Abstract
Objective
In this study, we aimed to determine drug-resistance mutations (DRMs) in HIV-1 patients with low-level viremia (LLV) and explored the performance of next-generation sequencing (NGS) in detecting HIV DRMs by using LLV samples.
Methods
Overall, 80 samples with LLV were amplified and sequenced using a commercial Sanger sequencing (SS) genotyping kit. Furthermore, 51 samples successfully sequenced using SS were simultaneously subjected to NGS. Genotyping success rates of various viremia categories by two sequencing methods were calculated. Stanford HIV-1 drug-resistance database (HIVdb version 8.9) was used to analyze the DRMs. In the NGS assay, a threshold of 5% was considered for reporting low-frequency variants, and the DRMs detected using SS and NGS were compared.
Results
The overall success rate of PR/RT regions was 88.1% (67/80) using SS and 86.3% (44/51) using NGS. Furthermore, a significant linear trend was noted between viral load and the genotyping success rate. A total of 38.8% (26/67) participants harbored at least one mutation, as revealed through SS. Moreover, the prevalence of DRMs in persistent LLV was significantly higher than that in intermittent LLV (62.1% vs. 21.1%; P < 0.05). A total of 69 DRMs were detected using the two sequencing methods at the threshold of 5%. Moreover, 10 DRMs missed by SS were detected using NGS, whereas 8 DRMs missed by NGS were detected by SS.
Conclusion
Our data suggested that the genotyping resistance testing is necessary to guide antiretroviral therapy optimization in LLV patients.
Keywords: HIV-1, low-level viremia, drug-resistance mutation, next-generation sequencing
Introduction
More than 75 million people have been infected with HIV-1, of which 32 million have died of AIDS-related diseases.1 The rapid antiviral therapy (ART) scale has effectively reduced HIV-related morbidity and mortality.2 ART can inhibit the viral replication and prolong the life span of HIV-infected patients. In China, low-level viremia (LLV) is detected in approximately 10–30% of patients.3,4 According to World Health Organization (WHO) guidelines, LLV is defined as the viral load (VL) between 50 and 999 copies/mL after 6 months of ART.5 In addition, two types of LLV have been described, namely intermittent LLV (iLLV/blip) and persistent LLV (pLLV).
Previous studies have confirmed that LLV has multiple risks such as virologic failure (VF),6–8 HIV transmission,9,10 emergence of new drug-resistance mutations (DRMs),11–13 enhanced immune activation,14 release of inflammatory factors,15 and accelerated clinical progression.16 Furthermore, according to data of a study, switching to second-line ART in patients with LLV resulted in a higher proportion of participants of virological suppression (VS).17 However, according to WHO guidelines, drug-resistance genotyping (RGT) and switching to second-line ART are recommended only when the VL reaches 1000 copies/mL.18
As fewer genomic templates are available, the low amplification success rate of VL < 1000 copies/mL is a major concern, and managing emerging DRMs in LLV remains a clinical challenge.19 Without the data of RGT, clinicians can maintain the regimen when DRMs have already appeared or can empirically switch to a new regimen when the virus is still sensitive.20 However, the accumulation of DRMs and cross-resistance would significantly reduce future therapeutic options. Therefore, understanding the drug-resistance profiles in patients with LLV is necessary.
It is well known that only variants with frequency > 20% can be detected using Sanger sequencing (SS).21 Increasing evidence suggests that low-frequency HIV-1 drug-resistance mutations detected using next-generation sequencing (NGS) may impair treatment outcomes.22–24 Considering the ability of NGS to reveal low-prevalence DRMs, a 5% cut-off threshold of minority variants for NGS seemed to be a good compromise.22,25 However, it is unclear whether the cases of LLV would benefit from this new technology.
In this study, we aimed to describe DRMs in patients with LLV and to precisely profile the prevalence of DRMs in iLLV and pLLV. Furthermore, we explored the performance of NGS in detecting HIV DRMs by using LLV samples and compared the results with those of SS.
Patients and Methods
Selection of Patients
This observational, retrospective, and single-centered study was conducted at Beijing Ditan Hospital from October 2, 2020, to May 10, 2022. The inclusion criteria were as follows: (i) age ≥ 18 years, (ii) treatment-experienced with first- or second-line ART, and (iii) experienced LLV in ART. In total, 80 participants who met the definition of LLV were enrolled. Their EDTA plasma samples during LLV were collected and stored at −80°C for genotyping.
Definition and Data Collection
According to WHO guidelines, LLV was defined as the VL between 50 and 999 copies/mL after 6 months of ART.5 iLLV refers to an independent LLV with previous and subsequent VL < 50 copies/mL.8 pLLV was defined as two or more consecutive VL between 50 and 999 copies/mL.11
Demographic and clinical data, including age, gender, HIV transmission route, HIV-1 RNA VL at baseline, VL at genotyping, CD4+ T-cell counts at baseline, and ART regimen, were collected from the database of the national free antiretroviral treatment plan.
HIV RNA Extraction
At least 500 µL plasma samples were concentrated using ultracentrifugation at 20,000 g and 4°C for 2 h to enhance the sensitivity for detecting mutations at low VL. A portion of the supernatant was removed, and the remaining 200 µL supernatant with the concentrated virus was used for RNA extraction using the Viral RNA Extraction Kit (Guangzhou Life Technologies Daan Diagnostics Co., Ltd.) according to the manufacturer’s instructions.
Sanger Sequencing
The amplification of the entire pol gene containing reverse transcriptase, protease, and integrase regions was performed using a commercial Sanger genotyping kit (Guangzhou Life Technologies Daan Diagnostics Co., Ltd.). The positive PCR products were purified and sequenced using the 3500XL DX genetic analyzer. HIV-1 subtypes were determined using the COMET online tool (http://comet.retrovirology.lu). Subsequently, HIV DRMs and resistance interpretations were confirmed using the Stanford HIVdb algorithm version 9.0. Low-, intermediate-, and high drug-resistance were defined as resistant.
Next-Generation Sequencing
A total of 51 samples successfully sequenced using SS were simultaneously subjected to NGS. Extracted RNA was used for the amplification of the PR region (4–99 amino acids) and partial RT region (1–251 amino acids) by using an in-house method (Promega, Madison, WI, USA). Second-round amplicons were cleaned using KAPA PureBeads (Roche, Basel, Switzerland) and quantified using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Carlsbad, CA, USA). Sequencing libraries were prepared using the 96-sample KAPA HyperPlus Kit (Roche, Basel, Switzerland). The samples were pooled in each run with a 40% Phix control library (v3, Illumina, San Diego, CA, USA) to increase the diversity of libraries and further sequenced on the Illumina Miseq system by using a v3600-cycle reagent kit (Illumina, San Diego, CA, USA). The raw data were analyzed using the HyDRA Web tool (http://hydra.canada.ca/, accessed on May 20, 2022) according to the HyDRA Web User Guide,26 producing lists and frequencies of DRMs, which were interpreted using the HIVdb algorithm version 9.0. The NGS assay used a threshold of 5% for reporting low-frequency variants.
Data Analysis
Continuous variables are described as the median and interquartile range (IQR), whereas categorical variables are presented as the percentage. The amplification success rates of PR/RT and IN regions were evaluated at 50–99, 100–199, 200–399, and 400–999 copies/mL. Correlation between VL and genotyping was assessed using logistic regression. The Chi-square test was used to compare the prevalence of DRMs in the pLLV and iLLV groups. Binary logistic regression analysis was used to evaluate the predictive factors for LLV patients who had DRMs. A P value of <0.1 was considered significant. Statistical analysis was performed using SPSS 22.0 (SPSS, Chicago, IL, USA) and GraphPad 7 (GraphPad Software, La Jolla, CA, USA) software.
Ethics
This study was approved by the Ethics Committee of Beijing Ditan Hospital of Capital Medical University (Approval number: 2021-022-01) and complied with the Declaration of Helsinki. All participants provided written informed consent to use their plasma samples and clinical data.
Results
Patient Characteristics
Overall, 80 patients met the definition of LLV, of which 62.5% (50/80) and 37.5% (30/80) were iLLV and pLLV, respectively. In total, 90% participants (72/80) were men, with a median age of 35 years (IQR: 27–43 years) at HIV diagnosis. At baseline, the median HIV-1 RNA VL and CD4+ T-cell counts were 297,107 copies/mL (IQR: 43,652–424,165 copies/mL) and 263 cells/µL (IQR: 62–402 cells/µL), respectively. The median VL at genotyping was 196 copies/mL (IQR: 68–253 copies/mL). In total, 83.8% (n = 67) patients received non-nucleoside reverse transcriptase inhibitor (NNRTI)-based regimen, and 8.8% (n = 7) received protease inhibitor (PI)-based regimen. The most common regimen was lamivudine and tenofovir combined with efavirenz (78.8%). Patient characteristics are given in Table 1.
Table 1.
Demographic Characteristics of the Study Participants
| Variable | Patients (n=80) |
|---|---|
| Age (years) | |
| <30 | 26 (32.5%) |
| 30–50 | 45 (56.2%) |
| >50 | 9 (11.3%) |
| Gender | |
| Male | 72 (90%) |
| Female | 8 (10%) |
| HIV transmission route | |
| Homosexual | 67 (83.8%) |
| Heterosexual | 11 (13.8%) |
| Others | 2 (2.5%) |
| CD4 count, cells/µL, at baseline, median (25th–75th) | 263 (62–402) |
| HIV-1 RNA copies/mL at baseline, median (25th–75th) | 297,107 (43,652–424,165) |
| Plasma viral load at LLV, copies/mL | |
| 50–99 | 43 (53.7%) |
| 100–199 | 14 (17.5%) |
| 200–399 | 12 (15%) |
| 400–999 | 11 (13.8%) |
| ART regimen | |
| 3TC+TDF+EFV | 63 (78.8%) |
| 3TC+TDF+LPV/r | 5 (6.3%) |
| TAF+EVG/c/FTC | 3 (3.8%) |
| AZT+3TC+EFV | 2 (2.5%) |
| 3TC+TDF+DTG | 2 (2.5%) |
| Others | 5 (6.3%) |
| Type of LLV | |
| iLLV | 50 (62.5%) |
| pLLV | 30 (37.5%) |
Abbreviations: 3TC, lamivudine; TDF, tenofovir; EFV, efavirenz; LPV/r, lopinavir/ritonavir; TAF, tenofovir alafenamide; EVG/c/FTC, elvitegravir; FTC, emtricitabine; AZT, zidovudine; DTG, dolutegravir; LLV, low-level viremia; iLLV, intermittent low-level viremia; pLLV, persistent low-level viremia.
Genotyping Success Rate of SS
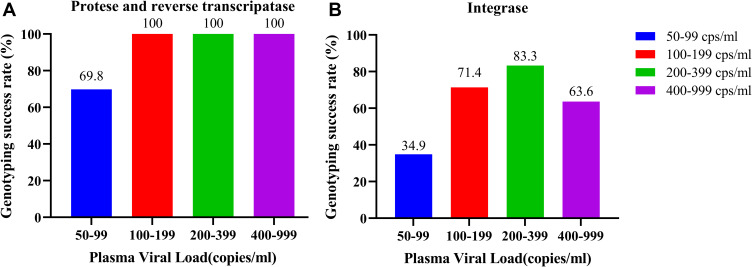
The overall success rates of SS in PR/RT regions and IN region were 83.8% (67/80) and 51.3% (41/80), respectively. For the VL of 50–99, 100–199, 200–399, and 400–999 copies/mL, the success rates of PR and RT regions were 69.8% (30/43), 100% (14/14), 100% (12/12), and 100% (11/11), respectively, whereas those of the IN region were 34.9% (15/43), 64.3% (9/14), 83.3% (10/12), and 63.6% (7/11), respectively. A significant linear trend existed between VL and the genotyping success rate (PR and RT: P = 0.021; IN: P = 0.019). Genotyping success rates of various VL categories by SS are detailed in Figure 1.
Figure 1.
Genotyping success rates of different viremia categories. (A) Genotyping success rates of different viremia categories at protease and reverse transcriptase regions. (B) Genotyping success rates of different viremia categories at integrase region.
HIV Subtypes and DRMs Detected by SS
Subtypes and DRMs were analyzed in 67 participants with successful amplification of the PR and RT regions. CRF01_AE was the most frequently occurring genotype (44.8%; 30/67), followed by CRF07_BC (25.4%; 17/67), B (17.9%; 12/67), and A (3%; 2/67). Overall, 38.8% (26/67) participants harbored at least one mutation. A total of 28.4% (19/67), 22.4% (15/67), and 4.5% (3/67) participants were resistant to NNRTIs, nucleoside reverse transcriptase inhibitors (NRTIs), and PIs, respectively. Overall, 20.9% (14/67) participants were resistant to both NRTIs and NNRTIs, and 1.5% (1/67) were resistant to NRTIs, NNRTIs, and PIs.
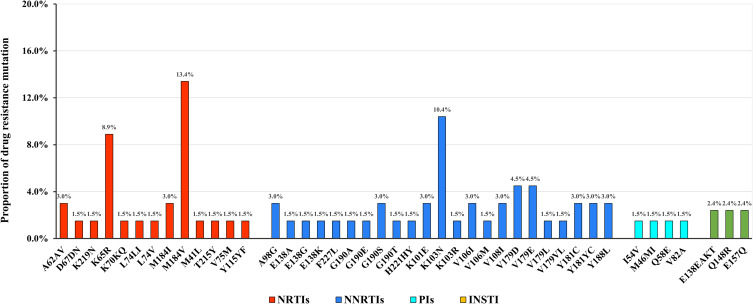
The most common NRTI-associated mutation was M184V, observed in 13.4% (9/67) participants, and the most common NNRTI-associated mutation was K103N, observed in 10.4% (7/67) participants. Three PI major mutations including M46MI, I54V, and V82A were observed in three participants. Two major IN-associated mutations (E138EAKT and Q148R) were observed in one participant, and one IN accessory mutation (E157Q) was observed in another participant. Patterns of DRMs in HIV-infected patients with LLV are depicted in Figure 2.
Figure 2.
Patterns of DRMs in HIV-infected patients with LLV.
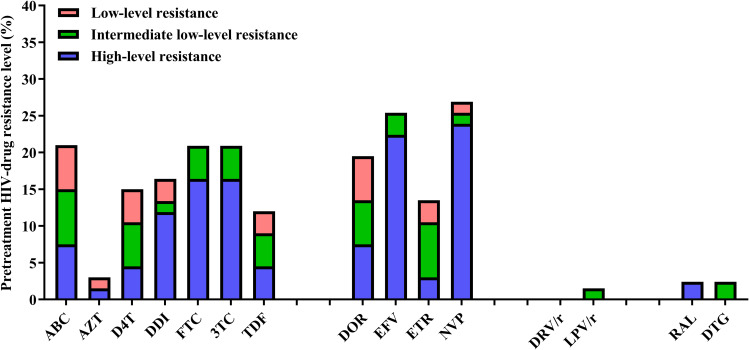
Regarding the NRTI regimen, 20.9% of the participants exhibited low- to high-level resistance to abacavir, lamivudine, and emtricitabine. Furthermore, 18 (26.9%) and 17 (25.4%) participants exhibited low- to high-level resistance to nevirapine and efavirenz, respectively. Additionally, 1.5% and 2.4% participants were resistant to PIs and INSTIs, respectively. The effect of resistance mutations on drug susceptibility is illustrated in Figure 3.
Figure 3.
Different levels of drug resistance in HIV-infected patients with LLV.
Comparison of the Prevalence of DRMs Between pLLV and iLLV
In our study, 50 participants were iLLV; among them, samples of 38 participants were successfully sequenced. Overall, 21.1% (8/38) participants had at least one mutation with a median DRMs count of 1 (IQR: 1–3). DRMs of participants with iLLV are given in Table 2. Furthermore, 30 participants were pLLV; among them, samples of 29 participants were successfully sequenced. Overall, 62.1% (18/29) of the patients with pLLV had at least one mutation with a median DRMs count of 4 (IQR: 1–5). The prevalence of DRMs in pLLV was significantly higher than that in iLLV (P < 0.001). DRMs of participants with pLLV are detailed in Table 3.
Table 2.
Drug Resistance Mutations in 8 Patients with Intermittent LLV
| Case ID | Gender | Age | TR | VL | Subtype | ART at the Time of DRM Test | Drug-Resistant Mutation | |||
|---|---|---|---|---|---|---|---|---|---|---|
| NNRTIs | NRTIs | PIs | INSTIs | |||||||
| F3821 | Male | 32 | Heterosexual | 53 | CRF07_BC | 3TC+TDF+EFV | – | – | Q58E | – |
| F4301 | Male | 27 | Homosexual | 60 | A | 3TC+TDF+DTG | E138K, F227L, V179L | M184V | – | – |
| F8115 | Male | 33 | Homosexual | 70 | CRF55_01B | 3TC+TDF+EFV | V179E | – | – | – |
| F8805 | Male | 27 | Homosexual | 73 | B | 3TC+TDF+EFV | V106I | – | – | – |
| F8830 | Male | 29 | Homosexual | 517 | B | 3TC+TDF+LPV/r | K103N | – | – | – |
| F9271 | Male | 23 | Homosexual | 82 | CRF01_AE | 3TC+AZT+LPV/r | Y188L | M184I, V75M | – | – |
| g1793 | Female | 31 | Heterosexual | 63 | CRF01_AE | 3TC+TDF+EFV | G190T | A62AV, K65R | – | – |
Abbreviations: VL, viral load; TR, transmission route; NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors; INSTIs, integrase inhibitors.
Table 3.
Drug Resistance Mutations in 18 Patients with Persistent LLV
| Case ID | Gender | Age | TR | VL | Subtype | ART at the Time of DRM Test | Drug-Resistant Mutation | |||
|---|---|---|---|---|---|---|---|---|---|---|
| NNRTIs | NRTIs | PIs | INSTIs | |||||||
| F3848 | Male | 35 | Homosexual | 142 | CRF01_AE | 3TC+TDF+EFV | – | – | M46MI | – |
| F4947 | Male | 45 | Homosexual | 124 | CRF07_BC | 3TC+TDF+EFV | E138G | – | – | – |
| F5603 | Male | 28 | Homosexual | 71 | CRF07_BC | 3TC+TDF+EFV | V179D | – | – | – |
| F5756 | Male | 50 | Homosexual | 342 | CRF01_AE | 3TC+TDF+EFV | – | D67DN | – | – |
| F6332 | Male | 31 | Heterosexual | 443 | CRF01_AE | 3TC+TDF+EFV | K101E, G190S | K65R | – | – |
| F6370 | Male | 30 | Homosexual | 85 | A | 3TC+TDF+EFV | K101E, G190S, Y181C, M184I | A62V, K65R, M184I | – | – |
| F6856 | Male | 28 | Homosexual | 388 | B | 3TC+TDF+EFV | A98G, K103N | M41L, T215Y, M184V | – | – |
| F7282 | Male | 50 | Homosexual | 867 | CRF01_AE | 3TC+TDF+EFV | Y181C | L74V, M184V | I54V, V82A | – |
| F7343 | Male | 36 | Homosexual | 811 | CRF55_01B | 3TC+TDF+EFV | Y181YC, G190A, H221HY, V179E | L74LI, M184V, Y115YF | – | – |
| F7492 | Male | 35 | Homosexual | 66 | B | 3TC+TDF+EFV | K103N, Y181YC, V108I | K65R, M184V | – | – |
| F7570 | Male | 56 | Homosexual | 371 | CRF07_BC | 3TC+TDF+EFV | A98G, K103N, V179E | M184V | – | E138EAKT, Q148R |
| F8103 | Male | 26 | Heterosexual | 111 | CRF01_AE | 3TC+TDF+EFV | V106I, Y188L, V179VL | K70KQ, M184V | – | – |
| F8355 | Male | 28 | Homosexual | 805 | CRF01_AE | 3TC+TDF+EFV | V106M, K103R, V179D | M184V | – | – |
| F9710 | Male | 23 | Homosexual | 323 | CRF01_AE | 3TC+TDF+EFV | K103N | – | – | – |
| g622 | Male | 44 | Homosexual | 223 | CRF01_AE | 3TC+TDF+EFV | V108I | – | – | – |
| g3789 | Male | 28 | Heterosexual | 180 | CRF07_BC | 3TC+TDF+EFV | G190E, E138A | K65R, K219N | – | – |
| g3914 | Male | 20 | Homosexual | 166 | CRF01_AE | 3TC+TDF+EFV | V108I, K103N | K65R, M184V | – | – |
| g4391 | Male | 30 | Homosexual | 301 | CRF07_BC | 3TC+TDF+EFV | – | K103N | – | – |
Abbreviations: VL, viral load; TR, transmission route; NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors; INSTIs, integrase inhibitors.
Comparison of DRMs and Drug-Resistance Interpretations Between NGS and SS
The overall success rates of NGS in PR/RT regions and IN region were 86.3% (44/51) and 88.2% (45/51), respectively. The PR/RT regions of 44 samples were successfully sequenced using SS and NGS simultaneously. Furthermore, we considered a 5% threshold to compare the results of the two sequencing methods. These two sequencing methods simultaneously detected 69 DRMs. A total of 10 DRMs, including 2 NRTI-related, 5 NNRTI-related, and 3 PI-related DRMs, detected using NGS were missed by SS. The frequency of 6 DRMs was <20%, whereas 4 DRMs was >20%. DRMs detected by NGS but missed by SS at the threshold of 5% are shown in Table 4. Furthermore, 8 DRMs were only detected using SS but missed by NGS, including 2 NRTI-related, 5 NNRTI-related, and 1 PI-related DRMs. DRMs detected by SS but missed by NGS at the threshold of 5% are shown in Table 5.
Table 4.
DRMs Detected by NGS but Missed by SS at the Threshold of 5%
| Case ID | LLV | VL | Sanger | NGS | ||||
|---|---|---|---|---|---|---|---|---|
| PIs | NRTIs | NNRTIs | PIs | NRTIs | NNRTIs | |||
| f6462 | pLLV | 95 | None | None | None | N88S (5.1) | None | None |
| f6332 | pLLV | 443 | None | K65R | K101E, G190S | None | K65R (70.31) | K101E (69.52), G190S (69.72), Y181C (7.82), |
| f7282 | pLLV | 867 | I54V, V82A | L74V, M184V | Y181C | M46I (15.15), I54V (98.66), V82A (97.71) | L74V (98.19), M184V (98.71) | Y181C (97.99), E138K (6.81), P236L (5.36) |
| G682 | pLLV | 89 | None | None | None | None | None | E138G (97.95) |
| G1128 | Blip | 986 | None | None | None | I47V (6.29) | None | None |
| GMg-3789 | pLLV | 180 | None | K65R, K219N | G190E, E138A | None | K65R (98.44), K219N (98.48), D67G (98.81) | G190E (98.76), E138A (97.62) |
| GMg-3914 | pLLV | 166 | None | K65R, M184V | V108I, K103N | None | K65R (98.94), M184V (98.12), K219E (41.44) | V108I (96.25), K103N (97.52) |
| GMg-4391 | pLLV | 301 | None | None | K103N | None | None | K103N (97.84), K101E (33.39) |
Note: The red-colored text represents DRMs detected by NGS but missed by SS at the threshold of 5%.
Abbreviations: LLV, low-level viremia; VL, viral load; NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; PI, protease inhibitors.
Table 5.
DRMs Detected by SS but Missed by NGS at the Threshold of 5%
| Case ID | LLV | VL | Sanger | NGS | ||||
|---|---|---|---|---|---|---|---|---|
| PIs | NRTIs | NNRTIs | PIs | NRTIs | NNRTIs | |||
| f3848 | pLLV | 142 | M46MI | None | None | None | None | None |
| f4947 | pLLV | 124 | None | None | E138G | None | None | None |
| f5756 | pLLV | 342 | None | D67DN | None | None | None | None |
| f8103 | pLLV | 111 | None | K70KQ, M184V | Y188L, V106I, V179VL | None | M184V (98.09) | Y188L (97.81) |
| f8355 | pLLV | 805 | None | M184V | V106M, V179D, K103R, | None | M184V (98.22) | V106M (97.49), V179D (96.68) |
| f8805 | Blip | 73 | None | None | V106I | None | None | None |
Note: The red-colored text represents DRMs detected by SS but missed by NGS at the threshold of 5%.
Abbreviations: LLV, low-level viremia; VL, viral load; NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors.
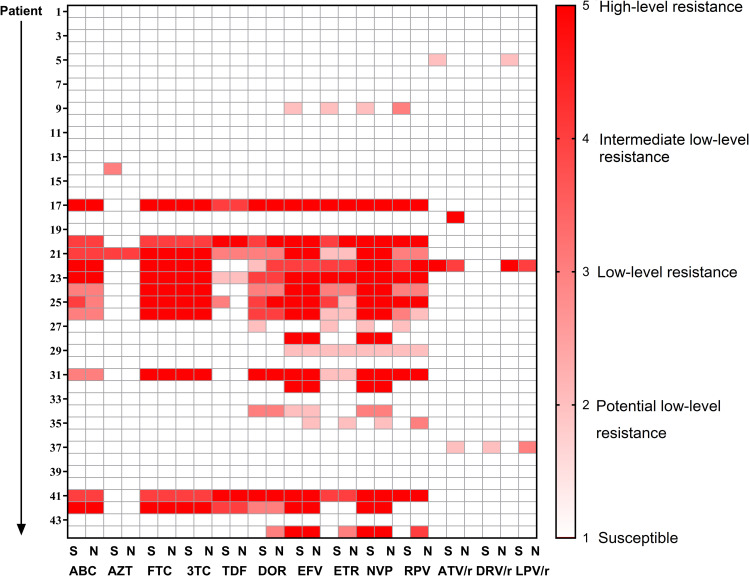
For 44 samples, NGS detected higher levels of NRTI, NNRTI, and PI resistance for 0% (n = 0), 9.1% (n = 4), and 6.8% (n = 3) of samples, respectively. By contrast, SS detected higher levels of NRTI, NNRTI, and PI resistance for 4.4% (n = 2), 9.1% (n = 4), and 2.3% (n = 1) of samples, respectively. Comparison of drug resistance interpretation between SS and NGS is shown in Figure 4.
Figure 4.
Comparison of drug resistance interpretation between SS and NGS.
Risk Factors of LLV Patients Having Mutations
We further focused on the risk factors associated with LLV patients who had DRMs. Binary logistic regression analysis was performed to introduce all dependent variables into the regression equation (Supplementary Table 1). In univariate analysis, VL at LLV and pLLV were observed to be associated with LLV patients who had DRMs. Moreover, in multivariate analysis, we observed that only pLLV [aOR: 5.621 (1.822–17.341)] was a risk factor for LLV patients who had DRMs. This suggested that the DRMs are more likely to occur when the variants continue replicating.
Discussion
To the best of our knowledge, this is the first study examining the feasibility of genotyping in LLV in China. We obtained a high genotyping success rate for the samples with low copy numbers. Furthermore, the results revealed that DRMs were common in patients with LLV, and the prevalence of DRMs in pLLV was significantly higher than that in iLLV. At the threshold of 5%, both NGS and SS detected new DRMs, reflecting that the two sequencing methods could complement each other rather than replace each other.
Using the same commercial Sanger genotyping kit, the success rate of samples with VL > 1000 copies/mL was 95%. In our study, the overall success rate of specimens with VL between 50 and 999 copies/mL using SS was 83.8%, which is consistent with the results of a previous study that included samples with VL between 200 and 999 copies/mL.27 Furthermore, the volume of most of the plasma specimens in our study was less than 1 mL, unlike previous studies that required a high plasma input of more than 1 mL for samples with VL < 1000 copies /mL.11,20
Studies in developed countries have reported that the prevalence of DRMs in patients with LLV ranges from 17% to 75%, depending on distinct study designs and the definition of LLV.28–30 In our study, the overall prevalence of HIV DRMs in patients with LLV was 38.8%, and further detailed analysis revealed that these patients were mainly resistant to NNRTIs and NRTIs. Due to the limitation of economic conditions, 3TC + TDF + EFV are still the mainstream first-line antiviral drugs in China, and fewer patients use INSITs. Overall, 61.2% cases with LLV did not have DRMs to the current ART regimen. This could be because of several reasons, including technical errors, the influence of the type of blood collection tube used,31 infections,32 vaccinations,33 and low drug concentrations in blood because of poor adherence or absorption of antiretroviral drugs.34
Consistent with the results of previous studies, both iLLV and pLLV were found to be associated with emerging DRMs, which decreases further therapeutic options.35,36 Contrary to the findings of a previous study, the prevalence of DRMs in the pLLV cohort was significantly higher than that in the iLLV cohort, and we observed that pLLV was a risk factor for LLV patients who had DRMs. This suggested that the DRMs are more likely to occur when variants continue replicating.27 Our previous study reported that blip is not associated with VF,3 and existing evidence suggests that blip can be detected with DRMs. Our results are consistent with those of a Spanish study that assessed the emergence of new DRMs during blip but reported no association between blip and VF.37
Most participants with LLV in this study were CRF01_AE, the predominant circulating subtype in China.38,39 DRMs were prevalent in patients with LLV; 28.4% patients had at least 1 NNRTI mutation, and 22.4% had at least 1 NRTI mutation. Predictably, the patterns of DRMs observed in our study are comparable with those reported in previous studies in which M184V/I and K103N were the most commonly detected mutations.12,27,28 Furthermore, three major PI-associated mutations, including M46MI, I54V, and V82A, were revealed in three participants; however, the participants with these mutations had no known exposure to the PI-related regimen. Similarly, one participant had two major IN-associated mutations; however, he did not receive IN-related regimen. The low prevalence of PI- and IN-related DRMs is mainly because of the small number of people using the related regimen and the high genetic barrier.
However, in some studies, only the variants with frequency >20% could be detected using SS.40,41 Low-frequency DRMs are associated with an increased risk of VF.42–44 Therefore, we evaluated the performance of NGS in detecting HIV-1 DRMs by using LLV samples and compared the results with those of SS. PR/RT regions of 44 LLV samples were successfully sequenced through NGS simultaneously. In our study, NGS missed 8 DRMs detected by SS; however, in another study, NGS detected all DRMs using the same sequencing platform at 5% threshold, although the study included samples with VL > 1000 copies/ mL.37 We considered that the missing of DRMs by NGS was due to the low VL and a low number of amplified cDNA templates.25
Studies in the developed and developing countries have reported that even patients with LLV and a low VL have the risk of VF.7,8,21 Moreover, a previous study demonstrated that switching to second-line ART in patients with LLV resulted in VS in most of the patients.12 However, the threshold for VF is 1000 copies/mL and LLV are non-failure which no management is required according to WHO guidelines.4 Our study demonstrated the influence of pLLV on VF and the high prevalence of DRMs in patients with pLLV, which warrants updating the guidelines in the future.
The strengths of this study are that to the best of our knowledge, this study is the first on the prevalence of DRMs in patients with LLV in China, and the results are clinically significant. Second, we observed that both patients with pLLV and iLLV had DRMs. Still, the prevalence of DRMs was significantly higher in patients with pLLV than in those with iLLV, which could remarkably reduce the therapeutic options for other regimens. Finally, we explored the performance of NGS in detecting HIV DRMs by using LLV samples and compared the results with those of SS.
Our study also has some limitations. First, the amplification efficiency for the IN region was significantly lower than that for the PR and RT regions, which may be related to primer specificity. Second, the DRMs were only described when LLV occurred, and we did not perform genotypic resistance testing at baseline compared with LLV. Finally, because of COVID-19 pandemic and incomplete follow-up data of patients, we could not further follow up. However, we will continue to pay attention to these patients after the pandemic eases.
Conclusion
In our study, we achieved 83.8% genotyping success rate for PR and RT regions from samples with relatively low copy numbers. Both patients with pLLV and iLLV had DRMs; however, the prevalence of DRMs in pLLV (62.1%) was significantly higher than that in iLLV (21.1%). At the threshold of 5%, both NGS and SS detected new DRMs, reflecting that the two sequencing methods could complement each other rather than replace each other. Our findings endorse that lowering the WHO threshold for VF and RGT is necessary to guide ART optimization in this setting.
Acknowledgments
We thank the staff from the HIV genotyping laboratory at Beijing Ditan Hospital. We also thank all the implementing partners of Beijing CDC for providing technical guidance on second-generation sequencing.
Funding Statement
This work was partly funded by The National 13th Five-Year Grand Program on Key Infectious Disease Control (2018ZX10302-102 to F.Z., 2018ZX10715-005 to H.Z.) and the R&D Program of Beijing Municipal Education Commission (KM202210025004).
Disclosure
The authors have no conflict of interest to declare in this work.
References
- 1.World Health Organization. HIV/AIDS; 2020. Available from: http://www.who.int/mediacentre/factsheets/fs360/en/. Accessed November 7, 2022.
- 2.Zhang F, Dou Z, Ma Y, et al. Five-year outcomes of the China National Free Antiretroviral Treatment Program. Ann Intern Med. 2009;151:241–251. doi: 10.7326/0003-4819-151-4-200908180-00006 [DOI] [PubMed] [Google Scholar]
- 3.Zhang T, Ding H, An M, et al. Factors associated with high-risk low-level viremia leading to virologic failure: 16-year retrospective study of a Chinese antiretroviral therapy cohort. BMC Infect Dis. 2020;201:147. doi: 10.1186/s12879-020-4837-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, Chen M, Zhao H, et al. Persistent low-level viremia is an independent risk factor for virologic failure: a retrospective cohort study in China. Infect Drug Resist. 2021;14:4529–4537. doi: 10.2147/IDR.S332924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Consolidated Guidelines on HIV Prevention, Diagnosis, Treatment and Care for Key Populations. World Health Organization; 2016. [PubMed] [Google Scholar]
- 6.Joya C, Won SH, Schofield C, et al. Persistent low-level viremia while on antiretroviral therapy is an independent risk factor for virologic failure. Clin Infect Dis. 2019;69:2145–2152. doi: 10.1093/cid/ciz129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermans LE, Moorhouse MM, Carmona S, et al. Effect of HIV-1 low-level viraemia during antiretroviral therapy on treatment outcomes in WHO-guided South African treatment programmes: a multicentre cohort study. Lancet Infect Dis. 2018;18:188–197. doi: 10.1016/S1473-3099(17)30681-3 [DOI] [PubMed] [Google Scholar]
- 8.Grennan JT, Loutfy MR, Su D, et al. Magnitude of virologic blips is associated with a higher risk for virologic rebound in HIV-infected individuals: a recurrent events analysis. J Infect Dis. 2012;8:1230–1238. doi: 10.1093/infdis/jis104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Abrams EJ, Ammann A, et al. Perinatal transmission of human immunodeficiency virus type 1 by pregnant women with RNA virus loads <1000 copies/mL. J Infect Dis. 2001;183:539–545. doi: 10.1086/318530 [DOI] [PubMed] [Google Scholar]
- 10.Hughes JP, Baeten JM, Lingappa JR, et al. Determinants of per-coital-act HIV-1 infectivity among African HIV-1-serodiscordant couples. J Infect Dis. 2012;205:358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taiwo B, Gallien S, Aga E, et al. Antiretroviral drug resistance in HIV-1-infected patients experiencing persistent low-level viremia during first-line therapy. J Infect Dis. 2011;204:515–520. doi: 10.1093/infdis/jir353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JA, Amstutz A, Nsakala BL, et al. Extensive drug resistance during low-level HIV viraemia while taking NNRTI-based ART supports lowering the viral load threshold for regimen switch in resource-limited settings: a pre-planned analysis from the SESOTHO trial. J Antimicrob Chemother. 2021;76:1294–1298. doi: 10.1093/jac/dkab025 [DOI] [PubMed] [Google Scholar]
- 13.Kantor R, Delong A, Schreier L, et al. HIV second-line failure and drug resistance at high- and low-level viremia in Western Kenya. AIDS. 2018;32:2485–2496. doi: 10.1097/QAD.0000000000001964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson AC, Younger SR, Martin JN, et al. Immunologic and virologic evolution during periods of intermittent and persistent low-level viremia. AIDS. 2004;18:981–989. doi: 10.1097/00002030-200404300-00005 [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Ma SS, Zhang WY, et al. Changes in peripheral blood inflammatory factors (TNF-alpha and IL-6) and intestinal flora in AIDS and HIV-positive individuals. J Zhejiang Univ Sci B. 2019;20:793–802. doi: 10.1631/jzus.B1900075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernal E, Gómez JM, Jarrín I, et al. Low-level viremia is associated with clinical progression in HIV-infected patients receiving antiretroviral treatment. J Acquir Immune Defic Syndr. 2018;78(3):329–337. doi: 10.1097/QAI.0000000000001678 [DOI] [PubMed] [Google Scholar]
- 17.Amstutz A, Nsakala B, Vanobberghen F, et al. Switch to second-line versus continued first-line antiretroviral therapy for patients with low-level HIV-1 viremia: an open-label randomized controlled trial in Lesotho. PLoS Med. 2020;17(9):e1003325. doi: 10.1371/journal.pmed.1003325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. Guidelines for the Diagnosis, Prevention and Management of Cryptococcal Disease in HIV-Infected Adults, Adolescents and Children: Supplement to the 2016 Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection. World Health Organization; 2018. [PubMed] [Google Scholar]
- 19.Vandamme A-M, Houyez F, Banhegyi D, et al. Laboratory guidelines for the practical use of HIV drug resistance tests in patient follow-up. Antivir Ther. 2001;6(1):21–39. doi: 10.1177/135965350100600103 [DOI] [PubMed] [Google Scholar]
- 20.Delaugerre C, Gallien S, Flandre P, et al. Impact of low-level-viremia on HIV-1 drug-resistance evolution among antiretroviral treated-patients. PLoS One. 2012;7:e36673. doi: 10.1371/journal.pone.0036673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esber A, Polyak C, Kiweewa F, et al. Persistent low-level viremia predicts subsequent virologic failure: is it time to change the third 90? Clin Infect Dis. 2019;69:805–812. doi: 10.1093/cid/ciy989 [DOI] [PubMed] [Google Scholar]
- 22.Inzaule SC, Hamers RL, Julian MN, et al. Clinically relevant thresholds for ultrasensitive HIV drug resistance testing: a multi-country nested case-control study. Lancet HIV. 2018;58:446–449. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JA, Li JF, Wei XR, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balduin M, Oette M, Däumer MP, et al. prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naïve patients and their impact on the virological failure. J Clin Virol. 2009;45(1):34–38. doi: 10.1016/j.jcv.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 25.Tzou PL, Ariyaratne P, Varghese V, et al. Comparison of an in vitro diagnostic next-generation sequencing assay with sanger sequencing for HIV-1Genotypic resistance testing. J Clin Microbiol. 2018;56:e00105–e00118. doi: 10.1128/JCM.00105-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nykoluk M, Taylor T, Enns E, et al. HyDRA web user guide; 2016. Available from: https://hydra.canada.ca/pages/about?lang=en-CA. Accessed March 20, 2022.
- 27.Bangalee A, Hans L, Steegen K. Feasibility and clinical relevance of HIV-1 drug resistance testing in patients with low-level viraemia in South Africa. J Antimicrob Chemother. 2021;76:2659–2665. doi: 10.1093/jac/dkab220 [DOI] [PubMed] [Google Scholar]
- 28.Nettles RE, Kieffer TL, Simmons RP, et al. Genotypic resistance in HIV-1–infected patients with persistently detectable low-level viremia while receiving highly active antiretroviral therapy. Clin Infect Dis. 2004;39:1030–1037. doi: 10.1086/423388 [DOI] [PubMed] [Google Scholar]
- 29.Serna AZ, Min JE, Woods C, et al. Performance of HIV-1 drug resistance testing at low-level viremia and its ability to predict future virologic outcomes and viral evolution in treatment-naive individuals. Clin Infect Dis. 2014;58:1165–1173. doi: 10.1093/cid/ciu019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao SW, Liu ZH, Wu TS, et al. Prevalence of drug resistance mutations in HIV-infected individuals with low-level viraemia under combination antiretroviral therapy: an observational study in a tertiary hospital in Northern Taiwan, 2017–19. J Antimicrob Chemother. 2021;76:722–728. doi: 10.1093/jac/dkaa510 [DOI] [PubMed] [Google Scholar]
- 31.Stosor V, Palella FJ, Berzins B, et al. Transient viremia in HIV-infected patients and use of plasma preparation tubes. Clin Infect Dis. 2005;41(11):1671–1674. doi: 10.1086/498025 [DOI] [PubMed] [Google Scholar]
- 32.Jones LE, Perelson AS. Opportunistic infection as a cause of transient viremia in chronically infected HIV patients under treatment with HAART. Bull Math Biol. 2005;67(6):1227–1251. doi: 10.1016/j.bulm.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 33.Gunthard HF, Wong JK, Spina CA, et al. Effect of influenza vaccination on viral replication and immune response in persons infected with human immunodeficiency virus receiving potent antiretroviral therapy. J Infect Dis. 2000;181(2):522–531. doi: 10.1086/315260 [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Serna A, Swenson LC, Watson B, et al. A single untimed plasma drug concentration measurement during low-level HIV viremia predicts virologic failure. Clin Microbiol Infect. 2016;22(12):1004–e9. [DOI] [PubMed] [Google Scholar]
- 35.Cohen Stuart JW, Wensing AM, Kovacs C, et al. Transient relapses (“blips”) of plasma HIV RNA levels during HAART are associated with drug resistance. J Acquir Immune Defic Syndr. 2001;28:105–113. doi: 10.1097/00126334-200110010-00001 [DOI] [PubMed] [Google Scholar]
- 36.Li JZ, Gallien S, Do TD, et al. Prevalence and significance of HIV-1 drug resistance mutations among patients on antiretroviral therapy with detectable low-level viremia. Antimicrob Agents Chemother. 2012;56:5998–6000. doi: 10.1128/AAC.01217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macias J, Palomares JC, Mira JA, et al. Transient rebounds of HIV plasma viremia are associated with the emergence of drug resistance mutations in patients on highly active antiretroviral therapy. J Infect. 2005;51:195–200. doi: 10.1016/j.jinf.2004.11.010 [DOI] [PubMed] [Google Scholar]
- 38.Yu F, Li Q, Wang L, et al. Drug resistance to HIV-1 integrase inhibitors among treatment-naive patients in Beijing, China. Pharmgenomics Pers Med. 2022;15:195–203. doi: 10.2147/PGPM.S345797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan HB, Liu ZQ, Wu XF, et al. Prevalence of transmitted HIV-1 drug resistance among treatment-naïve individuals in China, 2000–2016. Arch Virol. 2021;166:2451–2460. doi: 10.1007/s00705-021-05140-9 [DOI] [PubMed] [Google Scholar]
- 40.Chen NY, Kao SW, Liu ZH, et al. Shall I trust the report? Variable performance of Sanger sequencing revealed by deep sequencing on HIV drug resistance mutation detection. Int J Infect Dis. 2020;93:182–191. doi: 10.1016/j.ijid.2020.02.004 [DOI] [PubMed] [Google Scholar]
- 41.Li M, Liang S, Zhou C, et al. HIV drug resistance mutations detection by next-generation sequencing during antiretroviral therapy interruption in China. Pathogens. 2021;10:264. doi: 10.3390/pathogens10030264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;52:569–573. doi: 10.1097/QAI.0b013e3181ba11e8 [DOI] [PubMed] [Google Scholar]
- 43.Kyeyune F, Gibson RM, Nankya I, et al. Low-frequency drug resistance in HIV-infected Ugandans on antiretroviral treatment is associated with regimen failure. Antimicrob Agents Chemother. 2016;60:3380–3397. doi: 10.1128/AAC.00038-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mbunkah HA, Bertagnolio S, Hamers RL, et al. Low-abundance drug-resistant HIV-1 variants in antiretroviral drug-naive individuals: a systematic review of detection methods, prevalence, and clinical impact. J Infect Dis. 2020;221:1584–1597. doi: 10.1093/infdis/jiz650 [DOI] [PMC free article] [PubMed] [Google Scholar]