Abstract
Lasiodiplodia (family Botryosphaeriaceae) is a widely distributed fungal genus that causes a variety of diseases in tropical and subtropical regions. During 2020–2021, a routine survey of fruit tree plants was conducted in five Egyptian Governorates, and fresh samples exhibiting dieback, decline, leaf spot and root rot symptoms were collected. Collection from eight different symptomatic leaves, twigs, branches and roots of fruit trees yielded 18 Lasiodiplodia-like isolates. The sequencing data from the internal transcribed spacer region (ITS), partial translation elongation factor 1-alpha (tef1-a) and β-tubulin (tub2) were used to infer phylogenetic relationships with known Lasiodiplodia species. Two isolates obtained from black necrotic lesions on Phoenix dactylifera leaves were identified as a putative novel species, L. newvalleyensis sp. nov., and were thus subjected to further morphological characterization. The results of isolation and molecular characterization revealed that L. theobromae (n = 9) was the most common species on Mangifera indica, Citrus reticulata, C. sinensis, Ficus carica, Prunus persica, Prunus armeniaca and Pyrus communis trees. Lasiodiplodia pseudotheobromae (n = 5) was isolated from M. indica, Prunus persica and C. sinensis. Lasiodiplodia laeliocattleyae (n = 2) was isolated from C. reticulata. Pathogenicity test results suggested that all Lasiodiplodia species were pathogenic to their hosts. The present study is considered the first to characterize and decipher the diversity of Lasiodiplodia species associated with fruit trees in Egypt, using the multi-locus ITS, tef1-a and tub2 sequence data, along with morphological and pathogenic trials. To our knowledge, this is the first report of L. newvalleyensis on Phoenix dactylifera and L. laeliocattleya on C. reticulata in Egypt and worldwide.
Keywords: elongation factor 1-alpha, β-tubulin, ITS, morphological characterization, pathogenicity phylogenetic analysis
1. Introduction
The family Botryosphaeriaceae encompasses several fungal species that are found in all environmental and climatic zones of the world as endophytes or saprophytes pathogens [1]. Lasiodiplodia (family Botryosphaeriaceae) is a pluralistic genus distributed in tropical and subtropical areas that causes a variety of diseases, including cankers, dieback, fruit or root rot, branch blight, stem end rot and gummosis on a wide range of woody and fruit trees [1,2,3,4,5]. Since 2004 and until 2017, 43 species of Lasiodiplodia have been described [1,3,4,6]. Nonetheless, five new Lasiodiplodia species associated with blueberries have recently been discovered in China [7], bringing the genus Lasiodiplodia to forty-eight species. Members of the genus Lasiodiplodia exhibit diverse lifestyles on a wide range of host plants, ranging from endophytes, which cause asymptomatic infection on different plant tissues, pathogens, which cause diseases and saprophytes [1,8]. Among the Lasiodiplodia species, L. theobromae is a well-known plant pathogen associated with up to 500 hosts [9]. Diseases caused by species in the Botryosphaeriaceae have been reported since 1971 when Botryodiplodia theobromae was isolated from fruit rot and dieback of mango in Egypt. The fungal agent was later synonymized under L. theobromae and regarded as a causal pathogen for dieback on mango [3,10,11]; root rot on sugar beet dieback [12]; and canker and soft rot on other hosts, such as grapevine [13], walnut [14], maize [15], citrus [16], Annona spp. [17], Phoenix dactylifera [18], pome, stone fruit [19], Citrus sinensis, C. aurantifolia [20] and ornamental Ficus trees [21].
Characterization of Lasiodiplodia species has primarily relied on cultural and conidial characteristics and phylogenetic data [3,5,8,22,23,24,25]. Cultural and conidial characterization are often misleading and result in inaccurate identification due to overlapping in morphology [25,26]. Therefore, molecular characterization based on multi-locus sequence data has widely been applied to identify the Lasiodiplodia species, especially the L. theobromae species complex, which is difficult to distinguish based on morphology [1,8,23]. Recent multi-locus phylogenetic approaches using DNA sequence data of the internal transcribed spacers (ITS) of genomic rDNA [27], along with protein-coding genes such as translation elongation factor 1-alpha (tef1-a) and β-tubulin (tub2) [1,5,7,23], have aided in the identification of Lasiodiplodia species with strong phylogenetic support.
Based on the cosmopolitan presence of Lasiodiplodia species on various hosts and a very recent study [20], the distribution and prevalence of this fungal agent could be extended to other hosts in Egypt. In this sense, Lasiodiplodia species considered as a major pathogens occurring on a variety of hosts causing stem-end rot, fruit rot, decline, cankers and dieback. The current study was aimed at characterizing and deciphering the diversity of Lasiodiplodia species associated with wider fruit tree hosts in Egypt, using the ITS, tef1-a and tub2 sequence data, together with morphological and pathogenic trials.
2. Materials and Methods
2.1. Sampling and Isolation
During 2020–2021, surveys of fruit tree plants, including Mangifera indica, Citrus reticulata, Citrus sinensis, Ficus carica, Prunus persica, Prunus armeniaca, Pyrus communis and Phoenix dactylifera, were conducted across five Egyptian Governorates: Beheira, Giza, Kaliobyia, Sharkia and New Valley (Table S1). A total of fifty-seven symptomatic leaves, twigs, branches and roots of plants exhibiting leaf spot, dieback, decline and root rot symptoms were collected. Samples were subjected to pathogen isolation, as previously described [22]. The obtained Lasiodiplodia-like isolates and other associated fungi were cultured on potato dextrose agar (PDA) and stored at 5 °C in a refrigerator. The cultures were maintained in the culture collection facility at the Vegetable Diseases Research Department, Plant Pathology Research Institute, Agricultural Research Center (ARC), Giza, Egypt.
2.2. DNA Extraction and PCR Amplification
Genomic DNA was extracted from 5-day-old cultures of isolated fungi [28]. PCR amplification and sequencing of the ITS region of rDNA, including 5.8S, was performed using the primers ITS4 and ITS5 [27]. Part of the tef1-α region was amplified using EF1-728F and EF1-986R [29], and the tub2 region was amplified using Bt1a and Bt1b primers [30]. PCR amplifications were carried out in an ESCO Swift Maxi Thermal Cycler [31]. The resultant PCR amplicons were gel purified using the CloneJet PCR cloning kit (ThermoFisher Scientific, Waltham, MA, USA) and sequenced in both directions using Sanger sequencing at Macrogen Inc. (Seoul, Korea). Sequences obtained in this study were deposited in GenBank database, and their accession numbers were obtained (Table 1).
Table 1.
Lasiodiplodia sequences and their accession numbers used in the phylogenetic analyses.
| Species | Strain | Host | Country | GenBank Accession Numbers | ||
|---|---|---|---|---|---|---|
| ITS | tef1-α | tub2 | ||||
| L. aquilariae | GuoLD01961 * | Aquilaria crassna | Laos | KY783442 | KY848600 | - |
| L. avicenniae | CMW 41467 * | Avocennia marina | South Africa | KP860835 | KP860680 | KP860758 |
| L. avicenniae | LAS 199 | Avocennia marina | South Africa | KU587957 | KU587947 | KU587868 |
| L. americana | CERC 1961 = CFCC 50065 * | Pistachia vera | China | KP217059 | KP217067 | KP217075 |
| L. brasiliensis | GuoLD01736 | Carica papaya | Brazil | KY783475 | KY848612 | KY848556 |
| L. brasiliensis | CMW35884 |
Adansonia
madagascariensis |
Madagascar | KU887094 | KU886972 | KU887466 |
| L. bruguierae | CMW41470 * |
Bruguiera
gymnorrhiza |
South Africa | KP860833 | KP860678 | KP860756 |
| L. bruguierae | CMW42480 * |
Bruguiera
gymnorrhiza |
South Africa | KP860832 | KP860677 | KP860755 |
| L. caatinguensis | CMM1325 * | Citrus sinensis | Brazil | KT154760 | KT008006 | KT154767 |
| L. caatinguensis | IBL381 * | Spondias purpurea | Brazil | KT154757 | KT154751 | KT154764 |
| L. chinensis | CGMCC3.18066 * | Hevea brasiliensis | China | KX499899 | KX499937 | KX500012 |
| L. chinensis | CGMCC3.18067 |
Sterculia
lychnophora |
China | KX499901 | KX499939 | KX500014 |
| L. chonburiensis | MFLUCC 16-0376 * | Pandanaceae | Thailand | MH275066 | MH412773 | MH412742 |
| L. cinnamomi | CFCC 51997 * | Cinnamomum camphora | China | MG866028 | MH236799 | MH236797 |
| L. citricola | IRAN1521C * | Citrus sp. | Iran | GU945353 | GU945339 | KU887504 |
| L. citricola | IRAN1522C * | Citrus sp. | Iran | GU945354 | GU945340 | KU887505 |
| L. clavispora | CGMCC 3.19594 * |
Vaccinium
uliginosum |
China | MK802166 | OL773697 | MK816339 |
| L. clavispora | CGMCC 3.19595 |
Vaccinium
uliginosum |
China | MK802165 | OL773696 | MK816338 |
| L. crassispora | CMW 13488 |
Eucalyptus
urophylla |
Venezuela | DQ103552 | DQ103559 | KU887507 |
| L. crassispora | WAC12533 | Santalum album | Australia | DQ103550 | DQ103557 | - |
| L. curvata | GuoLD01755 | Aquilaria crassna | Laos | KY783443 | KY848601 | KY848532 |
| L. curvata | GuoLD01906 | Aquilaria crassna | Laos | KY783437 | KY84859 | KY848529 |
| L. euphorbicola | CMW36231 * | Adansonia digitata | Botswana | KU887187 | KU887063 | KU887494 |
| L. euphorbicola | CMW 3609 * | Adansonia digitata | Zimbabwe | KF234543 | KF226689 | KF254926 |
| L. endophytica | MFLUCC 18-1121 | Magnolia acuminata | China | MK501838 | MK584572 | MK550606 |
| L. exigua | IBL 104 = CBS 137785 * | Retama raetam | Tunisia | KJ638317 | KJ638336 | KU887509 |
| L. fujianensis | CGMCC3.19593 | Vaccinium uliginosum | China | MK802164 | MK887178 | MK816337 |
| L. gilanensis | IRAN 1501C | Unknown | Iran | GU945352 | GU945341 | KU887510 |
| L. gilanensis | IRAN 1523C * | Unknown | Iran | GU945351 | GU945342 | KU887511 |
| L. gonubiensis | CMW 14077 * |
Syzygium
cordatum |
South Africa | AY639595 | DQ103566 | DQ458860 |
| L. gonubiensis | CMW 14078 * |
Syzygium
cordatum |
South Africa | AY639594 | DQ103567 | EU673126 |
| L. gravistriata | CMM 4564 * | Anacardium humile | Brazil | KT250949 | KT250950 | - |
| L. gravistriata | CMM 4565 * | Anacardium humile | Brazil | KT250947 | KT266812 | - |
| L. henanica | XCN6 = CGMCC 3.19176 | Vaccinium uliginosum | China | MH729351 | MH729357 | MH729360 |
| L. hormozganensis | IRAN 1498C * | Mangifera indica | Iran | GU945356 | GU945344 | KU887514 |
| L. hormozganensis | IRAN 1500C * | Olea sp. | Iran | GU945355 | GU945343 | KU887515 |
| L. hyalina | CGMCC 3.17975 * | Acacia confusa | China | KX499879 | KX499917 | KX499992 |
| L. iraniensis | CMW 36237 * | Adansonia digitata | Mozambique | KU887121 | KU886998 | KU887499 |
| L. iraniensis | CMW 36239 * | Adansonia digitata | Mozambique | KU887123 | KU887000 | KU887501 |
| L. iraniensis | IRAN 1502C * | Juglans sp. | Iran | GU945347 | GU945335 | KU887517 |
| L. iraniensis | IRAN 1520C * | Salvadora persica | Iran | GU945348 | GU945336 | KU887516 |
| L. irregularis | GuoLD01673 | Aquilaria crassna | Laos | KY783472 | KY848610 | KY848553 |
| L. laeliocattleyae | CBS 130992 * | Mangifera indica | Egypt | JN814397 | JN814424 | KU887508 |
| L. laeliocattleyae | EGY2033 | Citrus reticulata | Egypt | ON392181 | OP080238 | OP080255 |
| L. laeliocattleyae | EGY2038 | Citrus reticulata | Egypt | ON392185 | OP080242 | OP080259 |
| L. laosensis | GuoLD01818 | Aquilaria crassna | Laos | KY783471 | KY848609 | KY848552 |
| L. laosensis | GuoLD01963 | Aquilaria crassna | Laos | KY783450 | KY848603 | KY848536 |
| L. lignicola | CBS 134112 * | dead wood | Thailand | JX646797 | KU887003 | JX646845 |
| L. macroconidica | GuoLD01752 * | Aquilaria crassna | Laos | KY783438 | KY848597 | KY848530 |
| L. macrospora | CMM3833 * | Jatropha curcas | Brazil | KF234557 | KF226718 | KF254941 |
| L. magnoliae | MFLUCC18-0948 * | Magnolia candolii | China | MK499387 | MK568537 | MK521587 |
| L. mahajangana | CMW 27801 * | Terminalia catappa | Madagascar | FJ900595 | FJ900641 | FJ900630 |
| L. mahajangana | CMW 27818 * | Terminalia catappa | Madagascar | FJ900596 | FJ900642 | FJ900631 |
| L. margaritacea | CBS 122519 * | Adansonia gibbosa | Australia | EU144050 | EU144065 | KU887520 |
| L. mediterranea | CBS 137783 * | Quercus ilex | Italy | KJ638312 | KJ638331 | KU887521 |
| L. mediterranea | CBS 137784 * | Vitis vinifera | Italy | KJ638311 | KJ638330 | KU887522 |
| L. microcondia | GuoLD01889 | Aquilaria crassna | Laos | KY783441 | KY848614 | - |
| L. missouriana | UCD 2193MO * | Vitis vinifera | USA | HQ288225 | HQ288267 | HQ288304 |
| L. missouriana | UCD 2199MO * | Vitis vinifera | USA | HQ288226 | HQ288268 | HQ288305 |
| L. nanpingensis | CGMCC3.19597 |
Vaccinium
uliginosum |
China | MK802168 | OL773699 | MK816341 |
| L. nanpingensis | CGMCC319596 |
Vaccinium
uliginosum |
China | MK802168 | OL773698 | MK816340 |
| L. newvalleyensis | EGY20113 * | Phoenix dactylifera | Egypt | ON392175 | OP080253 | OP080271 |
| L. newvalleyensis | EGY20114 * | Phoenix dactylifera | Egypt | ON392180 | OP080254 | OP080272 |
| L. pandanicola | MFLUCC 16-0265 * | Pandanaceae | Thailand | MH275068 | MH412774 | - |
| L. paraphysoides | CGMCC 3.19174 = QD7 | Vaccinium uliginosum | China | MH729349 | MH729355 | MH729358 |
| L. paraphysoides | CGMCC 3.19175 = QD8 | Vaccinium uliginosum | China | MH729350 | MH729356 | MH729359 |
| L. parva | CBS 456.78 * | Cassava field-soil | USA | EF622083 | EF622063 | KU887523 |
| L. parva | CBS 494.78 | Cassava field-soil | USA | EF622084 | EF622064 | EU673114 |
| L. plurivora | STE-U 4583 */CBS 121103 | Vitis vinifera | South Africa | AY343482 | EF445396 | KU887525 |
| L. pontae | IBL12 = CMM1277 * | Spondias purpurea | Brazil | KT151794 | KT151791 | KT151797 |
| L. pseudotheobromae | CBS 116459 * | Gmelina arborea | Costa Rica | EF622077 | EF622057 | EU673111 |
| L. pseudotheobromae | CGMCC 3.18047 |
Pteridium
aquilinum |
China | KX499876 | KX499914 | KX499989 |
| L. pseudotheobromae | EGY2041 | Citrus sinensis | Egypt | ON392168 | OP080243 | OP080260 |
| L. pseudotheobromae | EGY2043 | Mangifera indica | Egypt | ON392170 | OP080245 | OP080262 |
| L. pseudotheobromae | EGY2048 | Prunus persica | Egypt | ON392172 | OP080247 | OP080264 |
| L. pseudotheobromae | EGY2049 | Mangifera indica | Egypt | ON392173 | OP080248 | OP080265 |
| L. pseudotheobromae | EGY20101 | Mangifera indica | Egypt | ON392179 | OP080252 | OP080270 |
| L. pyriformis | CBS 121770 * | Acacia mellifera | Namibia | EU101307 | EU101352 | KU887527 |
| L. pyriformis | CBS 121771 * | Acacia mellifera | Namibia | EU101308 | EU101353 | KU887528 |
| L. rubropurpurea | WAC 12535 * | Eucalyptus grandis | Australia | DQ103553 | DQ103571 | EU673136 |
| L. rubropurpurea | WAC 12536 * | Eucalyptus grandis | Australia | DQ103554 | DQ103572 | KU887530 |
| L. sterculiae | CBS342.78 * | Sterculia oblonga | Germany | KX464140 | KX464634 | KX464908 |
| L. subglobosa | CMM3872 * | Jatropha curcas | Brazil | KF234558 | KF226721 | KF254942 |
| L. subglobosa | CMM4046 * | Jatropha curcas | Brazil | KF234560 | KF226723 | KF254944 |
| L. tenuiconidia | GuoLD01857 | Aquilaria crassna | Laos | KY783466 | KY848619 | KY848586 |
| L. thailandica | CPC22795 * | Albizia chinensis | China | KJ193637 | KJ193681 | KY751301 |
| L. theobromae | CBS 111530 * | Unknown | Unknown | EF622074 | EF622054 | KU887531 |
| L. theobromae | CBS 164.96 | Fruit on coral reef coast |
Papua New Guinea | AY640255 | AY640258 | KU887532 |
| L. theobromae | EGY2035 | Citrus reticulata | Egypt | ON392182 | OP080239 | OP080256 |
| L. theobromae | EGY2036 | Citrus reticulata | Egypt | ON392183 | OP080240 | OP080257 |
| L. theobromae | EGY2037 | Citrus reticulata | Egypt | ON392184 | OP080241 | OP080258 |
| L. theobromae | EGY2042 | Mangifera indica | Egypt | ON392169 | OP080244 | OP080261 |
| L. theobromae | EGY2046 | Pyrus communis | Egypt | ON392171 | OP080246 | OP080263 |
| L. theobromae | EGY2050 | Pyrus communis | Egypt | ON392174 | OP080249 | OP080266 |
| L. theobromae | EGY2082 | Mangifera indica | Egypt | ON392176 | OP080237 | OP080267 |
| L. theobromae | EGY2083 | Ficus carica | Egypt | ON392177 | OP080250 | OP080268 |
| L. theobromae | EGY20100 | Prunus armeniaca | Egypt | ON392178 | OP080251 | OP080269 |
| L. tropica | GuoLD01846 | Aquilaria crassna | Laos | KY783454 | KY848616 | KY848540 |
| L. venezuelensis | WAC 12539 * | Acacia mangium | Venezuela | DQ103547 | DQ103568 | KU887533 |
| L. venezuelensis | WAC 12540 * | Acacia mangium | Venezuela | DQ103548 | DQ103569 | KU887534 |
| L. viticola | UCD 2553AR * | Vitis sp. | USA | HQ288227 | HQ288269 | HQ288306 |
| L. viticola | UCD 2604MO * | Vitis sp. | USA | HQ288228 | HQ288270 | HQ288307 |
| L. vitis | CBS 124060 * | Vitis vinifera | Italy | KX464148 | KX464642 | KX464917 |
| Diplodia mutila | CMW 7060 * | Fraxinus excelsior | Netherlands | AY236955 | AY236904 | AY236933 |
* Isolates represent ex-type. The isolates obtained in this study are boldfaced, and those new species are in red boldface.
2.3. Phylogenetic Analyses
MEGA XI (version 11.0.8) was used to trim and edit the obtained ITS, tef1-α and tub2 sequences to remove ambiguous ends from both directions [32]. MAFFT version 7 was used to assemble and align the sequences with the closely related Lasiodiplodia spp. [33]. Sequences were retrieved from the NCBI GenBank database (http://www.ncbi.nlm.nih.gov, accessed on 25 July 2022). Phylogenetic analysis was conducted using PAUP version 4.0a [34]. Maximum parsimony (MP) analysis was conducted using the heuristic search option with random stepwise addition based on 1000 replicates, tree bisection and reconnection (TBR) as branch swapping algorithms, and random taxon addition sequences for the construction of MP trees. Branches of zero length were collapsed, and all multiple equally parsimonious trees were saved. MAXTREES was set to 10,000. In the analysis, all characters were unordered and had equal weight; gaps were treated as missing data. Tree length (TL), consistency index (CI), rescaled consistency index (RC), retention index (RI) and the homoplasy index (HI) were calculated for parsimony [35]. The phylogenetic relationship was inferred with 1000 bootstrap replicates and included 104 sequences, representing 103 of Lasiodiplodia species, and a Diplodia mutila (CMW 7060) sequence as an outgroup taxon (Table 1). Bayesian analysis was performed using MrBayes v3.2.7a [36] on Cipres Science Gateway (www.phylo.org, accessed on 25 July 2022) [37], on the combined, partitioned dataset with the substitution models, calculated for each partition, by ModelFinder on IQ-TREE multicore version 2.2.0 [38,39]. Bayesian analysis was run in duplicate with four Markov chain Monte Carlo (MCMC) chains, with random trees for 10,000,000 generations, sampled every 1000 generations. The temperature value was lowered to 0.10, burn-in was set to 0.25 and the run was automatically stopped when the average standard deviation of split frequencies ended up below 0.01. A total of 4222 trees were read in the two runs, 2111 each, and 25% of trees were discarded in each run as the burn-in phase of the analysis. Posterior probabilities were determined from a consensus tree generated from the remaining 1584 trees of each run. Maximum likelihood (ML) analysis was computed with IQ-TREE multicore version 2.2.0, setting ModelFinder + tree reconstruction + ultrafast bootstrap based on 10,000 replicates [39,40,41]. The phylogenetic trees of the MP, ML and BP were viewed in FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree, accessed on 25 July 2022).
2.4. Morphological Examination
Fungal structures were examined by inducing sporulation on 2% water agar (WA) medium supplemented with double-autoclaved pine needles, as described by Ismail et al. [3]. A 5-mm mycelial plug from each isolate was placed in the center of WA plates and incubated for 10–20 days at 25 ± 2 °C near direct light with a 12 h photoperiod. Sections were made through conidiomata using Leica CM1100 microtome and mounted in lactic acid. Measurements were done for 30 conidiogenous cells, 30 paraphyses and 50 conidia from material mounted in water. Fungal structures were imaged with a Nikon Coolpix 995 digital camera connected to a Leica, DM 25,000 LED microscope. Colony morphology was observed on PDA medium after 7 days of incubation at 25 °C in the dark.
2.5. Evaluation of Temperature’s Effect on the Mycelial Growth
The effects of different temperature on the mycelial growth of L. newvalleyensis were investigated. Three plates for each temperature were inoculated with 6-mm plugs from the actively margins of 5-days-old cultures in the center of the 85-mm PDA. Petri dishes were incubated in the dark at 6 different temperatures (10, 15, 20, 25, 30 and 35 °C). After 3 days, colony diameters were determined, and the data were converted to radial growth in millimeters.
2.6. Pathogenicity Test on Seedlings and Leaves
Lasiodiplodia isolates were tested for their pathogenicity against their hosts of origin. Pathogenicity was determined in 6–10-month-old seedlings of Citrus reticulata, M. indica, Prunus persica, Prunus armeniaca and Pyrus communis. Apparently healthy leaves of Citrus reticulata, F. carica, M. indica and Phoenix dactylifera were selected for pathogenicity. Three replicates were used, and each replicate consisted of three leaves, meaning a total of 12 leaves were used for each isolate. Lasiodiplodia isolates were plated on PDA for 5-days at 25 ± 2 °C in the dark prior to inoculation [3,22]. Inoculations of seedlings and leaves were performed according to Ismail et al. [3,22]. Three replicates were used per isolate, and each replicate comprised three plants with a total of 12 seedlings for each isolate. The inoculated plants were maintained under greenhouse conditions at 25 ± 2 °C and 70–80% relative humidity, and examined periodically for symptom development. The trials were arranged in a completely randomized factorial design, and the trials were repeated once. After 30 days, the pathogenicity of the tested isolates was terminated, and the results were recorded as the extent of necrotic lesions (in centimeters) developed around the inoculation sites for seedlings and leaves. The dimensions of the inoculated wounds were not subtracted from final measurements. Values were transformed by Log2 for analysis and separation of means. Re-isolation of the tested isolates was performed from the margins of the necrotic lesions on PDA medium amended with streptomycin sulfate (0.1g L−1) and incubated in the dark at 25 ± 2 °C.
2.7. Data Analysis
The obtained data were subjected to one-way ANOVA [42]. The data of lesion lengths were not normally distributed and were then log transformed. Mean values of the transformed lesion diameters (cm) and mycelial growth (mm) were compared using the least-significant difference (LSD) test at (p < 0.05). The statistical program SPSS 8.0 was used to analyse the data.
3. Results
3.1. Symptoms, Isolation and Frequency
Several symptom patterns on different fruit tree organs were observed, but the most prevalent disease phenotype was dieback and decline. On mango trees, stem cracking symptoms with black liquid oozing from infected tissues were also observed (Figure 1A). Other symptoms observed included dieback of the young twigs starting from the tip and extending downward (Figure 1B), infected twigs (cross section) showing brown vascular discoloration of tissues on one side (Figure 1C), brown to black lesions on the leaf margins (Figure 1D), root rot of mango seedlings (Figure 1E), black lesions under cambium tissues of the crown area (Figure 1F) and apical part of roots (cross section) showing brown discoloration of internal tissues (Figure 1G).
Figure 1.
Symptoms observed on M. indica plants included stem cracking and gummosis (A) and dieback of the young twigs starting from the tip and extending downward (B). Cross-section of infected twigs showing the brown vascular discoloration of tissues in one side (C). Brown to black lesions on the leaf margins of the affected leaves (D). Root rot of mango seedlings (E). Black lesions under cambium tissues of the crown area (F), and cross-section of an apical part of roots showing brown discoloration of internal tissues (G).
On Prunus persica trees, the observed symptoms were dieback of the young twigs and branches starting from the tip and extending downward (Figure 2A); infected twigs (cross and longitudinal sections) showing the brown vascular discoloration of tissues in one side (Figure 2B–D); brown and root rot, especially on old trees (Figure 2E); and brown discoloration under cambium tissues of the crown area (Figure 2F). The symptom on Pyrus communis trees was dieback of the young twigs starting from the tip and extending downward (Figure 3A). Cross-sections of infected twigs to compare the infected and healthy tissues also showed brown vascular discoloration of tissues on one side (Figure 3B,C). It was possible to observe dieback and decline of the young twigs and branches of Prunus armeniaca starting from the tip extending downward (Figure 3D) and dieback of the branches on one side, giving V-shape symptoms (Figure 3E,F). Lesions with different appearances were observed on C. reticulata: large necrotic black lesions starting from the leaf margins and inside the leaf blades (Figure 4A,B). In addition, dieback symptoms were observed on young twigs of C. reticulata (Figure 4C) and C. sinensis (Figure 4D). Furthermore, brown to black lesions were recorded on the young leaves of F. carica (Figure 4E,F) as well as on the leaves of Phoenix dactylifera (Figure 4G,H). A total of 18 Lasiodiplodia-like isolates (growing fast on medium, with a greenish brown to dark greyish blue mycelium) and other associated fungi (4 isolates of Alternaria spp., 2 isolates of Cladosporium spp. and 2 isolates of Pestalotiopsis spp.) were isolated from eight different fruit trees from five Egyptian Governorates. In total, 18 Lasiodiplodia-like isolates were isolated—4 from branches, 7 from leaves, 4 from twigs, 2 from roots and 1 from stem cracking (Table S2). All isolates were included in the phylogenetic study.
Figure 2.
Symptoms observed on Prunus persica plants included dieback of the young twigs and branches starting from the tip and extending downward (A); cross (B,C) and longitudinal (D) sections of infected twigs showing the brown vascular discoloration of tissues in one side, crown and root rot (E); brown discoloration under cambium tissues of the crown area (F).
Figure 3.
Symptoms on Pyrus communis plants included dieback of the young twigs starting from the tip and extending downward (A); cross-sections of infected twigs showing the brown vascular discoloration of tissues on one side (B,C); dieback and decline of the young twigs and branches of Prunus armeniaca starting from the tip and extending downward (D); dieback of the branches on one side, giving V-shape symptoms (E,F).
Figure 4.
Symptoms on C. reticulata: large necrotic black lesions that start from the leaf margins and inside leaf blade (A,B); dieback on young twigs of C. reticulata (C) and C. sinensis (D); brown to black lesions on F. carica (E,F) and on Phoenix dactylifera (G,H).
3.2. Phylogenetic Analyses
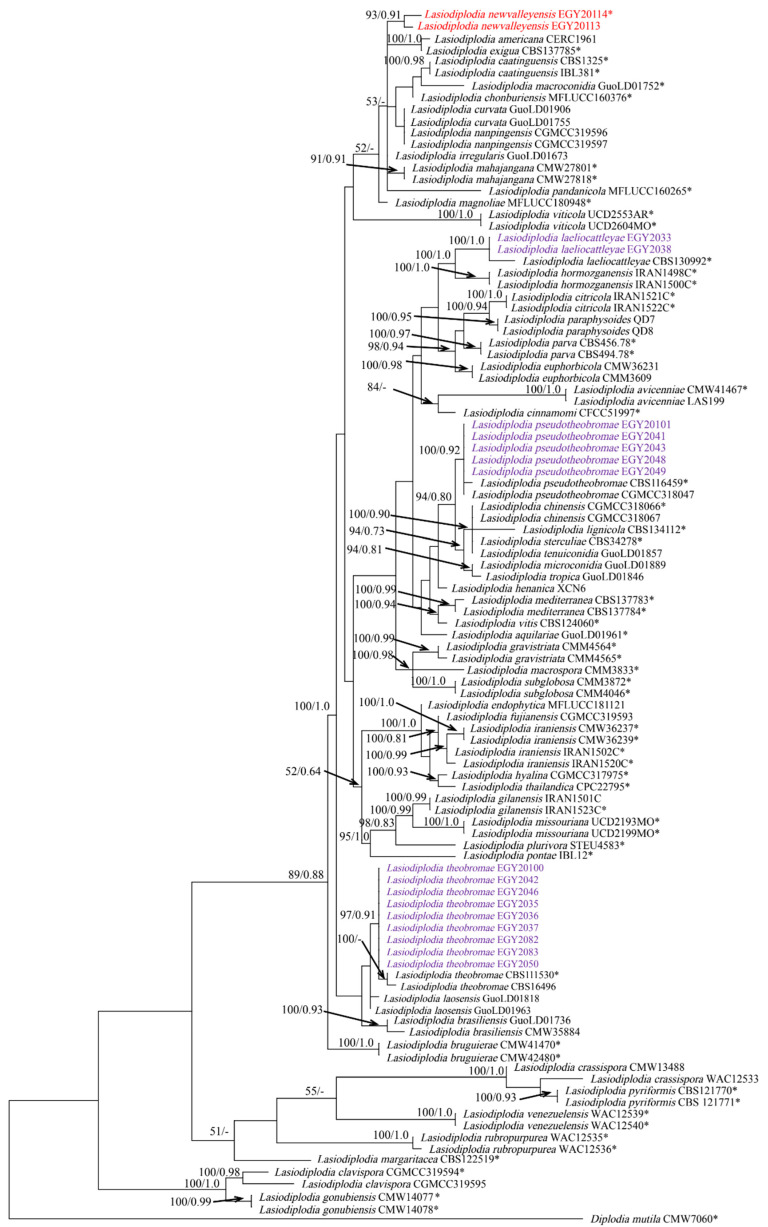
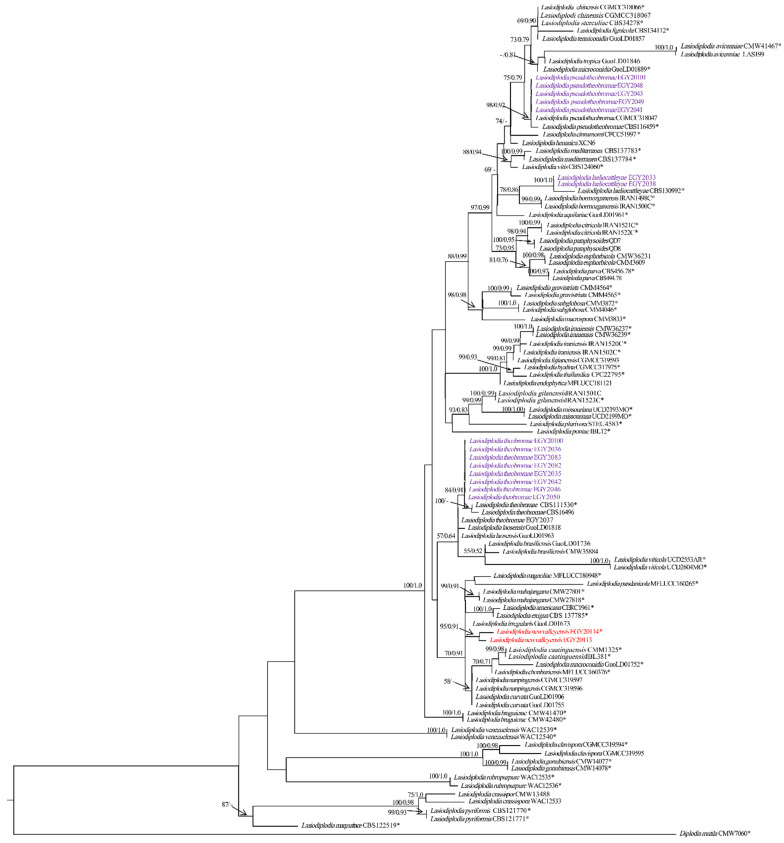
The sequences of the three gene regions were combined, yielding a dataset consisting of 1114 characters (ITS: 482 bps; tef-1α: 274 bps; tub2: 358 bps), including gaps of 104 Lasiodiplodia taxa (Table S3). Of these characters, 72 characters were parsimony-uninformative, 156 were parsimony-informative and 886 (proportion = 0.795) were constant. Heuristic search with the random addition of taxa (1000 replicates) resulted in the phylogenetic tree (TL = 445 steps, CI = 0.633, RI = 0.868, RC = 0.550, HI = 0.366) and the most parsimonious tree is presented in Figure 5. The topology of the tree generated by MP analysis was congruent with the 50% majority-rule consensus tree. The phylogenetic tree generated by ML analysis based on the combined ITS, tef-1α and tub2 sequence alignments is presented in Figure 6. Based on the ITS, tef-1α and tub2 dataset, ML analysis revealed that Lasiodiplodia isolates can be grouped into five major clades. Among all, five isolates belong to clade containing L. pseudotheobromae (CBS116459 and CGMCC3 18047), as highly supported by the bootstrap (BS)/posterior probability (PP) values of 98/0.92%. Most of the isolates (nine isolates) grouped with L. theobromae (CBS111530 and CBS164.96) in a clade, which was strongly supported with BS/PP values of 84/0.91% (Figure 6). Additionally, two isolates clustered with L. laeliocattleyae (CBS130992) in a clade, which was supported with strong values of BS/PP, 100/1.0%. Notably, two isolates, EGY20113 and EGY20114, of L. newvalleyensis, representing a potential novel species grouped together in an distant clade, which was supported with BS/PP 93/0.91%, sister to a clade containing L. exigua BL104 and L. americana CERC1961, that highly supported with BS/PP 100/1.0% and to a clade containing L. mahajangana CMW27801 and CMW27818, which was supported with BS/PP 99/1.0%.
Figure 5.
Phylogenetic tree based on maximum parsimony analysis (MP) through heuristic searches of the combined ITS, tef-1α and tub2 dataset of Lasiodiplodia species. Branches are shown on nodes with bootstrap values (BS %) and Bayesian posterior probabilities (PP). Branches not supported with BS or PP are marked with –, and isolates representing ex-type are marked with *. Diplodia mutila CMW 7060 was used as an outgroup taxon to validate the tree. The isolates obtained in this study are blue, and those newly described and ex-type species are in red boldface.
Figure 6.
Phylogenetic tree based of maximum likelihood analyses (ML) based on the combined ITS, tef-1α and tub2 dataset of Lasiodiplodia species. Branches are shown on nodes with bootstrap values (BS %) and Bayesian posterior probabilities (PP). Branches not supported with BS or PP are marked with –, and isolates representing ex-type are marked with *. Diplodia mutila CMW 7060 was used as an outgroup taxon to validate the tree. The isolates obtained in this study are blue, and those newly described and ex-type species are in red boldface.
3.3. Taxonomy
Lasiodiplodia newvalleyensis A.M. Ismail, S.M. El-Ganainy and E.S Elshewy, sp. nov (Figure 6).
MycoBank: MB843771.
The etymology refers to the place New Valley Governorate from where this species was isolated.
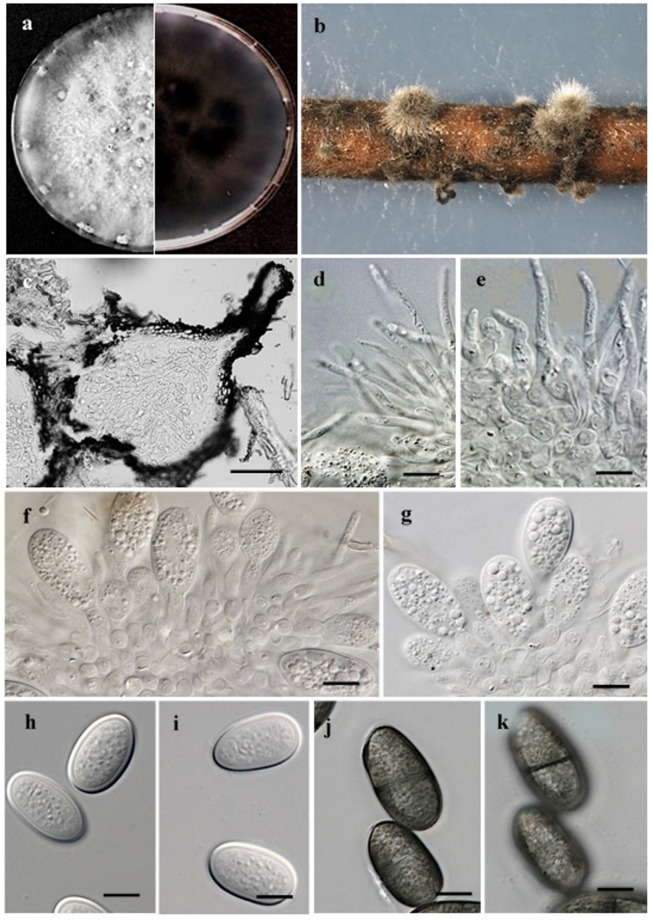
Sexual morph: Absent. Asexual morph; Conidiomata (Figure 7b) produced on pine needles on WA within 10–15 days; mostly solitary or in aggregates; dark-grey to black; globose to subglobose; covered with dense hairy mycelium; semi-immersed; becomes erumpent when mature. A vertical section through pycnidia shows outer layers of pycnidia composed of approximately 4–8 dark-brown, thick-walled cells layers of textura angularis, followed by hyaline thin-walled cells towards the centre (Figure 7c). Paraphyses (Figure 7d,e), hyaline and subcylindrical, arise between the conidiogenous cells. They are aseptate, wider at the base, slightly swollen at the apex, 14.9–44.5 µm long and 1.9–3.7 µm wide. Conidiophores reduced to conidiogenous cells. Conidiogenous cells (Figure 7f,g) are holoblastic, thin-walled hyaline, cylindrical and sometimes swollen slightly at the base. They have a rounded apex, proliferate recurrently to produce 1–2-minute annelations, are 4.6–10.5 µm long and are 3.2–5 µm wide. Conidia (Figure 7h–k) are initially hyaline, smooth, thick-walled, aseptate and obovoid to ellipsoid, contain granular contents and are mostly round at both ends; they have the same form when mature. Conidia become brown, are septate with 1-septum, have longitudinal striations and measure 17.2–26.7 × 10.5–13.3 µm (av. of 50 conidia ± SD = 22 ± 1.8 µm long, 11.7 ± 0.7 µm wide, L/W ratio = 1.8).
Figure 7.
Lasiodiplodia newvalleyensis holotype EGY H-240483. (a) Colony morphology, front and reverse sides; (b) conidiomata formed on pine needles on WA; (c) vertical section through pycnidia; (d,e) hyaline septate paraphyses formed between conidiogenous cells; (f,g) conidiogenous cells; (h,i) hyaline immature thick-walled conidia; and (j,k) dark mature conidia at two different focal planes to show longitudinal striation. Scale bars: (c) = 20 µm; (d–k) = 10 µm.
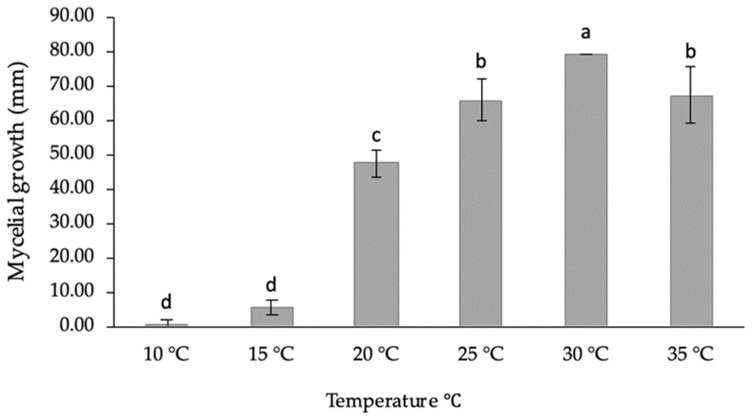
Cultural characteristics (Figure 7a): Colonies raised on a mycelium mat were moderately dense, and initially white to smoke-grey but turned greenish grey on the front side and greenish grey on the reverse side. The colour becomes dark slate blue with age. Pycnidia was produced on PDA after 7 days under the above-mentioned conditions. Colonies reached the edge of the Petri plate, 85 mm, after 3-days in the dark at 30 °C. Cardinal temperature requirements for growth: minimum, 15 °C; maximum, 35 °C; and optimum, 30 °C (Figure 8). No growth was observed at 10 °C. Isolates produced a pink pigment in PDA medium at 35 °C.
Figure 8.
The effect of temperature on the mycelial growth of L. newvalleyensis after 3-days on PDA medium. Means followed by the same letter are not significantly different according to LSD test (p < 0.05).
Materials examined: Egypt, New Valley Governorate—large dark-brown lesions on leaves of date palm trees (Phoenix dactylifera), May 2020, A.M. Ismail, (holotype; a dry culture on pine needles: EGY H-240483); living culture ex-type: EGY20114.
Notes:Lasiodiplodia newvalleyensis is phylogenetically distinct from other species of Lsiodiplodia. It forms a basal clade comprised of L. nanpingensis, L. mahajangana, L. curvata, L. irregularis, L. pandanicola, L. magnoliae, L. chonburiensis, L. caatinguensis, L. exigua and L. americana. Morphologically, the unbranched and shorter paraphyses (14.9– 44.5 × 1.9–3.7 µm) of L. newvalleyensis make the latter distinct from L. nanpingensis (102 × 3.5 µm) [7], L. caatinguensis (31.1–60.2 × 2.1–5.0 μm) [5] and L. exigua (66 × 5 µm) [43]. Furthermore, the aseptate paraphyses of L. newvalleyensis distinguished it from 1-septate L. irregularis [44] and from L. mahajangana [45]. The curved shape of conidia of L. curvata distinguished it from L. newvalleyensis [44]. Moreover, L. newvalleyensis have longer conidia (17.2–26.7 × 10.5–13.3 µm) than L. caatinguensis (13–20.2 × 10.1–12.5 μm) [5]. In addition, the conidia dimensions of L. newvalleyensis (17.2–26.7 × 10.5–13.3 µm) are distinguishable from those of L. pandanicola (14–38 × 9–22 µm) [46] and L. magnoliae (24–30 × 11–15 μm) [47]. The conidia shape (obovoid to ellipsoid) and dimensions (17.2–26.7 × 10.5–13.3 µm) of L. newvalleyensis are also distinguishable from those of L. chonburiensis that has subglobose to oval conidia with dimensions 23 × 12 µm [46]. Lasiodiplodia newvalleyensis and L. americana share almost the same conidia characteristics; however, the later differs by its longer (90 × 2–3.5 µm) and 1–3-septate paraphyses [48].
3.4. Pathogenicity Tests on Seedlings and Leaves
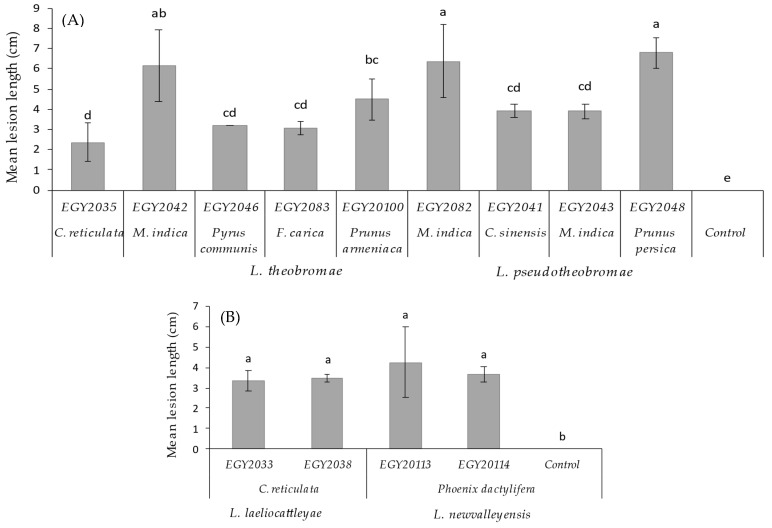
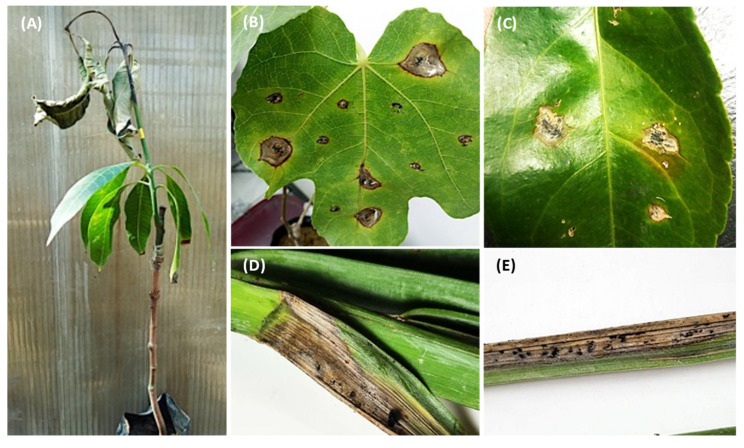
Pathogenicity tests revealed that all isolates were pathogenic to their hosts of origin to different degrees of severity. The control plants exhibited small zones of necrotic tissues due to wound reaction. Not all Lasiodiplodia isolates from the same species reacted in the same manner on the tested hosts. There was significant (p < 0.05) variation between isolates of L. theobromae and L. pseudotheobromae in terms of lesion length (Figure 9A). Out of all L. theobromae isolates, only EGY2082 and EGY2042 were aggressive on Mangifera indica, producing the largest lesions measuring 6.33 and 5.65cm (Figure 9A). EGY2048 was the most aggressive among L. pseudotheobromae isolates, causing lesions of 6.26 cm on Prunus persica (Figure 9A). The remaining L. theobromae and L. pseudotheobromae isolates induced smaller lesions that were not significantly different according to the LSD test (p < 0.05). Some isolates (EGY2048, EGY2082 and EGY20100) induced typical dieback symptoms on Mangifera indica in the early stage of infection, which progressed further with the fungal growth (upward and downward) and led to wilting and drying of the apical part and the terminal leaves, giving the scorched appearance (Figure 10A). The L. theobromae isolate (EGY2082) was pathogenic to F. carica and induced necrotic tissues similar to those observed on the origin host (Figure 10B). Both L. laeliocattleyae isolates (EGY2033 and EGY2038) were pathogenic to C. reticulata leaves (Figure 10B) with average lesion lengths of 3.27 and 3.49 cm, respectively, and were not statistically different (p < 0.05) from each other (Figure 9B). Additionally, the two isolates (EGY20113 and EGY20114) of the novel L. newvalleyensis species were highly pathogenic to Phoenix dactylifera leaves (Figure 10D,E) and produced lesions with average diameters of 4.44 and 3.91 cm, respectively (Figure 9B).
Figure 9.
Mean lesion size (mm) (y-axis) on stems (A) and leaves (B) of fruit trees inoculated with 9 isolates (6 of L. theobromae and 3 of L. pseudotheobromae) and 4 isolates (2 of L. laeliocattleyae and 2 of L. newvalleyensis) (x-axis). Data in these columns are the means of n = 9 lesions. Bars above the columns represent standard deviation of the mean. Columns bearing the same letters are not significantly different according to the LSD test (p < 0.05).
Figure 10.
Typical dieback symptoms on mango seedlings after 30 days of inoculation (A): necrotic lesions developed around the inoculated tissues of F. carica (B), C. reticulata (C) and Phoenix dactylifera (D), and black pycnidia developed on the necrotic area of Phoenix dactylifera (E).
4. Discussion
Based on the results of the current study, four species of Lasiodiplodia associated with diseases on different fruit trees were isolated and characterized. These were identified as L. theobromae, L. pseudotheobromae, L. laeliocattleya and the newly recognized species L. newvalleyensis. The new species was distinguished from other taxa in Lasiodiplodia based on the phylogenetic inferences of the ITS, tef1-α and tub2 and morphological characteristics. To our knowledge, this is the first report of L. newvalleyensis causing leaf lesions on Phoenix dactylifera in Egypt and worldwide.
Lasiodiplodia species do not only occur as latent endophytes in asymptomatic plants, but are also associated with different symptoms occurring on a variety of hosts, including stem-end rot, fruit rot, decline, cankers and dieback [3,49]. In Egypt, L. theobromae, previously known as Botryodiplodia theobromae, was considered as the main causal agent of fruit rot and dieback of mango [10]. In the current work, L. theobromae was the most commonly isolated species causing different kinds of symptoms on M. indica, C. reticulata, C. sinensis, F. carica, Prunus persica and Pyrus communis trees. This finding is supported by previous studies which showed that L. theobromae has the ability to target a wide variety of fruit and woody trees plants in Egypt [18,19], along with ornamental Ficus trees [21]. Lasiodiplodia theobromae was also reported to cause gummosis and dieback of Prunus persica in Egypt [50]. Very recently, L. theobromae was reported as a causal agent of dieback, branch cankers and gummosis on C. sinensis and C. aurantifolia in Egypt [20]. Similar results were reported, and L. theobromae was the most frequently isolated from M. indica in Western Australia and Brazil [51,52].
In our study, L. pseudotheobromae ranked second in terms of isolation frequency and was associated with leaf lesions and dieback of M. indica and C. sinensis, along with root rot on Prunus persica. This species has a worldwide distribution and causes mainly stem-end rot, dieback and cankers on a wide range of hosts [3,4,5,24,25,49,53,54,55,56]. It was reported to cause dieback in only mango trees in Egypt [3]. However, the current study reported the presence of L. pseudotheobromae on other trees in Egypt. Reports on various hosts in different geographical areas suggested that L. pseudotheobromae has a wide host range and that its distribution might extend to other plant hosts and areas [45]. The low frequently with which L. laeliocattleya was isolated from C. reticulata suggests that this species has a limited geographical distribution. However, it has previously been reported to be on mango trees in Egypt [3] and Peru [57] and on coconut and mango trees in Brazil [52,58].
The extensive phylogenetic Inference based on multiple gene sequences has played an important role in delimiting novel species in the genus Lasiodiplodia [7,25,59]. In this study, the use of combined ITS, tef1-α and tub2 sequence data enabled us to resolute the single cryptic species within L. theobromae species complex and provide novel clues into taxonomic novelties. The newly identified species was named as L. newvalleyensis, and its morphological description is supplemented. Several studies have demonstrated that using a single gene region is insufficient to delimit cryptic species [60,61,62], and therefore, to resolve species boundaries in the genus Lasiodiplodia, more than one gene region is required. This approach has revealed the presence of cryptic species in several genera in the family Botryosphaeriaceae. The multi-locus sequence data of ITS, tef1-α and tub2 were used to separate Lasiodiplodia species in this study. Several studies have relied on morphological characteristics such as conidia dimensions, morphology and morphology; the sizes of paraphyses; and DNA sequence data for identifying Lasiodiplodia species [7,44,46,47,48]. However, several morphological features can overlap [25,26,63] but are still complimentary tools when combined with DNA phylogeny to distinguish new species in Botryosphaeriaceae. In this study, the shapes and lengths of paraphyses were used to differentiate L. newvalleyensis from the closely related species (Figure 7). Burgess et al. relied on the septation of paraphyses to discriminate between Lasiodiplodia spp. and indicated that L. crassispora, L. gonubiensis and L. venezuelensis have septate paraphyses, whereas other species are aseptate [64]. However, in this study, septate paraphyses were observed for L. pseudotheobromae, as previously reported by Alves et al. [56]. Using a similar approach, Damm et al. distinguished L. plurivora from L. crassispora and L. venezuelensis based on the morphology of the paraphyses [65]. This was also followed by a study of Abdollahzadeh et al. who distinguished L. gilanensis from L. plurivora and L. hormozganensis from L. parva and L. citricola using the morphology of the paraphyses [25]. In addition, Ismail et al. relied on the morphology of the paraphyses to distinguish L. laeliocattleya from the phylogenetically related L. hormozganensis [3].
Culture characteristics have also played a role in distinguishing Lasiodiplodia species. Alves et al. discriminated L. parva and L. pseudotheobromae from L. theobromae based on the production of a pink pigment in culture [56]. In contrast, the findings of Abdollahzadeh et al. revealed that L. theobromae and other Lasiodiplodia species, with the exception of L. hormozganensis, produce pink pigment on PDA at 35 °C [25]. In the present study, L. newvalleyensis produced a dark-pink pigment in PDA after 4 days at 35 °C; the color become darker with age. Colonies of L. newvalleyensis covered the 90 mm plates after 3 d at the optimum temperature of 30 °C. This finding is supported by those reported in previous studies that the optimum growth temperature for Lasiodiplodia species ranges between 25 and 30 °C [66,67]. Moreover, L. newvalleyensis could not grow at 10 °C, which is in contrast with the observations made by Alves et al. [56] and those of Abdollahzadeh et al., who found that all studied Lasiodiplodia isolates grow at the same temperature [25]. Our results are corroborated by those of a study on the mycelial growth of L. viticola, which could not grow at 10 °C [68]. However, the recently described novel species L. guilinensis, L. huangyanensis, L. linhaiensis and L. ponkanicola showed the ability to grow at 10 °C [67]. Thus, culture characteristics are of limited value in species determination due to their variation between isolates of a given species.
All Lasiodiplodia species showed the ability to spread through the internal tissues above and below the points of inoculation, causing brown to black necrotic lesions (Figure 10). The upward and downward progression inside the apparently healthy tissues reflected the well-known endophytic nature of these fungi [68,69,70,71]. In our study, we could not compare the severity of certain species on their hosts due to the low number of isolates recovered from the same hosts. This was evident for the single isolates of L. theobromae obtained from Pyrus communis, M. indica, Prunus armeniaca, C. reticulata and F. carica. There was significant (p < 0.05) variation within isolates of L. pesudotheobromae and L. theobromae in terms of severity. Variation in severity among L. theobromae and L. brasiliensis was also reported [72]. Recent findings confirmed that isolates of L. theobromae are more virulent than D. seriata on grapevines in Mexico [73]. Our results indicated that L. theobromae was more aggressive than L. pesudotheobromae, which induced the largest lesions and severe dieback symptoms on M. indica. These results are in contrast with those obtained by Ismail et al., who demonstrated that L. pesudotheobromae was highly pathogenic to M. indica than L. theobromae [3]. Furthermore, Leala et al. confirmed that L. pesudotheobromae and L. theobromae are pathogenic to acid lime and valencia orange [20]. Therefore, the high-frequency isolation, together with the results of pathogenicity, led us to consider that L. pesudotheobromae and L. theobromae are important fungal pathogens in Egypt. The low incidence, together with the fact that the only two isolates of L. laeliocattleya induced the smallest lesions on C. reticulata, suggest that this species is of a little importance and does not contribute significantly to citrus diseases. Our implications are based on earlier reports which demonstrated that L. mahajagana was not a primary pathogen due to its low incidence and virulence on Terminalia catappa [45], and Fusicoccum bacilliforme is a weak pathogen on mango plants due to its low isolation frequency and the small lesions it produces on mango plants [74]. A recent study also confirmed our suggestion that only L. pesudotheobromae and L. theobromae have been reported on citrus in Egypt [20]. Likewise, it was stated that species of Lasiodiplodia were more virulent against citrus, L. pesudotheobromae being the most widely distributed in China [73]. The two isolates of the newly described species L. newvalleyensis showed pathogenic ability on the leaves of Phoenix dactylifera, and there was no significant (p < 0.05) difference among them in terms of severity [66].
To conclude, the studies demonstrated here added a new species and two new host records to the list of Lasiodiplodia species. Therefore, this is the first report of L. laeliocattleya on C. reticulata and L. newvalleyensis on Phoenix dactylifera in Egypt and worldwide. The L. laeliocattleya and the newly described species L. newvalleyensis might pose a major threat to citrus and date palm cultivations and other fruit trees in the reported area. Therefore, further studies are needed, including extensive surveys and pathogenicity assays to clarify the ecology and to highlight their relative roles in causing diseases on other hosts. The external and internal symptoms developed by Lasiodiplodia species can evidently reflect the capacity of inoculated fungi to cause diseases and to spread rapidly throughout the vascular tissues, even if their hosts are not subjected to stress factors.
Acknowledgments
Authors extend their appreciation to the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia, for supporting this research work through grant number GRANT488.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8111203/s1, Table S1: Best-fit model of evolution according to BIC; Table S2: Isolates obtained in this study and their origins; Table S3: SNP positions of ITS, tef, and tub2 genes.
Author Contributions
Conceptualization, A.M.I., D.M., E.S.E. and S.M.E.-G.; Investigation, E.S.E.; methodology, E.S.E., S.M.E.-G. and D.M.; M.I.A.; K.A.A.; software, A.M.I. and D.M.; writing—original draft preparation, A.M.I. and Z.I.; writing—review and editing, S.M.E.-G., A.M.I. and D.M. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All the data related to this study is mentioned in the manuscript.
Conflicts of Interest
There is no conflict of interest among the authors.
Funding Statement
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (GRANT488).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Phillips A., Alves A., Abdollahzadeh J., Slippers B., Wingfield M.J., Groenewald J., Crous P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2006;55:53–63. doi: 10.3114/sim0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freire F., Cardoso J., Viana F., Martins M. Status of Lasiodiplodia theobromae as a plant pathogen in Brazil. Essentia. 2011;12:53–71. [Google Scholar]
- 3.Ismail A., Cirvilleri G., Polizzi G., Crous P., Groenewald J., Lombard L. Lasiodiplodia species associated with dieback disease of mango (Mangifera indica) in Egypt. Australas. Plant Pathol. 2012;41:649–660. doi: 10.1007/s13313-012-0163-1. [DOI] [Google Scholar]
- 4.Netto M.S., Assunção I.P., Lima G.S., Marques M.W., Lima W.G., Monteiro J.H., de Queiroz Balbino V., Michereff S.J., Phillips A.J., Câmara M.P. Species of Lasiodiplodia associated with papaya stem-end rot in Brazil. Fungal Divers. 2014;67:127–141. doi: 10.1007/s13225-014-0279-4. [DOI] [Google Scholar]
- 5.Coutinho I., Freire F., Lima C., Lima J., Gonçalves F., Machado A., Silva A., Cardoso J. Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in northeastern Brazil. Plant Pathol. 2017;66:90–104. doi: 10.1111/ppa.12565. [DOI] [Google Scholar]
- 6.Machado A.R., Pinho D.B., Pereira O.L. Phylogeny, identification and pathogenicity of the Botryosphaeriaceae associated with collar and root rot of the biofuel plant jatropha curcas in Brazil, with a description of new species of Lasiodiplodia. Fungal Divers. 2014;67:231–247. doi: 10.1007/s13225-013-0274-1. [DOI] [Google Scholar]
- 7.Wang Y., Zhang Y., Bhoyroo V., Rampadarath S., Jeewon R. Multigene phylogenetics and morphology reveal five novel Lasiodiplodia species associated with blueberries. Life. 2021;11:657. doi: 10.3390/life11070657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crous P.W., Slippers B., Wingfield M.J., Rheeder J., Marasas W.F., Philips A.J., Alves A., Burgess T., Barber P., Groenewald J.Z. Phylogenetic lineages in the Botryosphaeriaceae. Stud. Mycol. 2006;55:235–253. doi: 10.3114/sim.55.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Punithalingam E. Plant Diseases Attributed to Botryodiplodia theobromae Pat. J. Cramer; Vaduz, Liechtenstein: 1980. [Google Scholar]
- 10.Ragab M., Sabet K., Dawood N. Botryodiplodia theobromae Pat. The cause of fruit rot and dieback of mango in A.R.E. Agrc. Res. Rev. Cairo. 1971;49:81–97. [Google Scholar]
- 11.Abdalla M., Safie M., El-Boghdady M., Soltan H. Fruit coating with certain plant oils for controlling post-harvest diseases of mangoes with special reference to stem end rot. Egypt. J. Appl. Sci. 2003;18:116–136. [Google Scholar]
- 12.Abd-El Ghani H.S., Fatouh H.M. First record of sugar beet root rot disease caused by Botryodiplodia theobromae in Egypt. Egypt. J. Phytopathol. 2005;33:107–108. [Google Scholar]
- 13.El-Goorani M.A., El-Meligi M.A., Elarosi H.M., Wasfy E.H. Ceratocystis black peduncle rot of banana in Egypt/il marciume bruno del peduncolo della banana da ceratocystis in Egitto. Phytopathol. Mediter. 1972;11:193–194. [Google Scholar]
- 14.Haggag W., Abou Rayya M., Kasim N. First report of a canker disease of walnut caused by Botryodiplodia theobromae in Egypt. Plant Dis. 2007;91:226. doi: 10.1094/PDIS-91-2-0226B. [DOI] [PubMed] [Google Scholar]
- 15.Diab M., Khalil I., Dawood N., El-Assiuty E. Ear and grain rot of maize caused by Botryodiplodia theobromae pathogens in Egypt. Minufiya J. Agric. Res. 1984;9:129–138. [Google Scholar]
- 16.Abo-El-Dahab M., El-Kazazz S., Shoeib A., El-Sheikh M. Biochemical changes in citrus fruits infected with Botryodiplodia theobromae. J. Agric. Sci. Mansoura Univ. 1992;7:3524–3532. [Google Scholar]
- 17.Haggag W.M., Nofal M. Improving the biological control of Botryodiplodia disease on some annona cultivars using single or multi-bioagents in Egypt. Biol. Control. 2006;38:341–349. doi: 10.1016/j.biocontrol.2006.02.010. [DOI] [Google Scholar]
- 18.Kamhawy M.A., Mahrous H.A., Shalaby M.S., El-Sharabasy S.M. Histopathology and control of Botryodiplodia theobromae rot of date palm off-shoot (Phoenix dactylifera L.) variety Zaghloul. Egypt. J. Agric. Res. 2005;83:1533–1546. [Google Scholar]
- 19.Barakat F., Abdel Salam A., Abada K., Korra A. In Pathogenicity of fungi associated with die-back of peach and some pome and stone fruit trees; Proceedings of the Sixth Congress of Phytopathology; Cairo, Egypt. 5–7 March 1990; pp. 311–323. [Google Scholar]
- 20.Leala M., El–Shahawy I., Tolba I. Characterization of Lasiodiplodia isolates obtained from acid lime and Valencia orange in Egypt. Azhar J. Agric. Res. 2021;46:55–67. doi: 10.21608/ajar.2021.245614. [DOI] [Google Scholar]
- 21.Rehab M., Rashed M., Ammar M., El-Morsy S. Dieback and sooty canker of ficus trees in Egypt and its control. Pak. J. Biol. Sci. 2014;17:364–371. doi: 10.3923/pjbs.2014.364.371. [DOI] [PubMed] [Google Scholar]
- 22.Ismail A., Cirvilleri G., Lombard L., Crous P., Groenewald J., Polizzi G. Characterisation of Neofusicoccum species causing mango dieback in Italy. J. Plant Pathol. 2013;95:549–557. [Google Scholar]
- 23.Slippers B., Boissin E., Phillips A., Groenewald J.Z., Lombard L., Wingfield M.J., Postma A., Burgess T., Crous P.W. Phylogenetic lineages in the Botryosphaeriales: A systematic and evolutionary framework. Stud. Mycol. 2013;76:31–49. doi: 10.3114/sim0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou Z., He W., Zhang Y. Does morphology matter in taxonomy of Lasiodiplodia? An answer from Lasiodiplodia hyalina sp. nov. Mycosphere. 2017;8:1014–1027. doi: 10.5943/mycosphere/8/2/5. [DOI] [Google Scholar]
- 25.Abdollahzadeh J., Javadi A., Goltapeh E.M., Zare R., Phillips A. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Pers.: Mol. Phylogeny Evol. Fungi. 2010;25:1–10. doi: 10.3767/003158510X524150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slippers B., Crous P.W., Denman S., Coutinho T.A., Wingfield B.D., Wingfield M.J. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia. 2004;96:83–101. doi: 10.1080/15572536.2005.11833000. [DOI] [PubMed] [Google Scholar]
- 27.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 28.Dellaporta S.L., Wood J., Hicks J.B. A plant DNA minipreparation: Version ii. Plant Mol. Biol. Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 29.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 30.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the pcr to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alhudaib K.A., El-Ganainy S.M., Almaghasla M.I., Sattar M.N. Characterization and control of Thielaviopsis punctulata on date palm in Saudi Arabia. Plants. 2022;11:250. doi: 10.3390/plants11030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K., Stecher G., Kumar S. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K., Standley D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swofford D.L. Paup: Phylogenetic Analysis Using Parsimony (and Other Methods), 4.0 Beta. 2002. [(accessed on 25 July 2022)]. Available online: http://paup.csit.fsu.edu/
- 35.Hillis D.M., Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993;42:182–192. doi: 10.1093/sysbio/42.2.182. [DOI] [Google Scholar]
- 36.Ronquist F., Huelsenbeck J.P. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 37.Miller M., Pfeiffer W., Schwartz T. Cipres Science Gateway Survey. [(accessed on 25 July 2022)]. Available online: http://www.phylo.org.
- 38.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. Iq-tree 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalyaanamoorthy S., Minh B.Q., Wong T.K., Von Haeseler A., Jermiin L.S. Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chernomor O., Von Haeseler A., Minh B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snedecor G., Cochran W. Statistical Methods. 7th ed. Iowa State University Press; Ames, IA, USA: 1980. [Google Scholar]
- 43.Linaldeddu B.T., Deidda A., Scanu B., Franceschini A., Serra S., Berraf-Tebbal A., Zouaoui Boutiti M., Ben Jamâa M., Phillips A.J. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Divers. 2015;71:201–214. doi: 10.1007/s13225-014-0301-x. [DOI] [Google Scholar]
- 44.Wang Y., Lin S., Zhao L., Sun X., He W., Zhang Y., Dai Y.-C. Lasiodiplodia spp. Associated with Aquilaria Crassna in Laos. Mycol. Prog. 2019;18:683–701. doi: 10.1007/s11557-019-01481-7. [DOI] [Google Scholar]
- 45.Begoude B., Slippers B., Wingfield M.J., Roux J. Botryosphaeriaceae associated with Terminalia catappa in cameroon, south africa and madagascar. Mycol. Prog. 2010;9:101–123. doi: 10.1007/s11557-009-0622-4. [DOI] [Google Scholar]
- 46.Tibpromma S., Hyde K.D., McKenzie E.H., Bhat D.J., Phillips A.J., Wanasinghe D.N., Samarakoon M.C., Jayawardena R.S., Dissanayake A.J., Tennakoon D.S. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018;93:1–160. doi: 10.1007/s13225-018-0408-6. [DOI] [Google Scholar]
- 47.de Silva N.I., Phillips A.J., Liu J.-K., Lumyong S., Hyde K.D. Phylogeny and morphology of Lasiodiplodia species associated with Magnolia forest plants. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-50804-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S., Li G., Liu F., Michailides T.J. Novel species of Botryosphaeriaceae associated with shoot blight of pistachio. Mycologia. 2015;107:780–792. doi: 10.3852/14-242. [DOI] [PubMed] [Google Scholar]
- 49.Trakunyingcharoen T., Lombard L., Groenewald J.Z., Cheewangkoon R., To-Anun C., Crous P. Caulicolous Botryosphaeriales from Thailand. Pers. Mol. Phylogeny Evol. Fungi. 2015;34:87–99. doi: 10.3767/003158515X685841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khalid I.A. Pathological Studies on Lasiodiplodia theobromae the Causal Agent of Gummosis and Die-Back of Peach. Cairo University; Giza, Egypt: 2012. [Google Scholar]
- 51.Sakalidis M.L., Ray J.D., Lanoiselet V., Hardy G.E., Burgess T.I. Pathogenic Botryosphaeriaceae associated with Mangifera indica in the Kimberley region of Western Australia. Eur. J. Plant Pathol. 2011;130:379–391. doi: 10.1007/s10658-011-9760-z. [DOI] [Google Scholar]
- 52.Marques M.W., Lima N.B., de Morais M.A., Michereff S.J., Phillips A.J., Câmara M.P. Botryosphaeria, Neofusicoccum, Neoscytalidium and Pseudofusicoccum species associated with mango in Brazil. Fungal Divers. 2013;61:195–208. doi: 10.1007/s13225-013-0258-1. [DOI] [Google Scholar]
- 53.Li G., Liu F., Li J., Liu Q., Chen S. Characterization of Botryosphaeria dothidea and Lasiodiplodia pseudotheobromae from English Walnut in China. J. Phytopathol. 2016;164:348–353. doi: 10.1111/jph.12422. [DOI] [Google Scholar]
- 54.Dissanayake A.J., Zhang W., Mei L., Chukeatirote E., Yan J.Y., Li X., Hyde K.D. Lasiodiplodia pseudotheobromae causes pedicel and peduncle discolouration of grapes in China. Australas. Plant Dis. Notes. 2015;10:1–5. doi: 10.1007/s13314-015-0170-5. [DOI] [Google Scholar]
- 55.Phillips A., Alves A., Pennycook S., Johnston P., Ramaley A., Akulov A., Crous P. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Pers. Mol. Phylogeny Evol. Fungi. 2008;21:29–55. doi: 10.3767/003158508X340742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alves A., Crous P.W., Correia A., Phillips A. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 2008;28:1–13. [Google Scholar]
- 57.Rodríguez-Gálvez E., Guerrero P., Barradas C., Crous P.W., Alves A. Phylogeny and pathogenicity of Lasiodiplodia species associated with dieback of mango in Peru. Fungal Biol. 2017;121:452–465. doi: 10.1016/j.funbio.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 58.Rosado A.W.C., Machado A.R., Freire F.d.C.O., Pereira O.L. Phylogeny, identification, and pathogenicity of Lasiodiplodia associated with postharvest stem-end rot of coconut in Brazil. Plant Dis. 2016;100:561–568. doi: 10.1094/PDIS-03-15-0242-RE. [DOI] [PubMed] [Google Scholar]
- 59.Phillips A.J., Hyde K.D., Alves A., Liu J.-K.J. Families in Botryosphaeriales: A phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019;94:1–22. doi: 10.1007/s13225-018-0416-6. [DOI] [Google Scholar]
- 60.Huda-Shakirah A.R., Mohamed Nor N.M.I., Zakaria L., Leong Y.-H., Mohd M.H. Lasiodiplodia theobromae as a causal pathogen of leaf blight, stem canker, and pod rot of Theobroma cacao in Malaysia. Sci. Rep. 2022;12:8966. doi: 10.1038/s41598-022-13057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berraf-Tebbal A., Mahamedi A.E., Aigoun-Mouhous W., Špetík M., Čechová J., Pokluda R., Baranek M., Eichmeier A., Alves A. Lasiodiplodia mitidjana sp. nov. and other Botryosphaeriaceae species causing branch canker and dieback of Citrus sinensis in Algeria. PLoS ONE. 2020;15:e0232448. doi: 10.1371/journal.pone.0232448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang L., Li H., Zhou L., Chen F. Lasiodiplodia pseudotheobromae causes stem canker of chinese hackberry in China. J. For. Res. 2020;31:2571–2580. doi: 10.1007/s11676-019-01049-x. [DOI] [Google Scholar]
- 63.Pennycook S., Samuels G. Botryosphaeria and Fusicoccum species associated with ripe fruit rot of Actinidia deliciosa (kiwifruit) in New Zealand. Mycotaxon. 1985;24:445–458. [Google Scholar]
- 64.Burgess T.I., Barber P.A., Mohali S., Pegg G., de Beer W., Wingfield M.J. Three new Lasiodiplodia spp. From the tropics, recognized based on DNA sequence comparisons and morphology. Mycologia. 2006;98:423–435. doi: 10.1080/15572536.2006.11832677. [DOI] [PubMed] [Google Scholar]
- 65.Damm U., Crous P.W., Fourie P.H. Botryosphaeriaceae as potential pathogens of Prunus species in South africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia. 2007;99:664–680. doi: 10.1080/15572536.2007.11832531. [DOI] [PubMed] [Google Scholar]
- 66.Xiao X., Wang W., Crous P., Wang H., Jiao C., Huang F., Pu Z., Zhu Z., Li H. Species of Botryosphaeriaceae associated with citrus branch diseases in China. Pers. Mol. Phylogeny Evol. Fungi. 2021;47:106–135. doi: 10.3767/persoonia.2021.47.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kenfaoui J., Lahlali R., Mennani M., Radouane N., Goura K., El Hamss H., El Ghadraoui L., Fontaine F., Tahiri A., Barka E.A. Botryosphaeria dieback (Lasiodiplodia viticola): An imminent emerging threat to the moroccan vineyards. Plants. 2022;11:2167. doi: 10.3390/plants11162167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moubasher H.A., Balbool B.A., Helmy Y.A., Alsuhaibani A.M., Atta A.A., Sheir D.H., Abdel-Azeem A.M. Insights into asparaginase from endophytic fungus Lasiodiplodia theobromae: Purification, characterization and antileukemic activity. Int. J. Environ. Res. Public Health. 2022;19:680. doi: 10.3390/ijerph19020680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salvatore M.M., Alves A., Andolfi A. Secondary metabolites of Lasiodiplodia theobromae: Distribution, chemical diversity, bioactivity, and implications of their occurrence. Toxins. 2020;12:457. doi: 10.3390/toxins12070457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slippers B., Wingfield M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007;21:90–106. doi: 10.1016/j.fbr.2007.06.002. [DOI] [Google Scholar]
- 71.Pérez C.A., Wingfield M.J., Slippers B., Altier N.A., Blanchette R.A. Endophytic and canker-associated Botryosphaeriaceae occurring on non-native eucalyptus and native Myrtaceae trees in Uruguay. Fungal Divers. 2010;41:53–69. doi: 10.1007/s13225-009-0014-8. [DOI] [Google Scholar]
- 72.Nur-Shakirah A.O., Khadijah M.S., Kee Y.J., Huda-Shakirah A.R., Mohd Hafifi A.B., Nurul-Aliyaa Y.A., Chew B.L., Zakaria L., Mohamed Nor N.M.I., Sreeramanan S. Characterization of Lasiodiplodia species causing leaf blight, stem rot and fruit rot of fig (Ficus carica) in Malaysia. Plant Pathol. 2022;71:1594–1605. doi: 10.1111/ppa.13580. [DOI] [Google Scholar]
- 73.Úrbez-Torres J., Leavitt G., Guerrero J., Guevara J., Gubler W. Identification and pathogenicity of Lasiodiplodia theobromae and Diplodia seriata, the causal agents of bot canker disease of grapevines in Mexico. Plant Dis. 2008;92:519–529. doi: 10.1094/PDIS-92-4-0519. [DOI] [PubMed] [Google Scholar]
- 74.Jacobs R. Characterisation of Botryosphaeria Species from Mango in South Africa. University of Pretoria; Pretoria, South Africa: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data related to this study is mentioned in the manuscript.