolar macrophages. The expression of pknB in alveolar macrophages taken from a patient with tuberculosis was examined. RT-PCR was performed on cDNA prepared from alveolar macrophages obtained by lung lavage of a patient suffering from tuberculosis. Figure 5C shows RT-PCR amplification of pknB in three of four preparations from this patient. No detectable amplification of pknB was observed with cDNA prepared from alveolar macrophages obtained from patients suffering from conditions other than tuberculosis.
DISCUSSION
This study characterized PknB, a member of the newly described eukaryotic-like serine/threonine kinase family from M. tuberculosis (3, 6). M. tuberculosis encodes in its genome 11 putative serine/threonine kinases. In contrast, E. coli and other bacteria whose genomes have been completely sequenced thus far do not contain this family of proteins. Growth and survival of M. tuberculosis inside host macrophages together with its long quiescent dormant state represent unique characteristics of this pathogen that may explain the need for a large number of these unique regulatory proteins. An important objective is to identify the roles of each of these protein kinases in the metabolic processes unique to M. tuberculosis. Perhaps the question of greatest importance is whether the PSTK genes encode active protein kinases. The findings of the present study show that PknB is indeed a functional protein kinase able to phosphorylate itself as well as model substrates such as MBP. PknB kinase activity and its properties are similar to those first described for the M. tuberculosis PSTK, PknD, a putative transmembrane kinase encoded by a gene located in the vicinity of the phosphate-specific transport operon (27).
As emphasized earlier, prokaryotic PSTKs fall into two categories, (i) those that are involved in pathogenicity and (ii) those that are involved in control of developmental processes. The best-studied example of a kinase involved in pathogenicity is provided by yopO (ypkA) of Y. pseudotuberculosis (12). YopO is secreted and targeted to the inner surface of the host cell plasma membrane (12) and is proposed to interfere with the host response by the disruption of cell signaling events. This interference probably occurs by phosphorylation of eukaryotic substrates. We have shown that PknB cross-reacts with certain eukaryotic antiphosphoprotein antibodies such as anti-MKK3/6 and anti-P38. However, there is no evidence for PknB secretion from M. tuberculosis, and therefore it seems unlikely that it will interact with eukaryotic signaling elements. Cross-recognition of PknB and eukaryotic kinases suggests that PknB has limited structural homology with these proteins, as suggested by their sequence similarities (3). Nevertheless, given that both P38 and MKK3/6 phosphoproteins are part of a cascade controlling cellular responses to stress and cytokines, it is also possible that PknB and these eukaryotic signaling elements are evolutionary related.
As shown in Fig. 1A, PknB is encoded by an open reading frame (Rv 0014c) that is in a cluster with pknA, the protein phosphatase ppp, and two other open reading frames that are predicted to encode the penicillin-binding protein PbpA and the cell division protein RodA. These open reading frames appear to form an operon structure. In E. coli, both rodA and pbpA are involved in cell structure determination and constitute a single transcriptional unit (20). Clusters of peptidoglycan biosynthesis and cell division genes were identified and sequenced in both gram-positive and gram-negative bacteria (1, 11, 20, 24, 38). These gene clusters are involved in the switching mechanism between cell elongation and division (4, 10, 14, 32). Cell division genes are not usually found coordinately expressed with kinases or phosphatases. The presence of pknB, pknA, and ppp genes within the cell division gene cluster may indicate an unusual regulatory cascade controlling M. tuberculosis cell growth.
By using the Kyte-Doolittle algorithm (16) to calculate hydrophilicity and transmembrane regions, PknB is predicted to contain a transmembrane domain between amino acids 332 and 350. Furthermore, fingerprint scan analysis (2) revealed that PknB possesses three of four known domains of the bacterial sensor C-terminal signature. In prokaryotes, histidine protein kinases function as sensors for various external signals (33). However, it was shown recently that Myxobacteria and Streptomyces contain serine/threonine kinases that are transmembrane proteins that may also serve as receptors for external signals (13, 25, 36). For example, Streptomyces AfsK and AfsR are suggested to be a two-component regulatory system involving serine protein kinases (19). Thus, it is reasonable to propose that PknB may serve as a sensor for external signals that translate into cell morphology changes such as division and elongation.
Most of the bacterial eukaryotic-like protein kinases that have been characterized so far have been shown to be involved in the regulation of different developmental states of the bacterium (35, 36, 40, 43). From this point of view, the best-studied organisms are the highly developed bacteria. For example, the Ser/Thr kinase Pkn1 of Myxococcus xanthus is expressed exclusively during sporulation and its inactivation inhibits spore formation in this bacterium (40). In a similar manner, inactivation of the Streptomyces granaticolor PSTK pkg2 resulted in changes in the formation of bacterial aerial hyphae (25). Both myxobacteria and Streptomyces spp. display developmental characteristics comparable to those of multicellular organisms. In contrast, the unicellular mycobacteria lack a complex developmental life cycle. However, it has been proposed that the M. tuberculosis “dormant state” could be considered as a new developmental growth state that is associated with prolonged latent infection and may be analogous to sporulation (9, 26). Interestingly, induction of the mycobacterial stationary phase by low oxygen tension has been shown to result in cell wall thickening (8). Thus, it is possible that PknB may be involved in M. tuberculosis progression into the late stationary or dormant stages. The finding that PknB expression is constitutive and present under both in vitro and in vivo growth conditions supports the hypothesis that PknB is a bacterial sensor that transduces signals through the mycobacterial membrane to an undefined accessory protein. Efforts to identify the candidate ligands for PknB are now in progress as well as gene knockout experiments that will attempt to demonstrate a role for PknB in the regulation of M. tuberculosis growth and pathogenicity.
ACKNOWLEDGMENTS
We thank Julian Davies for being our mentor and for his generous help and support. We thank Siobhan Cowley for assistance in preparation of the manuscript. We also thank Neil Reiner for helpful comments on the manuscript and John Belisle and his team from Colorado State University for kindly providing materials for this study.
This research was funded by Glaxo Wellcome Action TB program and the British Columbia TB Veterans Association.
REFERENCES
- 1.Asoh S, Matsuzawa H, Ishino F, Strominger J L, Matsuhashi M, Ohta T. Nucleotide sequence of the pbpA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K12. Eur J Biochem. 1986;160:231–238. doi: 10.1111/j.1432-1033.1986.tb09961.x. [DOI] [PubMed] [Google Scholar]
- 2.Attwood T K, Beck M E, Flower D R, Scordis P, Selley J N. The PRINTS protein fingerprint database in its fifth year. Nucleic Acids Res. 1998;26:304–308. doi: 10.1093/nar/26.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Av-Gay Y, Davies J. Components of eukaryotic-like protein signaling pathways in Mycobacterium tuberculosis. Microb Comp Genomics. 1997;2:63–73. [Google Scholar]
- 4.Begg K J, Donachie W D. Division planes alternate in spherical cells of Escherichia coli. J Bacteriol. 1998;180:2564–2567. doi: 10.1128/jb.180.9.2564-2567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brozna J P, Horan M, Rademacher J M, Pabst K M, Pabst M J. Monocyte responses to sulfatide from Mycobacterium tuberculosis: inhibition of priming for enhanced release of superoxide, associated with increased secretion of interleukin-1 and tumor necrosis factor alpha, and altered protein phosphorylation. Infect Immun. 1991;59:2542–2548. doi: 10.1128/iai.59.8.2542-2548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Cozzone A J. ATP-dependent protein kinases in bacteria. J Cell Biochem. 1993;51:7–13. doi: 10.1002/jcb.240510103. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMaio J, Zhang Y, Ko C, Young D B, Bishai W R. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donachie W D, Addinall S, Begg K. Cell shape and chromosome partition in prokaryotes or why E. coli is rod-shaped and haploid. Bioessays. 1995;17:569–576. doi: 10.1002/bies.950170616. [DOI] [PubMed] [Google Scholar]
- 11.Fsihi H, De Rossi E, Salazar L, Cantoni R, Labo M, Riccardi G, Takiff H E, Eiglmeier K, Bergh S, Cole S T. Gene arrangement and organization in a approximately 76 kb fragment encompassing the oriC region of the chromosome of Mycobacterium leprae. Microbiology. 1996;142:3147–3161. doi: 10.1099/13500872-142-11-3147. [DOI] [PubMed] [Google Scholar]
- 12.Hakansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanlon W A, Inouye M, Inouye S. Pkn9, a Ser/Thr protein kinase involved in the development of Myxococcus xanthus. Mol Microbiol. 1997;23:459–471. doi: 10.1046/j.1365-2958.1997.d01-1871.x. [DOI] [PubMed] [Google Scholar]
- 14.Henriques A O, Glaser P, Piggot P J, Moran C P., Jr Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol. 1998;28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- 15.Kamps M P, Sefton B M. Acid and base hydrolysis of phosphoproteins bound to Immobilon facilitates the analysis of phosphoamino acids in gel-fractionated proteins. Anal Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R F. A simple method for displaying the hydrophathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Landman O, Shiffman D, Av-Gay Y, Aharonowitz Y, Cohen G. High level expression in Escherichia coli of isopenicillin N synthase genes from Flavobacterium and Streptomyces, and recovery of active enzyme from inclusion bodies. FEMS Microbiol Lett. 1991;68:239–244. doi: 10.1016/0378-1097(91)90362-e. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto A, Hong S K, Ishizuka H, Horinouchi S, Beppu T. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene. 1994;146:47–56. doi: 10.1016/0378-1119(94)90832-x. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzawa H, Asoh S, Kunai K, Muraiso K, Takasuga A, Ohta T. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J Bacteriol. 1989;171:558–560. doi: 10.1128/jb.171.1.558-560.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonough K A, Kress Y, Bloom B R. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect Immun. 1993;61:2763–2773. doi: 10.1128/iai.61.7.2763-2773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta P K, King C H, White E H, Murtagh J J, Jr, Quinn F D. Comparison of in vitro models for the study of Mycobacterium tuberculosis invasion and intracellular replication. Infect Immun. 1996;64:2673–2679. doi: 10.1128/iai.64.7.2673-2679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno C, Mehlert A, Lamb J. The inhibitory effects of mycobacterial lipoarabinomannan and polysaccharides upon polyclonal and monoclonal human T cell proliferation. Clin Exp Immunol. 1988;74:206–210. [PMC free article] [PubMed] [Google Scholar]
- 24.Murray T, Popham D L, Setlow P. Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A. J Bacteriol. 1997;179:3021–3029. doi: 10.1128/jb.179.9.3021-3029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadvornik R, Vomastek T, Janecek J, Technikova Z, Branny P. Pkg2, a novel transmembrane protein Ser/Thr kinase of Streptomyces granaticolor. J Bacteriol. 1999;181:15–23. doi: 10.1128/jb.181.1.15-23.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrish N M, Dick J D, Bishai W R. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 27.Peirs P, De Wit L, Braibant M, Huygen K, Content J. A serine/threonine protein kinase from Mycobacterium tuberculosis. Eur J Biochem. 1997;244:604–612. doi: 10.1111/j.1432-1033.1997.00604.x. [DOI] [PubMed] [Google Scholar]
- 28.Potts M, Sun H, Mockaitis K, Kennelly P J, Reed D, Tonks N K. A protein-tyrosine/serine phosphatase encoded by the genome of the cyanobacterium Nostoc. J Biol Chem. 1993;268:7632–7635. [PubMed] [Google Scholar]
- 29.Russell D G, Dant J, Sturgill-Koszycki S. Mycobacterium avium- and Mycobacterium tuberculosis-containing vacuoles are dynamic, fusion-competent vesicles that are accessible to glycosphingolipids from the host cell plasmalemma. J Immunol. 1996;156:4764–4773. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Sibley L D, Hunter S W, Brennan P J, Krahenbuhl J L. Mycobacterial lipoarabinomannan inhibits gamma interferon-mediated activation of macrophages. Infect Immun. 1988;56:1232–1236. doi: 10.1128/iai.56.5.1232-1236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Signoretto C, Di Stefano F, Canepari P. Modified peptidoglycan chemical composition in shape-altered Escherichia coli. Microbiology. 1996;142:1919–1926. doi: 10.1099/13500872-142-8-1919. [DOI] [PubMed] [Google Scholar]
- 33.Stock J B, Ninfa A J, Stock A. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturgill-Koszycki S, Schlesinger P H, Chakraborty P, Haddix P L, Collins H L, Fok A K, Allen R D, Gluck S L, Heuser J, Russell D G. Lack of acidification in mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 35.Udo H, Inouye M, Inouye S. Biochemical characterization of Pkn2, a protein Ser/Thr kinase from Myxococcus xanthus, a gram-negative developmental bacterium. FEBS Lett. 1997;400:188–192. doi: 10.1016/s0014-5793(96)01384-1. [DOI] [PubMed] [Google Scholar]
- 36.Udo H, Munoz-Dorado J, Inouye M, Inouye S. Myxococcus xanthus, a gram-negative bacterium, contains a transmembrane protein serine/threonine kinase that blocks the secretion of beta-lactamase by phosphorylation. Genes Dev. 1995;9:972–983. doi: 10.1101/gad.9.8.972. [DOI] [PubMed] [Google Scholar]
- 37.Urabe H, Ogawara H. Cloning, sequencing and expression of serine/threonine kinase-encoding genes from Streptomyces coelicolor A3(2) Gene. 1995;153:99–104. doi: 10.1016/0378-1119(94)00789-u. [DOI] [PubMed] [Google Scholar]
- 38.Wada A, Watanabe H. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J Bacteriol. 1998;180:2759–2765. doi: 10.1128/jb.180.10.2759-2765.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J Y, Li C H, Yang C J, Mushegian A, Jin S G. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J Bacteriol. 1998;180:6764–6768. doi: 10.1128/jb.180.24.6764-6768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C C. Bacterial signalling involving eukaryotic-type protein kinases. Mol Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C C, Friry A, Peng L. Molecular and genetic analysis of two closely linked genes that encode, respectively, a protein phosphatase 1/2A/2B homolog and a protein kinase homolog in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1998;180:2616–2622. doi: 10.1128/jb.180.10.2616-2622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C C, Libs L. Cloning and characterisation of the pknD gene encoding an eukaryotic-type protein kinase in the cyanobacterium Anabaena sp. PCC7120. Mol Gen Genet. 1998;258:26–33. doi: 10.1007/s004380050703. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, Inouye M, Inouye S. Reciprocal regulation of the differentiation of Myxococcus xanthus by Pkn5 and Pkn6, eukaryotic-like Ser/Thr protein kinases. Mol Microbiol. 1996;20:435–447. doi: 10.1111/j.1365-2958.1996.tb02630.x. [DOI] [PubMed] [Google Scholar]