Abstract
Germline and tumor BRCA testing constitutes a valuable tool for clinical decision-making in the management of epithelial ovarian cancer (EOC) patients. Tissue testing is able to identify both germline (g) and somatic (s) BRCA variants, but tissue preservation methods and the widespread implementation of NGS represent pre-analytical and analytical challenges that need to be managed. This study was carried out on a multicenter prospective GEICO cohort of EOC patients with known gBRCA status in order to determine the inter-laboratory reproducibility of tissue sBRCA testing. The study consisted of two independent experimental approaches, a bilateral comparison between two reference laboratories (RLs) testing 82 formalin-paraffin-embedded (FFPE) EOC samples each, and a Ring Test Trial (RTT) with five participating clinical laboratories (CLs) evaluating the performance of tissue BRCA testing in a total of nine samples. Importantly, labs employed their own locally adopted next-generation sequencing (NGS) analytical approach. BRCA mutation frequency in the RL sub-study cohort was 23.17%: 12 (63.1%) germline and 6 (31.6%) somatic. Concordance between the two RLs with respect to BRCA status was 84.2% (gBRCA 100%). The RTT study distributed a total of nine samples (three commercial synthetic human FFPE references, three FFPE, and three OC DNA) among five CLs. The median concordance detection rate among them was 64.7% (range: 35.3–70.6%). Analytical discrepancies were mainly due to the minimum variant allele frequency thresholds, bioinformatic pipeline filters, and downstream variant interpretation, some of them with consequences of clinical relevance. Our study demonstrates a wide range of concordance in the identification and interpretation of BRCA sequencing data, highlighting the relevance of establishing standard criteria for detecting, interpreting, and reporting BRCA variants.
Keywords: ovarian cancer, BRCA mutations, NGS, BRCA testing, Ring Test Trial
1. Introduction
According to Globocan’s 2020 projections, by 2040, the number of women around the world diagnosed with ovarian cancer will rise by almost 37% up to 428,966. The number of women dying from ovarian cancer each year is projected to increase up to 313,617, which is a 50% increase from 2020 to 2022 [1]. Fortunately, the treatment of ovarian cancer has improved significantly in recent years. Today, the use of poly (ADP-Ribose) polymerase inhibitors (PARPis) has been shown to be particularly efficient in epithelial ovarian cancer (EOC) patients harboring germline or somatic BRCA mutations (gBRCA and sBRCA, respectively) [2,3], in addition to tumors with high genomic instability caused by defects in key genes within the homologous recombination (HR) DNA repair machinery [4].
In addition to other genes, both BRCA1 and BRCA2 genes play a key role within the HR DNA repair system [5]. Indeed, germline and somatic deleterious mutations in HR genes are identified in approximately 30% of EOC patients, but up to 75% of them are BRCA1 and BRCA2 gene mutations [5,6]. Furthermore, sBRCA mutations have been described in up to 7% of EOC patients in the first-line or platinum-sensitive relapsed clinical setting [5,7,8].
Consequently, BRCA testing in EOC plays a key role in the clinical decision-making process, not only for the identification of familial cancer predisposition but also to personalize therapeutic treatment [9]. The germline testing of BRCA is widespread in medical genetics laboratories, but not the identification of patients with sBRCA tumors that can benefit from PARPi therapy. Tumor BRCA testing on formalin-fixed paraffin-embedded (FFPE) tissue is key to identifying patients with sBRCA tumors, which in addition has the advantage of the simultaneous assessment of both somatic and germline mutations using an easily accessible material that is routinely available in any pathology laboratory worldwide. Currently, there is no consensus, but it is recommended to perform both germline and tumor BRCA characterization in EOC patients [10,11]. Indeed, sBRCA testing is now required to support treatment decisions in many countries, so it is essential that testing is robust [12].
Therefore, high-throughput next-generation sequencing (NGS)-based FFPE-derived DNA sequencing approaches are being implemented, given that they permit fast multiplex testing on small quantities of DNA, improving both the capacity and the cost-effectiveness of mutational analysis compared with traditional methods such as Sanger sequencing [13].
However, not all NGS-based sequencing technology is equal. Both the sequencing error and capacity of the sequencer are important to be considered, as well as data analysis software and the experience that the testing laboratory or center has. For this reason, the Spanish Group of Research on Ovarian Cancer (GEICO) conducted a study to evaluate a range of tumor BRCA testing approaches. First, we evaluated the inter-laboratory reproducibility of two Spanish reference laboratories identifying both gBRCA and sBRCA mutations in 100 FFPE EOC samples. Additionally, second, we determined the efficacy of a spectrum of tumor BRCA testing workflows within five Spanish clinical laboratories (CL) to accurately identify sBRCA variants in clinical practice in the context of a Ring Test Trial (RTT).
2. Materials and Methods
2.1. Study Design
The study consisted of a prospective, observational, multicenter study developed at the national level with 30 reference sites in Spain, during a data collection period of 60 months (including follow-up).
The study included 400 patients that met the following inclusion criteria:
Written informed consent signed by all patients participating in the study.
Histological diagnosis of non-mucinous epithelial ovarian, primary peritoneal, or tubal carcinoma confirmed not more than 2 years prior to the date of ICF signature.
Adult women (18 years old or older at the time of diagnosis).
Patients may have had other malignancies and have received or are receiving any anticancer therapy, including investigational drugs.
Availability of FFPE tumor blocks from the primary tumor for genetic analysis and willingness (100 valid cases).
Patients with gBRCA testing performed at the site according to current clinical guidelines or willing to be tested centrally if local testing is not available.
Exclusion criteria:
Patients without an available medical record (lost, empty, or unretrievable clinical information).
This study has been conducted in accordance with the ethical principles that have their origin in the Declaration of Helsinki (18th World Medical Association General Assembly, Helsinki, Finland) and according to the Spanish Order SAS/3470/2009, and Law 14/2007, of July 3rd, on Biomedical Research. Ethical committee approval (expedient number 025-19), informed consent, cancer family history, and clinical features were collected.
Tumor BRCA testing was performed in 100 EOC samples in two reference laboratories (RL1; RL2) to evaluate inter-laboratory reproducibility. The median age at diagnosis of the patients included in our series was 56 years (range 25–84 years). The main characteristics of the patients are included in Table 1. Approximately 16% were gBRCA mutation carriers.
Table 1.
Patient demographic and clinical characteristics.
| Clinicopathological Parameters | RL1 | RL2 |
|---|---|---|
| n = 100 | n = 82 | |
| Mean age at diagnosis (range) | 56 (25–84) years | 56 (25–84) years |
| Histology | ||
| High-grade serous | 83 (83%) | 69 (84.1%) |
| Low-grade serous | 4 (4%) | 2 (2.4%) |
| Endometrioid G1 | 3 (3%) | 3 (3.7%) |
| Endometrioid G2 | 4 (4%) | 3 (3.7%) |
| Endometrioid G3 | 0 | 0 |
| Clear cells | 4 (4%) | 3 (3.7%) |
| Seromucinous | 1 (1%) | 1 (1.2%) |
| Carcinosarcoma | 1 (1%) | 1 (1.2%) |
| FIGO Stage | ||
| I | 17 (17%) | 15 (18.3%) |
| II | 9 (9%) | 6 (7.3%) |
| III | 39 (39%) | 32 (39%) |
| IV | 21 (21%) | 17 (20.7%) |
| NA | 14 (14%) | 12 (14.6%) |
| gBRCA | ||
| wt-BRCA | 70 (70%) | 58 (70.7%) |
| VUS-BRCA | 13 (13%) | 10 (12.2%) |
| mt-BRCA | 16 (16%) | 13 (15.9%) |
| NA | 1 (1%) | 1 (1.2%) |
NA: not available; gBRCA: germline BRCA status; wt: wild type; VUS: variant of unknown significance; mt: mutated; RL: reference laboratory.
2.2. Tumor BRCA1/2 Testing Cross-Laboratory Validation
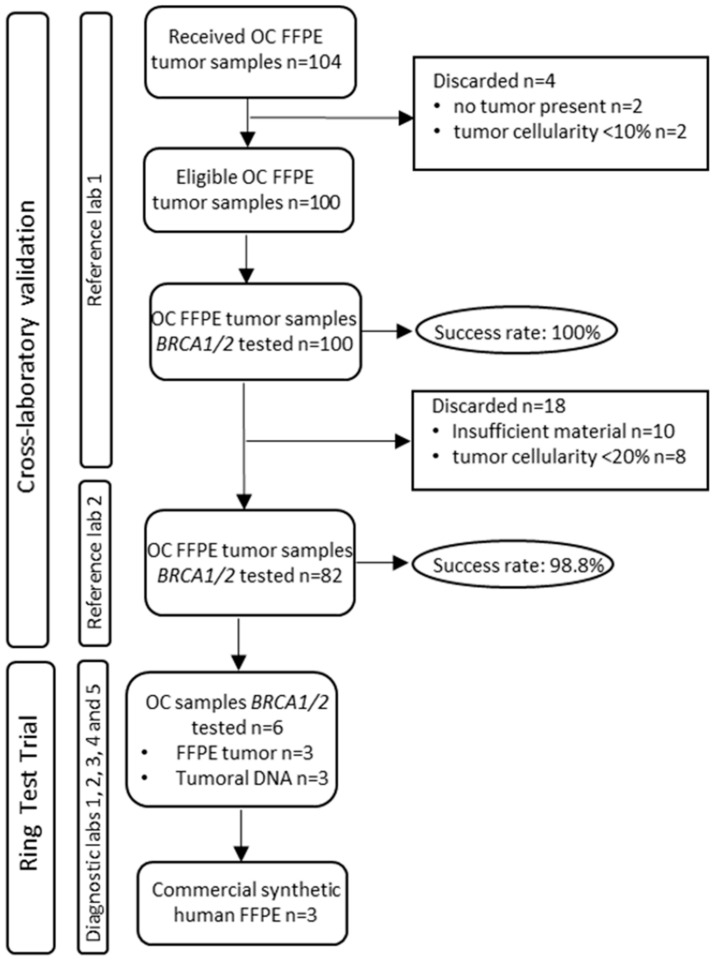
The first 100 valid FFPE tumor blocks from primary tumors were selected to be tested for tumor BRCA mutations at RL1. Of these, and after an initial quality control of the samples, 82 valid cases were sent to RL2 for further analysis of tumor mutations of BRCA. The workflow of the sample selection strategy is described in Figure 1.
Figure 1.
Sample selection workflow.
2.2.1. RL1 DNA Extraction
Hematoxylin and eosin (H&E) staining was performed to evaluate tumor cell percentage to guide the macrodissection of the solid tumor sample if ≤30%. A minimum of 10% tumor cellularity was required to be considered. For each sample, DNA was extracted from three unstained sections of 10µm thickness or three 0.6 mm needle biopsies using the QIAamp® DNA Investigator kit (QIAGEN, Hilden, Germany). DNA concentration was quantified using the Quant-iT™ PicoGreen™ dsDNA (ThermoFisher; Waltham, MA, USA) fluorimetric assay.
2.2.2. RL1 NGS
Sequencing was carried out with the Homologous Recombination Solution (HRS; Sophia Genetics) capture kit in the Illumina® MiSeq® sequencer (Illumina; San Diego, CA, USA). The study includes the analysis of the entire coding region and adjacent intronic regions (±25 pb) of 16 genes involved in homologous recombination repair. Bioinformatic analysis of the BRCA1/2 sequences was undertaken with the analysis software and algorithms developed by Sophia Genetics (Sophia DDM), with the support of other bioinformatic tools such as the Integrative Genome Viewer (IGV; www.broadinstitute.org (accessed on 20 September 2022). The reference genome used for the analysis was hg19. This analysis did not include large rearrangements. The sensitivity limit was set at 5% MAF for point variants and 10% MAF for insertion or deletion variants (indels). The minimum coverage to consider a region properly covered was 600 readings (600x). Pathogenic and likely pathogenic variants were visualized using the IGV software and classification was revised as described in Section 2.4.
2.2.3. RL2 DNA Extraction
Neoplastic content was assessed by H&E staining and a minimum of 20% tumor cellularity was required. For each sample, 5 × 10 µm FFPE sections were cut and genomic DNA (gDNA) was extracted using Maxwell® 16 FFPE Tissue LEV DNA Purification Kit (Promega). DNA was eluted in 60 µL of water and stored frozen at −20 °C.
The sample quality and DNA concentration were determined by fluorimetric quantification using Qubit Fluorimeter and Qubit dsDNA BR Assay Kit (Life Technologies, Carlsbad, CA, USA).
2.2.4. RL2 NGS
DNA libraries were generated from 500 ng of DNA per sample using the SureSelect XT Target Enrichment kit (Agilent Technologies Santa Clara, CA, USA). Briefly, gDNA was fragmented using a Covaris S2 sonicator (Covaris, Woburn, MA, USA) and QC was performed using an Agilent BioAnalyzer 2100 (Agilent Technologies) to ensure an average fragment size of 150 to 200 bp. Fragmentation was followed by end-repair, A-tailing, and sequencing adapter ligation. After 10 cycles of PCR amplification, PCR products were isolated on AMPure XP beads (Agencourt, Beckman Coulter; Pasadena, CA, USA). The prepared library was checked and quantified using an Agilent BioAnalyzer DNA1000 chip and 750 ng was hybridized to a biotinylated probe panel (VHIO-300) containing 430 cancer-related genes. The resulting library was sequenced using the Illumina sequencing by synthesis (SBS) technology (2 × 100 PE run). Sequencing reads were aligned (BWA v0.7.17, Samtools v1.9) against the hg19 reference genome, base recalibrated, indel realigned (GATK v3.7.0, abra2 v2.23) and variant called (VarScan2 v2.4.3, Mutect2 v4.1.0.0). Variants from both callers are reported. A minimum of 5 reads supporting the variant allele was required to identify a mutation. The sensitivity of the technique is 5% MAF for SNVs and 10% MAF for INDELs. Frequent single-nucleotide polymorphisms (SNPs) in the population were filtered based on the gnomAD database (allele frequency 0.0001) and copy number alterations (CNA) were calculated (CNVkit v0.9.6.dev0) but not included in the study. Variants were manually checked and the classification of identified variants was performed using publicly available databases as described in Section 2.4. A minimum 250x on-target coverage was required as part of sample QC.
2.3. Variant Analysis
Variant classification and interpretation were carried out using specific databases at the date of issuance of the report, including ClinVar, Breast Cancer Information Core NIH, BRCA Exchange, Leiden Open Variation Database (LOVD), Universal Mutation Database, Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff, COSMIC, cBioPortal, Oncokb and National Library of Medicine and National Institutes of Health (dbSNP). Variants were classified into five categories following the recommendations of the American College of Medical Genetics (ACMG) [14]. The nomenclature used to describe the genetic alterations was that proposed by the Society for Human Genome Variations (HGVS Variant Description Nomenclature) [15]. Reference sequences for BRCA1 and BRCA2 were NM007294.3 and NM000059.3, respectively.
2.4. Ring Test Trial (RTT)
Out of the 82 cases analyzed by the reference laboratories, 6 were selected by the RLs (based on the type(s) of mutation(s) detected) to conduct the RTT among five laboratories from GEICO hospitals. All laboratories received a total of 9 samples for the tumor testing of BRCA mutations, including commercial synthetic (CC_1-3) human FFPE from Horizon (Horizon Dx; Cambridge, UK) (n = 3), OC tumor tissue derived DNA (n = 3) (tDNA_1-3) and FFPE (n = 3) samples (FFPE_1-3) (Figure 1). Laboratories conducted the analyses using local tumor BRCA1/2 NGS testing approaches (Table 2) and reported to reference laboratories the list of identified BRCA1 and BRCA2 variants interpreted as variants of unknown significance (VUS), likely pathogenic (LP), or pathogenic (P). In order to evaluate the intra-lab RTT, RL1 and RL2 agreed upon a list of 17 genotype results (from the 9 supplied samples) along with their classification as VUS (n = 3), LP (n = 1) or P (n = 10). Four BRCA1/2 variants were present in tumor-derived samples (DNA1-3 and FFPE 1-3), ten in the reference samples (CC1-3), and three samples were wild-type (Table 3).
Table 2.
RTT tumor BRCA1/2 analysis strategies.
| Laboratory | Chemistry | NGS-Panel | NGS-Instrument |
|---|---|---|---|
| Lab1 | Multiplex-PCR | Oncomine Comprehensive Assay v3 (Thermo Fisher) | Ion S5™ System (Thermo Fisher Scientific) |
| Lab2 | Multiplex-PCR | BRCA MASTR Plus Dx (Multiplicom) | MiSeq (Illumina) |
| Lab3 | Hybrid Capture | Sure Select XT (Agilent) | Ion S5™ System (ThermoFisher Scientific) |
| Lab4 | Hybrid Capture | MiniHRS (Sophia Genetics) | MiSeq (Illumina) |
| Lab5 | Multiplex-PCR | Oncomine BRCA Research Assay (Thermo Fisher) | Ion S5™ System (Thermo Fisher Scientific) |
Table 3.
Summary of BRCA1/2 variants present in the samples used in the RTT.
| Variant Index | Sample | Variant | Clinical Classification |
Expected Allelic Frequency (%) |
|---|---|---|---|---|
| 1 | DNA_1 | BRCA1:c.3334G>T p.(Glu1112Ter) | P | 49.1 |
| 2 | DNA_2 | BRCA2:c.8802_8828del p.(Met2935_Gln2943del) | LP | 8.2 |
| 3 | DNA_3 | No pathogenic variant | ||
| 4 | FFPE_1 | BRCA1:c.80+6T>A | VUS | 47.4 |
| 5 | FFPE_2 | No pathogenic variant | ||
| 6 | FFPE_3 | BRCA2:c.5351dupA p.(Asn1784Lysfs) | P | 20.1 |
| 7 | CC_1 | BRCA2:c.5351del p.(Asn1784fs) | P | 40 |
| 8 | CC_1 | BRCA1:c.4327C>T p.(Arg1443Ter) | P | 32.5 |
| 9 | CC_1 | BRCA2:c.5073del p.(Lys1691fs) | P | 32.5 |
| 10 | CC_1 | BRCA2:c.8021dup p.(Ile2675fs) | P | 10 |
| 11 | CC_1 | BRCA1:c.1303G>T p.(Asp435Tyr) | VUS | 7.5 |
| 12 | CC_2 | BRCA2:c.5351del p.(Asn1784fs) | P | 10.2 |
| 13 | CC_2 | BRCA1:c.4327C>T p.(Arg1443Ter) | P | 3.9 |
| 14 | CC_2 | BRCA2:c.5073delA p.(Lys1691AsnfsTer15) | P | 3.1 |
| 15 | CC_2 | BRCA2:c.8021dup p.(Ile2675fs) | P | 4.5 |
| 16 | CC_2 | BRCA1:c.1303G>T p.(Asp435Tyr) | VUS | 8.2 |
| 17 | CC_3 | No pathogenic variant |
P: pathogenic; LP: likely pathogenic; VUS: variant of unknown significance. Expected allelic frequencies below 5% are in bold.
3. Results
3.1. Tissue BRCA Testing in the Bilateral RL Comparison
The rate of sequencing success for the 82 selected samples was 100% for RL1 and 98.8% for RL2 (Figure 1).
3.1.1. Pathogenic and Likely Pathogenic Findings (P and LP)
Of the complete series of 100 analyzed samples, RL1 identified twenty-three (23%) P/LP BRCA mutations, 15 (65.2%) germline, and 7 (30.4%) somatic. Germline information was missing for one of the cases (PID#72) (Table 4).
Table 4.
Pathogenic and likely pathogenic BRCA1/2 variants detected in EOC tumors.
| Patient ID | Description | RL1 | RL2 | Germline | ||||
|---|---|---|---|---|---|---|---|---|
| VAF (%) |
Reads | Class | VAF (%) |
Reads | Class | |||
| 7 | NM_007294.3(BRCA1):c.3627dupA p.(Glu1210Argfs) rs80357729 | 71 | 4287 | P | 46 | 361 | P | Yes |
| 11 | NM_007294.3(BRCA1):c.1674del p.(Gly559fs) rs80357600 | 59 | 13,641 | P | 52 | 362 | P | Yes |
| 13 | NM_007294(BRCA1): c.2843_2849del p.(Gly948Valfs*50) | 26 | 5823 | P | 13 | 313 | P | No |
| 14 | NM_007294.3(BRCA1):c.66_67AG p.(Glu23fs) rs80357914 | 70 | 13,914 | P | 64 | 342 | P | Yes |
| 15 | NM_000059.3(BRCA2):c.8802_8828del p.(Met2935_Gln2943del) | 8.2 | 6380 | VUS | 6 | 870 | P | No |
| 19 | NM_007294.3(BRCA1):c.115T>A p.(Cys39Ser) rs80357164 | 75 | 3148 | P | 78 | 125 | P | Yes |
| 21 | NM_007294.3(BRCA1):c.3331_3334del p.(Gln1111fs) rs80357701 | 68 | 557 | P | 57 | 65 | P | Yes |
| 28 | NM_007294.3(BRCA1):c.3648dupA p.(Ser1217Ilefs) rs80357902 | 80 | 5391 | P | 72 | 556 | P | No |
| 31 | NM_007294.3(BRCA1):c.3752_3755GTCT p.(Ser1253fs) rs80357868 | 55 | 4391 | P | 40 | 292 | P | Yes |
| 35 | NM_000059.3(BRCA2):c.715dup p.(Ser239fs) rs431825350 | 51 | 5313 | P | 52 | 257 | P | Yes |
| 48 | NM_007294.3(BRCA1): c.3334G>T p.(Glu1112Ter) | 49 | 18,219 | P | 53 | 704 | P | No |
| 62 | NM_000059.3(BRCA2):c.1128del p.(Phe376fs) rs80359263 | 73 | 4935 | P | 57 | 393 | P | Yes |
| 70 | NM_000059.3(BRCA2):c.5351dupA p.(Asn1784Lysfs) rs80359507 | 21 | 16,034 | P | 23 | 220 | P | No |
| 71 | NM_007294.3(BRCA1):c.845C>A p.(Ser282Ter) rs786203027 | 42 | 298 | P | 52 | 639 | P | Yes |
| 72 | NM_007294.3(BRCA1): c.5578del p.(His1860Thrfs*?) | 48 | 2399 | P | 36 | 110 | LP | NA |
| 79 | NM_007294.4(BRCA1):c.211A>G p.(Arg71Gly) rs80357382 | 71 | 6629 | P | 72 | 185 | P | Yes |
| 80 | NM_007294.4(BRCA1):c.211A>G p.(Arg71Gly) rs80357382 | 71 | 283 | P | 69 | 131 | P | Yes |
| 81 | NM_007294.3(BRCA1):c.3352C>T p.(Gln1118Ter) rs397507215 | 15 | 3636 | P | UR | No | ||
| 93 | NM_007294.3(BRCA1):c.3770_3771del p.(Glu1257fs) | 60 | 3503 | P | UR | Yes | ||
| 5 | NM_000059.3(BRCA2):c.9026_9030del p.(Tyr3009fs) | 45 | 994 | NT | Yes | |||
| 27 | NM_007294.3(BRCA1):c.3627dupA p.(Glu1210Argfs) | 45 | 3351 | NT | Yes | |||
| 36 | NM_000059.3(BRCA2):c.6275_6276del p.(Leu2092fs) | 53 | 21,450 | NT | Yes | |||
| 63 | NM_007294.3(BRCA1):c.1A>G p.(Met1Val) | 18 | 701 | NT | No | |||
| 110 | NM_000059.3(BRCA2): c.3022del p.(Ser1008Alafs*35) | 94 | 4321 | NT | No | |||
P: pathogenic; LP: likely pathogenic; VUS: variant of unknown significance; VAF: variant allelic frequency; UR: unreported; NT: non-tested; RL: reference laboratory; *: TERMINATION (Ter).
Focusing on the 82 samples tested in both RLs, RL1 reported 18 (21.9%) deleterious mutations and RL2 17 (21%) (Table 4). After agreeing on the results, a total of 19 P/LP mutations reached a consensus and three major discrepancies were detected between RL1 and RL2:
PID#81: BRCA1 c.3352C>T p.(Gln1118Ter), variant present at VAF < 5%, hence, below limit of reporting of RL2 and not called. Would not be a discrepancy, since RL1 and RL2 reported correctly according to their own specifications.
PID#93: BRCA1 c.3770_3771del p.(Glu1257fs), detection failure in RL2 as a consequence of sample handling and insufficient tumor material.
PID#15: BRCA2 c.8802_8828del p.(Met2935_Gln2943del), the different criterion in the interpretation, initially classified as VUS by the RL1.
Hence, the global BRCA mutation frequency in our cohort was 23.17%: 12 (63.1%) germline, 6 (31.6%) somatic, and 1 (5.3%) with no germline data available. No extra deleterious mutations in BRCA1/2 were detected in patients with germline mutations. Fifteen out of the nineteen mutations (79%) occurred in BRCA1 and four in BRCA2. Two of the BRCA1 mutations were detected in two different cases; hence, the number of different mutations was 17. Thirteen were frameshift alterations, including the four that occurred in BRCA2, three were nonsense (one recurrent), two were splicing alterations (recurrent) and one was missense (Table 4).
3.1.2. Variants of Uncertain Significance (VUS)
A total of 11 VUS were detected by RL1 in the 82 double-tested samples. RL2 described 8 VUS, seven of which were consistent with RL1. Discordant results had diverse underlying reasons:
PID#15: BRCA2 c.8802_8828del p.(Met2935_Gln2943del), an in-frame deletion initially considered as VUS by RL1 and after consensus was reported as LP (already mentioned in the previous section).
PID#13: BRCA2 c.2771A>T p.(Asn924Ile), an interpretation disagreement, classified as VUS and likely benign by RL1 and RL2, respectively.
PID#57 and PID#79: BRCA1 c.80+6T>A and c.4986+9A>C, both variants located in intronic regions called by RL1 that was removed by the RL2 intronic threshold setting (+/− 3) in the bioinformatic pipeline.
PID#73: BRCA2 c.353G>A p.(Arg118His), a missense change not reported by RL1 due to a bioinformatic error.
In summary, after consensus, VUS were detected in 11 out of 82 patients (13.4%), 7 in BRCA2 and 4 in BRCA1. Seven (64%) of the VUS alterations were missense; three were in intronic regions (BRCA1) and the remaining one corresponded to an in-frame deletion. Three of the patients with tumors presenting VUS carried a pathogenic mutation in BRCA1.
3.2. BRCA Ring Test Trial
Five laboratories participated in the RTT. Each of them employed different methodologies and data analysis pipelines to screen for BRCA1/2 alterations (Table 2 and Table 5). The participating labs had different minimum variant allele frequencies (VAF) and coverage specifications for testing (Table 5).
Table 5.
RTT NGS bioinformatic pipelines.
| RTT Laboratory | Data Analysis Tools/Pipelines | VAF | Minimum Coverage | Intron Flanking Region | Variant Annotation Databases |
|---|---|---|---|---|---|
| Lab1 | Ion ReporterTM Software Version 5.10 | 5% | 500× | ±10 bp | ClinVar, Varsome; COSMIC |
| Lab2 | MASTR Reporter 1.3.0 | 5% | 1000× | No | ClinVar; BRCA Exchange |
| Lab3 | novocraft V3.07.01, bamtools-2.4.1, VCFtools (0.1.15), bedtools v2.26.0-40, samtools 1.8, picardtools 2.8.3, ensembl vep release 94, CONTRA.v2.0.8, gatk-3.4.46 | 10% | 20× | NCBI, ClinVar, Ensembl, BRCA Exchange, cBioPortal | |
| Lab4 | Sophia DDM v3-Sophia Genetics | 5% | 500× | ClinVar, COSMIC, dbSNP, EXAC, g1000, ESP, EpiCov, GnomAD, | |
| Lab5 | Ion Reporter Software Version 5.16 | 5% | 100× | ±100 | dbSNP, BIC database, BRCA Exchange, BRCA Mutation Database |
RTT: Ring Test Trial; VAF: variant allelic frequency.
All of the results obtained in the different labs are summarized in Table 6. A total of 14 variants were evaluated. Three samples were wild-type for BRCA1/2 and their correct non-mutant genotype call was also included as a variant (see also Table 3). Individual sequencing analysis and clinical interpretation, as well as inter-laboratory detection and interpretation concordance for each variant, are included.
Table 6.
Summary of BRCA1/2 variants and results obtained in the RTT.
| Variant | Type of Variant | Sample | Lab1 | Lab2 | Lab3 | Lab4 | Lab5 | Detection Concordance to Reference Genotype (%) | Interpretation Concordance to Reference Genotype (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | nonsense | DNA_1 | * | 100% | 60% | ||||
| 2 | frameshift | DNA_2 | * | 80% | 75% | ||||
| 3 | wt | DNA_3 | 100% | 100% | |||||
| 4 | splicing | FFPE_1 | 40% | 100% | |||||
| 5 | wt | FFPE_2 | 100% | 100% | |||||
| 6 | frameshift | FFPE_3 | 40% | 100% | |||||
| 7 | frameshift | CC_1 | 20% | 100% | |||||
| 8 | nonsense | CC_1 | 100% | 100% | |||||
| 9 | frameshift | CC_1 | 60% | 100% | |||||
| 10 | frameshift | CC_1 | 20% | 100% | |||||
| 11 | missense | CC_1 | 20% | 100% | |||||
| 12 | frameshift | CC_2 | * | 50% | 50% | ||||
| 13 | nonsense | CC_2 | 50% | 100% | |||||
| 14 | frameshift | CC_2 | 25% | 100% | |||||
| 15 | frameshift | CC_2 | 0% | 100% | |||||
| 16 | missense | CC_2 | 25% | 100% | |||||
| 17 | wt | CC_3 | 100% | 100% |
Green: concordance in detection and interpretation; red: no reporting; orange: concordance in detection but not in interpretation; grey: no results delivered by the lab; *: discrepancy with clinical relevance.
The validated 17 genotypes were sent back to the participating laboratories to allow them to re-evaluate their results and determine the reason for miscalling. Variants 1–6 were present in the six tumor samples, whereas variants 7–17 corresponded to the three reference controls.
Variants present in tumor samples. No individual lab obtained a 100% variant calling success when reporting the results for variants 1–6. All the participants correctly identified the wild-type samples (variants 3 and 5) and variants 1 and 8, nonsense mutations in BRCA1. Fails in detection in tumor samples (samples DNA_1-3; FFPE_1-3) corresponded to two frameshift mutations in BRCA2 (variants 2 and 6) and a splicing alteration in BRCA1 (variant 4).
Variant 2: BRCA2 c.8802_8828del; p.(Met2935_Gln2943del), frameshift mutation not called in Lab2 as a consequence of bioinformatic filters.
Variant 4: BRCA1 c.80+6T>A, intronic VUS reported only by two laboratories; in the remaining laboratories it was missed due to the data analysis methods and filters applied for intronic sequences.
Variant 6: BRCA2 c.5351dupA; p.(Asn1784Lysfs), pathogenic mutation missed by three labs that used the Ion S5™ System (ThermoFisher Scientific) for sequencing (see Table 2). The alteration is an insertion located within a homopolymeric (polyA) region.
Variants present in synthetic human FFPE reference samples. The success in sample processing and sequencing was 100% for four out of the five participating labs. Lab4 reported low DNA yields from CC_1-3 local extraction. Due to this issue, CC_1 and CC_3 were processed using low DNA input amounts (below those recommended by the vendor) and no results were generated for CC_2.
Both CC_1 and CC_2 contained a total of five variants each, while CC_3 was found to be wild-type. The expected VAF for the synthetic variants presented a broad range of frequencies, some of them below the lower threshold (5%) for the majority of the laboratories (see Table 3 and Table 5). Regarding false positive results, an extra pathogenic nonsense mutation was reported in BRCA2 with a VAF of 5.6% by one of the labs (Lab2).
With regard to discrepancies in variant classification, a total of four discrepancies affecting three variants were identified (Table 6; orange). Importantly, three of them had clinical implications (Table 6; asterisk): BRCA1 c.3334G>T p.(Glu1112*), BRCA2 c.8802_8828del p.(Met2935_Gln2943del) and BRCA2 c.5351del p.(Asn1784fs) were classified as VUS instead of P/LP.
Overall, the intra-laboratory median concordance detection rate was 64.7% (35.3–70.6%) and 87.5% (75–100%) for variant classification.
Finally, reports of BRCA mutational tests by clinicians were also evaluated. The recommendations of Capoluongo et al. and Palacios et al. are taken as a reference [10,16]. The five report templates correctly included all data concerning patient identification and sample information (suitability of the tumor sample, neoplastic content, and statement of macro/micro-dissection). On the contrary, the methodology description sections presented high levels of heterogeneity, and relevant disclosures were missing in some cases, such as limitations in the detection of deep intronic variants or large indels, the sequencing of homopolymer regions, the description of areas of low coverage that may lead to false negative calls or the list of databases used for annotation. Concerning the interpretation of these findings and subsequent recommendations, in cases of pathogenic mutations emphasis should be placed on the need to recommend a germline study and the referral of the patient to genetic counseling.
4. Discussion
The approval of PARPi (olaparib, niraparib, and rucaparib) for patients with platinum-sensitive relapsed ovarian cancer (OC) has imposed the need for BRCA testing for proper patient management. The prevalence of gBRCA mutations is 18–25% in patients with high-grade OC, and an additional 8% presents sBRCA mutations [17,18,19,20]. In fact, both germline and somatic variants are included as predictive biomarkers of response in olaparib labeling in the EU [2,21]. Hence, the standard of gBRCA testing as the initial screening method has been challenged in favor of prioritizing tumor tissue DNA analysis. Tumor testing detects both germline and somatic mutations and is advantageous in terms of both time and cost, but in the absence of a paired normal sample, the variant origin may not be assigned. Therefore, any given patient with a deleterious mutation detected in a tumor sample should be referred to a genetic counseling unit so that a germline analysis of the detected variant based on observed VAF value can be performed. Several studies have evaluated the reliability of BRCA tumor testing compared with the germline in OC and concluded that this approach is efficient and feasible. The overall mutation rate in our study was within the range reported in other studies [6,22,23,24], as was the frequency of detected germline (63.1%) and somatic (31.6%) mutations [54–74 germline; 27–46 somatic] [5,6,23,25,26,27]. Concordance with the available gBRCA status data was 100%. Of note, no germline copy number variations (CNVs) were present in this particular cohort.
BRCA tumor testing presents relevant limitations affecting both pre-analytical (fixation process, storage conditions, age of the tumor block, tumor cellularity, tumor size, FFPE DNA extraction, and quantification, etc.) and analytical variables (library preparation, bioinformatic pipeline, variant interpretation, etc.).
This work was designed to evaluate the performance of routine real-life BRCA tumor testing in two settings: a two-sided cross-validation tumor BRCA mutation analysis of a series of 82 OC patients between two clinical reference laboratories and a ring trial involving five labs testing nine samples (six from EOC patients and three commercial controls). All labs used their own NGS methodology, data analysis, and interpretation pipelines. Pre-analytical conditions were highly homogeneous, since tumor paraffined slices were consecutively cut from a unique FFPE block per patient.
Discordance in the reported results for both sub-studies was variable. In the two-sided sub-study, discordant test results were discussed and a final genotype was agreed upon. This led to disagreement for only 3.4% of the total results, due to sample handling (one case), bioinformatic pipelines (two cases), and different variant classification criteria (two cases). In the ring trial, a perfect agreement between all participant labs was accomplished for only four genotypes (23.5%). At a more granular level, WT genotypes were correctly called by the five labs (hence, a 100% correct genotyping outcome), and point mutations and indels were correctly identified in 49% and 31% of instances, respectively.
The same NGS platform for sequencing was employed by both reference labs in the two-sided part of the study, possibly increasing the overall agreement of the obtained results. It is worth noting that BRCA genes include a significant amount of homopolymeric stretches, and some NGS approaches have limitations in analyzing these regions. Most of the non-reported variants in our study corresponded to indels located within low-complexity regions and this difficulty seemed to more frequently affect the labs that used an Ion S5™ System platform for sequencing (Thermo Fisher Scientific) [28,29].
Variants located within intronic regions were also a reason for non-concordance among different bioinformatic pipelines, since the distance from the canonical splicing sites cutoffs was not standardized between the labs.
The BRCA analysis of commercial reference samples highlighted the issues related to the limit of detection (LoD) and of reporting VAF thresholds (5–10%); for instance, CC_2 carried three variants with a VAF below 5% (BRCA1:c.4327C>T p.(Arg1443Ter); BRCA2:c.5073delA p.(Lys1691AsnfsTer15); and BRCA2:c.8021dup p.(Ile2675fs)). The value of LoD is particularly relevant for avoiding both false positive and false negative results. The tumor percentage of the starting sample is critical in order to maximize the detection of variants present at a frequency close to the LoD. An important limitation is a percentage of tumor cells in the sample [10,24]. In cases of low tumor cell content, pathologists should mark tumoral areas on hematoxylin-and-eosin-stained tumor slides to guide macro/micro-dissection.
Variant annotation and classification are also sources of differences among labs. A total of four clinically relevant discrepancies were detected, which could lead the oncologist to make different therapeutic decisions for the patient. This fact highlights the importance of standardizing not only the classification criteria, but also the use of specific databases and in silico prediction tools similar to those reported by the ENIGMA consortium (https://enigmaconsortium.org/ (accessed on 20 September 2022) for germline data [30]. Variants should be described as recommended by the Human Genome Variation Society (https://www.hgvs.org/ (accessed on 20 September 2022) [15] and classified according to the Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer into four tiers: tier I, variants with strong clinical significance; tier II, variants with potential clinical significance; tier III, variants with unknown clinical significance; and tier IV, variants that are benign or likely benign [31]. Regarding interpretation, discrepancies affecting non-reported variants in databases remain a challenge with clinically relevant implications.
An additional limitation associated with using tumor BRCA analysis as a universal screening method [22,32,33,34] is the possibility of missing deleterious large rearrangements that cause CNV changes at the sub-gene scale. Chandrasekaran et al. performed parallel germline and tumor testing in OC patients, and found that all germline P variants corresponding to CNVs (with a prevalence in their series of 5/303 (1.65%)) were missed in tumor analysis [22]. Vos et al. argued that exon deletions or duplications in BRCA genes are a minority of the deleterious variants in OC [35] and supported tumor BRCA testing as a prescreening for genetic predisposition if it was performed in accredited laboratories and using validated assays. In our study, labs did not include copy number assessment as part of their testing, confirming that these alterations remain under the radar in routine tumor testing approaches.
As mentioned above, our study has limitations, one of the main ones being the lack of analysis for major rearrangements. Additionally, the genomic regions examined in this study were limited to the coding exons and flanking intronic regions. Additionally, we also highlight the threshold within the detection sensitivity for variant calling.
In summary, our study portrayed a real-life routine testing setting in hospitals. We identified the stages during analytical processing that contributed the most to the relatively low agreement among labs: bioinformatic pipelines and their pre-established settings (minimum allele frequency, splice-site cutoff intronic position, false-positive call removal, etc.) as well as differences in the criteria for the classification of variants. In conclusion, the adoption of BRCA1/2 tumor testing will reduce the time and cost required to identify OC patients who could benefit from PARPi therapy, but critical aspects affecting the reported results are yet to be fully understood by the community, so that they may be managed to improve overall outcomes.
Acknowledgments
We thank all of the patients and their families and caregivers for their participation and hospital staff for their contributions to the study, particularly Tania Mazcuñan and Patricia Carretero for their technical assistance. We thank Patricio Ledesma, Aina Berbel, Isabel Cano and Maria Muntaner for operational support and Ana Levin for assistance in manuscript preparation.
Author Contributions
Design and conception of the study: I.R., A.O., J.A.L.-G. and A.V.; recruitment of patients and collection of clinical data: A.O., A.Y., R.M., A.G., A.B.S.-H., C.P.-S., P.B.-G., J.A., L.G., A.M., J.C., I.P., M.I., A.A., L.S.-L., E.G.-A. and I.R.; DNA extraction, NGS and interpretation of the variant data for the inter-laboratory cross-validation study and design of the sample strategy and analysis of the results for the RTT: Z.G.-C., J.A.L.-G., A.V. and J.M.; DNA extraction, NGS and interpretation of the variant data for the RTT: M.M., G.A.-A., J.P., J.R.A.-L. and G.M.-B.; writing—original draft preparation: Z.G.-C., A.V., I.R. and J.A.L.-G.; writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Instituto Valenciano de Oncologia (expedient number 025-19 (date of approval 07 March 2019)).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by Astra Zéneca Farmacéutica Spain SA (Grant Number GEICO60-0).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cabasag C.J., Fagan P.J., Ferlay J., Vignat J., Laversanne M., Liu L., van der Aa M.A., Bray F., Soerjomataram I. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. [(accessed on 11 May 2022)];Int. J. Cancer. 2022 151:1535–1541. doi: 10.1002/ijc.34002. Available online: https://pubmed.ncbi.nlm.nih.gov/35322413/ [DOI] [PubMed] [Google Scholar]
- 2.Ledermann J.A., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T., et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: An updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. [(accessed on 25 March 2022)];Lancet Oncol. 2016 17:1579–1589. doi: 10.1016/S1470-2045(16)30376-X. Available online: https://pubmed.ncbi.nlm.nih.gov/27617661/ [DOI] [PubMed] [Google Scholar]
- 3.Lheureux S., Lai Z., Dougherty B.A., Runswick S., Hodgson D.R., Timms K.M., Lanchbury J.S., Kaye S., Gourley C., Bowtell D., et al. Long-term responders on olaparib maintenance in high-grade serous ovarian cancer: Clinical and molecular characterization. [(accessed on 11 May 2022)];Clin. Cancer Res. 2017 23:4086–4094. doi: 10.1158/1078-0432.CCR-16-2615. Available online: https://pubmed.ncbi.nlm.nih.gov/28223274/ [DOI] [PubMed] [Google Scholar]
- 4.Vergote I., González-Martín A., Ray-Coquard I., Harter P., Colombo N., Pujol P., Lorusso D., Mirza M., Brasiuniene B., Madry R., et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. [(accessed on 11 May 2022)];Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2022 33:276–287. doi: 10.1016/j.annonc.2021.11.013. Available online: https://pubmed.ncbi.nlm.nih.gov/34861371/ [DOI] [PubMed] [Google Scholar]
- 5.Bell D., Berchuck A., Birrer M., Chien J., Cramer D.W., Dao F., Dhir R., Disaia P., Gabra H., Glenn P., et al. Integrated genomic analyses of ovarian carcinoma. [(accessed on 25 March 2022)];Nature. 2011 474:609–615. doi: 10.1038/nature10166. Available online: https://pubmed.ncbi.nlm.nih.gov/21720365/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennington K.P., Walsh T., Harrell M.I., Lee M.K., Pennil C.C., Rendi M.H., Thornton A., Norquist B.M., Casadei S., Nord A.S., et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. [(accessed on 25 March 2022)];Clin. Cancer Res. 2014 20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. Available online: https://pubmed.ncbi.nlm.nih.gov/24240112/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alsop K., Fereday S., Meldrum C., DeFazio A., Emmanuel C., George J., Dobrovic A., Birrer M.J., Webb P.M., Stewart C., et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian Ovarian Cancer Study Group. [(accessed on 11 May 2022)];J. Clin. Oncol. 2012 30:2654–2663. doi: 10.1200/JCO.2011.39.8545. Available online: https://pubmed.ncbi.nlm.nih.gov/22711857/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dann R.B., DeLoia J.A., Timms K.M., Zorn K.K., Potter J., Flake D.D., II, Lanchbury J.S., Krivak T.C. BRCA1/2 mutations and expression: Response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. [(accessed on 11 May 2022)];Gynecol. Oncol. 2012 125:677–682. doi: 10.1016/j.ygyno.2012.03.006. Available online: https://pubmed.ncbi.nlm.nih.gov/22406760/ [DOI] [PubMed] [Google Scholar]
- 9.Sánchez-Lorenzo L., Salas-Benito D., Villamayor J., Patiño-García A., González-Martín A. The BRCA gene in epithelial ovarian cancer. [(accessed on 11 May 2022)];Cancers. 2022 14:1235. doi: 10.3390/cancers14051235. Available online: https://pubmed.ncbi.nlm.nih.gov/35267543/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capoluongo E., Ellison G., López-Guerrero J.A., Penault-Llorca F., Ligtenberg M.J., Banerjee S., Singer C., Friedman E., Markiefka B., Schirmacher P., et al. Guidance statement on BRCA1/2 tumor testing in ovarian cancer patients. [(accessed on 25 March 2022)];Semin. Oncol. 2017 44:187–197. doi: 10.1053/j.seminoncol.2017.08.004. Available online: https://pubmed.ncbi.nlm.nih.gov/29248130/ [DOI] [PubMed] [Google Scholar]
- 11.Konstantinopoulos P.A., Norquist B., Lacchetti C., Armstrong D., Grisham R.N., Goodfellow P.J., Kohn E.C., Levine D.A., Liu J.F., Lu K.H., et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. [(accessed on 11 May 2022)];J. Clin. Oncol. 2020 38:1222–1245. doi: 10.1200/JCO.19.02960. Available online: https://pubmed.ncbi.nlm.nih.gov/31986064/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison G., Ahdesmäki M., Luke S., Waring P.M., Wallace A., Wright R., Röthlisberger B., Ludin K., Merkelbach-Bruse S., Heydt C., et al. An evaluation of the challenges to developing tumor BRCA1 and BRCA2 testing methodologies for clinical practice. [(accessed on 11 May 2022)];Hum. Mutat. 2018 39:394–405. doi: 10.1002/humu.23375. Available online: https://pubmed.ncbi.nlm.nih.gov/29215764/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegel V., Blatnik A., Škof E., Dragoš V.Š., Krajc M., Gregorič B., Škerl P., Strojnik K., Klančar G., Banjac M., et al. Real-world data on detection of germline and somatic pathogenic/likely pathogenic variants in BRCA1/2 and other susceptibility genes in ovarian cancer patients using next generation sequencing. [(accessed on 11 May 2022)];Cancers. 2022 14:1434. doi: 10.3390/cancers14061434. Available online: https://pubmed.ncbi.nlm.nih.gov/35326583/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. [(accessed on 25 March 2022)];Genet. Med. 2015 17:405–424. doi: 10.1038/gim.2015.30. Available online: https://pubmed.ncbi.nlm.nih.gov/25741868/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E., et al. HGVS recommendations for the description of sequence variants: 2016 update. [(accessed on 25 March 2022)];Hum. Mutat. 2016 37:564–569. doi: 10.1002/humu.22981. Available online: https://pubmed.ncbi.nlm.nih.gov/26931183/ [DOI] [PubMed] [Google Scholar]
- 16.Palacios J., De La Hoya M., Bellosillo B., De Juan I., Matias-Guiu X., Lázaro C., Palanca S., Osorio A., Rojo F., Rosa-Rosa J.M., et al. Mutational screening of BRCA1/2 genes as a predictive factor for therapeutic response in epithelial ovarian cancer: A consensus guide from the Spanish Society of Pathology (SEAP-IAP) and the Spanish Society of Human Genetics (AEGH) [(accessed on 25 March 2022)];Virchows Arch. 2020 476:195–207. doi: 10.1007/s00428-019-02709-3. Available online: https://pubmed.ncbi.nlm.nih.gov/31797087/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stratton J.F., Gayther S.A., Russell P., Dearden J., Gore M., Blake P., Easton D., Ponder B.A. Contribution of BRCA1 mutations to ovarian cancer. [(accessed on 25 March 2022)];N. Engl. J. Med. 1997 336:1125–1130. doi: 10.1056/NEJM199704173361602. Available online: https://pubmed.ncbi.nlm.nih.gov/9099656/ [DOI] [PubMed] [Google Scholar]
- 18.Norquist B.M., Harrell M.I., Brady M.F., Walsh T., Lee M.K., Gulsuner S., Bernards S.S., Casadei S., Yi Q., Burger R.A., et al. Inherited mutations in women with ovarian carcinoma. [(accessed on 25 March 2022)];JAMA Oncol. 2016 2:482–490. doi: 10.1001/jamaoncol.2015.5495. Available online: https://pubmed.ncbi.nlm.nih.gov/26720728/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrillo M., Marchetti C., De Leo R., Musella A., Capoluongo E.D., Paris I., Panici P.B., Scambia G., Fagotti A. BRCA mutational status, initial disease presentation, and clinical outcome in high-grade serous advanced ovarian cancer: A multicenter study. [(accessed on 25 March 2022)];Am. J. Obstet. Gynecol. 2017 217:334.e1–334.e9. doi: 10.1016/j.ajog.2017.05.036. Available online: https://pubmed.ncbi.nlm.nih.gov/28549976/ [DOI] [PubMed] [Google Scholar]
- 20.Zhang S., Royer R., Li S., McLaughlin J.R., Rosen B., Risch H.A., Fan I., Bradley L., Shaw P.A., Narod S.A. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. [(accessed on 25 March 2022)];Gynecol. Oncol. 2011 121:353–357. doi: 10.1016/j.ygyno.2011.01.020. Available online: https://pubmed.ncbi.nlm.nih.gov/21324516/ [DOI] [PubMed] [Google Scholar]
- 21.Weren R.D., Mensenkamp A.R., Simons M., Eijkelenboom A., Sie A.S., Ouchene H., van Asseldonk M., Gomez-Garcia E.B., Blok M.J., de Hullu J.A., et al. Novel BRCA1 and BRCA2 tumor test as basis for treatment decisions and referral for genetic counselling of patients with ovarian carcinomas. [(accessed on 25 March 2022)];Hum. Mutat. 2017 38:226–235. doi: 10.1002/humu.23137. Available online: https://pubmed.ncbi.nlm.nih.gov/27767231/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrasekaran D., Sobocan M., Blyuss O., Miller R.E., Evans O., Crusz S.M., Mills-Baldock T., Sun L., Hammond R.F.L., Gaba F., et al. Implementation of multigene germline and parallel somatic genetic testing in epithelial ovarian cancer: SIGNPOST study. [(accessed on 25 March 2022)];Cancers. 2021 13:4344. doi: 10.3390/cancers13174344. Available online: https://pubmed.ncbi.nlm.nih.gov/34503154/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mafficini A., Simbolo M., Parisi A., Rusev B., Luchini C., Cataldo I., Piazzola E., Sperandio N., Turri G., Franchi M., et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. [(accessed on 25 March 2022)];Oncotarget. 2016 7:1076–1083. doi: 10.18632/oncotarget.6834. Available online: https://pubmed.ncbi.nlm.nih.gov/26745875/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turashvili G., Lazaro C., Ying S., Charames G., Wong A., Hamilton K., Yee D., Agro E., Chang M., Pollett A., et al. Tumor BRCA testing in high grade serous carcinoma: Mutation rates and optimal tissue requirements. [(accessed on 25 March 2022)];Cancers. 2020 12:3468. doi: 10.3390/cancers12113468. Available online: https://pubmed.ncbi.nlm.nih.gov/33233347/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAlpine J.N., Porter H., Kobel M., Nelson B.H., Prentice L.M., Kalloger S.E., Senz J., Milne K., Ding J., Shah S.P., et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. [(accessed on 25 March 2022)];Mod. Pathol. 2012 25:740–750. doi: 10.1038/modpathol.2011.211. Available online: https://pubmed.ncbi.nlm.nih.gov/22282309/ [DOI] [PubMed] [Google Scholar]
- 26.Yang D., Khan S., Sun Y., Hess K., Shmulevich I., Sood A.K., Zhang W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. [(accessed on 25 March 2022)];JAMA. 2011 306:1557–1565. doi: 10.1001/jama.2011.1456. Available online: https://pubmed.ncbi.nlm.nih.gov/21990299/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennessy B.T., Timms K.M., Carey M.S., Gutin A., Meyer L.A., Flake D.D., 2nd, Abkevich V., Potter J., Pruss D., Glenn P., et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. [(accessed on 25 March 2022)];J. Clin. Oncol. 2010 28:3570–3576. doi: 10.1200/JCO.2009.27.2997. Available online: https://pubmed.ncbi.nlm.nih.gov/20606085/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bragg L.M., Stone G., Butler M.K., Hugenholtz P., Tyson G.W. Shining a light on dark sequencing: Characterising errors in Ion Torrent PGM data. [(accessed on 25 March 2022)];PLoS Comput. Biol. 2013 9:e1003031. doi: 10.1371/journal.pcbi.1003031. Available online: https://pubmed.ncbi.nlm.nih.gov/23592973/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeo Z.X., Wong J.C.L., Rozen S.G., Lee A.S.G. Evaluation and optimisation of indel detection workflows for ion torrent sequencing of the BRCA1 and BRCA2 genes. [(accessed on 25 March 2022)];BMC Genom. 2014 15:516. doi: 10.1186/1471-2164-15-516. Available online: https://pubmed.ncbi.nlm.nih.gov/24962530/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spurdle A.B., Healey S., Devereau A., Hogervorst F.B.L., Monteiro A.N.A., Nathanson K.L., Radice P., Stoppa-Lyonnet D., Tavtigian S., Wappenschmidt B., et al. ENIGMA—Evidence-based network for the interpretation of germline mutant alleles: An international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. [(accessed on 25 March 2022)];Hum. Mutat. 2012 33:2–7. doi: 10.1002/humu.21628. Available online: https://pubmed.ncbi.nlm.nih.gov/21990146/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. [(accessed on 30 March 2022)];J. Mol. Diagn. 2017 19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. Available online: https://pubmed.ncbi.nlm.nih.gov/27993330/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jonge M.M., Ruano D., van Eijk R., van der Stoep N., Nielsen M., Wijnen J.T., Ter Haar N.T., Baalbergen A., Bos M.E., Kagie M.J., et al. Validation and implementation of BRCA1/2 variant screening in ovarian tumor tissue. [(accessed on 25 March 2022)];J. Mol. Diagn. 2018 20:600–611. doi: 10.1016/j.jmoldx.2018.05.005. Available online: https://pubmed.ncbi.nlm.nih.gov/29936257/ [DOI] [PubMed] [Google Scholar]
- 33.Fumagalli C., Tomao F., Betella I., Rappa A., Calvello M., Bonanni B., Bernard L., Peccatori F., Colombo N., Viale G., et al. Tumor BRCA test for patients with epithelial ovarian cancer: The role of molecular pathology in the era of PARP inhibitor therapy. [(accessed on 25 March 2022)];Cancers. 2019 11:1641. doi: 10.3390/cancers11111641. Available online: https://pubmed.ncbi.nlm.nih.gov/31653094/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bekos C., Grimm C., Kranawetter M., Polterauer S., Oberndorfer F., Tan Y., Müllauer L., Singer C. Reliability of tumor testing compared to germline testing for detecting BRCA1 and BRCA2 mutations in patients with epithelial ovarian cancer. [(accessed on 25 March 2022)];J. Pers. Med. 2021 11:593. doi: 10.3390/jpm11070593. Available online: https://pubmed.ncbi.nlm.nih.gov/34202525/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vos J.R., Fakkert I.E., de Hullu J.A., van Altena A.M., Sie A.S., Ouchene H., Willems R.W., Nagtegaal I.D., Jongmans M.C., Mensenkamp A.R., et al. Universal tumor DNA BRCA1/2 testing of ovarian cancer: Prescreening PARPi treatment and genetic predisposition. [(accessed on 25 March 2022)];J. Natl. Cancer Inst. 2020 112:161–169. doi: 10.1093/jnci/djz080. Available online: https://pubmed.ncbi.nlm.nih.gov/31076742/ [DOI] [PMC free article] [PubMed] [Google Scholar]