Abstract
Mycobacterium tuberculosis H37Ra is an attenuated tubercle bacillus closely related to the virulent type strain M. tuberculosis H37Rv. Despite extensive study, the reason for the decreased virulence of M. tuberculosis H37Ra has not been determined. A genomic approach was therefore initiated to identify genetic differences between M. tuberculosis H37Rv and M. tuberculosis H37Ra as a means of pinpointing the attenuating mutation(s). Digestion with the rare-cutting restriction endonuclease DraI revealed two polymorphisms between the strains: a 480-kb fragment in M. tuberculosis H37Rv was replaced by two fragments of 220 and 260 kb in M. tuberculosis H37Ra, while there was a ∼7.9-kb DraI fragment in M. tuberculosis H37Ra that had no counterpart in M. tuberculosis H37Rv. As the M. tuberculosis insertion sequence IS6110 contains a single DraI restriction site, it was considered possible that these polymorphisms were the result of IS6110 transposition events in M. tuberculosis H37Ra, events that may have inactivated virulence genes. The 7.9-kb polymorphism was found to be due to the presence of the previously described H37Rv RvD2 deletion in M. tuberculosis H37Ra, with sequence analysis suggesting an IS6110-mediated deletion mechanism for loss of RvD2. Three other IS6110-catalyzed deletions from the M. tuberculosis H37Rv chromosome (RvD3 to RvD5) were also identified, suggesting that this mechanism plays an important role in genome plasticity in the tubercle bacilli. Comparative mapping and sequencing revealed that the 480-kb polymorphism was due to an IS6110 insertion in M. tuberculosis H37Ra near oriC. Complementation of M. tuberculosis H37Ra with a 2.9-kb restriction fragment from M. tuberculosis H37Rv that encompassed the IS6110 insertion did not increase the survival of recombinant M. tuberculosis H37Ra in mice. In conclusion, this study describes the presence and mechanisms of genomic variation between M. tuberculosis H37Ra and M. tuberculosis H37Rv, although the role that they play in the attenuation of M. tuberculosis H37Ra is unclear.
Identification of the virulence factors of Mycobacterium tuberculosis is a fundamental goal if new vaccines and antimycobacterial drugs against this pathogen are to be developed. Elucidation of such factors can be approached either by using molecular genetics to produce isogenic mutant strains that show decreased virulence or by studying attenuated strains that were generated by repeated subculture of virulent bacilli on laboratory media. Serial passage of M. tuberculosis H37 through media with different pHs allowed its dissociation into two forms: the virulent M. tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra (15). Although these strains are widely used in studies of mycobacterial virulence, the genetic reason for the decreased virulence of M. tuberculosis H37Ra has never been elucidated. As the original M. tuberculosis H37 was lost, we are left with comparison of M. tuberculosis H37Rv with M. tuberculosis H37Ra to identify the attenuating mutation(s).
The restriction endonuclease DraI has 38 sites on the 4.4-Mb M. tuberculosis H37Rv chromosome, with 16 of these due to the presence of IS6110, a 1.3-kb insertion sequence (IS) that carries a unique DraI site (3, 16). It has previously been reported that digestion of the genomes of M. tuberculosis H37Rv and M. tuberculosis H37Ra with the rare-cutting enzyme DraI and subsequent resolution by pulsed-field gel electrophoresis (PFGE) reveals a polymorphic site present only in M. tuberculosis H37Ra; a 480-kb DraI fragment present in the genome of M. tuberculosis H37Rv is replaced by fragments of 260 and 220 kb in M. tuberculosis H37Ra (4). It was tempting to speculate that this polymorphism was the result of an IS6110 insertion at this site in M. tuberculosis H37Ra, an insertion that may have inactivated virulence genes.
Other studies have previously tried to identify the genetic basis for the attenuation of M. tuberculosis H37Ra. Pascopella et al. (11) transformed a cosmid library of M. tuberculosis H37Rv into M. tuberculosis H37Ra and then selected for clones that were enriched for on passage through the mouse. A number of overlapping cosmid clones that gave enhanced growth and survival in the spleens of infected mice relative to that of wild-type M. tuberculosis H37Ra were identified. The locus that conferred this phenotype was termed ivg, for in vivo growth advantage. However, the ivg locus failed to restore full growth in the spleen, while no selective advantage was observed in the lung. Using probes generated from subtractive RNA hybridization, Kinger and Tyagi (8) isolated six clones that showed differential expression in M. tuberculosis H37Rv and M. tuberculosis H37Ra during in vitro culture. In a similar approach, Rindi et al. (13) described the use of mRNA differential display to identify genes that appeared to be down-regulated in M. tuberculosis H37Ra relative to M. tuberculosis H37Rv. However, in both cases it is unclear whether these genes play any role in vivo.
With the availability of the M. tuberculosis H37Rv sequence (3), a genomic approach was initiated to identify the molecular basis for the attenuation of M. tuberculosis H37Ra. A combination of clone mapping, sequence analysis, and comparative genomics was used to localize both the IS6110 insertion responsible for the 260- and 220-kb M. tuberculosis H37Ra polymorphism and a second DraI polymorphism. Complementation experiments were performed to assess the effects of the IS6110 insertion on virulence.
MATERIALS AND METHODS
DNA preparation, digestion, and PFGE.
M. tuberculosis H37Rv and M. tuberculosis H37Ra were grown in Middlebrook 7H9 medium (Difco) supplemented with albumin-dextrose-catalase (Difco) for 10 to 12 days at 37°C with mild shaking. Glycine was added to a final concentration of 0.2 M, and incubation was continued for a further 12 to 14 h before harvesting of cells at 4,000 × g for 30 min. The cells were washed once in 1× Tris-EDTA buffer (pH 7.6) and then encased in 0.5% low-melting-point agarose (Gibco BRL). The agarose blocks were then treated as previously described (12) to release genomic DNA. Prior to digestion, agarose plugs were washed for 2 to 4 h in 0.1% Triton X-100 (Serva) at 4°C and then in 1× medium buffer (10 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 50 mM NaCl) for 30 min at room temperature. Chromosomal DNA was digested overnight at 37°C with 20 U of DraI (Gibco BRL) in 1× medium buffer. Digestions were stopped by adding 1 ml of ice-cold 50 mM EDTA (pH 8.5).
DraI fragments were separated by migration through 1% agarose gels, at 8°C and 200 V, using a CHEF II Pulsaphor apparatus (Pharmacia) with a pulse ramp from 10 to 35 s for 24 h for fragments in the 0.2- to 1-Mb range.
Hybridizations.
DNA was transferred to Hybond C Extra nitrocellulose membranes (Amersham) by capillary transfer and fixed at 80°C for 2 h. Probes were labelled with [α-32P]dCTP (Amersham) by using the Prime-It II kit (Stratagene), and unincorporated nucleotides were removed by centrifugation through a P10 minicolumn (Bio-Rad). Hybridizations were carried out overnight at 37°C in 5× SSC (0.75 M NaCl, 0.75 M sodium citrate)–50% (vol/vol) formamide–10 mg of denatured salmon sperm DNA per ml (12). Membranes were washed at 37°C in 2× SSC–0.1% sodium dodecyl sulfate and 1× SSC–0.1% sodium dodecyl sulfate for 15 min each and then exposed to X-ray film (Amersham) at −80°C.
PCR.
Reaction mixtures contained 5 μl of 10× PCR buffer (100 mM β-mercaptoethanol, 600 mM Tris-HCl [pH 8.8]), 20 mM MgCl2, 170 mM (NH4)2SO4, 5 μl of 20 mM nucleotide mix, a 0.2 μM concentration of each primer, 10 to 50 ng of template DNA, 10% dimethyl sulfoxide, 0.5 U of Taq polymerase (Boehringer Mannheim), and sterile distilled water to 50 μl. Thermal cycling was performed on a PTC-100 amplifier (MJ Inc.) with an initial denaturation step of 90 s at 95°C, followed by 30 to 35 cycles of 30 s at 95°C, 1 min at 55°C, and 2 min at 72°C.
For expected products of greater than 3 kb, reactions were carried out with a GeneAmp XL PCR kit (Perkin Elmer). Reactions were set up according to the manufacturer's instructions, using 0.8 mM Mg acetate, a 0.2 μM concentration of each primer, and 10 to 30 ng of template DNA per reaction. Thermal cycles were 96°C for 1 min and then 15 two-step cycles at 94°C for 15 s and 70°C for 7 min, followed by 20 two-step cycles at 94°C for 15 s and 70°C for 8 min plus 15 s per cycle.
Sequencing reactions and computer analysis.
Sequencing reactions were performed with a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems) as previously described (1). Reaction mixtures were loaded onto 6% polyacrylamide gels, and electrophoresis was performed on a 373A or 377 automatic DNA sequencer (Applied Biosystems) for 10 to 12 h. Sequence data were transferred to a Digital Alpha station and analyzed by using the TED sequence analysis program from the Staden software package. The open reading frame (ORF) structure of the sequence was deduced by using the DIANA program (Sanger Centre, Cambridge, United Kingdom), with FASTA and BLAST used to compare sequences to those in in-house and public sequence databases.
Construction of recombinant M. tuberculosis H37Ra.
Digestion of the M. tuberculosis cosmid Y346 (12) with NaeI released a 2.9-kb fragment that encompassed the corresponding region of the H37Rv chromosome where IS6110 had inserted in M. tuberculosis H37Ra. The ORF structure of this region suggests that all necessary transcriptional signals for genes that may have been affected by the IS6110 insertion were present in the NaeI fragment. The 2.9-kb fragment was cloned into the mycobacterial attP-integrating shuttle vector pKINT (a gift from Douglas Young, Imperial College School of Medicine, London, United Kingdom) to generate pSG21. Electrocompetent M. tuberculosis H37Ra cells were generated as previously described (14). Four hundred microliters of electrocompetent bacilli and 5 to 10 μg of dialyzed pSG21 were mixed in 0.2-cm electroporation cuvettes (Bio-Rad) and electroporated by using a Bio-Rad Gene Pulser with settings of 2.5 kV, 25 μF, and 1,000 Ω. After electroporation and outgrowth, transformants were selected by plating on Middlebrook 7H11 medium (Difco) supplemented with oleic acid-albumin-dextrose-catalase (Difco) and 25 μg of kanamycin (Sigma) per ml. Kanamycin-resistant colonies appeared after 3 weeks and were checked for the presence of pSG21 by PCR.
Virulence studies.
Locally bred BALB/c mice (National Institute for Medical Research) were inoculated intravenously with approximately 106 CFU of M. tuberculosis H37Ra, M. tuberculosis H37Ra(pKINT), or M. tuberculosis H37Ra(pSG21). Mice were sacrificed at intervals, and their spleens and lungs were removed and weighed. The infected tissue was homogenized in 1 ml of chilled sterile saline by using a Beadbeater (Biospec Products) and 2.5-mm-diameter glass beads for 10 s at 5,000 beats/s. Serial 10-fold dilutions of the homogenate were made in saline, and 20-μl volumes were spotted onto Middlebrook 7H11 agar with oleic acid-albumin-dextrose-catalase (Difco) and incubated at 37°C for determination of viable counts. Counts were expressed as CFU per gram of infected tissue.
Nucleotide sequence accession number.
The nucleotide sequence of RvD2 from M. tuberculosis H37Ra has been deposited in the EMBL database under accession no. AJ242907.
RESULTS
Localization of DraI polymorphisms.
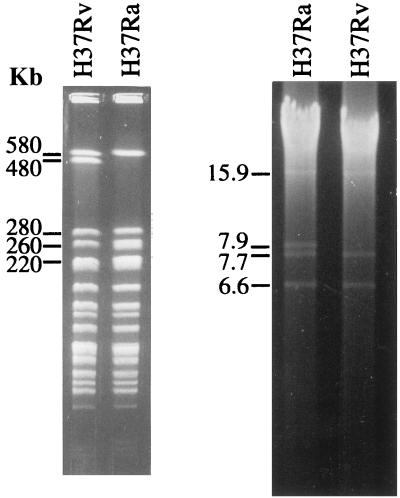
Digestion of the M. tuberculosis H37Rv and M. tuberculosis H37Ra chromosomes with DraI revealed two polymorphisms: a restriction fragment of 480 kb in M. tuberculosis H37Rv was replaced by two fragments of 260 and 220 kb in H37Ra, while a 7.9-kb DraI fragment that had no direct counterpart in M. tuberculosis H37Rv was present in M. tuberculosis H37Ra (Fig. 1).
FIG. 1.
DraI restriction profiles of M. tuberculosis H37Rv and M. tuberculosis H37Ra. (Left panel) Agarose-embedded DNA was digested with DraI and resolved by PFGE as described in Materials and Methods. The 480-kb fragment in M. tuberculosis H37Rv is replaced in M. tuberculosis H37Ra by bands of 260 and 220 kb. (Right panel) Digestion of DNA in solution with DraI and migration through a 0.7% agarose gel, revealing the 7.9-kb fragment present in M. tuberculosis H37Ra.
Since an integrated map showing the distribution of cosmid clones on the M. tuberculosis H37Rv chromosome had previously been determined (12), it was possible to select cosmids that could serve as linking clones between the 480-kb fragment in M. tuberculosis H37Rv and the 260- and 220-kb fragments in M. tuberculosis H37Ra. DraI digests of M. tuberculosis H37Rv and M. tuberculosis H37Ra were blotted onto nitrocellulose membranes and hybridized with selected cosmid clones, with Y346 (12) identified as binding both to the 480-kb and to the 260- and 220-kb fragments.
To determine where on the M. tuberculosis H37Rv chromosome the 7.9-kb DraI fragment was located, the fragment was gel purified, radiolabelled with [α-32P]dCTP, and used to probe DraI digests. The probe bound to all M. tuberculosis H37Rv DraI restriction fragments carrying IS6110, indicating that the 7.9-kb fragment contained at least one copy of this element. This complicated attempts to map the position of the fragment by hybridization.
Analysis of cosmid clone Y346.
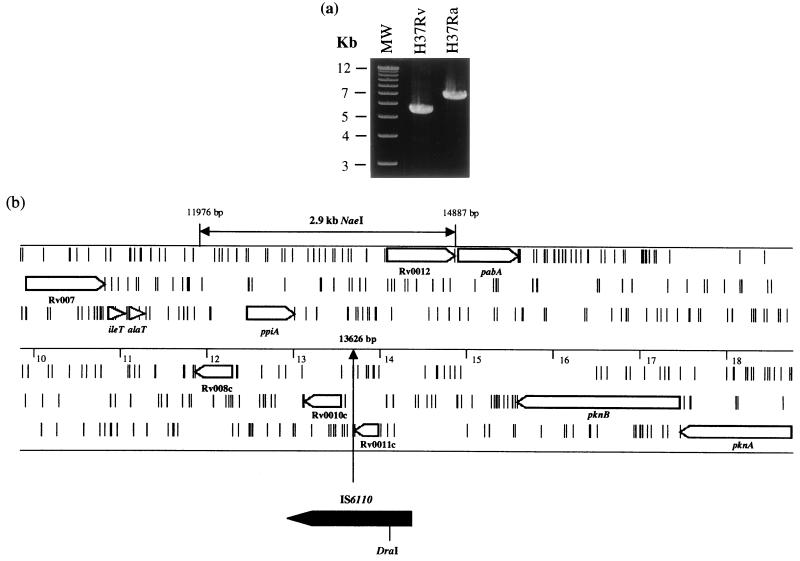
The insert ends of clone Y346 were sequenced, allowing the clone to be positioned on the M. tuberculosis genome between bp 6103 and 45420. PCR primers were designed to amplify overlapping fragments of ∼6 kb along the length of the cosmid. Reactions performed with the primers Y346.D1 and Y346.D2 (Table 1) generated a fragment in M. tuberculosis H37Ra that was ∼1.3 kb larger than that in M. tuberculosis H37Rv (Fig. 2a), indicating the presence of an internal IS6110 element. The larger M. tuberculosis H37Ra PCR product was gel purified and used as a template in sequencing reactions with primers that read out of IS6110. The relative genome position of the IS6110 insertion on the M. tuberculosis H37Rv genome (EMBL identification no. MTBH37RV) was determined to be bp 13626 (Fig. 2b).
TABLE 1.
Primers used in this study
| Primer | Description | Sequence | Expected product size (bp) |
|---|---|---|---|
| Y346.D1 | Flanks IS6110 insertion in M. tuberculosis H37Ra | CGAATGACCACGTTTGATCACAATCGC | 5,673 (H37Rv), 7,030 (H37Ra) |
| Y346.D2 | Flanks IS6110 insertion in M. tuberculosis H37Ra | CGATCCGATGTCTATTCCTTGGGCTG | |
| RvD2-17F | RvD2 internal primer | ACGGCAACGACAATCGTGGT | 1,327 (H37Ra) |
| RvD2-22R | RvD2 internal primer | AACTCTTGAACGCCTGCG |
FIG. 2.
(a) PCR products generated with primers that flank the IS6110 insertion at bp 13626. The product from M. tuberculosis H37Ra is 1.3 kb larger than that from M. tuberculosis H37Rv, due to the presence of IS6110. MW, molecular size markers. (b) The IS6110 insertion site in M. tuberculosis H37Ra. The region from ca. kb 10 to 19 of the M. tuberculosis H37Rv genome is shown, with ORFs present in all six reading frames and short vertical bars representing stop codons. The position on the M. tuberculosis H37Rv genome is marked on the middle scale, with the IS6110 insertion at bp 13626 indicated. The 2.9-kb NaeI fragment that was used in complementation experiments is marked. Genes are designated by their Rv designation or by their gene name.
Comparative genomics.
A comparative genomic study of the tubercle bacilli had revealed that M. tuberculosis H37Rv contains two deletions (RvD1 and RvD2) relative to other members of the complex (6). The RvD2 deletion was identified by comparing a local segment of the Mycobacterium bovis BCG genome to the corresponding region in M. tuberculosis H37Rv, disclosing a 6.5-kb locus that was present in M. bovis BCG but absent from M. tuberculosis H37Rv. Internal PCR primers, designed from the M. bovis BCG sequence of RvD1 and RvD2, were used to screen the M. tuberculosis H37Ra chromosome for the presence of the two deletions. While primers based on RvD1 did not produce an amplicon from M. tuberculosis H37Ra genomic DNA, RvD2 primers generated products of the expected size (Table 1). The RvD2 locus was therefore present in M. tuberculosis H37Ra.
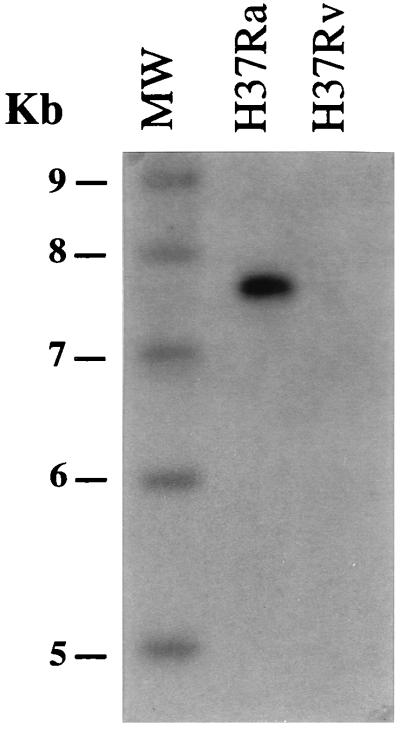
A PCR product generated with primers RvD2-17F and RvD2-22R (Table 1) was radiolabelled and used to probe DraI digests of M. tuberculosis H37Rv and M. tuberculosis H37Ra genomic DNAs. The internal RvD2 probe bound only to the 7.9-kb DraI fragment in M. tuberculosis H37Ra and gave no signal with M. tuberculosis H37Rv DNA (Fig. 3). Hence, the 7.9-kb DraI fragment in M. tuberculosis H37Ra corresponded to the RvD2 locus.
FIG. 3.
The gel shown in the right panel of Fig. 1 was blotted and hybridized with a radiolabelled internal RvD2 PCR product (Table 1) generated from M. bovis BCG Pasteur genomic DNA. The probe bound only to the 7.9-kb M. tuberculosis H37Ra DraI fragment and not to M. tuberculosis H37Rv DNA, indicating that the 7.9-kb DraI fragment in M. tuberculosis H37Ra is due to the presence of RvD2. MW, molecular size markers.
Sequence analysis of RvD2 from M. tuberculosis H37Ra.
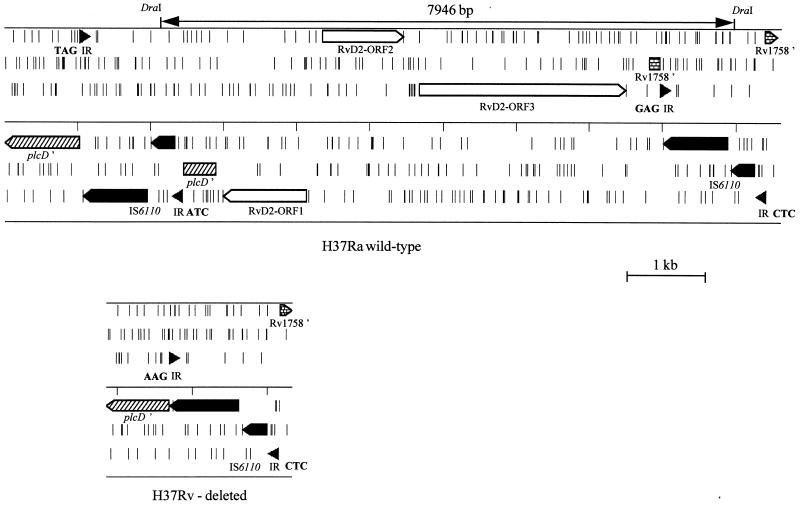
The entire RvD2 locus from M. tuberculosis H37Ra was sequenced by using primers based on the previously determined M. bovis BCG RvD2 sequence (6). The M. tuberculosis H37Ra RvD2 locus contained three ORFs (Fig. 4), with two flanking IS6110 elements that had interrupted two further ORFs. The sequences of the internal ORFs were almost identical to those of RvD2 ORF1 to ORF3 previously described for M. bovis BCG (6), which code for a putative sugar transferase, an oxidoreductase, and a membrane protein, respectively. However, ORF1 of M. tuberculosis H37Ra contained an extra base compared to those of M. bovis BCG Pasteur (sequence accession number Y18606) and M. bovis (sequence data were obtained from the M. bovis genome sequencing project [13a]). This introduced a frameshift in ORF1, changing the first part of the predicted amino acid sequence and shortening it by 13 amino acids. In total, four single-nucleotide differences were observed between ORF1 to ORF3 of M. tuberculosis H37Ra and those of M. bovis BCG. Apart from the IS6110 insertion, no differences were found between the M. bovis BCG and M. tuberculosis H37Ra sequences for the phospholipase C gene (plcD′) and the putative cutinase gene (Rv1758′).
FIG. 4.
Structures of the RvD2 loci in M. tuberculosis H37Ra and M. tuberculosis H37Rv. ORFs present in all six reading frames are shown, with short vertical bars representing stop codons. The ORFs and inverted repeats (IRs) of IS6110 are shown in black, with the 3-bp flanking DR sequences shown in boldface. Recombination between the IS6110 elements would lead to the deletion of the intevening sequence, generating the RvD2-deleted locus shown in M. tuberculosis H37Rv.
Comparison with the genome sequence of M. tuberculosis H37Rv revealed that the IS6110 insertion in the plcD′ gene (Rv1755c) is at a different position in M. tuberculosis H37Ra, with the IS6110 located 220 bp upstream of the site occupied in the virulent strain (Fig. 4). As M. tuberculosis H37Rv can have a second IS6110 located in the plcD gene (7), this indicates that this locus is a hot spot for IS6110 insertion.
In silico comparison of IS6110 elements with different DRs.
Transposition of IS6110 gives rise to the generation of 3-bp direct repeats (DRs) which flank the ends of the element (10). The absence of flanking DRs can be an indication of recombination between ISs (10), with deletion of the region between the elements. The observation that the RvD2 locus in M. tuberculosis H37Rv contains a copy of IS6110 lacking DRs indicated that this deletion was probably due to an IS6110-mediated mechanism. This led us to screen all 16 IS6110 insertions in the M. tuberculosis H37Rv genome sequence for the absence of DRs. In addition to RvD2 (at ca. kb 1987), three more IS6110 elements lacking DRs were located at ca. kb 1997, 2636, and 3711. PCR studies revealed that these three regions gave amplicons of the same size in M. tuberculosis H37Rv and M. tuberculosis H37Ra. In contrast, comparison of the flanking sequences with sequences in the M. bovis sequence database held at the Sanger Centre (13a) showed that in all three regions M. bovis contained additional DNA, ranging in size from 0.8 to 4 kb. The sizes of the deletions were confirmed by PCR with genomic DNAs from M. tuberculosis and M. bovis clinical isolates (data not shown). The deleted regions were named RvD3 (at ca. kb 1997; size, 1 kb), RvD4 (at ca. kb 2636; size, 0.8 kb), and RvD5 (at ca. kb 3711; size, 4 kb). It should be noted that RvD5 is identical to the deletion described by Fang et al. (5). Hence, these deletions appear to have occurred by recombination between two IS elements oriented in the same direction. As there are over 50 IS elements present in the genome of M. tuberculosis H37Rv (3, 7), this mechanism may be central to the generation of genomic variation. Analysis of RvD3 to RvD5 is ongoing and will be the subject of a future publication.
Complementation and virulence studies.
As the 7.9-kb polymorphism is due to a deletion from the M. tuberculosis H37Rv chromosome, it likely plays no role in the attenuation of M. tuberculosis H37Ra. Further studies therefore concentrated on the IS6110 insertion at bp 13626. To complement M. tuberculosis H37Ra with the wild-type locus from M. tuberculosis H37Rv, a 2.9-kb NaeI fragment that encompassed the region of the insertion was cloned into the mycobacterial integrating shuttle vector pKINT, generating the recombinant plasmid pSG21. Electrocompetent M. tuberculosis H37Ra was then transformed with pKINT or pSG21, and transformants were selected with kanamycin.
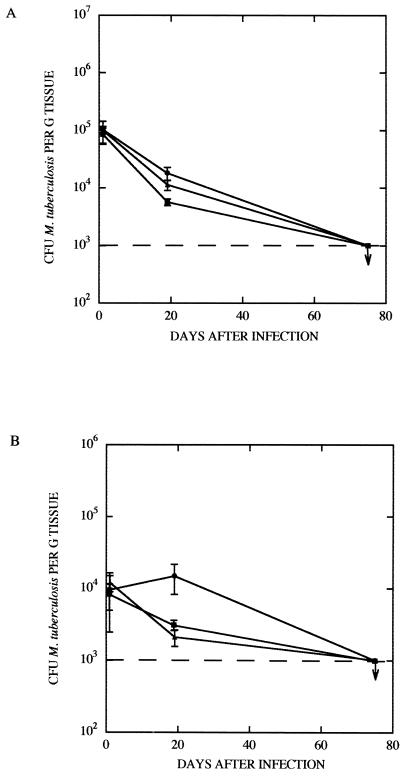
To assess the virulence of the recombinant bacilli, wild-type M. tuberculosis H37Ra, M. tuberculosis H37Ra(pKINT), and M. tuberculosis H37Ra(pSG21) were injected intravenously into BALB/c mice. Mice were sacrificed at days 1, 19, and 75, and bacterial counts in the spleens and lungs were determined. In the spleens (Fig. 5A) of infected mice, the bacterial counts for all three strains started at 105 CFU/g of infected tissue, but by day 19 the counts had fallen, with no significant difference between the H37Ra(pSG21) complemented strain and the two controls. By day 75 the counts for all three strains had fallen to ∼103 CFU/g. Counts in the lung (Fig. 5B) at day 1 were all approximately 104 CFU/g, but by day 19 the control M. tuberculosis H37Ra(pKINT) and the complemented M. tuberculosis H37Ra(pSG21) strains showed reduced growth compared to the wild-type M. tuberculosis H37Ra. However, by day 75 all counts had levelled out to ∼103 CFU/g. Hence, complementation of M. tuberculosis H37Ra with the 2.9-kb NaeI fragment imparted no in vivo selective advantage to the recombinant. In contrast, in control experiments performed with M. tuberculosis H37Rv, extensive bacterial growth was reflected in pronounced morbidity over the same period (data not shown).
FIG. 5.
Growth of bacilli in the spleens (A) and lungs (B) of BALB/c mice. The CFU per gram of infected tissue for wild-type M. tuberculosis H37Ra (●), M. tuberculosis H37Ra carrying the shuttle vector pKINT (■), or M. tuberculosis H37Ra harboring the complementing pSG21 (▴) are shown. Error bars indicate standard deviations.
DISCUSSION
Understanding of the basis of mycobacterial virulence depends on the identification of genes and gene products that contribute to pathogenesis. The mycobacterial strains M. bovis BCG and M. tuberculosis H37Ra are both attenuated derivatives of virulent tubercle bacilli that were generated by serial passage of the parent strain through laboratory media (2, 15). The nature of the isolation of these attenuated bacilli makes precise definition of the attenuating mutation(s) difficult to describe by classical genetic approaches. However, the genome sequence of M. tuberculosis H37Rv (3) offers the chance to use comparative genomics to identify variable loci between attenuated tubercle bacilli and the fully virulent M. tuberculosis H37Rv.
The observation that M. tuberculosis H37Ra showed two differences from the M. tuberculosis H37Rv DraI restriction pattern was intriguing. As insertion of IS6110 could have generated such DraI polymorphisms and in the process inactivated a virulence gene(s), it was imperative to define the nature of these variations. This study shows that IS6110 is the cause of both of these polymorphisms.
The RvD2 locus was originally detected as a deletion from the M. tuberculosis H37Rv genome (6). This region is present, however, in M. tuberculosis H37Ra, with evidence suggesting that RvD2 was lost from M. tuberculosis H37Rv following an IS6110 recombination event during its derivation from M. tuberculosis H37. Although the presence of RvD2 in M. tuberculosis H37Ra suggests that this locus might encode a repressive element, the existence of RvD2 in clinical isolates (6) indicates that this is not the case. The identification of three additional IS6110-mediated genomic deletions in M. tuberculosis H37Rv (RvD3 to RvD5) underlines the importance of this mechanism for the generation of genomic diversity in the tubercle bacilli. Our observation of IS-catalyzed genomic variation (RvD2 to RvD5) complements and extends the studies of Fang et al. (5), who have recently identified IS6110-mediated deletions of chromosomal segments in clinical isolates of M. tuberculosis. It also underlines the utility of in silico genomic comparison for rapid detection of variable regions in the genomes of mycobacteria. This way of identifying genomic particularities of mycobacterial strains will become increasingly important, especially with the imminent completion of the genome sequences of other tubercle bacilli (1a, 7a, 13a).
The loss of the 480-kb DraI fragment from the M. tuberculosis H37Ra restriction profile was due to the insertion of an IS6110 element that mapped to bp 13626 on the M. tuberculosis H37Rv sequence. Although the insertion did not disrupt any ORFs, it may have had polar effects on nearby genes, in particular Rv0010c, which codes for a protein of unknown function. Complementation of M. tuberculosis H37Ra with the wild-type locus from M. tuberculosis H37Rv [H37Ra(pSG21)] did not restore virulence, however, suggesting that this insertion is not the principal reason for the attenuation of M. tuberculosis H37Ra. Nevertheless, as the complementing DNA was cloned into pKINT, a vector that integrates at the chromosomal attB site (9), it is possible that only cis complementation of the locus at bp 13626 may restore virulence. Furthermore, Pascopella et al. (11) have proposed that multiple mutations, at different chromosomal loci, are responsible for the attenuation of the bacillus. It may therefore be necessary to complement M. tuberculosis H37Ra(pSG21) with the ivg locus described by Pascopella et al. (11), and possibly other loci, to achieve restoration of virulence.
The application of the genomic techniques described here successfully detected genetic variation between M. tuberculosis H37Rv and M. tuberculosis H37Ra. However, the basis for the decreased virulence of M. tuberculosis H37Ra remains unclear. The possibility remains that attenuation was caused by chromosomal rearrangements such as small duplications or inversions, variations that would have been difficult to detect by using the approach described above. Techniques that allow a more subtle examination of the genome may therefore need to be applied if we are to define the genetic basis of attenuation in M. tuberculosis H37Ra.
ACKNOWLEDGMENTS
We thank J. F. de La Palette for advice and discussion.
Financial support for this work was provided by the Wellcome Trust, the Institut Pasteur, and l'Association Française Raoul Follereau. S. V. Gordon was the recipient of a Wellcome Trust International Travelling fellowship.
REFERENCES
- 1.Brosch R, Gordon S V, Billault A, Garnier T, Eiglmeier K, Soravito C, Barrell B G, Cole S T. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Brosch, R., and S. T. Cole. 1999. [Online.] http://www.pasteur.fr/units/Lgmb/. [17 September 1999, last date accessed.]
- 2.Calmette A, Guerin C. Sur quelques propriétés du bacille tuberculeux d'origine, cultivé sur la bile de boeuf glycérinée. C R Acad Sci. 1909;149:716–718. [Google Scholar]
- 3.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T, Smith D. Toward mapping and sequencing the genome of Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection and control. Washington, D.C: American Society for Microbiology; 1994. pp. 227–238. [Google Scholar]
- 5.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole S T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 7.Gordon S V, Heym B, Parkhill J, Barrell B, Cole S T. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology. 1999;145:881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 7a.The Institute for Genomic Research. 1999. [Online.] http://www.tigr.org/tdb/mdb/mtdb.html. [1 September 1999, last date accessed.]
- 8.Kinger A K, Tyagi J S. Identification and cloning of genes differentially expressed in the virulent strain of Mycobacterium tuberculosis. Gene. 1993;131:113–117. doi: 10.1016/0378-1119(93)90678-v. [DOI] [PubMed] [Google Scholar]
- 9.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascopella L, Collins F M, Martin J M, Lee M H, Hatfull G F, Stover C K, Bloom B R, Jacobs W R., Jr Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect Immun. 1994;62:1313–1319. doi: 10.1128/iai.62.4.1313-1319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rindi L, Lari N, Garzelli C. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem Biophys Res Commun. 1999;258:94–101. doi: 10.1006/bbrc.1999.0591. [DOI] [PubMed] [Google Scholar]
- 13a.The Sanger Centre. 1999. [Online.] http://www.sanger.ac.uk/Projects/M_bovis. [17 September 1999, last date accessed.]
- 14.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 15.Steenken W, Oatway W H, Petroff S A. Biological studies of the tubercle bacillus. III. Dissociation and pathogenicity of the R and S variants of the human tubercle bacillus (H37) J Exp Med. 1934;60:515–540. doi: 10.1084/jem.60.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]