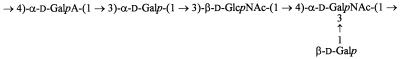Abstract
Shiga toxigenic Escherichia coli (STEC) strains are a diverse group of organisms capable of causing severe gastrointestinal disease in humans. Within the STEC family, eae-positive STEC strains, particularly those belonging to serogroups O157 and O111, appear to have greater virulence for humans. However, in spite of being eae negative, STEC strains belonging to serogroup O113 have frequently been associated with cases of severe STEC disease, including hemolytic-uremic syndrome (HUS). Western blot analysis with convalescent-phase serum from a patient with HUS caused by an O113:H21 STEC strain indicated that human immune responses were directed principally against lipopolysaccharide O antigen. Accordingly, the serum was used to isolate a clone expressing O113 O antigen from a cosmid library of O113:H21 DNA constructed in E. coli K-12. Sequence analysis indicated that the O113 O-antigen biosynthesis (rfb) locus contains a cluster of nine genes which may be cotranscribed. Comparison with sequence databases identified candidate genes for four glycosyl transferases, an O-acetyl transferase, an O-unit flippase, and an O-antigen polymerase, as well as copies of galE and gnd. Two additional, separately transcribed genes downstream of the O113 rfb region were predicted to encode enzymes involved in synthesis of activated sugar precursors, one of which (designated wbnF) was essential for O113 O-antigen synthesis, and so is clearly a part of the O113 rfb locus. Interestingly, expression of O113 O antigen by E. coli K-12 significantly increased in vitro adherence to both HEp-2 and Henle 407 cells.
Shiga toxigenic Escherichia coli (STEC) strains are an important cause of gastrointestinal disease in humans, particularly since such infections may result in life-threatening sequelae, such as hemolytic-uremic syndrome (HUS) (19, 30, 38). It has been recognized for a number of years that STEC strains causing human disease may belong to a broad range of O serogroups (19). However, a subset of these (particularly O157 and O111) appear to be responsible for the majority of serious cases (those complicated by HUS) (12, 19, 38). These STEC strains have the capacity to produce attaching-and-effacing (A/E) lesions on intestinal mucosa, a property encoded by a pathogenicity island termed the locus for enterocyte effacement (LEE) (7, 9). LEE encodes proteins with a range of functions, including a type III secretion system, various secreted effector proteins and their chaperons, the outer membrane protein intimin (the eae gene product), which mediates intimate attachment to the enterocyte cell surface, as well as the receptor for intimin (Tir) which is translocated into the plasma membrane of the enterocyte (6, 21). However, production of intimin is not essential for pathogenesis, because a significant minority of sporadic cases of HUS are caused by eae-negative STEC strains (38). One of the more common eae-negative STEC serogroups associated with human disease is O113 (particularly serotype O113:H21) (19, 38). Indeed, 2 of the 12 STEC strains originally isolated from patients with HUS by Karmali et al. (20) belonged to this serotype. An O113:H21 STEC strain was also responsible for a recent cluster of three HUS cases in Adelaide, South Australia (37); this was the first report of an apparent outbreak of HUS caused by an eae-negative STEC strain. Collectively, these findings indicate that an investigation of O113 STEC virulence factors may be warranted.
In previous studies, we have used Western immunoblot analysis and convalescent-phase HUS patient sera to examine the serological response to infection with eae-positive STEC strains belonging to serogroups O111 and O157 (32, 47). Antibody responses to intimin and Tir were detected, and convalescent-phase serum was also used to isolate clones containing a portion of the LEE from a cosmid library of O111 STEC DNA constructed in E. coli K-12 (47). However, the strongest immune response was directed against the lipopolysaccharide (LPS) O antigen. In the nonimmune host, LPS is believed to contribute to virulence by shielding the infecting organism from the bactericidal effects of serum (17, 39, 46). However, antibodies directed against LPS are likely to be highly protective (17), and anti-LPS seroconversion probably contributes to the sometimes rapid elimination of the causative STEC strain from the patient's gut during the course of HUS. Indeed, an O157-specific O-antigen–protein conjugate vaccine is currently being developed for prevention of infections caused by this STEC serogroup (22).
In the present study, we used Western immunoblot analysis to examine the antibody response of a patient with HUS due to an O113:H21 STEC strain. The convalescent-phase serum was also used to screen a cosmid gene bank of O113:H21 STEC DNA constructed in E. coli K-12, resulting in the isolation and characterization of the locus encoding biosynthesis of the O113 O antigen. The effect of expression of O113 O antigen on adherence of E. coli K-12 to epithelial cells was also investigated.
MATERIALS AND METHODS
Bacterial strains and cloning vectors.
The O113:H21 STEC strain 98NK2 was isolated from a patient with HUS at the Women's and Children's Hospital, South Australia, and has been described elsewhere (37). E. coli K-12 strains DH1 and JM109 have been described previously (13, 50). The cosmid vector pHC79 has also been described previously (16), and the phagemid pBC SK, which encodes chloramphenicol resistance, was obtained from Stratagene, La Jolla, Calif. All E. coli strains were routinely grown in Luria-Bertani (LB) medium (27) with or without 1.5% Bacto-Agar (Difco Laboratories, Detroit, Mich.). Where appropriate, ampicillin and chloramphenicol were added to growth media at concentrations of 50 and 25 μg/ml, respectively.
Western blot analysis.
Crude lysates of STEC or other E. coli strains were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described by Laemmli (24), and antigens were electrophoretically transferred onto nitrocellulose filters, as described by Towbin et al. (45). Filters were probed with convalescent-phase serum from the HUS patient from whom the O113:H21 STEC 98NK2 had been isolated (kindly provided by K. F. Jureidini and P. Henning, Renal Unit, Women's and Children's Hospital, North Adelaide, South Australia) (used at a dilution of 1:3,000), followed by goat anti-human immunoglobulin G (IgG) conjugated to alkaline phosphatase (Bio-Rad Laboratories, Hercules, Calif.). Alternatively, filters were probed with absorbed polyclonal rabbit E. coli O113-specific antiserum (obtained from the Institute of Medical and Veterinary Science, Adelaide, South Australia) (used at a dilution of 1:3,000), followed by goat anti-rabbit IgG–alkaline phosphatase conjugate (Bio-Rad). Immunoreactive bands were visualized with a chromogenic substrate (4-nitro blue tetrazolium and X-phosphate).
Construction of cosmid gene bank.
High-molecular-weight chromosomal DNA was extracted as described previously (34) from STEC 98NK2 and was digested partially with Sau3A1 in order to optimize the yield of fragments in the size range 35 to 40 kb. This DNA was ligated with a fivefold molar excess of pHC79 DNA, which had been digested with BamHI. Ligated DNA was packaged into lambda heads by using a Packagene kit (Promega Biotec, Madison, Wis.) and transfected into E. coli DH1, which had been grown in LB medium plus 2% maltose. The cells were then plated onto LB agar supplemented with ampicillin, and after incubation, clones were stored in LB medium plus ampicillin plus 15% glycerol in microtiter plates at −70°C.
Screening of cosmid clones by immunoblotting.
Cosmid clones were grown overnight at 37°C in 200 μl of LB medium plus ampicillin in microtiter plates and then spotted onto a nitrocellulose filter. Filters were then blocked, reacted with HUS patient serum (diluted 1:3,000), and then developed as described for Western blots above.
DNA sequencing.
Nested deletions of STEC DNA cloned into pBC SK were constructed by the method of Henikoff (14) with an Erase-a-base kit (Promega). This DNA was transformed into E. coli JM109, and the resulting plasmid DNA was characterized by restriction analysis. Double-stranded template DNA for sequencing was prepared as recommended in the Applied Biosystems sequencing manual. The sequence of both strands was then determined by using dye-labelled terminator chemistry on an Applied Biosystems model 373A automated DNA sequencer. The sequence was analyzed with DNASIS and PROSIS version 7.0 software (Hitachi Software Engineering, South San Francisco, Calif.). Comparison with sequence databases was carried out with the program BLASTX version 2.0 (1).
Cell culture and bacterial adherence assays.
The capacity of E. coli K-12 derivatives to adhere to either Henle 407 or HEp-2 cells was assessed essentially as described previously for adherence of STEC to Henle 407 cells (35). Briefly, Henle 407 or HEp-2 cells were grown in Dulbecco's modified Eagle's medium buffered with 20 mM HEPES and supplemented with 10% fetal calf serum, 2 mM l-glutamine, 50 IU of penicillin per ml, and 50 μg of streptomycin per ml. For bacterial adherence assays, cells were seeded into 24-well tissue culture trays and grown for 24 h before use, at which time the monolayer had attained confluence. The monolayers were washed twice with phosphate-buffered saline (pH 7.5) immediately prior to infection with bacteria.
For adherence assays, E. coli cells were grown overnight at 37°C in LB broth, and diluted to a density of 2 × 104 CFU/ml (confirmed by viable count) in the tissue culture medium described above supplemented with 50 μg of ampicillin per ml. Washed Henle 407 or HEp-2 monolayers were then infected with 1-ml aliquots of bacterial suspension. After incubation at 37°C for 3.5 h, the culture medium was removed, and the monolayers were washed four times with phosphate-buffered saline to remove nonadherent bacteria. The cell monolayers were then detached from the plate by treatment with 100 μl of 0.25% trypsin–0.02% EDTA. Cells were then lysed by addition of 400 μl of 0.025% Triton X-100, and 50-μl aliquots (and serial 10-fold dilutions thereof) were plated on LB agar to determine the total number of adherent bacteria. Assays were performed in quadruplicate, and the significance of differences between mean adherence values was analyzed with the unpaired Student's t test (two tailed).
Nucleotide sequence accession number.
The nucleotide sequence of the segment of 98NK2 DNA described in this study has been deposited with GenBank under accession no. AF172324.
RESULTS AND DISCUSSION
Western blot analysis of O113:H21 STEC and screening of cosmid gene bank using convalescent-phase HUS patient serum.
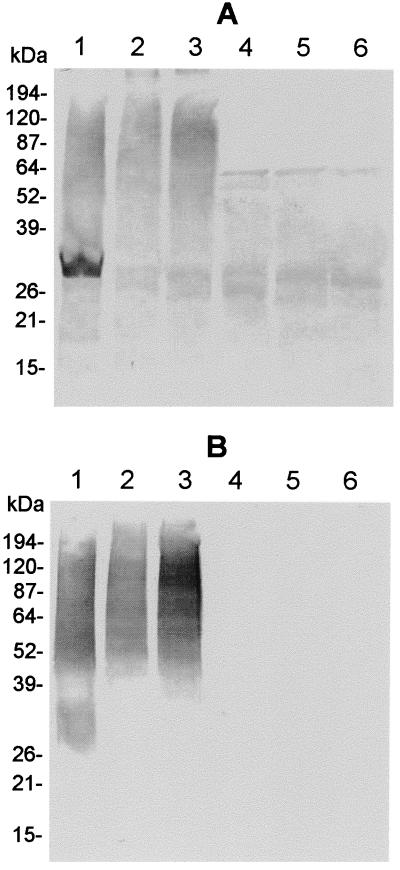
As a first step in characterization of O113 STEC antigens which might be involved in pathogenesis, a Western blot of STEC 98NK2 was probed with convalescent-phase serum from the HUS patient from whom this strain was isolated (used at a dilution of 1:3,000) (Fig. 1A, lane 1). The serum reacted principally with a smear of material migrating between the sizes of 50 and 150 kDa and with a discrete species of approximately 30 kDa. A similar labelling pattern was observed when two other O113:H21 STEC strains in our collection (97MW1 and MW10) were probed with the same serum or when 98NK2 lysates were probed with convalescent-phase sera from two other HUS patients with O113 STEC infection (results not presented). Predigestion of the 98NK2 lysate with proteinase K had no obvious impact on the intensity of labelling of the slower-migrating material, but removed the 30-kDa species completely (result not shown). This suggested that the higher-molecular-weight smear of immunoreactive material was nonproteinaceous, possibly LPS O antigen.
FIG. 1.
Western immunoblot analysis. Lysates of O113:H21 STEC strain 98NK2 (lane 1), or E. coli DH1 carrying pJCP590 (lane 2), pJCP591 (lane 3), pJCP592 (lane 4), pJCP593 (lane 5), or pHC79 (lane 6) were separated by SDS-PAGE, electroblotted, and probed with convalescent-phase HUS patient serum (A) or rabbit anti-E. coli O113 typing serum (B). The mobility of protein size markers is also indicated.
The convalescent-phase serum was then used to screen a 700-clone gene bank of 98NK2 DNA constructed in E. coli K-12 DH1 with cosmid pHC79, as described in Materials and Methods. Lysates from clones exhibiting immunoreactivity above background by dot immunoblotting were then retested by Western blotting. One of these was confirmed as positive and yielded a smear of high-molecular-weight immunoreactive material similar to the putative LPS seen in lysates of 98NK2 (Fig. 1A, lane 2). The recombinant cosmid from this clone (designated pJCP590) was found to contain an insert of approximately 35 kb of 98NK2 DNA. A series of deletion derivatives of pJCP590 were then generated by digestion with either SalI, EcoRI, or HindIII, followed by religation. These derivatives, designated pJCP591, pJCP592, and pJCP593, respectively, were then transformed into E. coli DH1. Unique EcoRI, HindIII, and SalI sites are located at nucleotide positions 0, 31, and 649, respectively, in pHC79, whereas the 98NK2 DNA had been cloned into the BamHI site at position 374. Thus, pJCP591 retains 98NK2 DNA from the opposite end of the original insert from that retained by the other two derivatives. Western blot analysis indicated that lysates of E. coli DH1(pJCP591) contained high-molecular-weight material that reacted with the convalescent-phase HUS patient serum, similar to that seen in lysates of E. coli DH1(pJCP590) (Fig. 1A, lane 3). This was not seen in lysates of E. coli DH1(pJCP592), DH1(pJCP593), or DH1(pHC79), which contained only weakly immunoreactive smaller species common to all strains (Fig. 1A, lanes 4 to 6). To confirm the identity of the high-molecular-weight immunoreactive material, Western blots of the various strains were probed with rabbit anti-O113 typing serum (Fig. 1B). This serum labelled high-molecular-weight material in the lysates of 98NK2, E. coli DH1(pJCP590), and DH1(pJCP591), but not any of the other strains. A culture of E. coli DH1(pJCP591) was also sent to a reference laboratory at the Institute of Medical and Veterinary Science, Adelaide, South Australia, and was confirmed as belonging to serogroup O113 on the basis of tube agglutination tests (result not shown). Thus, the 98NK2 DNA insert in pJCP591, which was approximately 14 kb in size, as judged by restriction analysis, was sufficient to direct biosynthesis of O113 LPS O antigen by E. coli DH1.
DNA sequence analysis.
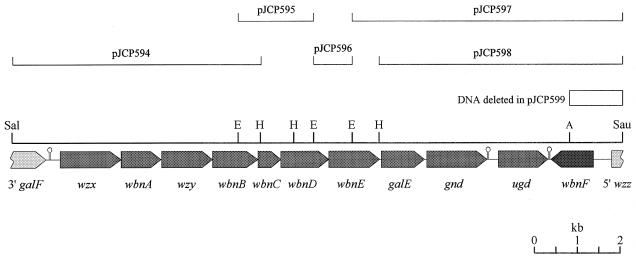
In order to determine the DNA sequence of the O113 O-antigen biosynthesis (rfb) locus, a series of EcoRI and HindIII restriction fragments from pJCP591 were subcloned into pBC SK and transformed into E. coli JM109 (Fig. 2). None of the subclones, designated pJCP594 to pJCP598, were capable of directing biosynthesis of O113 O antigen (result not shown). The various subclones were then subjected to sequence analysis, as described in Materials and Methods. Where subclones did not overlap (that is, between pJCP595 and pJCP596 and between pJCP596 and pJCP597), the sequence across the junction was determined with custom-designed oligonucleotide primers by using pJCP591 DNA as the template.
FIG. 2.
Map of the region of 98NK2 DNA cloned in pJCP591, showing the locations of the various subclones used to determine the DNA sequence. A, AatII; E, EcoRI; H, HindIII; Sal, SalI; Sau, Sau3A. The locations and direction of transcription of the various ORFs are shown as shaded pointed boxes below the map, and genes are named according to Reeves et al. (40). The locations of three putative transcription terminators are also indicated. The open box above the map indicates the region of the insert of pJCP591 deleted in pJCP599.
The positions of open reading frames (ORFs) within the compiled 14,263-bp sequence of the 98NK2 DNA insert in pJCP591 are shown in Fig. 2. Examination of the DNA sequence indicated that it contained 11 complete and 2 partial ORFs, and some features of the predicted protein products are listed in Table 1.
TABLE 1.
Summary of ORFs in pJCP591
| Gene name | Location in sequence | Predicted Mr | No. of aa | % G+C content | Similar protein(s) (reference or accession no.) | % Identical/ % similar (no. of aa) | Putative function of O113 protein |
|---|---|---|---|---|---|---|---|
| 3′galF | 1–801 | 52.2 | GalF, Escherichia coli K-12 (P78083) | 100/100 (262) | Regulation of UDP-Glc pyrophosphorylase | ||
| wzx | 1121–2542 | 54,495 | 473 | 29.8 | WzxC, Escherichia coli K-12 (41) | 21/42 (396) | Repeat unit transporter |
| GumJ, Xanthomonas campestris (U22511) | 23/42 (396) | ||||||
| PssL, Rhizobium leguminosarum (AF040104) | 21/42 (423) | ||||||
| wbnA | 2544–3464 | 34,881 | 306 | 28.6 | WbcG, Yersinia enterocolitica O:8 (51) | 39/55 (210) | Glycosyl transferase |
| Orf39x2, Vibrio cholerae O139 (44) | 35/51 (236) | ||||||
| wzy | 3480–4667 | 46,175 | 395 | 27.9 | O-Polymerase, Salmonella choleraesuis (25) | 21/39 (235) | O-Antigen polymerase |
| wbnB | 4670–5722 | 41,044 | 350 | 28.5 | YveP, Bacillus subtilis (Z71928) | 24/44 (355) | Glycosyl transferase |
| Orf224, Escherichia coli K-12 (42) | 28/44 (193) | ||||||
| wbnC | 5719–6258 | 20,194 | 179 | 28.5 | EpsH, Streptococcus thermophilus (43) | 49/69 (105) | O-Acetylase |
| ThgA, Escherichia coli (10) | 48/62 (102) | ||||||
| Cap1F, Streptococcus pneumoniae type 1 (29) | 38/58 (138) | ||||||
| wbnD | 6255–7388 | 44,519 | 377 | 26.3 | CapH, Staphylococcus aureus type 1 (26) | 27/49 (142) | Glycosyl transferase |
| WbcI, Yersinia enterocolitica O:8 (51) | 26/40 (183) | ||||||
| wbnE | 7388–8566 | 44,859 | 392 | 31.7 | EpsF, Streptococcus thermophilus (43) | 29/50 (271) | Glycosyl transferase |
| EpsG, Streptococcus thermophilus (43) | 24/43 (375) | ||||||
| RfbF, Campylobacter hyoilei (X91081) | 27/44 (323) | ||||||
| PglA, Neisseria meningitidis (18) | 26/41 (290) | ||||||
| galE | 8601–9614 | 37,859 | 337 | 35.4 | GalE, Yersinia enterocolitica (52) | 57/73 (335) | UDG-Glc-4-epimerase |
| gnd | 9674–11080 | 51,494 | 468 | 50.5 | Gnd, Escherichia coli (M64330, M63829) | 99/99 (468) | 6-Phosphogluconate dehydrogenase |
| ugd | 11329–12495 | 43,402 | 388 | 41.8 | Ugd, Escherichia coli O8:K40 (2) | 91/95 (388) | UDP-Glc-6-dehydrogenase |
| wbnF | 13565–12561a | 37,653 | 334 | 42.3 | Orf2, Escherichia coli O111 (3) | 85/90 (334) | Nucleotide sugar epimerase |
| NAD-dependent epimerase, Vibrio vulnificus (53) | 67/80 (332) | ||||||
| 5′wzz | 13991–14263 | 43.6 | Wzz, Escherichia coli O8:K40 (2) | 97/99 (91) | Chain length determinant | ||
| Wzz, Escherichia coli O157 (11) | 100/100 (86) |
Encoded on complementary strand.
BLASTX analysis indicated that the partial ORF at the 5′ end of the pJCP591 insert encoded a protein with 100% identity to the last 262 amino acids (aa) of GalF of E. coli K-12. GalF is not involved in O-antigen biosynthesis in E. coli, but is believed to regulate cellular levels of uridine diphosphoglucose (UDP-Glc) by interacting with the UDP-Glc pyrophosphorylase GalU (28). However, galF is known to be located just upstream of the rfb promoter region in E. coli O7 and K-12, as well as in a number of enteric microorganisms, including Salmonella typhimurium LT2, Salmonella enterica, Shigella dysenteriae, and Shigella flexneri (28). The galF gene was followed by a stem-loop structure (ΔG = −29.5 kcal/mol). Downstream of this, a copy of the JUMPstart sequence is located. This is a 39-bp element present just upstream of a number of polysaccharide gene clusters (15).
The JUMPstart sequence is followed by a cluster of nine genes exhibiting features common to other rfb loci, and these have been named in accordance with the system proposed by Reeves et al. (40). Each of the genes is preceded by a putative ribosome binding site, and most are tightly linked. Indeed, the genes named wbnB, wbnC, wbnD, and wbnE (Fig. 2) overlap each other by from 1 to 4 nucleotides (nt), and the only significant intergenic gaps are located between wbnE and galE (34 nt) and between galE and gnd (59 nt). These intergenic regions did not contain any potential stem-loop structures. However, a potential stem-loop transcription terminator (ΔG = −24.6 kcal/mol) was located immediately downstream of gnd. E. coli rfb loci are typically flanked by the JUMPstart element and gnd, and Wang and Reeves (48) have recently exploited this to amplify the rfb region of E. coli O157 by long-range PCR. Interestingly, with the exception of gnd, all of the genes in the putative O113 rfb cluster have very low G+C contents. Indeed, for six of the eight genes, the G+C content is <30%. This is markedly lower than that of the flanking E. coli O113 DNA sequences and that of the E. coli genome as a whole, suggesting that these rfb genes have been acquired from another bacterial species.
Predicted functions of proteins encoded by the major O113 rfb operon.
The structure of the E. coli O113 O-antigen chemical repeat unit has been shown to consist of a tetrasaccharide backbone with an additional galactose (Gal) side chain (31) and is shown in Fig. 3. The repeat unit also contains O-acetyl groups, but the position of these has not been determined. On the basis of this structure and the known mechanism of biosynthesis of other O-antigen polysaccharides (49), we predicted that the O113 rfb locus should contain genes encoding a total of five glycosyl transferases required for assembly of the oligosaccharide repeat unit, including one which initiates synthesis by linking the first sugar to a lipid carrier undecaprenyl phosphate (UndP) on the cytoplasmic face of the membrane. We would also expect to find at least one acetyl transferase, a repeat unit transporter (flippase) for translocation of the completed repeat unit across the cell membrane, and a polymerase to link repeat units together to form high-molecular-weight O antigen. We therefore conducted BLASTX analyses to identify candidates for these functions, the results of which are summarized in Table 1.
FIG. 3.
Structure of the oligosaccharide repeat unit of E. coli O113 O antigen, as determined by Parolis and Parolis (31). The position of O-acetyl groups was not determined. Sugars are abbreviated as follows: Gal, galactose; GalA, galacturonic acid; GlcNAc, N-acetyl glucosamine; and GalNAc, N-acetyl galactosamine.
The first gene in the major O113 rfb operon has been designated wzx, because it encodes a predicted protein with similarities to several putative polysaccharide repeat unit transporters (flippases), including WzxC from the colanic acid biosynthesis locus of E. coli K-12 (41). Wzx from E. coli O113 is predicted to be a highly hydrophobic integral membrane protein; hydrophobicity analysis conducted according to the method of Kyte and Doolittle (23) yielded a hydrophobicity index of 0.75 and indicated the presence of multiple potential membrane-spanning domains (result not presented).
The third ORF in the major O113 rfb operon encodes another highly hydrophobic protein (hydrophobicity index = 0.89) with 12 potential membrane-spanning domains (result not presented). This is a typical feature of polysaccharide polymerases, and the protein shows a modest, but significant degree of similarity to the O-antigen polymerase of Salmonella choleraesuis (25) (Table 1). Thus, this gene (designated wzy) is presumed to encode the O113 O-antigen polymerase. Polysaccharide polymerases from different bacteria typically exhibit only minimal sequence similarity to each other, because they must be specific for both the oligosaccharide repeat unit itself and the type of glycosidic linkage joining the repeat units.
The predicted products of the genes designated wbnA, wbnB, wbnD, and wbnE (see Fig. 2 for location) all exhibit significant sequence similarity to glycosyl transferases from a variety of gram-negative and gram-positive microorganisms (Table 1). However, in all of these cases, the specificity of the transferase is either unknown, or the degree of similarity is insufficient to assign substrate specificity to the putative O113 transferase with any confidence. None of the other O113 ORFs cloned in pJCP591 encode proteins with similarity to known glycosyl transferases, and so there appears to be one transferase gene too few to synthesize the O113 repeat unit. The gene most likely to be missing is that which encodes the transferase linking the first sugar to UndP. The enzyme responsible for this reaction would need to be capable of interaction with membrane lipid and would therefore be expected to contain hydrophobic membrane-spanning anchorage domains. However, none of the protein products of wbnA, wbnB, wbnD, or wbnE contain such regions. A similar situation was observed by Wang and Reeves (48) in the rfb locus of E. coli O157, which contained three rather than the predicted four putative glycosyl transferase genes. They explained this apparent deficiency by proposing that WecA (formerly Rfe), which initiates synthesis of enterobacterial common antigen by transferring N-acetyl glucosamine (GlcNAc) phosphate to UndP, could also initiate O-unit synthesis by transferring N-acetyl galactosamine (GalNAc) phosphate, as previously shown for Yersinia enterocolitica serotype O:8 (51). Similarly, WecA could also initiate O113 repeat unit synthesis by transferring GlcNAc phosphate (or possibly GalNAc phosphate) to UndP.
The predicted product of wbnC is a small protein (20.2 kDa) with significant similarity to members of the CysE-LacA-LpxA-NodL family of acetyl transferases from a wide range of bacteria (Table 1). Thus, WbnC is likely to be responsible for O acetylation of the O113 O-antigen polysaccharide, although the precise sugar(s) modified is yet to be determined. O acetylation is also likely to be responsible for the fact that the O113 O antigen migrates as a smear on SDS-PAGE (Fig. 1). Many O antigens migrate as a ladder-like pattern of discrete bands, each corresponding to a polysaccharide containing a different number of oligosaccharide repeat units. O acetylation, however, is often not stoichiometric, and so for O antigens with such modifications, polysaccharides containing a given number (n) of repeat units will vary in the extent to which each is O acetylated. Thus, the molecular weight will vary to the point where the mobility on SDS-PAGE overlaps that of polysaccharides containing either n − 1 or n + 1 repeat units.
The eighth ORF in the major O113 rfb operon encodes a protein with a very high degree of similarity to GalE proteins (UDP-Glc-4-epimerases) from a large number of gram-negative and gram-positive bacteria, the most closely related being that from Y. enterocolitica (57% identity and 73% similarity over 335 aa) (52). These enzymes convert UDP-Glc to UDP-Gal, and since the latter is required by E. coli for a number of purposes, a housekeeping copy of galE is located elsewhere in the chromosome (41). Thus, one might not have expected to find an additional copy of galE in the O-antigen biosynthesis locus of E. coli O113. However, the housekeeping galE gene in E. coli is subject to repression by Glc, and so under normal growth conditions, cellular levels of UDP-Gal may be low. Such levels may become limiting in E. coli O113, because its O antigen is rich in Gal. Thus, the presence of an additional galE gene, which may or may not be subject to catabolite repression, may be necessary to ensure adequate supplies of the activated precursor.
The final gene in the major O113 rfb operon is gnd, which as stated previously, is located at the 3′ end of numerous rfb loci (48). The product of this particular copy of gnd is 99% identical to several other E. coli Gnd proteins for which sequences have been deposited with GenBank (Table 1). Gnd is a 6-phosphogluconate dehydrogenase, but there is no evidence that this enzyme functions in O-antigen biosynthesis.
Predicted functions of proteins encoded by other genes in pJCP591.
Downstream of the major O113 rfb operon is a copy of ugd, which encodes UDP-glucose dehydrogenase. This gene is unlikely to be cotranscribed with the O113 rfb locus, because, as mentioned previously, there is a strong transcription terminator between gnd and ugd. The O113 Ugd protein exhibits 91% identity and 95% similarity to Ugd encoded by the his region of E. coli O8:K40 (2). UDP-Glc dehydrogenase converts UDP-Glc to UDP-glucuronic acid (UDP-GlcA), a precursor of UDP-galacturonic acid (UDP-GalA), which is one of the activated sugars required for O113 O-antigen biosynthesis. Thus, it is possible that this gene does actually function in O113 O-antigen biosynthesis. However, it is not possible to test this hypothesis by deletion mutagenesis of ugd in pJCP591, because E. coli K-12 contains another copy of ugd, the product of which exhibits 82% identity and 91% similarity (accession no. P76373).
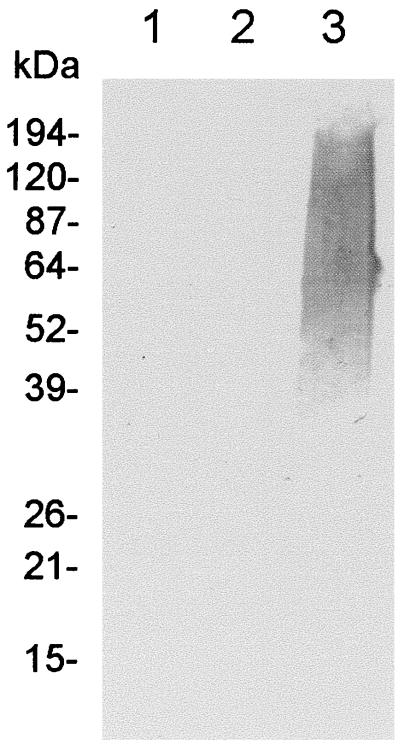
Conversion of UDP-GlcA to UDP-GalA would require the appropriate epimerase, and it is possible that the enzyme encoded by the galE homologue in the O113 rfb locus could fulfill this function. Interestingly, however, downstream of ugd, but on the opposite DNA strand, is a gene which we have designated wbnF. A putative transcription terminator sequence (ΔG = −29.2 kcal/mol) is also located between ugd and wbnF (Fig. 2). The predicted product of wbnF has significant homology to a variety of nucleotide sugar epimerases (Table 1). The most closely related protein is that encoded by a gene designated orf2, which is located in an analogous position downstream of the rfb region of E. coli O111 (3). This gene is also transcribed in the opposite direction from the rfb locus, but it is not known whether it is essential for synthesis of O111 O antigen. Thus, WbnF might be involved in synthesis of UDP-GalA; a possible alternative function could be conversion of UDP-GlcNAc to UDP-GalNAc, because the latter is also a precursor required for assembly of the O113 O-antigen repeat unit. BLAST analysis indicated that the E. coli K-12 genome does not contain a homologue of wbnF, and so its role in O113 O-antigen biosynthesis was examined by deleting the distal portion of the insert of pJCP591 by restriction with AatII, followed by religation. This removes the DNA between the AatII cleavage site at nucleotide position 13,004 in the pJCP591 insert (within wbnF) and that in the vector approximately 450 bp downstream from the original cloning site (Fig. 2). E. coli DH1 transformed with the religated deletion derivative (designated pJCP599) did not synthesize O113 O antigen, as judged by Western blot analysis with the O113-specific rabbit antiserum (Fig. 4) and was not agglutinated by the same serum. Thus, notwithstanding the uncertainty as to its precise function, wbnF is essential for O113 O-antigen biosynthesis in E. coli K-12; in spite of the fact that it is transcribed separately from the major O113 rfb operon, it should be considered part of the O113 rfb locus.
FIG. 4.
Western immunoblot analysis. Lysates of E. coli DH1 carrying pHC79 (lane 1), pJCP599 (lane 2), or pJCP591 (lane 3) were separated by SDS-PAGE, electroblotted, and probed with rabbit anti-E. coli O113 serum. The mobility of protein size markers is also indicated.
The final partial ORF in the insert of pJCP591 is the 5′ end of wzz. The predicted product has a very high degree of identity to the first 91 aa of Wzz proteins (polysaccharide chain length determinants) from various E. coli strains, S. dysenteriae, and S. flexneri. The sequence of the remainder of the O113 wzz gene was determined by direct analysis of PCR products obtained with primers based on that published for wzz from E. coli O8:K40 (2). The complete O113 Wzz was 94% identical to the homologue from E. coli O8:K40 and 99% identical to that from an E. coli O157 strain (11) (result not presented).
Clearly, a complete copy of the O113 wzz gene is not essential for expression of O113 O antigen in E. coli K-12 DH1(pJCP591), and presumably the chain length regulation function is carried out by the E. coli K-12 Wzz protein. However, the Western blot shown in Fig. 1 (lanes 2 and 3) suggests that the mean chain length of O113 O antigen expressed by E. coli K-12 carrying either pJCP590 or pJCP591 may be slightly higher than that expressed by the wild-type O113 strain 98NK2 (lane 1). Franco et al. (11) have recently reported that amino acid sequence variation in the Wzz proteins from E. coli strains belonging to different serogroups is directly responsible for variations in modal chain length of O antigen. A final feature worthy of mention is that examination of the noncoding O113 DNA between wbnF and wzz indicates that the region from nt 13737 to the wzz initiation codon (nt 13991) has 88% nucleotide sequence identity with E. coli O8:K40, including the last 106 bp of the O8:K40 ugd gene. Thus, it appears that the 3′ end of ugd has been duplicated in E. coli O113, perhaps as a consequence of a recombination event which resulted in insertion of wbnF; a homologue of this gene is not present in E. coli O8:K40.
Effect of expression of O113 O antigen on adherence to epithelial cells.
The O113 O antigen contains GalA, which is negatively charged at physiological pH. Thus, E. coli expressing this O-antigen type could have a significant net surface charge that could influence its capacity to interact with other structures, such as the surface of host cells during establishment of gut infection. To examine this possibility, the capacity of E. coli DH1(pJCP591) and DH1(pHC79) to adhere to either HEp-2 or Henle 407 cell monolayers was assessed, as described in Materials and Methods. At an initial dose of 2 × 104 CFU, the mean adherence ± standard error of E. coli DH1(pHC79) to Henle 407 cells after 3.5 h of incubation was (3.05 ± 0.42) × 104 CFU per well. This represented approximately 3.1% of the total number of bacteria present in the culture medium at the end of the assay. In contrast, the adherence of E. coli DH1(pJCP591) was (1.26 ± 0.13) × 105 CFU per well. Thus, expression of O113 O antigen increased adherence of E. coli DH1 to Henle 407 cells approximately fourfold (P < 0.001). A similar result was obtained with HEp-2 cells. Total adherence of E. coli DH1(pHC79) to HEp-2 cell monolayers after 3.5 h of incubation was (2.32 ± 0.38) × 104 CFU per well, compared with (4.80 ± 0.94) × 104 CFU per well for E. coli DH1(pJCP591) (P < 0.05). This enhancement of adherence due to expression of O113 O antigen contrasts with our previously reported finding that expression of O111 O antigen by E. coli K-12 had no effect on adherence to Henle 407 cells under the same experimental conditions as those used in the present study (36). O111 O antigen is a neutral polysaccharide, and so it is conceivable that net negative charge facilitates interaction between E. coli and the epithelial cell surface. However, analysis of the effects of additional O-antigen types is required before a definitive conclusion can be drawn. Interestingly, two previous studies have examined the role of O antigen in adherence of O157:H7 STEC by using TnphoA mutagenesis to construct strains deficient in O-antigen biosynthesis. In both studies, such mutants were found to be hyperadherent to HEp-2 cells in vitro (4, 5).
Conclusions.
This study demonstrates that O antigen is one of the principal targets of the immune responses of HUS patients infected with STEC belonging to serotype O113. Moreover, among a cosmid library of O113 STEC DNA constructed in E. coli K-12, a clone expressing O antigen was the only one to react strongly with convalescent-phase HUS patient serum. A deletion derivative of this cosmid (pJCP591) with a 14,263-bp O113 DNA insert was sufficient to direct biosynthesis of O113 O antigen in E. coli K-12. Sequence analysis indicated that this region contains a cluster of nine closely linked genes, including gnd at the 3′ end and with the JUMPstart sequence (15) at the 5′ end, as shown for rfb regions from other E. coli serogroups (48). Examination of the Bacterial Polysaccharide Gene Database (2a) indicates that to date only 7 of the 165 other E. coli O-antigen biosynthesis loci have been sequenced (either partially or in full). The O113 rfb region includes candidate genes for four glycosyl transferases, an O-acetyl transferase, an O-unit flippase, and an O-antigen polymerase, as well as a copy of galE. Assuming that WecA is capable of initiating O-antigen synthesis, as discussed above, these gene products are theoretically sufficient to assemble, export, and polymerize the O113 O-antigen repeat unit. Interestingly, two additional, separately transcribed genes downstream of the O113 rfb region were predicted to encode enzymes involved in synthesis of activated sugar precursors. One of these (wbnF) is on the complementary DNA strand, and deletion analysis indicated that it is essential for O113 O-antigen synthesis. WbnF has significant homology with nucleotide sugar epimerases from a variety of bacteria and may therefore be involved in synthesis of either UDP-GalA or UDP-GalNAc, both of which are present in the O113 repeat unit. Interestingly, however, the closest homologue of WbnF (85% identity) is encoded by a gene (orf2) located in a similar position and orientation downstream of the rfb region of E. coli O111. This serotype does not contain either of the sugars mentioned above in its O-antigen repeat unit. However, it is not known whether orf2 is essential for synthesis of O111 O antigen. Thus, it remains a possibility that orf2 is a remnant from an ancestral rfb locus, which has since undergone a serogroup-converting recombination event and that its retention in E. coli O111 has no functional significance. Such recombination events are likely to have played an important role in the evolution of the 166 distinct E. coli O serogroups.
The sequence data generated in this study also provide an opportunity to develop PCR assays for serogroup-specific portions of the O113 rfb locus. We have previously described a multiplex PCR assay for such regions from E. coli O111 and O157 (33), which are the most common STEC types responsible for HUS in Australia (38). We routinely use this assay, in combination with another specific for the genes encoding Shiga toxins 1 and 2, intimin, and a plasmid-encoded hemolysin, for direct detection and characterization of STEC in crude fecal culture extracts (33). STEC strains belonging to serogroup O113 were among the first STEC types to be associated with HUS in the landmark studies of Karmali et al. (20), and over the last 5 years have been the third most prevalent STEC strains associated with cases of HUS in South Australia. Thus, inclusion of a pair of primers specific for O113 in the rfb multiplex assay is likely to be a useful adjunct in the diagnosis of STEC disease and in epidemiological studies. A portion of the O113 wzy gene would be the preferred target for such an assay, because it showed the lowest degree of deduced amino acid sequence homology with any genes deposited with GenBank. Moreover, the fact that wzy encodes the putative O-antigen polymerase, which must exhibit absolute specificity for both the oligosaccharide repeat unit and the type of glycosidic linkage formed during polymerization, renders existence of homologous sequences in other organisms extremely unlikely.
The finding that expression of O113 O antigen significantly enhanced adherence of E. coli K-12 to epithelial cells of human origin was unexpected, given the results of studies with O157 and O111 strains discussed previously (4, 5, 36). However, the fact that in patients with HUS caused by O113 STEC, the major host immune response is directed against O antigen suggests an important role in the host-pathogen interaction. Unlike O111 and O157 STEC, O113 STEC strains are LEE negative and so lack the capacity to form A/E lesions on enterocytes (8). Thus, the molecular mechanisms whereby these bacteria interact with and attach to intestinal mucosa may be fundamentally different. The isolation of the O113 rfb region in the present study will facilitate future studies of the direct or indirect contribution of O antigen to intestinal colonization by STEC.
ACKNOWLEDGMENTS
We are grateful to Renato Morona and Peter Reeves for numerous helpful discussions and to Paul Henning and Fred Jureidini for providing convalescent-phase HUS patient sera.
This work was supported by grants from the National Health and Medical Research Council of Australia and the Women's and Children's Hospital Foundation.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor P A, Whitfield C. Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his-region containing gene clusters for multiple cell-surface polysaccharides. Mol Microbiol. 1997;26:145–161. doi: 10.1046/j.1365-2958.1997.5631930.x. [DOI] [PubMed] [Google Scholar]
- 2a.Bacterial Polysaccharide Gene Database 1998. [Online.] Microbiology Department, University of Sydney, Sydney, Australia. http://www.microbio.usyd.edu.au/BPGD. [17 September 1999, last date accessed.]
- 3.Bastin D A, Stevenson G, Brown P K, Haase A, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 4.Bilge S S, Vary J C, Jr, Dowell S F, Tarr P I. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cockerill F, III, Beebakhee G, Soni R, Sherman P. Polysaccharide side chains are not required for attaching and effacing adhesion of Escherichia coli O157:H7. Infect Immun. 1996;64:3196–3200. doi: 10.1128/iai.64.8.3196-3200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deibel C, Krämer S, Chakraborty T, Ebel F. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol Microbiol. 1998;28:463–474. doi: 10.1046/j.1365-2958.1998.00798.x. [DOI] [PubMed] [Google Scholar]
- 7.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 8.Dytoc M T, Ismaili A, Philpott D J, Soni R, Brunton J L, Sherman P M. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect Immun. 1994;62:3494–3505. doi: 10.1128/iai.62.8.3494-3505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y K, Lai L-C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 10.Fowler A V, Hediger M A, Musso R E, Zabin I. The amino acid sequence of thiogalactoside transacetylase of Escherichia coli. Biochimie. 1985;67:101–108. doi: 10.1016/s0300-9084(85)80235-2. [DOI] [PubMed] [Google Scholar]
- 11.Franco V A, Liu D, Reeves P R. The Wzz (Cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. J Bacteriol. 1998;180:2670–2675. doi: 10.1128/jb.180.10.2670-2675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R I, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–761. [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 16.Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 17.Hull S. Escherichia coli lipopolysaccharide in pathogenesis and virulence. In: Sussman M, editor. Escherichia coli: mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 145–167. [Google Scholar]
- 18.Jennings M P, Virji M, Evans D, Foster V, Srikhanta Y N, Steeghs L, van der Ley P, Moxon E R. Identification of a novel gene involved in pilin glycosylation in Neisseria meningitidis. Mol Microbiol. 1998;29:975–984. doi: 10.1046/j.1365-2958.1998.00962.x. [DOI] [PubMed] [Google Scholar]
- 19.Karmali M A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic uremic syndrome and infection by Verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 21.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 22.Konadu E Y, Parke J C, Jr, Tran H T, Bryla D A, Robbins J B, Szu S C. Investigational vaccine for Escherichia coli O157: phase I study of O157 O-specific polysaccharide-Pseudomonas aeruginosa recombinant exoprotein A conjugates in adults. J Infect Dis. 1998;177:383–387. doi: 10.1086/514203. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee S J, Romana L K, Reeves P R. Sequence and structural analysis of the rfb (O antigen) gene cluster from a group C1 Salmonella enterica strain. J Gen Microbiol. 1992;138:1843–1855. doi: 10.1099/00221287-138-9-1843. [DOI] [PubMed] [Google Scholar]
- 26.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.Marolda C L, Valvano M A. The GalF protein of Escherichia coli is not a UDP-glucose pyrophosphorylase but interacts with the GalU protein possibly to regulate cellular levels of UDP-glucose. Mol Microbiol. 1996;22:827–840. doi: 10.1046/j.1365-2958.1996.01531.x. [DOI] [PubMed] [Google Scholar]
- 29.Muñoz R, Mollerach M, López R, García E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 30.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parolis H, Parolis L A S. The structure of the O-specific polysaccharide from Escherichia coli O113 lipopolysaccharide. Carbohydr Res. 1995;267:263–269. doi: 10.1016/0008-6215(94)00303-w. [DOI] [PubMed] [Google Scholar]
- 32.Paton A W, Manning P A, Woodrow M C, Paton J C. Translocated intimin receptors (Tir) of Shiga-toxigenic Escherichia coli isolates belonging to serogroups O26, O111, and O157 react with sera from patients with hemolytic-uremic syndrome and exhibit marked sequence heterogeneity. Infect Immun. 1998;66:5580–5586. doi: 10.1128/iai.66.11.5580-5586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paton A W, Paton J C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paton A W, Paton J C, Heuzenroeder M W, Goldwater P N, Manning P A. Cloning and nucleotide sequence of a variant Shiga-like toxin II gene from Escherichia coli OX3:H21 isolated from a case of Sudden Infant Death Syndrome. Microb Pathog. 1992;13:225–236. doi: 10.1016/0882-4010(92)90023-h. [DOI] [PubMed] [Google Scholar]
- 35.Paton A W, Voss E, Manning P A, Paton J C. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun. 1997;65:3799–3805. doi: 10.1128/iai.65.9.3799-3805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paton A W, Voss E, Manning P A, Paton J C. Antibodies to lipopolysaccharide block adherence of Shiga toxin-producing Escherichia coli to human intestinal epithelial (Henle 407) cells. Microb Pathog. 1998;24:57–63. doi: 10.1006/mpat.1997.0172. [DOI] [PubMed] [Google Scholar]
- 37.Paton A W, Woodrow M C, Doyle R M, Lanser J A, Paton J C. Molecular characterization of a Shiga-toxigenic Escherichia coli O113:H21 strain Lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3357–3361. doi: 10.1128/jcm.37.10.3357-3361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paton J C, Paton A W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves P R. Role of O-antigen variation in the immune response. Trends Microbiol. 1995;3:381–386. doi: 10.1016/s0966-842x(00)88983-0. [DOI] [PubMed] [Google Scholar]
- 40.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stroeher U H, Parasivam G, Dredge B K, Manning P A. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J Bacteriol. 1997;179:2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valvano M A. Pathogenicity and molecular genetics of O-specific side-chain lipopolysaccharides of Escherichia coli. Can J Microbiol. 1992;38:711–719. doi: 10.1139/m92-117. [DOI] [PubMed] [Google Scholar]
- 47.Voss E, Paton A W, Manning P A, Paton J C. Molecular analysis of Shiga toxigenic Escherichia coli O111:H− proteins which react with sera from patients with hemolytic-uremic syndrome. Infect Immun. 1998;66:1467–1472. doi: 10.1128/iai.66.4.1467-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 50.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Radziejewska-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Toivanen P, Skurnik M. The gene cluster directing O-antigen biosynthesis in Yersinia enterocolitica serotype O:8: identification of the genes for mannose and galactose biosynthesis and the gene for the O-antigen polymerase. Microbiology. 1996;142:277–288. doi: 10.1099/13500872-142-2-277. [DOI] [PubMed] [Google Scholar]
- 53.Zuppardo A B, Siebeling R J. An epimerase gene essential for capsule synthesis in Vibrio vulnificus. Infect Immun. 1998;66:2601–2606. doi: 10.1128/iai.66.6.2601-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]