Summary Paragraph:
Receptor tyrosine kinase (RTK)-RAS signaling through the downstream mitogen activated protein kinase (MAPK) cascade regulates cell proliferation and survival. The SHOC2-MRAS-PP1C holophosphatase complex functions as a key regulator of RTK-RAS signaling by removing an inhibitory phosphorylation on RAF family proteins to potentiate MAPK signaling1. SHOC2 forms a ternary complex with MRAS and PP1C, and human germline gain-of-function mutations of this complex result in congenital RASopathy syndromes2-5. However, the structure and assembly of this complex are poorly understood. Here, we use cryogenic electron microscopy (cryo-EM) to resolve the structure of the SHOC2-MRAS-PP1C complex to 2.9Å resolution. Furthermore, we define the biophysical principles of holoenzyme interactions, elucidate the assembly order of the complex, and systematically interrogate the functional consequence of nearly all possible missense variants of SHOC2 through deep mutational scanning. We demonstrate that SHOC2 binds PP1C and MRAS through the concave surface of the leucine-rich repeat region and further engages PP1C through the N-terminal disordered region containing a cryptic RVxF motif. Initial complex formation is mediated by SHOC2-PP1C interactions and is stabilized by binding of GTP-loaded MRAS. These observations define how SHOC2 mutants in RASopathies and cancer stabilize interactions of complex members to enhance holophosphatase activity. Together, this integrative structure-function model comprehensively defines key binding interactions within the SHOC2-MRAS-PP1C holophosphatase complex to inform therapeutic development.
SHOC2 is a scaffold protein composed of leucine-rich repeats (LRRs) that binds directly to the PP1 catalytic subunit (PP1C). MRAS activation leads to membrane localization of the SHOC2-MRAS-PP1C (SMP) complex and dephosphorylation of RAF family proteins at key inhibitory phosphorylation sites, including S259 on CRAF (RAF1), S365 on BRAF, and S214 on ARAF (hereafter collectively referred to as ‘S259’), resulting in release of autoinhibition and potentiation of RAF activation1,6. The SMP holophosphatase plays a critical role in RAS-MAPK pathway signaling underlying normal developmental processes as well as oncogenic signaling in cancer. Activating mutations in SHOC2, PP1C, and MRAS are found in Noonan-like syndrome, a “RASopathy” syndrome characterized by congenital cardiac, skeletal, and cognitive deficits2,6-8. SHOC2 is essential for proliferation and survival of RAS-driven leukemia, non-small cell lung cancer models, and melanoma9,10. Moreover, SHOC2 depletion sensitizes RAS-driven cancers to MEK inhibition through disruption of RTK-mediated feedback signaling and MEK-inhibitor-induced RAF dimerization11,12. However, gaps in our knowledge of the biophysical underpinnings of complex assembly and function limit our understanding of mechanisms of disease and opportunities for therapeutic targeting of this ternary complex.
Structure of the SHOC2-MRAS-PP1C complex
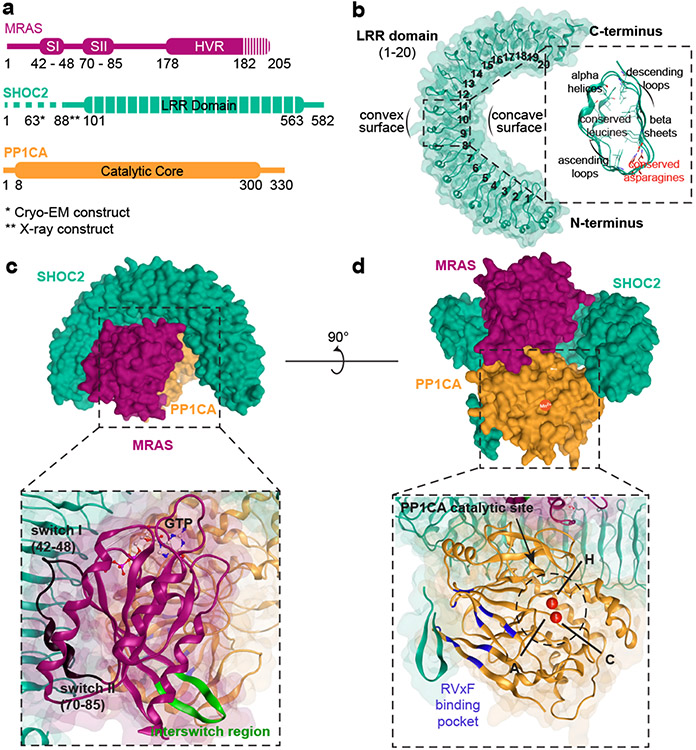
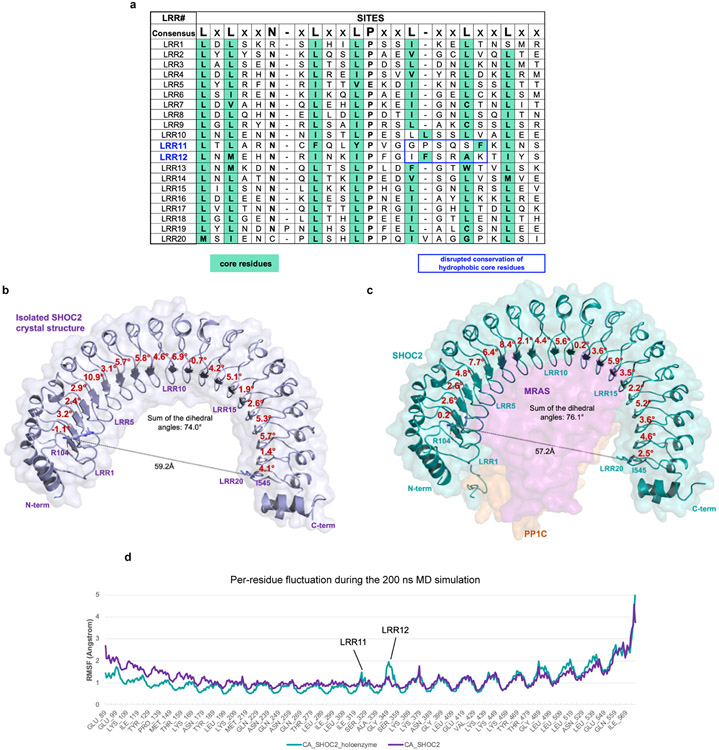
To understand the role of SHOC2 in RAS-MAPK pathway signaling, we solved the SHOC2 structure alone and in complex with PP1C and MRAS. Specifically, we developed an optimized expression systems to produce wild-type (WT) human SHOC2 and PP1C, as well as constitutively active GTP-bound MRASQ71L (Fig. 1a, Methods). We first used X-ray crystallography to solve the structure of WT SHOC2 apoprotein at 1.8 Å (Fig. 1b, Extended Data Table 1). The apo-SHOC2 structure is a canonical leucine-rich repeat (LRR) protein consisting of twenty tandem LRR domains. These LRR domains concatenate to form a conventional solenoid structure that has an average LRR corkscrew rotation of 4.0° (Fig. 1b) and is stabilized by an N-terminal flanking α-helix and C-terminal helix-turn-helix. Each LRR is composed of 22-24 amino acids containing a β-strand followed by a descending loop, α-helix, and ascending loop. The LRR β-strands assemble in parallel to generate the concave surface of the solenoid, while the α-helices form the majority of the convex surface. Internal to the structure, the conserved leucines of the LRR motif, LxxLxLxxN(x)1-2L, condense to form the hydrophobic core of the protein, while the conserved asparagine participates in a highly stabilizing “asparagine ladder” motif that has been shown to be critical to the LRR fold13. The SHOC2 structure does not demonstrate the presence of the flexible hinge within the medial LRRs that was predicted in previous computational models of SHOC214,15.
Figure 1: Structure of apo-SHOC2 and SHOC2-MRAS-PP1C holophosphatase complex.
a. Schematic diagram of complex members. Truncation of constructs is indicated in dashed lines. *Indicates 2-88 deletion SHOC2 construct utilized for X-ray crystallography and **2-63 deletion construct for cryo-EM. b. Overview of apo-SHOC2 crystal structure along with cross-sectional representation of LRR domain. c & d. Side views of Cryo-EM structure of SHOC2 complex with SHOC2 (teal), MRAS (maroon), and PP1C (yellow). A ribbon representation and view of MRAS (c) and PP1C (d) with relevant structural features annotated. d. Manganese ions (red), Hydrophobic (H), C-terminal (C), and Acidic (A) grooves are shown.
Since the conformation and binding interfaces of SHOC2 involved in complex formation with PP1C and MRAS cannot be inferred from the apoprotein structure alone, we used single particle cryo-EM to resolve the holophosphatase complex of an N-terminally truncated SHOC2, WT PP1C, and GTP-loaded MRASQ71L (Fig. 1c&d, Extended Data Fig. 1 & Extended Data Table 2). The resulting 2.9Å resolution structure unambiguously defined a ternary complex with SHOC2 engaging both PP1C and MRAS through its concave surface (Fig. 1d). The overall structure of SHOC2 observed in the holoenzyme complex is similar to the isolated apoprotein. Through protein-protein interactions, the SHOC2 complex buries a total of 5.934Å2 of solvent accessible surface area, with SHOC2-PP1C, SHOC2-MRAS, and MRAS-PP1C covering 3019Å2, 1729Å2, and 1186Å2, respectively (Extended Data Fig. 1e).
SHOC2 interacts with PP1C through two broad surfaces within the concave region of the LRR domains as well as a short stretch of its flexible N-terminal arm (residues 65-77). In this structure, the interaction between SHOC2 and PP1C does not lead to an appreciable change in PP1C conformation, in contrast to interactions of other holoenzyme partners with PP1C, such as MYPT1 and SDS2216,17. The most extensive SHOC2-PP1C interaction surface is found within the ascending loops of LRR2-5 and LRR7-11, which interfaces with a binding region of PP1C between the αG helix/loop and αA helix that resides proximal to but does not directly interact with the SILK binding region as previously suggested18. An additional contact surface on SHOC2 extends along the ascending loops of LRR13-16/18 with the positive charged surface of PP1C αF helix. These primary binding interactions between PP1C and SHOC2 occur on the face opposite from the PP1C catalytic site, leaving all three conventional PP1C binding grooves poised for substrate engagement. The SHOC2 LRR - PP1C interaction surface consists of 30 SHOC2 and 36 PP1C residues that primarily engage in a mixture of polar and pi-cation interactions with 1-2 residues per LRR (Fig. 2a). However, interactions through hydrogen and ionic bonds (SHOC2-PP1C: R203-E167; R182-E56, R203-E54; E155-R188) are predicted to create stabilizing interactions based on calculated residue interaction energies (Fig. 2a&b, Extended Data Fig. 2, Supplementary Table 1). We observed potential steric hinderance between the non-polar methyl-group of SHOC2 T411 and amine group of PP1C K147, which is anticipated to induce a modest destabilization of the interaction interface (Fig. 2b, Supplementary Table 1). The N-terminal region of SHOC2 exhibited low overall complexity and is generally predicted to be unstructured; however, this region has been shown to be necessary for MRAS and PP1C binding8. Within the holophosphatase structure, we observed that the N-terminal region of SHOC2 contains a cryptic RVxF motif (63GVAF66) that binds the RVxF interaction site of PP1C in a conventional manner19,20. In forming this interaction, SHOC2 residues 63-74 form a tight β-hairpin that extends the β-sandwich core of PP1C by engaging the RVxF binding motif composed primarily by β10 (Fig. 2a, Extended Data Fig. 2f). SHOC2 residues V64 and F66 embed within the hydrophobic RVxF binding pocket to enhance the affinity of SHOC2 for PP1C (Fig. 2a, Supplementary Table 1).
Figure 2: Detailed contacts between ternary SHOC2 complex members informs mechanism of assembly.
a. Enlarged images show surface contacting residues between SHOC2 LRR domains and PP1C (top left and middle) or MRAS (bottom), and residues of SHOC2 unstructured N-terminus contacting PP1C (top right). b. Energy contribution of key contact-residues between complex members (bars) and cumulative energy of interaction interface by Amber10 force field-based energy calculation (red line). c. Conformational comparison of MRAS switch I ‘open’ and ‘closed’ confirmation. d. Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC) analysis of SHOC2-MRAS-PP1C holoenzyme formation in the presence of MRAS-GDP (red line) MRAS-GppCp (blue line). Line trace represents n = 1 technical replicate and is representative of 3 biological replicates. Bio-Layer Interferometry (BLI) analysis of SHOC2 complex order of assembly for apo-SHOC2 with MRAS-GTP and PP1C (e) and SHOC2-PP1C activated engagement of MRAS-GTP binding (f). Line traces represents n = 1 technical replicate and is representative of 2 biological replicates. g. Schematic diagram of proposed model for SHOC2-MRAS-PP1C holophosphatase complex assembly whereby SHOC2 (teal) and PP1C (yellow) first engage in binding followed by MRAS-GTP (maroon) to stabilize and slow down dissociation of the complex. AUC and BLI experiments were repeated two or more times and representative is shown.
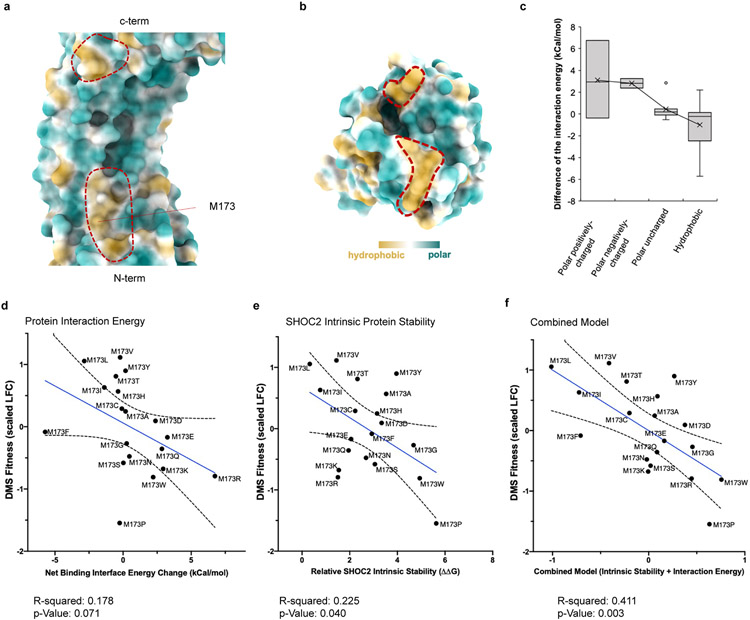
In contrast to the multiple interaction surfaces of SHOC2-PP1C, SHOC2 binds MRAS exclusively through the concave surface formed by the descending loop/beta-strands of SHOC2 LRR1-11. A total of 29 residues of SHOC2 engage in hydrophobic and polar contacts with 19 residues on MRAS between the switch I and II regions, which is found in its closed conformation (Supplementary Table 1). An additional minor hydrophobic contact surface occurs between the ascending loop of SHOC2 LRR 13-14 and the interstrand region of the MRAS G-domain, between the β5 strand and α4 helix (Fig. 2a, Extended Data Fig. 1). The N-terminal loop, switch I region, and α1 helix of MRAS also interact with PP1C on the acidic face that is proximal to the hydrophobic groove (Extended Data Fig. 1). Although the most stabilizing interactions between SHOC2 and MRAS involve selected ionic and hydrogen bonds (SHOC2-MRAS: R223-D43; R177-E47; K109-D64; R292-D41), most of the surface involves 20 hydrophobic interactions that further stabilize the complex (Fig. 2b, Supplementary Table 1). The SHOC2 M173I mutation has been previously reported to be a gain-of-function mutation found in Noonan-like Syndrome21. Consistent with this observation, M173 resides within the MRAS binding region of SHOC2, and comprehensive in silico structural analysis reveals a striking increase in overall energy stabilization when hydrophobic amino acid residues are substituted at the M173 position (Extended Data Fig. 3).
MRAS and PP1C interactions are primarily mediated by four hydrogen bonds (MRAS-PP1C: D48-R188; H53-D179; Q35-M190; K36-Q198) between the MRAS effector domain and a region near the hydrophobic groove of PP1C (residues 178-225) (Fig. 2a&b, Supplementary Table 1). Given the polar nature of this interaction and an inferred aggregate binding energy of only ~−63kCal/mol, this interaction was not anticipated to be stable in isolation. Indeed, using analytical ultracentrifugation experiments with isolated components of the complex demonstrate, we found that MRAS fails to bind PP1C in the absence of SHOC2 (Extended Data Fig. 2g). Previous studies have indicated that the SMP holophosphatase complex requires GTP-bound MRAS for complex formation and activation1. When complexed with GTP, RAS proteins are known to adopt an activated “closed” conformation wherein residues of their switch-I and switch-II domains (MRAS residues 42-48 and 70-85, respectively) undergo an essential conformational change to interact with the gamma phosphate of GTP, and in so doing generate a permissive RAS effector binding region6. Within the cryo-EM structure we observed that the same activating switch mechanism and RAS effector binding site occur in the assembly of the SMP holoenzyme complex. Indeed, assembly requires MRAS to be in the activated GTP-bound conformation to avoid extensive steric clashes with switch 1 and PP1C, permitting appropriate conformation for interactions between the MRAS effector binding region with PP1C and SHOC2 (Fig. 2c). We confirmed this requirement through analytical ultracentrifugation, in which MRAS bound to the non-hydrolyzable GTP analog GppCp drove assembly of the ternary complex as seen by a 52.1% shift in distinct sedimentation species to a Svedburg coefficient of 6.3 along with depletion of SHOC2/PP1c monomers and complex, whereas MRAS-GDP was unable to form a ternary complex (Fig. 2d).
Previous reports have suggested a potential flexible ‘hinge’ within the medial LRRs of SHOC214,15 due in part to non-conserved intervening residues at key leucine positions of the medial SHOC2 LLR domains 11 and 12 (Extended Data Fig. 4). Comparing our apo- SHOC2 and holoenzyme structures, we observed compression between LRR1 and LRR20 as well as an increased net corkscrew rotation of the LRR region when SHOC2 is bound to MRAS and PP1C compared to the unbound structure (Extended Data Fig. 4). Molecular dynamic (MD) simulation studies also revealed a high degree of flexibility/dynamic motion of LRR11 and 12 when in complex compared to in apo form (Extended Data Fig. 4c).
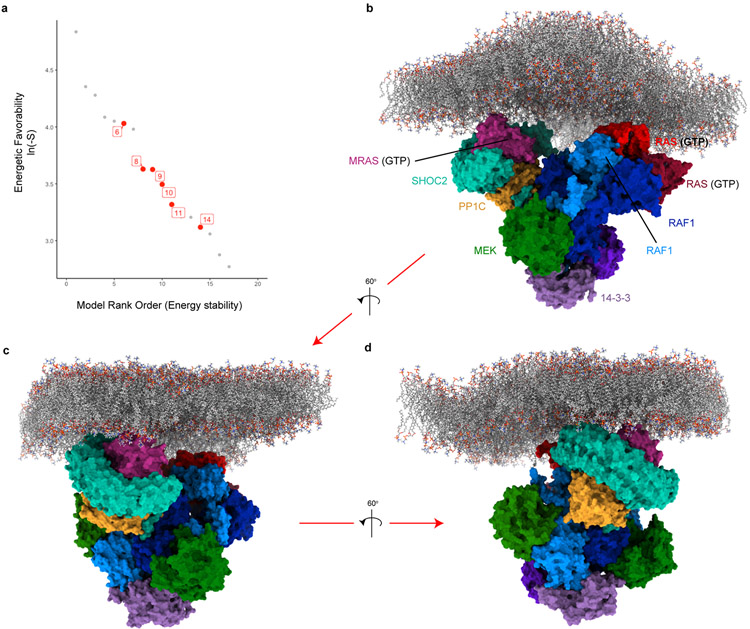
Finally, the SMP holophosphatase complex dephosphorylates RAF at the ‘S259’ position, inducing RAF release from its autoinhibited state when bound to 14-3-31. SHOC2-dependent PP1C dephosphorylation of RAF substrate is predicted to occur through proximal interactions at the plasma membrane for RAF-bound RAS proteins1, but the mechanism of this interaction has not been described. We systematically modeled the interaction between the SHOC2 ternary complex and RAF by merging the SHOC2 complex cryo-EM structure and a recently reported model of the RAS signalosome22 (Methods, Extended Data Fig. 5). This structural model of the multimeric complex reveals a spatially feasible and energetically favorable arrangement of membrane-bound SHOC2 ternary complex with the RAS signalosome, whereby S259 of RAF is engaged with the PP1C catalytic site. Furthermore, this model proposes that interactions between the SMP holophosphatase complex and RAF1 are mediated through PP1C, with no additional broad surfaces of SHOC2 in contact with any other member of the RAS-RAF complex.
Mechanism of SHOC2 complex assembly
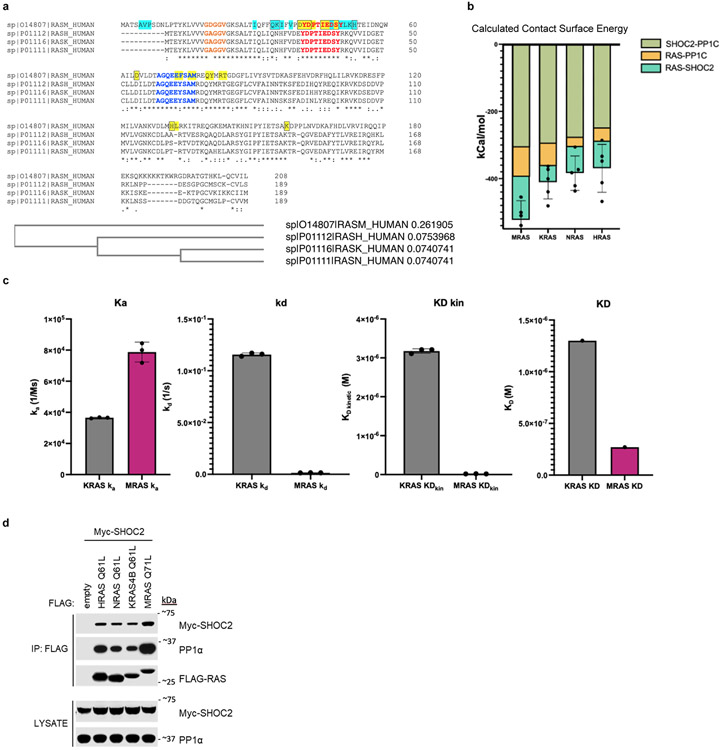
To define the mechanism of SMP holophosphatase complex assembly, we calculated interface contacting energy and observed that the SHOC2-PP1C interaction (−238 kCal/mol) demonstrates the greatest stability, primarily due to a greater number of electrostatic interactions compared to the other protein interfaces within the complex (Fig. 2b). To examine ternary complex assembly and dissociation, we performed Bio-layer interferometry (BLI) where SHOC2 was immobilized and sequentially exposed to either PP1C or activated MRAS bound to GppCp. These findings demonstrated clear engagement of SHOC2-PP1C in a fast on/off binding reaction with weaker affinity (2.7x104 Ms−1/0.39s−1 kd/ka) followed by kinetic locking via MRAS-GppCp binding (7.0x104 Ms−1/0.18s−1 kd/ka) (Fig. 2e&f). In contrast, SHOC2 sequentially exposed to MRAS-GTP followed by PP1C failed to form a complex. Consistent with observations from the cryo-EM structure, we confirmed the requirement of MRAS in the GTP-bound state to stably form the complete holoenzyme complex (Fig. 2g).
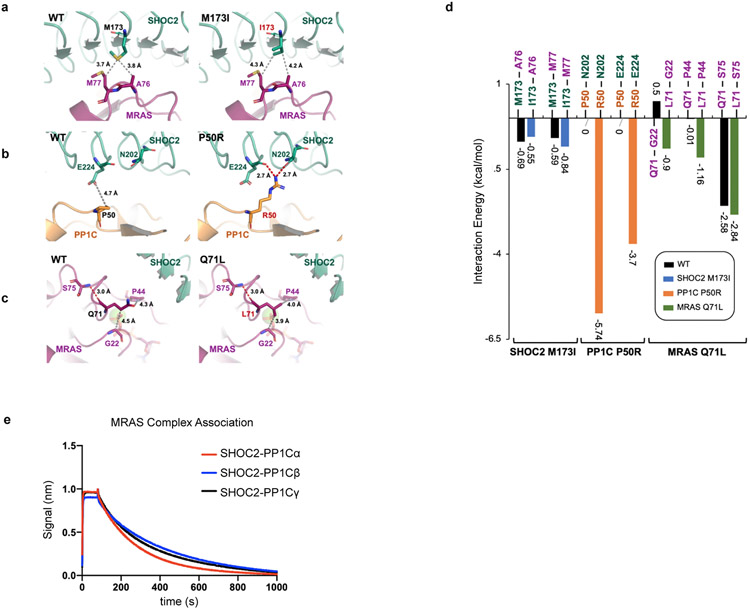
To further validate the cryo-EM model, we used BLI to assess the binding and kinetics of well-known gain of function (GOF) mutations in each complex member that have been reported in clinical cases of Noonan-Like Syndrome (PP1C P50R, MRAS Q71L, and SHOC2 M173I) (Supplementary Table 1, Extended Data Fig. 6). PP1C P50R resulted in an additional ionic interaction with E224 and hydrogen bond with N202 on the convex surface of SHOC2 LRR5, and an increased residue interaction energy of −9.4 kCal/mol. BLI studies demonstrated a 2-fold increase in Ka and 14.7-fold decrease in KD compared to PP1C wild type, with minimal difference in MRAS engagement (Extended Data Table 3). In contrast, we predicted that the SHOC2 M173I GOF mutation would further stabilize hydrophobic interaction with M77 found within the switch II domain of MRAS (Extended Data Table 3). Indeed, SHOC2 M173I induces relatively nominal changes in PP1C binding kinetics, but dramatically increases ka associated with MRAS (Extended Data Table 3). Finally, MRAS Q71L is a mutation with reduced intrinsic GTP hydrolysis and leads to a dramatic decrease in kd with the ternary complex. Combining all three mutant complex members, we observed a >33-fold increase in KD which further confirms the proposed model of complex assembly (Extended Data Table 3). SHOC2 is also known to engage various PP1C isoforms (PP1Cα/β/γ) and MRAS in an MRAS-GTP dependent manner18. In comparing the binding kinetics between SHOC2-PP1C isoforms and MRAS-GppCp, we found no substantial difference in binding kinetics (Extended Data Fig. 6e, Extended Data Table 3). Together, these studies implicate the GTP-bound, closed state of MRAS as a key requisite for stable complex formation.
Although MRAS has been reported to be the sole RAS isoform that binds the SHOC2 ternary complex, some studies suggest that additional RAS isoforms also bind SHOC223-25. Sequence alignment among RAS isoforms reveals conservation between MRAS and K/H/NRAS within several residue positions of switch I/II regions that engage in SHOC2/PP1C binding (Extended Data Fig. 7a). Modeling additional activated RAS isoforms into the complex, we observed a striking similarity in the overall orientation, and MD simulation revealed broadly stabilizing interactions between SHOC2/PP1C and the KRAS, NRAS, and HRAS isoforms (Extended Data Fig. 7b). To experimentally evaluate these in silico findings, we tested the ability of KRAS-GppCp to form a stable complex with the SHOC2 ternary complex. We observed a modestly higher kinetic on rate (ka) for active KRAS compared to MRAS binding to SHOC2/PP1C, but the interaction between SHOC2/KRAS/PP1C was transient due to a high dissociation rate constant in contrast to more stable engagement with MRAS (Extended Data Fig. 7c). To further complement these findings, we confirmed that SHOC2-PP1C interacts with RAS isoforms possessing Q61 mutations (H/K/NRAS), as assessed by immunoprecipitation studies in cells exogenously overexpressing RAS isoforms and SHOC2 (Extended Data Fig. 7d). These observations suggest that additional RAS isoforms beyond MRAS form SHOC2 ternary complexes.
Systematic structure-function analysis of SHOC2
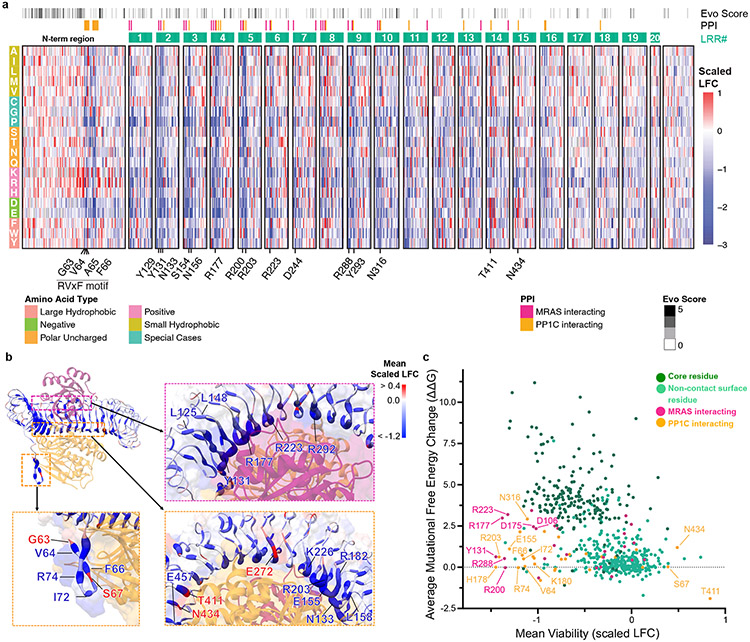
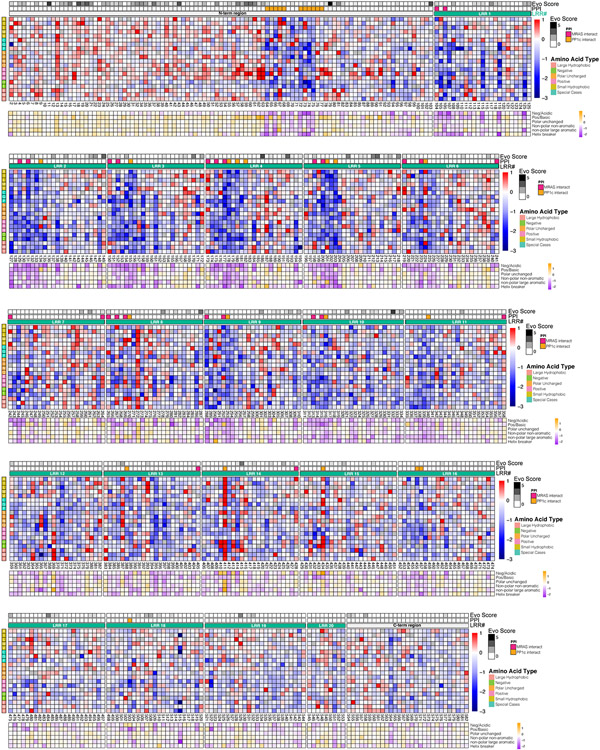
To complement the cryo-EM data and to define structure-function relationships within the SMP complex, we performed a deep mutational scanning (DMS) screen of the SHOC2 protein. Specifically, we created ~12,000 single residue variants of SHOC2 and assessed their function in KRAS-mutant cancer cells under SHOC2-dependent conditions and determined the scaled log2-fold change (LFC) of SHOC2 variants with a statistical threshold of gain- (GOF) and loss-of-function (LOF) set at >0.6 and <−0.6 respectively (Fig. 3a, Extended Data Fig. 8, Supplementary Table 2, Methods). We noted that at several residues within SHOC2, more than half of missense mutations scored as negatively selected LOF variants in the screen and refer to these positions as mutationally intolerant SHOC2 residue positions. These mutational intolerant positions occurred with a distinct periodicity and structurally reside within interaction surfaces between ternary complex members, with SHOC2 residues engaged in polar contacts exhibiting the greatest functional impact (Fig. 3b&c, Extended Data Fig. 9a). Indeed, we found that several SHOC2 residues that are predicted to form stabilizing interactions between SHOC2 and MRAS or PP1C were negatively selected in the screen upon mutation. Residues along the concave surface of SHOC2 that engage MRAS (E127, Y131, R177, R200, R223, R288) and PP1C (N156, H178, R203, Y293, N316) were found to be intolerant to mutation within the DMS screen (Fig. 3a&c). We validated representative LOF variants that scored in the DMS screen with mutations at positions involved in interactions with PP1C and MRAS in low attachment growth conditions (Extended Data Fig. 9b), an additional SHOC2-dependent functional assay18. Furthermore, we assessed downstream MAPK signaling and performed immunoprecipitation studies with select LOF mutants from the DMS screen (e.g. Y131E, R223F, E457K) and found that these variants displayed increased p-S259 RAF1, diminished MAPK activity, and disrupted SMP complex interactions compared to wildtype SHOC2 (Extended Data Fig. 9c-h).
Figure 3: Systematic deep mutational scanning (DMS) reveals structural constraints of SHOC2 complex function.
a. Heat map representation of log2-fold change (LFC) allele enrichment (red) and depletion (blue) between trametinib treatment and vehicle control, centered on SHOC2 wildtype (silent) and normalized to the mean of non-sense mutations (scaled LFC). SHOC2 positional evolutionary sequence variation (Evo Score) and protein-protein interacting residues (PPI) from cryo-EM data are indicated. b. Projections of observed DMS allele abundance on N-terminal unstructured region (bottom left), and MRAS (top right) and PP1C interface (bottom right) onto Cryo-EM structure. Color indicates mean positional Scaled LFC in the DMS fitness screen and size of residue indicates number of variants that score as GOF/LOF. c. Scatter plot showing position-level calculated, mean free-energy change upon mutation (intrinsic SHOC2 stability) and corresponding average scaled LFC for fitness in the SHOC2 DMS screen, with higher ddG values correspond to greater instability. Positive DMS scaled LFC: positive selection, GOF. Negative DMS Z-score: negative selection, LOF.
In agreement with the cryo-EM structure, SHOC2 alleles with mutations in the conserved hydrophobic residues of the cryptic N-terminal RVxF motif (V64 and F66) generally failed to functionally rescue viability in the DMS screen. Conversely, G63K/R/H substitutions scored as GOF in the screen (Extended Data Fig. 8&9). These GOF mutations change the WT 63GVAF66 sequence into a canonical RVxF motif, likely resulting in an increased stabilizing interaction through a new hydrogen bond formation with D242 on PP1C, similar to conventional RVxF motifs26. Indeed, SHOC2 G63R was found to enhance PP1C binding as well as increased MAPK activity, while V64 and F66 mutants displayed impaired binding and MAPK signaling compared to SHOC2 WT control (Extended Data Fig 9c-h). Positive charge substitutions at proximal positions (S57, A59, and P62) also resulted in GOF phenotypes within the DMS screen (Extended Data Fig. 9i, Supplementary Table 2). These mutations are anticipated to similarly enhance polar interactions between the N-terminal region of SHOC2 and the acidic surface of PP1C proximal to the hydrophobic tunnel of the RVxF binding pocket. Furthermore, substitutions replacing the hydroxyl sidechain of S67, which proceeds the RVxF (63GVAF66) motif, with bulkier hydrophobic side chains (S67W/I/V/F), resulted in a GOF phenotype (Fig. 3a, Extended Data Fig. 9i, Supplementary Table 2), likely by enhancing the hydrophobic interaction with the RVxF binding pocket on PP1C.
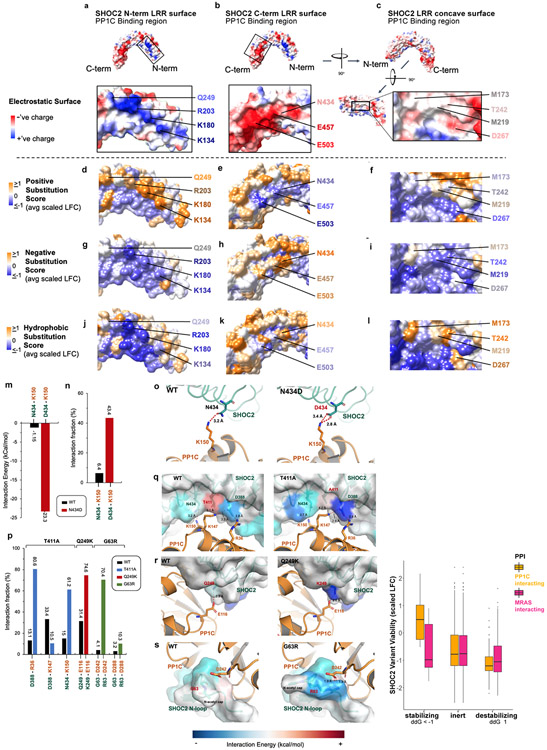
In addition, SHOC2-PP1C binding is also mediated through electrostatic interactions between PP1C and two key surfaces of within the LRR domains of SHOC2 (Extended Data Fig. 10a-b). The surface along the C-terminal LRRs of SHOC2 which binds PP1C is negatively charged and is functionally intolerant to positive charged amino acid substitutions, as exhibited by negative selection in the DMS screen (Extended Data Fig. 10e). Conversely, the basic surface within the N-terminal LRRs of SHOC2 that binds PP1C is intolerant to negative charged substitutions (Extended Data Fig. 10g). Furthermore, the substitution of uncharged SHOC2 residue N434, with negatively charged residues that match the overall local surface charge of the PP1C binding region of SHOC2 C-terminal LRRs exhibits a GOF phenotype in the DMS screen. Computational, structural analysis reveals N434D is predicted to enhanced ionic interactions resulting in a calculated interaction with K150 on PP1C, resulting in a calculated net-stabilization of < −20kCal/mol (Extended Data Fig. 10m-o). Indeed, SHOC2 N434D demonstrates enhanced binding to MRAS/PP1C in immunoprecipitation studies (Extended Data Fig. 9g).
SHOC2 M173I is clinically linked to a RASopathy phenotype in humans21. Indeed, M173I scores as a significant GOF in our screen and similar hydrophobic residue substitutions at M173V/L were also found to exhibit increased fitness above wildtype SHOC2 (Supplementary Table 2). These findings likely reflect further stabilization of the hydrophobic interaction between SHOC2 and MRAS surfaces as previously noted (Supplementary Table 1). Indeed, we corroborated these observations through detailed computational and structural analysis of the M173 position that revealed hydrophobic residue substitutions result in increased interprotein interaction energy as well as relatively lower intrinsic SHOC2 protein instability compared to other variants (Extended Data Fig. 3d-f).
As noted previously, steric hindrance at T411 has a destabilizing impact on the SHOC2-PP1C interaction interface (Fig. 2b, Supplementary Table 1). Mutations at T411 demonstrate strong positive selection in the DMS screen, suggesting GOF phenotypes conferred by these residues (Fig 2a, Supplementary Table 2). In the case of T411A, computational modeling suggests that the alanine mutation avoids the steric hindrance between the methyl group of WT SHOC2 T411 with the amine on K147 of PP1C (Extended Data Fig. 10p&q, Supplementary Table 1), and SHOC2 T411A allele was found to have enhanced complex member interactions and conferred an increase in MAPK activity (Extended Data Fig. 9c-h). Substitutions at T411 with positively charged residues were not enriched in the screen (Supplementary Table 2), likely due to enhanced electrostatic repulsion with K147 on PP1C. Collectively, integrative mapping of DMS and cryo-EM results suggest a high degree of structure-function concordance and define variants of SHOC2 that are critical for stabilizing or destabilizing SHOC2 interactions with complex members to mediate holophosphatase function (Extended Data Fig. 10t).
SHOC2-MRAS-PP1C complex in human disease
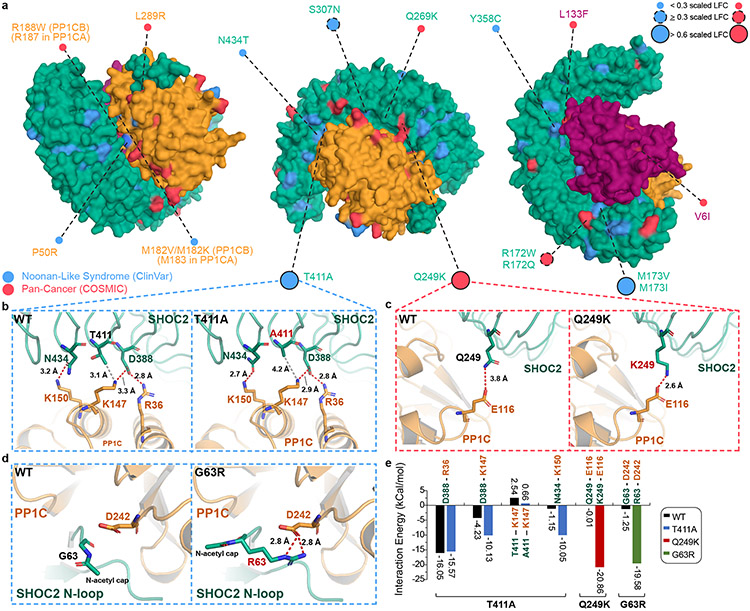
Germline SHOC2 gain-of-function mutations resulting in increased complex assembly or plasma membrane localization have been associated with Noonan-like Syndrome2,8. However, the functional consequences of only a small number of these mutations have been characterized. To investigate single-residue disease-associated mutations in SHOC2, we cross-referenced the DMS screen data with germline RASopathy mutations present in the ClinVar database and recurrent (>2 patients) mutations in human cancer using the COSMIC database (Supplementary Table 3). We specifically identified those mutations that reside within or proximal to protein-protein complex interfaces that also scored as GOF within our screen (scaled LFC >0.6) (Fig. 4a). Previously identified GOF RASopathy mutations include PP1C P50R and SHOC2 M173I/V2-5,21, located within interaction interfaces found in the cryo-EM structure (Fig 4a, Extended Data Fig. 8). Moreover, we identified previously uncharacterized variants in ClinVar for all three complex members that also reside in the protein interaction interfaces of the cryo-EM structure (Supplementary Table 3). Several additional SHOC2 RASopathy mutants score as GOF in the DMS screen (scaled LFC>0.6), including, G63R, T411A, Q249K, and Q269R (Fig. 4a-d). In addition to finding that T411A alleviates a solvation penalty and steric hindrance (Fig. 2b), this variant also enhanced adjacent contact sites between SHOC2 and PP1C (SHOC2-PP1C: D388-K147; N434-K150) through in silico modeling (Fig. 4e). In contrast, Q249K stabilizes the SHOC2-PP1C interface by creating a salt bridge with E116 of PP1C, further enhancing the binding energy by −22.67 kcal/mol (Fig. 4c&e, Extended Data Fig. 10p&r). We also identified and credentialed an uncharacterized RASopathy associated gain-of-function SHOC2 allele, G63R that completes a canonical RVxF motif, enabling tighter binding of the RVxF binding pocket on PP1C (Fig. 4d&e). Molecular modeling suggested that G63R further enhances this interaction by creating two additional hydrogen-bonds involving D242 on PP1C, releasing an additional interaction energy of 18.88 kcal/mol. These findings are in line with previous reports indicating a level of degeneracy in RVxF motifs where the conservation of VxF can be sufficient for PP1C binding20. In cells, all three novel SHOC2 pathogenic variants G63R, Q259K, and T411A demonstrated enhanced interactions with PP1C and MRAS as well as enhanced MAPK signaling (Extended Data Fig. 9) Thus, the integrated SMP holophosphatase structure and DMS screen data provide a valuable resource for future interpretation of pathogenic mutations in SHOC2, PP1C and MRAS that are observed in human RASopathy syndromes and cancer.
Figure 4: Structure-function analysis identifies disease-associated mutations.
a. Clinical missense mutations of SHOC2 complex members in Noonan-like Syndrome (NL-S) (ClinVar) and cancer (COSMIC database) with interface mutant alleles annotated. Lollipop size of interface mutants is proportional to DMS scaled LFC. b-c. Dynamic change in interaction surface between SHOC2 and PP1C in WT and novel NL-S (SHOC2 T411A) (b) or cancer associated mutations (SHOC2 Q249K) (c). d. Modeling of anticipated GOF G63R SHOC2 mutant. e. Contact surface energy of SHOC2 complex for novel functional pathogenic variants SHOC2 T411A, Q249K, and G63R, as predicted by Amber10 force field-based energy calculation.
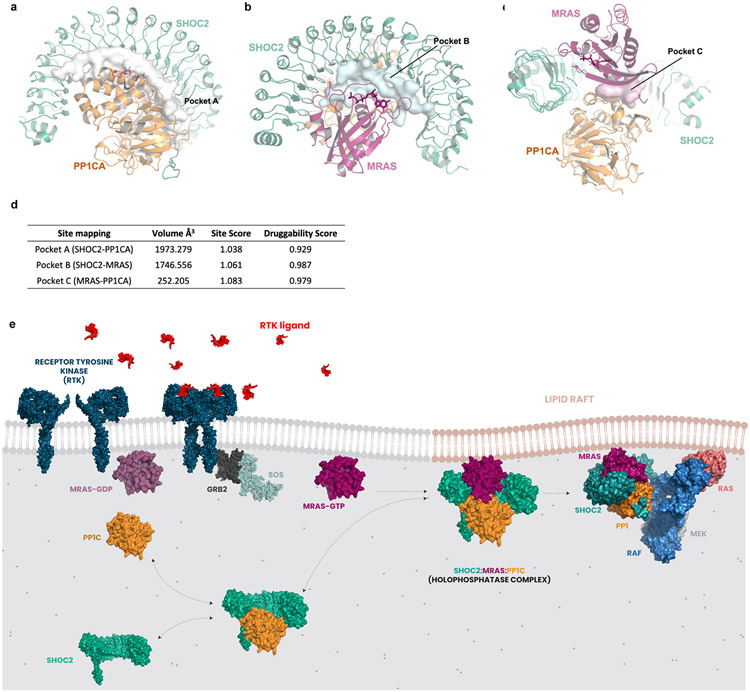
The SHOC2 ternary complex has a clear functional importance in RASopathies and RAS-driven cancers and remains an appealing therapeutic target. Using engineered degron systems, we have previously demonstrated proof-of-concept for small-molecule-mediated proteasomal degradation of SHOC211; however, small molecule ligands that bind to SHOC2 or the holophosphatase complex have not yet been reported. Enabled by insights from the cryo-EM structure and recent evidence demonstrating feasibility in developing selective inhibitors of PP1C holoenzymes27, we utilized the SiteMap algorithm (Methods) and identified three distinct pockets within the complex with favorable properties for binding of small molecule ligands (druggability scores >0.92) (Extended Data Fig. 11a-d). Notably, each pocket resides proximal to a critical stabilizing interface between each complex member interaction surface. At a combined volume of 3,961A3, these pockets present an attractive path toward discovery of small molecule ligands that disrupt holoenzyme formation or function.
Discussion
We have resolved the cryo-EM structure of the SMP holophosphatase demonstrating a ternary complex with PP1C and MRAS. Through integrative analysis of the SHOC2 DMS dataset, we developed a structure-function map of this complex, resolving the key functional contributions of residues within the N-terminal unstructured region and concave LRR surface to mediate complex stability and function. In addition, we provide a biophysical model suggesting that SHOC2 and PP1C exist in a relatively high affinity equilibrium between bound and unbound states (Extended Data Fig. 11e). Upon activation through GTP-loading, MRAS adopts an activated conformation and completes assembly of the SMP holophosphatase complex at the plasma membrane. We predict that sustained localization of the SHOC2 complex at the membrane mediates S259 RAF dephosphorylation in a RAS-GTP dependent manner. Moreover, the RAS-SHOC2-PP1C complex likely also localizes PP1C to lipid domains, where RAS has been known to cluster28, increasing local membrane concentration of PP1C and RAF1 substrate. Our finding that numerous pathogenic GOF variants of SHOC2 exhibit enhanced binding affinities for complex members further supports this model. Further studies are necessary to test whether SHOC2/RAS-GTP-dependent localization of PP1C at discrete lipid microdomains is necessary for its activity to dephosphorylate RAF or if the holophosphatase structurally confers substrate specificity. Furthermore, we provide structural, computational, and cell-based evidence that the canonical RAS isoforms (K/H/NRAS) can also mediate complex assembly, although with decreased stability compared with MRAS-mediated complex formation. However, our experimental evidence for complex member interactions was achieved using exogenously expressed, tagged-forms of SHOC2 and RAS proteins, and demonstration of endogenous interactions among these proteins will require further study.
These studies reveal disease mechanisms mediated by mutations in the SHOC2 ternary complex in congenital RASopathy syndromes and in cancer. We found that these mutations resulted in complex stabilization and enhanced interaction energy of SHOC2 with PP1C and/or MRAS. Collectively, this systematic structure-function map of SHOC2 provides a valuable resource for future interpretation of the functional significance of additional germline or tumor-associated mutations of SHOC2, as well as for inference of the functional implications for observed mutations in PP1C or MRAS. Moreover, SHOC2 is an essential mediator of RAS pathway signaling and RAS-driven cancer cells depend on SHOC2 for proliferation and survival, particularly in combination with MEK inhibition. The structural model of the SMP holophosphatase identifies key interaction interfaces that promote complex formation and function, and disruption of the SHOC2 complex represents an attractive therapeutic strategy to inhibit RAF kinase activation.
Finally, several recent studies have shed light on the autoinhibited structural state of RAF kinases when 14-3-3 is bound at two critical phosphorylation sites on RAF (S259 and S621)29,30, leading to a biophysical model of RAF activation through RBD/CRD engagement with RAS and phospholipids at the plasma membrane resulting in release RAF from 14-3-331. However, the structural basis by which the SMP holophosphatase functions within this stepwise activation model is yet to be determined. Our structural data does not support a SHOC2 ‘hinge’ within the medial LRRs; however, if such flexibility were to exist, it could serve to modulate access of RAF substrate to the PP1C hydrophobic grove and active site, which we expect are key to holoenzyme catalysis. In addition, our DMS screen revealed several mutationally sensitive SHOC2 surfaces, all which are interpreted as important for complex member binding; the lack of additional broad surfaces which are mutationally intolerant suggests that SHOC2 may not involved in direct binding or recruitment of RAF as a substrate. Further clarification of how the SHOC2 complex engages its sequestered RAF substrate will further inform and enable rational drug design.
Taken together, these studies provide a roadmap for characterizing disease-associated mutations in the SMP holophosphatase and yield insights that may unveil new avenues to therapeutically target this complex.
Methods
SHOC2 Production.
FL SHOC2 was cloned into pFastBac (ThermoFisher) along with sequence to produce an N’-terminally tagged 6XHis-GST-TEV SHOC2 protein. The protein was expressed in SF9 cells (Expression Systems) that had been transfected with baculovirus made from the relevant bacmid and then harvested by centrifugation after 72 hrs. Cell pellet was resuspended in 50 mM HEPES, pH 7.5, 500 mM NaCl, 10% glycerol, 0.5 mM TCEP containing protease inhibitors and lysozyme. The cells were lysed by microfluidizer and cell debris was removed by high speed centrifugation followed by filtration. The clarified lysate was supplemented with 20 mM Imidazole and passed over a HisTrap column (Cytiva), washed with 50 mM HEPES, pH 7.5, 500 mM NaCl, 10% glycerol, 0.5 mM TCEP and eluted over a 20 mM - 500 mM Imidazole gradient. The appropriate fractions were pooled and the 6XHis-GST tag was removed by TEV cleavage. The untagged protein was passed over an S200 size exclusion column (GE LifeSciences) that had been equilibrated in the wash buffer to further isolate purified protein. The pooled protein was concentrated by exploiting the slight affinity of the untagged SHOC2 to a HisTrap column. The protein was loaded onto the HisTrap column, the column was washed and protein was eluted in a single step 50 mM imidazole elution. The final protein was dialyzed into storage buffer of 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5 mM TCEP and stored at −80 C.
PP1C Production.
Full length PP1C (alpha isoform) was cloned into pET21b (ThermoFisher) along with sequence to generate an N’-terminally tagged 6XHis-SUMO-TEV PP1C protein. The construct was transformed into BL21DE3 cells (ThermoFisher) and cultured in TB media supplemented with 1 mM MnCl2. Protein expression was IPTG induced at an optical density of 0.8 (OD600) and incubated overnight at 18 C and the cells were harvested by ultracentrifugation. The cell pellet was resuspended for lysis in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.5 mM MnCl2, 10% glycerol, 0.5 mM TCEP supplemented with protease inhibitors and lysozyme. The cells were lysed by microfluidizer and the lysate was clarified by high speed centrifugation followed by filtration. The clarified lysate was supplemented with 20 mM Imidazole and passed over a HisTrap column (Cytiva), washed with 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.5 mM MnCl2, 10% glycerol, 0.5 mM TCEP and then eluted over a 20 mM - 500 mM Imidazole gradient. The appropriate fractions were pooled and the 6XHis-SUMO tag was removed by TEV cleavage. The protein was passed over the HisTrap column to separate the tag and collected from the fractionated flow through. The protein was pooled and concentrated using spin filters with a 10,000 MWCO to decrease the protein volume for the final SEC step. The protein was passed over an S75 column (GE LifeSciences) that was equilibrated with the final PP1C storage buffer 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.5 mM MnCl2, 10% glycerol, 0.5 mM TCEP. The purified protein was stored at −80 C.
MRAS Q71L Production.
MRAS Q71L (1-182) was cloned into pET21b (ThermoFisher) along with sequence to generate an N’-terminally tagged 6XHis-TEV MRAS protein. The construct was transformed into BL21DE3 cells (ThermoFisher) and cultured in TB media. Protein expression was IPTG induced at an optical density of 0.8 (OD600) and incubated overnight at 18 C and the cells were harvested by ultracentrifugation. The cell pellet was resuspended for lysis in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 0.5 mM TCEP, protease inhibitors and lysozyme and then lysed by microfluidizer. The lysate was clarified by high speed centrifugation followed by filtration. The clarified lysate was supplemented with 20 mM Imidazole and passed over a HisTrap column (Cytiva), washed with 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.5 mM TCEP and then eluted over a 20 mM - 500 mM Imidazole gradient. The appropriate fractions were pooled, treated with TEV and dialyzed at 4 C overnight into nucleotide exchange buffer, 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 0.5 mM TCEP. The dialyzed protein was treated with 1mM EDTA, 20 units of Alkaline Phosphatase (Sigma, P0114) per mg of protein, 4X molar excess of GppCp (Sigma, M3509) and incubated at room temperature for 2 hrs. Nucleotide exchange progress was monitored by UPLC. Post-exchange, 5 mM MgCl2 was added back to the protein and additional dialysis was performed to remove the EDTA. To remove the tags, the newly exchanged protein was passed over a His Trap column that was equilibrated in the wash buffer and collected from the fractionated flow through. The protein elutes in the flow through in two separate peaks, the first of which contains more contaminants and more aggregated protein than the second peak. The fractions for the second peak were pooled and concentrated to less than 12 ml volume. The protein was passed over an S75 (GE LifeSciences) sizing column that had been equilibrated with the final storage buffer, 25 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 10% glycerol and 0.5 mM TCEP. The pooled and purified protein was stored at −80 C.
Crystallization of apo-SHOC2, data collection, and structure determination.
All the crystallization trials were performed by hanging drop vapor diffusion method. Pure apo SHOC2 protein (truncated at position 88) was exchanged into the same final buffer but without glycerol and the concentration was adjusted to 6 mg/mL. Drops were set up by mixing the protein solution with crystallization solution at 2 μL : 2 μL and 2 μL : 3 μL drop ratios and let to equilibrate against 500 μL of well solution at 4°C. Large 3D crystals grew within 1-2 weeks from solutions comprising 350-600 mM MgNO3, 200 mM Tris pH 8.5 and 23-30% PEG 4000. Crystals were directly flash frozen in liquid nitrogen. X-ray data were collected at Brookhaven National Lab (NSLSII AMX). Data were indexed and scaled using iMosflm32. Structure was solved by molecular replacement using Phaser33 and a homology model. The program Coot34 and Phenix35 were used for structure refinement. Data collection and refinement statistics are reported in Extended Data Table 1.
Complex Formation.
The holoenzyme was formed by mixing individually purified proteins in a 1:2:2 stoichiometry of SHOC2:PP1C:MRAS Q71L and incubated overnight at 4 C. The formed complex was isolated by passing over an S200 (GE Lifesciences) that had been equilibrated with 50 mM HEPES, pH 7.4, 150 mM NaCl, 10% glycerol, 0.5 mM TCEP.
Cryo-EM sample preparation and data acquisition.
The complex was diluted to 2.75 mg/mL and supplemented with 0.025% (w/v) fluorinated octyl maltoside immediately prior to being applied to cryo-EM grids. Quantifoil 1.2/1.3 300 Au mesh grids (Quantifoil Micro Tools GmbH) were glow discharged for 30 s using a Gatan Solarus plasma cleaner (Gatan, Inc.) operating at 20W and using ambient air. Grid freezing was performed using a Vitrobot Mk IV (ThermoFisher, Inc.) with the blotting chamber held at 100% humidity and 18 °C. A 3.5 μL droplet of sample was applied to the grid, blotted for 5 s, and then plunged into liquid ethane. Data were collected at The University of Chicago Advanced Electron Microscopy Core Facility using a Titan Krios G3i electron microscope (ThermoFisher, Inc.) equipped with a BioQuantum K3 camera and energy filter. The camera was operated in CDS mode, and exposure movie data were recorded in super-resolution mode. A total of 4721 movies were collected. Data acquisition parameters are given in Extended Data Table.
Cryo-EM data processing.
All data processing steps were performed using Relion 4.0beta236 unless otherwise noted. Micrograph movies were summed and dose-weighted, and contrast transfer function (CTF) parameters were estimated using CTFFind 4.1.1437 on movie frame-averaged power spectra covering ~ 4 e-/Å2 dose. Micrographs showing extreme high outliers in corrected motion, poor power spectra Thon rings or CTF estimation fits, or large non-vitreous ice regions were removed, resulting in 4499 micrographs used for downstream data processing. A random subset of 1000 micrographs were processed first. Particles from this subset were picked using the ab initio particle picker from the CisTEM 1.0.0beta software package38 and filtered over multiple rounds of 2D classification. The resulting 85,761 particles were sufficient to generate an ab initio 3D model, and a single round of 3D classification was performed to further improve the particle stack. The resulting 40,069 particles were used to train a Topaz39 particle picking model. This model was then used to pick particles from the entire micrograph set, using a log-likelihood score of −3 as a cutoff. 644,140 particles remained after 2D and 3D classification, which were then used for iterative rounds of 3D refinement and CTF parameter (per-particle defocus, per-micrograph astigmatism, and per-image shift position beam-tilt, trefoil, and 4th order aberrations) refinement until no further improvement in resolution was observed. 3D classification with fixed particle poses was then used to remove remaining outlier particles. The resulting 450,317 particles were then subject to per-particle motion correction (Bayesian polishing), then further iterative 3D and CTF refinement until resolution improvements ceased. A focused 3D refinement using a mask excluding the distal/C-terminal tail of SHOC2 was used as the basis of a fixed-pose 3D classification to remove any remaining particles that were outliers specifically at the MRAS-PP1C-SHOC2 interface, but this resulted in the removal of only 933 particles. The remaining 449,384 particles were subject to a second round of Bayesian polishing and a final round of CTF refinement (additionally estimating CTF B-factors) before the final 3D refinement that was used as the basis for model building.
Cryo-EM model building.
The crystal structure of SHOC2 reported here and the publicly available crystal structures of MRAS and PP1C (PDB IDs 3E7A40 and 3KKO41, respectively) were rigid-body fit into the map using Chimera42 and used as the basis for model building. The model was refined by iterating between automated real-space refinement using Phenix35 and manual editing using Coot34. The map used for both manual and automate refinement was the result of sharpening by an automatically determined B-factor followed by filtering to local resolution using Relion 4.0beta2. Model geometry and map-model agreement statistics were calculated using Phenix and are given in Supplementary Table 2. The EMRinger43 score was calculated using the unsharpened map.
Bio-layer Interferometry (BLI).
All bio-layer interferometry was performed on a ForteBio Octet Red-384 system using SA sensors. C-terminally Avi-tagged full length SHOC2 that was biotinylated in vitro was loaded onto sensors for all experiments. All experiments were performed at 30 °C at a sensor height of 4 mm and an acquisition rate of 5 Hz in the following buffer: 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5 mM TCEP, 0.05% TWEEN-20. All experiments started with 60 s of sensor equilibration, followed by loading test sensors to 3 nm. Loaded sensors were then washed with buffer for 60 s. PP1c binding was evaluated at a maximum concentration of 10 μM, diluted 2x back (as limited by PP1c solubility) for 20 s. Formed complexes were then allowed to dissociate in buffer for 100 s. For MRAS binding, 3 nm of loaded SHOC2 was saturated with 10 μM of PP1c until signal equilibrium was reached (20 s). Sensors were then immediately dipped into wells containing up to 10 μM of MRAS and allowed to bind for 200 s, and dissociate for 1800 s until dissociation was complete or deemed too slow to continue. Data were then fitted to single-exponential models to obtain ka and kd (when applicable), as well as fittin response vs. concentration to fit for KD.
Sedimentation velocity – analytical ultracentrifugation (SV-AUC).
All SV-AUC was run using A280 migration as detection in a 2-slit cell. All proteins were sedimented in isolation and in all possible combinations across individual cells at the following concentrations: SHOC2, 7 μM, PP1c, 10 μM, MRAS, 20 μM. All spins were performed using a Beckman Optima XLA Ultracentrifuge 8-hole rotor at 20 C, 50k RPM. Data were then fitted in the program SEDFIT, fitting all scans until sedimentation was complete in c(s) mode from 0-15 Svedburgs, and identified peaks provided final S and MW values to attribute an identity to each sedimenting species, as well as relative percent of abundance.
Modeling and interface energy calculations.
SHOC2 complex Cyro-EM structure was prepared before modeling and simulations. The module of Protein Preparation in Schrödinger Maestro44-46 was applied to cap termini and repair residues. The minimization and optimization to the protein system was performed under Amber10:EHT force field (https://infoscience.epfl.ch/record/121435/files/Amber10i.pdf) to RMS gradient of the potential energy falls below 0.1 kcal/mol/Å using Molecular Operating Environment (MOE), (2019.01; Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2021). Default tether restraints from MOE were applied to the system. Protein models after in silico mutations underwent the same preparation procedure. The interface energy calculation between contacting residue pairs was processed using the module of Protein Contacts in MOE. Six types of contacts were identified: Hydrogen bonds (Hbond), Metal, Ionic, Arene, Covalent, and Van der Waals distance interactions (Distance). Proximity threshold was set to 4.5 Å. Atoms separated by greater than this distance were not considered to be interacting. Energy threshold was set to −0.5 kcal/mol for H-bond, H-pi, and Ionic bond.
MD simulations.
Schrödinger Desmond MD47 engine was used for simulations. An orthorhombic water box was applied to bury the prepared protein system with a minimum distance of 10 Å to the edges from the protein. Water molecules were described using the SPC model. Na+ ions were placed to neutralize the total net charge. The prepared system for simulation contained ~95k atoms. All simulations were performed following the OPLS4 force field48. The ensemble class of NPT was selected with simulation temperature set to 300K (Nose-Hoover chain) and pressure set to 1.01325 bar (Martyna-Tbias-Klein). A set of default minimization steps pre-defined in the Desmond protocol were adopted to relax the MD system. The simulation time was set to 200ns with 2000 frames evenly recorded (1 frame per 200 ps) during the sampling phase. Post simulation analysis of RMSD on α-carbon atoms was performed using Schrödinger simulation interaction diagram. Python-based analysis script analyze_trajectory_ppi.py was used to monitor contacting residue pairs during the MD course.
SHOC2 expression vector and mutagenesis library development.
Lentiviral vector pMT_025 was developed by Broad Institute Genetic Perturbation Platform (GPP). Open Reading Frames (ORF) can be cloned in through restriction/ligation at a multiple cloning site (MCS). The ORF expression is driven by EF1a promoter. A PAC gene is driven by SV40 promotor to confer puromycin resistance. The library was designed with the software tool and principles as previously described49. The full-length SHOC2 gene was mutagenized. At each codon position, except the start and stop codons, 19 missense changes, and 1 nonsense change were made, but due to the constraint of avoiding the development of restriction enzyme sites used for cloning in the body of the gene, some intended codon changes were not possible (10 variant positions missing in the designed SHOC2 library). In addition, we incorporated 342 silent changes scattered along the region of interest. It is important to note that in SHOC2 library variant designs, efforts were made to minimize codons that differ from the corresponding template codon by 1 nucleotide49. In all, the library had 11,952 variants. The cloning protocol was performed as previously described49. The mutagenesis library was synthesized by Twist BioScience, and the library created as a pool of linear fragments representing the full-length SHOC2 ORF with a short flank sequence, ~35 bases, at each end. The two flank sequences were designed to facilitate restriction/ligation cloning of the linear fragment library into pMT_025 expression vector. The linear fragment library and the vector were each digested with NheI and BamH1, and then ligated with pMT_025 that had been opened with NheI and BamH1. The ligation products were then transformed into Stbl4 competent cells (New England BioLabs). In order to achieve good library representation, approximately 1000 colonies per variant were cloned (e.g. 12 million colonies the entire SHOC2 variant library). Ultimately ~20 million colonies were obtained for the SHOC2 library. The colonies were harvested and plasmid DNA was extracted via maxi preparation kit (Qiagen). The resulting plasmid DNA library was sequenced via Illumina Nextera XT platform. The distribution of variants was computationally assessed.
SHOC2 variant library lentivirus production.
Lentivirus was created by Broad Institute Genetic Perturbation Platform (GPP). The detailed protocol is available at http://www.broadinstitute.org/rnai/public/resources/protocols/. Briefly, viral packaging cells 293T was transfected with pDNA library, a packaging plasmid containing gag, pol and rev genes (e.g. psPAX2, Addgene), and VSV-G expressing envelop plasmid (e.g. pMD2.G, Addgene), using TransIT-LT1 transfection reagent (Mirus Bio). Media was changed 6-8 hours post-transfection. Virus was harvested 30 hours post-transfection.
SHOC2 DMS Viability Screen.
Screening-scale infections were performed with virus in the 12-well format and infected wells were pooled 24 hr post-centrifugation. Infections were performed with 3 replicates per treatment arm, and a representation of at least 1000 cells per SHOC2 variant was achieved following puromycin selection. Approximately 3 days after infection and selection, all cells within a replicate were pooled and split into Falcon™ Cell Culture Multi Flasks flasks and treated in media with 10 nM trametinib or DMSO control. Cells were passaged in fresh media containing drugs or vehnicle control (DMSO) every 3-4 days. Cells were harvested 16 days after initiation of treatment and gDNA extracted (Genomic DNA Extraction Kit, Machery-Nagel). Twelve PCR reactions were performed for each gDNA sample. The volume of each PCR reaction was 100 μl and contained ~3 μg of gDNA. Herculase II (Agilent) was used as the DNA polymerase. All 12 PCR reactions for each gDNA sample were pooled, concentrated with a PCR cleanup kit (QIAGEN), loaded onto a 1% agarose gel, and separated by gel electrophoresis.
SHOC2 DMS PCR amplification and deconvolution.
The general screen deconvolution strategy and considerations were described in detail in49. The integrated ORF in genomic DNA was amplified by PCR. The PCR products were shotgun sheared with transposon, index labeled, and sequenced with next-generation sequencing technology. The PCR primers were designed in such a way that there is a ~100 bp extra sequence at each end leading up to the mutated ORF region. We used 2 primers: (Forward: 5’-ATTCTCCTTGGAATTTGCCCTT-3’; Reverse: 5’-CATAGCGTAAAAGGAGCAACA-3’). PCR reactions were performed for each gDNA sample with a reaction volume of 50 uL and with 1 ug gDNA. Q5 (New England BioLabs) was used as DNA polymerase. 1/3 of 96 PCR reactions of a gDNA sample were pooled, concentrated with Qiagen PCR cleanup kit, and then purified by 1% agarose gel. The excised bands were purified first by Qiagen Qiaquick kits, then by AMPure XP kit (Beckman Coulter). Following Illumina Nextera XT protocol, for each sample, we set up 6 Nextera reactions, each with 1 ng of purified ORF DNA. Each reaction was indexed with unique i7/i5 index pairs. After the limited-cycle PCR step, the Nextera reactions were purified with AMPure XP kit. All samples were then pooled and sequenced with Illumina Novaseq S4 platform. NovaSeq600 S4 data were processed with software AnalyzeSaturationMutagenesis, ASMv1.0 for short, which was developed by Broad Institute as previously described49. Typically, the pair-end reads were aligned to the ORF reference sequence. Multiple filters were applied and some reads were trimmed. The counts of detected variants were then tallied. The output files from ASMv1.0, one for each screening sample, were then parsed, annotated merged into a single .csv file that is ready for hit-calling utilizing software tools that are freely available49.
DMS analysis.
Abundance of each variant was calculated by the fraction of reads compared to the total reads of all variants in each endpoint, and Log2 Fold Change (LFC) was determined between Day 16, 10nM trametinib treatment compared to Day 16, vehicle (DMSO) control. To better appreciate our variant activity relative to wildtype SHOC2, the DMS LFC was centered to the mean of distribution of wildtype SHOC2 (silent mutations) and normalized against the mean of SHOC2 non-sense variants. The threshold for gain- (GOF) or loss-of function (LOF) has been determined based on two standard deviations above and below the mean of the SHOC2 wildtype (silent mutant) distribution (GOF >0.6 scaled LFC; and LOF < −0.6 scaled LFC). Evolutionary conservation score (Evo Score) was determined by Aminode: evolutionarily constrained regions and protein-protein interacting residues (PPI) from cryo-EM data are indicated.
In silico saturation mutagenesis with FoldX.
The in silico saturation mutagenesis studies on SHOC2 and SHOC2 complex that evaluate the protein stability from the perspective of free energy change (ΔΔG) upon mutations were performed using the FoldX50. MutateX51 was used for automation. The overall process was to systematically mutate each available residue within a protein or a protein complex to all other possible residue types and to predict ΔΔGs utilizing the FoldX energy calculation. The RepairPDB function of FoldX was first applied for energy minimization to modify the protein system to reasonable conformations. The BuildModel function was followed for the computational mutagenesis and reporting ΔΔG values. For the application on SHOC2 complex, the AnalyseComplex function was continued to further evaluate ΔΔG of interaction between protein chains upon mutation.
Generation of Expression Constructs.
SHOC2 ORF (SHOC2 transcript NM007373.3) containing wobble mutants was utilized to allow for SHOC2 protein expression in the presence of SHOC2 sgRNAs (both the NGG PAM sequence and the first amino acid in the guide sequence were mutated). SHOC2 variants were created using Q5® Site-Directed Mutagenesis Kit (NEB E0554S) and pDONR221-SHOC2(WT-wobble mutant) as template and mutations were sanger sequence confirmed. Variants were subsequently cloned into pLX307 utilizing Gateway™ LR Clonase™ II Enzyme mix (#11791020) and expression confirmed in mamallian cells.
Cell lines, Culturing, and Generation of SHOC2 KO and stable cell lines.
Cells (293T, PA-TU-8902, MIA PaCa-2) were grown in the following media supplemented with 2 mM glutamine, 50 U/mL penicillin, 50 U/mL of streptomycin (GIBCO), and 10% fetal bovine serum (Sigma): DMEM. PA-TU-8902 and MIA PaCa-2 cells were infected with virus expressing SHOC2 sgRNA or non-cutting control (sgCH2-2) made with plentiCRISPR v2-Blast vector (Addgene: 83480). Following blasticidin selection (2ug/ml) for three days, cells were serially diluted and single cell clones were selected and SHOC2 protein levels determined via western blot.
Growth In Low Attachment Assays.
MIA PaCa-2 with endogenous SHOC2 KO and stably expressing restored SHOC2 WT or various variants were seeded into 96 well Ultra-Low Attachment plates (Corning; 3904) at 5,000 cells/well. 7 days following seeding, cell viability was determined by Cell-Titer-Glo (CTG) (Promega; G7570) utilizing EnVision Plate Reader (PerkinElmer).
Immunoblot Analysis.
Cells were lysed using RIPA buffer (R0278; Sigma-Aldrich), quantified using BCA Protein Assay Kit (23227; Thermo Scientific), resolved on 4%–12% Bis-Tris gel, and transferred onto nitrocellulose membrane (IB23001; Thermo Scientific) utilizing iBlot 2 Dry Blotting System (IB21001; Thermo Scientific). All immunoblots were incubated with indicated primary antibodies and imaged using Odyssey CLx infrared imager (LICOR). Densitometry analysis was conducted using Fiji image-analysis software (Schindelin et al., 2012).
Immunoprecipitation studies.
4-7x106 cells were seeded in 10cm dish, transfected with either co-transfected with FLAG-K/H/N/MRAS expressing vector (3ug) and Myc-SHOC2 (0.5ug) for RAS pulldown studies, or co-transfected with SHOC2-V5 expressing vector (3ug) and HA-MRAS (3ug) for SHOC2 variant pulldown studies. 24hrs post-tranfection reagent addition to cells, media was changed, and cells were harvested after an additional 24hours. For RAS pulldown studies, cells were lysed in 1mL TNT-M lysis buffer (with 1mM DTT, protease/phosphatase inhibitor (Sigma cocktail 2+3). Following lysate harvest, 30ul was saved for input and 7 uL packed FLAG M2 beads (Sigma) were added to remaining lysate. Lysates with beads rotated 40C for 2 hours. Beads were washed (TNT-M wash buffer [50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton-X-100, 5 mM MgCl2]), 3 times, and beads were boiled in 1.5x LDS. Input and and eluted IP samples were immunoblotted for FLAG (Sigma F7425 1:4000), PP1 alpha (Upstate 06-221), and Myc (Abcam ab9106 1:2000). For SHOC2 variant pulldown studies, cells were lysed in 1mL IP lysis buffer (40mM Tris, pH 7.4, 150mM NaCl, 1% Triton-X-100, 5mM MgCl2, 10mM B-PG, 10mM pyroPP, 40mM HEPES). Lysates were quantified via Pierce™ BCA Protein Assay Kit, and 3.3mg lysate were equilibrated to 1.1mL volume. 100ul was saved as input, and 50ul of Anti-V5-tag mAb-Magnetic Beads (MBL, M215-11) were added to lysate, and incubated at 4C overnight rotating. Next day, beads were washed 3x rotating at 4C. Beads were boiled in 2x laemmli buffer, and input/eluted IP samples were immunoblotted for V5 (CST 13202S), HA (CST 3724TS), and blotted for PP1CB (Thermo Scientific PA5-78117).
In Silico modeling of SHOC2 complex interaction with RAS-RAF dimeric multimer unit.
Model incorporated referenced structural model of a RAS-RAF signalosome22. The module of protein-protein docking in MOE was used for this modeling. One signalosome unit that includes 2x RAS, 2x RAF, 2x MEK, and 2x 14-3-3 was extracted from the above-mentioned Ras-Raf signalosome structural model. The dephosphorylation pocket of PP1C in holoenzyme cryo-EM structure was defined as a pocket for gridding, and a S259 on RAF was pin-pointed for protein-protein docking. 100 possible docking conformations were sampled. Outcomes were ranked based on their energy profiles (van der Waals, electrostatic, and solvation energies). Manual structural inspection prioritized conformations that can have the RAS in SHOC2 holoenzyme also face the membrane.
Extended Data
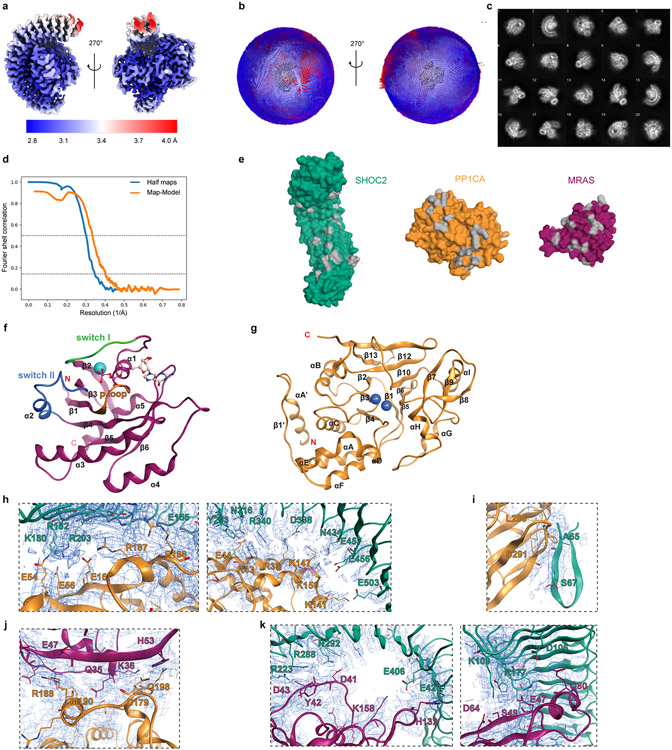
Extended Data Figure 1. Cryo-EM map of the SHOC2 holophosphatase complex, surface model of contact surfaces and secondary structure annotations.
a, The cryo-EM map used for modeling the complex, colored according to local resolution using the color map shown. The map was sharpened by an automatically determined B-factor of −90.7922 Å2 and filtered to local resolution, both determined by the methods implemented in Relion. b, A 3D histogram of the angular distribution in the final particle set as determined during the final 3D map refinement. Both the size and color of the bins correspond to particle counts. The views are the same as shown in (a), with the map itself rendered in the center of the histograms in grey. c, Reference-free 2D class averages generated from the final particle set. d, The fourier shell correlation (FSC) for the independently refined half maps from the final 3D map refinement (blue), and the full map and atomic model (orange). The “gold standard” half-maps FSC was calculated and corrected for masking effects using Relion; the map-model FSC was calculated by Phenix using a mask around the model based on the 2.9 Å global resolution. FSC=0.5 and 0.143 thresholds are marked by dashed lines. The half-maps FSC crosses the 0.143 threshold at 2.8925 Å resolution, and the map-model FSC crosses the 0.5 threshold at 3.01 Å resolution. e, Surface model of unbound SHOC2, PP1CA, and MRAS. Grey indicates the interacting surfaces. f, MRAS cartoon representation with secondary structured labeled. g, PP1CA cartoon representation with secondary structured labeled. h, SHOC2 LRR and PP1C interactions i, SHOC2 N-term region and PP1C j, PP1C and MRAS k, SHOC2 LRR and MRAS shown in local electron density map corresponding to protein-protein interaction sites in Figure 4. SHOC2 is shown in teal, PP1CA in yellow and MRAS in magenta. The map (2Fo-Fc) is at 4.5 sigma.
Extended Data Figure 2. 200ns MD simulation of the SHOC2 complex, cryo-EM maps of SHOC2 T411 and proximal interactions with PP1C, SHOC2 N-term region degenerate RVxF motif and PP1C RVxF binding pocket, and AUC analysis of PP1C pair-wise interactions with complex members.
a, An overview of the MD simulation system for the SHOC2 complex. b, Root-mean-square-deviation (RMSD) of the protein α-carbon throughout the simulation. c, Interaction fraction of contacting residue pairs between SHOC2 and PP1C. d, Interaction fraction of contacting residue pairs between SHOC2 and MRAS. e, Interaction fraction of contacting residue pairs between MRAS and PP1C. f, Local electron density map for T411 of SHOC2 (teal) and K147 of PP1CA (orange) and their neighboring residues (left) and SHOC2 N-terminal residues interacting with RVxF binding pocket of PP1c (right). The map (2Fo-Fc) is at 4.5 sigma. g, Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC) analysis of PP1C binding to SHOC2 or MRAS-GppCp compared to PP1C alone, and with the presence of SHOC2 and MRAS-GppCp.
Extended Data Figure 3. Surface view of SHOC2 M173 and in silico energy calculations of variant substitutions and correlation of intrinsic protein stability and interaction energies of SHOC2 M173 mutations with DMS functional scores.
Structural view of a, SHOC2 and b, MRAS with hydrophobicity (yellow) and polar (teal) surfaces colored, and red outlines indicate hydrophobic interaction surfaces. c, Box and whisker plot for calculated differences of SHOC2-MRAS interaction energy between wildtype M173 and models of variants. In silico mutagenesis modeling grouped based on their hydrophobicity and charge states: hydrophobic (n = 8 residue calculated energies: I, V, L, F, M, A, W, and P); polar uncharged group includes (n = 7 residue calculated energies: C, G, T, S, Y, N, and Q); polar negatively-charged (n = 2 residue calculated energies: D, and E); polar positively-charged (n = 3 residue calculated energies: H, K, and R). Center line represents median, whiskers represent the first and fourth quartiles, box edges represent the second and third quartiles. All other observed data points outside the boundary of the whiskers are plotted as outliers. Mean and outliers are shown in crosses and dots respectively. SHOC2 M173 variant fitness scores in the DMS screen are presented on the x-axes and d, calculated protein interaction energy between SHOC2 and complex members; e, impact on intrinsic protein stability; and f, combined multiple linear model (0.12*contact energy + 0.25*intrinsic protein stability − 0.75) are represented on the y-axes. Line (blue) represent linear regression model, 95% confidence interval of best fit line (dashed black lines), R2 (goodness of fit), and linear model p-value (analysis of regression coefficient significantly non-zero) indicated.
Extended Data Figure 4. The comparison of crystal SHOC2 and the SHOC2 in the Cryo-EM holoenzyme.
a, Primary sequence analysis of highly conserved SHOC2 LRRs with consensus sequence indicated above. The canonical highly conserved leucine-rich repeat motif is indicated above; boxes of hydrophobic residues based on structure data (teal); and disruption in core hydrophobic core residues within LRR11 and LRR12 are indicated (blue box). b, Measurements of dihedral angles between each two neighboring LRRs (red text) and the distance between alpha carbons of R104 at LRR1 and I545 on LRR20 for crystal SHOC2 (indicated by line), and c, the SHOC2 in the Cryo-EM holoenzyme. d, Per-residue fluctuation reflected from 200ns MD simulations for crystal SHOC2 and SHOC2-MRAS-PP1C holoenzyme.
Extended Data Figure 5. In Silico modeling of SHOC2 complex interaction with RAS-RAF dimeric multimer unit.
a, Rank order of 17 established models with preferred van der Waals, electrostatic, and solvation energies (natural log of negative S-score) were manually annotated for spatial accommodation of RAS members to be oriented/embedded within a plasma membrane (indicated red dots). The top energetically favorable model that accommodates RAS orientation within the membrane (model 6) was selected. b, Structural model of SHOC2 complex interacting with dimeric multimer unit (2x RAS, 2x RAF, 2x MEK, and 2x 14-3-3) is presented with c & d, additional rotational views of the docked complex. Individual protein units are colored and labeled.
Extended Data Figure 6. In silico modeling and energy calculation for SHOC2 M173I, PP1C P50R, and MRAS Q71L mutations and evaluation of PP1C isoforms in MRAS Complex Association.
a, Zoom-in views for SHOC2 M173 and modeled M173I mutation with distance measurement to contacting residues on MRAS. b, Zoom-in views for PP1C P50 and modeled P50R mutation with distance measurement to contacting residues on SHOC2. c, Zoom-in views for MRAS Q71 and modeled Q71L mutation with distance measurement to surrounding residues. d, Predicted interaction energy for the WT and mutated residues. e, Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC) analysis of SHOC2 holoenzyme with PP1C isoforms (PP1Cα/β/γ) formation in the presence of MRAS-GppCp. Line trace of 1 technical replicate, representative of 3 biological replicates.
Extended Data Figure 7. Evaluation of RAS isoforms in context of SHOC2 holophosphatase complex formation.
a, Multiple sequence alignment analysis (EMBL-EBI ClustalW) of RAS isoforms (MRAS, KRAS, HRAS, NRAS). Switch I (red), Switch II (blue), and P-loop (orange) are annotated. MRAS residues that interact with PP1C (cyan highlighted) and SHOC2 (yellow highlighted) are boxed if they contribute ≤ −1.5kcal/mol of calculated paired interaction. b, Mean interaction energy calculated through molecular dynamic simulation of RAS isoforms (n = average of 5 representative frames/RAS isoform) and error bars represent standard deviation of the mean. c, BLI experimentation of MRAS and KRAS complex with activated SHOC2-PP1C. Mean (n= 3 technical replicates, representative of 3 independent experiments) for binding constants (Ka, kd, KD kin) and error bars (standard deviation) are presented. d, Immunoprecipitation of various exogenously expressed oncogenic RAS isoforms from 293T cells co-transfected with FLAG-tagged RAS and Myc-tagged SHOC2 (representative of 3 biological replicates).
Extended Data Figure 8. High resolution heatmap of SHOC2 DMS screen.
High resolution heat map representation of log2-fold change (LFC) allele enrichment and depletion between trametinib treatment and vehicle control, centered on mean of wildtype (silent mutants) and scaled to mean of nonsense mutants (scaled LFC), providing relative enrichment (red) vs depletion (blue) relative to SHOC2 WT. SHOC2 positional evolutionary sequence variation (Evo Score; higher value = less conserved) and protein-protein interacting residues (PPI) from cryo-EM data are indicated (Methods). An additional heatmap is provided below which depicts the average scaled LFC score of residues that have been grouped according to biophysical characteristics (orange = GOF; purple = LOF), including negative-charge (D/E), positive-charge (K/R), and hydrophobic (G/A/V/L/I/M), polar uncharged (S/T/C/Y/N/Q), non-polar large aromatic (F/W/Y/H) and helix breaker (P/G).
Extended Data Figure 9. Analysis of mutational tolerance of SHOC2 residues based on deep mutational scanning and residue contact points within SHOC2 complex, Impact of SHOC2 variants on growth in low attachment, and Impact of SHOC2 variants on MAPK signaling, and Impact of SHOC2 variants on MAPK signaling in response to MEK inhibition.
a, Violin plot of SHOC2 mean positional viability for surface contacting residues between complex members, PP1C (green, n = 28 positions) and MRAS (maroon, n = 26 positions), compared to core-residues (brown, n = 198 positions) and surface non-contacting residues (yellow, n = 329). Center line represents median, whiskers represent the first and fourth quartiles, box edges represent the second and third quartiles. b, MIA PaCa-2 with knock-out of endogenous SHOC2 and stably re-expressing various SHOC2 gain-of-function (red) and loss-of-function (blue) variants were seeded in ultra-low attachment plates and cultured for 7 days. Viability endpoint via cell titer glow is presented on x-axis along scaled LFC from fitness screen with PaTu-8902. Error bars represent standard deviation of GILA CTG viability (n=6 technical replicates; representative of 3 biological replicates). Line (green) represent simple linear regression model, 95% confidence interval (black dashed lines), R2 (goodness of fit), and linear model p-value < 0.0001 (analysis of regression coefficient significantly non-zero) indicated. c, Wild type (WT) and gain- or loss-of-function (GOF/LOF) variants were stably expressed in KRAS mutant cell line MIA PaCa-2 with knock-out of endogenous SHOC2. d, Densitometry quantification of P-S259 RAF1 relative to total RAF1 and normalized to levels from wildtype expressing cells. Center line represents median and whiskers represent interquartile range. ***p<0.001, two-sided t-test between LOF (n = 5 variants) and GOF (n = 6 variants) SHOC2 alleles, representative of 3 biological replicates. e, Wild type (WT) and gain- or loss-of-function (GOF/LOF) variants were stably expressed in KRAS mutant cell line MIA PaCa-2 with knock-out of endogenous SHOC2. Cells were treated with the MEK1/2 inhibitor trametinib (10nM) for 24 hours prior to Western blot. f, Densitometry quantification of P-S259 RAF1 relative to total RAF1 and normalized to levels from wildtype expressing cells. Center line represents median and whiskers represent interquartile range. ***p<0.001, two-sided t-test between LOF (n = 6 variants) and GOF (n = 6 variants) SHOC2 alleles, representative of 3 biological replicates. g, Immunoprecipitation of V5-tagged SHOC2 variants in 293T cells co-transfected with HA-MRAS. h, Densitometry analysis of relative prey including endogenous PP1CB (yellow) and MRAS (maroon) normalized to V5 bait (y-axis) and DMS fitness score (LFC Z-score) (x-axis). Lines represent simple linear regression model, R2 (goodness of fit), and linear model p-value < 0.0001 (analysis of regression coefficient significantly non-zero) indicated, representatitve of 3 biological replicates. i, Deep mutational scanning results for N-terminal region of SHOC2 (residues 60-68) depicted via sequence logo plot per amino acid substitution at respective positions (ggseqlogo).
Extended Data Figure 10. Functional consequence of mutations in SHOC2 LRR surface based on biophysical attributes of amino acid substitutions, In silico mutagenesis study of N434D, 200ns MD simulations for SHOC2 T411A, Q249K, and G63R mutations.
Three major regions of SHOC2 LRR that mediated complex member binding: (1) C-term PP1C binding region - left; (2) N-term PP1C binding region - middle; (3) Concave MRAS binding surface - right are presented in columns. Electrostatic surface depiction of SHOC2 (red = negative; blue = positive) for a & b, SHOC2 LRR region surfaces that bind PP1C and c, MRAS are presented (1st row), along with select protein-protein interacting residues of SHOC2 labeled. Subsequently, the SHOC2 Deep Mutational Scanning (DMS) screen functional score (Scaled LFC) was averaged for every surface residue of SHOC2 based on the biophysical characteristics of substituted residues at each given surface position and projected onto the SHOC2 surface with colorimetric scale (orange = GOF; purple = LOF). The average functional impact (mean scaled LFC) of positively charged residues (K/R) are presented (2nd row) for d, C-term PP1C binding region - left; e, N-term PP1C binding region - middle; f, Concave MRAS binding surface – right. The average functional impact of negatively charged residues (D/E) are presented (3rd row) for g, C-term PP1C binding region - left; h, N-term PP1C binding region - middle; i, Concave MRAS binding surface – right. The average functional impact of hydrophobic residues - non-polar, non-aromatic (G/A/V/L/I/M) are presented (4th row) for j, C-term PP1C binding region - left; k, N-term PP1C binding region - middle; l, Concave MRAS binding surface. m, Predicted interaction energy towards the K150 on PP1C for the SHOC2 WT and N434D mutation. n, Interaction fraction of contacting residue pairs for WT and the N434D mutation during the 200ns MD simulation. Zoom-in views for SHOC2 N434 o, and modeled N434D mutation with distance measurement to PP1C K150. p, Interaction fraction of contacting residue pairs for WT and mutations. r, Zoom-in views for SHOC2 T411 and modeled T411A mutation with distance measurement to contacting residues on PP1C. s, Zoom-in views for SHOC2 Q249 and modeled Q249K mutation with distance measurement to contacting residues on PP1C. t, Zoom-in views for modeled SHOC2 G63 and modeled G63R mutation with distance measurement to contacting residues on PP1C. The calculated interaction energy is colored to the SHOC2 protein surface for visualization. u, Boxplot of SHOC2 variants with mutations at protein interaction sites that are stabilizing (ddG ≤ −1), inert (ddG: >−1 and <1), destabilizing (ddG ≥ 1) by FoldX computations. Center line represents median, whiskers represent the first and fifth quartiles, box edges represent the second and fourth quartiles of SHOC2 variants with mutations in residues interacting with PP1C that are stabilizing (n = 18 variants), inert (n = 912 variants), destabilizing (n = 134 variants) or interacting with MRAS that are stabilizing (n = 13 variants), inert (n = 779 variants), and destabilizing (n = 196 variants) that were functionally evalulated in the DMS screen.
Extended Data Figure 11. Druggability analysis of SHOC2 holophosphatase complex and schematic diagram of proposed model for SHOC2 holophosphatase complex assembly.
a, SiteMap analysis of SHOC2 complex identifying druggable binding pockets between SHOC2-PP1C, b, SHOC2-MRAS and c, PP1C-MRAS. d, SiteScore is capped at 1.0 to limit the impact of hydrophilicity in charged and highly polar sites. A SiteScore of 0.80 has been found to accurately distinguish between drug-binding and non-drug-binding sites. For Dscore, the hydrophilic score is not capped. This one of the keys for distinguishing “difficult” and “undruggable” targets from “druggable” ones. e, Hypothesized model of the SHOC2 holophosphatase complex. MRAS is GDP bound and PP1C and SHOC2 exist in bound/unbound equilibrium in cytoplasm. Upon RTK stimulation and MRAS-GTP activation, the SHOC2-PP1C complex binds with MRAS at the plasma membrane to produce stable complex formation, and likely localizes the SHOC2 holophosphatase to lipid domains with concentrated RAS-bound RAF1 to dephosphorylate ‘S259’ on RAF and enable MAPK signaling.
Extended Data Table 1. X-ray crystallography data collection and refinement statistics.
Metrics from X-ray crystal structure data collection and processing are indicated, including Fourier shell correlation (FSC). *Values in parentheses are for the highest-resolution shell.
| APO-SHOC2 (PDB ID: 7T7A) |
|
|---|---|
| Data collection | |
| Space group | C 121 |
| Cell dimensions | |
| a, b, c (Å) | 91.93, 103.03, 121.09 |
| α, β, γ (°) | 90.00, 101.92, 90.00 |
| Resolution (Å) | 48.5-1.79(1.85-1.79)* |
| Rsym or Rmerge | 0.091(0.46) |
| I / σI | 11.73(1.3) |
| Completeness (%) | 96.82(94.26) |
| Redundancy | 4.4 (4.5) |
| Refinement | |
| Resolution (Å) | 48.5*1.79 |
| No. reflections | 99833(9677) |
| Rwork / Rfree | 0.20(0.30)/0.22(0.32) |
| No. atoms | |
| Protein | 7748 |
| Ligand/ion | 5 |
| Water | 550 |
| B-factors | 40.57 |
| Protein | 40.41 |
| Ligand/ion | 56.69 |
| Water | 42.70 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.01 |
Extended Data Table 2. Cryo-EM data collection, refinement, and validation statistics.
Metrics from cryo-EM structure data collection and processing are indicated, including Fourier shell correlation (FSC).
| SHOC2-PPIC-MRAS complex (EMDB-26667) (PDB 7UPI) |
|
|---|---|
| Data collection and processing | |
| Magnification | 81,000x |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 60 |
| Defocus range (μm) | −2.84 to −0.29 |
| Pixel size (Å) | 1.068 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 1,388,538 |
| Final particle images (no.) | 449,384 |
| Map resolution (Å) | 2.8925 |
| FSC threshold | @ FSC = 0.143 |
| Map resolution range (Å) | 2.7967 to 4.8389 |
| Refinement | |
| Initial model used (PDB code) | 3KKO, 3E7A |
| Model resolution (Å) | 2.98 |
| FSC threshold | @ FSC = 0.5 |
| Map sharpening B factor (Å2) | −90.7922 |
| Model composition | |
| Non-hydrogen atoms | 7778 |
| Protein residues | 972 |
| Ligands | GTP: 1 |
| Mg: 1 | |
| Mn: 2 | |
| C1: 1 | |
| B factors (Å2) (min/max/mean) | |
| Protein | 18.03/167.15/66.13 |
| Ligand | 22.16/82.04/42.97 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.435 |
| Validation | |
| MolProbity score | 1.04 |
| Clashscore | 2.56 |
| Poor rotamers (%) | 0.00 (0) |
| Ramachandran plot | |
| Favored (%) | 98.13 |
| Allowed (%) | 1.87 |
| Disallowed (%) | 0.00 |
Extended Data Table 3. Bio-layer interferometry analysis of SHOC2 ternary complex.
Binding kinetic values for step-specific complex member binding (Interaction) are indicated. Association rate constant (ka), dissociation rate constant (kd), steady-state dissociation constant (KD), and least squares fit (LSF) are shown. Values represent the mean of three replicates ± standard deviation. *Kinetics too irreversible to be determined.
| Interaction | SHOC2 | PP1C | MRAS | ka (1/Ms) x104 |
kd (1/s) x10−3 |
KD kinetic (M) x10−8 |
KD (M) x10−8 |
LSF (KD) x10−7 |
|---|---|---|---|---|---|---|---|---|
| SHOC2 >► PP1CA | WT | WT | — | 2.7 ± 0.1 | 390 ± 22 | 1,413 ± 135 | 670 | 5 |
| SHOC2 > PP1CAP50R | WT | P50R | — | 5.5 ± 1.4 | 182 ±79 | 96 ± 141 | 180 | 2.4 |
| SHOC2M1731 >■ PP1CA | M173I | WT | — | 5.0 ±0.7 | 384 ± 39 | 775 ± 137 | 450 | 3.2 |
| SHOC2M1731 >► PP1CAP50R | M173I | P50R | — | 7.0 ± 0.4 | 194 ± 46 | 279 ± 13 | 190 | 1.5 |
| SHOC2-PP1CA >■ MRAS | WT | WT | WT | 7.2 ± 0.9 | 5.2 ± 3.3 | 7.4 ± 0.7 | 20 | 0.6 |
| SHOC2-PP1CA >► MRASQ71L | WT | WT | Q71L | 8.1 ± 1.5 | 0.3 ± 0.1 | 0.4 ± 0.1 | 6.7 | 0.2 |
| SHOC2M173I-PP1CA >► MRAS | M173I | WT | WT | 6.4 ± 1.2 | 1.5 ± 1.3 | 2.4 ± 0.3 | 8.5 | 0 |
| SHOC2-PP1CAP50R >► MRAS | WT | P50R | WT | 7.9 ± 0.6 | 1.4 ± 0.8 | 1.8 ± 0.2 | 27 | 0.9 |
| SHOC2M173I-PP1CAP50R > MRAS | M173I | P50R | WT | 9.3 ± 1.4 | 0.5 ± 0.1 | 0.6 ± 0.1 | 20 | 1 |
| SHOC2M173I-PP1CAP50R > MRASQ71L | M173I | P50R | Q71L | * | * | * | 0.6 | 0.3 |
| SHOC2 >► PP1CB | WT | WT | — | 1.0 ± 4.4 | 7.5 ± 0.8 | 4,180 ± 2,570 | 1,100 | 6.2 |
| SHOC2-PP1CB >► MRAS | WT | WT | WT | 5.7 ± 6.9 | 2.8 ± 0.2 | 4.9 ± 0.5 | 20 | 0.6 |
| SHOC2 >► PP1CC | WT | WT | — | 3.5 ± 1.0 | 377 ± 41 | 1130 ± 112 | 650 | 12 |
| SHOC2-PP1CC >► MRAS | WT | WT | WT | 5.5 ± 5.6 | 3.7 ± 0.3 | 6.9 ± 0.6 | 19 | 0.4 |
Supplementary Material
Acknowledgments:
This work was funded in part by NIH/NCI (F32CA243290, J.J.K.), the Lustgarten Foundation, the Doris Duke Charitable Foundation, Pancreatic Cancer Action Network, NIH-NCI K08 CA218420-02, P50CA127003 (A.J.A.) and the Dana-Farber Cancer Institute Hale Center for Pancreatic Cancer Research (A.J.A., W.C.H.), U01 CA176058 (W.C.H.), U01 CA224146 (A.J.A., W.C.H.), and U01 CA250549 (A.J.A., W.C.H.). This work was funded in part by the Deerfield-Broad Discovery Research Collaboration. Deerfield Management Company, L.P. is a healthcare focused investment management firm.
Footnotes
Competing Interest: D.E.R. receives research funding from members of the Functional Genomics Consortium (Abbvie, BMS, Jannsen, Merck, Vir), and is a director of Addgene, Inc. D.L.M consults for Legal consulting Shook, Hardy & Bacon. F.M. is a consultant for the following companies: Amgen; Daiichi Ltd., Frontiers Med, Exuma Biotech, Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Inc., PellePharm, Pfizer Inc., PMV Pharma and Quanta Therapeutics. F.M. is a consultant for and cofounder of the following companies (with ownership interest including stock options): BridgeBio; DNAtrix Inc., Olema Pharmaceuticals, Inc., and Quartz. F.M. is the scientific director of the National Cancer Institute RAS Initiative at the Frederick National Laboratory for Cancer Research/Leidos Biomedical Research, Inc. F.M. has been a recipient of research grants from Daiichi Sankyo and Gilead Sciences and has a current grant from Boehringer-Ingelheim. W.C.H. is a consultant for Thermo Fisher, Solasta Ventures, MPM Capital, KSQ Therapeutics, Tyra Biosciences, Jubilant Therapeutics, Function Oncology, RAPPTA Therapeutics, Frontier Medicine and Calyx. A.J.A. has consulted for Oncorus, Inc., Arrakis Therapeutics, Syros Pharmaceuticals, Boehringer Ingelheim, T-knife Therapeutics, AstraZeneca, Mirati Therapeutics and Merck & Co., Inc, and has research funding from Mirati Therapeutics, Syros Pharmaceuticals, Bristol Myers Squibb, Revolution Medicines, Novartis, and Novo Ventures that is unrelated to this work. W.C.H. and A.J.A. have funding from Deerfield, Inc. that is related to the work described here.
Supplementary Information: Supplementary Information is available for this paper.
Data availability:
The coordinates and structure factors for apo-SHOC2 and cryo-EM structures are deposited in the protein databank (Apo SHOC2 X-ray structure PDB ID: 7T7A; Complex cryo-EM structure PDB ID: 7UPI). The variant information for disease associated mutations for complex members are publicly available (Clinvar: https://www.ncbi.nlm.nih.gov/clinvar/ and COSMIC: https://cancer.sanger.ac.uk/cosmic). Primary data are provided with this paper, and additional data relating to this study are available at:
Code availability:
Molecular Operating Environment (MOE) and Schrödinger software are publicly available for commercial and non-commercial use. Custom code for DMS analyses are available at: https://github.com/jkwonbio/Structure-function-analysis-of-the-SHOC2-MRAS-PP1C-holophosphatase-complex.git
References
- 1.Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M & McCormick F A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol. Cell 22, 217–230 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Cordeddu V et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet 41, 1022–1026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins EM et al. Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight 2, e91225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gripp KW et al. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am. J. Med. Genet. A 170, 2237–2247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L et al. De novo missense variants in PPP1CB are associated with intellectual disability and congenital heart disease. Hum. Genet 135, 1399–1409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simanshu DK, Nissley DV & McCormick F RAS Proteins and Their Regulators in Human Disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gripp KW & Lin AE Costello syndrome: a Ras/mitogen activated protein kinase pathway syndrome (rasopathy) resulting from HRAS germline mutations. Genet. Med 14, 285–292 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Young LC et al. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc. Natl. Acad. Sci. U. S. A 115, E10576–E10585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behan FM et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Sulahian R et al. Synthetic Lethal Interaction of SHOC2 Depletion with MEK Inhibition in RAS-Driven Cancers. Cell Rep. 29, 118–134.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boned Del Río I et al. SHOC2 complex-driven RAF dimerization selectively contributes to ERK pathway dynamics. Proc. Natl. Acad. Sci. U. S. A 116, 13330–13339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein SA, Majumdar A & Barrick D A Second Backbone: The Contribution of a Buried Asparagine Ladder to the Global and Local Stability of a Leucine-Rich Repeat Protein. Biochemistry 58, 3480–3493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motta M et al. SHOC2 subcellular shuttling requires the KEKE motif-rich region and N-terminal leucine-rich repeat domain and impacts on ERK signalling. Hum. Mol. Genet 25, 3824–3835 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Kwon JJ & Hahn WC A Leucine-Rich Repeat Protein Provides a SHOC2 the RAS Circuit: a Structure-Function Perspective. Mol. Cell. Biol 41, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terrak M, Kerff F, Langsetmo K, Tao T & Dominguez R Structural basis of protein phosphatase 1 regulation. Nature 429, 780–784 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Choy MS et al. SDS22 selectively recognizes and traps metal-deficient inactive PP1. Proc. Natl. Acad. Sci. U. S. A 116, 20472–20481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young LC et al. An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol. Cell 52, 679–692 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Egloff MP et al. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16, 1876–1887 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S & Lee EY A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J. Biol. Chem 272, 28368–28372 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Hannig V, Jeoung M, Jang ER, Phillips JA 3rd & Galperin E A Novel SHOC2 Variant in Rasopathy. Hum. Mutat 35, 1290–1294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mysore VP et al. A structural model of a Ras-Raf signalosome. Nat. Struct. Mol. Biol 28, 847–857 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieburth DS, Sun Q & Han M SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell 94, 119–130 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Matsunaga-Udagawa R et al. The scaffold protein Shoc2/SUR-8 accelerates the interaction of Ras and Raf. J. Biol. Chem 285, 7818–7826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Han M & Guan KL The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev. 14, 895–900 (2000). [PMC free article] [PubMed] [Google Scholar]
- 26.Wakula P, Beullens M, Ceulemans H, Stalmans W & Bollen M Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem 278, 18817–18823 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Krzyzosiak A et al. Target-Based Discovery of an Inhibitor of the Regulatory Phosphatase PPP1R15B. Cell 174, 1216–1228.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parton RG & Hancock JF Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol. 14, 141–147 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Park E et al. Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed E, Symons M, Macdonald SG, McCormick F & Ruggieri R Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science 265, 1713–1716 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Tran TH et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun 12, 1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battye TGG, Kontogiannis L, Johnson O, Powell HR & Leslie AGW iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimanius D, Dong L, Sharov G, Nakane T & Scheres SHW New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J (2021) doi: 10.1042/BCJ20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant T, Rohou A & Grigorieff N cisTEM, user-friendly software for single-particle image processing. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bepler T et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelker MS, Page R & Peti W Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J. Mol. Biol 385, 11–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shima F et al. Structural basis for conformational dynamics of GTP-bound Ras protein. J. Biol. Chem 285, 22696–22705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Barad BA et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu K et al. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins 82, 1646–1655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salam NK, Adzhigirey M, Sherman W & Pearlman DA Structure-based approach to the prediction of disulfide bonds in proteins. Protein Eng. Des. Sel 27, 365–374 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Beard H, Cholleti A, Pearlman D, Sherman W & Loving KA Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS One 8, e82849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowers KJ et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. in SC ’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing 43–43 (2006). [Google Scholar]
- 48.Lu C et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput 17, 4291–4300 (2021). [DOI] [PubMed] [Google Scholar]
- 49. https://www.biorxiv.org/content/10.1101/2021.06.16.448102v1.
- 50.Schymkowitz J et al. The FoldX web server: an online force field. Nucleic Acids Res. 33, W382–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiberti M et al. MutateX: an automated pipeline for in-silico saturation mutagenesis of protein structures and structural ensembles. bioRxiv 824938 (2019) doi: 10.1101/824938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for apo-SHOC2 and cryo-EM structures are deposited in the protein databank (Apo SHOC2 X-ray structure PDB ID: 7T7A; Complex cryo-EM structure PDB ID: 7UPI). The variant information for disease associated mutations for complex members are publicly available (Clinvar: https://www.ncbi.nlm.nih.gov/clinvar/ and COSMIC: https://cancer.sanger.ac.uk/cosmic). Primary data are provided with this paper, and additional data relating to this study are available at:
Molecular Operating Environment (MOE) and Schrödinger software are publicly available for commercial and non-commercial use. Custom code for DMS analyses are available at: https://github.com/jkwonbio/Structure-function-analysis-of-the-SHOC2-MRAS-PP1C-holophosphatase-complex.git