Abstract
Among the medically most important snakes in the world, the species belonging to the genus Daboia have been attributed to the highest number of human envenomings, deaths and disabilities. Given their significant clinical relevance, the venoms of Russell’s vipers (D. russelii and D. siamensis) have been the primary focus of research. In contrast, the composition, activity, ecology and evolution of venom of its congener, the Palestine viper (D. palaestinae), have remained largely understudied. Therefore, to unravel the factors responsible for the enhanced medical relevance of D. russelii in comparison to D. palaestinae, we comparatively evaluated their venom proteomes, biochemical activities, and mortality and morbidity inflicting potentials. Furthermore, the synthesis and regulation of venom in snakes have also remained underinvestigated, and the relative contribution of each venom gland remains unclear. We address this knowledge gap by sequencing the tissue transcriptomes of both venom glands of D. palaestinae, and comparatively evaluating their contribution to the secreted venom concoction. Our findings highlight the disparity in the venom composition, function and toxicities of the two Daboia species. We also show that toxin production is not partitioned between the two venom glands of D. palaestinae.
Keywords: Daboia palaestinae, Daboia russelii, snake venoms, venomics, transcriptomes, preclinical assessment
1. Introduction
The genus Daboia is currently constituted by four snake species that are distributed across Asia, the Middle East and Africa: Russell’s viper (D. russelii), Siamese or the eastern Russell’s viper (D. siamensis), Palestine viper (D. palaestinae), and the Moorish viper (D. mauritanica). Among them, D. russelii is arguably the medically most important species, being responsible for the largest number of human envenomings, deaths and disabilities globally [1]. Considering their relatively greater medical relevance, the two species of Russell’s vipers, D. russelii and D. siamensis, have been extensively investigated [2,3,4,5,6]. In contrast, the composition, activity, ecology, and evolution of D. mauritanica and D. palaestinae venom have remained largely uninvestigated. This is despite the fact that the latter is the most common venomous snake species in Israel that causes hundreds of envenomations in humans and livestock. It has also been reported that D. palaestinae is responsible for a significant number of snakebites in Lebanon, northwestern Jordan and the Palestinian territories [7,8,9]. Moreover, understanding the mechanisms involving venom production and regulation in snakes has remained the least studied. For instance, despite the presence of a pair of venom glands in snakes, their relative contribution to the secreted venom cocktail remains unclear.
To address this knowledge gap and unravel the influence of phylogenetic divergence and biogeography in shaping the composition and activity of Daboia venoms, we comparatively investigated the venoms of D. russelii from western India and D. palaestinae from Israel, whose range distributions are separated by over 5500 km. Comparative proteomics and biochemical assessment provided fascinating insights into the venom constitution and function. Toxicity profiling and the preclinical evaluation of venom-induced morbidity revealed the underlying factors responsible for the enhanced medical importance of D. russelii venoms in the Indian subcontinent. Furthermore, comparative transcriptomics of both venom glands of D. palaestinae provided insights into the production and regulation of venom toxins in this species.
2. Results
2.1. Venom Proteomics
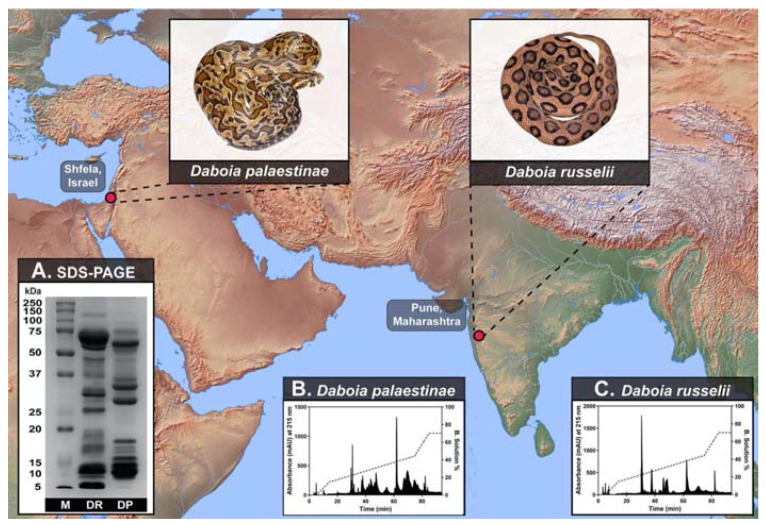
To determine the differences in the proteomic composition of D. russelii and D. palaestinae venoms, we subjected them to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and Reversed-phase high-performance liquid chromatography (RP-HPLC). Despite their close phylogenetic relationship, the venom composition of the two Daboia species was very distinct (Figure 1A). SDS-PAGE and RP-HPLC profiles of D. russelii and D. palaestinae venoms unveiled significant differences in patterns and intensities of protein bands (between 10–20 kDa and 50–75 kDa) (Figure 1A; Supplementary Materials Figure S1) and RP-HPLC peaks (between the retention time of 40 to 80 min) (Figure 1B,C; Supplementary Materials Figure S2), highlighting the considerable differences in their venom compositions. Moreover, searching the mass spectrometric data from individual gel-excised bands of D. palaestinae venom against NCBI-NR Serpentes databases (taxid: 8570) and D. palaestinae venom gland transcriptome identified 91 non-redundant protein families (Figure 2A; Supplementary Materials Table S1; Supplementary Materials Files S1). Among these were 16 toxin families, including cysteine-rich secretory proteins (CRISP), disintegrin, phospholipase A2 (PLA2), cystatin, vascular endothelial growth factor (VEGF), L-amino acid oxidase (LAAO), nerve growth factor (NGF), Kunitz-type serine protease inhibitor (Kunitz), 5′-nucleotidase (5′-NT), lectin, hyaluronidase (HYL), phosphodiesterase (PDE), phospholipase B (PLB), serpin, snake venom serine protease (SVSP), and snake venom metalloproteinase (SVMP) (Figure 2A; Supplementary Materials Table S1).
Figure 1.
Sampling locations of D. palaestinae (Israel) and D. russelii (India), along with their representative photographs, are shown. Panel (A) depicts the comparative SDS-PAGE profiles of D. palaestinae (DP) and D. russelii (DR) venoms (M: protein marker), while their respective RP-HPLC profiles are shown in panels (B,C).
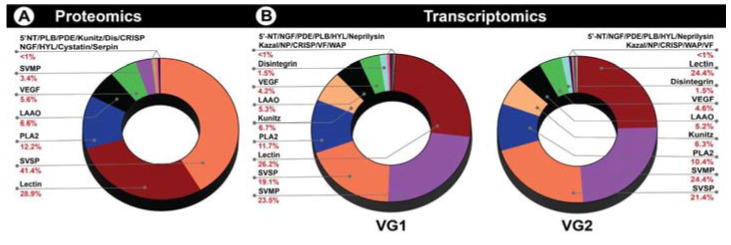
Figure 2.
Venom proteome and venom gland transcriptomes of D. palaestinae. Here, the doughnut charts indicate the relative abundance of various toxins in the (A) venom profile of D. palaestinae and (B) tissue transcriptomes of the left and right venom glands of this species.
Tandem mass spectrometry further revealed that D. palaestinae venom is predominantly constituted by SVSP (41%), lectins (29%) and PLA2s (12.2%; Figure 2A; Supplementary Table S1). Additionally, CRISP, VEGF, SVMP, LAAO, 5′-NT, PDE, NGF, HYL, Kunitz, PLB, serpin, cystatin and disintegrin were also identified as the minor components of the venom proteome (Figure 2A; Supplementary Materials Table S1).
2.2. Venom Gland Transcriptomics
Tissue transcriptomes of both venom glands from a male D. palaestinae were sequenced on an Illumina HiSeq 2500 platform. A total of 41,875,666 and 43,278,432 sequences were retrieved from the left (VG1) and right (VG2) venom glands, respectively (Supplementary Materials Table S2). A combined de novo transcriptome from both glands was assembled using Trinity v2.11.0 [10] and a total of 191,425 transcripts were identified. The assembly was characterised by an N50 statistics of 1812 based on all transcripts. The annotation of the resultant venom gland transcripts identified multiple toxin-encoding genes from both glands (Supplementary Materials Table S3). D. palaestinae venom gland transcriptome profile (VG1 and VG2, respectively) showed the abundance of SVMP (23.5% and 24.3%), lectin (26.2% and 24.4%), SVSP (19.0% and 21.4%) and PLA2 (11.7% and 10.4%) transcripts (Figure 2B, Supplementary Table S3). Transcripts encoding LAAO, disintegrin, VEGF, Kunitz, PLB, hyaluronidase, PDE, 5′-NT, natriuretic peptide, Kazal-type serine protease inhibitors, CRISPs, cobra venom factor (CVF), waprin and NGF were also retrieved from these glands. Interestingly, we recovered a large number of transcripts encoding snake venom metalloproteinase inhibitors (SVMPI) from the D. palaestinae venom glands. Although the role of SVMPIs in venom remains poorly understood, the active-site tripeptide region of a synthetic SVMPI identified from D. siamensis has been shown to inhibit SVMPs under experimental conditions. Therefore, SVMPIs may putatively be involved in the inhibition of SVMPs in the venom glands to prevent autotoxicity [11]. SVMPI transcripts are also known to code the precursors of Bradykinin-potentiating peptides (BPP) and natriuretic peptides (NP) in Viperidae snakes [12,13]. Manual inspection of these transcripts in VG1 and VG2, respectively, revealed that most of them encode for SVMPI and BPP (62.49% and 62.49%), followed by SVMPI, BPP and NP (24.99% and 24.99%) and SVMPI only (12.50% and 12.50%).
2.3. Venom Biochemistry
2.3.1. PLA2 assay
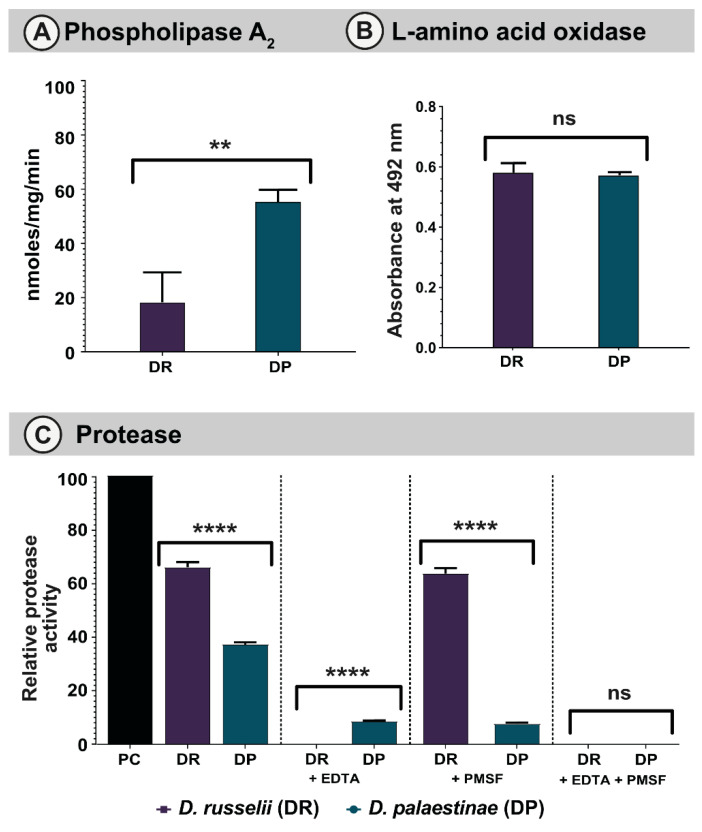
Phospholipases are among the major classes of snake venom toxins that exhibit numerous pharmacological effects, including cytotoxicity, neurotoxicity, myotoxicity and the perturbation of haemostasis. Viperidae snakes are known to possess both catalytic and non-catalytic forms of PLA2s [14]. However, in our PLA2 assay, both D. russelii and D. palaestinae venoms exhibited very low PLA2 activities (18.27 nmoles/mg/min to 55.27 nmoles/mg/min, respectively; p = 0.0058; Figure 3A).
Figure 3.
Biochemical activities of Daboia venoms. This figure depicts (A) PLA2, (B) LAAO, and (C) proteolytic (with and without SVMP and SVSP inhibitors) activities of Daboia venoms. Here, the standard deviation is represented as error bars (PC: positive control; DR: D. russelii; DP: D. palaestinae). The statistical significance is represented as follows: p < 0.01 and 0.0001 are indicated as ** and ****, respectively. ns indicates statistical insignificance.
2.3.2. LAAO Assay
LAAOs are flavoproteins that catalyse the stereospecific oxidative deamination of L-amino acids to α-keto acids. Snake venom LAAOs exhibit diverse pharmacological effects, such as oedema, haemorrhage, myotoxicity, apoptosis and necrosis [15,16,17]. In LAAO assays, both Daboia venoms showed notable L-amino acid oxidation, albeit the difference between the two was not statistically significant (p = 0.6581, Figure 3B).
2.3.3. Snake Venom Protease Assay
Snake venom proteases, such as SVSPs and SVMPs, are known to significantly contribute to the clinical manifestations of snakebite victims [18]. Viperid venoms, in particular, are enriched with these proteolytic enzymes. As a result, they inflict fibrinolysis, inhibition of platelet aggregation, and degradation of the capillary basement membrane [19,20]. When the proteolytic potentials of D. russelii and D. palaestinae venoms were evaluated in comparison to a bovine pancreatic protease (positive control), both venoms exhibited significant proteolysis, with the D. russelii venom exhibiting relatively increased activity than D. palaestinae (70% vs. 38%, respectively; p < 0.001; Figure 3C). We further assessed the relative contributions of SVMP and SVSP towards proteolysis using ethylenediamine tetraacetic acid (EDTA) and phenylmethylsulfonyl fluoride (PMSF), a metal chelator and protease inhibitor which are known to inhibit these toxins, respectively. While the addition of EDTA completely inhibited the proteolysis of azocasein by D. russelii venom, PMSF did not seem to diminish this effect (Figure 3C). Interestingly, both PMSF and EDTA were documented to inhibit proteolytic effects of D. palaestinae venom to a similar extent. When both PMSF and EDTA were added to these reaction mixtures, the proteolytic activities of Daboia venoms were completely suppressed.
2.3.4. DNase Assay
ETosis is a unique mechanism wherein cells, such as neutrophils, monocytes, or macrophages, release their DNA and granular contents to restrict the movement of venom toxins in the bloodstream. As toxins are trapped at a particular site, the rate of local tissue damage accelerates significantly [21]. Venoms that show DNase activity cause the cleavage of these extracellular traps, leading to the diffusion of venom toxins. Interestingly, certain populations of D. russelii in India were found to cleave DNA [22]. However, consistent with the absence of DNase activity in the majority of Daboia populations [22], the venoms of D. russelii (Maharashtra) and D. palaestinae did not exhibit this activity (Supplementary Materials Figure S3).
2.3.5. Fibrinogenolytic Assay
Fibrinogen, a glycoprotein that forms a hexameric complex (Aα/Bβ/γ)2, is cleaved by thrombin when released into the blood. This reaction results in the formation of fibrin monomers and a mesh-like network that ultimately leads to blood clots [23]. Snake venoms can prolong clotting time by cleaving fibrinogen in blood [5,24]. Therefore, we assessed the abilities of D. russelii and D. palaestinae venoms in cleaving the human fibrinogen. Compared to the control–human fibrinogen with three distinct bands–D. palaestinae venom was found to cleave Aα and Bβ components (Supplementary Materials Figure S4A,B), whereas the D. russelii venom exhibited a partial cleavage of Aα subunit retaining the other two components (verified with densitometric analyses of bands; Supplementary Materials Figure S5A,B). Moreover, when D. russelii venom was incubated with EDTA (SVMP inhibitor), all three bands were observed. While the treatment with PMSF (SVSP inhibitor) did not completely inhibit the cleavage of Aα band, the addition of both inhibitors showed all three bands, suggesting the complete inhibition of fibrinogenolytic activity (Supplementary Figures S4 and S5).
2.4. Preclinical Assessments
2.4.1. Venom Toxicity
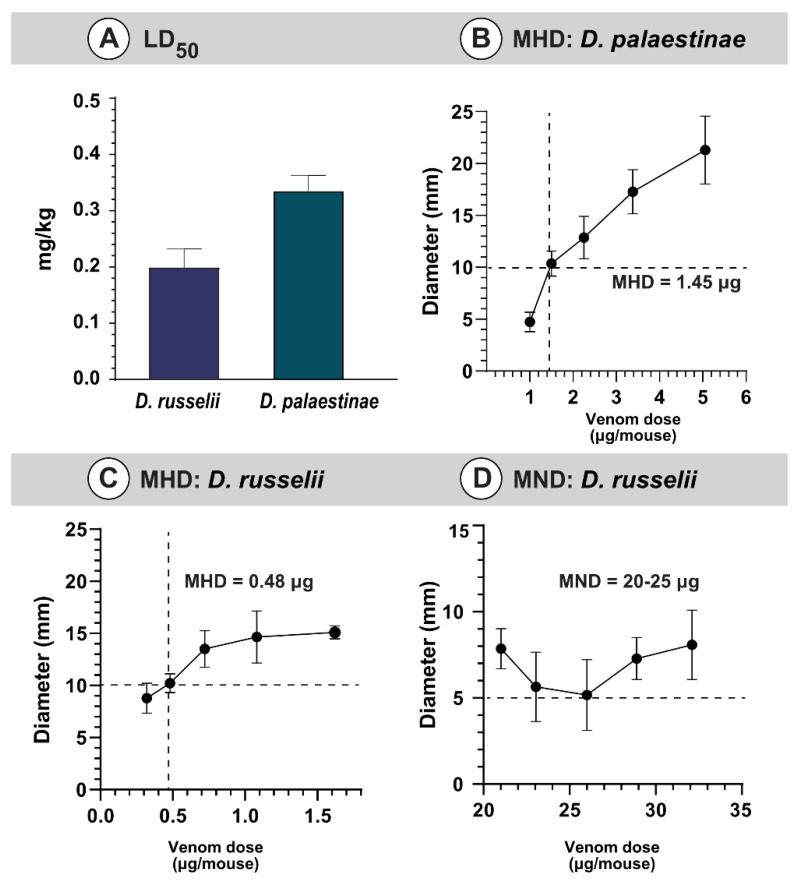
Toxicity profiles of Daboia venoms were evaluated in the mouse model of envenoming. In these experiments, the venom collected from the western Indian population of D. russelii was found to be relatively more toxic (0.20 mg/kg) than the venom of D. palaestinae (0.34 mg/kg) from Israel (Figure 4A). Previous studies have highlighted the remarkable biogeographic variation in the lethal potency of D. russelii venoms across the Indian subcontinent, ranging from 0.1 mg/kg to 0.4 mg/kg [6,22,25]. Interestingly, toxicities of D. russelii and D. palaestinae venoms were very similar to those of D. siamensis (0.3–0.6 mg/kg) and D. mauritanica (0.33 mg/kg) [2,26].
Figure 4.
Preclinical assessment of Daboia venoms. This figure depicts the (A) LD50 of D. russelii and D. palaestinae venoms, MHDs of (B) D. palaestinae and (C) D. russelii venom, and the (D) MND of D. russelii venom.
2.4.2. Venom-Induced Morbidity
Envenomations caused due to vipers are also known to inflict morbid symptoms [27]. For instance, over 50% of Daboia bite victims are documented to suffer from various forms of morbidities [28]. Considering this, we performed preclinical experiments using D. russelii and D. palaestinae venoms. Haemorrhagic abilities were observed for both venoms under investigation, and the diameter of lesions was directly proportional to the increasing venom concentrations (Figure 4B,C, Supplementary Materials Figure S6A,B). The Russell’s viper venom from western India (MHD of 0.48 μg/mouse) was thrice as haemorrhagic as its conger from the Middle East (MHD of 1.48 μg/mouse; Figure 4B,C). Preliminary dose-finding investigations using a single mouse per venom dose did not identify necrotising activities for the D. palaestinae venom, even up to 35 μg. Hence, given animal ethics, complete experiments involving five mice per venom dose were not carried out for this species. In contrast, between 20 to 25 μg of the D. russelii venom produced significant necrotic lesions in mice (5 mm diameter; Figure 4D; Supplementary Materials Figure S6).
2.4.3. Venom Induced Nephrotoxicity
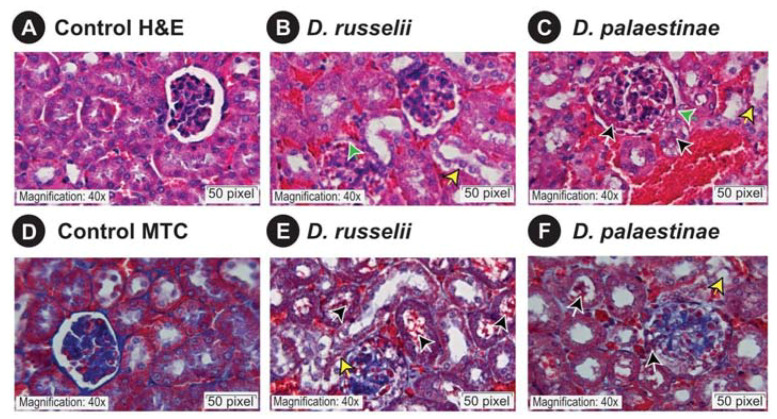
As Russell’s vipers from certain regions in India and Israel have been documented to inflict kidney injury in human snakebite victims and livestock [29,30,31], we evaluated the nephrotoxic potentials of Daboia venoms using the mouse model. Microscopic examinations of kidney sections of venom-injected mice showed significant pathologies relative to those of the control mice which received normal saline alone. While there was no evidence of inflammation documented in kidneys of the control group, and the morphologies of the renal tubules and glomeruli were also normal (Figure 5A,D), the administration of 10 μg of D. russelii (Figure 5B,E) and D. palaestinae (Figure 5C,F) venom in the treatment group resulted in shrinkage of the bowman’s capsular space and the formation of haemorrhagic casts. Additionally, changes were also observed in the proximal tubule of the treatment group, such as the loss of proximal brush borders, cytoplasmic vacuolation and formation of interlumen haemorrhagic casts.
Figure 5.
Microscopic observations of mouse kidney sections. This figure shows hematoxylin-eosin and Masson’s trichrome stained kidney sections of the control mice (A,D), and D. russelii (B,E) and (D). palaestinae (C,F) venom-injected mice. A scale bar of 20 μm is shown, along with green, yellow and black arrows that indicate glomerular capsular space shrinkage, tubular injury and cellular debris, respectively.
3. Discussion
3.1. Disparate Venoms of D. russelii and D. palaestinae
Despite the vast spatial and biogeographic separation, the venoms of the Daboia species across the world share several medically important toxins, such as SVMP, SVSP, lectins, PLA2s and Kunitz [3,6,22,32,33,34]. However, the relative abundance of these toxins has been documented to vary significantly between the western Mediterranean and the eastern tropical lineages. Previously, it has been theorised that the venoms of western Mediterranean Daboia species, including D. mauritanica and D. palaestinae, are enriched with SVMPs and lectins, while venoms of eastern tropical Daboia snakes are rich in PLA2s [32,35]. In line with this hypothesis, we recovered a greater number of SVMP and lectin transcripts from the venom glands of D. palaestinae (Figure 2B; Supplementary Materials Table S3). However, as evidenced by SDS-PAGE and mass spectrometry profiles, the venom proteome of D. palaestinae was found to be rich in SVSPs, lectins and PLA2s, rather than SVMPs (Figure 1A and Figure 2A; Supplementary Materials Figure S1 and Table S1). Such a discrepancy between the snake venom gland transcriptome and proteome has also been previously reported [13,22,36,37,38]. However, in the case of D. russelii, varying degrees of SVMPs, ranging from 3 to 22%, have also been recorded across the Indian subcontinent [39,40]. Intraspecific differences in relative toxin constituents have been attributed to a range of ecological and environmental factors [22].
Venoms of D. palaestinae and D. russelii also varied in their biochemical profiles. For example, the venom of D. palaestinae exhibited relatively higher PLA2 activity compared to D. russelii, whereas the latter species was found to exhibit significantly higher SVMP-mediated proteolytic activity than the former (Figure 3). The prominent role of D. russelii SVMPs in inflicting proteolysis was evidenced by the addition of EDTA [41,42]. While EDTA completely inhibited proteolysis of the azocasein substrate, PMSF did not diminish this activity, suggesting that SVMPs are responsible for the proteolytic effects of D. russelii venom. Interestingly, the addition of either EDTA or PMSF reduced the overall proteolytic activity of D. palaestinae venom to a similar extent, suggesting a synergy between these toxin types. Consistently, when both EDTA and PMSF were added to the reaction, the proteolytic activity of D. palaestinae venom was completely suppressed (Figure 3C). Moreover, the fibrinogenolytic effect of D. palaestinae venom was inhibited by a combination of EDTA and PMSF, whereas EDTA alone completely inhibited the fibrinogenolytic activity of D. russelii venom (Supplementary Materials Figures S4 and S5). It should be noted that given the significant differences documented in the venoms of the pan-Indian populations of D. russelii [6,22,25,43], it is likely that the profiles described here do not fully represent the profiles of their species, and variations in the abundance and functions of toxins are expected in accordance with the literature.
3.2. Venom Production in D. palaestinae Is Not Partitioned between the Venom Glands
Venom, a complex biochemical cocktail of proteins, carbohydrates, salts and amino acids, is a metabolically taxing trait. To capitalise on the evolutionary advantage of this unique molecular innovation, venomous animals have optimised pathways associated with venom production, storage and delivery. Previous reports have highlighted ingenious adaptations of certain venomous organisms to overcome constraints associated with venom storage and target-specific deployment. Sea anemones and other cnidarians have evolved phyletically unique cells called cnidocytes, or stinging cells, for resource specialisation and partitioning of venom storage [44,45]. Research on the starlet sea anemone, Nematostella vectensis, revealed that the early stages of developing cnidocytes exhibit high levels of transcription and translation of venom and structural protein-coding genes [44]. However, since the mature cells have space constraints, given a large centralised capsule, they are marked by decreased levels of both these processes [44]. The geographer cone snail (Conus geographus) provides an example of the spatial distinction in venom production to ensure target-specific deployment of venom. The distal region of the venom duct is shown to produce a cocktail exclusively deployed for defensive purposes, while the proximal end is responsible for producing predation-specific venoms [46]. Similarly, in the assassin bug, Pristhesancus plagipennis, three unique venom gland lumens have been documented to produce distinct venom cocktails [47]. The venom produced in the posterior main gland has been shown to be responsible for inducing prey paralysis, while the anterior main gland produces defensive venoms [47].
In contrast to the aforementioned venomous animals, advanced snakes with a pair of prominent venom glands, perhaps, do not require partitioning of venom production. To shed light on venom production and resource partitioning, we sequenced transcriptomes of both venom glands of Daboia palaestinae (VG1 and VG2, respectively). In line with our hypothesis, a highly similar abundance of venom transcripts across the two venom glands was recovered. The abundance of medically most important toxins, including SVMP (23.5% and 24.3%), lectin (26.2% and 24.4%), PLA2 (11.6% and 10.4%), SVSP (19.0% and 21.4%) and Kunitz (6.7% and 6.3%) were comparable across VG1 and VG2 (Figure 2B, Supplementary Materials Table S3). Similar chromatography profiles have also been reported for the secretory proteomes of the left and right Duvernoy’s gland from the false coral snake, Rhinobothryum bovallii [48]. Similarly, venom fractions collected from the left and right venom glands of Naja siamensis exhibited similarities in their composition and receptor-binding activities [49]. A shared venom production strategy between the two glands decreases the metabolic stress on either of the glands while also facilitating the rapid replenishment of their toxin reserve.
3.3. The Role of Compositional and Activity Differences in Determining the Clinical Relevance of Daboia Species
Russell’s vipers (D. russelii and D. siamensis) are amongst snakes capable of delivering the most life-threatening bites to humans. Envenoming by these snakes results in innumerable deaths and immutable morbidities. In India, over 40% of snakebite mortalities have been attributed to D. russelii envenoming [50]. Similarly, the western Mediterranean congener of Russell’s viper, the Palestine viper (D. palaestinae), is endemic to parts of the Levant and is considered the medically most important snake in Israel. Despite both D. palaestinae and D. russelii thriving closer to farmlands and human residential habitats, only a few hundred D. palaestinae bites are documented in Israel, corresponding to a fraction of D. russelii envenoming’s in India [50,51]. This discrepancy in snakebite burden is despite the two species exhibiting a similar toxicity profile against mammals (D. palaestinae: 0.34 mg/kg vs. D. russelii: 0.20 mg/kg; Figure 4A; Supplementary Materials Table S4), and both being capable of injecting nearly 200 mg of the venom in a single bite [22,52].
Envenoming by Daboia snakes is typically characterised by local oedema, tissue necrosis, haemorrhage and coagulopathy [6,53,54]. Moreover, these snakes are also known to inflict nephrotoxicity in snakebite victims [29]. In our preclinical assays involving the administration of Daboia venoms into mice, both D. russelii and D. palaestinae venoms were found to cause considerable kidney injury. While the control group that only received normal saline showed no signs of pathology, the administration of minuscule amounts of Daboia venom (10 μg/mouse) resulted in significant deformation of the tubular region and glomeruli within 15 min of injection. These alarming findings highlight the severe clinical repercussions of Daboia envenoming.
Interestingly, in our preclinical assays, while both D. russelii and D. palaestinae venoms exhibited haemorrhagic effects, necrotizing activity was only documented in the former species (Figure 4B–D, Supplementary Materials Figures S6 and S7). Clinically, however, D. russelii is also known to induce extreme systemic manifestations, such as disseminated intravascular coagulation, capillary leakage syndrome, and intracerebral and subarachnoid haemorrhage [55,56,57]. These differences in the abilities to inflict local and systemic complications could largely explain the relatively greater proportion and severity of morbidities caused by D. russelii, in comparison to D. palaestinae. Moreover, such differences in venom activities could be dictated by local ecological and environmental factors, including prey availability, predator density, and seasonal and ontogenetic shifts. Consistent with this hypothesis, the amount of venom injected, and the clinical manifestations of D. palaestinae bite incidents have been recorded to vary across seasons, with slightly severe symptoms observed in patients bitten during spring or immediately after brumation [8,52].
Moreover, the role of demographics in determining the extent of the human–animal conflict cannot be overstated. With over 1.4 billion people, India is the second-most populous country in the world. It is also home to a vast diversity of snake species that could deliver clinically severe bites to humans. Snakebite is a persistent occupational hazard for farmers and cattle herders of poor agricultural subsistence [58]. Coincidentally, around 60% of the Indian population, who are involved in agriculture and allied activities, reside in rural areas, as opposed to a mere 7% in Israel [59]. The sheer difference in the densities of the human population, and that of the medically relevant snake species in these regions, could explain the significantly disproportionate burden of snakebites in India and Israel. Therefore, in addition to research on snake venom composition, activity and ecology, investigations into the human and snake demography are imperative for the effective management of the prevailing global snakebite crisis.
4. Conclusions
In this study, we report a comparative analysis of the venoms of D. palaestinae from Israel and D. russelii from India, whose distribution ranges are separated by over 5500 km. While the venom of D. russelii assessed here was enriched with high molecular weight, haemorrhage-inducing toxins such as SVMPs, the D. palaestinae venom was enriched by SVSPs, PLA2s and lectins. However, we show that despite the overall similarity in venom functions, the differences in their abilities to inflict local and systemic complications, and the demographics of the vulnerable human populations in the two regions, could explain the disproportionate medical relevance of these snake species. Moreover, this is the first holistic characterisation of the venom of D. palaestinae–the most common and medically relevant venomous snake in the State of Israel–using an ‘omics’ approach. Furthermore, comparative tissue transcriptomics revealed the absence of partitioning of venom production between the pair of venom glands in D. palaestinae.
5. Materials and Methods
5.1. Sampling Permits, and the Collection of Venom and Venom Gland
A male Palestine viper (Daboia palaestinae) was sourced from the Southern Shfela region of Israel in compliance with the Nature and National Parks Protection Authority (Permit #: 2015/41135). Venom from an adult Russell’s viper (Daboia russelii) was collected from the Pune district of Maharashtra in India with prior permission from the state forest department (#Desk-22 (8)/WL/CR-60/(17-18)/2708). The freshly extracted venoms from these individuals were flash-frozen, lyophilised and stored at −80° C until further use. Post milking, D. palaestinae was maintained in captivity for three days and humanely euthanised on the fourth day, when the transcriptional activity in the venom glands is believed to be the highest [60]. Euthanisation of the animal was carried out via intramuscular injection of sodium pentobarbital (0.6–0.8 mL/kg) by a licensed veterinarian. After the complete cessation of vital signs and physiological reflexes, both venom glands were dissected, flash-frozen and stored at −80 °C until further processing.
5.2. Ethical Statements
The venom toxicity and morbidity assays in mice were performed using the standard protocols recommended by the World Health Organisation (WHO) [61]. The Institutional Animal Ethics Committee (IAEC), Indian Institute of Science (IISc), Bangalore (CAF/Ethics/769/2020; approved on: 16 October 2020), reviewed and approved these protocols. Experiments were also designed adhering to the guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA).
5.3. Proteomic Analyses
5.3.1. Protein Estimation, One-Dimensional Gel Electrophoresis and In-Gel Digestion
Protein concentrations of venoms were determined using the Bradford method and the Bovine serum albumin (BSA) standard [62]. Variations in the proteomic profiles of D. palaestinae and D. russelii venoms were assessed using SDS-PAGE. The reduced venom samples (20 µg) were subjected to 12.5% polyacrylamide gel electrophoresis at a constant voltage (80 V) [63]. The Precision Plus Dual Color (Bio-Rad Laboratories, Hercules, CA, USA) protein ladder was used as a reference for determining the molecular weight of proteins. Post-separation, the gels were stained overnight with Coomassie Brilliant Blue R-250 (Sisco Research Laboratories Pvt. Ltd., Mumbai, India) and destained the following day. The protein bands were visualised using an iBright CL1000 gel documentation system (Thermo Fisher Scientific, MA, USA), and densitometric analysis of individual bands was performed using the ImageJ software [64].
The individual protein bands were excised and collected separately for mass spectrometric analyses. Briefly, the gel bands were destained and dehydrated with 50% acetonitrile. Following destaining, proteins were reduced with 10 mM dithiothreitol (DTT) at 56 °C for 1 h and alkylated using 30 mM iodoacetamide (IAA), in the dark at room temperature, for 45 min. Then, the bands were washed with 25 mM ammonium bicarbonate in water and acetonitrile solution (1:1, v/v), and excess solvent was removed using a vacuum concentrator (Thermo Fisher Scientific, MA, USA). The samples were then digested with trypsin (0.2 µg/µL) overnight at 37 °C, and the peptides were extracted the next day into 50 μL of 50% acetonitrile solution.
5.3.2. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
The proteomic composition of individually excised D. palaestinae venom bands was characterised using tandem mass spectrometry. Samples (40 µg) were first reduced with 10 mM dithiothreitol (DTT), alkylated using 30 mM iodoacetamide (IAA) and further digested with trypsin (0.2 µg/µL) overnight at 37 °C. The digested samples were run through a C18 nano-LC column (50 cm × 75 µm, 3 µm particle size and 100 Å pore size) with a Thermo EASY nLC 1200 series system (Thermo Fisher Scientific, MA, United States) at a constant flow rate of 300 nl/min for 120 min by varying the concentrations of buffer A (0.1% formic acid in HPLC grade water) and buffer B (0.1% formic acid in 80% acetonitrile) as 10–45% over 98 min, 45–95% over 4 min and 95% over 18 min. Samples were then subjected to tandem mass spectrometry on a Thermo Orbitrap Fusion Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The following parameters were defined for the MS scans: range (m/z) of 375–1700 with a resolution of 120,000 and maximum injection time of 50 ms. Furthermore, an ion trap detector with high collision energy fragmentation (30%) was used to perform the fragment scans (MS/MS) with a scan range (m/z) of 100–2000 and a maximum injection time of 35 ms.
The identities of the individual toxins in each of the excised bands were determined by searching the raw MS/MS spectra against the National Center for Biotechnology Information non-redundant (NCBI-NR) Serpentes database (taxid: 8570; with 410,049 entries as of December 2021) and the D. palaestinae venom gland transcriptome generated in this study, using PEAKS Studio X Plus (Bioinformatics Solutions Inc., Waterloo, ON, Canada). The following parameters were defined for the search: the parent and fragment mass error tolerance limits were set to 10 ppm and 0.6 Da, respectively. A ‘monoisotopic’ precursor ion search type with ‘semispecific’ trypsin digestion with a maximum of three missed cleavages, cysteine carbamidomethylation (+57.02) as a fixed modification; methionine oxidation (+15.99) as a variable modification was specified. For match acceptance, the filtering parameters were set to a False Discovery Rate (FDR) of 0.1%, detection of ≥1 unique peptide, and a-10lgP protein score of ≥50. The raw mass spectrometry data have been made available at the ProteomeXchange Consortium via the PRIDE partner repository [65] with the data identifier PXD031190 (Reviewer account details: Username: reviewer_pxd031190@ebi.ac.uk; Password: YtCME8qx). Hits with at least one unique matching peptide were considered for downstream analyses. The redundant protein hits from each protein family were removed manually. The relative abundance of each toxin hit in a fraction was determined by estimating the area under the spectral curve (AUC). These AUC values, which represent mean peak intensities, were obtained from PEAKS Studio analyses, and were then normalised across the gel bands using the densitometric estimates from the SDS-PAGE profiles [66]. The relative abundance of a protein family hit (X) was estimated using the equation below, where ‘N’ indicates the number of bands in the SDS-PAGE profile.
5.3.3. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
Lyophilised Daboia venoms (200 μg) were reconstituted in molecular grade water and loaded onto a 4.6 × 250 mm, C18 (5 μm, 300 Å) reversed-phase column attached to a Shimadzu LC-20AD series HPLC system (Kyoto, Japan). The column was equilibrated with solution A [0.1% trifluoroacetic acid (TFA) in water (v/v)] and the fractions obtained were eluted at a flow rate of 1 mL/min using the graded concentrations of solution B [0.1% TFA in 100% acetonitrile (v/v)]: 5% for 5 min, 5–15% for 10 min, 15–45% for 60 min, and 45–70% for 10 min and 70% for 5 min at a flow rate of 1 mL/min. The absorbance was monitored at 215 nm.
5.4. Transcriptomics Analyses
5.4.1. RNA Isolation and Sequencing
The total RNA was extracted from freshly preserved venom gland tissues of D. palaestinae using the TRIzol™ Reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. Isolated RNA samples were treated with Turbo DNase (Thermo Fisher Scientific, Waltham, MA, USA) to remove DNA contamination, followed by another round of extraction with TRIzol™. The concentration and purity of the isolated RNA were measured using an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA), and the integrity was assessed on a bioanalyzer Agilent 4200 TapeStation system using RNA HS ScreenTape (Cat# 5067-5579; Agilent Technologies, Santa Clara, CA, USA). RNA samples that had an RNA Integrity Number (RIN) of greater than 8 were used to generate cDNA library using the NEBNext® Ultra™ RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) for Illumina® and subsequently sequenced on an Illumina HiSeq 2500 platform, with a sequencing depth of 20 million reads (2 × 150 bp paired-end). Raw sequencing results are deposited to NCBI’s Sequence Read Archive (SRA) repository: Bioproject: PRJNA800175; SRA: SRR17903474 and SRR17903475.
5.4.2. Quality Assessment, De Novo Assembly and Annotation
The acquired raw data were screened for high-quality reads using Trimmomatic v0.39 [67]. The filtering process involved the removal of adapter sequences, trimming leading and trailing low-quality bases (<3), and discarding short (<20 nucleotides) and low-quality reads (<25; sliding window of 4). The quality of the processed data was then assessed with FASTQC v0.11.9 [68], before and after trimming. The curated reads were de novo assembled into contigs using Trinity v2.11.0 [69] with the following parameters: k-mer = 25, minimum k-mer coverage = 1, minimum contig length = 200, pair distance = 500 and the maximum number of reads per graph = 200,000. Reads were aligned back onto the transcriptome using bowtie v2.4.2 [70] to evaluate the quality of the assembly. TransDecoder v5.5.0 [10] was used to predict the coding regions in transcripts that code for a contiguous stretch of over 30 amino acids. The coding transcripts were annotated with BLAST searches [71] against the SwissProt (December 2021) and NCBI’s non-redundant protein databases.
5.4.3. Quantification and Differential Expression Analyses
The abundance of transcripts in Fragments per kilobase of exon per million fragments mapped (FPKM) was quantified using RSEM v1.3.3 [69]. Venom gland-specific expression was determined by pairwise differential expression analysis performed using the EdgeR package within the Bioconductor tool [72]. A predefined cut-off value for fold change in expression (≥2) and probability of p ≥ 0.9 was used to identify significantly differentially expressed transcripts.
5.5. Biochemical Characterisation
5.5.1. Colourimetric Phospholipase A2 (PLA2) Assay
The phospholipase activity of venom PLA2 was assessed using a chromogenic lipid substrate, 4-nitro-3-[octanoyloxy] benzoic acid (NOB; Enzo Life Sciences, New York, NY, USA). Briefly, 5 µg of the venom was added to 500 mM NOB substrate dissolved in a 200 µL reaction buffer (10 mM Tris–HCl, 10 mM CaCl2, 100 mM NaCl, pH 7.8). The mixture was incubated at 37 °C for 40 min, and the kinetics of the assay was monitored by measuring the absorbance at 425 nm every 10 min using an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA). A blank containing only the chromogenic substrate and the buffer without any enzyme was included in the experiment. The endpoint reading at the 40th min was considered for calculating the specific PLA2 activity after subtracting the blank. An identical protocol was followed, and a standard curve with varying concentrations of the NOB substrate (4 nanomoles to 130 nanomoles) and 4M NaOH was plotted. The amount of the phospholipid substrate in nmol cleaved per minute per mg of the venom was calculated by extrapolation from the standard curve [73,74].
5.5.2. L-amino Acid Oxidase (LAAO) Assay
The LAAO activity of D. russelii and D. palaestinae venoms was evaluated using a previously described endpoint assay [22]. The L-leucine substrate solution (5 mM L-leucine, 50 nM Tris-HCl buffer, 5 IU/mL horseradish peroxidase, 2 mM o-phenylenediamine dihydrochloride) was incubated with 10 microlitres of the crude venom at 37 °C. After one hour of incubation, the reaction was terminated by the addition of 2M H2SO4 solution. The absorbance of the solution was recorded at 492 nm using an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA).
5.5.3. Snake Venom Protease Assay
The snake venom protease activity was assessed using previously established protocols [75], wherein a known amount of the crude venom (10 µg) was incubated with the azocasein substrate at 37 °C for 90 min. Post-incubation, the reaction was stopped by adding trichloroacetic acid (200 µL). Before the addition of azocasein, both venoms were incubated at 37 °C for 15 min with 0.1 M EDTA and 0.04 M PMSF to assess the contribution of SVMP and SVSP, respectively, towards the overall proteolytic activity [41,42]. This mixture was then subjected to centrifugation at 1000× g for 5 min. The supernatant was mixed with equal volumes of 0.5 M NaOH, and the absorbance was measured in an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA) at 440 nm. Purified bovine pancreatic protease (Sigma-Aldrich, Burlington, MA, USA) was used as a positive control, and the relative proteolytic activities of Daboia venoms were calculated.
5.5.4. DNase Assay
The DNase assay was conducted on D. russelii and D. palaestinae venoms using a previously described protocol [76]. A known concentration of the crude venom was added to the purified calf thymus DNA (Sigma-Aldrich, Burlington, MA, USA) dissolved in phosphate buffer saline (PBS; pH 7.4), and the reaction mixture was incubated at 37 °C for 60 min. This mixture was thereafter run on a 0.8% agarose gel electrophoresis and imaged using an iBright CL1000 (Thermo Fisher Scientific, MA, USA). Intact calf thymus DNA, and DNase I from the bovine pancreas (15 U), were used as negative and positive controls, respectively.
5.5.5. Fibrinogenolytic Assay
The fibrinogenolytic activity of Daboia venoms was visualised electrophoretically [77]. Briefly, 1.5 µg of crude venom in phosphate buffer saline (PBS, pH 7.4) was incubated with 15 µg of human fibrinogen (Sigma-Aldrich, Burlington, MA, USA) at 37 °C for 60 min. We also preincubated the venom with 0.1 M EDTA (SVMP inhibitor) and/or 0.04 M PMSF (SVSP inhibitor) at 37 °C for 15 min to assess the respective contributions of SVMP and SVSP towards proteolysis [78]. The reaction was stopped by adding an equal volume of sample loading buffer (1 M Tris-HCl, pH 6.8; 50% Glycerol; 0.5% Bromophenol blue; 10% SDS; and 20% β-mercaptoethanol) and heated at 70 °C for 10 min. A 15% polyacrylamide gel was run, and the banding patterns of the fibrinogen cleavage products were observed by comparing them to an untreated human fibrinogen control. Densitometric analyses of the bands were carried out using the ImageJ tool [64].
5.6. Preclinical Assessments
5.6.1. The Median lethal Dose (LD50)
The median lethal dose or the LD50 of the venom, which is defined as the minimum amount of venom that can kill 50% of the test population, was determined using the murine model of envenoming [61]. Five distinct concentrations of D. russelii and D. palaestinae venoms, prepared in the physiological saline (0.9% NaCl), were administered intravenously into the caudal vein of male CD-1 mice (200 µL/mouse). Death and survival patterns were recorded for each venom dose group (n = 5), 24 h post-venom injection. Finally, using Probit analysis, the LD50 values were calculated with 95% confidence intervals [79].
5.6.2. The Minimum Haemorrhagic Dose (MHD)
MHD is defined as the amount of venom in μg that induces a 10 mm haemorrhagic lesion within three hours of intradermal injection in mice [61,80,81]. To determine the MHD of D. russelii and D. palaestinae venoms, five graded venom concentrations were dissolved in the physiological saline (50 µL) before being intradermally injected into a group of five male mice (CD-1 mice; 18–22 gm). The group, where only the physiological saline was administered, served as the negative control. Three hours post-venom injection, mice were humanely euthanised with CO2 asphyxiation, and the diameter of the haemorrhagic lesion on the dorsal skin patch was measured using a vernier calliper.
5.6.3. The Minimum Necrotic Dose (MND)
MND is defined as the amount of venom in μg that induces a 5 mm necrotic lesion within 72 h of intradermal injection in mice [61,80]. To determine the MND of Daboia venoms, five graded venom concentrations were dissolved in physiological saline (50 µL) and intradermally injected into a group of five male mice (CD-1 mice; 18–22 gm). The group receiving physiological saline alone served as the negative control. Mice were euthanised humanely, 72 h post-venom injection, and the dorsal skin patch was examined for necrotic lesions.
5.7. Nephrotoxic Potentials of Daboia Venoms
The nephrotoxic effects of D. russelii and D. palaestinae venoms were assessed by injecting 10 μg of the venom into the caudal vein of 4 male CD-1 mice (18–22 g). Both kidneys were harvested immediately after the death of the animal and were washed with 1X PBS, fixed in 10% buffered formalin for 24 h, and dehydrated with ascending concentrations of ethyl alcohol (70 and 95% for 30 min; 100% for 2 h), and cleared in xylene (Thermofisher, Waltham, MA, USA). Tissues were embedded in paraffin (Thermofisher, Waltham, MA, USA) at 58 °C, following which 3 μm sections were prepared using a Leica microtome (RM2245, Wetzlar, Germany). The slides were then stained with hematoxylin (Leica, Wetzlar, Germany), eosin (Leica, Wetzlar, Germany) and Masson’s trichrome staining (MTS; Path Stains, Bengaluru, India). The slides obtained were visualised using an Olympus light microscope (Ix81, Olympus, Shinjuku, Japan) at a 40× magnification, and images were acquired and analysed using CellSens dimension imaging software (Olympus, Shinjuku, Japan). The histological structure of renal tubules and glomeruli of the treatment group (10 μg of venom) was assessed in comparison to the control that received 200 μL of normal saline [82].
5.8. Statistical Analysis
Statistical comparisons between samples were carried out using an unpaired t-test and one-way ANOVA in GraphPad Prism (GraphPad Software 9.0, San Diego, CA USA, www.graphpad.com, accessed on 22 September 2022).
Acknowledgments
The authors are thankful to the Forest Departments of Maharashtra for the sampling permits and the logistic support during venom collection, as well as to the Nature and National Parks Protection Authority, Israel. The authors are also thankful to Ashok Captain, Anil Khaire (Indian Herpetological Society), Aditya Maladi (IISc) and Vivek Suranse (IISc) for their assistance with sampling, and to Matthieu Berroneau (www.matthieu-berroneau.fr, accessed on 22 September 2022) and Ajinkya Unawane for sharing photographs of D. palaestinae and D. russelii, respectively, in Figure 1.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/toxins14110725/s1, Figure S1: SDS PAGE profile of D. palaestinae venom; Figure S2: RP-HPLC profiles of Daboia palaestinae and Daboia russelii venoms; Figure S3: The DNase activity of Daboia venoms; Figure S4: The fibrinogenolytic activity of D. palaestinae venom; Figure S5: The fibrinogenolytic activity of the D. russelii venom; Figure S6: The hemorrhagic potential of Daboia venoms; Figure S7: The necrotic potential of D. russelii venom. Table S1: The proteomic composition of D. palaestinae venom; Table S2: Venom gland transcriptome assembly statistics; Table S3: Comparative transcriptomics of the left and right venom glands of D. palaestinae; Table S4: Toxicity profiles of Daboia venoms. File S1: Mass spectrometry of D. palaestinae venom.
Author Contributions
Conceptualisation: K.S. and Y.M.; Formal analysis: R.R.S.L., S.K., S.A., N.K., P.J., N.Y.S., R.A., N.P. and K.S.; Funding acquisition: K.S. and Y.M.; Investigation: R.R.S.L., S.K., S.A., N.K., P.J., N.Y.S., K.S., R.A. and N.P., Sample collection: N.P. and D.H.; Supervision: K.S.; Visualisation: S.A., S.K. and K.S.; original draft: R.R.S.L., N.K., N.Y.S., S.K. and K.S.; Review and editing: Y.M. and K.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The venom toxicity and morbidity assays in mice were performed using the standard protocols recommended by the World Health Organisation (WHO) [60] The Institutional Animal Ethics Committee (IAEC), Indian Institute of Science (IISc), Bangalore (CAF/Ethics/769/2020; approved on: 16th October 2020), reviewed and approved these protocols. Experiments were also designed adhering to the guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA).
Data Availability Statement
The raw proteomics data generated for this study can be found at PRIDE Database (Accession No: PXD031190). The transcriptomics data presented in this study can be openly accessed via Sequence Read Archive (SRA) at NCBI (Bioproject: PRJNA800175; SRA: SRR17903474 and SRR17903475).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Key Contribution
The first comprehensive investigation of the venom constitution of the Palestine viper (Daboia palaestinae) using venom proteomics and venom gland transcriptomics. Comparative transcriptomics of both venom glands of D. palaestinae reveals equal contribution by both glands to the secreted venom arsenal. The venom of D. palaestinae and the closely related Indian counterpart, D. russelii, exhibit notable compositional and functional differences. Variations in venom-induced toxicity and morbidities perhaps contribute to the enhanced clinical relevance of D. russelii in the Indian subcontinent.
Funding Statement
KS was supported by the Wellcome Trust DBT India Alliance Fellowship (IA/I/19/2/504647). This research was also supported by DST-FIST (SR/FST/LS-II/2018/233) and the Marie Skłdowska-Curie Individual Fellowship (654294), awarded to K.S., Y.M. acknowledges support from the Israel Science Foundation grant (636/21). S.L. was supported by the Prime Minister’s Research Fellowship, Government of India (0201058).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Warrell D.A. Snake venoms in science and clinical medicine 1. Russell’s viper: Biology, venom and treatment of bites. Trans. R. Soc. Trop. Med. Hyg. 1989;83:732–740. doi: 10.1016/0035-9203(89)90311-8. [DOI] [PubMed] [Google Scholar]
- 2.Chaisakul J., Alsolaiss J., Charoenpitakchai M., Wiwatwarayos K., Sookprasert N., Harrison R.A., Chaiyabutr N., Chanhome L., Tan C.H., Casewell N.R. Evaluation of the geographical utility of Eastern Russell’s viper (Daboia siamensis) antivenom from Thailand and an assessment of its protective effects against venom-induced nephrotoxicity. PLoS Negl. Trop. Dis. 2019;13:e0007338. doi: 10.1371/journal.pntd.0007338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faisal T., Tan K.Y., Sim S.M., Quraishi N., Tan N.H., Tan C.H. Proteomics, functional characterization and antivenom neutralization of the venom of Pakistani Russell’s viper (Daboia russelii) from the wild. J. Proteom. 2018;183:1–13. doi: 10.1016/j.jprot.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Senji Laxme R.R., Khochare S., De Souza H.F., Ahuja B., Suranse V., Martin G., Whitaker R., Sunagar K. Beyond the ‘big four’: Venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Negl. Trop. Dis. 2019;13:e0007899. doi: 10.1371/journal.pntd.0007899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee A.K. The pro-coagulant fibrinogenolytic serine protease isoenzymes purified from Daboia russelii russelii venom coagulate the blood through factor V activation: Role of glycosylation on enzymatic activity. PLoS ONE. 2014;9:e86823. doi: 10.1371/journal.pone.0086823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pla D., Sanz L., Quesada-Bernat S., Villalta M., Baal J., Chowdhury M.A.W., León G., Gutiérrez J.M., Kuch U., Calvete J.J. Phylovenomics of Daboia russelii across the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. J. Proteom. 2019;207:103443. doi: 10.1016/j.jprot.2019.103443. [DOI] [PubMed] [Google Scholar]
- 7.Abu Baker M.A., Al-Saraireh M., Amr Z., Amr S.S., Warrell D.A. Snakebites in Jordan: A clinical and epidemiological study. Toxicon. 2022;208:18–30. doi: 10.1016/j.toxicon.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Agajany N., Kozer E., Agajany N., Trotzky D., Kishk I.A., Youngster I. Is severity of Daboia (Vipera) palaestinae snakebites influenced by season of exposure? Toxicon. 2021;206:51–54. doi: 10.1016/j.toxicon.2021.12.016. [DOI] [PubMed] [Google Scholar]
- 9.El Zahran T., Kazzi Z., Chehadeh A.A.-H., Sadek R., El Sayed M.J. Snakebites in Lebanon: A descriptive study of snakebite victims treated at a tertiary care center in Beirut, Lebanon. J. Emergencies Trauma Shock. 2018;11:119. doi: 10.4103/JETS.JETS_141_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yee K.T., Pitts M., Tongyoo P., Rojnuckarin P., Wilkinson M.C. Snake venom metalloproteinases and their peptide inhibitors from Myanmar Russell’s viper venom. Toxins. 2017;9:15. doi: 10.3390/toxins9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higuchi S., Murayama N., Saguchi K., Ohi H., Fujita Y., Camargo A.C., Ogawa T., Deshimaru M., Ohno M. Bradykinin-potentiating peptides and C-type natriuretic peptides from snake venom. Immunopharmacology. 1999;44:129–135. doi: 10.1016/S0162-3109(99)00119-8. [DOI] [PubMed] [Google Scholar]
- 13.Leonardi A., Sajevic T., Pungerčar J., Križaj I. Comprehensive study of the proteome and transcriptome of the venom of the most venomous european viper: Discovery of a new subclass of ancestral snake venom metalloproteinase precursor-derived proteins. J. Proteome Res. 2019;18:2287–2309. doi: 10.1021/acs.jproteome.9b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kini R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Costa T.R., Burin S.M., Menaldo D.L., de Castro F.A., Sampaio S.V. Snake venom L-amino acid oxidases: An overview on their antitumor effects. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014;20:23. doi: 10.1186/1678-9199-20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paloschi M.V., Pontes A.S., Soares A.M., Zuliani J.P. An update on potential molecular mechanisms underlying the actions of snake venom L-amino acid oxidases (LAAOs) Current medicinal chemistry. 2018;25:2520–2530. doi: 10.2174/0929867324666171109114125. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro P.H., Zuliani J.P., Fernandes C.F., Calderon L.A., Stábeli R.G., Nomizo A., Soares A.M. Mechanism of the cytotoxic effect of l-amino acid oxidase isolated from Bothrops alternatus snake venom. Int. J. Biol. Macromol. 2016;92:329–337. doi: 10.1016/j.ijbiomac.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Slagboom J., Kool J., Harrison R.A., Casewell N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017;177:947–959. doi: 10.1111/bjh.14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asega A.F., Menezes M.C., Trevisan-Silva D., Cajado-Carvalho D., Bertholim L., Oliveira A.K., Zelanis A., Serrano S.M.T. Cleavage of proteoglycans, plasma proteins and the platelet-derived growth factor receptor in the hemorrhagic process induced by snake venom metalloproteinases. Sci. Rep. 2020;10:12912. doi: 10.1038/s41598-020-69396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Q., Clemetson J., Clemetson K.J. Snake venoms and hemostasis. J. Thromb. Haemost. 2005;3:1791–1799. doi: 10.1111/j.1538-7836.2005.01358.x. [DOI] [PubMed] [Google Scholar]
- 21.Katkar G.D., Sundaram M.S., NaveenKumar S.K., Swethakumar B., Sharma R.D., Paul M., Vishalakshi G.J., Devaraja S., Girish K.S., Kemparaju K. NETosis and lack of DNase activity are key factors in Echis carinatus venom-induced tissue destruction. Nat. Commun. 2016;7:11361. doi: 10.1038/ncomms11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senji Laxme R., Khochare S., Attarde S., Suranse V., Iyer A., Casewell N.R., Whitaker R., Martin G., Sunagar K. Biogeographic venom variation in Russell’s viper (Daboia russelii) and the preclinical inefficacy of antivenom therapy in snakebite hotspots. PLoS Negl. Trop. Dis. 2021;15:e0009247. doi: 10.1371/journal.pntd.0009247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kattula S., Byrnes J.R., Wolberg A.S. Fibrinogen and fibrin in hemostasis and thrombosis. Arterioscler. Thromb. Vasc. Biol. 2017;37:e13–e21. doi: 10.1161/ATVBAHA.117.308564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazaki Y., Morita T. Snake venom components affecting blood coagulation and the vascular system: Structural similarities and marked diversity. Curr. Pharm. Des. 2007;13:2872–2886. doi: 10.2174/138161207782023775. [DOI] [PubMed] [Google Scholar]
- 25.Prasad N.B., Uma B., Bhatt S.K., Gowda V.T. Comparative characterisation of Russell’s viper (Daboia/Vipera russelli) venoms from different regions of the Indian peninsula. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1999;1428:121–136. doi: 10.1016/S0304-4165(99)00053-7. [DOI] [PubMed] [Google Scholar]
- 26.Oukkache N., Lalaoui M., Ghalim N. General characterization of venom from the Moroccan snakes Macrovipera mauritanica and Cerastes cerastes. J. Venom. Anim. Toxins Incl. Trop. Dis. 2012;18:411–420. doi: 10.1590/S1678-91992012000400009. [DOI] [Google Scholar]
- 27.Alirol E., Sharma S.K., Bawaskar H.S., Kuch U., Chappuis F. Snake bite in South Asia: A review. PLoS neglected tropical diseases. 2010;4:e603. doi: 10.1371/journal.pntd.0000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kularatne S. Epidemiology and clinical picture of the Russell’s viper (Daboia russelii russelii) bite in Anuradhapura, Sri Lanka: A prospective study of 336 patients. Southeast Asian J. Trop. Med. Public Health. 2003;34:855–862. [PubMed] [Google Scholar]
- 29.Vikrant S., Jaryal A., Parashar A. Clinicopathological spectrum of snake bite-induced acute kidney injury from India. World J. Nephrol. 2017;6:150. doi: 10.5527/wjn.v6.i3.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ariga K., Dutta T.K., Haridasan S., Puthenpurackal P.S.P., Harichandrakumar K., Parameswaran S. Chronic Kidney Disease after Snake Envenomation Induced Acute Kidney Injury. Saudi J. Kidney Dis. Transplant. 2021;32:146. doi: 10.4103/1319-2442.318516. [DOI] [PubMed] [Google Scholar]
- 31.Tirosh-Levy S., Solomovich-Manor R., Comte J., Nissan I., Sutton G.A., Gabay A., Gazit E., Steinman A. Daboia (Vipera) palaestinae Envenomation in 123 Horses: Treatment and Efficacy of Antivenom Administration. Toxins. 2019;11:168. doi: 10.3390/toxins11030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damm M., Hempel B.-F., Süssmuth R.D. Old World Vipers—A Review about Snake Venom Proteomics of Viperinae and Their Variations. Toxins. 2021;13:427. doi: 10.3390/toxins13060427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makran B., Fahmi L., Pla D., Sanz L., Oukkache N., Lkhider M., Ghalim N., Calvete J.J. Snake venomics of Macrovipera mauritanica from Morocco, and assessment of the para-specific immunoreactivity of an experimental monospecific and a commercial antivenoms. J. Proteom. 2012;75:2431–2441. doi: 10.1016/j.jprot.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 34.Risch M., Georgieva D., von Bergen M., Jehmlich N., Genov N., Arni R.K., Betzel C. Snake venomics of the Siamese Russell’s viper (Daboia russelli siamensis)—Relation to pharmacological activities. J. Proteom. 2009;72:256–269. doi: 10.1016/j.jprot.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Momic T., Arlinghaus F.T., Arien-Zakay H., Katzhendler J., Eble J.A., Marcinkiewicz C., Lazarovici P. Pharmacological aspects of Vipera xantina palestinae venom. Toxins. 2011;3:1420–1432. doi: 10.3390/toxins3111420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durban J., Pérez A., Sanz L., Gómez A., Bonilla F., Rodríguez S., Chacón D., Sasa M., Angulo Y., Gutiérrez J.M., et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee A.K., Kalita B., Mackessy S.P. A proteomic analysis of Pakistan Daboia russelii russelii venom and assessment of potency of Indian polyvalent and monovalent antivenom. J. Proteom. 2016;144:73–86. doi: 10.1016/j.jprot.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Sunagar K., Khochare S., Senji Laxme R., Attarde S., Dam P., Suranse V., Khaire A., Martin G., Captain A. A wolf in another wolf’s clothing: Post-genomic regulation dictates venom profiles of medically-important cryptic kraits in India. Toxins. 2021;13:69. doi: 10.3390/toxins13010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalita B., Mackessy S.P., Mukherjee A.K. Proteomic analysis reveals geographic variation in venom composition of Russell’s Viper in the Indian subcontinent: Implications for clinical manifestations post-envenomation and antivenom treatment. Expert Rev. Proteom. 2018;15:837–849. doi: 10.1080/14789450.2018.1528150. [DOI] [PubMed] [Google Scholar]
- 40.Kalita B., Patra A., Das A., Mukherjee A.K. Proteomic analysis and immuno-profiling of eastern India Russell’s Viper (Daboia russelii) venom: Correlation between RVV composition and clinical manifestations post RV bite. J. Proteome Res. 2018;17:2819–2833. doi: 10.1021/acs.jproteome.8b00291. [DOI] [PubMed] [Google Scholar]
- 41.Meléndez-Martínez D., Plenge-Tellechea L.F., Gatica-Colima A., Cruz-Pérez M.S., Aguilar-Yáñez J.M., Licona-Cassani C. Functional mining of the Crotalus Spp. venom protease repertoire reveals potential for chronic wound therapeutics. Molecules. 2020;25:3401. doi: 10.3390/molecules25153401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita K.M., Alves A.F., Barbaro K.C., Santoro M.L. Bothrops jararaca venom metalloproteinases are essential for coagulopathy and increase plasma tissue factor levels during envenomation. PLoS Negl. Trop. Dis. 2014;8:e2814. doi: 10.1371/journal.pntd.0002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma M., Gogoi N., Dhananjaya B., Menon J.C., Doley R. Geographical variation of Indian Russell’s viper venom and neutralization of its coagulopathy by polyvalent antivenom. Toxin Rev. 2014;33:7–15. doi: 10.3109/15569543.2013.855789. [DOI] [Google Scholar]
- 44.Sunagar K., Columbus-Shenkar Y.Y., Fridrich A., Gutkovich N., Aharoni R., Moran Y. Cell type-specific expression profiling unravels the development and evolution of stinging cells in sea anemone. BMC Biol. 2018;16:108. doi: 10.1186/s12915-018-0578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Columbus-Shenkar Y.Y., Sachkova M.Y., Macrander J., Fridrich A., Modepalli V., Reitzel A.M., Sunagar K., Moran Y. Dynamics of venom composition across a complex life cycle. eLife. 2018;7:e35014. doi: 10.7554/eLife.35014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dutertre S., Jin A.-H., Vetter I., Hamilton B., Sunagar K., Lavergne V., Dutertre V., Fry B.G., Antunes A., Venter D.J., et al. Evolution of separate predation-and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014;5:3521. doi: 10.1038/ncomms4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker A.A., Mayhew M.L., Jin J., Herzig V., Undheim E.A., Sombke A., Fry B.G., Meritt D.J., King G.F. The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat. Commun. 2018;9:755. doi: 10.1038/s41467-018-03091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calvete J.J., Bonilla F., Granados-Martínez S., Sanz L., Lomonte B., Sasa M. Venomics of the Duvernoy’s gland secretion of the false coral snake Rhinobothryum bovallii (Andersson, 1916) and assessment of venom lethality towards synapsid and diapsid animal models. J. Proteom. 2020;225:103882. doi: 10.1016/j.jprot.2020.103882. [DOI] [PubMed] [Google Scholar]
- 49.Harris R.J., Zdenek C.N., Nouwens A., Sweeney C., Dunstan N., Fry B.G. A symmetry or asymmetry: Functional and compositional comparison of venom from the left and right glands of the Indochinese spitting cobra (Naja siamensis) Toxicon: X. 2020;7:100050. doi: 10.1016/j.toxcx.2020.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suraweera W., Warrell D., Whitaker R., Menon G., Rodrigues R., Fu S.H., Begum R., Sati P., Piyasena K. Trends in snakebite deaths in India from 2000 to 2019 in a nationally representative mortality study. eLife. 2020;9:e54076. doi: 10.7554/eLife.54076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abd Rabou A. On the occurrence and health risks of the venomous Palestine Viper (Vipera palaestinae Werner, 1938) in the Gaza Strip-Palestine. Biomed. J. Sci. Tech. Res. Mini Rev. 2019;18:13934–13937. doi: 10.26717/BJSTR.2019.18.003219. [DOI] [Google Scholar]
- 52.Kochva E. A quantitative study of venom secretion by Vipera palaestinae. Am. J. Trop. Med. Hyg. 1960;9:381–390. doi: 10.4269/ajtmh.1960.9.381. [DOI] [PubMed] [Google Scholar]
- 53.Bentur Y., Cahana A. Unusual local complications of Vipera palaestinae bite. Toxicon. 2003;41:633–635. doi: 10.1016/S0041-0101(02)00367-7. [DOI] [PubMed] [Google Scholar]
- 54.Pivko-Levy D., Munchnak I., Rimon A., Balla U., Scolnik D., Hoyte C., Voliovitch Y., Glatstein M. Evaluation of antivenom therapy for Vipera palaestinae bites in children: Experience of two large, tertiary care pediatric hospitals. Clin. Toxicol. 2017;55:235–240. doi: 10.1080/15563650.2016.1277233. [DOI] [PubMed] [Google Scholar]
- 55.Adhikari R.B., Gawarammana I.B., De Silva D., Dangolla A., Mallawa C., Premarathna A., Silva I.D. Clinico-epidemiology and management of Russell’s viper (Daboia russelii) envenoming in dogs in Sri Lanka. Toxicol. Rep. 2019;6:809–818. doi: 10.1016/j.toxrep.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warrell D.A. Handbook of: Clinical Toxicology of Animal Venoms and Poisons. CRC Press; Boca Raton, FL, USA: 2017. Clinical toxicology of snakebite in Asia; pp. 493–594. [Google Scholar]
- 57.Warrell D.A., Gutiérrez J.M., Calvete J.J., Williams D. New approaches & technologies of venomics to meet the challenge of human envenoming by snakebites in India. Indian J. Med. Res. 2013;138:38–59. [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison R.A., Hargreaves A., Wagstaff S.C., Faragher B., Lalloo D.G. Snake envenoming: A disease of poverty. PLoS Negl. Trop. Dis. 2009;3:e569. doi: 10.1371/journal.pntd.0000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.United Nations, Department of Economics and Social Affairs, Population Division . World Urbanization Prospects: The 2018 Revision (ST/ESA/SER/420) United Nations; New York, NY, USA: 2018. [Google Scholar]
- 60.Leary S.L., Underwood W., Anthony R., Cartner S., Corey D., Grandin T., Greenacre C., Gwaltney-Brant S., McCrackin M.A., Meyer R., et al. AVMA Guidelines for the Euthanasia of Animals: 2013 Edition. American Veterinary Medical Association; Schaumburg, IL, USA: 2013. [Google Scholar]
- 61.WHO . World Health Organisation Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins. World Health Organisation; Geneva, Switzerland: 2018. [Google Scholar]
- 62.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 63.Smith B. SDS Polyacrylamide Gel Electrophoresis of Proteins. Springer; Berlin/Heidelberg, Germany: 1984. pp. 41–55. [DOI] [PubMed] [Google Scholar]
- 64.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan N.H., Wong K.Y., Tan C.H. Venomics of Naja sputatrix, the Javan spitting cobra: A short neurotoxin-driven venom needing improved antivenom neutralization. J. Proteom. 2017;157:18–32. doi: 10.1016/j.jprot.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 67.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrews S. Babraham Bioinformatics-FastQC a Quality Control Tool for High Throughput Sequence Data. [(accessed on 22 October 2022)]. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc.2010.
- 69.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature biotechnology. 2011;29:644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 72.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freitas-de-Sousa L.A., Nachtigall P.G., Portes-Junior J.A., Holding M.L., Nystrom G.S., Ellsworth S.A., Guimarães N.C., Tioyama E., Ortiz F., Silva B.R., et al. Size matters: An evaluation of the molecular basis of ontogenetic modifications in the composition of Bothrops jararacussu snake venom. Toxins. 2020;12:791. doi: 10.3390/toxins12120791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tasoulis T., Lee M.S., Ziajko M., Dunstan N., Sumner J., Isbister G.K. Activity of two key toxin groups in Australian elapid venoms show a strong correlation to phylogeny but not to diet. BMC Evol. Biol. 2020;20:9. doi: 10.1186/s12862-020-1578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chowdhury M., Miyoshi S., Shinoda S. Purification and characterization of a protease produced by Vibrio mimicus. Infect. Immun. 1990;58:4159–4162. doi: 10.1128/iai.58.12.4159-4162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gerceker D., Karasartova D., Elyürek E., Barkar S., KIyan M., Özsan T.M., Calgin M.K., Sahin F. A new, simple, rapid test for detection of DNase activity of microorganisms: DNase Tube test. J. Gen. Appl. Microbiol. 2009;55:291–294. doi: 10.2323/jgam.55.291. [DOI] [PubMed] [Google Scholar]
- 77.Teng C.-M., Ouyang C., Lin S.-C. Species difference in the fibrinogenolytic effects of α-and β-fibrinogenases from Trimeresurus mucrosquamatus snake venom. Toxicon. 1985;23:777–782. doi: 10.1016/0041-0101(85)90008-X. [DOI] [PubMed] [Google Scholar]
- 78.op den Brouw B., Ghezellou P., Casewell N.R., Ali S.A., Fathinia B., Fry B.G., Bos M.H.A., Ikonomopoulou M.P. Pharmacological characterisation of Pseudocerastes and Eristicophis viper venoms reveal anticancer (Melanoma) properties and a potentially novel mode of fibrinogenolysis. Int. J. Mol. Sci. 2021;22:6896. doi: 10.3390/ijms22136896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finney D.J. Probit Analysis. 3rd ed. Cambridge University Press; London, UK: 1971. [Google Scholar]
- 80.Gutiérrez J.M., Solano G., Pla D., Herrera M., Segura Á., Vargas M., Villalta M., Sánchez A., Sanz L., Lomonte B., et al. Preclinical evaluation of the efficacy of antivenoms for snakebite envenoming: State-of-the-art and challenges ahead. Toxins. 2017;9:163. doi: 10.3390/toxins9050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kondo H., Kondo S., Ikezawa H., Murata R., Ohsaka A. Studies on the quantitative method for determination of hemorrhagic activity of Habu snake venom. Jpn. J. Med. Sci. Biol. 1960;13:43–51. doi: 10.7883/yoken1952.13.43. [DOI] [PubMed] [Google Scholar]
- 82.Hedrich H. The Laboratory Mouse. Academic Press; Cambridge, MA, UK: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw proteomics data generated for this study can be found at PRIDE Database (Accession No: PXD031190). The transcriptomics data presented in this study can be openly accessed via Sequence Read Archive (SRA) at NCBI (Bioproject: PRJNA800175; SRA: SRR17903474 and SRR17903475).