Abstract
The filamentous bacteriophage CTXΦ, which encodes cholera toxin (CT) in toxigenic Vibrio cholerae, is known to propagate by infecting susceptible strains of V. cholerae by using the toxin coregulated pilus (TCP) as its receptor and thereby causing the origination of new strains of toxigenic V. cholerae from nontoxigenic progenitors. Besides V. cholerae, Vibrio mimicus strains which are normally TCP negative have also been shown to occasionally produce CT and cause diarrhea in humans. We analyzed nontoxigenic V. mimicus strains isolated from surface waters in Bangladesh for susceptibility and lysogenic conversion by CTXΦ and studied the expression of CT in the lysogens by using genetically marked derivatives of the phage. Of 27 V. mimicus strains analyzed, which were all negative for genes encoding TCP but positive for the regulatory gene toxR, 2 strains (7.4%) were infected by CTX-KmΦ, derived from strain SM44(P27459 ctx::km), and the phage genome integrated into the host chromosome, forming stable lysogens. The lysogens spontaneously produced infectious phage particles in the supernatant fluids of the culture, and high titers of the phage could be achieved when the lysogens were induced with mitomycin C. This is the first demonstration of lysogenic conversion of V. mimicus strains by CTXΦ. When a genetically marked derivative of the replicative form of the CTXΦ genome carrying a functional ctxAB operon, pMSF9.2, was introduced into nontoxigenic V. mimicus strains, the plasmid integrated into the host genome and the strains produced CT both in vitro and inside the intestines of adult rabbits and caused mild-to-severe diarrhea in rabbits. This suggested that in the natural habitat infection of nontoxigenic V. mimicus strains by wild-type CTXΦ may lead to the origination of toxigenic V. mimicus strains which are capable of producing biologically active CT. The results of this study also supported the existence of a TCP-independent mechanism for infection by CTXΦ and showed that at least one species of Vibrio other than V. cholerae may contribute to the propagation of the phage.
Vibrio cholerae belonging to the O1 or O139 serogroup is the etiologic agent of cholera, an acute dehydrating diarrhea which is caused principally by the potent enterotoxin cholera toxin (CT) produced by these organisms during pathogenesis (30). The ctxAB operon, which encodes the A and B subunits of CT, resides in the genome of CTXΦ, a filamentous bacteriophage which exists as a prophage in the chromosome of toxigenic V. cholerae (39). We have previously studied the induction of CTX prophage and demonstrated that the propagation of CTXΦ is associated with the origination of new toxigenic strains of V. cholerae from nontoxigenic progenitors (12, 13). The receptor for CTXΦ for invading V. cholerae cells is the toxin coregulated pilus (TCP), the genes for which are located on a large DNA region referred to as the TCP-ACF pathogenicity island, which includes the tcp and acf gene clusters (20, 22). However, recent studies involving molecular analysis of naturally occurring strains of toxigenic V. cholerae have shown that although most toxigenic strains carry the TCP pathogenicity island, a small proportion of toxigenic strains which are negative for genes encoding TCP also exist (15, 33).
Besides toxigenic strains of V. cholerae which carry the CTX prophage, Vibrio mimicus has also been implicated in diarrheal disease, and some strains of V. mimicus have been demonstrated to produce CT (7, 18, 31, 34). The mechanism involved in the acquisition of ctxAB genes by V. mimicus, however, has not been clearly demonstrated. In the present study we analyzed environmental strains of V. mimicus, isolated from surface waters in Bangladesh, for their susceptibility and lysogenic conversion by CTXΦ and tested the toxigenicity of the lysogens under laboratory conditions and inside the intestines of adult rabbits, using genetically marked derivatives of the phage. The study was designed to investigate whether CT-negative V. mimicus strains are also infected and converted to toxigenic strains by the phage and to evaluate the possible role of V. mimicus in the propagation of CTXΦ in the natural habitat.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
Strains of V. mimicus analyzed in the present study were isolated from surface water samples collected in Dhaka between January and December 1997. The strains were stored either in lyophilized form or in sealed deep nutrient agar at room temperature until they were used for the present study. Before use, the identities of the cultures were confirmed by biochemical methods (8), and the presence or absence of genes encoding CT, TCP, and ToxR, as well as the CTXΦ attachment sequence attRS, was tested with specific DNA probes or PCR assays (9, 13). Details of the strains are presented in Table 1. Relevant characteristics of reference bacterial strains and properties of phages and plasmids used in this study are listed in Table 2.
TABLE 1.
Analysis of 27 V. mimicus strains isolated from environmental surface waters in Bangladesh for rRNA gene restriction patterns (ribotype), for presence of genes encoding CT, TCP, and ToxR as well as the attRS sequence, and for susceptibility of the strains to genetically marked derivatives of CTXΦ
| rRNA gene restriction pattern | Presence ofa:
|
No. of strains (total strains tested)b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | zot | tcpA | tcpI | acfB | toxR | attRS | Infected with CTX-KmΦ | Lysogenizedc with CTX-KmΦ | Transformed with pMSF9.2 | Chromosomalc integration of pMSF9.2 | |
| A | − | − | − | − | − | + | + | 1 (5) | 1 | 2 (2) | 2 |
| B | − | − | − | − | − | + | + | 1 (6) | 1 | 3 (3) | 3 |
| C | − | − | − | − | − | + | + | 0 (8) | 0 | 2 (2) | 2 |
| D | − | − | − | − | − | + | − | 0 (3) | 0 | NDd | |
| E | − | − | − | − | − | + | − | 0 (5) | 0 | ND | |
| Total | 2 (27) | 2 | 7 (7) | 7 | |||||||
Presence of different genes was detected with DNA probes and PCR assays (see the text for details). +, present; −, absent.
Susceptibility and lysogenic conversion of V. mimicus strains by CTXΦ was determined with two genetically marked derivatives of the phage, i.e., CTX-KmΦ and pMSF9.2 (see the text for details).
Integration of the phage genome into the host chromosome was determined by Southern blot analysis of genomic and plasmid DNA isolated from native and infected strains.
ND, not done.
TABLE 2.
Characteristics of V. cholerae reference strains, plasmids, and phages used in the study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| SM44 | Derivative of El Tor strain P27459, in which the CTX genetic element was marked with a kanamycin resistance determinant by marker exchange disrupting the ctxAB operon | 16 |
| RV508 | Derivative of classical biotype strain 569B, which is known to constitutively express CT, TCP, and other toxR-regulated gene products | 39 |
| 569B | V. cholerae O1 classical Inaba strain | Laboratory collection |
| P-27457 | V. cholerae O1 El Tor Inaba strain | Laboratory collection |
| SA-406 | Nontoxigenic TCP-positive V. cholerae O1 El Tor biotype strain | 13 |
| O395 | Classical Ogawa streptomycin-resistant strain | Laboratory collection |
| TCP2 | Derivative of strain O395, which carries deletions in the tcpA and ctxA genes | 36 |
| O395(pCTX-Km) | Strain O395 carrying the RF of CTX-KmΦ derived from strain SM44 | 13 |
| pRT41 | Derivative of pBR322 carrying both the ctxA and ctxB genes, including the wild-type promoter | 38 |
| pMSF9 | Derivative of pCTX-Km in which a 0.4-kb deletion was introduced in the zot gene in order to stop the morphogenesis of CTX-KmΦ | This study |
| pMSF9.2 | Derivative of pMSF9 in which the ctxAB operon was reinstated from pRT41; this construct carried both a functional ctxAB operon and the Kmr cassette | This study |
Recombinant DNA procedures.
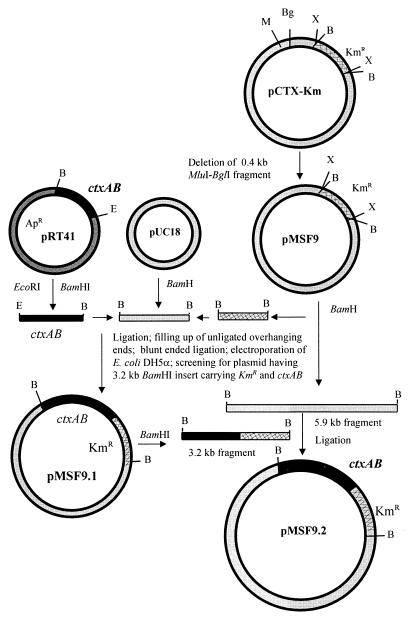
For in vitro DNA manipulations, pUC18, chromogenic substrate (X-Gal-[5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]), and DNA restriction and modifying enzymes were purchased from Bethesda Research Laboratories (BRL) (Gaithersburg, Md.) and used in accordance with the manufacturer's suggestions. The strategy for the construction of a genetically marked derivative of the replicative form (RF) of the CTXΦ genome carrying a functional ctxAB operon is shown in Fig. 1. CTX-KmΦ, isolated from strain SM44 (16, 39), was used to infect the classical biotype strain O395. The RF of the phage genome pCTX-Km isolated from strain O395(pCTX-Km) carried a kanamycin resistance (Kmr) determinant in place of the ctxAB genes. Before reinstating the ctxAB genes into the RF DNA, a mutation was introduced into the zot gene to stop phage morphogenesis (39) by deleting a 0.4-kb MluI-BglI fragment of the gene, and the resulting plasmid was designated pMSF9. This was done by digesting pCTX-Km simultaneously with MluI and BglI to excise a 0.4-kb portion of the DNA. The remaining 7.2-kb fragment was purified, the overhanging termini were filled by the Klenow fragment of Escherichia coli DNA polymerase I and ligated with T4 DNA ligase, and the DNA was electroporated into strain O395. Kanamycin-resistant colonies were selected and tested for the presence of the plasmid and for their inability to produce CTX-KmΦ particles.
FIG. 1.
Strategy for the construction of pMSF9.2, a derivative of pCTX-Km carrying a functional ctxAB operon. The following letters denote restriction sites on the maps: B, BamHI; X, XbaI; E, EcoRI; Bg, BglI; M, MluI. See the text for details.
The entire ctxAB operon was obtained from another recombinant plasmid, pRT41 (38), and reinstated into pMSF9 by a number of cloning steps (Fig. 1) to construct pMSF9.2. Briefly, pMSF9 was digested with BamHI, and a 1.3-kb fragment encoding Kmr and the remaining 5.9-kb fragment of the phage genome were isolated. The 1.9-kb BamHI-EcoRI insert carrying the entire ctxAB operon, including the wild-type promoter, was isolated from pRT41. The DNA fragments carrying the ctxAB operon and Kmr were sequentially ligated to BamHI-cleaved dephosphorylated pUC18. The ligated DNA was isolated and subjected to filling up of protruding ends with the Klenow fragment of E. coli DNA polymerase I. The resulting blunt ends were then ligated, and the ligation mixture was used to electroporate E. coli DH5α. Colonies which were simultaneously resistant to kanamycin and ampicillin were screened for the presence of a pUC18 derivative with a 3.2-kb BamHI insert carrying the ctxAB genes and the gene encoding Kmr, and this plasmid was designated pMSF9.1. The 3.2-kb BamHI fragment of pMSF9.1 was isolated and ligated with the 5.9-kb BamHI fragment of pMSF9. The ligated DNA was used to electroporate V. cholerae O395, and colonies were selected for resistance to kanamycin. The ultimate plasmid construct designated pMSF9.2 thus consisted of a functional ctxAB operon and a Kmr cassette and most of the RF DNA of CTXΦ but was unable to support the morphogenesis of infectious phage particles.
Preparation of phage.
CTX-KmΦ used in this study was prepared from a culture of strain O395 carrying the RF of the phage genome as described by us previously (13). Briefly, the culture supernatant was sterilized by filtration through 0.22-μm-pore-size filters (Millipore Corporation, Bedford, Mass.). To confirm that the filtrate did not contain any bacterial cells, aliquots of the filtrate were streaked on Luria agar plates and incubated overnight at 37°C. The filtrate was titrated for infectious phage particles by incubating aliquots of the supernatants with strain RV508 for 30 min at 30°C and then selecting for colonies resistant to kanamycin.
Probes and PCR assays.
The gene probes used in this study to detect the CTXΦ genome were a 0.5-kb EcoRI fragment of pCVD27 (19) carrying part of the ctxA gene and an 840-bp region internal to the zot gene amplified by PCR from the recombinant plasmid pBB241 as described previously (1, 11). The toxR gene probe was a 2.4-kb BamHI fragment of pVM7 (25), which is a pBR322-derived plasmid carrying the entire toxR gene. The 18-bp attRS sequence was identified by using a synthetic oligonucleotide corresponding to the attRS sequence (28). The rRNA gene probe consisted of a 7.5-kb BamHI fragment of the E. coli rRNA clone pKK3535 (2, 10). Colony blots or Southern blots were prepared with nylon filters (Hybond; Amersham International plc., Ayelesbury, United Kingdom) and processed by standard methods (24). The polynucleotide probes were labeled by random priming (14) with a random-primer DNA-labeling kit (BRL) and [α-32P]deoxycytidine triphosphate (3,000 Ci/mmol; Amersham), and oligonucleotide probes were labeled by 3′ tailing with terminal deoxynucleotidyl transferase (BRL) and [α-32P]dCTP (Amersham). Southern blots and colony blots were hybridized with the labeled probes, and autoradiographs were developed as described by us previously (9–12).
The presence of the TCP pathogenicity island was detected by PCR assays specific for the tcpA, tcpI, and acfB genes. The presence of tcpA genes specific for the classical and El Tor biotypes was detected by a multiplex PCR assay (21), and the tcpI and acfB genes were detected by specific PCR assays described by us previously (9, 13). The PCR-negative strains were further confirmed for the absence of the relevant genes by colony blot hybridization with the corresponding PCR-generated amplicons derived from a positive control strain, 569B, as specific probes.
Infection of recipient strains.
The susceptibility of V. mimicus strains to CTXΦ was assayed under laboratory conditions by previously described methods (12, 13) with CTX-KmΦ. Briefly, the recipient strains were grown in Luria-Bertani (LB) medium (1% Bacto Tryptone, 0.5% Bacto Yeast Extract, 1% NaCl) or in AKI medium (1.5% Bacto Peptone, 0.4% yeast extract, 0.5% NaCl, 0.3% NaHCO3, pH 7.4) at 30°C; the cells were precipitated by centrifugation and washed in fresh LB or AKI medium. The recipient cells and phage particles were mixed in the appropriate medium to a make an approximate final concentration of 106 bacterial cells and 106 phage particles per ml. The mixture was incubated for 16 h at 30°C, and aliquots of the culture were diluted and plated on Luria agar plates containing kanamycin (50 μg/ml) to select for kanamycin-resistant colonies and on plates devoid of kanamycin to determine the total number of colonies. The results were expressed as a percentage of the total colonies that became resistant to kanamycin. In each round of assay, the V. cholerae O1 classical strains O395, RV508, and TCP2, a derivative of strain O395 with tcpA deleted, and an El Tor biotype strain, SA-406, were included as controls. To allow optimal expression of the phage receptor TCP in the control strains, classical strains were grown in LB medium whereas the El Tor strain was grown in AKI medium. The V. mimicus strains were tested for susceptibility to the phage in both LB and AKI media separately.
Analysis of infected strains.
Representative colonies of V. mimicus which were either infected by CTX-KmΦ or electroporated with pMSF9.2 were grown in LB medium containing kanamycin (50 μg/ml) and were analyzed for the presence of the phage genome. Total DNA or plasmid DNA was extracted from overnight cultures by standard methods (24) and purified with microcentrifuge filter units (Ultrafree-Probind; Sigma). Integration of the phage genome into the chromosome of the recipient cells was studied by comparative Southern blot analysis of total DNA and plasmid preparations from infected and native strains (see Fig. 2). Production of extracellular phage particles by V. mimicus strains infected with CTX-KmΦ was assayed by growing the strains in the presence of mitomycin C (20 ng/ml) or without mitomycin C and titrating the supernatant fluids of the cultures for the presence of phage particles with strain RV508 as the recipient as described by us previously (12, 13).
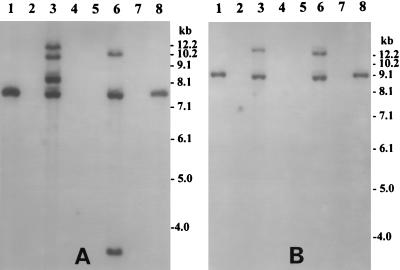
FIG. 2.
Southern hybridization analysis of genomic DNA or plasmids isolated from V. mimicus strains infected with CTX-KmΦ or electroporated with pMSF9.2 and the corresponding native strains. (A) Total DNA or plasmid isolated from strains infected with CTX-KmΦ was digested with BglI and probed with an 850-bp PCR-generated probe for the zonula occludens toxin gene. Lanes 1 and 8, pCTX-Km linearized with BglI; lanes 2 and 5, total DNA from native strains 2V8 and 88V114, respectively; lanes 3 and 6, total DNA from infected strains 2V8 (CTX-Km) and 88V114 (CTX-Km), respectively; lanes 4 and 7, plasmid preparations from strain 2V8 (CTX-Km) and 88V114 (CTX-Km). (B) Total DNA or plasmid isolated from strains electroporated with pMSF9.2 was digested with BglII and hybridized with the ctxA probe. Lanes 1 and 8, pMSF9.2 linearized with BglII; lanes 2 and 5, total DNA from native strains 2V8 and 88V114, respectively; lanes 3 and 6, total DNA from transformed strains 2V8 (MSF9.2) and 88V114 (MSF9.2), respectively; lanes 4 and 7, plasmid preparations from strain 2V8 (MSF9.2) and 88V114 (MSF9.2). The numbers indicating the molecular sizes of bands correspond to a 1-kb DNA ladder (BRL). The absence of bands in the lanes containing plasmid preparations and the positive hybridization of digested chromosomal DNA of the transductants demonstrate the integration of CTX-KmΦ DNA or pMSF9.2 into the chromosomes of recipient V. mimicus strains.
Assay for CT production.
The ability of V. mimicus strains carrying the integrated form of pMSF9.2 to produce CT was determined by the GM1-ganglioside-dependent enzyme-linked immunosorbent assay (GM1-ELISA) and the rabbit ileal loop assay as described previously (6, 9, 32). To study the expression of CT in LB as well as in AKI medium, each strain was tested separately with either of the media. For each round of CT assay, 5 ml of LB (pH 6.5) or AKI (pH 7.4) medium was inoculated with approximately 103 bacterial cells and grown for 16 h at 30°C with shaking. The culture was centrifuged at 4,000 × g for 5 min, and the supernatant was collected and filtered through 0.22-μm-pore-size Millipore filters. Aliquots of the undiluted supernatant, 10-fold and 100-fold dilutions of the supernatant, and dilutions of purified CT (Sigma) were used for the toxin assay. Briefly, 100 μl of the samples was added into each well of microtiter plates precoated with GM1 and incubated at room temperature for 90 min. After the plates were washed, with phosphate-buffered saline containing 0.5% Tween-20, the GM1-bound CT was reacted with rabbit anti-CT antibody (Sigma). Antibody binding to CT was detected by reaction with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (whole-molecule) antibody (Sigma), and the substrates o-phenyldiamine and hydrogen peroxide. Quantification of CT production was done with a standard curve prepared for each batch of assay. The amount of CT produced by each strain was the mean value of five different assays with the same strain and culture conditions. The classical strain 569B, El Tor strain P27457, and V. mimicus strains lysogenized with CTX-KmΦ were used as positive and negative control strains in each round of assay.
Ileal loop assay.
Culture filtrates of the lysogens and the corresponding native strains, along with the control strains, were tested in ileal loops of adult New Zealand White rabbits obtained from the breeding facilities of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B). A maximum of six ileal loops approximately 10 cm in length were made in each rabbit, which had previously been fasting for 48 h. Culture filtrates prepared for the ELISA test were also used for the ileal loop assay, and 1 ml of the filtrate was inoculated into each loop as described previously (6). Culture supernatant from each strain was tested in at least five rabbits. After 18 h, the rabbits were sacrificed and the loops were examined for fluid accumulation. The results were expressed as the volume of fluid accumulated in milliliters per centimeter of the loop.
Assay for diarrhea in rabbits.
The diarrheal response of rabbits to V. mimicus strains carrying the integrated form of pMSF9.2 and to the corresponding native strains as well as the control strains were assayed in adult rabbits with the removable intestinal tie-adult rabbit diarrhea (RITARD) model (35). Adult New Zealand White rabbits weighing 1.5 to 2.7 kg were used to prepare the RITARD model. The rabbits were starved for the previous 24 h, and surgery was done under a local anesthetic. The cecum of each animal was ligated to prevent it from retaining fluid secreted by the small intestine, and a temporary removable tie of the small bowel was introduced at the time of challenge. Strains were grown in Casamino acid-yeast extract broth as described previously (35), and cells were precipitated by centrifugation and resuspended in 10 mM phosphate-buffered saline, pH 7.4, at a concentration of approximately 109 per ml. One ml of the suspension was injected into the lumen of the anterior jejunum. The removable tie in the intestine was removed after 2 h of inoculation. Each strain was inoculated in at least 5 different rabbits. The rabbits were observed for overt diarrhea and for death, and stools or rectal swabs were cultured on gelatin agar plates containing kanamycin whenever appropriate to monitor shedding of the challenge organisms. Observations were made at 6-h intervals during the following 5 days of inoculation, the number of rabbits developing moderate-to-severe diarrhea was arbitrarily scored, and the number of deaths was recorded. Rabbits that died with or without diarrhea were subjected to postmortem examinations to check for the presence of fluid in the intestine.
RESULTS AND DISCUSSION
Species of the genus Vibrio have been regarded as a group of organisms whose major habitats are aquatic ecosystems, although several of the species are pathogenic in humans and produce putative colonization factors, enterotoxins, hemolysins, hemagglutinins, and possibly other virulence-associated factors. V. cholerae, Vibrio parahaemolyticus, and V. mimicus are known to cause diarrheal illness in humans (8). It has been suggested that acquisition of virulence-associated genes has allowed specific vibrios to adapt to the human intestinal environment (13, 20). The most potent enterotoxin, cholera toxin (CT) produced by toxigenic strains of V. cholerae, is encoded by the CTX bacteriophage (39), and propagation of the phage in its natural habitat is associated with the origination of new toxigenic strains (13). The factors associated with the propagation of the phage and the evolutionary significance of its interaction with the host bacteria have yet to be clearly elucidated. Preliminary studies by us and others have shown that CTXΦ infects V. cholerae by using the pilus colonization factor TCP as the receptor and lysogenizes the susceptible bacteria (13, 39). This implies that CTXΦ should specifically infect V. cholerae strains which are capable of expressing TCP. However, we have shown previously that a V. cholerae non-O1 strain which was negative for TCP was also infected by CTXΦ, although at a low efficiency (13). This indicated that there may be an alternative TCP-independent mechanism for CTXΦ infection and thus raised questions as to whether CTXΦ is also able to infect and utilize strains belonging to other species of the genus for its propagation. In the present study we tested the susceptibility of environmental V. mimicus strains to the phage and the expression of CT in these strains, since V. mimicus has been reported to occasionally produce CT (7, 34). In addition, we tested the ability of infected strains to produce extracellular phage particles in order to understand whether environmental strains of V. mimicus may also contribute to the propagation of CTXΦ.
Distribution of virulence genes and attRS sequence.
Naturally occurring toxigenic strains of V. cholerae carry the CTXΦ genome integrated in the chromosome at a site specified by the attachment sequence attRS. In addition, these strains normally possess genes for TCP and the transcriptional regulator ToxR (17, 37). We screened the V. mimicus strains in the present study for the presence of these genes with either DNA probes or PCR assays. All V. mimicus strains studied were negative for the tcpA, tcpI, and acfB genes and presumably the entire TCP pathogenicity island. To our knowledge, V. mimicus strains have never been shown previously to carry genes encoding TCP, although the species has close similarity with V. cholerae (4, 8). However, all strains tested in the present study carried the toxR gene and 19 of the 27 strains (70.3%) carried the attRS sequence (Table 1). The toxR gene has also been found to be widely distributed among nontoxigenic V. cholerae strains (13). This is probably because ToxR is involved in the regulation of a number of genes in addition to those encoding TCP and CT (5, 25–27) and may be part of a common regulatory mechanism possessed by different species of the genus. The high prevalence of the attRS sequence in V. mimicus strains suggested that these strains were capable of integrating the phage genome once it was inside the cell by whatever means and thus that they can possibly act as a reservoir of the CTXΦ genome in the aquatic environment. This was further confirmed by the observed integration of the phage derivative pMSF9.2 in the chromosomes of V. mimicus strains which were electroporated with the plasmid (Table 1).
Infection and analysis of recipient strains.
We used two genetically marked derivatives of CTXΦ or its RF DNA, namely, CTX-KmΦ, and pMSF9.2 (Table 2) to study the susceptibility of the strains and their ability to express CT. The Kmr marker present in the genome of the phage allowed us to conveniently detect and analyze infected cells. Of 27 V. mimicus strains exposed to CTX-KmΦ, 2 strains (7.4%) were infected. These two V. mimicus strains were distinctly susceptible to the phage in repeated assays, producing between 12 and 56 infected colonies per 107 live cells (susceptibility, between [1.2 × 10−6]% and [5.6 × 10−6]%). No significant difference was noted in the phage susceptibility of these two V. mimicus strains whether the strains were tested in LB or AKI medium. The remaining 25 strains were completely resistant to infection by the phage, and no kanamycin-resistant transductant was detected in these strains when they were assayed under identical conditions. The susceptibility of V. mimicus strains was significantly low compared to that of the control V. cholerae classical biotype strains RV508 and O395 (mean, 89.1 and 45.2%, respectively) but was comparable to that of the nontoxigenic El Tor biotype strain SA-406 (mean susceptibility, [6.5 × 10−6]%), even when the strain was grown in AKI medium to allow expression of TCP (the receptor for CTXΦ). This was probably because the classical biotype strains expressed TCP more adequately than the El Tor strain under in vitro conditions, and moreover, strain RV508 is a mutant that constitutively expresses TCP and other toxR-regulated genes. It was, however, interesting to note that 2 of 27 V. mimicus strains which did not carry genes for TCP were infected at an efficiency comparable to that of the TCP-positive El Tor strain of V. cholerae O1. The present study thus showed for the first time that besides V. cholerae some V. mimicus strains can also be infected by the phage and confirmed our previous speculation (13) that in addition to the TCP-mediated mechanism there may be a second mechanism for CTXΦ infection. However, the control strain, TCP2, which is a TCP-negative derivative of the classical strain O395, was not infected by the phage in repeated assays, although the parent strain was infected at high frequency. Thus the TCP-independent infection by CTXΦ was strain specific. However, it was not clear from this study what determined the phage sensitivity of the two V. mimicus strains.
We analyzed the rRNA gene restriction patterns (ribotype) of the V. mimicus strains to determine clonal relationships among the phage-susceptible and phage-resistant strains. Ribotyping was done with the restriction enzyme BglI, which has been previously used by us and others to study clonal relationships among V. cholerae strains (9, 10, 29). V. mimicus strains analyzed in the present study revealed five different BglI restriction patterns (patterns A through E) of their rRNA genes. The two strains which were susceptible to the phage produced restriction patterns A and B, but nine other strains which produced either of these patterns were not susceptible (Table 1). Thus, there was no association between ribotype and susceptibility to the phage. All five ribotype patterns were widely different from previously described ribotype patterns produced by toxigenic V. cholerae strains (9, 10, 29). In the present study, V. mimicus strains were identified and differentiated from V. cholerae based primarily on biochemical reactions (8) and then serology to specifically exclude V. cholerae O1 and O139 strains. Analysis of rRNA gene restriction patterns allowed us to further verify whether particular mutants of nontoxigenic V. cholerae O1 impaired in sucrose fermentation or in the expression of serotype-specific antigens were wrongly classified as V. mimicus. Comparison of ribotype patterns of the V. mimicus strains with previously reported ribotype patterns of toxigenic V. cholerae (9, 10, 29) ruled out this possibility.
In all V. mimicus strains which were infected by CTX-KmΦ or electroporated with pMSF9.2, the phage genome integrated into the chromosome of the host forming lysogens, as was evidenced by Southern blot analysis of plasmid and chromosomal DNA preparations from representative infected cells (Fig. 2). Although plasmid preparations from the freshly infected cells showed the presence of the phage genome, the lysogens spontaneously lost the RF of the phage, as confirmed by subsequent plasmid preparations (data not shown). To analyze strains infected with CTX-KmΦ, which did not carry the ctxA gene, chromosomal DNA was digested with BglI to cleave within the zot gene (1), and a zot probe was used to determine the number of fragments produced. Since, in pMSF9.2, a deletion was introduced in the zot gene, we used a different enzyme, BglII, to digest chromosomal DNA derived from the electroporated strains and used the ctxA probe. Since, there is no BglII site within the ctxA gene, the number of fragments produced represented the approximate number of copies of the CTXΦ genome which integrated into the chromosome of the infected cells. Thus, hybridization of BglI- or BglII-digested genomic DNA from infected or electroporated strains with the zot or ctxA probe and interpretation of the resulting restriction patterns as described previously (9) confirmed that the phage genome integrated into the chromosome of V. mimicus cells and two or three copies of the phage genome were present in tandem repeats in different host chromosomes (Fig. 2). The absence of a detectable level of the RF DNA of the phage was evidenced by the absence of hybridization signals in lanes containing plasmid preparations from the infected strains (Fig. 2).
Expression of CT.
Unlike CTX-KmΦ (39), the newly constructed derivative pMSF9.2 in the present study carried a functional ctxAB operon, which allowed us to study the expression of CT in the V. mimicus cells. The introduction of a deletion mutation in the zot gene (Fig. 1) while constructing pMSF9.2 was primarily a safety precaution to avoid the creation of a live phage carrying both a wild-type ctxAB operon and a gene encoding kanamycin resistance.
Production of CT by the lysogens was initially studied in vitro by GM1-based ELISA, using an antibody against the B subunit of CT (Table 3). To ascertain whether the strains produced both the A and B subunits of CT and whether the toxin was biologically active, we used the ligated-ileal-loop assay in rabbits and observed fluid accumulation. Finally we studied the diarrheal response of adult rabbits challenged with the toxigenic lysogens (Table 4). These assays confirmed that in contrast to the corresponding native strains the lysogens produced biologically active CT both in vivo and under in vitro conditions (Tables 3 and 4). Production of CT by the toxigenic V. mimicus strains in vitro was less than that by the classical V. cholerae strain 569B but was comparable to that produced by the El Tor strain P-27457. Rabbits challenged with native V. mimicus strains and with V. mimicus strains lysogenized with CTX-KmΦ did not show a diarrheal response (Table 4), but V. mimicus strains carrying integrated pMSF9.2, which carried a functional ctxAB operon, produced diarrhea in rabbits. This suggested that the diarrhea was in response to CT and not due to other possible factors, such as zonula occludens toxin or accessory cholera enterotoxin, which are also known to be encoded by the CTXΦ genome (1, 39). Several previous studies have shown that certain naturally occurring V. mimicus strains produce enterotoxins identical to CT (7, 34). Hence, the lysogenic conversion of V. mimicus strains by CTXΦ demonstrated in the present study under laboratory conditions may have been occurring in the ecological habitat as well.
TABLE 3.
Production of CT by environmental V. mimicus strains with integrated form of pMSF9.2, a genetically marked derivative of the RF of the CTXΦ genome carrying a functional ctxAB operon and encoding resistance to kanamycin
| Strain | Characteristics | CT productiona (GM1 ELISA) | Fluid accumulationb in rabbit ileal loops (ml/cm of ileal loop) |
|---|---|---|---|
| 569B | Toxigenic classical biotype strain of V. cholerae O1 | 6.13 ± 1.02 | 3.61 ± 0.51 |
| P-27457 | Toxigenic El Tor biotype strain of V. cholerae O1 | 2.87 ± 0.73 | 2.39 ± 0.47 |
| 2V-8 | Wild-type CTX-negative V. mimicus strain | UD (<0.01)c | 0 |
| 2V-8(MSF9.2) | Strain 2V-8 carrying integrated pMSF9.2 | 2.36 ± 0.42 | 2.16 ± 0.61 |
| 2V-8(CTX-Km) | Strain 2V-8 lysogenized with CTX-KmΦ | UD (<0.01) | 0 |
| 88V-114 | Wild-type CTX-negative, V. mimicus strain | UD (<0.01) | 0 |
| 88V-114(MSF9.2) | Strain 88V-114 carrying integrated pMSF9.2 | 2.95 ± 0.72 | 2.45 ± 0.65 |
| 88V-114(CTX-Km) | Strain 88V-114 lysogenized with CTX-KmΦ | UD (<0.01) | 0 |
Toxin amounts are expressed in micrograms per unit of optical density of the culture at 600 nm. The values represent the averages of five independent observations in LB medium for the classical biotype strain and in AKI medium for the El Tor and V. mimicus strains. No significant difference in toxin production was noted when the El Tor and V. mimicus strains were tested in LB or AKI medium.
Values represent averages of five independent observations made in different rabbits.
UD, undetectable. The toxin amounts were less than 0.01 μg/ml, which was the lowest concentration of purified toxin used as a control (see the text for details).
TABLE 4.
Diarrheal response of adult rabbits to V. mimicus strains lysogenized with genetically marked derivatives of CTXΦ
| Strain | Characteristics | No. of animals challenged | No. responding with
|
||
|---|---|---|---|---|---|
| Fatal diarrheaa | Nonfatal diarrheabc | No symptomsc | |||
| P-27457 | Toxigenic clinical strain of V. cholerae O1, El Tor | 5 | 4 | 1 | 0 |
| 2V-8 | Wild-type CTX-negative V. mimicus strain | 6 | 0 | 0 | 6 |
| 2V-8(MSF9.2) | Strain 2V-8 carrying integrated pMSF9.2 | 7 | 0 | 6 | 1 |
| 2V-8(CTX-Km) | Strain 2V-8 lysogenized with CTX-KmΦ | 5 | 0 | 0 | 5 |
| 88V-114 | Wild-type CTX-negative V. mimicus strain | 6 | 0 | 0 | 6 |
| 88V-114(MSF9.2) | Strain 88V-114 carrying integrated pMSF9.2 | 7 | 2 | 5 | 0 |
| 88V-114(CTX-Km) | Strain 88V-114 lysogenized with CTX-KmΦ | 5 | 0 | 0 | 5 |
This category includes two animals with fluid-filled intestines but no diarrhea at the time of death, as determined by postmortem examinations.
The duration of diarrhea was 5 days for the rabbit challenged with the El Tor strain and between 2 and 5 days for the animals challenged with V. mimicus strains.
Animals belonging to these categories were positive for the challenge organism either in stool or rectal-swab cultures for 1 to 6 days.
Regulation of CT expression in V. mimicus strains.
Expression of critical virulence genes in V. cholerae is known to be coordinately regulated so that multiple genes respond in a similar fashion to environmental conditions (5). Coordinate expression of virulence genes results from the activity of a cascading system of regulatory factors. ToxR, a 32-kDa transmembrane protein, is the master regulator which is itself regulated by environmental signals. ToxR regulates not only the expression of ctxAB and the TCP colonization factor, but also at least 17 distinct genes that constitute the ToxR regulon (5). The ToxR regulon is controlled through another regulatory factor called ToxT, a 32-kDa protein. ToxR controls the transcription of the toxT gene, and the resulting increased expression of the ToxT protein leads to activation of other genes in the ToxR regulon. In the present study, the V. mimicus strains were negative for the entire TCP pathogenicity island, which includes the toxT gene (22), although these strains carried the toxR gene. The expression of CT in these strains thus was independent of toxT. It may be mentioned that V. cholerae has ToxT-dependent and ToxT-independent branches of the ToxR regulon (3, 5). The ToxR protein binds to a tandemly repeated 7-bp DNA sequence found upstream of the ctxAB structural gene and directly increases transcription of ctxAB, resulting in higher levels of CT expression. In view of the absence of toxT in the V. mimicus strains, it is likely that ToxR directly regulated the expression of CT from the integrated form of the CTXΦ genome in the V. mimicus lysogens. It may be mentioned that CT can also be expressed from the RF of the CTXΦ genome independently of ToxR (23). However, in the present study, the V. mimicus lysogens did not carry any detectable level of the RF DNA, as evidenced by Southern blot hybridization of plasmid preparations (Fig. 2).
Propagation of CTXΦ.
The V. mimicus lysogens of CTX-KmΦ spontaneously produced infectious phage particles in the supernatant fluids of culture (Table 5), and high titers of the phage (mean, 4.2 × 105 particles/ml) could be obtained when induced with mitomycin C, as determined by titration with strain RV508 as the recipient. The demonstration of the presence of V. mimicus strains in the environment which were capable of harboring the CTXΦ genome and producing infectious phage particles indicated that some V. mimicus strains may have a role in supporting the replication and propagation of CTXΦ in the environmental habitat. The discovery of CTXΦ is beginning to provide an understanding of the role of bacteriophages in the evolution of vibrios and the mutual benefit imparted. Although the ctxAB genes do not have a direct role in the morphogenesis of CTXΦ, the toxigenic property imparted by the phage to its host provides greater evolutionary fitness to the bacterium, since toxigenic strains are selectively enriched in the intestinal environment. Moreover, acquisition of the diarrheagenic property allows rapid spread of the bacteria and thus of the bacteriophage. In the present study, although the number of rabbits that developed fatal diarrhea in response to the V. mimicus strains was less than that in response to the control El Tor strain, the surviving rabbits shed the organisms in the diarrheal stools for 2 to 6 days. In previous studies the specificity of the phage towards infecting V. cholerae was assumed to be a result of the expression by V. cholerae of the pilus colonization factor, TCP, which is also known to be the receptor for CTXΦ. In the present study we demonstrated that CTXΦ can also infect selective strains of V. mimicus and convert them to toxigenic strains capable of producing diarrhea in the rabbit model. This study thus showed that at least one Vibrio species other than V. cholerae can contribute to the propagation of CTXΦ. Our efforts are at present directed towards understanding the mechanism that allows TCP-independent infection by CTXΦ and to further studying the regulation of the ctxAB operon in V. mimicus.
TABLE 5.
Production of cell-free phage particles by V. mimicus and V. cholerae strains carrying a genetically marked derivative of CTXΦ prophage
| Strain | Description | Titer of CTX-KmΦ in supernatants of culture (particles/ml)a:
|
|
|---|---|---|---|
| Induced with mitomycin C | Without mitomycin C | ||
| 2V-8(CTX-Km) | Environmental V. mimicus strain 2V-8 lysogenized with CTX-KmΦ | 3.6 × 105 | 0.5 × 102 |
| 88V-114(CTX-Km) | Environmental V. mimicus strain 88V-114 lysogenized with CTX-KmΦ | 4.2 × 105 | 1.2 × 102 |
| SM44(P27459 ctx::km) | El Tor lysogen of CTXΦ marked with a Kmr determinant disrupting the ctxAB operon | 4.3 × 105 | 0.6 × 102 |
Values are averages of three independent observations.
ACKNOWLEDGMENTS
This research was funded by the United States Agency for International Development (USAID) under grant HRN-5986-A-00-6005-00 with the ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries.
We thank V. I. Mathan for helpful discussions and suggestions. We thank Manujendra N. Saha for helping with this study and Afjal Hossain for secretarial assistance.
REFERENCES
- 1.Baudry B, Fasano A, Ketley J, Kaper J B. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60:428–434. doi: 10.1128/iai.60.2.428-434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rRNB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 3.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of Tox T mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 4.Davis B R, Fanning G R, Madden J M, Steigerwalt A G, Bradford H B, Jr, Smith H L, Jr, Brenner D J. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus. J Clin Microbiol. 1981;14:631–639. doi: 10.1128/jcm.14.6.631-639.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De S N, Chatterje D N. An experimental study of the mechanisms of action of Vibrio cholerae on the intestinal mucous membrane. J Pathol Bacteriol. 1953;46:559–562. doi: 10.1002/path.1700660228. [DOI] [PubMed] [Google Scholar]
- 7.Dotevall H, Jonson-Stormberg G, Sanyal S, Holmgren J. Characterization of enterotoxin and soluble hemagglutinin from Vibrio mimicus; identity with V. cholerae O1 toxin and hemagglutinin. FEMS Microbiol Lett. 1985;27:17–22. [Google Scholar]
- 8.Farmer J J, III, Brenner F W H, Kelly T M. Vibrio. In: Lennete E H, editor. Manual of clinical microbiology. 4th ed. Washington, D.C: American Society for Microbiology; 1985. pp. 281–301. [Google Scholar]
- 9.Faruque S M, Ahmed K M, Alim A R M A, Qadri F, Siddique A K, Albert M J. Emergence of a new clone of toxigenic Vibrio cholerae biotype El Tor displacing V. cholerae O139 Bengal in Bangladesh. J Clin Microbiol. 1997;35:624–630. doi: 10.1128/jcm.35.3.624-630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque S M, Roy S K, Alim A R M A, Siddique A K, Albert M J. Molecular epidemiology of toxigenic V. cholerae in Bangladesh studied by numerical analysis of rRNA gene restriction patterns. J Clin Microbiol. 1995;33:2833–2838. doi: 10.1128/jcm.33.11.2833-2838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque S M, Comstock L, Kaper J B, Albert M J. Distribution of zonula occludens toxin (zot) gene among clinical isolates of Vibrio cholerae O1 from Bangladesh and Africa. J Diarrhoeal Dis Res. 1994;12:222–224. [PubMed] [Google Scholar]
- 12.Faruque S M, Asadulghani, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic V. cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque S M, Asadulghani, Saha M N, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for the origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg A, Volgelstein B. A technique for radio labelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh C, Nandi R K, Dasgupta S K, Nair G B, Hall R H, Ghose A C. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb Pathog. 1997;22:199–208. doi: 10.1006/mpat.1996.0105. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg I, Mekalanos J J. Effect of a recA mutation on cholera toxin gene amplification and deletion events. J Bacteriol. 1986;165:723–731. doi: 10.1128/jb.165.3.723-731.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili and ToxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda J M, Powers C, Bryant R G, Abbott S L. Current perspective on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–267. doi: 10.1128/cmr.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaper J B, Morris J G, Jr, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious disease. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 65–77. [Google Scholar]
- 20.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keasler S P, Hall R H. Detection and biotyping of Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 22.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 23.Lazar S, Waldor M K. ToxR-independent expression of cholera toxin from the replicative form of CTXΦ. Infect Immun. 1998;66:394–397. doi: 10.1128/iai.66.1.394-397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsot C, Mekalanos J J. Expression of the Vibrio cholerae gene encoding aldehyde dehydrogenase is under control of ToxR, the cholera toxin transcriptional activator. J Bacteriol. 1991;173:2842–2851. doi: 10.1128/jb.173.9.2842-2851.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popovic T, Bopp C, Olsvic O, Wachsmuth K. Epidemiological application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabbani G H, Greenough W B. Cholera. In: Lebenthal E, Duffy M, editors. Textbook of secretory diarrhea. New York, N.Y: Raven Press Inc.; 1990. pp. 233–253. [Google Scholar]
- 31.Ramamurthy T, Albert M J, Huq A, Colwell R R, Takeda Y, Takeda T, Shimada T, Mandal B K, Nair G B. Vibrio mimicus with multiple toxin types isolated from human and environmental sources. J Med Microbiol. 1994;40:194–196. doi: 10.1099/00222615-40-3-194. [DOI] [PubMed] [Google Scholar]
- 32.Sack D A, Huda S, Neogi P K B, Daniel R R, Spira W M. Microtiter ganglioside enzyme-linked immunosorbent assay for Vibrio and Escherichia coli heat labile enterotoxins and antitoxins. J Clin Microbiol. 1980;1:35–40. doi: 10.1128/jcm.11.1.35-40.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Said B, Smith H R, Scotland S M, Rowe B. Detection and differentiation of the gene for toxin co-regulated pili (tcpA) in Vibrio cholerae non-O1 using the polymerase chain reaction. FEMS Microbiol Lett. 1995;125:205–210. doi: 10.1111/j.1574-6968.1995.tb07359.x. [DOI] [PubMed] [Google Scholar]
- 34.Spira W M, Fedorka-Cray P J. Purification of enterotoxins from Vibrio mimicus that appear to be identical to cholera toxin. Infect Immun. 1984;45:679–684. doi: 10.1128/iai.45.3.679-684.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spira W M, Sack R B, Froehlich J L. Simple adult rabbit model for Vibrio cholerae and enterotoxigenic Escherichia coli diarrhea. Infect Immun. 1981;32:739–747. doi: 10.1128/iai.32.2.739-747.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor R, Shaw C, Peterson K, Spears P, Mekalanos J. Safe, live Vibrio cholerae vaccines? Vaccine. 1988;6:151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- 37.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]