Abstract
The Hedgehog (Hh) cascade is central to development, tissue homeostasis, and cancer. A pivotal step in Hh signal transduction is the activation of GLI transcription factors by the atypical G protein-coupled receptor (GPCR) SMOOTHENED (SMO). How SMO activates GLI remains unclear. Here we show that SMO employs a decoy substrate sequence to physically block the active site of the PKA catalytic subunit (PKA-C) and extinguish its enzymatic activity. As a result, GLI is released from phosphorylation-induced inhibition. Using a combination of in vitro, cellular, and organismal models, we demonstrate that interfering with SMO / PKA pseudosubstrate interactions prevents Hh signal transduction. The mechanism uncovered echoes one utilized by the Wnt cascade, revealing an unexpected similarity in how these two essential developmental and cancer pathways signal intracellularly. More broadly, our findings define a mode of GPCR-PKA communication that may be harnessed by a range of membrane receptors and kinases.
The Hh signaling cascade is fundamental to embryogenesis, controlling the development of nearly every vertebrate organ1–4. Insufficient Hh pathway activity underlies birth defects affecting the nervous, cardiovascular, and musculoskeletal systems5–7. On the other hand, Hh pathway overactivation drives several common cancers, including basal cell carcinoma of the skin (the most common cancer in North America) and medulloblastoma (the most common pediatric brain tumor)8,9.
The Hh pathway utilizes an unusual signal transduction mechanism involving layers of repressive interactions1–4. In the pathway “off” state, PKA-C phosphorylates GLI, stimulating its proteolysis into a truncated transcriptional repressor that inhibits target gene expression4,10. In the pathway “on” state, Hh ligands bind to and inactivate the 12-transmembrane sterol transporter PATCHED1 (PTCH1), which releases SMO from PTCH1-mediated inhibition4. This process allows SMO to access its endogenous sterol ligands and undergo an activating conformational change11–14. Once activated, SMO blocks PKA-C-mediated phosphorylation of GLI15–18, a key step that likely occurs within a solitary microtubule-based organelle known as the primary cilium19–21. This tiny cell-surface compartment is thought to facilitate the essential protein-protein interactions and biochemical reactions in the Hh pathway, owing to its high surface-area-to-volume ratio, compartmentalized structure, and unique protein and lipid repertoire. Following the loss of PKA-C-mediated phosphorylation, GLI becomes activated and consequently controls the expression of proliferative or differentiative genes10. SMO regulation of PKA-C is thus a critical event in transducing Hh signals from the cell surface to the nucleus. However, the underlying mechanism has remained mysterious for decades1–4.
GPCRs classically inhibit PKA-C via well-characterized signaling cascades involving heterotrimeric G protein-mediated effects on cAMP. Lowering cAMP reduces PKA-C activity, either by promoting interactions with PKA regulatory (PKA-R) subunits to form inactive PKA holoenzymes22–24 or by affecting the conformational states of pre-formed PKA-C / PKA-R complexes25–28. In contrast, we recently found that SMO prevents PKA-C from phosphorylating substrates via a noncanonical mechanism. SMO directly interacts with PKA-C subunits, recruiting them to the ciliary membrane and thereby restricting their access to soluble GLI proteins. SMO / PKA-C interactions are triggered by GPCR kinases 2 and 3 (GRK2/3), which recognize the SMO active state and phosphorylate the cytoplasmic tail (C-tail) of SMO to promote PKA-C binding. Based on these observations, we proposed that active, phosphorylated SMO binds to and sequesters PKA-C within the cilium, which prevents phosphorylation of GLI and thereby promotes GLI activation29.
Here we uncover a critical and unexpected component of the SMO / PKA-C regulatory mechanism in which SMO physically blocks PKA-C enzymatic activity. We show that the SMO proximal C-tail (pCT) acts as a decoy PKA-C substrate that binds to and occludes the kinase active site, thereby preventing phosphorylation of PKA-C targets. SMO is, to our knowledge, the first example of a GPCR that functions as a direct PKA-C inhibitor. However, this decoy substrate mechanism appears to apply more generally to transmembrane receptors and kinases in other signaling pathways.
RESULTS
SMO binds and inhibits PKA-C as a pseudosubstrate
The principal regulators of PKA-C activity within cells are PKA-R subunits30 and the heat-stable protein kinase inhibitor (PKI) proteins31. PKI proteins and type I (PKA-RI) regulatory subunits operate as pseudosubstrates that bind within the PKA-C active site but cannot undergo phosphorylation. As a result, the enzyme’s phosphoryl transfer and substrate turnover cycle is interrupted30–33. PKA-R / PKA-C holoenzymes dissociate upon cAMP binding to PKA-R, releasing catalytically active PKA-C30. In contrast, PKI proteins interact with PKA-C independently of cAMP levels31. Despite their divergent regulatory influences, PKI and PKA-RI engage the PKA-C active site cleft using similar sequence motifs30,33.
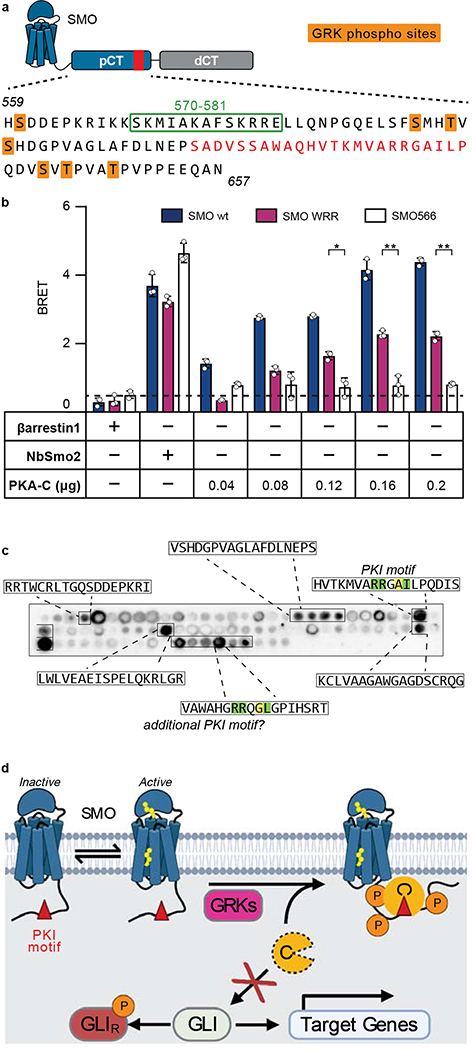
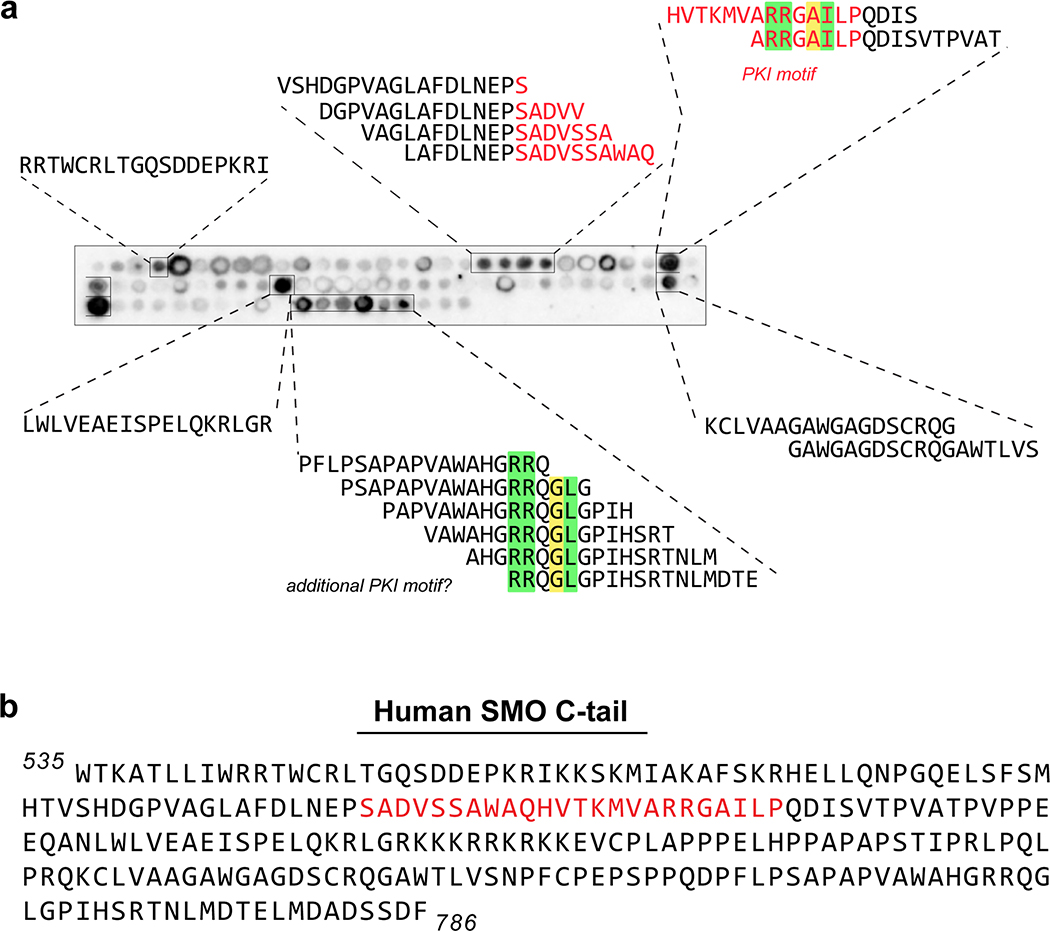
Inspection of the mouse SMO sequence revealed a region within the pCT (residues 615–638) that bears the hallmarks of a PKA-C pseudosubstrate (Fig. 1a). First, in contrast to canonical PKA-C substrates, which contain a serine or threonine at a canonical phosphorylation site (P-site)34, SMO possesses a non-phosphorylatable residue, alanine, at this position. Second, SMO contains arginines at the P−2 and P−3 positions that are essential in other pseudosubstrates (PKI and PKA-RI) for binding the PKA-C active site35–38. Third, SMO contains a hydrophobic residue, isoleucine, at P+1 aromatic and a bulkier residue, tryptophan, at P−13, both of which contribute to high-affinity PKI interactions with PKA-C39–41. Finally, SMO harbors a predicted α-helical sequence N-terminal to the pseudos region (Fig. 1a), resembling a domain in PKI that is required for high-affinity interactions with PKA-C42,43. Based on these observations and the studies described below, we refer to SMO residues 615–638 as the “SMO PKI motif.”
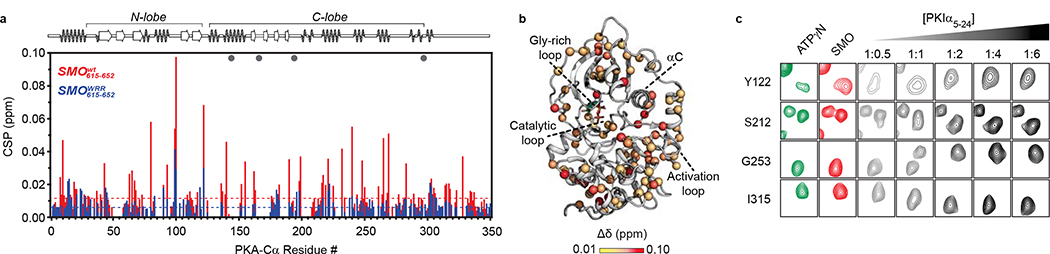
Fig. 1: SMO binds PKA-C as a pseudosubstrate.
a, CLUSTAL alignment of the mouse SMO pCT with the PKIɑ pseudosubstrate region. Additional PKA-C pseudosubstrate and substrate sequences are provided for comparison32,65,98. P-site is yellow; other key conserved residues are green. Spiral cartoon above alignment indicates predicted SMO helical region. Standard (615–638) and extended (615–652) SMO peptides used for in vitro assays are colored red or black, respectively. Inset, structure of PKA-Cɑ bound to PKIɑ(5–24) (PDB: 3FJQ), with ATP colored orange and key PKI residues colored as described above. b, Top, fluorescence polarization assay employing FAM-labeled SMO peptide, 1 mM ATP, and varying concentrations of human PKA-Cɑ. Points represent the mean from two separate experiments composed of quadruplicates. Bottom, the same assay except with 3 μM PKA-Cɑ and varying concentrations of ATP. Quadruplicates representative of two independent trials are plotted. c, Overlay of purified mouse PKA-Cɑ onto an array of SMO peptides containing the indicated substitutions in the P−13, P−3, and P−2 positions. d, SPR sensorgram for binding of PKA-Cɑ, in concentrations ranging from 5 nM to 2.5 μM, to GST-tagged wild-type SMO pCT. Buffer contained 1 mM ATP and 10 mM MgCl2. e, As d, but with SMO pCT harboring the WRR mutation. Note that the negative signal results from the subtraction of the non-specific binding of PKA-Cɑ to a GST control surface.
We hypothesized that SMO utilizes its PKI motif as a pseudosubstrate to bind and inhibit PKA-C, thereby activating GLI. Consistent with this hypothesis, residues 615–637 of mouse SMO are essential for communication with GLI44, although the structure, function, and interacting partner(s) of this SMO region are all undefined. Furthermore, the P, P+1, P−2, P−3, and P−13 residues critical for PKA-C pseudosubstrate function in bona fide PKI proteins are highly conserved among SMO orthologs (Extended Data Fig. 1), suggestive of a central role in SMO functionality.
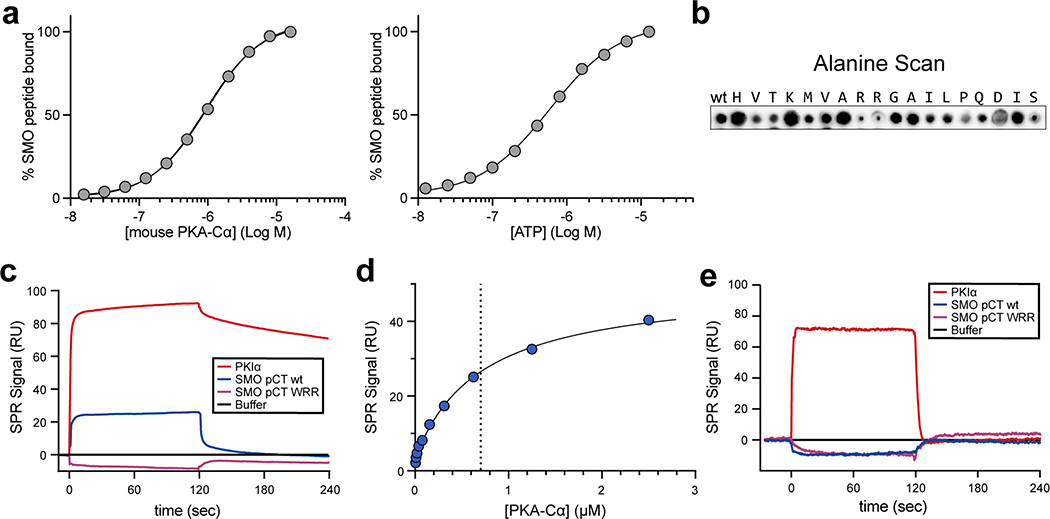
We measured the affinity of the SMO PKI motif for PKA-Cα, the best-studied and most ubiquitously expressed PKA-C isoform32, using fluorescence polarization assays. A fluorescently labeled peptide encompassing the SMO PKI motif (standard SMO peptide, Fig. 1a, red) bound saturably to purified human (KD = 823 nM, Fig. 1b (top)) or mouse (KD = 911 nM, Extended Data Fig. 2a (left)) PKA-Cα. These interactions showed pseudosubstrate characteristics: they were strongly ATP-dependent45 (EC50 for ATP = 19.2 μM (human) or 0.59 μM (mouse), Fig. 1b (bottom) and Extended Data Fig. 2a (right)) and were blocked by alanine substitution of the P−2 / P−3 arginines and the P−13 tryptophan, hereafter referred to as the “WRR mutation” (Fig. 1c, Extended Data Fig. 2b). Along similar lines, direct surface plasmon resonance (SPR) binding studies revealed that a recombinant protein encompassing the entire 93 amino-acid SMO pCT bound specifically to mouse PKA-Cα (Fig. 1d). These interactions displayed fast on- and off-rates with transient kinetics, and steady-state analysis of the binding data revealed a KD of 752 nM (Extended Data Fig. 2c,d). Consistent with our peptide binding studies, PKA-C interactions with the SMO pCT depended strictly on ATP and MgCl2 (Extended Data Fig. 2e), and no PKA-C binding was observed for the WRR mutant (Fig. 1e, Extended Data Fig. 2c,e). Interestingly, the above SMO PKI binding assays revealed weaker interactions with PKA-C than observed for conventional pseudosubstrates (KD = 1.1 nM for PKIα(5–24) peptide46). We discuss the biochemical basis and biological significance of these affinity differences below.
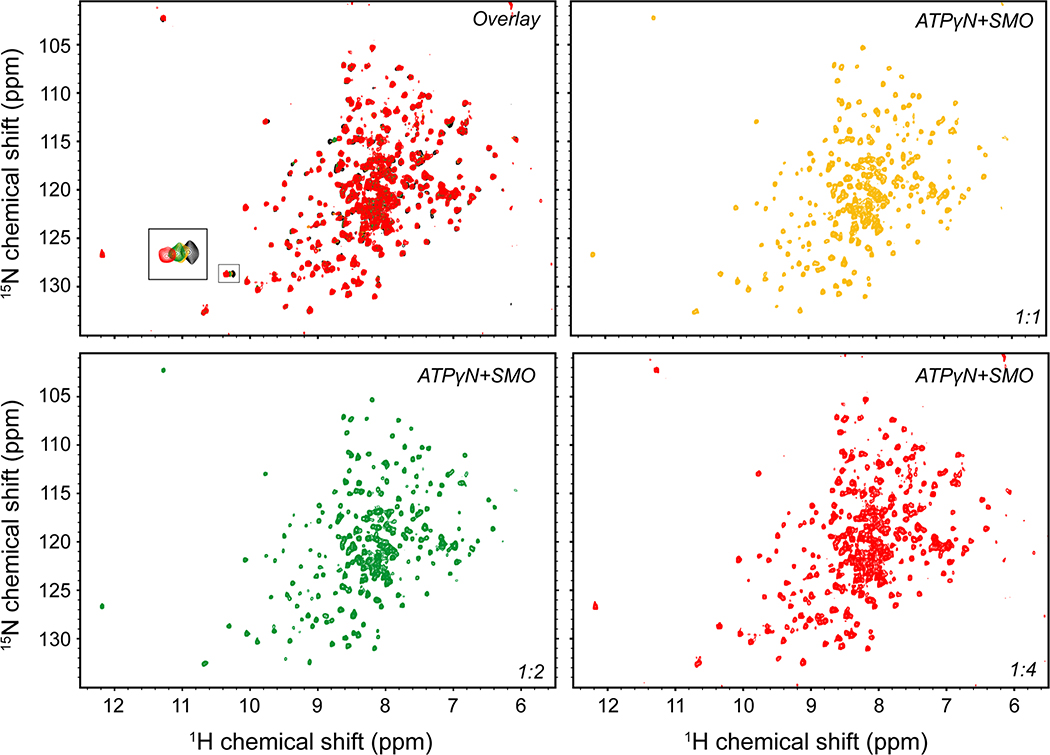
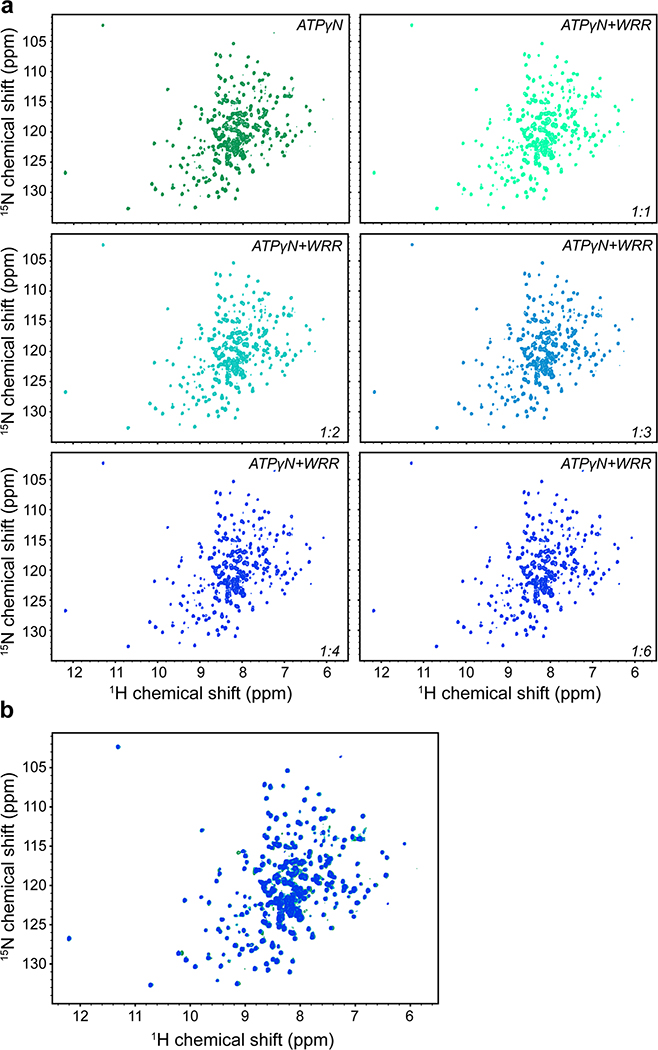
We next studied the interaction between PKA-C and the SMO PKI motif at the atomic level using nuclear magnetic resonance (NMR) spectroscopy. Binding of a peptide encompassing the SMO PKI motif induced chemical shift perturbations (CSP) in the amide fingerprint of the enzyme, especially within key domains throughout the PKA-C kinase core (Fig. 2a, Extended Data Fig. 3–5). The SMO peptide CSPs reflect specific effects on PKA-C, based on several lines of evidence. First, the CSPs involve regions throughout the kinase N-lobe critical for pseudosubstrate peptide binding and subsequent reorientation of the kinase domain, such as the αA, αF, and αG helices as well as the catalytic loop (Fig. 2a,b). Second, a SMO peptide harboring the WRR mutation displayed consistently smaller CSPs than its wild-type counterpart, particularly in the kinase N-lobe including the peptide-binding groove (Fig. 2a, Extended Data Fig. 4). Third, several of the SMO peptide CSPs exhibit a linear dependence on SMO peptide concentration (Extended Data Fig. 3), indicative of a specific interaction with the kinase. Finally, PKIα(5–24) readily displaced the SMO peptide when added as a competitor in our NMR samples, demonstrating that both peptides compete for overlapping sites within the PKA-C peptide-binding groove (Fig. 2c, Extended Data Fig. 5). Thus, the SMO peptide biases the kinase toward an inactive conformation, although to a lesser extent than does PKIα(5–24). This conclusion is consistent with the divergent biological functions and regulatory mechanisms of PKI vs. SMO (see “Discussion”).
Fig. 2: SMO induces changes in the amide fingerprint of PKA-C.
a, Mapping of amide backbone chemical shift perturbations (CSP) for [1H, 15N]-labeled PKA-Cɑ bound to nucleotide (ATPɣN) and either an extended SMO wild-type (red) or WRR mutant (blue) peptide, provided at a 1:6 PKA-Cɑ:peptide ratio, and calculated relative to ATPɣN-bound PKA-Cɑ without peptide. Key secondary structural elements within the N-lobe and C-lobe of PKA-Cɑ are determined via assignment of resonances in the kinase spectrum as previously described43, and are indicated along the X-axis. Gray spheres designate PKA-Cɑ residues that typically show a signal for PKIɑ(5–24)43,99,100 but are broadened out upon binding SMO. Dashed lines indicate one standard deviation from baseline for each dataset. b, CSP values from a were mapped onto the PKA-Cɑ structure (PDB: 4WB5) and displayed as a heatmap. c, Competition-induced changes in the indicated residues observed in the NMR spectrum upon titration of PKIɑ(5–24) peptide (1:0.5, 1:1, 1:2, 1:4, or 1:6 ratios, relative to PKA-Cɑ) into a SMO peptide:ATPɣN:PKA-Cɑ complex (1:6 PKA-Cɑ:SMO peptide ratio). Spectrum for ATPɣN-bound PKA-Cɑ (green) and SMO peptide:ATPɣN:PKA-Cɑ complex (red) are shown for reference.
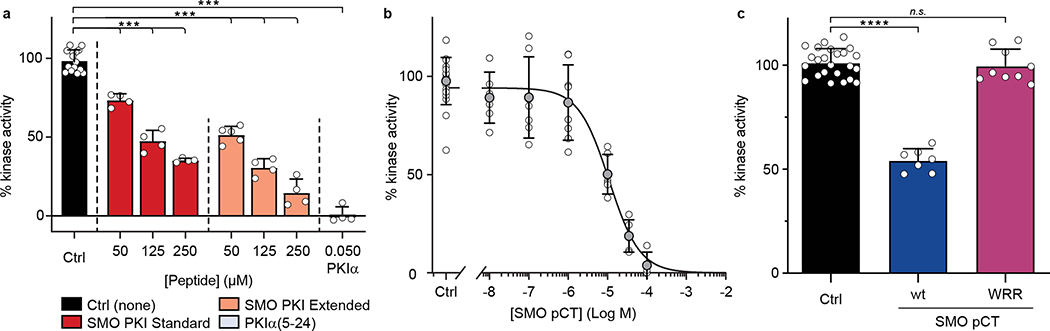
SMO PKI peptides also blocked PKA-C enzymatic activity in vitro, as assessed via a PKA-C substrate phosphorylation assay47, with an extended length peptide (Fig. 1a) inhibiting PKA-C more profoundly (Fig. 3a). In keeping with this trend, the recombinant SMO pCT (Supplementary Fig. 1a,b) required 5–10-fold lower concentrations to efficiently block PKA-C activity compared to SMO PKI peptides (IC50 = 10.9 μM for pCT vs. 50–125 μM for SMO PKI peptides, Fig. 3b). Consistent with a pseudosubstrate mode of inhibition, introducing the WRR mutation into the SMO pCT restored PKA-C activity (Fig. 3c). These data indicate that the SMO PKI motif is sufficient to inhibit PKA-C substrate phosphorylation, with additional sequences in the pCT enhancing the efficiency of inhibition.
Fig. 3: SMO inhibits PKA-C enzymatic activity.
a, Spectrophotometric assay of PKA-Cɑ substrate phosphorylation, in the presence of standard (red) or extended (coral) SMO peptides (see Fig. 1a) or a control PKIα(5–24) peptide (grey). b, Concentration dependent inhibition of PKA-Cɑ with recombinant SMO pCT. c, As a, but comparing wild-type vs. WRR mutant versions of the recombinant SMO pCT.Inhibition in a-c is calculated relative to a control without SMO or PKIα(5–24) peptide (Ctrl.). Data in a-c is shown as the mean +/− standard deviation of two independent experiments. P < 0.001 (***); P < 0.0001 (****), n.s. = not significant. See Supplementary Table 1 for full statistical analysis.
The SMO PKI motif is required for Hh signal transduction
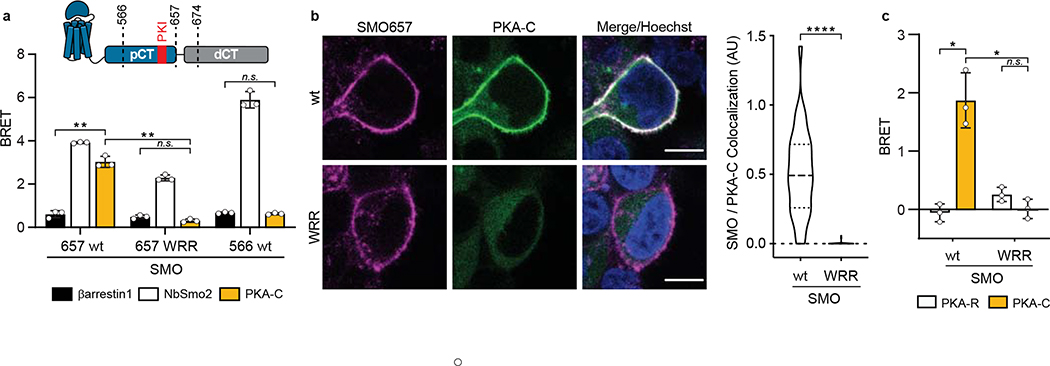
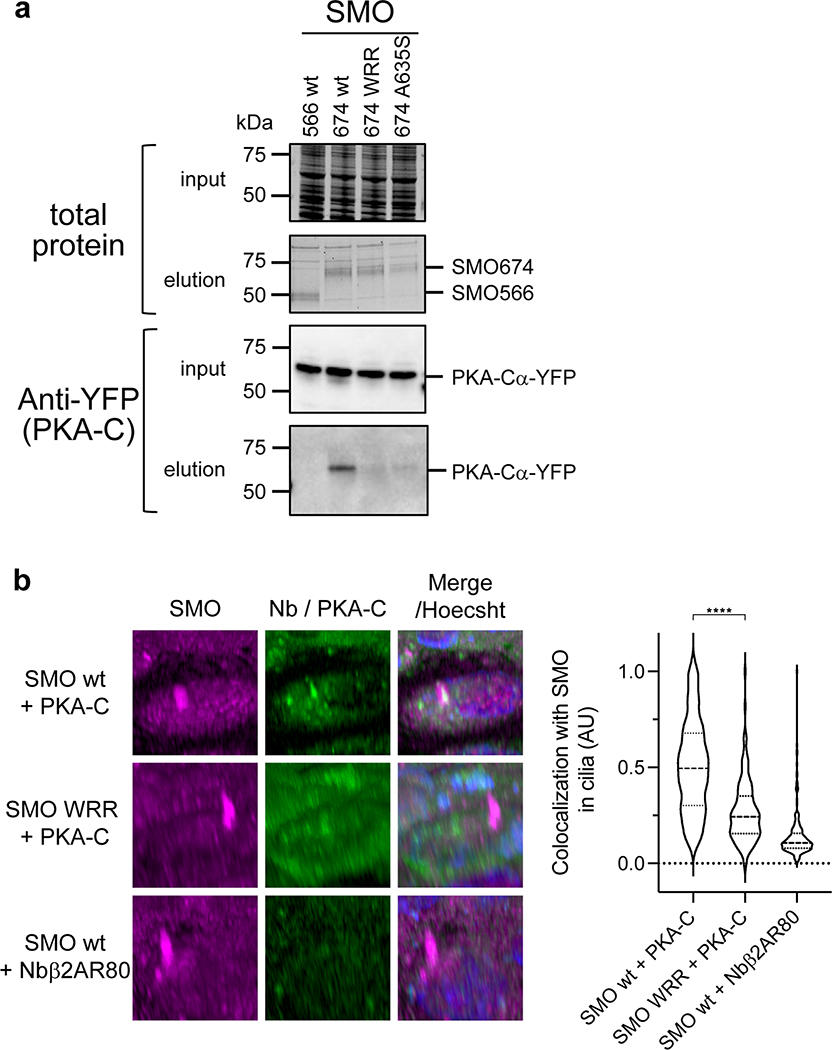
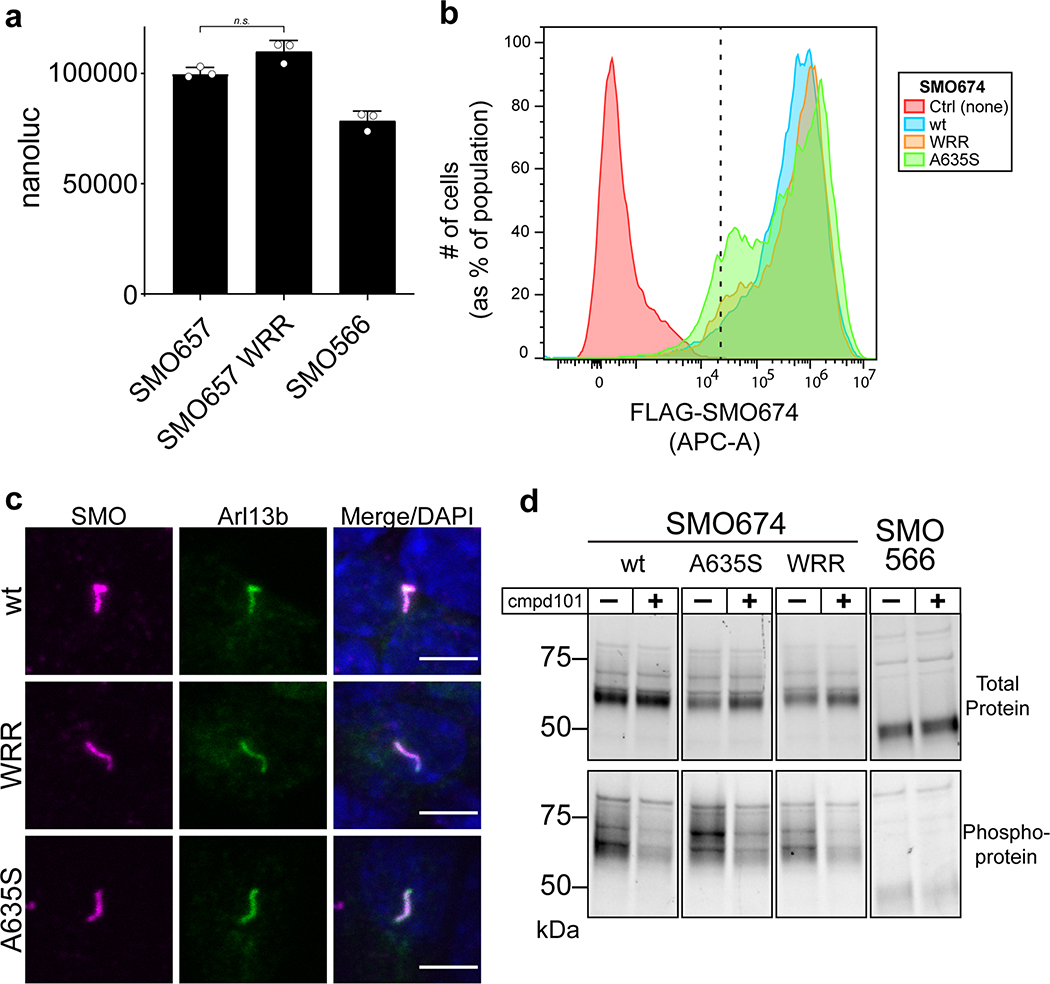
We next asked whether the SMO PKI motif contributes to PKA-C binding and inhibition in living systems. Our initial studies involved a simplified HEK293 cell model for SMO / PKA-C regulation and employed truncated SMO constructs (either SMO657 or SMO674, with the number indicating the C-terminal-most residue) that exhibit improved expression and biochemical stability compared to full-length SMO29. We used bioluminescence resonance energy transfer (BRET)48,49 to detect interactions between nanoluciferase (nanoluc)-tagged SMO and YFP-tagged PKA-C constructs in a cellular environment29. In this assay, PKA-C exhibited strong, specific BRET with wild-type SMO657, but not with SMO657 harboring the WRR mutation (Fig. 4a). Coimmunoprecipitation assays confirmed these results (Extended Data Fig. 6a). Similarly, the WRR mutation prevented SMO674 from colocalizing with PKA-C at the plasma membrane29, as determined by live-cell confocal microscopy (Fig. 4b).
Fig. 4: The SMO PKI motif is required for Hh signal transduction.
a, Top, schematic diagram of truncated SMO expression constructs. Bottom, BRET analysis of SMO / PKA-C interactions in HEK293 cells expressing nanoluc-tagged wild-type SMO657 (SMO657 wt) or SMO657 harboring the WRR mutation (SMO657 WRR), along with YFP-tagged PKA-Cɑ. SMO566 (which lacks the C-tail) serves as a negative control donor. YFP-tagged βarrestin1 (which exhibits minimal binding to SMO) and NbSmo2 (which binds the intracellular surface of the SMO 7TM domain) serve as negative and positive control acceptors, respectively29. Data represent mean +/− standard deviation, n = three biologically independent samples. b, Left, confocal images of live HEK293 cells coexpressing GFP-tagged PKA-Cɑ with FLAG-tagged wild-type or mutant SMO674. Cells were treated with SMO agonist (SAG21k). Scale bar = 10 μm. Right, quantification of SMO / PKA-C colocalization with the median represented by a dashed line and the upper and lower quartiles indicated by dotted lines (n=34 or 48 cells for “wt” or “WRR”, respectively, examined over two independent experiments). c, BRET analysis of SMO / PKA-C interactions in IMCD3 cells expressing nanoluc-tagged wild-type or mutant SMO, along with low levels of PKA-Cɑ-YFP. PKA-RIɑ-YFP serves as a negative control. Under these conditions, PKA-Cɑ-YFP is expressed at substantially lower levels than PKA-RIɑ-YFP29. Data represent mean +/− standard deviation, n = three biologically independent samples. P < 0.05 (*); P < 0.01 (**), P < 0.0001 (****), n.s. = not significant. See Supplementary Table 1 for full statistical analysis.
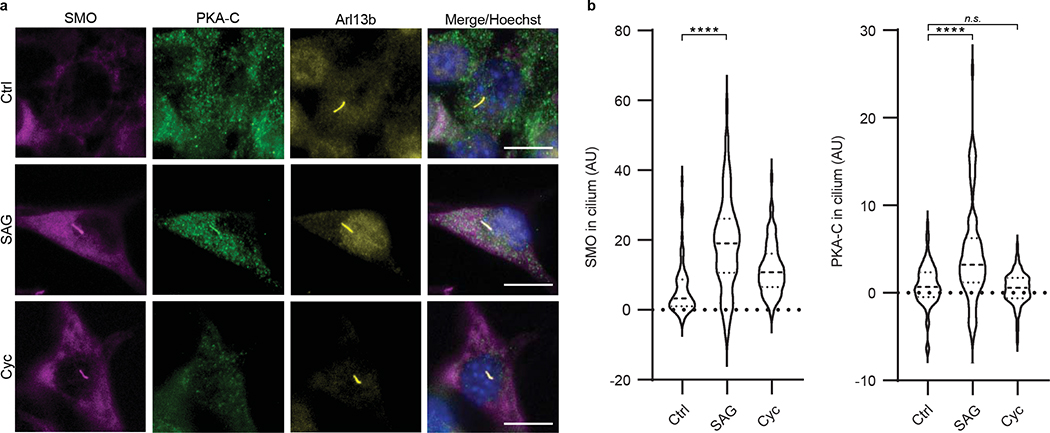
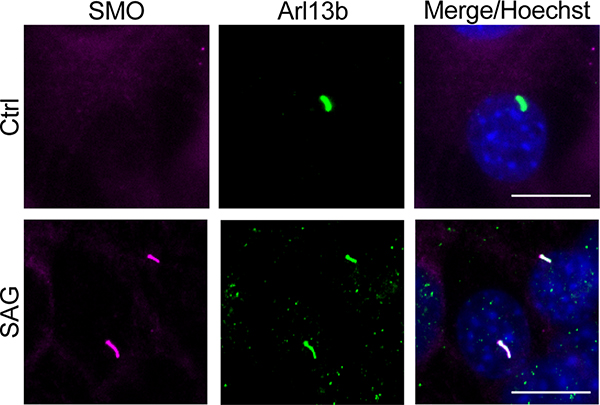
We extended the above findings to more physiological cellular models of Hh signaling in primary cilia, using SMO constructs with full-length unmodified C-termini. We studied SMO / PKA-C colocalization in cilia using inner medullary collecting duct (IMCD3) cells, a robustly ciliated kidney cell line that is used extensively in studies of ciliary Hh signal transduction29,50–52. Upon Hh pathway activation, SMO accumulates in IMCD3 primary cilia50–52, where it colocalizes with PKA-C29. In contrast, PKA-C displayed dramatically reduced colocalization in cilia with SMO containing the WRR mutation (Extended Data Fig. 6b). The WRR mutation also prevented interaction between SMO and PKA-C (expressed at near-endogenous levels29) in these ciliated cells (Fig. 4c). We also observed that SMO colocalizes with endogenous PKA-C in the primary cilium of NIH3T3 fibroblasts (Fig. 5, Extended Data Fig. 7), another widely utilized cultured cell model of Hh signal transduction. Colocalization only occurred when SMO was in an active, agonist-bound state; it did not occur when SMO was bound to the inverse agonist cyclopamine (Fig. 5), which induces SMO to accumulate in cilia but traps it in an inactive conformation53–55. We conclude the SMO colcocalization with endogenous PKA-C in NIH3T3 cilia is highly specific, as it only occurs when SMO is in an active state.
Fig. 5: SMO colocalizes with endogenous PKA-C in primary cilia.
a, Left, immunofluorescence microscopy images of endogenous PKA-C (green) in NIH3T3 cells stably expressing a V5-TurboID-tagged full-length SMO construct at low levels (see Extended Data Fig. 7 and “Supplementary Note 1”), and treated with SAG, cyclopamine (“Cyc”), or vehicle control (“Ctrl”) for 8 hours. SMO is marked by a fluorescent streptavidin conjugate (magenta). Cilium is marked by Arl13b (yellow). Scale bar = 10 μm. b, Quantification of SMO or PKA-C signal intensity in cilia, with the median represented by a dashed line and the upper and lower quartiles indicated by dotted lines (n=74, 81, or 78 cells per condition for “Ctrl”, “SAG”, or “Cyc”, respectively, examined over two independent experiments). P < 0.0001 (****), n.s. = not significant. See Supplementary Table 1 for full statistical analysis.
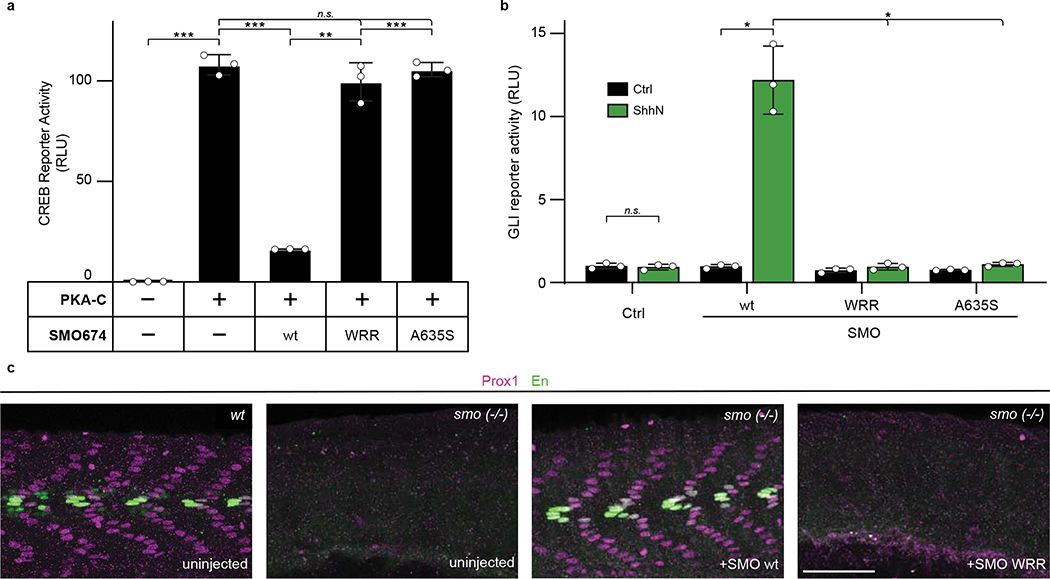
We next assessed the impact of the SMO PKI motif on PKA-C activity in cells using a model PKA-C substrate, the cyclic AMP response element binding protein (CREB) transcription factor56, and a CREB transcriptional reporter assay. This experimental paradigm entails overexpression of PKA-C at levels exceeding those of endogenous PKA-R, thereby minimizing potentially confounding contributions from heterotrimeric G protein- and cAMP-containing cascades29. Under these conditions, wild-type SMO674 blocked PKA-C-mediated CREB reporter activation while SMO674 harboring the WRR mutation failed to do so (Fig. 6a). We observed similar effects with serine substitution of the P-site alanine (A635S, Fig. 6a and Extended Data Fig. 6a), which converts pseudosubstrates to substrates and thereby promotes their dissociation from the PKA-C active site38,57.
Fig. 6: The SMO PKI motif is required for Hh signal transduction.
a, CREB transcriptional reporter assay, reflecting PKA-C mediated substrate phosphorylation, in HEK293 cells transfected with PKA-Cɑ and the indicated SMO674 constructs. Data represent the mean +/− standard deviation, n = three biologically independent samples. b, GLI transcriptional reporter assay in Smo−/− MEFs transfected with a GFP negative control (Ctrl), or the indicated wild-type (wt) or mutant SMO constructs. Cells were treated with conditioned medium containing the N-terminal signaling domain of Sonic hedgehog (ShhN, green), or control, non-ShhN-containing conditioned medium (Ctrl, black). Data represent the mean +/− standard deviation, n = three biologically independent samples. c, Wild-type or smo−/− zebrafish injected with the indicated mRNA constructs were stained for Prox1 (magenta) or Engrailed (En, green) to mark muscle fiber nuclei. n=12 (uninjected), n=41 (SMO wt), n= 47 (SMO WRR). P < 0.05 (*); P < 0.01 (**), P < 0.001 (***), n.s. = not significant. See Supplementary Table 1 for full statistical analysis of Fig. 6a,b.
To capture the entire process of Hh signal transduction from the cell surface to the nucleus, we used mouse embryonic fibroblasts (MEFs) and a GLI transcriptional reporter assay11,58,59. This model strictly requires SMO, PKA-C, GLI, and an intact cilium58,60,61. In Smo−/− MEFs, transfection of wild-type SMO enabled strong GLI transcriptional responses to Hh ligands, whereas the WRR and A635S mutants were almost completely devoid of activity (Fig. 6b).
Mutations in the SMO PKI motif specifically affect binding to and inhibition of PKA-C, based on the following control studies. First, the expression levels and electrophoretic mobilities of these mutants were similar to wild-type controls, and the mutants localized correctly to primary cilia (Extended Data Fig. 6a, 6b, and 8). In addition, the SMO mutations only minimally affected BRET with nanobody 2 (NbSmo2, Fig. 4a), which selectively binds the active state of SMO via the cytoplasmic face of its seven-transmembrane (7TM) domain29. Finally, the mutants underwent normal SMO activity-dependent phosphorylation by GRK2/3 kinases29,62 (Extended Data Fig. 8d). These data rule out possible nonspecific effects of the mutations on SMO expression, trafficking, conformational activation, or ability to serve as a GRK2/3 substrate.
To examine the role of the SMO PKI motif in Hh signal transduction in vivo, we studied the specification of slow muscle cell types in zebrafish embryos, a widely utilized model of morphogenetic Hh signal transduction during vertebrate development29,63,64. While expression of wild-type SMO restored correct muscle specification to smo−/− zebrafish, the WRR or A635S mutants did not (Fig. 6c, Supplementary Table 2).
Taken together, our studies demonstrate a requirement for PKA-C pseudosubstrate interactions in SMO inhibition of PKA-C, and activation of GLI, during Hh signal transduction in both cultured cells and organisms.
An avidity-based mechanism for SMO inhibition of PKA-C
PKA-C interactions with the SMO PKI motif, while essential for Hh signal transduction (Fig. 6), appear weaker (KD = 823–911 nM for SMO PKI peptides, Fig. 1b, Extended Data Fig. 2a) than those with a canonical PKA-C pseudosubstrate, (KD = 1.1 nM for PKIα(5–24) peptide46). SMO / PKA-C interactions, however, are also influenced by sequences outside the PKI motif, indicating that they are more favorable in vivo than the in vitro measurements suggest. Two lines of evidence support this proposal. First, SMO mutations known to reduce PKA-C interactions in vivo, including deletion of a predicted amphipathic helix spanning residues 570–581 or alanine substitution of GRK2/3 phosphorylation sites29, map to non-PKI regions of the pCT (summarized in Fig. 7a). Second, SMO pCT interactions with PKA-C are enhanced by the distal SMO C-tail (dCT, residues 658–793)29, demonstrating that this region harbors additional PKA-C binding determinants. Accordingly, in HEK293 cells, PKA-C interactions with SMO containing its full-length C-tail are only partially blocked by the WRR mutation (Fig. 7b), in contrast to the near-complete loss of interaction with SMO lacking the dCT (Fig. 4a),
Fig. 7: An avidity-based mechanism for SMO inhibition of PKA-C.
a, Annotated sequence of the mouse SMO pCT. PKI motif is indicated in red, along with GRK2/3 phosphorylation sites (orange) and residues 570–581 (green box), previously shown to influence SMO / PKA-C interactions and Hh signal transduction29,54. b, BRET analysis of SMO / PKA-C interactions in HEK293 cells transfected with full-length wild-type SMO (wt, navy) WRR mutant (purple), or C-terminally truncated (SMO566, white) versions of SMO as donor, and the indicated DNA amounts of PKA-Cɑ as acceptor. Nonspecific signal is indicated by the negative control BRET acceptor βarrestin1 (dashed line). Data represent the mean +/− standard deviation, n = three biologically independent samples.). P < 0.05 (*); P < 0.01 (**), See Supplementary Table 1 for full statistical analysis. c, Representative image of a tiled array of 18mer peptides covering the complete C-tail of human SMO, probed with PKA-Cɑ as in Fig. 1c. Peptides that bind are boxed and their sequences indicated. d, Proposed model for Hh signal transduction, as described in “Discussion.”
In light of the above findings, we searched for additional PKA-C binding sequences in the SMO C-tail using a tiled peptide microarray. While this approach is not ideal for detecting binding interactions involving extensive secondary structure, it is well-suited to uncover short linear PKA-C binding motifs within protein domains predicted to be largely unstructured or disordered65,66, such the SMO C-tail14,29. Indeed, our microarray identified several PKA-C-binding sequences in addition to the PKI motif, including one in the dCT with PKI-like attributes (Fig. 7c, Extended Data Fig. 9).
Taken together, our data support an avidity-based mechanism for SMO / PKA-C binding. A set of ancillary interactions cooperate to stabilize the core binding events between the SMO PKI motif and the PKA-C active site. This avidity-based mechanism may provide the essential link between SMO activation and disruption of PKA-C phosphoryl transfer during Hh signal transduction.
DISCUSSION
SMO inhibition of PKA-C is a central aspect of Hh signal transduction in development and disease. Instead of obeying the existing paradigms for GPCR-PKA signaling, SMO enjoys a private signaling pathway whereby it directly binds PKA-C as a pseudosubstrate. This both restricts access of PKA-C to soluble targets and extinguishes its enzymatic activity. A PKI motif encoded by the SMO cytoplasmic domain is central to this mechanism in two ways. First, the PKI motif sequesters PKA-C away from GLI by promoting SMO / PKA-C interactions, which recruits PKA-C to the ciliary membrane. Second, the PKI motif occludes the PKA-C active site, interrupting catalysis. By utilizing this two-pronged strategy, SMO can efficiently block PKA-C-mediated phosphorylation, activate GLI, and promote transcription of Hh pathway target genes.
Based on our findings, we propose a revised model for Hh signal transduction downstream of SMO (Fig. 7d). In the pathway “off” state, SMO is in an inactive conformation11,53,59,67. As a result, SMO cannot be phosphorylated efficiently by GRK2/329,62, and exists at low levels in the ciliary membrane54,68,69. We expect interactions between PKA-C and the SMO PKI motif to be unfavorable under these conditions due to their distinct subcellular localizations and relatively weak affinity. In the pathway “on” state, Hh-mediated inactivation of PTCH1 enables SMO to bind its endogenous sterol ligands and adopt an active conformation11–13,59; alternatively, oncogenic PTCH1 loss-of-function mutations or SMO gain-of-function mutations (i.e., SMOM2) constitutively activate SMO by stabilizing the active conformation of the SMO 7M domain12,67,70,71. Consequently, active SMO is recognized by GRK2/3, undergoes phosphorylation of its pCT, and accumulates in the cilium, bringing SMO in close proximity to ciliary PKA-C. This allows SMO to bind PKA-C via its PKI motif, which both sequesters PKA-C at the ciliary membrane29 and blocks PKA-C-mediated catalysis.
Along with the PKI motif, our study identified additional PKA-C-binding sequences in the SMO pCT and dCT (Fig. 7); while such sequences cannot support Hh signal transduction in the absence of an intact SMO PKI motif (Fig. 6b,c), they may nevertheless contribute to pathway activation in a more peripheral way, such as by stabilizing the interaction between the PKI motif and the kinase active site. We expect that binding of the SMO PKI motif to PKA-C is also enhanced by the membrane itself, which restricts diffusion of proteins to two dimensions and can thereby dramatically increase effective concentrations relative to soluble compartments72. Additionally, SMO activation could conceivably decrease ciliary cAMP via coupling to inhibitory (Gαi/o/z) G protein cascades59,73,74 or by triggering the ciliary exit of GPR161, a constitutively active GPCR that couples to stimulatory (Gαs) G proteins50,52,75. These processes, while not absolutely required for Hh signal transduction75–80, may reduce levels of free (non-PKA-R-bound) PKA-C in cilia81, and thereby aid the SMO PKI motif in binding and inhibiting the ciliary pool of PKA-C. Thus, activation of SMO prevents PKA-C substrate phosphorylation via a host of auxiliary interactions and regulatory influences that assist the essential pseudosubstrate action of the SMO PKI motif.
Our study helps explain a crucial but enigmatic aspect of Hh signal transduction, namely how PKA-C is inhibited, and GLI activated, only when the Hh pathway is in an “on” state. Indeed, our findings are consistent with a number of prior observations, such as that inactivation of GRK2/3 decreases SMO / PKA-C interactions29, blocks SMO / PKA-C colocalization29, and abolishes Hh signal transduction64,82,83. Based on our model, these effects can be explained by GRK2/3 inhibition weakening the SMO / PKA-C interaction below a critical threshold such that the PKI motif cannot efficiently engage the enzyme’s active site. Furthermore, our findings suggest that SMO / PKA-C interactions are appropriately tuned: If the SMO PKI motif were to bind PKA-C with high affinity, the Hh pathway could misfire, as PKA-C would be inhibited (and GLI dephosphorylated) even when SMO is inactive, similar to organismal phenotypes observed upon loss of PKA-C function60,84–86. This proposal is consistent with the relatively modest effects of the SMO pseudosubstrate peptide on the PKA-C amide fingerprint in our NMR studies. Unlike PKI, which functions as a constitutive PKA-C inhibitor, only the active state of SMO is expected to efficiently bind and inhibit PKA-C under physiological conditions. Thus, short peptides containing only the minimal SMO pseudosubstrate sequence but missing the key structural determinants that specify the SMO active state (such as the 7TM domain and GRK2/3 phosphorylation sites) have only a limited capacity to bias PKA-C toward an inactive conformation. Conversely, reducing the affinity of PKA-C for the SMO PKI motif, as occurs with the SMO WRR or A635S mutations, prevents the SMO active state from inhibiting PKA-C and thereby hinders GLI transcription. A similar effect on Hh signaling occurs when PKA-C is ectopically activated84–86; although the PKA-C binding site on SMO remains intact under these conditions, the pool of active PKA-C is expanded beyond the capacity for SMO-mediated sequestration and inhibition. Future structural studies of full-length SMO / PKA-C complexes will provide a more detailed mechanistic understanding of how the SMO pCT engages PKA-C, and may also reveal precisely how GRK2/3 facilitates these interactions.
Our model predicts that SMO and PKA-C will interact in cilia under physiological conditions, but only when SMO is in an active conformation. In support of this prediction, we found that endogenous PKA-C colocalizes in NIH3T3 cilia with agonist- but not inverse agonist-bound SMO. These studies focused on endogenous PKA-C subunits, thereby minimizing potential artifactual effects of PKA-C overexpression on endogenous PKA regulatory complexes (see also “Supplementary Note 1”). Our model is consistent with a recent report that stimulation of the Hh pathway decreased PKA-C kinase activity in cilia, as measured by a live-cell fluorescent PKA-C activity reporter 79. While the authors interpreted their result in terms of potential cAMP-dependent processes downstream of SMO, an alternative and non-mutually exclusive explanation is that SMO directly inhibits PKA-C enzymatic activity via the pseudosubstrate mechanism described here. Going forward, an important goal is to determine the relative stoichiometry of SMO and free (i.e., non-PKA-R-bound) PKA-C in cilia (see “Supplementary Note 2”), as well as these proteins’ activity states within the various ciliary subcompartments where each biochemical step of the pathway occurs. This remains extremely challenging at present given the relatively limited signal-to-noise and spatial resolution of currently available tools. However, new advances in ciliary proteomics and live-cilium biosensors may enable such studies in the future.
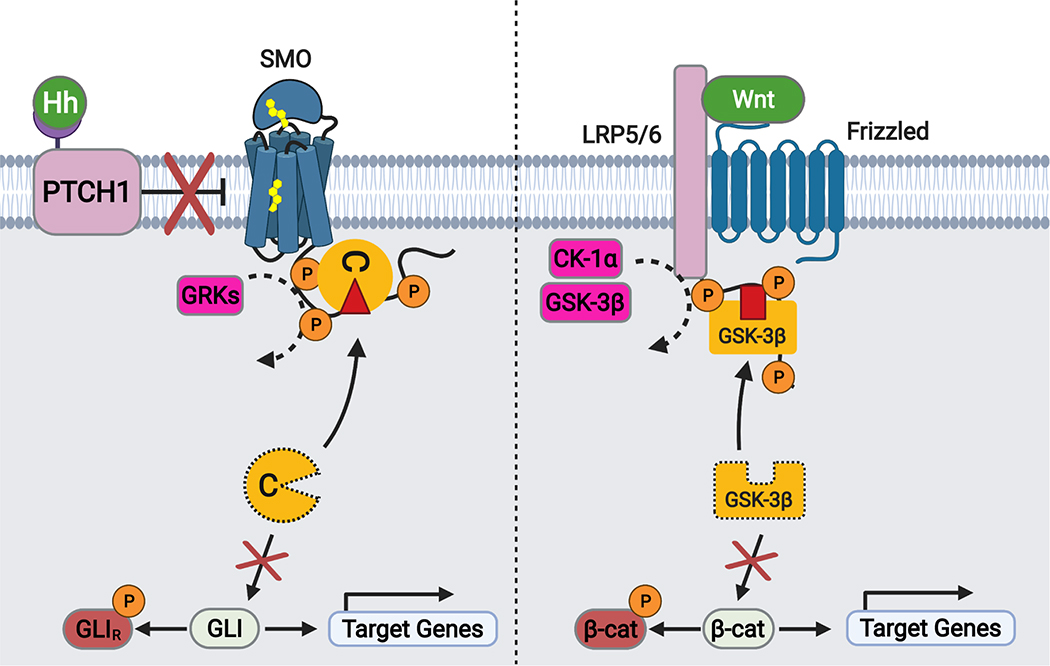
The mechanism described above represents, to our knowledge, an unprecedented mode of GPCR-kinase communication. It is, however, remarkably similar to a strategy used by another critical developmental and cancer pathway: the Wnt cascade87–90 (Extended Data Fig. 10). During the initial steps of Wnt signal transduction at the plasma membrane, the cytoplasmic tail of LDL receptor-related proteins 5/6 (LRP5/6) serves as a pseudosubstrate inhibitor of GSK-3β kinases. This blocks phosphorylation and degradation of the transcriptional coactivator β-catenin91–94. Moreover, the LRP5/6 tail binds GSK-3β with low affinity, but Wnt pathway activation enhances this interaction through phosphorylation of the LRP5/6 tail as well as recruitment of GSK-3β-containing protein complexes to the plasma membrane91–93. Thus, in both the Hh and Wnt cascades, a phosphorylated receptor tail sequesters and competitively inhibits a kinase via an avidity-based mechanism. It was unexpected that these two pathways, which utilize distinct molecular components and rely on disparate subcellular environments (cilium vs. plasma membrane)21,95, nevertheless share a common mechanism for intracellular signal transduction. Why such a mechanism evolved to carry out central steps in these pathways is presently unclear. One possibility is that it serves to spatially restrict the kinase-inhibiting effects of upstream receptors and thereby avoid pleiotropic effects of inhibiting these kinases globally. This may help to ensure signaling specificity and reduce untoward crosstalk with other pathways88,91,94. It is also possible that such a mechanism is particularly well-suited to the embryogenesis- and homeostasis-related functions of Hh and Wnt signaling. In any event, the use of a similar strategy during Hh and Wnt signal transduction hints at a more general applicability to a range of transmembrane receptors and kinases. Given the ubiquity of these proteins in metazoan physiology and disease22,23,33, and their prevalence as therapeutic targets96,97, this mechanism represents an exciting area for future study.
METHODS:
Cell culture and zebrafish husbandry.
HEK293FT cells (gift of P. Beachy, Stanford University), IMCD3 Flp-in cells (gift of P. Jackson, Stanford University), Sf9 cells (gift of K.C. Garcia, Stanford University), Smo−/− MEFs (gift of P. Beachy, Stanford University), and HEK293-Freestyle cells (gift of R. Majeti, Stanford University) were grown as previously described29. NIH3T3 Flp-in cells (gift of R. Rohatgi, Stanford University) were grown according to the manufacturer’s instructions in DMEM (MT10013CM, Fisher Scientific) with 10%FBS (MT35010CV, Fisher Scientific). Stably transfected HEK293 Flp-in T-rex cells (gift of D. Julius, UCSF) were constructed and maintained as previously described11. IMCD3 stable lines coexpressing FLAG-SMO + PKAC-mNG or FLAG-SMO + β2ARNb80-mNG were previously described, and the stable line expressing FLAG-SMO(WRR) + PKACmNG was produced as previously described. Zebrafish were maintained as previously described29.
Antibodies, small molecules, and other reagents.
SAG21k was a gift from P. Beachy. Cmpd101 was obtained from Hello Bio. Rabbit anti-GFP (which also detects YFP) was obtained from Thermo Fisher Scientific (A11122). SAG (S7779) and Cyclopamine (S1146) were obtained from Selleckchem. Poly-D-Lysine (A003E) and Biotin (B4501-1G) were obtained from Sigma. 20% PFA was obtained from Electron Microscopy Science (15713). Hygromycin B (10687010) and Lipofectamine 2000 reagent (52887) were from Invitrogen. Mouse anti-V5 (R960-25) was obtained from Thermo Fisher. Mouse anti-Arl13b was obtained from Antibodies Inc (75–287, IMCD3 studies), and rabbit anti-Arl13b was obtained from ProteinTech (17711-1-AP, NIH3T3 studies). Mouse anti-Prox1 was obtained from the Monoclonal antibody facility, University of Oregon (4D9), and rabbit anti-engrailed was obtained from AngioBio (11-002P). Purified mouse anti-PKA-C was from BD Biosciences (610980). Rhodamine anti-rabbit IgG (711025152), Alexa 488 anti-mouse IgG (715545150), Alexa 647 streptavidin (016600084) were obtained from Jackson ImmunoResearch. Alexa 647-conjugated M1 FLAG antibody and M1 FLAG affinity resin were prepared in-house. Control or ShhN conditioned medium was prepared from stably transfected HEK293 cells as previously described29. Dual-luciferase assay was obtained from Promega. Coelenterazine h was obtained from NanoLight Technology (301–500). Furimazine was obtained from AOBIOUS (AOB36539). For transfection studies, TransIT 2020 and TransIT 293 were obtained from Mirus Bio.
DNA constructs.
For expression and purification of the SMO pCT in E. coli, the ShhN gene was excised from ShhN / pHTSHP59 and replaced with residues 565–657 of mouse SMO (565KRIKK…PEEQAN657), downstream of the MBP-His8-SUMO tags in the vector pHTSHP, or for GST-constructs in the vector pGexKG. The resulting construct was termed SMO pCT(565–657). GST-PKI construct in pGexKG was previously described101. The SMO655-nanoluc, SMO674, and SMO-nanoluc constructs (all in pVLAD6, with N-terminal hemagglutinin signal sequence and FLAG tag) were previously described29. Full-length SMO (in pGEN, with C-terminal myc tag) was previously described102. Mouse PKA-Cα-YFP, PKA-RIα-YFP, and NbSmo2-YFP constructs in pVLAD6 were previously described29. Barrestin1-YFP in pCDNA3.1-zeo was previously described29. To construct IMCD3 Flp-in stable lines coexpressing FLAG-tagged mutant SMO with mNeonGreen-tagged PKA-Cα (using an IRES element), the previously described construct SMO-IRES-PKACαmNG / pEF5-FRT29 was modified to introduce the WRR mutation into SMO. To construct stable Flp-in HEK293 cell lines coexpressing wild-type or mutant SMO674 (which includes an N-terminal hemagglutinin signal sequence and FLAG tag) along with GFP-tagged PKACα, we cloned a SMO674-IRES-PKACαGFP cassette into the pCDNA5-FRT-TO vector. Untagged human and mouse PKACα constructs in the vector pRSETb were previously described101. TurboID is a gift from Alice Ting’s lab at Stanford. mSmo-V5-TurboID was inserted into the pEF5-FRT expression vector following the vendor’s instruction (ThermoFisher #V602020). Sequences of all oligonucleotides are provided in Supplementary Table 3. All mutant DNA sequences were prepared via PCR-based mutagenesis, cloned into their respective vector backbones via Gibson assembly, and verified by Sanger sequencing.
Peptide synthesis.
SMO PKI peptides (standard = SADVSSAWAQHVTKMVARRGAILP; extended = SADVSSAWAQHVTKMVARRGAILPQDVSVTPVATPVPP) were prepared via standard solid-phase synthesis by either GenScript or Elim Bio, purified via reversed-phase HPLC to ≥85% purity, and the sequence / molecular mass confirmed by mass spectrometry. A WRR mutant version of the extended SMO peptide was synthesized and validated in the same manner. For fluorescence polarization assays, a FAM fluorophore and 3xPEG linker were added to the N-terminus of the standard SMO PKI peptide during synthesis. PKIα(5–24) Peptide (TTYADFIASGRTGRRNAIHD) was synthesized as described for the SMO-PKI peptides by GeneCust (Boynes, France).
Fluorescence polarization assays.
Human or mouse PKA-Cα subunits were purified as previously described103. In addition, the protein obtained from the S200 was further purified via cation exchange chromatography. For this, the S200 peak fractions were pooled and dialyzed overnight into MonoS buffer (20 mM KH2PO4 pH 6.5, 5 mM DTT) before loading onto a MonoS cation exchange column. PKA-Cα was eluted with a gradient of 0–300mM KCl in MonoS buffer. Binding of a SMO peptide containing the PKI motif was investigated by fluorescence polarization (FP). Assay buffer for all FP experiments consisted of 50 mM MOPS pH 7, 35 mM NaCl, 10 mM MgCl2, 1 mM DTT and 0.005% Triton X-100 with the addition of 1 mM ATP in the PKA-C titrations. Experiments were performed by adding 50 μl of the titration component (PKA-C or ATP) to 150 μl of FAM-SMO containing solution. For SMO/PKA-C binding experiments, a two-fold dilution series from 16uM to 0uM of either mouse or human PKA-Cα was added to 40 nM FAM-SMO. To assess the ATP dependence of SMO binding to PKA-C, a titration with varying the ATP concentration from 0 to 1 mM (human PKA-Cα) or 12.8 μM (mouse PKA-Cα) was used with keeping PKA-C at 3 μM and FAM-SMO at 25 nM. Readings were taken with a Tecan Genios plate reader using black flat bottom 96-well Costar plates. Each experiment was repeated in at least 2 independent trials with each condition representing triplicate measurements with FAM readings at 485 nm excitation and 535 nm emission.
Peptide arrays.
Peptide arrays were synthesized with a MultiPep Flexible Parallel Peptide Synthesizer (Intavis Bioanalytical Instruments, Germany). The blots were probed with mouse or human PKA Cα protein, purified as described above for “Fluorescence polarization assays.” Unless noted otherwise all steps were carried out at room temperature. The blots were initially soaked with 100% ethanol for 5 minutes followed by 5 times 5-minute washes with water. All subsequent washes were carried out for 5 minutes 5 times with TTBS. The membranes were washed with TTBS, blocked with 5% milk in TTBS for 1 hour and washed again with TTBS. They were then soaked with TTBS containing 2 μg PKA-Cα, 5% milk, 10 mM MgCl2 and 1mM ATP overnight at 4° C. The next day the blots were washed, blocked with 5% milk in TTBS and washed again before incubation overnight at 4° C with TTBS containing 5% milk and a primary PKA-C antibody (generated in-house previously). The next day the blots were washed, then incubated with an HRP-conjugated anti-rabbit antibody (Prometheus™) in 5% milk TTBS for one hour followed by a final round of washes with TTBS. The membranes were finally covered with SuperSignal West Pico PLUS Chemiluminescent Substrate for detection of HRP (Thermo Scientific # 34580) and imaged with a ChemiDoc MP Imaging System from BIO RAD. A representative array image (representative of two separate trials) is shown; we excluded from analysis any spots that were not observed consistently across replicates.
Surface plasmon resonance (SPR).
Mouse PKA-Cα was overexpressed in E. coli BL21(DE3) cells after induction with 0.4 mM IPTG for 16 h at RT using the expression vector pRSETb-mPKACα and then purified by affinity chromatography using an IP20-resin as described earlier104. GST-PKI, GST-SMO pCT wt and WRR mutant were overexpressed in E. coli BL21(DE3) for 16 hr at room temperature, and the GST fusion proteins were purified using Protino gluthathione agarose 4B according to the manufacturer’s instruction (Macherey-Nagel). Protein-rich fractions were pooled followed by a buffer exchange via PD-10 desalting column (Cytiva, Marlborough, MA, USA) against low salt buffer (20 mM Tris-HCl, pH 8.0, 25 mM NaCl). The protein was further purified by loading onto a 1 mL ResourceQ anion exchange column (Cytiva, Marlborough, MA, USA) pre-equilibrated in low-salt buffer. Column was washed extensively with low salt buffer and protein was eluted with a gradient 0% to 100% high-salt buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl). Fractions containing the wild-type and WRR mutant GST-SMO pCT were pooled, respectively. SPR interaction studies were performed according to previous studies57,101. Briefly, the interaction studies were performed in running buffer (20 mM MOPS, pH 7.0, 150 mM NaCl, 50 μM EDTA, 0.005% P20 surfactant) at 25 °C using a Biacore T200 instrument (GE Healthcare). For measurements involving ATP/MgCl2, the buffer was supplemented with 1 mM ATP and 10 mM MgCl2. Polyclonal anti-GST antibody (Carl Roth, 3998.1) was covalently immobilized to all four flow cells of a CM5 sensorchip (Cytiva, Marlborough, MA, USA) to a level of 8,000 response units (RU) via standard NHS/EDC amine coupling. Each measurement cycle started with the sequential capture of 40−60 RU of GST-PKI and 125–200 RU of free GST. GST-SMO-RLG wt and GST-SMO-RLG WRR mutant on separate flow cell (flow rate 10 μL/min). Interaction analysis was then initiated by the injection of increasing concentrations of PKA Cα (5 nM – 2.5 μM) at a flow rate of 30 μL/min for 120 s (association) followed by 120 s dissociation with buffer without PKA Cα. Nonspecific binding was removed by subtracting SPR signals from the flow cell with only GST protein captured and additional buffer blank runs without PKA Cα (double referencing) employing Biacore T200 Evaluation Software 3.0 (Cytiva, Marlborough, MA, USA). After each cycle, the sensorchip was regenerated by injecting up to 5 times with 10 mM glycine, pH 1.9 or 2.2, to remove the GST-fusion proteins from the antibody surfaces until the baseline level was reached. Steady-state analysis was performed with Biacore T200 Evaluation Software 3.0.
NMR spectroscopy.
Recombinant human PKA-Cα (encoded by the PRKACA gene, uniprot P17612) was expressed and purified as reported100,105. Briefly, transformed E.coli BL21 (DE3) pLysS (Agilent) cells were cultured in deuterated (2H) M9 minimal medium supplemented with 15NH4Cl. Protein overexpression was initiated by adding 0.4 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) and carried out for 12 hours at 20 °C. The collected cell pellet was then resuspended in 50 mM Tris-HCl pH 8.0, 30 mM KH2PO4, 200 mM NaCl, 5 mM β-mercaptoethanol, 0.15 mg/mL lysozyme, 200 μM ATP, DnaseI, and protease inhibitor (Sigma) and passed through a French press (2 times). The cell resuspension was then cleared by centrifugation (40.000 × g for 45 minutes), and the supernatant was incubated overnight with Ni2+-NTA resin (Thermo Fisher). His-tagged PKA-Cα was eluted using 50 mM Tris-HCl pH 8.0, 30 mM KH2PO4, 100 mM NaCl, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF) supplemented with 200 mM of imidazole. The His-tag was removed during an overnight dialysis step performed in 20 mM KH2PO4 (pH 6.5), 25 mM KCl, 5 mM β-mercaptoethanol, 0.1 mM PMSF, using stoichiometric quantities of recombinant tobacco etch (TEV) protease. Finally, cationic exchange chromatography was performed to separate the three different isoforms of PKA-Cα, representing the three different phosphorylation states of the kinase, using a linear gradient of KCl in 20 mM KH2PO4 at a pH of 6.5 (HiTrap HP SP column, GE Healthcare Life Science). All the NMR experiments were performed using PKA-Cα isoform II, corresponding to phosphorylation at S10, T197, and S338 residues106. The purity of the protein preparation was tested using SDS-PAGE electrophoresis and Mass spectrometry analysis (purity >97%).
A portion of the lyophilized extended SMO PKI peptide (see above) was resuspended in 20 mM KH2PO4 (pH 6.5), 90 mM KCl, 10 mM MgCl2, 10 mM DTT, 1 mM NaN3, to a final concentration of 4 mM. The NMR experiments were performed on 100 μM uniformly 2H, 15N-labeled PKA-Cα sample in 20 mM KH2PO4, 90 mM KCl, 10 mM MgCl2, 10 mM DTT, 1 mM NaN3 at pH of 6.5 and saturated with 12 mM of a non-hydrolyzable ATP analog (ATPγN). Modified [1H-15N]-TROSY-HSQC spectra were acquired on a Bruker Advance III spectrometer operating at a proton frequency of 850 MHz and on a 600 MHz Bruker Avance NEO spectrometer, equipped with a TCI cryoprobe, at an acquisition temperature of 300K. First, a spectrum of ATPγN-saturated PKA-Cα complex (PKA-Cα/ATPγN) was recorded with 2048 (proton) and 128 (nitrogen) complex points. Stoichiometric amounts of SMO PKI peptide (hereafter referred to as SMO-PKI) were then added to the PKA-Cα/ATPγN complex until saturation (1:0, 1:1, 1:2, and 1:4 SMO-PKI:PKA-C/ATPγN molar ratio), with a concentration of SMO-PKI ranging from 0.1 to 0.4 mM. The same set of experiments was repeated for the PKA-C titrations with the WRR mutant. Briefly, lyophilized SMO PKI WRR mutant was resuspended in the NMR buffer to a final concentration of 5 mM. Stochiometric amount of this solution was added to the ATPγN-bound PKA-Cα (100 μM) until saturation, with 1:0, 1:1, 1:2, 1:4, and 1:6 PKA-Cα/ATPγN:SMO-WRR molar ratios. A [1H, 15N]-TROSY spectrum was recorded after each addition. All the spectra were processed using NMRPipe107, and visualized using NMRFAM-SPARKY software108. Combined chemical shift perturbation (CSPs) were calculated using the 1H and the 15N chemical shift derived from the PKA-Cα/ATPγN complex and the 1:4 SMO-PKI:PKA-C/ATPγN complex. The CSPs was calculated using the following equation109:
The PKIα(5–24) CSPs were originally published earlier100 and are included here for reference.
For the displacement experiments, stochiometric quantities of PKIα(5–24) ranging from 0.05 to 0.6 mM total concentration (1:0, 1:0.5, 1:0, 1:1, 1:2, 1:4, and 1:6 PKA-C/ATPγN:PKIα molar ratio) were added to a SMO-PKI-bound PKA-Cα/ATPγN complex (1:6 PKA-Cα:SMO-PKI molar ratio). All the experiments were carried out under the same conditions used for the titration of SMO PKI WT.
Purification of SMO pCT for PKA-C activity assays.
MBP-His8-SUMO-tagged SMO pCT (SMO pCT(565–657) / pHTSHP) was transformed into BL21 (DE3) E coli and grown to OD600= ~0.5–1.0 in 1 L terrific broth (TB) with ampicillin at 37°C in 2800ml baffled Fernbach flasks. IPTG was then added to 0.4mM and the temperature lowered to 18°C for 18 hours. Cells were harvested by centrifugation (5000 × g), snap-frozen in liquid nitrogen, and stored at −80°C. Cell pellets were resuspended in 20ml binding buffer (50mM Tris pH 8.0, 300mM NaCl, 10mM imidazole, protease inhibitors (Pierce)) at 4°C with stirring. Lysozyme was added at 1mg/ml as well as benzonase (10,000X, Sigma), and samples were lysed by sonication. Lysates were clarified by centrifugation at 40,000 × g, 30 min, 4°C. Supernatant was run by gravity over a column of 6ml NiNTA affinity resin (Qiagen) pre-equilibrated in binding buffer. The column was washed twice with 5 column volumes of wash buffer (50mM Tris pH 8.0, 300mM NaCl, 25mM imidazole) and eluted with four successive 1 column volumes of elution buffer (50mM Tris pH 8.0, 300mM NaCl, 250mM imidazole). Protein content of fractions was estimated by Quickstart Bradford assay and purity determined by SDS-PAGE. Protein-rich fractions were pooled and cleaved by the addition of 1.4mg Ulp1 enzyme (prepared in-house) and 1mM DTT for 1 hour at room temperature. After incubation, the samples were dialyzed overnight against dialysis buffer (50mM HEPES pH 8.0, 150mM NaCl, 7mM β-mercaptoethanol. The completed digestion reaction was applied to NiNTA resin, washed twice with 3 column volumes of dialysis buffer, and eluted with 3 column volumes of elution buffer. The flow-through and wash fractions were pooled, centrifugated at 20,000 × g, 5 min, 4°C to pellet any insoluble material, and further purified by loading onto a 1ml HiTrap HP SP cation exchange column pre-equilibrated in low-salt buffer (50mM HEPES pH8.0, 150mM NaCl). Column was washed extensively with low salt buffer and protein was eluted with a gradient to high-salt buffer (50mM HEPES pH 8.0, 1.2M NaCl). Fractions containing the cleaved SMO pCT were pooled, concentrated, and injected onto a Superdex 200 (10/300) gel filtration column equilibrated in gel filtration buffer (50 mM HEPES pH 7.5, 300 mM NaCl). The peak fractions were collected and analyzed by SDS-PAGE. Fractions containing intact pCT were pooled, snap-frozen in liquid nitrogen, and stored at −80°C. For the experiments in Fig. 3c, wild-type or mutant pCT proteins were purified by NiNTA affinity as described above and used directly in PKA-C activity assays.
In vitro PKA-C activity assays.
For in vitro PKA-C activity assays, recombinant mouse PKA catalytic subunit (Cα) was overexpressed in E. coli BL21(DE3) cells after induction with 0.4 mM IPTG for 14 hr at room temperature using the expression vector pRSETb / mPKACα and then purified by affinity chromatography using an IP20-resin as described earlier104. PKA catalytic activity was assayed using a coupled spectrophotometric assay as described previously47. Briefly, the reaction mixture contained 100 mM MOPS (pH 7), 10 mM MgCl2, 100 μM ATP, 1 mM phosphoenolpyruvate, 15 U/mL lactate dehydrogenase, 70 U/mL pyruvate kinase, 200 mM reduced nicotinamide adenine dinucleotide, 5 mM β-mercaptoethanol with 15–30 nM Cα and 260 μM Kemptide (LRRASLG; GeneCust) as a substrate. Formation of PKA-C:SMO complex (concentrations of SMO peptide or recombinant SMO pCT protein are indicated in the figures) was carried out for 3 minutes at room temperature in the assay mixture. The apparent IC50 for SMO pCT were determined by fitting the concentration-dependent activity to a sigmoid dose-response model. All data were plotted as means of at least two independent experiments measured in duplicate each with standard deviation (SD) (n = 4–24 data points per condition).
MALS.
Purified SMO pCT was concentrated to 5 mg/ml and analyzed via SEC-MALS using a Superdex 75 gel chromatography column (GE Healthcare) equilibrated in gel filtration buffer (50 mM HEPES pH 7.5, 300 mM NaCl) with an in-line DAWN MALS detector (Wyatt Technology.)
BRET.
BRET in HEK293 or IMCD3 cells was performed as previously described29. Briefly, HEK293 or IMCD3 cells were transiently transfected with 1.2 μg DNA made up of nanoluc-tagged SMO (0.3 μg) along with YFP-tagged βarrestin1(0.3 μg), NbSmo2 (0.9 μg), PKA-Cα (0.1 μg), or PKA-RIα (0.1 μg) plasmids. The amount of DNA was kept constant in each condition by the addition of empty pV1392 plasmid. We used these amounts of plasmid for all experiments with the following two exceptions: (1) Fig. 4b which examined a range of PKA-Cα DNA amounts as indicated in the figure legend; (2) Fig 3d which used 0.3 μg of PKA-Cα-YFP (corresponding to a 0.33–2.68% increase in the size of the endogenous PKA-C pool, as determined by quantitative immunoblotting29) or PKA-RIα-YFP (which displays no BRET with SMO even though it expresses at substantially higher levels than does PKA-Cα-YFP29). Cells were replated in poly-D-lysine coated white opaque 96-well plates, loaded with 5 μM coelenterazine h (HEK293) or 10 μM furimazine (IMCD3), and analyzed for BRET on a Tecan Spark multimode plate reader. The background signal from cells expressing nanoluc-tagged SMO without BRET acceptor was subtracted from all measurements. For all BRET assays, data represent mean ± SD from triplicate wells, and data are representative of at least two independent experiments.
Flow cytometry.
Cell surface expression of wild-type and mutant SMO constructs was analyzed via flow cytometry as previously described29. Briefly, HEK293-Freestyle cells were infected with BacMam viruses encoding the indicated wild-type or mutant SMO674 constructs. 1–2 days later, live cells were stained with Alexa 647-conjugated M1 anti-FLAG antibody (1:1000, 5 minutes) and analyzed on a Cytoflex flow cytometer (Beckman Coulter). To determine the percentage of positive cells, a gate was established and the percent of cells falling within that gate was calculated. This was repeated on two independent occasions.
CREB reporter assays.
CREB transcriptional reporter assays were performed as previously described29. Briefly, HEK293FT cells were transfected in 24-well plates with a 30:1 mixture of CRE-Firefly reporter (pGL4.29[luc2p/CRE/Hygro]) and constitutively expressing SV40-Renilla plasmids (20%(w/w)), PKA-Cα (0.625%(w/w)), along with the indicated wild-type or mutant SMO674 plasmids (24%(w/w)). A GFP expression plasmid was used to bring the total amount of DNA in each well to 250 ng. Two days later, reporter activity was measured via dual luciferase assay. Reporter activity is expressed as a ratio of Firefly/Renilla (relative luciferase units (RLU)). For all CREB assays, data represent mean ± SD from triplicate wells, and data are representative of at least two independent experiments.
Coimmunoprecipitation assay.
SMO / PKA-C coimmunoprecipitation was performed as previously described29, with minor modifications. Briefly, 3 ml HEK293-Freestyle cells were infected with BacMam viruses encoding PKA-Cα-YFP the indicated wild-type or mutant SMO674 constructs and solubilized in low-salt solubilization buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.1 mM TCEP, 0.5% GDN, 1 mM CaCl2•6H2O, protease inhibitor tablet) to prepare a whole-cell lysate. A portion of the lysate was reserved for SDS-PAGE analysis, and the remainder was incubated with FLAG affinity resin (10 μl settled resin per condition). After a one-hour incubation, resin was washed three times in low-salt wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.1 mM TCEP, 0.05% GDN, 1 mM CaCl2•6H2O), and protein eluted in 40 μl of the same buffer supplemented with 5 mM EDTA and 0.2 mg/ml FLAG peptide. Proteins were separated by SDS-PAGE on a 4–20% Stain-Free TGX gel (BioRad), and total protein in lysate or eluate was visualized via Stain Free imaging. To detect PKA-Cα-YFP, inputs and eluates were transferred to PVDF membranes and processed for immunoblotting with anti-GFP antibodies (1:5000) as previously described29. In-gel Pro-Q Diamond assay to detect phosphoprotein was performed according to the manufacturer’s instructions, as previously described29. Presented data are representative of experiments repeated in at least two independent trials.
HEK293 imaging.
Imaging and quantification of SMO / PKA-C colocalization in HEK293 cells were performed as previously described29, with minor modifications. Briefly, HEK293 Flp-in T-rex stable cell lines coexpressing GFP-tagged PKA-Cα with either wild-type or mutant FLAG-tagged SMO674 were plated onto μ–slide 8-well glass chamber slides (ibidi), and treated overnight with 1 μg/ml doxycycline (to induce SMO expression) and 1 μM SAG21k (to induce SMO activation). Live cells were subsequently stained for 5 min with an Alexa647-conjugated M1 anti-FLAG antibody (1:2000) followed by washing in HBSS, mounting, and visualization. Images were collected on a Leica SP8 laser scanning confocal microscope, using a 40x water immersion lens. All images were acquired with identical zoom / exposure / gain settings and processed identically in Fiji. SMO / PKA-C colocalization was determined by measuring the background-subtracted PKA-C (green) fluorescence at the membrane (GFPmembrane − GFPcytoplasm) and the background-subtracted SMO (red) fluorescence at the membrane (Alexa647membrane − Alexa647cytoplasm), using four independent line-scans across the membrane in Fiji. The background-subtracted green fluorescence divided by the background-subtracted red fluorescence is referred to as “SMO / PKA-C colocalization” and reported in arbitrary units (AU). The imaging was repeated on multiple occasions and all data were pooled together to generate the plots/quantification in the relevant figure panels.
IMCD3 imaging.
Imaging and quantification of SMO ciliary accumulation and SMO / PKA-C colocalization in IMCD3 cells were performed as previously described29. Briefly, to assess ciliary accumulation of wild-type or mutant SMO, IMCD3 cells were transiently transfected on coverslips with the indicated myc-tagged wild-type or mutant SMO / pGEN constructs, grown to confluency, fixed, and permeabilized. Coverslips were then stained with anti-myc (SMO) and anti-Arl13b (cilia) antibodies, along with DAPI to mark the nucleus. For quantitative assessment of SMO / PKA-C colocalization in live IMCD3 cilia, Flp-in IMCD3 cell lines coexpressing mNeonGreen-tagged PKA-Cα with either wild-type or mutant FLAG-tagged SMO were plated onto μ–slide 8-well glass chamber slides (ibidi), grown to confluency, and treated overnight in low-serum medium with 1 μM SAG21k (to induce SMO activation and ciliary accumulation.) A previously described cell line coexpressing mNeonGreen-tagged Nbβ2AR80 with wild-type FLAG-tagged SMO29 was used as a negative control. Live cells were subsequently stained for 5–10 min with an Alexa647-conjugated M1 anti-FLAG antibody (1:1000) and Hoechst counterstain, followed by washing in HBSS, mounting, and visualization. Cells were imaged on a Leica SP8 laser scanning confocal using a 40x water immersion lens. Three-dimensional reconstructions of Z-stacks were performed in Fiji using the 3D Viewer plugin. Quantification of SMO (red) and PKA-C/Nbβ2AR80 (green) staining was performed using CiliaQ110 and reported as a ratio of the green fluorescence in each cilium, normalized to the red fluorescence (“Colocalization with SMO in cilia (AU)”), as previously described29. All images were acquired with identical zoom / exposure / gain settings. The imaging was repeated on multiple occasions and all data were pooled together to generate the plots/quantification in the relevant figure panels.
Immunofluorescence microscopy in NIH3T3 Flp-in cells.
NIH3T3 cell lines stably expressing mSmo-V5-TurboID were generated using Flp-In system following manufacturer’s instructions. Briefly, Flp-In NIH3T3 cells were co-transfected with pEF5-FRT-mSmo-V5-TurboID and Flp recombinase expressing plasmid pOG44 with Lipofectamine 2000 reagent. Cells were selected with 0.2 mg/ml hygromycin. Single colonies were expanded and protein expression was confirmed by immunoblotting, immunofluorescence microscopy and qPCR. A cell clone that expressed low levels of SMO and displayed strong SAG-induced accumulation of SMO in cilia was chosen and expanded for further study.
mSmo-V5-TurboID stable cells were grown on Poly-D-Lysine-coated 12mm #1.5 coverslips. Once reaching 80–90% confluency, cells were treated with low-serum medium (DMEM + 0.5% FBS) or low-serum medium + 100nM SAG / 5μM Cyclopamine for indicated times. Live cells were subsequently incubated with 500μM Biotin for 15min at 37°C, 5% CO2, and washed once with DPBS before fixation in 4% PFA. Cells were blocked with blocking buffer (0.2% Triton X-100, 2% Donkey serum in PBS) for 1h at room temperature. Cells were first stained with rabbit anti-ARL13B (1:1000) and mouse anti-PKA-C (1:200) overnight at 4°C. Subsequently, cells were incubated with Rhodamine anti-Rabbit (1:100), Alexa 488 anti-mouse (1:200) and Alexa 647 streptavidin (1:200) for 1h at room temperature followed by Hoechst 33342 staining and mounting.
Imaging was performed with a LEICA DMi8 system with 63X oil-immersion lens. Quantification of SMO and PKA-C staining was performed in FIJI. Briefly, the contour of the cilium was outlined in Arl13b channel, and the intensity within the area of interest was measured in SMO and PKA-C channels. After that the contour was dragged to the nearby area immediately adjacent to the cilium to measure the background values, which were then subtracted from the cilium intensity. The intensities are reported in arbitrary units (AU).
GLI reporter assays.
GLI transcriptional reporter assays were performed as previously described29. Briefly, Smo−/− MEFs were transfected with a 30:1 mixture of 8xGli-Firefly and SV40-Renilla plasmids (50% (w/w)), along with the indicated full-length wild-type or mutant SMO constructs (in a pGEN vector, with C-terminal myc tag) (2%(w/w)), and GFP to adjust the amount of DNA to 250 ng/well. Cells were cultured to confluency, shifted to 0.5% FBS-containing medium, and treated with control or ShhN conditioned medium (1:20 dilution) for 2 days. Reporter activity was then measured via dual luciferase assay. For all GLI assays, data represent mean ± SD from triplicate wells, and data are representative of at least two independent experiments.
Zebrafish embryological studies.
Zebrafish mRNA injection, fixing, mounting, and staining of muscle cell markers were all performed on Danio rerio embryos (both males and females) derived from Tubingen strain adult zebrafish (<26 hours post-fertilization) kept on a light–dark cycle of 14 hours in light and 10 hours in dark at 27°C, using anti-Prox1 (1:1000) and anti-Engrailed (1:5) as previously described29, except that a far-red secondary antibody (donkey anti-rabbit 649, Jackson Laboratory, 1:500) was used instead of a red secondary antibody during immunohistochemistry. Zebrafish (D. rerio) were maintained in accordance with approved institutional protocols at the University of Utah. Specifically, all experimental protocols were approved by the University of Utah Institutional Animal Care and Use Committee and were in accordance with the guidelines from the National Institutes of Health (Approval number: 19-11006). The numbers of animals used in each experiment are indicated in the corresponding figure or table legends.
Software.
Microscopy data were acquired using Application Suite X 3.5.7 (Leica) and analyzed using Fiji 2.3.0. SDS-PAGE and Western blots were acquired using Image Lab 6.1.0 (BioRad). BRET measurements were acquired using SparkControl 2.2 (Tecan). SPR data were acquired and analyzed using Biacore T200 Evaluation Software 3.0 (Cytiva). MALS data were acquired using Astra 8.0 (Wyatt). Flow cytometry data were acquired using CytExpert 2.3.1.22 (Beckman Coulter) and analyzed using FlowJo 10.8.1. NMR data were analyzed using NMRpipe 8 and NMRFAM-SPARKY 1.3. PDB files were viewed using Pymol 2.4.2. Graphs, curve fitting, and statistical analysis were performed in GraphPad Prism 9.3.1. Alignments were generated using CLUSTAL Omega 1.2.2. Graphics were generated using BioRender or Adobe Illustrator 2021.
Statistics and reproducibility.
All statistics used GraphPad Prism (v.9.3.1) unless stated otherwise, using t-tests or ANOVA as indicated in Supplementary Table 1. No statistical method was used to predetermine sample size. However, we used similar numbers of cells and animals to previous studies in our field that employed these experimental models. All experiments shown were reproduced on at least two independent occasions. For all statistical analyses, n reflects the number of independent experimental/biological replicates. No data were excluded from the analyses. Randomization and blinding methods were not used.
DATA AVAILABILITY STATEMENT:
Source data, including uncropped gels and Western blots, are provided with this paper. All unique biological materials are available upon request from the authors.
Extended Data
Extended Data Fig. 1. Sequence alignment of SMO PKI motif.
Extended alignment of a portion of the pCT from the indicated SMO orthologs, with key PKI motif residues colored as in Fig. 1a.
Extended Data Fig. 2. Additional binding and peptide array studies, SPR sensorgrams, and SMO pCT purification strategy.
a, Fluorescence polarization assays using mouse PKA-Cɑ, performed as in Fig. 1b. Triplicate points from representative experiments are shown. b, Peptide array, performed as in Fig. 1c, but with individual residues in the human SMO PKI motif mutated to alanine. c, SPR sensorgram for 625 nM PKA-Cɑ binding to GST-tagged wild-type (blue) or WRR mutant (purple) SMO pCT, or a PKIɑ positive control (red), in the presence of ATP and MgCl2. d, Exemplary steady-state analysis of binding interactions between human PKA-Cɑ and a recombinant wild-type SMO pCT, with a KD of 703 +/− 0.003 nM (dotted line) as assessed by SPR. This measurement was made three times, resulting in a mean KD value of 752 +/− 34 nM. e, SPR sensorgram, performed as in c, but with ATP and MgCl2 omitted from the buffer. PKA-Cɑ was present at 2.5 μM. Note that although removal of ATP and MgCl2 does not completely eliminate steady-state binding to the PKIɑ positive control, it dramatically accelerates the dissociation rate, as expected.
Extended Data Fig. 3. Raw NMR spectra for wild-type SMO peptide binding to PKA-Cα at varying kinase:peptide ratios.
2D NMR spectra from an experiment in which SMO peptide was titrated into ATPγN-bound PKA-Cα at the indicated kinase:peptide ratios. Upper left panel represents an overlay of all three spectra to highlight the concentration dependence of the SMO peptide-induced changes observed in each spectrum. Black spectrum represents ATPγN-bound PKA-Cα without SMO peptide (see also Extended Data Fig. 4, 5). In the overlay plot, a box denotes one example of a peak (likely corresponding to an unassigned tryptophan residue) that changes linearly according to SMO peptide concentration (magnified in the inset at left).
Extended Data Fig. 4. Raw NMR spectra for WRR mutant peptide binding to PKA-Cα, as shown in Fig 2a.
a, The WRR mutant SMO peptide was titrated into ATPγN-bound PKA-Cα at the indicated kinase:peptide ratios. ATPγN-bound PKA-Cα without peptide (dark green spectrum, upper left) is shown for reference. See Extended Data Fig. 5 for raw NMR spectrum corresponding to PKA-Cα:wild-type SMO peptide at 1:6 ratio. b, Overlay of the spectra in a.
Extended Data Fig. 5. Raw NMR spectra for wild-type SMO peptide binding to PKA-Cα, as shown in Fig. 2a, and PKIα(5–24)-induced displacement of SMO peptide from PKA-Cα, as shown in Fig. 2c.
a, 2D NMR spectra for ATPγN-bound PKA-Cα, either alone (left) or with SMO peptide added at a 1:6 kinase:peptide ratio (right). b, 2D NMR spectra for titration of PKIα(5–24) peptide into the SMO peptide:ATPγN:PKA-Cα complex at the indicated kinase: PKIα(5–24) ratios. Upper left panel represents an overlay of the individual spectra to highlight the concentration dependence of the PKIα(5–24) peptide-induced effects.
Extended Data Fig. 6. Coimmunoprecipitation studies and ciliary colocalization studies to assess SMO / PKA-C interactions.
a, Coimmunoprecipitation of PKA-Cɑ-YFP with the indicated FLAG-tagged wild-type or mutant SMO constructs was assessed using FLAG chromatography from lysates of transfected HEK293 cells. Data shown are representative of two independent experiments. b, Left, Colocalization of FLAG-tagged wild-type or mutant SMO674 (magenta) with mNeonGreen-tagged PKA-Cɑ (green) in ciliated IMCD3 cells stably expressing both constructs and treated with the SMO agonist SAG21k. Cilia are marked by the SMO (FLAG-647) stain. mNeonGreen-tagged Nbβ2AR80 (which does not bind SMO29) serves as a negative control. 3D reconstructions from Z-stacks of confocal live-cell images are shown. Right, quantification of microscopy studies with the median represented by a dashed line and the upper and lower quartiles indicated by dotted lines (n=142–244 cilia per condition). P < 0.0001 (****). See Supplementary Table 1 for full statistical analysis.
Extended Data Fig. 7. Characterization of NIH3T3 cell line expressing epitope-tagged SMO.
The NIH3T3 cell line used in Fig. 5 exhibited trace amounts of SMO in cilia under vehicle (“Ctrl”) -treated conditions, and a dramatic accumulation of SMO in cilia under SAG-treated conditions. SMO (magenta) is visualized using an anti-V5 antibody, and cilia are visualized using an anti-Arl13b antibody (green). Scale bar = 10 μm. Data shown are representative of two independent experiments.
Extended Data Fig. 8. Controls for SMO / PKA-C binding, colocalization, and signaling studies.
a, Expression levels of SMO constructs in Fig. 3a, assessed by whole-cell nanoluc measurements. Data represent the mean +/− standard deviation, n = three biologically independent samples. n.s. = not significant. b, Surface levels of N-terminally FLAG-tagged wild-type or mutant SMO674 constructs were quantified via expression in HEK293 cells followed by FLAG staining and flow cytometry. Mock-infected cells stained with FLAG antibody (red) serve as a negative control. A representative histogram is shown. The % of FLAG-positive cells (i.e., those to the right of the vertical dashed line) are: 0.3 +/− 0.3% (Ctrl); 95.9 +/− 0.9% (wt), 96.0 +/− 2.5% (WRR); 93.4 +/− 4.2% (A635S); values represent the mean +/− standard deviation from two biologically independent samples. See Supplementary Table 1 for full statistical analysis, and Supplementary Figure 2 for gating strategy.. c, Ciliary localization in IMCD3 cells of myc-tagged wild-type or mutant SMO proteins (magenta). Cilia were visualized with Arl13b antibody (green). Scale bar = 5 μm. Data shown are representative of two independent experiments. d, GRK2/3-dependent phosphorylation of FLAG-tagged wild-type or mutant SMO674 constructs was determined via expression in HEK293 cells treated with or without the GRK2/3 inhibitor cmpd101, followed by FLAG purification. Levels of total and phosphorylated SMO were assessed by Stain Free imaging and ProQ Diamond fluorescence, respectively. SMO566, which is not phosphorylated by GRK2/3 (as it does not contain the C-tail and therefore lacks all previously mapped physiological GRK2/3 phosphorylation sites), serves as a negative control. Data shown are representative of two independent experiments.
Extended Data Fig. 9. Complete data set from SMO C-tail peptide array studies.
a, The same SMO tiled peptide array from Fig. 7c, but including the sequences of all positive hits in each array cluster. b, Complete human SMO C-tail sequence used to create the peptide array. In a,b, the SMO PKI motif identified in the pCT is indicated in red. Key residues in this PKI motif, along with ones in the candidate PKI motif in the dCT, are colored as in Fig. 1a.
Extended Data Fig. 10. Similarity between signal transduction mechanisms in the Hh and Wnt pathways.
Schematic diagram of transmembrane signal transduction in the Hh (left) and Wnt (right) pathways. During Hh signal transduction, active SMO is phosphorylated on its cytoplasmic tail by GRK2/3, triggering membrane sequestration and inhibition of PKA-C, and ultimately stabilization and activation of GLI. During Wnt signal transduction, active LRP5/6 is phosphorylated on its cytoplasmic tail by glycogen synthase kinase (GSK)-3β and casein kinase (CK)-1ɑ, triggering membrane sequestration and inhibition of GSK-3β, and ultimately stabilization and activation of β-catenin. Note that this is a simplified and highly schematized diagram and is not intended to be comprehensive; many other components of both pathways (for example, the destruction complex in which GSK-3β and β-catenin reside) are omitted in order to highlight mechanistic similarities between the underlying transmembrane signaling mechanisms.
Supplementary Material
ACKNOWLEDGMENTS:
We thank J. Zalatan for making us aware of the parallels between SMO / PKA-C regulation in the Hh pathway and LRP / GSK-3β regulation in the Wnt pathway. We thank S. Lusk and K. Kwan (University of Utah) for providing smo-null zebrafish (smohi1640), and D. Klatt Shaw and D. Grunwald (University of Utah) for sharing advice and reagents regarding zebrafish immunohistochemistry. We thank J. Müller and S. Kasten (Univ of Kassel) for excellent technical assistance. We thank the Johnson Foundation Structural Biology and Biophysics Core at the Perelman School of Medicine (University of Pennsylvania; Philadelphia, PA) for performing SEC-MALS analyses. We thank D. Julius, S. Nakielny, A. Manglik, K. Basham, and M. He for providing feedback on this manuscript. B.R.M. acknowledges support from the 5 for the Fight Foundation (award # 6000-32705) and a Cancer Center Support Grant Pilot Project Fund from the Huntsman Cancer Institute (award #200206). This work was supported by the DFG grant GRK2749/1 (F.W.H.), and NIH grants R01GM100310-08 (G.V.), 1R35GM130389 (S.S.T.), 1R03TR002947 (S.S.T.) and 1R35GM133672 (B.R.M.).
Footnotes
COMPETING FINANCIAL INTERESTS:
The authors declare no competing financial interests.
REFERENCES:
- 1.Briscoe J & Therond PP The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 14, 416–29 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW & McMahon AP Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15, 3059–87 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Ingham PW, Nakano Y & Seger C Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 12, 393–406 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Kong JH, Siebold C & Rohatgi R Biochemical mechanisms of vertebrate hedgehog signaling. Development 146(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muenke M & Beachy PA Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev 10, 262–9 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Digilio MC et al. Atrioventricular canal defect and genetic syndromes: The unifying role of sonic hedgehog. Clin Genet 95, 268–276 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Anderson E, Peluso S, Lettice LA & Hill RE Human limb abnormalities caused by disruption of hedgehog signaling. Trends Genet 28, 364–73 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Pak E & Segal RA Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev Cell 38, 333–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F, Zhang Y, Sun B, McMahon AP & Wang Y Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem Biol 24, 252–280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui CC & Angers S Gli proteins in development and disease. Annu Rev Cell Dev Biol 27, 513–37 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Deshpande I et al. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571, 284–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang P et al. Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell 174, 312–324 e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrne EFX et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi X, Friedberg L, De Bose-Boyd R, Long T & Li X Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling. Nat Chem Biol 16, 1368–1375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Fallon JF & Beachy PA Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–34 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Humke EW, Dorn KV, Milenkovic L, Scott MP & Rohatgi R The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev 24, 670–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewiadomski P et al. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep 6, 168–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J et al. PKA-mediated Gli2 and Gli3 phosphorylation is inhibited by Hedgehog signaling in cilia and reduced in Talpid3 mutant. Dev Biol 429, 147–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nachury MV & Mick DU Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol 20, 389–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiter JF & Leroux MR Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gigante ED & Caspary T Signaling in the primary cilium through the lens of the Hedgehog pathway. Wiley Interdiscip Rev Dev Biol 9, e377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefkowitz RJ The superfamily of heptahelical receptors. Nat Cell Biol 2, E133–6 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Pierce KL, Premont RT & Lefkowitz RJ Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3, 639–50 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Walker-Gray R, Stengel F & Gold MG Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc Natl Acad Sci U S A 114, 10414–10419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DA, Leathers VL, Martinez AM, Walsh DA & Fletcher WH Fluorescence resonance energy transfer within a heterochromatic cAMP-dependent protein kinase holoenzyme under equilibrium conditions: new insights into the conformational changes that result in cAMP-dependent activation. Biochemistry 32, 6402–10 (1993). [DOI] [PubMed] [Google Scholar]
- 26.Yang S, Fletcher WH & Johnson DA Regulation of cAMP-dependent protein kinase: enzyme activation without dissociation. Biochemistry 34, 6267–71 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Kopperud R et al. Formation of inactive cAMP-saturated holoenzyme of cAMP-dependent protein kinase under physiological conditions. J Biol Chem 277, 13443–8 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Smith FD et al. Local protein kinase A action proceeds through intact holoenzymes. Science 356, 1288–1293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arveseth CD et al. Smoothened transduces hedgehog signals via activity-dependent sequestration of PKA catalytic subunits. PLoS Biol 19, e3001191 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor SS, Ilouz R, Zhang P & Kornev AP Assembly of allosteric macromolecular switches: lessons from PKA. Nat Rev Mol Cell Biol 13, 646–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton GD & Dewey WL Protein kinase inhibitor peptide (PKI): a family of endogenous neuropeptides that modulate neuronal cAMP-dependent protein kinase function. Neuropeptides 40, 23–34 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Soberg K & Skalhegg BS The Molecular Basis for Specificity at the Level of the Protein Kinase a Catalytic Subunit. Front Endocrinol (Lausanne) 9, 538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SS, Zhang P, Steichen JM, Keshwani MM & Kornev AP PKA: lessons learned after twenty years. Biochim Biophys Acta 1834, 1271–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemp BE & Pearson RB Protein kinase recognition sequence motifs. Trends Biochem Sci 15, 342–6 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Scott JD, Fischer EH, Demaille JG & Krebs EG Identification of an inhibitory region of the heat-stable protein inhibitor of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A 82, 4379–83 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng HC, van Patten SM, Smith AJ & Walsh DA An active twenty-amino-acid-residue peptide derived from the inhibitor protein of the cyclic AMP-dependent protein kinase. Biochem J 231, 655–61 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corbin JD et al. Studies on the properties and mode of action of the purified regulatory subunit of bovine heart adenosine 3’:5’-monophosphate-dependent protein kinase. J Biol Chem 253, 3997–4003 (1978). [PubMed] [Google Scholar]
- 38.Scott JD, Glaccum MB, Fischer EH & Krebs EG Primary-structure requirements for inhibition by the heat-stable inhibitor of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A 83, 1613–6 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glass DB, Cheng HC, Mende-Mueller L, Reed J & Walsh DA Primary structural determinants essential for potent inhibition of cAMP-dependent protein kinase by inhibitory peptides corresponding to the active portion of the heat-stable inhibitor protein. J Biol Chem 264, 8802–10 (1989). [PubMed] [Google Scholar]
- 40.Knighton DR et al. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 414–20 (1991). [DOI] [PubMed] [Google Scholar]
- 41.Knighton DR et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–14 (1991). [DOI] [PubMed] [Google Scholar]
- 42.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M & Taylor SS Dynamics of cAMP-dependent protein kinase. Chem Rev 101, 2243–70 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Olivieri C et al. Multi-state recognition pathway of the intrinsically disordered protein kinase inhibitor by protein kinase A. Elife 9(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J et al. The role of ciliary trafficking in Hedgehog receptor signaling. Sci Signal 8, ra55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitehouse S & Walsh DA Mg X ATP2-dependent interaction of the inhibitor protein of the cAMP-dependent protein kinase with the catalytic subunit. J Biol Chem 258, 3682–92 (1983). [PubMed] [Google Scholar]
- 46.Knape MJ et al. Divalent metal ions control activity and inhibition of protein kinases. Metallomics 9, 1576–1584 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Cook PF, Neville ME Jr., Vrana KE, Hartl FT & Roskoski R Jr. Adenosine cyclic 3’,5’-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry 21, 5794–9 (1982). [DOI] [PubMed] [Google Scholar]
- 48.Marullo S & Bouvier M Resonance energy transfer approaches in molecular pharmacology and beyond. Trends Pharmacol Sci 28, 362–5 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Machleidt T et al. NanoBRET--A Novel BRET Platform for the Analysis of Protein-Protein Interactions. ACS Chem Biol 10, 1797–804 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Mick DU et al. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell 35, 497–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desai PB, Stuck MW, Lv B & Pazour GJ Ubiquitin links smoothened to intraflagellar transport to regulate Hedgehog signaling. J Cell Biol 219(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pal K et al. Smoothened determines beta-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol 212, 861–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohatgi R, Milenkovic L, Corcoran RB & Scott MP Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci U S A 106, 3196–201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Kato M & Beachy PA Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci U S A 106, 21666–71 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson CW, Chen MH & Chuang PT Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS One 4, e5182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaywitz AJ & Greenberg ME CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 68, 821–61 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Knape MJ et al. Divalent Metal Ions Mg(2)(+) and Ca(2)(+) Have Distinct Effects on Protein Kinase A Activity and Regulation. ACS Chem Biol 10, 2303–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varjosalo M, Li SP & Taipale J Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell 10, 177–86 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Myers BR, Neahring L, Zhang Y, Roberts KJ & Beachy PA Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci U S A 114, E11141–E11150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuson M, He M & Anderson KV Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development 138, 4921–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lipinski RJ, Bijlsma MF, Gipp JJ, Podhaizer DJ & Bushman W Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC Cell Biol 9, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W et al. Activity-dependent internalization of smoothened mediated by beta-arrestin 2 and GRK2. Science 306, 2257–60 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Wolff C, Roy S & Ingham PW Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol 13, 1169–81 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Zhao Z et al. An essential role for Grk2 in Hedgehog signalling downstream of Smoothened. EMBO Rep 17, 739–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eccles RL et al. Bimodal antagonism of PKA signalling by ARHGAP36. Nat Commun 7, 12963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sastri M, Barraclough DM, Carmichael PT & Taylor SS A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci U S A 102, 349–54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taipale J et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005–9 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Corbit KC et al. Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–21 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Rohatgi R, Milenkovic L & Scott MP Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–6 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Xie J et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–2 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Sweeney RT et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat Genet 46, 722–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yogurtcu ON & Johnson ME Cytosolic proteins can exploit membrane localization to trigger functional assembly. PLoS Comput Biol 14, e1006031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riobo NA, Saucy B, Dilizio C & Manning DR Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A 103, 12607–12 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen F, Cheng L, Douglas AE, Riobo NA & Manning DR Smoothened is a fully competent activator of the heterotrimeric G protein G(i). Mol Pharmacol 83, 691–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mukhopadhyay S et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 152, 210–23 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Hwang SH et al. The G protein-coupled receptor Gpr161 regulates forelimb formation, limb patterning and skeletal morphogenesis in a primary cilium-dependent manner. Development 145(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shimada IS et al. Basal Suppression of the Sonic Hedgehog Pathway by the G-Protein-Coupled Receptor Gpr161 Restricts Medulloblastoma Pathogenesis. Cell Rep 22, 1169–1184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Regard JB et al. Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat Med 19, 1505–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moore BS et al. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci U S A 113, 13069–13074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Low WC et al. The decoupling of Smoothened from Galphai proteins has little effect on Gli3 protein processing and Hedgehog-regulated chick neural tube patterning. Dev Biol 321, 188–96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tschaikner P, Enzler F, Torres-Quesada O, Aanstad P & Stefan E Hedgehog and Gpr161: Regulating cAMP Signaling in the Primary Cilium. Cells 9(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pusapati GV et al. G protein-coupled receptors control the sensitivity of cells to the morphogen Sonic Hedgehog. Sci Signal 11(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meloni AR et al. Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Mol Cell Biol 26, 7550–60 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hammerschmidt M, Bitgood MJ & McMahon AP Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev 10, 647–58 (1996). [DOI] [PubMed] [Google Scholar]
- 85.Concordet JP et al. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835–46 (1996). [DOI] [PubMed] [Google Scholar]
- 86.Barresi MJ, Stickney HL & Devoto SH The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development 127, 2189–99 (2000). [DOI] [PubMed] [Google Scholar]
- 87.MacDonald BT, Tamai K & He X Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17, 9–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Metcalfe C & Bienz M Inhibition of GSK3 by Wnt signalling--two contrasting models. J Cell Sci 124, 3537–44 (2011). [DOI] [PubMed] [Google Scholar]
- 89.Nusse R & Clevers H Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Steinhart Z & Angers S Wnt signaling in development and tissue homeostasis. Development 145(2018). [DOI] [PubMed] [Google Scholar]
- 91.Piao S et al. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One 3, e4046 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cselenyi CS et al. LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3’s phosphorylation of beta-catenin. Proc Natl Acad Sci U S A 105, 8032–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu G, Huang H, Garcia Abreu J & He X Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One 4, e4926 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stamos JL, Chu ML, Enos MD, Shah N & Weis WI Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6. Elife 3, e01998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ocbina PJ, Tuson M & Anderson KV Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS One 4, e6839 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB & Gloriam DE Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oprea TI et al. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov 17, 317–332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han P, Sonati P, Rubin C & Michaeli T PDE7A1, a cAMP-specific phosphodiesterase, inhibits cAMP-dependent protein kinase by a direct interaction with C. J Biol Chem 281, 15050–7 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Kim J et al. Dysfunctional conformational dynamics of protein kinase A induced by a lethal mutant of phospholamban hinder phosphorylation. Proc Natl Acad Sci U S A 112, 3716–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Olivieri C et al. Defective internal allosteric network imparts dysfunctional ATP/substrate-binding cooperativity in oncogenic chimera of protein kinase A. Commun Biol 4, 321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES:
- 101.Zimmermann B, Schweinsberg S, Drewianka S & Herberg FW Effect of metal ions on high-affinity binding of pseudosubstrate inhibitors to PKA. Biochem J 413, 93–101 (2008). [DOI] [PubMed] [Google Scholar]
- 102.Myers BR et al. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev Cell 26, 346–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu TW et al. Two PKA RIalpha holoenzyme states define ATP as an isoform-specific orthosteric inhibitor that competes with the allosteric activator, cAMP. Proc Natl Acad Sci U S A 116, 16347–16356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Olsen SR & Uhler MD Affinity purification of the C alpha and C beta isoforms of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem 264, 18662–6 (1989). [PubMed] [Google Scholar]
- 105.Walker C et al. Cushing’s syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity. Sci Adv 5, eaaw9298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walsh DA & Ashby CD Protein kinases: aspects of their regulation and diversity. Recent Prog Horm Res 29, 329–59 (1973). [DOI] [PubMed] [Google Scholar]
- 107.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–93 (1995). [DOI] [PubMed] [Google Scholar]
- 108.Lee W, Tonelli M & Markley JL NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Williamson MP Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 73, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- 110.Hansen JN, Rassmann S, Stuven B, Jurisch-Yaksi N & Wachten D CiliaQ: a simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur Phys J E Soft Matter 44, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data, including uncropped gels and Western blots, are provided with this paper. All unique biological materials are available upon request from the authors.