Abstract
129sv mice functionally deleted of the antimicrobial resistance gene, Nramp1, were found to be as resistant as wild-type mice to infection with the virulent H37Rv strain of Mycobacterium tuberculosis, as determined by monitoring bacterial growth in major organs and recording host survival times. Death of infected mice of both types was associated with extensive infection-induced pathology in the lungs but not in other major organs. These findings are in keeping with the view that Nramp1 is of limited importance in resistance to tuberculosis in mice.
Nramp1 is a gene involved in determining the ability of mice to resist infection with certain unrelated intracellular pathogens, including Salmonella typhimurium, Leishmania donovani, the attenuated bacillus Calmette-Guérin (BCG) strain of Mycobacterium bovis, as well as other species of Mycobacterium (reviewed in references 5, 6, and 24). The gene is located at the Ity/Lsh/Bcg locus of mouse chromosome 1 and encodes a transmembrane protein that translocates to the macrophage phagocytic vacuole following phagocytosis of microorganisms (5, 6). The gene has two alleles, a dominant resistance one and a recessive susceptibility one, which results in inbred mouse strains being assignable to one or other of two nonoverlapping groups according to their resistance to the aforementioned intracellular pathogens (15). The susceptibility allele differs from the resistance allele by a substitution of adenine for guanine at base position 596 of the gene, resulting in replacement of glycine with aspartic acid at amino acid position 169 in a predicted transmembrane domain of the protein (15). Because Nramp1D169 is unstable and rapidly degraded (14), mice homozygous for this allele are as susceptible to infection as mice functionally deleted of the resistance allele by homologous recombination (26).
The importance of Nramp1 in natural resistance to infection with certain intracellular pathogens, including several species of Mycobacterium, is indisputable. However, the belief that the gene is also important in determining resistance of mice to infection with virulent Mycobacterium tuberculosis has been mainly inferred from the results of resistance studies with other Mycobacterium species, especially BCG. The belief has been recently challenged by the finding (16, 17, 19) that mouse strains homozygous for the Nramp1 resistance allele are equally, or even less resistant to M. tuberculosis infection than strains homozygous for the Nramp1 susceptibility allele. It has also been demonstrated (18) that Nramp1 alleles segregate independently of resistance to M. tuberculosis in F2 generation progeny of homozygous Nramp1 resistance and Nramp1 susceptibility parental mouse strains. Even so, according to one study (4), Nramp1 is involved in resistance to infection initiated with very small intravenous (i.v.) inocula of M. tuberculosis. Therefore, the subject would appear to be controversial and deserving of additional investigation. It would seem important to determine, for instance, whether mice that bear a homozygous null allele at the Nramp1 locus generated by homologous recombination are less resistant to M. tuberculosis infection than their isogenic wild-type counterparts. This report shows that mice bearing a homozygous deletion at Nramp1 are not more susceptible than their wild-type counterparts to infection initiated with small inocula of M. tuberculosis.
MATERIALS AND METHODS
Mice.
The 129sv mouse stock was initially obtained from Taconic Farms (Germantown, N.Y.) and subsequently bred in the Animal Care Center at McGill University. The 129sv strain bears a resistance, wild-type allele at Nramp1 (G169). The procedures used to generate, and the characterization of, mutant 129sv mice bearing a null mutation at Nramp1 (Nramp1−/− mice) were described previously (26). Wild-type and null Nramp1−/− mutant 129sv mice were maintained by brother-sister mating and were housed under pathogen-free conditions and used in experiments at 10 weeks of age.
Bacteria.
The H37Rv strain of M. tuberculosis was obtained from the Trudeau Institute Culture Collection as a frozen (−150°C) log-phase (2 × 108/ml) culture in Proskauer and Beck medium containing 0.01% Tween 80. The method used for growing M. tuberculosis in suspension culture was described previously (9). For infection, a vial of the culture was thawed, diluted 1 in 10 in phosphate-buffered saline (PBS) containing 0.01% Tween 80 (PBS-Tween), subjected to two 5-s pulses of ultrasound to break up clumps, and diluted appropriately in PBS-Tween for inoculation via a lateral tail vein. Infection via the respiratory route with 2 × 102 CFU of H37Rv was achieved by subjecting the mice to aerosolized bacilli in an aerosol infection apparatus (Tri Instruments, Jamaica, N.Y.), as described previously (17). The course of infection was monitored by enumerating M. tuberculosis in major organs at progressive times after inoculation. This involved plating 10-fold serial dilutions of whole organ homogenates on nutrient agar and enumerating bacterial colonies after 3 weeks of incubation as described previously (9). Homogenization of organs was performed in the cold (10°C) PBS-Tween in glass tubes with close-fitting motorized Teflon pestles.
Photography.
The lungs, livers, spleens, and kidneys of wild-type and Nramp1−/− mice were harvested on day 200 of infection and photographed with a Nikon 35-mm camera.
Statistics.
Where warranted, the significance of differences between the geometric means of CFU per organ at particular times was determined by Student's t test. The significance of differences between survival times of groups of 10 mice was determined with the log-rank test for comparisons of survival curves.
RESULTS AND DISCUSSION
Previous studies (11) have shown that the effect of Nramp1 on resistance to BCG infection is most obvious when small i.v. inocula (<105 CFU) are used to initiate infection. Therefore, in the study reported here, a 104 i.v. inoculum of H37Rv was used to monitor resistance to bacterial growth in major organs of Nramp1−/− and wild-type (Nramp1+/+) mice and to determine host survival times. The decision not to use a smaller i.v. inoculum for this experiment was dictated by the wish to use host survival time as a measure of resistance to infection-induced lung disease and the need, therefore, for mice to succumb to infection in a reasonable period of time. It is known (19) in this regard that a 105 i.v. inoculum of H37/Rv can take over 300 days to kill mice of resistant strains and about half as long to kill mice of susceptible strains, such as 129sv.
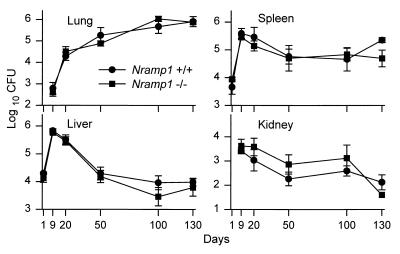
The lack of effect of Nramp1 deletion on susceptibility of 129sv mice to M. tuberculosis replication in major organs (Fig. 1) was paralleled by a study of host survival (Fig. 2). It can be seen that there was no significant difference in the ability of the two types of mice to control bacterial growth in lungs, kidneys, livers, and spleens over a 130-day period of infection. In both types of mice, infection progressed in the liver and spleen for about 10 days and then underwent partial resolution before entering an approximate stationary phase. In the lungs, however, infection progressed for an additional 10 days before its rate of progression was significantly reduced, although not halted. It is known (3, 21) that resolution of infection in the liver and spleen, and reduction in the rate of lung infection, is mediated predominantly by CD4 Th1 cells, with a contribution from CD8 T cells. It can be seen in Fig. 1 that no bacilli were detected in the lung on day 1 of infection. This is because only about 0.1% of an i.v. inoculum implants in this organ (9), which means that the lungs of both types of mice would have received only about 10 bacilli, a number below the detection limits of the plating assay. The kidneys likewise received too small a fraction of the i.v. inoculum to be detected on day 1. It is apparent that resolution of infection in the kidneys occurred in concert with resolution of infection in the liver and spleen.
FIG. 1.
Growth of M. tuberculosis H37Rv in lungs, livers, spleens, and kidneys of Nramp1−/− and wild-type (Nramp1+/+) mice inoculated i.v. with 104 CFU of bacilli. There was no significant difference in the ability of mutant and wild-type mice to control bacterial replication over 130 days of infection except for day 130 in the spleen, where the differences between the means was significant (P < 0.05). Means ± standard errors of five mice per group per time point are shown.
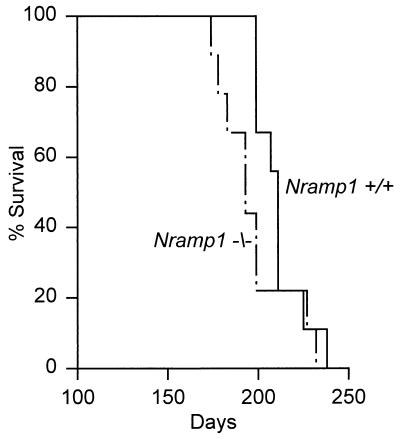
FIG. 2.
Survival times of 10 Nramp1−/− and 10 wild-type mice inoculated i.v. with 104 CFU of H37Rv. There was no significant difference (P > 0.2) between the survival times of mutant and wild-type mice.
The foregoing evidence that Nramp1 is not important in the resistance of 129sv mice to H37Rv infection is supported by the results of the host survival study shown in Fig. 2. It can be seen that there was no significant difference (P > 0.27) between the survival times of wild-type and Nramp1−/− mice infected with 104 bacilli i.v. It should be pointed out that the survival times of wild-type and Nramp1−/− 129sv mice as shown in Fig. 2 is shorter than the median survival times published for mice of certain other inbred strains infected with 10 times more bacilli (19). This is because the 129sv strain, although homozygous for the resistance allele of Nramp1, is an M. tuberculosis-susceptible strain (19). Therefore, the forgoing results serve to show that the absence of Nramp1 does not make a susceptible strain more susceptible to tuberculosis. In addition, lung infection in 129sv mice as shown here, like lung infection in another M. tuberculosis-susceptible mouse strain, is progressive, in contrast to lung infection in resistant mouse strains, which is stabilized by immunity and caused to become approximately stationary (17, 20). The possibility that the genetic susceptibility of 129sv mice in some way might mask a relatively small contribution of Nramp1 because of the presence of susceptibility genes should be considered. Even so, the contribution would be very small, given the results of a previous study (17) showing that M. tuberculosis-resistant BALB/c mice homozygous for the susceptibility allele of Nramp1 and congenic BALB/c mice homozygous for the resistance allele (Ity/Lsh/Bcg locus) are identical in resistance to M. tuberculosis infection.
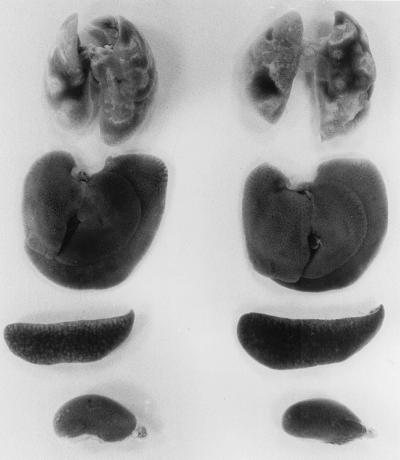
That compromised lung function was the most likely cause of death of mutant and wild-type mice is suggested by the macroscopic appearance of major organs harvested on day 200 of infection, as shown in Fig. 3. It can be seen at this stage of infection that whereas infection-induced disease was very extensive in the lungs of Nramp1−/− and wild-type mice, there was no macroscopic evidence of disease in other organs. This is in keeping with the results of published histological study of the organs of resistant and susceptible mouse strains (20), which shows that M. tuberculosis-induced histopathology develops progressively only in the lungs. This study also showed (20) that whereas H37Rv in the liver and spleen is confined to compact granulomas that remain small in number and size over a protracted period of time, H37Rv in the lungs was associated with the development of extensive pathology and organ consolidation. Because tuberculosis is confined to the lungs of more than 85% of humans who develop the disease (10), it seems reasonable to suggest, in view of the foregoing discussion, that mouse tuberculosis is a relevant model of the human disease. However, because BCG does not cause progressive disease in any organ of mice, it is perhaps not surprising that a resistance mechanism capable of reducing the rate of BCG growth is not capable of slowing infection with the virulent pathogen. It is apparent that the virulence of M. tuberculosis is determined to some extent by its ability to resist an Nramp1-dependent defense mechanism that enables mouse macrophages to control intracellular growth of several other Mycobacterium species in vitro (2, 7, 8, 12) and enables mice to better defend against infection with these species (13, 23). One possibility is that M. tuberculosis inhibits fusion of the phagosome to Nramp1-positive vesicles (7).
FIG. 3.
Macroscopic appearance of major organs of wild-type (left) and Nramp1−/− (right) mice harvested on day 200 of infection initiated with 104 CFU of H37Rv i.v. Shown from top to bottom are the lungs, liver, spleen, and left kidney. Only the lungs of both types of mice show disease.
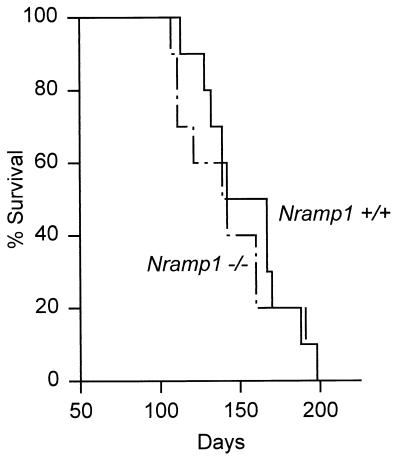
To determine whether the foregoing results also apply to mice infected via the respiratory route, we infected 10 Nramp1−/− and 10 Nramp1+/+ mice by aerosol with 2 × 102 H37Rv CFU and monitored their survival times. The results in Fig. 4 show that their was no significant difference (P > 0.6) between the survival times of mutant and wild-type mice. This result is in keeping with the results of a previous study (7) that compared the survival times of BALB/c (Nramp1 susceptibility) and DBA/2 (Nramp1 resistance) mice infected via the respiratory route.
FIG. 4.
Survival times of 10 Nramp1−/− and 10 wild-type mice infected via the respiratory route with 2 × 102 CFU of H37Rv. There was no significant difference (P > 0.6) between the survival times.
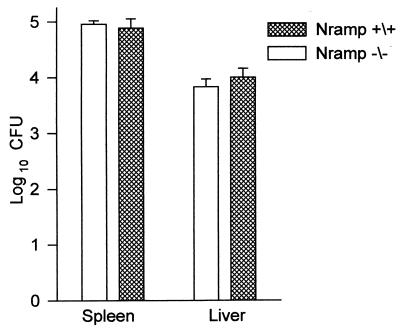
The possibility that a contribution of Nramp1 to resistance to M. tuberculosis infection, as seen by others (4), would be revealed if smaller i.v. inoculum of virulent M. tuberculosis were used to initiate infection was tested by inoculating wild-type and Nramp1−/− mice i.v. with 103 CFU of H37Rv and determining the number of CFU in lungs, livers, kidneys, and spleens 30 days later. The results in Fig. 5 clearly show, however, that the livers and spleens of both types of mice contained the same number of bacilli at day 30 of infection. No bacilli were detected in the lungs or kidneys on day 30, indicating that these organ may not have become infected. This would not be surprising given that only 0.1% of the i.v. inoculum would be expected to implant in the lung (9, 16). With regard to this result, moreover, it was shown earlier (16, 26) that an even smaller inoculum (102) of M. tuberculosis given via the respiratory route failed to reveal a contribution of Nramp1 to resistance to tuberculosis.
FIG. 5.
Number of H37Rv CFU in livers and spleens of Nramp1−/− and Nramp1+/+ mice given 103 CFU of H37Rv i.v. 30 days earlier. No bacteria were detected in the lungs. Means ± standard errors of five mice per group are shown.
Last, the difficulty of showing a protective role for Nramp1 in resistance to tuberculosis in mice must be reconciled with the results of a recent study showing a role for the human NRAMP1 homologue in resistance to the disease in humans. It was demonstrated (2) in a case-control study in Gambia, West Africa, that of 410 smear-positive patients and 410 ethnically matched controls, patients with polymorphism in intron 4 and in the 3′ untranslated region of NRAMP1 were particularly overrepresented (P < 0.001) in the tuberculous population, as opposed to those with most common NRAMP1 genotypes. This was interpreted as evidence that the gene plays a role in susceptibility to tuberculosis in this African population. However, the authors point out that their methodology does not allow them to exclude the involvement of a gene closely linked to the NRAMP1 locus but suggest that this is unlikely because of the importance of Nramp1 in resistance to tuberculosis in mice. However, recent segregation and linkage analysis of multicase tuberculosis families in a region of Brazil (22) failed to show evidence that NRAMP1 is involved to any significant extent in susceptibility and resistance to pulmonary tuberculosis. The evidence that NRAMP1 is involved in resistance to leprosy is suggestive, in that a study in Vietnam shows that NRAMP1, or a closely linked gene, is involved in susceptibility to this disease in Vietnamese families, although no evidence was found that it is involved in susceptibility of Chinese families (1).
The discrepancy between the apparent lack of Nramp1 deletion on susceptibility to experimental tuberculosis infection of 129sv mice with M. tuberculosis as described here, and the positive association between certain NRAMP1 polymorphisms and susceptibility to tuberculosis and leprosy as noted in certain human studies (see above) has yet to be explained. One possibility is that human NRAMP1 does not play an important role per se in susceptibility to tuberculosis, but that a gene nearby and in linkage disequilibrium is important and responsible for the apparent allelic association. In this connection, attention was focused on NRAMP1 because of the belief that it is important in resistance to mouse tuberculosis. The second possibility is that NRAMP1 is important for susceptibility to tuberculosis in humans but does not play a role in the susceptibility of mice. If it turns out that polymorphisms at human NRAMP1 are important in determining resistance or susceptibility to tuberculosis in certain human populations, it will be important to develop an animal model with which to analyze the role of this gene in more detail.
ACKNOWLEDGMENTS
This work was supported by grants to R.J.N. (AI-37844 and AI-40071) from the National Institutes of Health and to P.G. from the Howard Hughes Medical Institute. P.G. is supported by a Senior Scientist Salary Award from the Medical Research Council of Canada.
REFERENCES
- 1.Abel L, Sanchez F O, Oberti J. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;77:133–145. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
- 2.Bellamy R, Ruwende C, Corra T, McAdam K P W J, Whittle H C, Hill A V S. Variations in the NRAMP1 susceptibility gene and susceptibility to tuberculosis in West Africa. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 3.Boom W H. The role of T cell subsets in Mycobacterium tuberculosis infection. Infect Agents Dis. 1996;5:73–81. [PubMed] [Google Scholar]
- 4.Brown D H, Miles B H, Zwilling B S. Growth of Mycobacterium tuberculosis in BCG-resistant and -susceptible mice: establishment of latency and reactivation. Infect Immun. 1995;63:2243–2247. doi: 10.1128/iai.63.6.2243-2247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buschman E, Vidal S, Skamene E. Nonspecific resistance to mycobacteria: the role of the Nramp1 gene. Behring Inst Mitt. 1997;99:51–57. [PubMed] [Google Scholar]
- 6.Cellier M, Belouchi A, Gros P. Resistance to intracellular infections: comparative genomic analysis of Nramp1. Trends Genet. 1996;12:201–204. doi: 10.1016/0168-9525(96)30042-5. [DOI] [PubMed] [Google Scholar]
- 7.De Chastellier C, Frehel C, Offreso C, Skamene E. Implication of phogosome-lysosome fusion in restriction of Mycobacterium avium growth in bone marrow macrophages from genetically resistant mice. Infect Immun. 1993;61:3775–3784. doi: 10.1128/iai.61.9.3775-3784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis M, Forget A, Pelletier M, Gervais F, Skamene E. Killing of Mycobacterium smegmatis by macrophages from genetically susceptible and resistant mice. J Leukoc Biol. 1990;47:25–30. doi: 10.1002/jlb.47.1.25. [DOI] [PubMed] [Google Scholar]
- 9.Dunn P L, North R J. Virulence ranking of some Mycobacterium tuberculosis and Mycobacterium bovis strains according to their ability to multiply in the lungs, induce lung pathology, and cause mortality in mice. Infect Immun. 1995;63:3428–3437. doi: 10.1128/iai.63.9.3428-3437.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farer L S, Lowell A M, Meador M P. Extrapulmonary tuberculosis in the United States. Am J Epidemiol. 1979;109:205–210. doi: 10.1093/oxfordjournals.aje.a112675. [DOI] [PubMed] [Google Scholar]
- 11.Forget A, Skamene E. Genetic regulation of early host defense mechanisms controlling mycobacterial infections. In: Skamene E, editor. Genetic control of host resistance to infection and malignancy. New York, N.Y: Allen R. Liss Inc.; 1985. pp. 265–277. [Google Scholar]
- 12.Gomes M S, Appelberg R. Evidence for a link between iron metabolism and Nramp1 gene function in innate resistance against Mycobacterium avium. Immunology. 1998;95:165–168. doi: 10.1046/j.1365-2567.1998.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotto Y, Buscham E, Skamene E. Regulation of host resistance to Mycobacterium intracellulare in vivo and in vitro by the Bcg gene. Immunogenetics. 1984;30:218–221. doi: 10.1007/BF02421210. [DOI] [PubMed] [Google Scholar]
- 14.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bsg/Ity/Lsh locus: genetic transfer of resistance to infection in C57BL/6 transgenic mice for the Nramp1gly169 allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P. Haplotype mapping and sequence analysis of the mouse Nramp gene predicts susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 16.Masu S H, Kim Y, Hashim R, Wang G-Z, Dimmer C, Smith D. Response of inbred mice to aerosol challenge with Mycobacterium tuberculosis. Infect Immun. 1987;55:1862–1866. doi: 10.1128/iai.55.8.1862-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina E, North R J. Evidence inconsistent with a role for the Bcg gene (Nramp1) in resistance of mice to infection with Mycobacterium tuberculosis. J Exp Med. 1996;183:1045–1051. doi: 10.1084/jem.183.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina E, Rogerson B J, North R J. The Nramp1 antimicrobial resistance gene segregates independently of resistance to virulent Mycobacterium tuberculosis. Immunology. 1996;88:479–481. doi: 10.1046/j.1365-2567.1996.d01-700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medina E, North R J. Resistance ranking of some common inbred mouse strains to Mycobacterium tuberculosis and relationship to major histocompatibility haplotype and Nramp1 genotype. Immunology. 1998;93:270–274. doi: 10.1046/j.1365-2567.1998.00419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medina E, North R J. Genetically susceptible mice remain proportionally more susceptible to tuberculosis after vaccination. Immunology. 1999;96:16–21. doi: 10.1046/j.1365-2567.1999.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orme I A, Anderson P, Boom W H. T cell responses to Mycobacterium tuberculosis. J Infect Dis. 1993;167:1481–1497. doi: 10.1093/infdis/167.6.1481. [DOI] [PubMed] [Google Scholar]
- 22.Shaw M A, Collins A, Peakock C S, Miller E N, Black G F, Sibthorpe D, Lins-Lainson Z, Shaw J J, Ramos F, Silveira F, Blackwell J M. Evidence that genetic susceptibility to Mycobacterium tuberculosis in a Brazilian population is under oligogenic control: linkage study of the candidate genes NRAMP1 and TNFA. Tuberc Lung Dis. 1997;78:35–45. doi: 10.1016/s0962-8479(97)90014-9. [DOI] [PubMed] [Google Scholar]
- 23.Skamene E, Gros P, Forget A, Patel P J, Nesbitt M. Regulation of resistance to leprosy by chromosome 1 locus in the mouse. Immunogenetics. 1984;19:117–120. doi: 10.1007/BF00387854. [DOI] [PubMed] [Google Scholar]
- 24.Skamene E, Schurr E, Gros P. Infection genomics: Nramp1 as a major determinant of natural resistance to intracellular infections. Annu Rev Med. 1998;49:275–287. doi: 10.1146/annurev.med.49.1.275. [DOI] [PubMed] [Google Scholar]
- 25.Stokes R W, Orme I, Collins F M. Role of mononuclear phagocytes in expression of resistance and susceptibility to Mycobacterium avium infections in mice. Infect Immun. 1986;54:811–819. doi: 10.1128/iai.54.3.811-819.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vidal S, Tremblay M L, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]