Abstract
Fruit rot of cucurbits caused by several pathogenic fungi has become an important postharvest disease worldwide. In 2022, fruit rot on watermelon (Citrullus lanatus) and muskmelon (Cucumis melo) was observed during the postharvest storage phase in the Chiang Mai and Phitsanulok Provinces of northern Thailand. These diseases can lead to significant economic losses. This present study was conducted to isolate the causal agent of fungi in lesions of fruit rot. A total of four fungal isolates were obtained, of which two isolates (SDBR-CMU422 and SDBR-CMU423) were obtained from rot lesions of watermelons, while the remaining isolates (SDBR-CMU424 and SDBR-CMU425) were obtained from rot lesions of muskmelons. All fungal isolates were identified using both morphological characteristics and molecular analyses. Morphologically, all isolated fungal isolates were classified into the genus Fusarium. Multi-gene phylogenetic analyses of a combination of the translation elongation factor 1-alpha (tef-1), calmodulin (cam), and RNA polymerase second largest subunit (rpb2) genes reveled that four fungal isolates belonged to the Fusarium incarnatum–equiseti species complex and were distinct from all other known species. Thus, we have described them as two new species, namely F. citrullicola (SDBR-CMU422 and SDBR-CMU423) and F. melonis (SDBR-CMU424 and SDBR-CMU425). A full description, illustrations, and a phylogenetic tree indicating the position of both new species have been provided. Moreover, pathogenicity tests were subsequently performed and the results showed that F. citrullicola and F. melonis caused symptoms of fruit rot on inoculated watermelon and muskmelon fruits, respectively. Notably, this outcome was indicative of the symptoms that appeared during the postharvest storage phase. To our knowledge, two new pathogenic fungi, F. citrullicola and F. melonis, are new causal agents of watermelon and muskmelon fruit rot, respectively. Importantly, these findings provide valuable information for the development of effective strategies for the monitoring and prevention of these diseases.
Keywords: cucurbit, fruit rot, fungal disease, new species, pathogen identification, taxonomy
1. Introduction
Watermelon [Citrullus lanatus (Thunb.) Mats. & Nakai] and muskmelon (Cucumis melo L.) are both cucurbit species that belong to the family Cucurbitaceae [1]. Both crops are extensively cultivated in temperate, subtropical, and tropical regions throughout the world [2,3]. Both fruits have been described as healthy food choices for human consumption [4,5]. The primary nutritional components found in watermelon and muskmelon fruits include amino acids, ascorbic acid, β-carotene, carbohydrates, fatty acids, flavonoids, minerals, potassium, sugars, vitamins, and a number of bioactive compounds [2,5,6,7,8]. They also possess beneficial medicinal properties such as analgesic, anticancer, anti-inflammatory, antimicrobial, antioxidant, antiulcer, and hepatoprotective properties [9,10]. More interestingly, the watermelon fruit is a great natural source of lycopene and also principally contains about 93% water [2]. In 2020, China is known to be the largest producer of watermelon and melon (including muskmelon) fruits in the global market, followed by Turkey, India, and Iran [11]. In Southeast Asia, Indonesia led in melon production in 2020, followed by the Lao People’s Democratic Republic and the Philippines [11]. In addition, Vietnam led Southeast Asia in watermelon production in 2020, followed by Indonesia, the Lao People’s Democratic Republic, and Thailand [11]. Currently, muskmelon has emerged as one of the economically important crops in Thailand.
Several diseases caused by microorganisms (bacteria, fungi, and viruses) can have negative effects on cucurbit plants throughout both the growing period and the postharvest period [4,12,13,14]. Fruit rot disease is one of the most typical preharvest and postharvest diseases of cucurbit fruits in Thailand and worldwide [4,15,16,17,18,19]. This disease has caused huge losses through reductions in harvest yields and lowered standards of quality, both of which can have a significant negative economic impact [20,21,22]. Previous studies indicate that the fungal species belonging to the genera Alternaria, Didymella, Epicoccum, Fusarium, Lasiodiplodia, Myrothecium, Penicillium, Phomopsis, Phytophthora, Pythium, Rhizoctonia, and Sclerotium have been reported as causal agents of fruit rot in cucurbits (cantaloupes, cucumbers, melons, pumpkins, squashes, and watermelons) [4,15,17,23,24,25,26]. The demand for watermelon and muskmelon fruits has risen due to the rapid growth of the world’s population and an increased interest in pursuing a healthy lifestyle. Therefore, plantation areas dedicated to the cultivation of both watermelon and other melon plants have increased significantly. Conversely, the prevalence and severity of some fungal-based diseases have also increased when plants have been grown in sub-optimal locations [4,27,28]. In Thailand, the major areas for watermelon and melon cultivation are located in the northern region of the country, including Chiang Mai, Kamphaeng Phet, Phayao, Phichit, Phitsanulok, and Sukhothai Provinces. However, there have been relatively few studies on incidences of postharvest fruit rot of watermelon and muskmelon in Thailand. In 2022, fruit rot caused by fungi was observed on watermelon and muskmelon during the postharvest storage phase in Chiang Mai and Phitsanulok Provinces, respectively. The incidence of this disease ranged from 20 to 30% according to the number of fruits in each pallet box (100 fruits per pallet box). Therefore, this study aimed to isolate the causal fungal agents of these fruit rot diseases. The obtained fungi were identified and described by their morphological characteristics and through multigene phylogenetic analysis. Subsequently, a pathogenicity test was subsequently performed and Koch’s postulates were employed to evaluate the asymptomatic fruits of watermelon and muskmelon using the isolated fungi.
2. Materials and Methods
2.1. Sample Collection
Fruit rot disease on watermelon (C. lanatus) and muskmelon (C. melo) fruits was observed during the postharvest storage phase in Chiang Mai and Phitsanulok Provinces of northern Thailand in 2022. Ten fruits of each symptomatic watermelon and muskmelon from different locations were randomly collected, maintained in sterile plastic boxes, and carried to the laboratory within 48 h of collection. After being transferred to the laboratory, symptomatic fruits were examined using a stereo microscope (Nikon H55OS, Tokyo, Japan) and then stored in a plastic box with wet filter paper in order to stimulate sporulation.
2.2. Fungal Isolation
Fruits samples were processed for the isolation of fungal causal agents. Causal fungi were isolated from lesions using a single conidial isolation on 1.0% water agar containing 0.5 mg/l streptomycin under a stereo microscope according to the method described by Choi et al. [29]. The isolated plates were incubated at 25 °C in darkness for 24–48 h, and the germinated conidia were then transferred onto potato dextrose agar (PDA; Conda, Madrid, Spain) containing 0.5 mg/L streptomycin. Pure fungal isolates were deposited in the culture collection of the Sustainable Development of Biological Resources, Faculty of Science, Chiang Mai University (SDBR-CMU), Chiang Mai Province, Thailand.
2.3. Fungal Identification
2.3.1. Morphological Study
The morphological characteristics of fungal isolates were determined according to methods established by Crous et al. [30] and Wang et al. [31,32]. Colony characteristics, including colony morphology, pigmentation, and odor, were observed on PDA, oatmeal agar (OA; Difco, Le Pont de Claix, France), and synthetic nutrient-poor agar (SNA) after one week of incubation in the dark at 25 °C. Color notations were rated according to the color charts of Kornerup and Wanscher [33]. Micromorphological characteristics were identified using sterile water as a mounting medium under a light microscope (Nikon Eclipse Ni-U, Tokyo, Japan). Anatomical structure related to size data (e.g., chlamydospores, conidiophores, conidiogenous cells, conidia, and phialides) was based on at least 50 measurements of each structure using the Tarosoft (R) Image Frame Work program.
2.3.2. DNA Extraction and PCR Amplification and Sequencing
Genomic DNA of each fungal isolate was extracted from the fungal mycelia grown on PDA in darkness at 25 °C for five days using the Fungal DNA Extraction Kit (FAVORGEN, Ping-Tung, Taiwan) following the manufacturer’s protocol. The translation elongation factor 1-alpha (tef-1), calmodulin (cam), and RNA polymerase second largest subunit (rpb2) genes were amplified by the polymerase chain reaction (PCR) using EF1/EF2 primers [34], CAL-228F/CAL-2Rd primers [35] and RPB2-5F2/RPB2-7cR primers [36], respectively (Table 1). The amplification program of three genes was conducted in separate PCR reactions. The amplification process consisted of an initial denaturation step at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing steps at 60 °C for 50 s (tef-1), 59 °C for 30 s (cam) and 52 °C for 1 min (rpb2), and a final extension step at 72 °C for 1 min on a peqSTAR thermal cycler (PEQLAB Ltd., Fareham, UK). PCR products were checked on 1% agarose gel electrophoresis.
Table 1.
Details of primers and the obtained PCR product in this study.
| Gene | Primer Name | Primer Sequence | The Obtained Length (bp) | |||
|---|---|---|---|---|---|---|
| SDBR-CMU422 | SDBR-CMU423 | SDBR-CMU424 | SDBR-CMU425 | |||
| tef-1 | EF1 EF2 |
5′-ATGGGTAAGGARGACAAGAC-3′ 5′-GGARGTACCAGTSATCATG-3′ |
692 | 691 | 691 | 686 |
| cam | CAL-228F CAL-2Rd |
5′-GAGTTCAAGGAGGCCTTCTCCC-3′ 5′-TGRTCNGCCTCDCGGATCATCTC-3′ |
606 | 603 | 601 | 597 |
| rpb2 | RPB2-5F2 RPB2-7cR |
5′-GGGGWGAYCAGAAGAAGGC-3′ 5′-CCCATRGCTTGYTTRCCCAT-3′ |
1152 | 1148 | 1141 | 1131 |
2.3.3. Sequencing
PCR products were purified using the PCR Clean-Up Gel Extraction NucleoSpin® Gel (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. The purified PCR products were directly sequenced. Sequencing reactions were performed and the sequences were then automatically determined in a genetic analyzer at the 1st Base Company Co., Ltd., (Kembangan, Malaysia) using EF1/EF2, CAL-228F/CAL-2Rd and RPB2-5F2/RPB2-7cR primers for tef-1, cam, and rpb2, respectively.
2.3.4. Sequence Alignment and Phylogenetic Analyses
An analysis of the tef-1, cam, and rpb2 sequences was carried out with the use of similarity searches using the BLAST program available at NCBI (https://blast.ncbi.nlm.nih.gov, accessed on 5 August 2022). The sequences from this study, along with those obtained from previous studies and the GenBank database (with ≥60% query coverage and ≥85–100% sequence similarity) were selected and are listed in Table 2. Multiple sequence alignment was performed with MUSCLE [37] and improved where necessary using BioEdit v. 6.0.7 [38]. Phylogenetic analysis was carried out using combination datasets of tef-1, cam, and rpb2. Fusarium camptoceras CBS 193.65 and F. neosemitectum CBS 115476 within the F. camptoceras species complex (FCAMSC) were used as the outgroup. A phylogenetic tree was constructed under the maximum likelihood (ML) and Bayesian inference (BI) methods. The ML analysis was carried out using RAxML v7.0.3 on the GTRCAT model with 25 categories and 1000 bootstrap (BS) replications [39,40] via the online portal CIPRES Science Gateway v. 3.3 [41]. BI analysis was performed with MrBayes v3.2.6 software for Windows [42]. The best substitution models for BI and ML analyses were estimated using the jModelTest 2.1.10 [43] by employing the Akaike information criterion (AIC). Both ML and BI analyses were based on the GTR + I + G model. For BI analysis, six simultaneous Markov chains were run for one million generations with random initial trees, wherein every 1000 generations were sampled. A burn-in phase was employed to discard the first 2000 trees, while the remaining trees were used to construct the 50% majority-rule consensus phylogram with calculated Bayesian posterior probabilities (PP). The tree topologies were visualized in FigTree v1.4.0 [44].
Table 2.
Details of sequences used in the molecular phylogenetic analysis.
| Fungal Taxa | Strain/Isolate | GenBank Accession Number | Reference | ||
|---|---|---|---|---|---|
| tef-1 | cam | rpb2 | |||
| Fusarium aberrans | CBS 131385 T | MN170445 | MN170311 | MN170378 | [45] |
| F. aberrans | CBS 131387 | MN170446 | MN170312 | MN170379 | [45] |
| F. arcuatisporum | LC12147 T | MK289584 | MK289697 | MK289739 | [31] |
| F. arcuatisporum | LC11639 | MK289586 | MK289658 | MK289736 | [31] |
| F. brevicaudatum | NRRL 43638 T | GQ505665 | GQ505576 | GQ505843 | [46] |
| F. brevicaudatum | NRRL 43694 | GQ505668 | GQ505579 | GQ505846 | [46] |
| F. bubalinum | CBS 161.25 T | MN170448 | MN170314 | MN170381 | [45] |
| F. caatingaense | URM 6779 T | LS398466 | – | LS398495 | [47] |
| F. caatingaense | URM 6778 | LS398465 | – | LS398494 | [47] |
| F. cateniforme | CBS 150.25 T | MN170451 | MN170317 | MN170384 | [45] |
| F. citri | LC6896 T | MK289617 | MK289668 | MK289771 | [31] |
| F. citri | LC4879 | MK289615 | MK289665 | MK289768 | [31] |
| F. citrullicola | SDBR-CMU422 T | OP020920 | OP020924 | OP020928 | This study |
| F. citrullicola | SDBR-CMU423 | OP020921 | OP020925 | OP020929 | This study |
| F. clavum | CBS 126202 T | MN170456 | MN170322 | MN170389 | [45] |
| F. clavum | NRRL 34032 | GQ505635 | GQ505547 | GQ505813 | [46] |
| F. coffeatum | CBS 635.76 T | MN120755 | MN120696 | MN120736 | [48] |
| F. coffeatum | CBS 430.81 | MN120756 | MN120697 | MN120737 | [48] |
| F. compactum | CBS 186.31 ET | GQ505648 | GQ505560 | GQ505826 | [46] |
| F. compactum | CBS 185.31 | GQ505646 | GQ505558 | GQ505824 | [46] |
| F. croceum | CBS 131777 T | MN170463 | MN170329 | MN170396 | [45] |
| F. croceum | NRRL 3020 | GQ505586 | GQ505498 | GQ505764 | [46] |
| F. duofalcatisporum | CBS 384.94 T | GQ505652 | GQ505564 | GQ505830 | [46] |
| F. duofalcatisporum | CBS 264.50 | GQ505651 | GQ505563 | GQ505829 | [46] |
| F. equiseti | CBS 307.94 NT | GQ505599 | GQ505511 | GQ505777 | [46] |
| F. equiseti | CBS 245.61 | GQ505594 | GQ505506 | GQ505772 | [46] |
| F. fasciculatum | CBS 131382 T | MN170473 | MN170339 | MN170406 | [45] |
| F. fasciculatum | CBS 131383 | MN170474 | MN170340 | MN170407 | [45] |
| F. flagelliforme | CBS 162.57 T | GQ505645 | GQ505557 | GQ505823 | [46] |
| F. flagelliforme | CBS 259.54 | GQ505650 | GQ505562 | GQ505828 | [46] |
| F. gracilipes | NRRL 43635 T | GQ505662 | GQ505573 | GQ505840 | [46] |
| F. guilinense | LC12160 T | MK289594 | MK289652 | MK289747 | [31] |
| F. guilinense | NRRL 32865 | GQ505614 | GQ505526 | GQ505792 | [46] |
| F. hainanense | LC11638 T | MK289581 | MK289657 | MK289735 | [31] |
| F. hainanense | LC12161 | MK289595 | MK289648 | MK289748 | [31] |
| F. humuli | CQ1039 T | MK289570 | MK289712 | MK289724 | [31] |
| F. humuli | CQ1032 | MK289568 | MK289710 | MK289722 | [31] |
| F. incarnatum | CBS 132.73 NT | MN170476 | MN170342 | MN170409 | [45] |
| F. incarnatum | NRRL 32866 | GQ505615 | GQ505527 | GQ505793 | [46] |
| F. ipomoeae | LC12165 T | MK289599 | MK289704 | MK289752 | [31] |
| F. ipomoeae | LC12166 | MK289600 | MK289706 | MK289753 | [31] |
| F. irregulare | LC7188 T | MK289629 | MK289680 | MK289783 | [31] |
| F. irregulare | LC12146 | MK289583 | MK289682 | MK289738 | [31] |
| F. lacertarum | NRRL 20423 T | GQ505593 | GQ505505 | GQ505771 | [46] |
| F. lacertarum | LC7942 | MK289643 | MK289696 | MK289797 | [31] |
| F. longicaudatum | CBS 123.73 T | MN170481 | MN170347 | MN170414 | [45] |
| F. longifundum | CBS 235.79 T | GQ505649 | GQ505561 | GQ505827 | [46] |
| F. luffae | LC12167 T | MK289601 | MK289698 | MK289754 | [31] |
| F. luffae | NRRL 32522 | GQ505612 | GQ505524 | GQ505790 | [46] |
| F. melonis | SDBR-CMU424 T | OP020922 | OP020926 | OP020930 | This study |
| F. melonis | SDBR-CMU425 | OP020923 | OP020927 | OP020931 | This study |
| F. monophialidicum | NRRL 54973 T | MN170483 | MN170349 | MN170416 | [45] |
| F. mucidum | CBS 102395 T | MN170485 | MN170351 | MN170418 | [45] |
| F. mucidum | CBS 102394 | MN170484 | MN170350 | MN170417 | [45] |
| F. multiceps | CBS 130386 T | GQ505666 | GQ505577 | GQ505844 | [46] |
| F. nanum | LC12168 T | MK289602 | MK289651 | MK289755 | [31] |
| F. nanum | LC1384 | MK289611 | MK289661 | MK289764 | [31] |
| F. neoscirpi | CBS 610.95 T | GQ505601 | GQ505513 | GQ505779 | [46] |
| F. pernambucanum | URM 7559 T | LS398489 | – | LS398519 | [47] |
| F. pernambucanum | URM 6801 | LS398483 | – | LS398513 | [47] |
| F. persicinum | CBS 479.83 T | MN170495 | MN170361 | MN170428 | [45] |
| F. persicinum | CBS 131780 | MN170496 | MN170362 | MN170429 | [45] |
| F. scirpi | CBS 447.84 NT | GQ505654 | GQ505566 | GQ505832 | [46] |
| F. scirpi | CBS 448.84 | GQ505592 | GQ505504 | GQ505770 | [46] |
| F. serpentinum | CBS 119880 T | MN170499 | MN170365 | MN170432 | [45] |
| F. sulawesiense | InaCC F940 T | LS479443 | LS479422 | LS479855 | [49] |
| F. sulawesiense | Indo186 | LS479449 | LS479426 | LS479864 | [49] |
| F. tanahbumbuense | InaCC F965 T | LS479448 | LS479432 | LS479863 | [49] |
| F. tanahbumbuense | NRRL 34005 | GQ505629 | GQ505541 | GQ505807 | [46] |
| F. toxicum | CBS 406.86 T | MN170508 | MN170374 | MN170441 | [45] |
| F. toxicum | CBS 219.63 | MN170507 | MN170373 | MN170440 | [45] |
| F. camptoceras | CBS 193.65 ET | MN170450 | MN170316 | MN170383 | [45] |
| F. neosemitectum | CBS 189.60 T | MN170489 | MN170355 | MN170422 | [45] |
Note: species obtained in this study are in bold. Superscript “T”, “ET”, and “NT” represents ex-type, epi-type, and neotype species, respectively. “–” represents the absence of sequence data in GenBank.
2.4. Pathogenicity Tests
Asymptomatic commercial watermelon and muskmelon fruits were carefully washed, and the surfaces were disinfected by immersion in 1.5% (v/v) sterile sodium hypochlorite solution for 5 min. They were then subsequently washed three times with sterile distilled water. The surface disinfected fruits were then air-dried at room temperature (25 ± 2 °C) for 10 min [50]. After being air-dried, a uniform wound (5 pores, 1 cm in depth and 1 mm in width) was made at the equator of each fruit using aseptic needles [4]. Conidial suspensions of all fungal isolates were prepared from each fungal culture grown on PDA at 25 °C for two weeks and suspended in sterile distilled water. The suspension was filtered through two layers of sterile cheesecloth, diluted in distilled water with 0.05% (v/v) Tween 20, and adjusted to 1 × 106 conidia/mL using a hemacytometer. Five hundred microliters of the conidial suspension was dropped onto the wounded fruits. Accordingly, control fruits were also wounded and treated with sterile distilled water. Each fruit was then placed in a separate sterile plastic box (26 cm × 35.5 cm × 20 cm) at conditions of 80% relative humidity. The plastic boxes were stored in a growth chamber at 25 °C under a 12 h period of light for one week. Ten replications were conducted for each treatment. The experiments were independently repeated twice. The disease severity score was employed to evaluate the specimens following the method described by Safari et al. [51] with mild (1–25%), moderate (26–50%), severe (51–75%), and very severe (76–100%) degrees of infection for the damaged fruit areas. To authenticate the causal agent, the fungi were re-isolated from the lesions following the method described by Bika and Baysal-Gurel [52].
3. Results
3.1. Sample Collection and Disease Symptoms
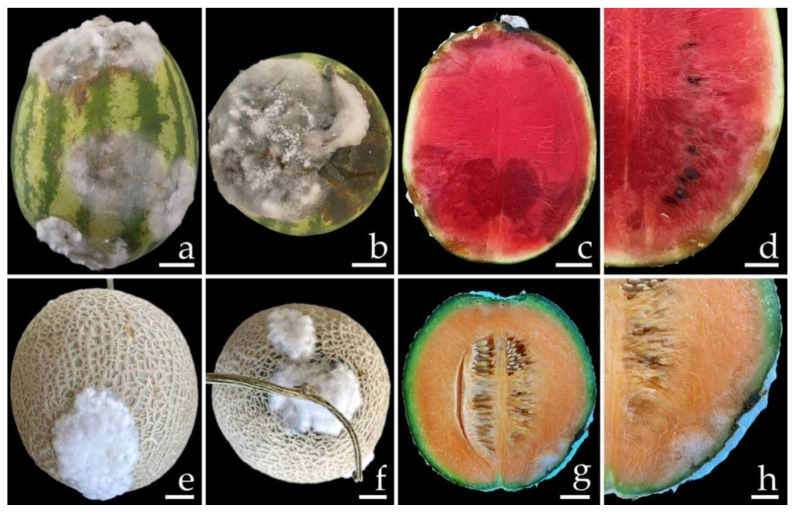
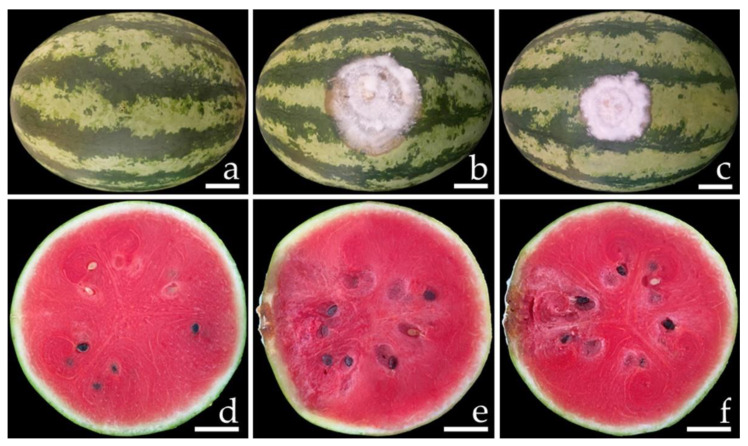
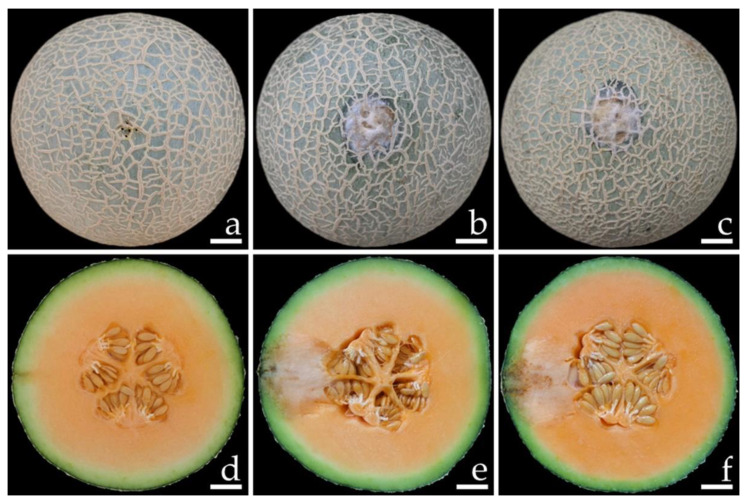
Samples of fruit rot on specimens of watermelon (C. lanatus) and muskmelon (C. melo) were collected from postharvest storage pallet boxes located in Chiang Mai and Phitsanulok Provinces of northern Thailand, respectively. The incidence of this disease ranged from 20 to 30% according to the number of fruits in each pallet box (100 fruits per pallet box). Symptoms on watermelon were characterized by the initial presence of small light-brown spots. These spots then expanded into irregular brown spots, and the epidermal tissue was covered with white mycelia tissue (Figure 1a,b). Disease symptoms on the muskmelon started at the top and base of the fruit appearing as brown spots surrounded by a bruise margin. Eventually, white mycelial masses covered the advanced lesions (Figure 1e,f). Lesions of both the watermelon and muskmelon fruit finally became widened and merged to cover the entire fruit, causing both of the infected fruits to appear bruised, ruptured, and decayed. The internal area of decay appeared to be clearly rotten and was surrounded by water-soaked tissue (Figure 1c,d,g,h).
Figure 1.
Natural symptoms of fruit rot disease on watermelon (a–d) and melon (e–h). The infected watermelon (a) and melon (e) fruits covered with white mycelium in the epidermal tissue. The top view of infected watermelon (b) and melon (f) fruits. A cross-section of a mature lesion of infected watermelon (c,d) and melon (g,h) fruits revealed the internal decayed area. Scale bars: (a–c) = 30 mm; (d) = 15 mm; (e–g) = 20 mm; (h) = 10 mm.
3.2. Fungal Isolation
A total of four fungal isolates were obtained in this study. Two fungal isolates, CMU422 and CMU423, were isolated from watermelon fruits rot collected from Chiang Mai Province and two isolates, CMU424 and CMU425, were isolated from muskmelon fruits rot collected from Phitsanulok Province. All fungal isolates were deposited at the SDBR-CMU under the accession numbers SDBR-CMU422, SDBR-CMU423, SDBR-CMU424, and SDBR-CMU425, respectively.
3.3. Morphological Study
Fungal colonies of each isolate were observed on three different agar media including PDA, OA, and SNA at 25 °C. After being incubated for one week, OA was found to be the best media by displaying the highest colony diameter of all four isolates. All four fungal isolates produced conidiophores, conidiogenous cells, chlamydospores, phialides, and conidia in all of the agar media. Based on the morphological characteristics, all fungal isolates were initially identified as belonging to the genus Fusarium [30,31,32,45]. The results obtained from morphological observation of the fungal colony and the micromorphological characters revealed that the isolate SDBR-CMU422 was similar to the isolate SDBR-CMU423, and that the isolate SDBR-CMU424 was similar to the isolate SDBR-CMU425. The fungal identification was then further confirmed through multi-gene phylogenetic analysis of a combination of the translation elongation factor 1-alpha (tef-1), calmodulin (cam), and the RNA polymerase second largest subunit (rpb2) genes.
3.4. Phylogenetic Results
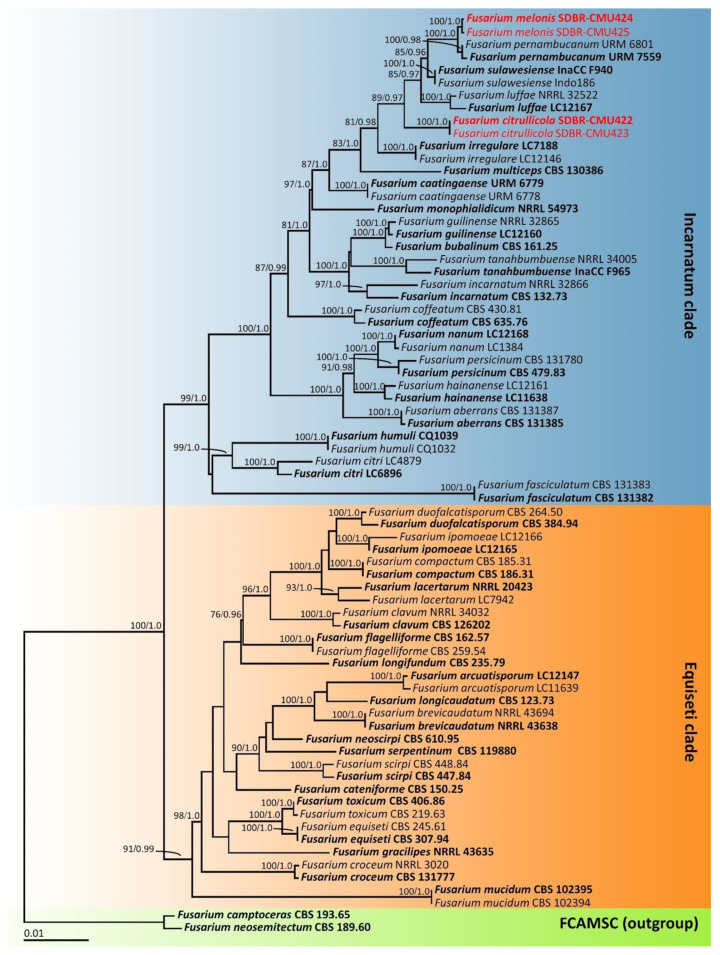
The tef-1, cam, and rpb2 sequences of each fungal isolate were amplified, sequenced, and deposited in the GenBank database (Table 2). The combined tef-1, cam, and rpb2 sequence dataset consisted of 73 taxa, while the aligned dataset was comprised of 2096 characters including gaps (tef-1: 1–669, cam: 670–1231 and rpb2: 1232–2096). ML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −9306.5763. The matrix contained 555 distinct alignment patterns with 5.24% undetermined characters or gaps. Estimated base frequencies were recorded as follows: A = 0.2810, C = 0.2397, G = 0.2704, T = 0.2089; while substitution rates were established as AC = 0.6704, AG = 5.9889, AT = 0.7165, CG = 0.8948, CT = 19.6030, GT = 1.0000. The gamma distribution shape parameter alpha value was equal to 0.2311 and the tree-length value was equal to 0.6021. In addition, the final average standard deviation of the split frequencies at the end of the total MCMC generations was calculated as 0.00708 through BI analysis. In terms of topology, the phylograms of the ML and BI analyses were found to be similar (data not shown). Therefore, the phylogram obtained from the ML analysis was selected and is presented in Figure 2. Our phylogenetic tree was constructed concordantly and is supported by previous studies [30,31,32,45]. A phylogram clearly separated the four fungal isolates obtained in this study into two monophyletic clades within the Incarnatum clade of the Fusarium incarnatum–equiseti species complex. The results indicate that the sequences of two fungal isolates, SDBR-CMU422 and SDBR-CMU423 (introduced as F. citrullicola), were clearly separated from the previously known Fusarium species in the Incarnatum clade with a high support value (100% BS and 1.0 PP). Moreover, two fungal isolates, SDBR-CMU424 and SDBR-CMU425 (introduced as F. melonis), formed a sister taxon to F. pernambucanum with high BS (100%) and PP (1.0) supports.
Figure 2.
Phylogram derived from maximum likelihood analysis of 73 taxa of the combined tef-1, cam, and rpb2 sequences. Fusarium camptoceras CBS 193.65 and F. neosemitectum CBS 115476 were used as the outgroup. The numbers above branches represent bootstrap percentages (left) and Bayesian posterior probabilities (right). Bootstrap values ≥ 75% and Bayesian posterior probabilities ≥ 0.95 are shown. The scale bar represents the expected number of nucleotide substitutions per site. Sequences of fungal species obtained in this study are in red. Type species are in bold.
3.5. Taxonomic Description
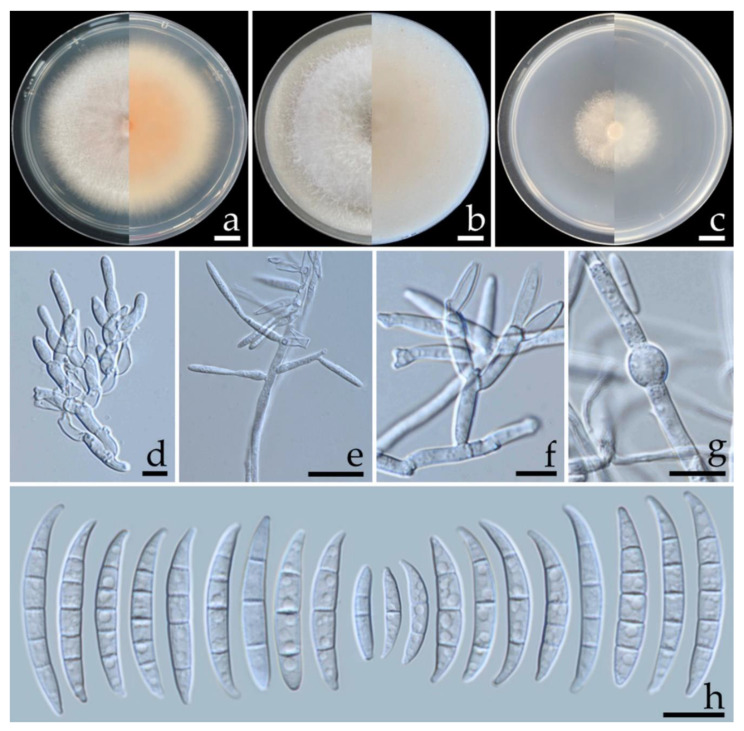
Fusarium citrullicola S. Khuna, J. Kumla & N. Suwannarach, sp. nov. (Figure 3).
Figure 3.
Fusarium citrullicola (SDBR-CMUS422; holotype). Colonies incubated at 25 °C for one week on PDA (a), OA (b), and SNA (c) (left, surface view and right, reverse view). Conidiophores on aerial mycelium (d). Lateral monophialides on aerial mycelium (e). Polyphialides on aerial mycelium (f). Chlamydospores (g). Aerial conidia (h). Scale bars: (a–c) = 10 mm; (d–h) = 10 µm.
MycoBank No.: 845955.
Etymology: ‘citrullicola’ referring to the Citrullus-inhibitor.
Holotype: THAILAND, Chiang Mai Province, Mueang District, 18°45′31″N, 98°58′20″E, on fruit rot lesion of Citrullus lanatus, 18 May 2022, S. Khuna, ex-type culture: SDBR-CMU422.
Description: Colonies on PDA, OA, and SNA were described at 25 °C after seven days of incubation. Colonies on PDA grew to 68.0–74.5 mm in diameter, slightly raised, aerial mycelia dense, colony margin entire, orange white (6A2) in the center, white at the margin; reverse light orange (6A5) in the center, white at the margin. Colonies on OA reached 75.0–85.0 mm in diameter, umbilicate, aerial mycelia dense, colony margin entire, surface white; reverse pale orange (5A3) in the center, white at the margin. Colonies on SNA attained a diameter of 45.5–51.0 mm, flat, aerial mycelia scant, colony margin entire, surface white; reverse white. Pigment and odor absent. No sporodochia were observed in all agar media. Conidiophores borne on aerial mycelium, 10–120 × 1.8–3.2 µm, unbranched, sympodial, or irregularly branched, bearing terminal or lateral phialides, with apical whorls of 1–3 phialides. Phialides mono- and polyphialidic, subulate to subcylindrical, sometimes proliferating percurrently, smooth and thin-walled, hyaline, 8.4–30.4 × 2.0–4.7 µm (av. ± SD: 16.8 ± 5.3 × 2.9 ± 0.5 µm). Chlamydospores abundant, intercalarily or terminal, globose, ellipsoid, smooth, thick-walled, hyaline, 0–4-septate, 4.7–15.6 × 4.6–14.4 µm (av. ± SD: 10.0 ± 2.7 × 7.7 ± 2.5 µm). Conidia falcate, curved dorsiventrally, sometimes straight, tapering towards both ends, smooth to slightly rough, hyaline, apical cell pointed to blunt, basal cell blunt to barely notched, 1–5-septate; 1-septate conidia 8.0–21.0 × 2.0–3.8 µm (av. ± SD: 14.9 ± 2.7 × 2.8 ± 0.4 µm); 2-septate conidia 13.9–24.6 × 2.1–3.9 µm (av. ± SD: 19.3 ± 2.4 × 3.1 ± 0.4 µm); 3-septate conidia 17.7–34.4 × 2.3–3.9 µm (av. ± SD: 26.6 ± 3.8 × 3.1 ± 0.4 µm); 4-septate conidia 26.7–35.3 × 2.4–4.4 µm (av. ± SD: 31.0 ± 2.3 × 3.7 ± 0.5 µm); 5-septate conidia 28.0–39.0 × 2.4–4.9 µm (av. ± SD: 32.7 ± 2.4 × 3.9 ± 0.4 µm).
Additional specimen examined: THAILAND, Chiang Mai Province, Mueang District, 18°45′31″N, 98°58′20″E, on fruit rot lesion of Citrullus lanatus, 18 May 2022, S. Khuna, SDBR-CMU423.
GenBank accession numbers: holotype SDBR-CMUS422 (tef-1: OP020920, cam: OP020924, rpb2: OP020928); additional specimen SDBR-CMUS423 (tef-1: OP020921, cam: OP020925, rpb2: OP020929).
Note: The colony characteristics of F. citrullicola on PDA were similar to those of F. sulawesiense that were found to have caused crown rot on banana fruit [31,49] and black rot on papaya fruit [53]. However, the growth of F. citrullicola displayed faster growth than F. sulawesiense (36.4–42.0 mm) on PDA at 25 °C [49]. The phylogenetic analyses of the combined tef-1, cam, and rpb2 sequences confirmed that F. citrullicola was clearly distinguishable from F. sulawesiense and the previously known Fusarium species in the Incarnatum clade with a high support value (Figure 2).
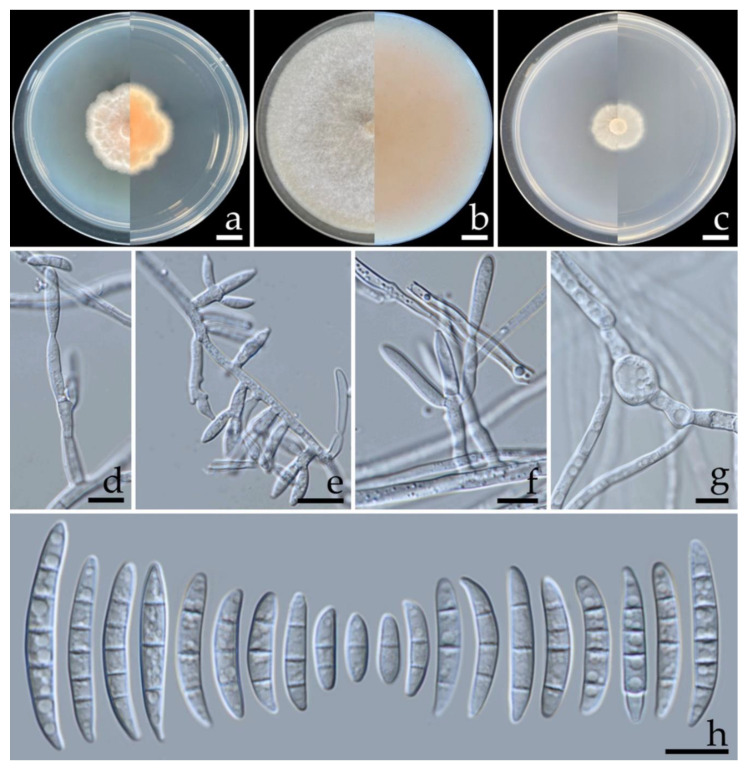
Fusarium melonis S. Khuna, J. Kumla & N. Suwannarach, sp. nov. (Figure 4).
Figure 4.
Fusarium melonis (SDBR-CMUS424; holotype). Colonies incubated at 25 °C for one week on PDA (a), OA (b), and SNA (c) (left, surface view and right, reverse view). Conidiophores on aerial mycelium (d). Mono- and polyphialides on aerial mycelium (e). Polyphialides on aerial mycelium (f). Chlamydospores (g). Aerial conidia (h). Scale bars: (a–c) = 10 mm; (d–h) = 10 µm.
MycoBank No.: 845956.
Etymology: ‘melonis’ referring to the host plant, Cucumis melo.
Holotype: THAILAND, Phitsanulok Province, Wang Thong District, 16°50′37″N, 100°36′00″E, on fruit rot lesion of Cucumis melo, 17 March 2022, S. Khuna, ex-type culture: SDBR-CMU424.
Description: Colonies on PDA, OA, and SNA were described at 25 °C after seven days of incubation. Colonies on PDA were 32.5–38.0 mm in diameter, flat, aerial mycelia scant, colony margin undulate, light orange (6A4) in the center, white at the margin; reverse orange (6A7) in the center, white at the margin. Colonies on OA grew to 85.0 mm in diameter, flat, aerial mycelia dense, colony margin entire, surface white; reverse light orange (6A4) in the center, white at the margin. Colonies on SNA reached a diameter of 15.0–20.5 mm, flat, aerial mycelia scant, colony margin entire, white; reverse white. Pigment and odor absent. No Sporodochia were observed in all agar media. Conidiophores borne on aerial mycelium, 13–85 × 1.9–4.2 µm, unbranched, sympodial branched, bearing terminal or lateral phialides, with apical whorls of 1–3 phialides. Phialides mono- and polyphialidic, subulate to sub-cylindrical, smooth and thin-walled, hyaline, 10.2–35.3 × 2.3–3.8 µm (av. ± SD: 18.6 ± 5.2 × 3.0 ± 0.4 µm). Chlamydospores abundant, intercalarily or terminal, globose, ellipsoid, smooth, thick-walled, hyaline, 0–2-septate, 5.3–15.3 × 4.7–12.8 µm (av. ± SD: 9.5 ± 2.2 × 7.9 ± 1.9 µm). Conidia ellipsoidal to falcate, slightly curved, sometimes straight, smooth to slightly rough, hyaline, apical cell pointed to blunt, basal cell obtuse to papillate, non-foot shaped, 1–5-septate; 1-septate conidia 11.7–22.8 × 2.6–4.0 µm (av. ± SD: 16.4 ± 2.1 × 3.2 ± 0.3 µm); 2-septate conidia 15.6–23.4 × 2.9–4.3 µm (av. ± SD: 18.8 ± 1.6 × 3.5 ± 0.3 µm); 3-septate conidia 17.9–31.3 × 3.1–4.6 µm (av. ± SD: 25.9 ± 3.2 × 3.6 ± 0.3 µm); 4-septate conidia 26.2–34.0 × 3.2–4.6 µm (av. ± SD: 30.4 ± 1.6 × 3.8 ± 0.3 µm); 5-septate conidia 26.7–45.8 × 3.2–4.8 µm (av. ± SD: 33.6 ± 4.0 × 4.0 ± 0.4 µm).
Additional specimen examined: THAILAND, Phitsanulok Province, Wang Thong District, 16°50′37″N, 100°36′00″E, on fruit rot lesion of Cucumis melo, 17 March 2022, S. Khuna, SDBR-CMU425.
GenBank accession numbers: holotype SDBR-CMUS424 (tef-1: OP020922, cam: OP020926, rpb2: OP020930); additional specimen SDBR-CMUS425 (tef-1: OP020923, cam: OP020927, rpb2: OP020931).
Note: Morphologically, F. melonis was similar to F. pernambucanum. However, the colony diameter of F. melonis on PDA at 25 °C (32.5–38.0 mm in diameter) was clearly smaller than F. pernambucanum (52.5–84 mm in diameter) [47]. The multi-gene phylogenetic analyses indicated that F. melonis clearly distinguished it from the other previously known F. incarnatum–equiseti species complexes and formed a sister clade to F. pernambucanum. However, a pairwise nucleotide comparison of tef-1 data also indicated that F. melonis differed from F. pernambucanum by 2.2% (15/690 bp). Furthermore, F. pernambucanum was isolated from fruit rot disease of muskmelons (C. melo) grown in China [54], fruit rot disease on melons collected from Brazil [55], leaf blight disease found on plum trees (Prunus salicina) grown in China [56] and insects (Aleurocanthus woglumi and Dactylopius opuntiae) indigenous to Brazil [47].
3.6. Pathogenicity Test
Conidia of all fungal isolates were used in this experiment. The initial symptoms were observed on both inoculated watermelon and muskmelon fruits two days after inoculation. Initially, small light-brown to brown spots appeared on the fruits. The lesions then enlarged rapidly and developed into brown to dark brown spots on the watermelon fruit and green bruised spots on the melon fruit, both of which were covered with white mycelia surrounding each lesion. After one week of incubation, the lesion diameters on the inoculated fruits were within the ranges of 6.0–6.5 and 2.5–3.0 cm on the watermelons (Figure 5b,c) and muskmelons (Figure 6b,c), respectively. The inoculated watermelons exhibited moderate infections indicated by symptoms of rot, whereas the muskmelons exhibited mild infections. A cross-section revealed that the internal lesion area appeared to be rotting and was surrounded by water-soaked tissue (Figure 5e,f and 6e,f). The diameters of the internal lesions on the watermelon and muskmelon fruits ranged from 6.5–7.5 and 4.0–4.5 cm, respectively. The lesions then spread to the entire fruit and coalesced within 12 and 14 days on the watermelon and muskmelon specimens, respectively, after the occurrence of necrosis. After that, the fruits became completely soft and rotten. These disease symptoms were similar to those seen during the postharvest storage phase. However, no symptoms of plant disease were observed in the inoculation treatments involving sterile distilled water among both wounded watermelon (Figure 5a,d) and muskmelon (Figure 6a,d) fruits. The fungi were re-isolated from symptomatic fruit tissue and then cultured on PDA in order to fulfill Koch’s postulates. The re-isolated fungi were identified as F. citrullicola and F. melonis.
Figure 5.
Pathogenicity test using F. citrullicola SDBR-CMU422 and SDBR-CMU423 on watermelon (Citrullus lanatus) fruits after one week of inoculation. Control fruit treated with water instead of inoculum (a,d). Fruit rot after inoculation of isolate SDBR-CMU422 (b,e) and isolate SDBR-CMU423 (c,f). Scale bars = 30 mm.
Figure 6.
Pathogenicity test using F. melonis SDBR-CMU424 and SDBR-CMU425 on melon (Cucumis melo) fruits after one week of inoculation. Control fruit treated with water instead of inoculum (a,d). Fruit rot after inoculation of isolate SDBR-CMU424 (b,e) and isolate SDBR-CMU425 (c,f). Scale bars = 20 mm.
4. Discussion
With regard to the genus Fusarium (Nectriaceae, Hypocreales), currently, there are more than 400 accepted species that have been divided into 23 species complexes [32,57]. The Fusarium species is known to cause several diseases including crown rot, root rot, wilt, fruit rot, leaf spot, stem rot, and seedling blight among cucurbits (cucumbers, muskmelons, pumpkins, squash, and watermelons) worldwide [14,15,23,54,58,59,60]. Traditionally, Fusarium species are mainly identified by macromorphological characteristics (colony morphology, pigmentation, and type of aerial mycelium), and micromorphological characteristics (the shapes and sizes of conidiophores, conidiogenous cells, macroconidia, microconidia, and the presence or absence of chlamydospores) [30,61,62]. However, morphological characteristics cannot be used to distinguish between the closely related species of Fusarium due to the wide range of morphological variations [30,61]. Therefore, it is essential to identify Fusarium species by applying a molecular approach. Ribosomal DNA [the internal transcribed spacer (ITS) and the large subunit (LSU) regions] and protein-coding [cam, tef-1, β-tubulin (tub2), RNA polymerase largest subunit (rpb1 and rpb2)] genes have provided researchers with a powerful tool in the identification of the Fusarium species [30,31,36,49,63,64,65]. Nevertheless, using only the ribosomal DNA gene did not resolve the identification of Fusarium at the species level [66,67]. Currently, a combination of morphological characteristics and multi-gene molecular phylogeny are being used for the accurate identification of the Fusarium species [30,31,32,45,47,36,49,63,64,65]. In this study, two new Fusarium species, namely F. citrullicola and F. melonis, were obtained from the rot lesions of watermelon and muskmelon fruits, respectively, that were collected from northern Thailand. All fungal species were identified according to their morphological and molecular characteristics in accordance with the identification methods established for the identification approach of Fusarium [30,31,32,47].
Both of our new fungal species belong to the F. incarnatum–equiseti species complex (FIESC). FIESC is a highly diverse group that is widely distributed. Accordingly, the majority of them are saprobes that are recognized as pathogens of plants, humans, and animals, and have been found in various environmental habitats [4,17,18,19,31,46,68,69]. Generally, identification based only on the morphological characteristics of FIESC is difficult because many species have similar outward appearances and display overlapping microscopic characteristics [61,70]. Therefore, molecular multi-gene phylogenetic analysis would be essential to accurately identify the FIESC species [31,45]. Prior to this study, this species complex comprised of 38 recognized phylogenetic species and has been separated into two main clades including the Equiseti clade (19 species) and the Incarnatum clade (19 species) [31,45,47,65,70]. Our multi-gene phylogenetic analyses revealed that two new species, F. citrullicola and F. melonis, formed distinct lineages from previously known species within the Incarnatum clade of FIESC. The different colony characteristics between the two new species indicate that F. citrullicola could more effectively grow on PDA and SNA when compared with F. melonis. On the OA medium, F. citrullicola generated an umbilicate colony, but F. melonis established a flat colony. Micromorphological characteristics indicate that F. citrullicola presents falcate conidia with tapering towards both ends, while F. melonis presents ellipsoidal to falcate conidia that are slightly curved. Additionally, F. citrullicola presents four septate chlamydospores, whereas F. melonis presents two septate chlamydospores. Phylogenetic analyses confirmed that F. citrullicola and F. melonis are actually different species. In addition, F. melonis formed a sister clade to F. pernambucanum. However, a nucleotide comparison of the tef-1 gene showed that F. melonis differed from F. pernambucanum by 2.2% (15/690 bp). Jeewon and Hyde [71] suggested that the nucleotide comparisons of reliable genes should be more than 1.5% different in order to justify the existence of a novel species. Therefore, F. melonis and F. pernambucanum can be considered different species.
To fulfill Koch’s postulates, pathogenicity tests were conducted on all isolates of F. citrullicola and F. melonis that had manifested the same symptoms as those observed during the postharvest storage phase. Therefore, F. citrullicola and F. melonis can be considered causal agents for watermelon and muskmelon fruit rot, respectively. Our results are supported by the findings of several previous studies, which indicated that Fusarium is an economically significant plant pathogen. Accordingly, some species of FIESC have been reported to cause fruit rot disease in various cucurbit plants worldwide [4,18]. For example, F. equiseti caused fruit rot disease on watermelon specimens collected in China [72], Malaysia [18], and the United States [24]. Ezrari et al. [16] found that F. equiseti caused pre- and postharvest fruit rot on zucchini plants (Cucurbita pepo) in Morocco. Notably, F. equiseti has been reported as a causal agent of postharvest fruit rot on both oriental melons and cantaloupes grown in Korea [73] and Thailand [4], respectively. Fusarium incarnatum was found as a causal agent of fruit rot on cucumbers grown in Mexico [17] and muskmelons cultivated in Thailand [19]. Fusarium pernambucanum caused fruit rot disease on muskmelons grown in China [54]. In Brazil, F. pernambucanum and F. sulawesiense were found to cause fruit rot on melons [55]. Additionally, other Fusarium species in the F. chlamydosporum species complex (F. chlamydosporum), the F. fujikuroi species complex (F. annulatum, F. moniliforme, F. proliferatum, and F. verticillioides), the F. solani species complex (F. falciforme, F. petroliphilum, and F. solani), the F. oxysporum species complex (F. kalimantanense), the F. sambucinum species complex (F. asiaticum, F. culmorum, F. graminearum, and F. sambucinum) and the F. tricinctum species complex (F. acuminatum) were also found to cause fruit rot on numerous cucurbits (cucumbers, melons, pumpkins, squashes, and watermelons) [22,25,55,73,74,75,76,77,78,79].
In Thailand, F. equiseti and F. incarnatum were found to be causal agents of rot among cantaloupes [4] and muskmelons [19], respectively. In addition, F. equiseti has been reported as a causal agent of muskmelon wilt disease [80]. In the current study, the disease symptoms observed in incidences of watermelon and muskmelon fruit rot caused by F. citrullicola and F. melonis, respectively, are similar to those that were caused by those known pathogens. Therefore, we have proposed that F. citrullicola and F. melonis be added as the causal agents of fruit rot on watermelons and muskmelons, respectively. However, there have been no prior reports of fruit rot disease on watermelons grown in Thailand. Thus, this study was determined to be the first investigative report on watermelon fruit rot in Thailand. Generally, watermelons and muskmelons are cultivated and harvested in Thailand throughout the cool to early wet seasons (November to June). Thus, during these seasons, fruit rot in watermelons and muskmelons can be found. Follow-up studies are needed to clarify the timing of the infections that occur in these fruits via fungal pathogens. This can be accomplished by monitoring the presence of the disease causal agents in these fruits at different stages of development in cultivation areas during both the pre-and postharvest processes, as well as during the postharvest storage period. Additional investigations will also be necessary to determine the disease’s inoculum source and the meteorological conditions that influence infection and disease development.
5. Conclusions
Fruit rot disease caused by the Fusarium species is one of the most important postharvest diseases of cucurbits in the world. In this study, two new pathogenic Fusarium species, namely F. citrullicola and F. melonis, were isolated from infected watermelon and melon fruits, respectively. Their identification was based on morphological characteristics and multi-gene phylogenetic analyses. The pathogenicity test of F. citrullicola and F. melonis revealed the same symptoms under artificial inoculation conditions as those observed during the postharvest storage phase. Thus, F. citrullicola and F. melonis have been proposed as new causal agents of watermelon and muskmelon fruit rot, respectively. Consequently, further studies involving the distribution of these diseases in other regions of Thailand, and the control of these diseases, will need to be conducted. In order to address the significant economic losses caused by this disease, it will be essential to develop effective monitoring and preventative strategies in the future.
Acknowledgments
The authors are grateful to Russell Kirk Hollis for his kind help in the English correction.
Author Contributions
Conceptualization, N.S. and J.K.; methodology, S.K., J.K., W.N. and N.S.; software, S.K., T.T. and J.K.; validation, S.K., J.K. and N.S.; formal analysis, S.K., J.K. and N.S.; investigation, S.K., T.T., J.K. and N.S.; resources, S.K., J.K. and N.S.; data curation, S.K., T.T., J.K. and N.S.; writing—original draft, S.K., J.K. and N.S.; writing—review and editing, S.K., J.K., T.T., W.N., S.L. and N.S.; supervision, N.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA sequence data obtained from this study have been deposited in GenBank under accession numbers; tef-1 (OP020920, OP020921, OP020922, OP020923), cam (OP020924, OP020925, OP020926, OP020927), and rpb2 (OP020928, OP020929, OP020930, OP020931).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The authors gratefully acknowledge the financial supported by Chiang Mai University Junior Research Fellowship Program (No. JRCMU2565_056), Chiang Mai University, Thailand.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saediman H., Alwi L.O., Rianse I.S., Taridala S.A.A., Salahuddin S., Indarsyih Y., Astuti R.W. Comparative profitability of melon and watermelon production in South Konawe District of Southeast Sulawesi. WSEAS Trans. Bus. Econ. 2020;17:933–939. doi: 10.37394/23207.2020.17.91. [DOI] [Google Scholar]
- 2.Assefa A.D., Hur O.S., Ro N.Y., Lee J.E., Hwang A.J., Kim B.S., Rhee J.H., Yi J.Y., Kim J.H., Lee H.S., et al. Fruit morphology, citrulline, and arginine levels in diverse watermelon (Citrullus lanatus) germplasm collections. Plants. 2020;9:1054. doi: 10.3390/plants9091054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kesh H., Kaushik P. Advances in melon (Cucumis melo L.) breeding: An update. Sci. Hortic. 2021;282:110045. doi: 10.1016/j.scienta.2021.110045. [DOI] [Google Scholar]
- 4.Nuangmek W., Aiduang W., Suwannarach N., Kumla J., Kiatsiriroat T., Lumyong S. First report of fruit rot on cantaloupe caused by Fusarium equiseti in Thailand. J. Gen. Plant Pathol. 2019;85:295–300. doi: 10.1007/s10327-019-00841-1. [DOI] [Google Scholar]
- 5.Manivannan A., Lee E.S., Han K., Lee H.E., Kim D.S. Versatile nutraceutical potentials of watermelon—A modest fruit loaded with pharmaceutically valuable phytochemicals. Molecules. 2020;25:5258. doi: 10.3390/molecules25225258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perkins-Veazie P., Davis A., Collins J.K. Watermelon: From dessert to functional food. Isr. J. Plant Sci. 2013;60:395–402. [Google Scholar]
- 7.Lester G.E., Hodges D.M. Antioxidants associated with fruit senescence and human health: Novel orange-fleshed non-netted honey dew melon genotype comparisons following different seasonal productions and cold storage durations. Postharv. Biol. Technol. 2008;48:347–354. doi: 10.1016/j.postharvbio.2007.11.008. [DOI] [Google Scholar]
- 8.Parle M., Singh K. Musk melon is eat-must melon. Int. Res. J. Pharm. 2011;2:52–57. [Google Scholar]
- 9.Maoto M.M., Beswa D., Jideani A.I.O. Watermelon as a potential fruit snack. Int. J. Food Prop. 2019;22:355–370. doi: 10.1080/10942912.2019.1584212. [DOI] [Google Scholar]
- 10.Vella F.M., Cautela D., Laratta B. Characterization of polyphenolic compounds in cantaloupe melon by-products. Foods. 2019;8:196. doi: 10.3390/foods8060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food and Agriculture Organization of the United Nations. [(accessed on 23 August 2022)]. Available online: https://www.fao.org/faostat/en/#home.
- 12.Keinath A.P. From native plants in Central Europe to cultivated crops worldwide: The emergence of Didymella bryoniae as a cucurbit pathogen. HortScience. 2011;46:532–535. doi: 10.21273/HORTSCI.46.4.532. [DOI] [Google Scholar]
- 13.Li P.F., Ren R.S., Yao X.F., Xu J.H., Babu B., Paret M.L., Yang X.P. Identification and characterization of the causal agent of gummy stem blight from muskmelon and watermelon in East China. J. Phytopathol. 2015;163:314–319. doi: 10.1111/jph.12277. [DOI] [Google Scholar]
- 14.Nuangmek W., Aiduang W., Suwannarach N., Kumla J., Lumyong S. First report of gummy stem blight caused by Stagonosporopsis cucurbitacearum on cantaloupe in Thailand. Can. J. Plant Pathol. 2018;40:306–311. doi: 10.1080/07060661.2018.1424038. [DOI] [Google Scholar]
- 15.Babadoost M., Zitter T.A. Fruit rots of pumpkin: A serious threat to the pumpkin industry. Plant Dis. 2009;93:772–782. doi: 10.1094/PDIS-93-8-0772. [DOI] [PubMed] [Google Scholar]
- 16.Ezrari S., Lahlali R., Radouane N., Tahiri A., Lazraq A. First report of Fusarium equiseti causing pre- and postharvest fruit rot on zucchini in Morocco. J. Plant Pathol. 2020;102:251. doi: 10.1007/s42161-019-00389-1. [DOI] [Google Scholar]
- 17.García-Estrada R.S., Márquez-Zequera I., Tovar-Pedraza J.M., Cruz-Lachica I. First report of cucumber fruit rot caused by Fusarium incarnatum in Mexico. Plant Dis. 2020;105:497. doi: 10.1094/PDIS-07-20-1533-PDN. [DOI] [PubMed] [Google Scholar]
- 18.Rahman M.Z., Ahmad K., Siddiqui Y., Saad N., Hun T.G., Hata E.M., Rashed O., Hossain M.I. First report of Fusarium equiseti, causing fruit rot disease of watermelon in Malaysia. Plant Dis. 2022;106:326. doi: 10.1094/PDIS-05-21-1027-PDN. [DOI] [PubMed] [Google Scholar]
- 19.Wonglom P., Sunpapao A. Fusarium incarnatum is associated with postharvest fruit rot of muskmelon (Cucumis melo) J. Phytopathol. 2020;168:204–210. doi: 10.1111/jph.12882. [DOI] [Google Scholar]
- 20.Li Y.G., Zhang R., Meng L., Ali E., Ji P., Zhang Q.F., Cui G.W. Occurrence of fruit rot of cantaloupe caused by Fusarium equiseti in China. Plant Dis. 2019;103:2683. doi: 10.1094/PDIS-03-19-0671-PDN. [DOI] [Google Scholar]
- 21.Lima E.N., Oster A.H., Bordallo P.N., Araújo A.A.C., Silva D.E.M., Lima C.S. A novel lineage in the Fusarium incarnatum-equiseti species complex is one of the causal agents of fusarium rot on melon fruits in Northeast Brazil. Plant Pathol. 2021;70:133–143. doi: 10.1111/ppa.13271. [DOI] [Google Scholar]
- 22.Oyedeji E.O., Arogundade O., Tairu F.M., Elum C.G. Identification and characterization of fungi pathogen causing fruit rot disease of watermelon (Citrullus lanatus) Arch. Phytopathol. Plant Prot. 2022;55:344–354. doi: 10.1080/03235408.2021.2019990. [DOI] [Google Scholar]
- 23.Tuttle McGrath M. Diseases of Cucurbits and their Management. In: Naqvi S.A.M.H., editor. Diseases of Fruits and Vegetables. Volume 1. Springer; Dordrecht, The Netherlands: 2004. pp. 455–510. [Google Scholar]
- 24.Li Y., Ji P. First report of fruit rot of watermelon caused by Fusarium equiseti in Georgia in the United States. Plant Dis. 2015;99:1272. doi: 10.1094/PDIS-10-14-1074-PDN. [DOI] [Google Scholar]
- 25.Rivedal H.M., Stone A.G., Johnson K.B. First report of Fusarium culmorum causing fruit rot of winter squash (Cucurbita maxima) in Oregon. Plant Dis. 2018;102:2659. doi: 10.1094/PDIS-06-18-0922-PDN. [DOI] [Google Scholar]
- 26.Suwannarach N., Khuna S., Kumla J., Tanruean K., Lumyong S. First report of Lasiodiplodia theobromae causing fruit rot on melon (Cucumis melo) in Thailand. Plant Dis. 2019;104:280. doi: 10.1094/PDIS-07-19-1454-PDN. [DOI] [Google Scholar]
- 27.Wilkinson K., Grant W.P., Green L.E., Hunter S., Jeger M.J., Lowe P., Medley G.F., Mills P., Phillipson J., Poppy G.M., et al. Infectious diseases of animals and plants: An interdisciplinary approach. Philos. Trans. R. Soc. B. 2011;366:1933–1942. doi: 10.1098/rstb.2010.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suwannarach N., Khuna S., Kumla J., Cheewangkoon R., Suttiprapan P., Lumyong S. Morphology characterization, molecular identification, and pathogenicity of fungal pathogen causing kaffir lime leaf blight in northern Thailand. Plants. 2022;11:273. doi: 10.3390/plants11030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi Y.W., Hyde K.D., Ho W.H. Single spore isolation of fungi. Fungal Divers. 1999;3:29–38. [Google Scholar]
- 30.Crous P.W., Lombard L., Sandoval-Denis M., Seifert K.A., Schroers H.-J., Chaverri P., Gené J., Guarro J., Hirooka Y., Bensch K., et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021;98:100116. doi: 10.1016/j.simyco.2021.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M.M., Chen Q., Diao Y.Z., Duan W.J., Cai L. Fusarium incarnatum-equiseti complex from China. Persoonia. 2019;43:70–89. doi: 10.3767/persoonia.2019.43.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M.M., Crous P.W., Sandoval-Denis M., Han S.L., Liu F., Liang J.M., Duan W.J., Cai L. Fusarium and allied genera from China: Species diversity and distribution. Persoonia. 2022;48:1–53. doi: 10.3767/persoonia.2022.48.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornerup A., Wanscher J.H. Methuen Handbook of Colour. 3rd ed. Eyre Methuen; London, UK: 1978. 252p [Google Scholar]
- 34.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 36.O’Donnell K., Sutton D.A., Rinaldi M.G., Sarver B.A.J., Balajee S.A., Schroers H.-J., Summerbell R.C., Robert V.A.R.G., Crous P.W., Zhang N., et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010;48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall T. Bioedit Version 6.0.7. 2004. [(accessed on 20 August 2022)]. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html.
- 39.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Miller M.A., Pfeiffer W., Schwartz T. Creating the cipres science gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; Manhattan, NY, USA: IEEE; pp. 1–8. [Google Scholar]
- 42.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambaut A. FigTree Tree Figure Drawing Tool Version 131. Institute of Evolutionary 623 Biology, University of Edinburgh; Edinburgh, Scotland: 2019. [(accessed on 10 August 2022)]. Available online: http://treebioedacuk/software/figtree/ [Google Scholar]
- 45.Xia J.W., Sandoval-Denis M., Crous P.W., Zhang X.G., Lombard L. Numbers to names—Restyling the Fusarium incarnatum-equiseti species complex. Persoonia. 2019;43:186–221. doi: 10.3767/persoonia.2019.43.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Donnell K., Sutton D.A., Rinaldi M.G., Gueidan C., Crous P.W., Geiser D.M. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 2009;47:3851–3861. doi: 10.1128/JCM.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos A.C.S., Trindade J.V.C., Lima C.S., Barbosa R.N., Costa A.F., Tiago P.V., Oliveira N.T. Morphology, phylogeny, and sexual stage of Fusarium caatingaense and Fusarium pernambucanum, new species of the Fusarium incarnatum-equiseti species complex associated with insects in Brazil. Mycologia. 2019;111:244–259. doi: 10.1080/00275514.2019.1573047. [DOI] [PubMed] [Google Scholar]
- 48.Lombard L., van Doorn R., Crous P.W. Neotypification of Fusarium chlamydosporum—A reappraisal of a clinically important species complex. Fungal Syst. Evol. 2019;4:183–200. doi: 10.3114/fuse.2019.04.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maryani N., Sandoval-Denis M., Lombard L., Crous P.W., Kema G.H.J. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia. 2019;43:48–69. doi: 10.3767/persoonia.2019.43.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Oliveira M.J., Laranjeira D., Câmara M.P.S., Laranjeira F.F., Armengol J., Michereff S.J. Effects of wounding, humidity, temperature, and inoculum concentrations on the severity of corky dry rot caused by Fusarium semitectum in melon fruits. Acta Sci. Agron. 2014;36:281–289. doi: 10.4025/actasciagron.v36i3.17656. [DOI] [Google Scholar]
- 51.Safari Z.S., Ding P., Nakasha J.J., Yuso S.F. Combining chitosan and vanillin to retain postharvest quality of tomato fruit during ambient temperature storage. Coatings. 2020;10:1222. doi: 10.3390/coatings10121222. [DOI] [Google Scholar]
- 52.Bika R., Baysal-Gurel F. Identification of Fusarium commune, the causal agent of postharvest zinnia meltdown disease in Tennessee. HortTechnology. 2021;31:432–439. doi: 10.21273/HORTTECH04795-21. [DOI] [Google Scholar]
- 53.Yi R.H., Lian T., Su J.J., Chen J. First report of internal black rot on Carica papaya fruit caused by Fusarium sulawesiense in China. Plant Dis. 2022;106:319. doi: 10.1094/PDIS-04-21-0721-PDN. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X.P., Xia J.W., Liu J.K., Zhao D., Kong L.G., Zhu X.P. First report of Fusarium pernambucanum causing fruit rot of muskmelon in China. Plant Dis. 2022;106:1997. doi: 10.1094/PDIS-07-21-1520-PDN. [DOI] [PubMed] [Google Scholar]
- 55.Araújo M.B., Moreira G.M., Nascimento L.V., Nogueira G.A., Nascimento S.R.C., Pfenning L.H., Ambrósio M.M.Q. Fusarium rot of melon is caused by several Fusarium species. Plant Pathol. 2021;70:712–721. doi: 10.1111/ppa.13328. [DOI] [Google Scholar]
- 56.Lu M., Zhang Y., Li Q., Huang S., Tang L., Chen X., Guo T., Mo J., Ma L. First report of leaf blight caused by Fusarium pernambucanum and Fusarium sulawesiense on plum in Sichuan, China. Plant Dis. 2022;106:2759. doi: 10.1094/PDIS-12-21-2672-PDN. [DOI] [PubMed] [Google Scholar]
- 57.Laraba I., McCormick S.P., Vaughan M.M., Geiser D.M., O’Donnell K. Phylogenetic diversity, trichothecene potential, and pathogenicity within Fusarium sambucinum species complex. PLoS ONE. 2021;16:e0245037. doi: 10.1371/journal.pone.0245037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pavlou G.C., Vakalounakis D.J., Ligoxigakis E.K. Control of root and stem rot of cucumber, caused by Fusarium oxysporum f. sp. radicis-cucumerinum, by grafting onto resistant rootstocks. Plant Dis. 2002;86:379–382. doi: 10.1094/PDIS.2002.86.4.379. [DOI] [PubMed] [Google Scholar]
- 59.Shanmugam V., Veena K.H., Jain S., Tripathi M., Aggarwal R., Singh A.K. First report of seedling blight caused by Fusarium solani on cucumber from India. J. Plant Pathol. 2016;98:677–697. [Google Scholar]
- 60.Gao X., Wang Y., Liu Y., Zhang M., Zhang W., Li Y. First report of leaf spot on cucumber caused by Fusarium incarnatum in China. Plant Dis. 2020;104:973. doi: 10.1094/PDIS-10-19-2102-PDN. [DOI] [Google Scholar]
- 61.Leslie J.F., Summerell B.A. In: The Fusarium Laboratory Manual. 1st ed. Ames I.A., editor. Blackwell Publishing Professional; New York, NY, USA: 2006. pp. 8–240. [Google Scholar]
- 62.Rahjoo V., Zad J., Javan-Nikkhah M., Mirzadi Gohari A., Okhovvat S.M., Bihamta M.R., Razzaghian J., Klemsdal S.S. Morphological and molecular identification of Fusarium isolated from maize ears in Iran. J. Plant Pathol. 2008;90:463–468. [Google Scholar]
- 63.Geiser D.M., Jiménez-Gasco M.M., Kang S., Makalowska I., Veeraraghavan N., Ward T.J., Zhang N., Kuldau G.A., O’Donnell K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004;110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 64.Nitschke E., Nihlgard M., Varrelmann M. Differentiation of eleven Fusarium spp. isolated from sugar beet, using restriction fragment analysis of a polymerase chain reaction-amplified translation elongation factor 1α gene fragment. Phytopathology. 2009;99:921–929. doi: 10.1094/PHYTO-99-8-0921. [DOI] [PubMed] [Google Scholar]
- 65.Jedidi I., Jurado M., Cruz A., Trabelsi M.M., Said S., González-Jaén M.T. Phylogenetic analysis and growth profiles of Fusarium incarnatum-equiseti species complex strains isolated from Tunisian cereals. Int. J. Food Microbiol. 2021;353:109297. doi: 10.1016/j.ijfoodmicro.2021.109297. [DOI] [PubMed] [Google Scholar]
- 66.O’Donnell K., Ward T.J., Robert V.A.R.G., Crous P.W., Geiser D.M., Kang S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica. 2015;43:583–595. doi: 10.1007/s12600-015-0484-z. [DOI] [Google Scholar]
- 67.Balajee S.A., Borman A.M., Brandt M.E., Cano J., Cuenca-Estrella M., Dannaoui E., Guarro J., Haase G., Kibbler C.C., Meyer W., et al. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: Where are we and where should we go from here? J. Clin. Microbiol. 2009;47:877–884. doi: 10.1128/JCM.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akram W., Ahmad A., Luo W., Yasin N.A., Wu T., Guo J., Wang Q., Li G. First report of stem and root rot of Chinese kale caused by Fusarium incarnatum-equiseti species complex in China. Plant Dis. 2019;103:1781. doi: 10.1094/PDIS-02-19-0261-PDN. [DOI] [Google Scholar]
- 69.Ismail S.I., Noor Asha N.A., Zulperi D. First report of Fusarium incarnatum-equiseti species complex causing leaf spot on rockmelon (Cucumis melo) in Malaysia. Plant Dis. 2021;105:1197. doi: 10.1094/PDIS-06-20-1380-PDN. [DOI] [PubMed] [Google Scholar]
- 70.Villani A., Moretti A., Saeger S.D., Han Z., Mavungu J.D.D., Soares C.M.G., Proctor R.H., Venâncio A., Lima N., Stea G., et al. A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatum-equiseti species complex. Int. J. Food Microbiol. 2016;234:24–35. doi: 10.1016/j.ijfoodmicro.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 71.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic am-biguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 72.Li Y.G., Song X.L., Wang X.Q., Zhang H., Tian S., Ji P. First report of fruit rot of watermelon caused by Fusarium equiseti in China. Plant Dis. 2018;102:1852. doi: 10.1094/PDIS-01-18-0199-PDN. [DOI] [Google Scholar]
- 73.Kim J.W., Kim H.J. Fusarium fruit rot of posthavest oriental melon (Cucumis melo L. var. makuwa Mak.) caused by Fusarium spp. Res. Plant Dis. 2004;10:260–267. doi: 10.5423/RPD.2004.10.4.260. [DOI] [Google Scholar]
- 74.Ikediugwu F.E.O., Ogieva W.O. Fruit rot of Citrullus lanatus in Nigeria caused by Fusarium solani. Trans. Br. Mycol. Soc. 1978;71:209–213. doi: 10.1016/S0007-1536(78)80100-4. [DOI] [Google Scholar]
- 75.Rampersad S.N. First report of Fusarium solani fruit rot of pumpkin (Cucurbita pepo) in Trinidad. Plant Dis. 2009;93:547. doi: 10.1094/PDIS-93-5-0547B. [DOI] [PubMed] [Google Scholar]
- 76.González V., Armengol J., Garcés-Claver A. First report of Fusarium petroliphilum causing fruit rot of butternut squash in Spain. Plant Dis. 2018;102:1662. doi: 10.1094/PDIS-11-17-1740-PDN. [DOI] [Google Scholar]
- 77.Li Y.G., Jiang W.Y., Jiang D., Wang R.T., Tian S., Ji P., Jiang B.W. First report of fruit rot on postharvest pumpkin caused by Fusarium acuminatum in China. Plant Dis. 2019;103:1035. doi: 10.1094/PDIS-11-18-1957-PDN. [DOI] [Google Scholar]
- 78.Hao F., Zang Q., Ding W., Ma E., Huang Y., Wang Y. First report of fruit rot of melon caused by Fusarium asiaticum in China. Plant Dis. 2021;105:1225. doi: 10.1094/PDIS-08-20-1857-PDN. [DOI] [PubMed] [Google Scholar]
- 79.Parra M.Á., Gómez J., Aguilar F.W., Martínez J.A. Fusarium annulatum causes Fusarium rot of cantaloupe melons in Spain. Phytopathol. Mediterr. 2022;61:269–277. doi: 10.36253/phyto-13454. [DOI] [Google Scholar]
- 80.Nuangmek W., Aiduang W., Kumla J., Lumyong S., Suwannarach N. Evaluation of a newly identified endophytic fungus, Trichoderma phayaoense for plant growth promotion and biological control of gummy stem blight and wilt of muskmelon. Front. Microbiol. 2021;12:634772. doi: 10.3389/fmicb.2021.634772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DNA sequence data obtained from this study have been deposited in GenBank under accession numbers; tef-1 (OP020920, OP020921, OP020922, OP020923), cam (OP020924, OP020925, OP020926, OP020927), and rpb2 (OP020928, OP020929, OP020930, OP020931).