Abstract
We previously reported on the existence of a family of lipoprotein genes, designated 2.9 lipoprotein genes, encoded in at least seven versions on the circular (supercoiled) cp32 and cp18 plasmids of Borrelia burgdorferi 297. A distinguishing feature of the 2.9 lipoproteins were highly similar signal sequences but variable mature polypeptides that segregated into two antigenic classes. Further screenings of B. burgdorferi 297 genomic libraries led to the identification of three additional 2.9 lipoprotein genes, renamed herein mlp, for multicopy lipoprotein genes. Computer analyses and immunoblotting revealed that Mlp-9 segregated with the antigenic class I lipoproteins, whereas Mlp-8 and Mlp-10 were members of class II. Northern blotting showed that all three of the mlp genes were expressed when B. burgdorferi was cultivated in vitro at 34°C, although mlp-9 and mlp-10 transcripts were expressed at very low levels. Additional combined immunoblotting and comparative reverse transcription-PCR analyses performed on borreliae cultivated in vitro at 23, 34, or 37°C indicated that although Mlp-8 was substantially more abundant than Mlp-9 or Mlp-10, all three of the mlp genes were upregulated during B. burgdorferi replication at 37°C. Expression of the same three lipoproteins was further enhanced upon growth of the spirochetes within dialysis membrane chambers (DMCs) implanted intraperitoneally in rats (i.e., spirochetes in a mammalian host-adapted state), suggesting that temperature alone did not account for maximal upregulation of the mlp genes. That certain mlp genes are likely expressed during the growth of B. burgdorferi in mammalian tissues was supported by findings of antibodies against all three Mlp lipoproteins in mice after challenge with Ixodes scapularis nymphs harboring B. burgdorferi 297. The combined data suggest that as opposed to being differentially expressed in any reciprocal fashion (e.g., OspA/OspC), at least three mlp genes are simultaneously upregulated by temperature (37°C) and some other mammalian host factor(s). The findings have importance not only for understanding alternative modes of differential antigen expression by B. burgdorferi but also for assessing whether one or more of the Mlp lipoproteins represent new candidate vaccinogens for Lyme disease.
Lyme disease, a multisystem infectious disorder caused by the spirochetal bacterium Borrelia burgdorferi (34), is the most prevalent arthropod-borne disease in the United States (7). In 1996, almost 17,000 cases of Lyme disease were reported to the Centers for Disease Control and Prevention, an increase of 41% above 1995 and a record high (7). Thus, Lyme disease continues to represent a significant public health problem (7).
The zoonotic life cycle of B. burgdorferi is complex and depends on horizontal transmission between immature ticks and mice; humans are accidental hosts (34). During nymphal feeding, profound changes occur in the antigenic repertoire of B. burgdorferi as it migrates from the midgut and salivary glands of the tick into mammalian tissue (13, 32). The paradigm for this phenomenon is the inverse relationship between the expression of OspA and OspC (27, 32). Such findings coincide with additional observations that other B. burgdorferi antigens are expressed predominantly during growth either in vitro (ostensibly analogous to the tick environment) or in vivo (i.e., during mammalian infection) (1, 3, 4, 8, 10, 12, 15, 16, 25, 29, 30, 38, 40, 42). Interestingly, these differentially expressed antigens are virtually all plasmid encoded, underscoring the importance of plasmids for the organism's zoonotic cycle and for virulence expression.
The 2.9 locus, a ca. 3-kb segment of DNA encoding a number of genes on the circular (supercoiled) cp32 and cp18 plasmids, was first reported to exist in at least seven versions in B. burgdorferi 297 (29). As originally described (29), the 5′ end of each 2.9 locus contains four tandem open reading frames (ORFs), designated ABCD. Just downstream of these four ORFs is another ORF encoding a highly repeated (rep) region; the putative directions of transcription for rep were designated rep+ and rep−. In at least one 2.9 locus, the rep+ region is replaced by an ORF in the opposite orientation designated rev. Further downstream of the rep/rev region is usually a single lipoprotein gene, except in one instance where two tandem lipoprotein genes have been noted (29). The 2.9 lipoprotein genes encode highly similar signal sequences but variable mature polypeptides that segregate into two antigenic classes based on size, hydrophilicity, sequence similarities, and reactivity with polyclonal antisera. These lipoproteins are renamed herein Mlp, for multicopy lipoproteins.
While physiological functions have not been ascribed to any of the proteins encoded within the 2.9 loci, Guina and Oliver (19) reported on a comparable orfA (blyA) gene within an ABCD operon of B. burgdorferi B31 that encodes a hemolysin-like protein. The same authors (19) also described an orfB (blyB) gene that encodes a 13-kDa protein which stabilizes the hemolytic activity of the orfA gene product. Regarding lipoprotein genes, Theisen (41) reported on the existence in B. afzelii, B. garinii, and B. burgdorferi sensu stricto DK7 of a 33-kDa lipoprotein (NlpH), encoded on a supercoiled plasmid, which binds Congo red, a property associated with bacterial virulence and the binding of hemin; the nlpH orthologs are highly homologous with the class I Mlp lipoproteins of B. burgdorferi 297. Furthermore, the DNA sequences flanking nlpH include a repetitive region (i.e., rep) remarkably similar to the genetic organization of the 2.9 loci of B. burgdorferi 297. Finally, protease sensitivity experiments suggested that NlpH is surface-exposed in B. afzelii. These combined findings suggest that (i) one or more of the 2.9 lipoproteins may be potential targets for bactericidal antibodies (i.e., vaccine candidates) and (ii) the 2.9 lipoprotein genes may be widely distributed among the various B. burgdorferi sensu lato genospecies. Regarding other 2.9 locus genes, Gilmore and Mbow (18) identified in B. burgdorferi B31 a rev gene product that is expressed in infected (tick-inoculated) mice as well as in the sera of human Lyme disease patients, suggesting that a rev gene product may play an important role for spirochete survival during the mammalian phase of infection.
Herein we describe the identification and characterization of three additional 2.9 loci and provide evidence that their mlp genes are all upregulated during the growth of B. burgdorferi in vitro at 37°C as well as when B. burgdorferi replicates in dialysis membrane chambers implanted intraperitoneally in rats (i.e., during a mammalian host-adapted state). The findings have importance not only for understanding the regulation of differential antigen expression by B. burgdorferi but ultimately for assessing whether one or more of the 2.9 lipoproteins represent new candidate vaccinogens for Lyme disease.
MATERIALS AND METHODS
Bacterial strains.
Low-passage, virulent B. burgdorferi strain 297 was described previously (20). For in vitro cultivation, spirochetes from not more than three serial passages were cultivated at 34°C in complete BSK-H medium (Sigma Chemical Co., St. Louis, Mo.) (28). For temperature shift experiments, spirochetes were first cultivated at 23°C until cell densities reached approximately 5 × 106 cells per ml. The culture then was divided and diluted 1,000-fold with fresh (prewarmed) BSK-H medium; cultures were further incubated at either 23, 34, or 37°C. Spirochetes were harvested at the mid-logarithmic phase (ca. 107 cells per ml) for further analyses. To obtain B. burgdorferi in the mammalian host-adapted state, spirochetes were cultivated in dialysis membrane chambers (DMCs) implanted in rat peritoneal cavities as described by Akins et al. (1) except that BSK-H medium was used in place of BSK II medium. To standardize total B. burgdorferi proteins among differing spirochete samples, total protein was determined by the bicinchoninic acid assay (Pierce Chemical Co., Rockford, Ill.). Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was used as the cloning host and was cultivated either in yeast-tryptone broth or on yeast-tryptone agar supplemented with 100 μg of ampicillin per ml.
Tick rearing, infection, and infestation.
Four BALB/c mice each were inoculated with 2.5 × 105 B. burgdorferi 297 spirochetes. Half of the cells were administered by intraperitoneal injection, and the remaining half were given via subcutaneous injection over the sternum. To ensure infection, 2 weeks later, ear punch biopsies were obtained (33) and cultured in BSK II medium supplemented with rifampin (50 μg/ml) and amphotericin B (25 μg/ml); cultures were examined by dark-field microscopy daily after the third day for the presence of spirochetes.
Pathogen-free Ixodes scapularis nymphs were derived from a colony maintained in the laboratory of S. K. Wikel. Larvae and nymphs obtained blood meals from laboratory mice, whereas fertilized adult females fed on sheep. All tick life cycle stages were stored in 16-ml glass vials (Wheaton Glass, Millville, N.J.) with screen lids. Ticks were maintained at 22°C with a 16 h:8 h light:dark photoperiod in a desiccator over a super-saturated K2SO4 solution, providing a relative humidity of 97%. I. scapularis larvae were infected with B. burgdorferi by permitting them to obtain a blood meal from the infected mice. Engorged larvae were allowed to molt to the nymphal stage. Digestive tracts were removed from 30 representative nymphs, suspended in 0.1 M phosphate-buffered saline (pH 7.2), and examined by dark-field microscopy. Nymphs derived only from feedings that resulted in ≥90% infection were used in this study.
To infect mice with B. burgdorferi 297, naive mice were each infested with 10 I. scapularis nymphs (a number shown in earlier titration experiments to result in 100% infection of mice) confined within a capsule placed on the back. Each capsule consisted of the top portion of a 1.5-ml polypropylene microcentrifuge tube secured to closely clipped fur by a mixture (weight/weight) of 4 parts colophonium and 1 part beeswax. Nymphs, on average, fed on mice for approximately 4 days. Ear punch biopsies were taken 2 weeks after animal infestation and cultured in BSK-H medium with antibiotics (Sigma catalog number A-1956) to confirm that all mice were infected.
Identification of E. coli clones harboring B. burgdorferi 2.9 loci.
A genomic DNA library of low-passage (virulent) B. burgdorferi 297 DNA (29) was screened by hybridization with a 32P-labeled 2.9orfD probe as previously described (29). Plasmid DNAs were then isolated from hybridizing clones and were subjected to DNA sequence analysis (below).
Pulsed-field gel electrophoresis and Southern hybridization analyses.
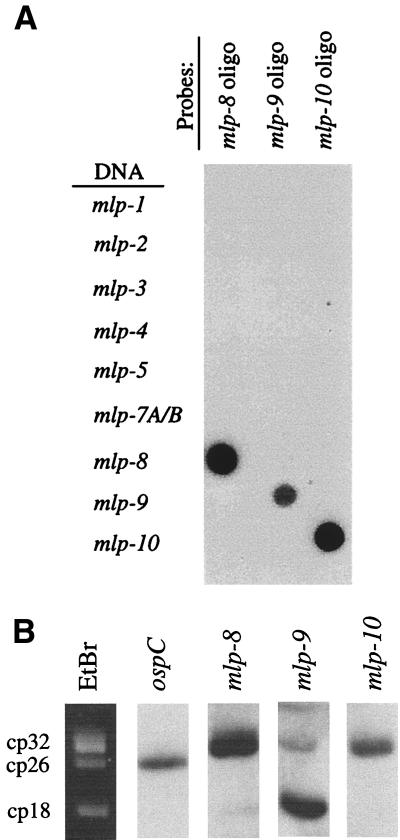
Pulsed-field gel electrophoresis and Southern hybridization analyses were performed as described previously (20, 29). Methods for the isolation of B. burgdorferi genomic DNA and supercoiled plasmid DNA were as reported elsewhere (29). Hybridization probes for ospC and lipoprotein genes mlp-8, mlp-9, and mlp-10 are listed in Table 1. The specificities of the mlp gene probes were confirmed by performing dot blot Southern hybridization analyses (Fig. 3A). In these assays, 50-ng aliquots of purified plasmid DNAs harboring each mlp gene were first loaded into a 96-well transfer device; plasmid DNAs were UV cross-linked to nylon membranes. Nylon membranes were then cut into strips such that each strip contained all 10 mlp genes for Southern hybridizations with individual radioactive oligonucleotide probes.
TABLE 1.
Oligonucleotide primers and probes used in this study
| Designation | Sequence (5′-3′) | Purpose |
|---|---|---|
| mlp-8-oligo | GTCCCCTTCAAGTGCCCCTTGAAC | mlp-8 probe used in Fig. 3 and 4 |
| mlp-9-oligo | CCATGTTTATTATCATTACATTTCTTTGGATTATT | mlp-9 probe used in Fig. 3 and 4 |
| mlp-10-oligo | TTAAGAGATTCTTTAACTAAAGTTTTGTATGTGG | mlp-10 probe used in Fig. 3 and 4 |
| ospC-oligo | CTGCCACAATAGGACTTGTAAGCTCTTTAACTG | ospC probe used in Fig. 3 |
| flaB-oligo | CAATACCAGAAGCGCCCGAATATACATAATC | flaB probe used in Fig. 4 |
| Primer A | CTGTAAATTTATCTATGTCCCCTTC | Primer used in Fig. 5 |
| Primer B | GAGCAATGTCTATAACAATGATAG | Primer used in Fig. 5 |
| Primer C | TTTACAAAAAGATATATCTAATTTAGATAC | Primer used in Fig. 5 |
| Primer D | TGGGCATGGAACAATTAATAGC | Primer used in Fig. 5 |
| Primer E | CTTGCAAGAAATTCTCCTTTAC | Primer used in Fig. 5 |
| mlp-cons-5′ | TAACAATGAAAATYATCAACATATTATTTTG | 5′ primer for mlp-8, mlp-9, and mlp-10 used in Fig. 8 |
| mlp-8-3′ | Same as primer A | 3′ mlp-8 primer used in Fig. 8 |
| mlp-9-3′ | CATGTTTATTATCATTACATTTGTTTG | 3′ mlp-9 primer used in Fig. 8 |
| mlp-10-3′ | Same as mlp-10-oligo | 3′ mlp-10 primer used in Fig. 8 |
| ospA-5′ | TGTAAGCAAAATGTTAGCAGCCTT | 5′ ospA primer used in Fig. 8 |
| ospA-3′ | CAGCAGTTAGAGTTCCTTCAAG | 3′ ospA primer used in Fig. 8 |
| ospC-5′ | GCTGATGAGTCTGTTAAAGGG | 5′ ospC primer used in Fig. 8 |
| ospC-3′ | GCATCTCTTTAGCTGCTTTTG | 3′ ospC primer used in Fig. 8 |
| flaB-5′ | ATGATTATCAATCATAATACATCAGC | 5′ flaB primer used in Fig. 8 |
| flaB-3′ | GTTGTCTGAATAAAATTAATAGCC | 3′ flaB primer used in Fig. 8 |
| Mlp-cons-Bam-5′ | CGGGATCCAATTCTAATGATAATGACAC | 5′ primer for full-length GST-mlp fusion |
| Mlp-8-Eco-3′ | CGGAATTCTTATTAGGAATTACCACCACCAC | 3′ primer for full-length GST-Mlp-8 fusion |
| Mlp-9-Eco-3′ | CGGAATTCTTATTAGTTTTGCCAATTATCTGTAAG | 3′ primer for full-length GST-Mlp-9 fusion |
| Mlp-10-Eco-3′ | CGGAATTCTTATTAGGACCCGTTGCAGGGAC | 3′ primer for full-length GST-Mlp-10 fusion |
| Mlp-8-BglII-5′ | GGAAGATCTGAAGGGGACATAGATAAATTT | 5′ primer for epitopic fusion protein of Mlp-8 |
| Mlp-8-HindIII-3′ | CACAAGCTTGGAATTACCACCACCACATGT | 3′ primer for epitopic fusion protein of Mlp-8 |
| Mlp-10-BglII-5′ | GGAAGATCTACTTTAGTTAAAGAATCTCTTAAAG | 5′ primer for epitopic fusion protein of Mlp-10 |
| Mlp-10-HindIII-3′ | CACAAGCTTGGGACTATTTGTTTGTGCTGT | 3′ primer for epitopic fusion protein of Mlp-10 |
FIG. 3.
Specificities of oligonucleotide probes for the mlp genes and localization of mlp-8, mlp-9, and mlp-10 to supercoiled plasmids of B. burgdorferi 297. (A) Dot blot hybridization assay; target (plasmid) DNAs encoding each mlp gene were spotted onto nylon membranes, cross-linked, and hybridized with radioactively labeled oligonucleotide probes for mlp-8, mlp-9, or mlp-10. Designations at the left denote each target DNA. (B) Supercoiled plasmids isolated from B. burgdorferi 297 were separated by electrophoresis on 0.4% agarose and hybridized with probes for ospC, mlp-8, mlp-9, or mlp-10. The left-most lane shows the ethidium bromide (EtBr)-stained plasmid profile. Circular plasmids are denoted at the left of the ethidium bromide-stained gel; cp26 encodes ospC. cp18 identity was confirmed by hybridization with probes specific for mlp-3 (29) and p21 (2) (not shown).
Two-dimensional pulsed-field gel electrophoresis was used for the separation of supercoiled plasmid DNA, linear plasmid DNA, and linear chromosomal DNA of B. burgdorferi (25, 29); separated DNAs were transferred to nylon membranes for Southern hybridization analyses. To localize each individual mlp gene to a specific species of supercoiled plasmid, 200 ng of purified supercoiled plasmid DNA was loaded per well of a 0.4% agarose gel (29). After separation by electrophoresis, DNAs were transferred to nylon membranes and subjected to Southern hybridization analysis (29).
Northern blot analysis.
Total RNA was isolated from B. burgdorferi 297 by using an Ultraspec RNA Isolation System (Biotecx, Houston, Tex.) according to the protocol of the manufacturer. Northern blot analysis was carried out as described previously (29). The hybridization probes for the flagellin gene (flaB) and the mlp-8, mlp-9, and mlp-10 genes are listed in Table 1.
RT-PCR.
Spirochetes were harvested at the mid-logarithmic phase of growth (107 cells per ml) from either in vitro cultures or DMCs in rats. Total RNA was isolated as for Northern blot experiments. Extracted RNA (10 μg) was incubated with 10 U of RQ1 DNase I (Promega Corp., Madison, Wis.) at 37°C for 3 h. RNA was extracted once with phenol-chloroform and precipitated with cold 100% ethanol. RNA pellets were then suspended in 50 μl of diethyl pyrocarbonate-treated water. DNase treatment was repeated if DNA contamination was detected after PCR analysis. One-step reverse transcription-PCRs (RT-PCRs) reactions were performed with the Titan RT-PCR system (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer's recommendations. The 20-μl reaction mixtures included reaction buffer, 5 U of RNase inhibitor, 5 mM dithiothreitol, 0.8 mM deoxynucleoside triphosphates, 0.3 μM each oligonucleotide primer, and 1 μl of enzyme mixture. For results shown in Fig. 4, 100 ng of DNA-free total RNA was used; for results shown in Fig. 8, 10-fold serial dilutions of RNA were used in the reactions. The concentrations of total RNA used varied depending on the relative abundances of the transcripts; for detection of flaB and ospA transcripts, 1 pg to 1 ng of total RNA was used, whereas 100-fold-greater amounts of RNA (100 pg to 100 ng) were used in reactions to detect mlp-8, mlp-9, and mlp-10 transcripts. cDNA syntheses were performed by incubating all reaction mixtures at 50°C for 30 min. Amplifications were carried out in a Perkin-Elmer GeneAmp 2400 Thermocycler set for the following parameters: 94°C for 3 min; 30 cycles of 94°C for 15 s, 50°C for 15 s, and 72°C for 30 s, with increments of 5 s for each cycle. For all RT-PCRs, a negative control was performed by omitting reverse transcriptase in the reaction mixtures; positive controls were carried out with 50 ng of B. burgdorferi genomic DNA as template in place of total RNA. Primers used for RT-PCRs are listed in Table 1.
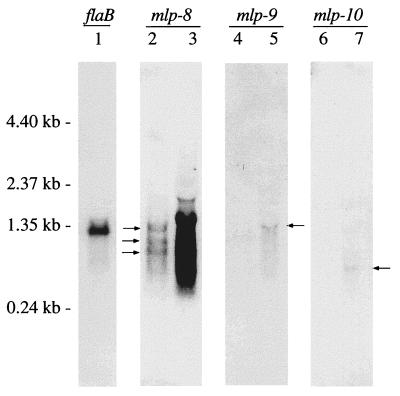
FIG. 4.
Northern blot analysis of the mlp genes. RNA from B. burgdorferi 297 was hybridized with probes (indicated above the lanes) specific for flaB, mlp-8, mlp-9, and mlp-10. Lanes 1, 2, 4, and 6 contain 3 μg of RNA; lanes 3, 5, and 7 contain 30 μg of RNA. Arrows beside lane 2 show 0.6-, 0.9-, and 1.3-kb transcripts of mlp-8. Arrows beside lanes 5 and 7 show transcripts of 1.3 and 0.4 kb, respectively. Molecular size markers are shown at the left.
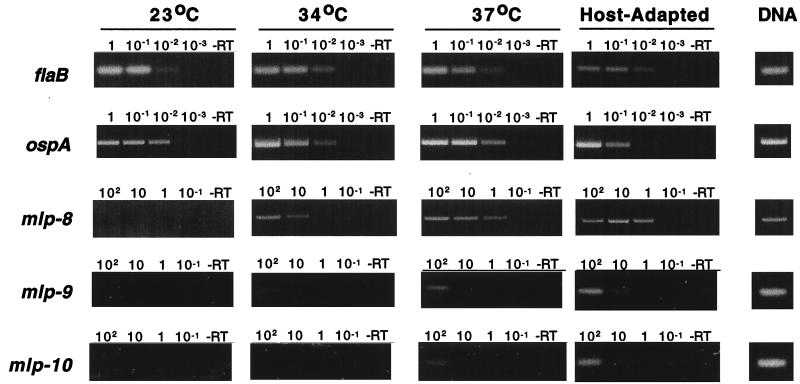
FIG. 8.
Comparative RT-PCR analysis of mlp-8, mlp-9, and mlp-10 gene expression in spirochetes cultivated in vitro or within DMCs in rat peritoneal cavities. Total RNA was isolated from spirochetes grown in vitro either at 23, 34, or 37°C or within DMCs in rat peritoneal cavities (host adapted). Tenfold serial dilutions of RNA were used for amplification of each mRNA. The final amounts (nanograms) of RNA used in each reaction is indicated above each lane. -RT denotes reaction mixtures lacking reverse transcriptase. The column at the right (DNA) corresponds to reactions using B. burgdorferi genomic DNA as the template in place of RNA. Note that for the analyses of flaB and ospA, 100-fold-lower quantities of RNA were required to visualize endpoints.
DNA sequencing and computer analyses.
Nucleotide sequencing was performed with an Applied Biosystems model 373A automated DNA sequencer and PRISM ready reaction DyeDeoxy terminator cycle sequencing kits as instructed by the manufacturer (Applied Biosystems Inc., Foster City, Calif.). Nucleotide and deduced amino acid sequences were analyzed and manipulated by using the University of Wisconsin Computer Genetics Group Wisconsin Package version 7.3 (GenBank database release 82.0) (13), Lasergene (DNASTAR, Madison, Wis.), and MacVector version 4.1.1 (International Biotechnologies Inc.-Kodak, New Haven, Conn.) software packages. Promoter prediction analyses were carried out with the Promoter Prediction by Neural Network software (29a).
Fusion proteins.
A glutathione S-transferase (GST) fusion protein of OspC was described previously (9). GST fusions of lipoproteins Mlp-8, Mlp-9, and Mlp-10 (lacking acylation sites) were generated by PCR amplification of DNAs from lambda clones encoding the corresponding predicated mature portions of each protein; the oligonucleotide primers used for PCR are listed in Table 1. PCR products were restriction enzyme digested and ligated into the corresponding polylinker sites of pGEX4T-2 (Pharmacia Biotech, Inc.). The resultant fusion proteins were purified by affinity chromatography as described previously (25).
To construct a partial fusion protein (epitope-specific version) of Mlp-8, a DNA fragment encoding amino acid residues 130 to 149 was amplified by PCR from the DNA of its lambda clone (see Table 1 for oligonucleotide primers). This partial fusion protein was designed to contain less than five consecutive amino acids identical to any other Mlp lipoprotein. The PCR product was restriction enzyme digested and ligated into the corresponding polylinker sites of pQE40 (Qiagen, Inc.). The partial fusion protein also contained a His6 tag and a murine dihydrofolate reductase protein. A comparable protocol (see Table 1 for oligonucleotide primers) was used for construction of a partial fusion protein of Mlp-10, which contained amino acid residues 123 to 143. Epitope-specific partial fusion proteins were purified by affinity chromatography on a nickel-nitrilotriacetic acid matrix as instructed by the manufacturer (Qiagen). Protein concentrations were determined by the bicinchoninic acid assay (Pierce).
Antibodies and antisera.
Rat polyclonal antisera directed against fusion proteins were prepared according to a previously published protocol (25). Polyclonal antisera against OspC, monoclonal antibody 14D2-27 against OspA, and monoclonal antibody 8H3-33 against FlaB of B. burgdorferi B31 were previously described (1). Monoclonal antibody 17C3-73 directed against Mlp-9 was generated by immunizing BALB/c mice with the full-length fusion protein according to previously published protocols (1, 31). Sera from groups of B. burgdorferi-infected C3H/HeJ mice were obtained at 0, 2, 4, 8, and 16 weeks after infestation with I. scapularis nymphs harboring B. burgdorferi 297; like sera were pooled and stored at −70°C until use.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Samples for protein analysis were boiled for 10 min in final sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol [vol/vol], 5% [vol/vol] 2-mercaptoethanol, 2.0% SDS, 0.001% [vol/vol] bromophenol blue) prior to electrophoresis through 2.4% stacking and 12.5 or 15% polyacrylamide resolving gels. Gels were then stained with Coomassie brilliant blue. Alternatively, proteins were transferred electrophoretically to a 0.45-μm-pore-size nitrocellulose filter (Schleicher & Schuell, Keene, N.H.) for immunoblotting. Immunoblots were incubated with primary antibodies at the following dilutions: 1:500 for rat Mlp epitope-specific antisera and sera from B. burgdorferi-infected mice; 1:2,000 for all other rat polyclonal antisera; and 1:50 for hybridoma clone supernatants. This was followed by incubations with 1:2,000 dilutions of either goat anti-mouse or goat anti-rat immunoglobulin G (heavy- and light-chain-specific)-horseradish peroxidase conjugates and then, in some experiments (Fig. 7), rabbit anti-goat immunoglobulin G-horseradish peroxidase conjugates (all from Jackson ImmunoResearch, West Grove, Pa.). Immunoblots were developed with 4-chloro-1-naphthol as the substrate.
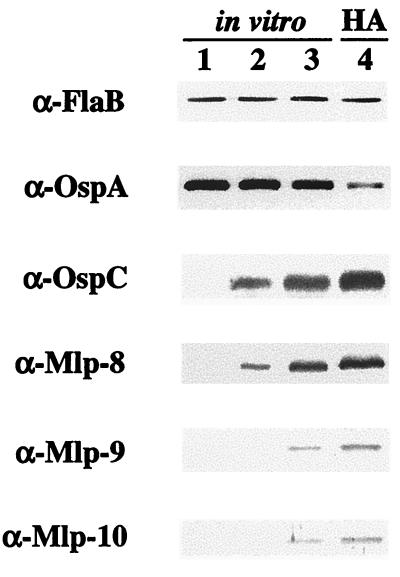
FIG. 7.
Immunoblot analysis of B. burgdorferi. Whole-cell lysates from spirochetes cultivated in vitro at 23°C (lane 1), 34°C (lane 2), or 37°C (lane 3) or within DMCs in rat peritoneal cavities (lane 4) were probed with antibodies directed against the indicated antigens (left). Five micrograms of total protein per gel lane was loaded, except for OspA and OspC, in which case 0.1 μg of protein was used. Antibodies (indicated at the left) used to detect Mlp-8, Mlp-9, and Mlp-10 are those used for Fig. 6; α-Mlp-9 is monoclonal antibody 17C3-73. HA, mammalian host-adapted spirochetes from DMCs implanted in rat peritoneal cavities.
Nucleotide sequence accession numbers.
Nucleotide sequences for the mlp-8, mlp-9, and mlp-10 genes of strain 297 were submitted to GenBank under the accession no. AF046998, AF046999, and AF047000, respectively.
RESULTS
Identification of three new 2.9 loci.
We initially reported on the presence of seven 2.9 loci in B. burgdorferi 297 (29) distributed among two species of circular (supercoiled) plasmids (cp32 and cp18). Based upon the fact that none of the previously reported 2.9 orfD sequences of the seven 2.9 loci completely matched the original orfD probe used initially to identify each locus (29), we assumed that one or more additional 2.9 loci were present in B. burgdorferi 297. In the present study, further screening of a lambda genomic library by using a 32P-labeled probe from a highly conserved region of orfD (29) led to the identification of three other loci, designated 2.9-8, 2.9-9, and 2.9-10 (Fig. 1). Hundreds of additional 2.9 locus-encoding DNA fragments from B. burgdorferi 297 genomic libraries screened via high-throughput automated DNA sequencing did not yield any other unique 2.9 loci.
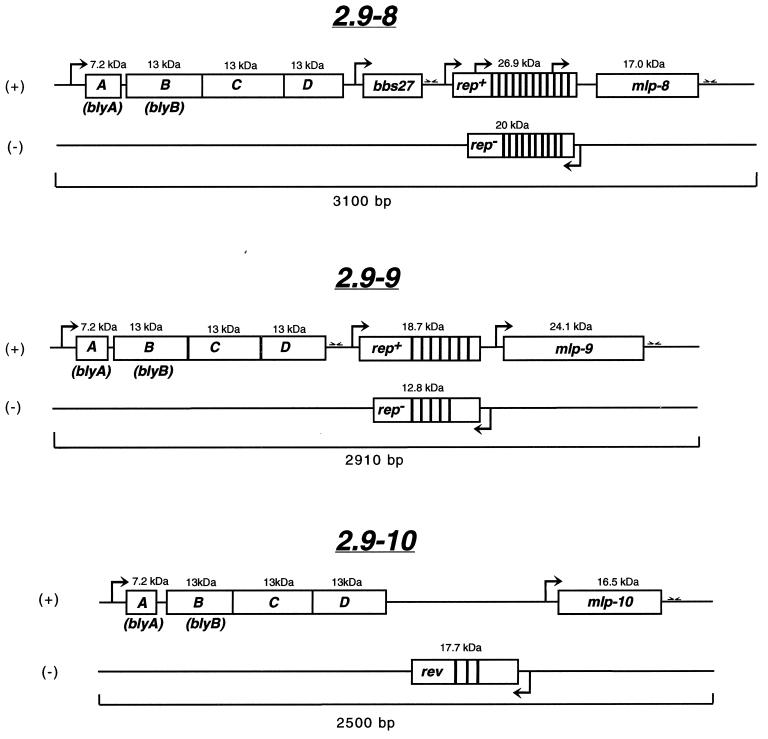
FIG. 1.
Schematic representation of three new 2.9 loci of B. burgdorferi 297. ORFs are shown as boxed regions; the number above each box indicates the size of the polypeptide encoded by each ORF. Promoter-like elements are delineated as arrows in the direction of transcription; stem-loop structures (putative rho-independent terminators) are shown as inverted half arrows. Areas of close vertical lines represent repeat motifs. Positive and negative DNA strands are designated by + and −, respectively. rep, repeat-containing gene; mlp, multicopy lipoprotein; rev, an ORF in the opposite orientation as rep.
As depicted in Fig. 1, like the other 2.9 loci described previously (29), each new 2.9 locus harbored an operon of four highly conserved ORFs (ABCD). Of note, the 2.9-8 and 2.9-10 orfD sequences matched the original orfD sequence first used to detect the 2.9 loci (29). Downstream of the ABCD genes, the 2.9-8 and 2.9-9 loci both contained rep+/rep− genes, whereas the 2.9-10 locus contained a rev gene rather than rep+/rep− genes. Upstream of the rep+ gene in the 2.9-8 locus was a unique ORF not present in any other 2.9 locus; this ORF is identical to a hypothetical ORF, BBS27, which has been localized to the circular plasmid cp32-3 in B. burgdorferi B31 (4a). Further downstream of the rep+/rep− and rev genes of each 2.9 locus were the mlp lipoprotein genes.
The three new Mlp lipoproteins segregate into two classes.
Our interests have been focused largely on characterizing the 2.9 (Mlp) lipoproteins encoded downstream of the rep+/rep− regions (29). We reported previously that these lipoproteins segregate into two distinct classes (I and II) based on size, hydrophilicity, sequence similarities, and reactivity with polyclonal antisera (29). Lipoproteins within each class tend to share 65 to 86% sequence identity, whereas members between the two classes are only 20 to 30% identical (29). In the present study, sequence analysis revealed that Mlp-9 segregated most closely with the class I lipoproteins (70 to 80% identity with other class I lipoproteins), whereas Mlp-8 and Mlp-10 appeared to fall into class II (70 to 86% identity with other class II lipoproteins). This theoretical distinction was supported by immunoblot analyses which showed that polyclonal anti-Mlp-9 antiserum cross-reacted with the class I lipoproteins (Mlp-1, -4, -5, -7B, and -9), whereas polyclonal anti-Mlp-8 and anti-Mlp-10 antisera cross-reacted with the class II lipoproteins (Mlp-2, -3, -7A, -8, and -10) (Fig. 2). Finally, BLAST searches revealed that lipoproteins Mlp-8, -9, and -10 have extensive amino acid homologies with the analogous ORFs in B. burgdorferi B31 (paralogous family 113) (4a, 17); Mlp-9 was 72% identical to ORF BBQ35, and Mlp-8 and Mlp-10 were 69 to 83% identical to ORFs BBL28, BBM28, BBN28, BBO28, BBP28, BBR28, and BBS30. Members of the paralogous family 113 (4a, 17) were noted to be related to the Mlp lipoproteins (29).
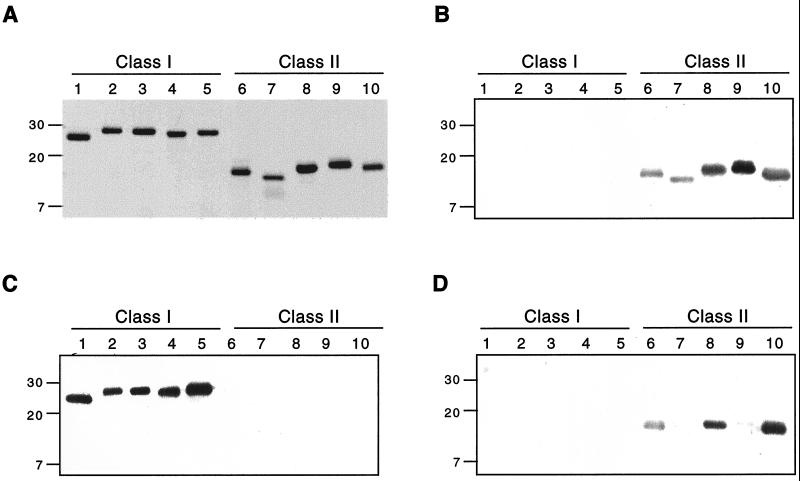
FIG. 2.
Lipoproteins Mlp-8, Mlp-9, and Mlp-10 segregate into two distinct antigenic classes. Purified recombinant versions of the 10 lipoproteins were separated by SDS-PAGE (0.5 μg per gel lane). Numbers at the left represent protein molecular masses in kilodaltons. Lanes 1 through 5, Mlp-1, -4, -5, -7B, and -9 (antigenic class I), respectively; lanes 6 through 10, Mlp-2, -3, -7A, -8, and -10 (antigenic class II), respectively. (A) Gel stained with Coomassie brilliant blue; (B to D) immunoblots of panel A, using rat polyclonal antisera raised against recombinant Mlp-8, Mlp-9, and Mlp-10, respectively. Note that these antisera are cross-reactive with other Mlp members of the same antigenic class.
The three new 2.9 loci are encoded on circular plasmids cp32 and cp18.
We showed previously that the original 2.9 loci were distributed among circular plasmids cp32 and cp18 (29). To localize the new 2.9 loci among the genetic contents of B. burgdorferi 297, Southern blot analyses were performed with radioactive oligonucleotide probes specific for each of the three new mlp genes. Given the high DNA homologies among all of the mlp genes, it first was necessary to confirm the specificities of the oligonucleotide probes used for detecting and localizing individual mlp genes. To accomplish this, a standard dot blot hybridization was carried out; as shown in Fig. 3A, each probe hybridized only with its corresponding template. When used to probe borrelial DNA separated by two-dimensional pulsed-field gel electrophoresis (29), the three probes hybridized to the circular plasmids but not to the linear plasmids (not shown). To identify the circular plasmid(s) containing these genes, supercoiled plasmids of B. burgdorferi were first isolated by two successive rounds of CsCl density gradient centrifugation and then used in Southern blot assays with the probes specific for each mlp gene. As shown in Fig. 3B, probes for mlp-8 and mlp-10 hybridized strongly with the cp32 plasmid(s), whereas the probe for mlp-9 hybridized most intensely with cp18. Some residual hybridization of the mlp-9 probe with cp32, however, also was detectable, possibly due to minor trapping of cp18 within cp32 DNA.
Analyses of gene transcripts.
To assess whether the three new mlp genes were transcribed in B. burgdorferi 297 during in vitro cultivation, total RNA was isolated and quantities of 3 and 30 μg were subjected to Northern blot hybridization. The same specific hybridization probes (Fig. 3A) used to localize the lipoprotein genes to the cp32 and cp18 plasmids (Fig. 3B) were used in these experiments. Probes for mlp-9 and mlp-10 yielded hybridization signals that were only barely detectable even when 30 μg of RNA was probed (Fig. 4). The probe for mlp-10 appeared to hybridize with a single mRNA species of 0.4 kb, consistent with the predicted size of the transcript (Fig. 4, lane 7). The Northern blot for mlp-9, however, revealed a 1.3-kb mRNA species (Fig. 4, lane 5) which did not match the predicted size but rather corresponded to the size of the mRNA if the lipoprotein gene were cotranscribed with its upstream rep+ gene.
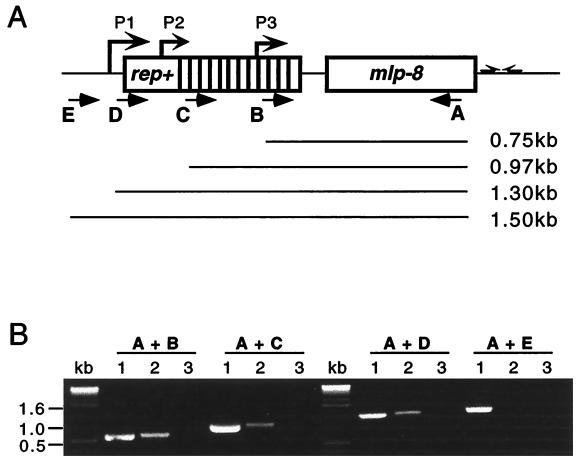
In contrast to mlp-9 and mlp-10, hybridization signals for mlp-8 were strong (Fig. 4), with the lesser amount of RNA (3 μg) clearly revealing more than one mRNA species of about 0.6, 0.9, and 1.3 kb. Interestingly, computer-assisted promoter analysis revealed three potential promoters (P1, P2, and P3 [Fig. 5A]) at about 680, 890, and 1,300 bp upstream of the mlp-8 gene, locations which coincided precisely with results of the Northern blot analyses (Fig. 4). However, because of the high degrees of DNA sequence homologies within the 5′ regions of all of the mlp genes (29), assigning transcriptional initiation sites via primer extension analyses to discern which of these might serve as promoters was not feasible. We thus used RT-PCR as an alternative strategy to corroborate the existence of a mlp-8 transcript(s) larger than 680 bp. The method used one fixed 3′ oligonucleotide primer (primer A) specific for mlp-8 (Table 1); the specificity of primer A was determined by performing PCR with each mlp gene (not shown). Four different 5′ primers (B, C, D, and E), positioned strategically either within or just upstream of the 2.9-8 rep+ region (Fig. 5A), served as 5′ primers; these primers were designed with maximum specificity for the 2.9-8 locus by making them complementary to areas between the repeats of rep+ (Table 1). Using paired combinations of primer A and the other 5′ primers, three of the four RT-PCRs yielded amplification products (Fig. 5B); the exception was the one using primer E positioned 5 bp upstream of the −10 region of the theoretical P1 promoter. These results suggest that the mlp-8 gene may be cotranscribed with its upstream rep+ gene from the theoretical P1 promoter or that multiple promoters are involved; although Northern blotting implied that the mlp-8 gene may be transcribed from three promoters, results of RT-PCR experiments were unable to discern this because the smaller amplicons could have derived from a larger (i.e., A plus D) mRNA template.
FIG. 5.
Evidence that mlp-8 is cotranscribed with its upstream rep+ gene. (A) Schematic of the RT-PCR strategy used for assessing the sizes of mlp-8 transcripts. Four pairs of oligonucleotide primers were used; the 5′ primer for each pair was either primer B, C, D, or E, positioned downstream of putative promoter P3, P2, or P1 or upstream of P1, respectively. The 3′ primer for all amplifications was primer A, which was positioned near the end of the mlp-8 gene. Prospective transcripts are indicated by the solid lines below the schematic. (B) Ethidium bromide-stained amplicons. Lanes: 1, PCR products from B. burgdorferi genomic DNA (positive controls); 2, RT-PCR products from the four RT-PCR reaction combinations (i.e., primers A plus B, A plus C, A plus D, or A plus E); lanes 3, RT-PCRs lacking reverse transcriptase (negative controls).
Expression of the Mlp lipoproteins.
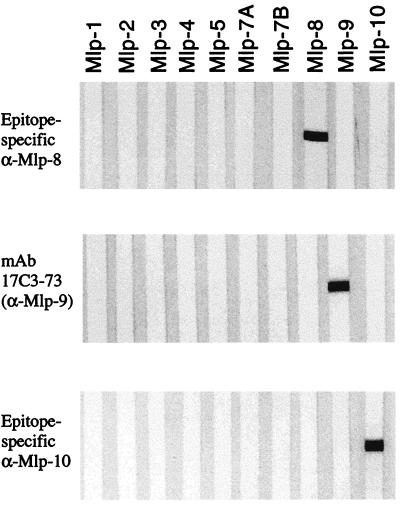
Recently a new animal model was described for studying differential antigen expression by B. burgdorferi as it replicates in a mammalian host-adapted state (1). Using this model, we examined the influence of various B. burgdorferi cultivation conditions on expression (as detected by immunoblotting) of one or more of its Mlp lipoproteins. However, because polyclonal antiserum directed against an individual Mlp tends to be cross-reactive with other Mlp lipoproteins (particularly those within the same antigenic class [Fig. 2]), the first objective was to generate monoclonal or polyclonal antibodies with absolute specificity for each lipoprotein. In the case of Mlp-9, this was accomplished by producing a monoclonal antibody (17C3-73) (Fig. 6). For Mlp-8 and Mlp-10, specific antisera generated against epitope-specific versions of the polypeptides were produced (Fig. 6).
FIG. 6.
Specificities of antibodies directed against Mlp-8, Mlp-9, and Mlp-10. Ten individual purified recombinant Mlp proteins (0.5 μg of each per gel lane) were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed either with rat antisera directed against epitope-specific versions (see text) of Mlp-8 and Mlp-10 or with a monoclonal antibody (mAb; 17C3-73) directed against Mlp-9.
Each of these antibody reagents was used in immunoblots of whole-cell lysates derived from B. burgdorferi cultivated either under various in vitro temperatures (23, 34, or 37°C) or in DMCs implanted in rat peritoneal cavities. Under all conditions examined, the expression of FlaB remained constant and was used as an internal standard for protein loading among the various gel lanes (Fig. 7). Spirochetes cultivated in vitro at 23°C did not display any appreciable amounts of the three Mlp lipoproteins (Fig. 7, lane 1). When these organisms were shifted to 34°C, expression of Mlp-8 was readily apparent, whereas Mlp-9 and Mlp-10 were only barely detectable (lane 2), findings which were all consistent with earlier Northern blot data (Fig. 4). Spirochetes shifted from 23 to 37°C exhibited levels of expression of all three Mlp lipoproteins greater than those observed for organisms grown at 23 or 34°C. These results suggest that increased temperature is a component in the induction of the Mlp lipoproteins. However, expression of all three Mlp lipoproteins by organisms cultivated in DMCs (lane 4) was uniformly higher than in spirochetes cultivated in vitro at 37°C (lane 3); this differential was particularly striking for Mlp-9 and Mlp-10. Thus, elevated temperature alone did not account for the more abundant expression of these lipoproteins in organisms grown in DMCs. As expected (1), OspA, but not flagellin (FlaB), was downregulated within B. burgdorferi replicating in DMCs, whereas OspC was markedly upregulated.
Comparative RT-PCR (Fig. 8) also was performed on 10-fold serial dilutions of RNA as a more sensitive method for assessing gene expression and for correlation with protein expression data (Fig. 7). For the mlp genes, a conserved oligonucleotide primer (mlp-cons-5′) (Table 1) served as the 5′ primer in all RT-PCRs. The 3′ primers (Table 1) were derivatives of the same three oligonucleotides shown to be specific for each mlp gene (Fig. 3A), but some adjustments were made to make the melting temperatures of the primer pairs compatible. The specificity of each primer pair for its corresponding mlp gene was confirmed in separate PCRs using the cloned genes as templates (not shown). At the various quantities of RNA assayed, RT-PCR analyses did not detect transcripts for mlp-8, mlp-9, and mlp-10, using RNA derived from spirochetes cultivated in vitro at 23°C; these RT-PCR results were consistent with the negative immunoblot results of Fig. 7. When spirochetes were cultivated at 34°C, transcripts for mlp-9 and mlp-10 still could not be detected, whereas transcript levels for mlp-8 increased at least 100-fold, findings again consistent with immunoblot results (Fig. 7). Low levels of transcripts, however, for mlp-9 and mlp-10 could be detected when higher amounts of RNA (e.g., 1.0 μg) were used in RT-PCRs (data not shown), consistent with the positive Northern blot results shown in Fig. 4. RT-PCR assays additionally revealed that elevated temperature (37°C) or mammalian host adaptation (organisms cultivated in DMCs) increased the mRNA levels for mlp-8, mlp-9, and mlp-10 at least 1 order of magnitude or more in comparison with levels demonstrable for spirochetes cultivated at 34°C (Fig. 8). Under the same conditions, mRNA levels for flaB and ospA were unchanged and markedly reduced, respectively, as would be predicted from prior studies (1).
B. burgdorferi-infected mice produce antibodies against the Mlp lipoproteins.
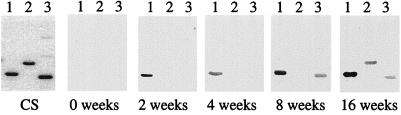
Results from immunoblot and RT-PCR experiments described above implied that one or more of the Mlp lipoproteins should be expressed as spirochetes replicate in mammalian tissues. To examine this, antibody responses of B. burgdorferi-infected mice were used as surrogate markers of lipoprotein expression during spirochetal growth in vivo. Groups of C3H/HeJ mice were infected by infestation with I. scapularis nymphs harboring B. burgdorferi 297. After various intervals postinfection, sera were collected; like sera were pooled and used in immunoblotting experiments with each of the three Mlp lipoproteins. As shown in Fig. 9, antibody reactivity with Mlp-8 was detectable as early as 2 weeks postinfection and became even more intense as the infection progressed to 16 weeks. In contrast, antibodies directed against Mlp-9 and Mlp-10 did not appear until 16 and 8 weeks postinfection, respectively.
FIG. 9.
Tick-inoculated mice infected with B. burgdorferi 297 produce antibodies against the Mlp lipoproteins. Recombinant Mlp-8, -9, and -10 (lanes 1, 2, and 3, respectively) (0.5 μg of protein per gel lane) were separated by SDS-PAGE. The gel was either stained with Coomassie brilliant blue (CS) or transferred to nitrocellulose membranes for immunoblotting. Membranes were immunoblotted with sera harvested from B. burgdorferi-infected (tick-inoculated) C3H/HeJ mice at 0, 2, 4, 8, or 16 weeks post-infection. Note that antibodies directed against Mlp-8 were detectable as early as 2 weeks postinfection, whereas antibodies against Mlp-10 and Mlp-9 were detected at 8 and 16 weeks postinfection, respectively.
DISCUSSION
Elucidating the temporal patterns of antigen expression as B. burgdorferi cycles between its arthropod and mammalian hosts is of paramount importance in understanding the pathogenesis of Lyme disease; differential antigen expression likely underlies events associated with tissue invasion, dissemination, and bacterial chronicity. Moreover, differential antigen expression patterns provide a potential window into the genetics of B. burgdorferi and have profound importance in the selection of new vaccine candidates and the development of new serodiagnostic reagents for Lyme disease. The redundancy of the 2.9 locus (29) led us to hypothesize that one or more of the Mlp lipoproteins may undergo differential antigen expression in B. burgdorferi. As a prelude to exploring this possibility, it first was necessary to identify virtually all of the 2.9 loci in B. burgdorferi 297. Porcella et al. (29) reported on seven 2.9 loci in B. burgdorferi 297; subsequent DNA sequence analyses revealed that the 2.9-6 locus actually was identical to 2.9-1 (unpublished data). Given the presence of two tandem mlp-7A and mlp-7B genes on the 2.9-7 locus (29), a total of six distinct 2.9 loci and seven mlp genes actually were identified in our original study (29). That the 2.9 loci are encoded on as many as nine cp32 plasmids and one or more cp18 plasmid (a naturally occurring truncated species of cp32 [37]) (4a, 5, 39) prompted us to seek and identify the additional 2.9 loci described in this report. Two of these contained orfD genes that were precise matches for the orfD probe used in the initial study of Porcella et al. (29). While this does not preclude the possibility that other, as yet unidentified 2.9 loci exist in B. burgdorferi 297, that extensive screenings yielded the same loci many times over suggests that virtually all of the 2.9 loci of B. burgdorferi 297 were identified in the present study.
Herein we have renamed the 10 lipoprotein genes of the 2.9 loci of B. burgdorferi strain 297 as mlp-1, mlp-2, mlp-3, mlp-4, mlp-5, mlp-7A, mlp-7B, mlp-8, mlp-9, and mlp-10. There is no mlp-6 gene because the original 2.9-6 locus is identical to 2.9-1. Seven of mlp genes are encoded on cp32 plasmids, whereas two (mlp-3 and mlp-9) are on cp18. Five of the corresponding lipoproteins fall into each of the two antigenic classes (in class I, Mlp-1, Mlp-4, Mlp-5, Mlp-7B, and Mlp-9; in class II, Mlp-2, Mlp-3, Mlp-7A, Mlp-8, and Mlp-10). Interestingly, analysis of available DNA sequence information suggests that seven of the Mlp paralogs in B. burgdorferi B31, segregate into antigenic class II whereas only one Mlp paralog (BBQ35; encoded on lp56) (4a) belongs to antigenic class I.
As in the study of Porcella et al. (29), Northern blot analysis herein established that the basal levels of transcription among the mlp-8, mlp-9, and mlp-10 genes varied considerably. Immunoblots using antibodies specific for each Mlp confirmed that such variation also existed at the protein level. These findings suggest that despite structural relatedness, members of the Mlp family may not function equivalently in the zoonotic life cycle of B. burgdorferi, or they may even carry out disparate function(s). Regardless, the mechanism(s) for variability in the levels of Mlp expression remains unclear but is somewhat paradoxical given that all three Mlp genes were uniformly inducible by either temperature shift in vitro or adaptation during growth of B. burgdorferi in DMCs. The theoretical 5′ promoter regions for all of the mlp genes are highly similar, often differing by only one or a few nucleotides (29). Such similarities have precluded using transcriptional initiation approaches (e.g., primer extension analyses) as a strategy to assist with mlp promoter assignments. In this study, combined Northern blot and RT-PCR data suggested that the mlp-8 and mlp-9 genes may utilize multiple promoters and/or may be cotranscribed with the upstream rep+ gene. The significantly greater nucleotide sequence diversity of the rep+ region than of the 5′ regions of the mlp genes may engender different promoter strengths (11, 26) that account for variable basal expression levels of the mlp genes. Additional work will be required to address this possibility.
The mlp genes overall were less efficiently expressed when B. burgdorferi was cultivated at lower temperature (i.e., 23°C), a condition likely analogous to spirochetes within ticks. When the in vitro growth temperature for B. burgdorferi was shifted to 34 or 37°C, conditions which ostensibly mimic temperature transitions during tick feeding on a mammalian host, there were marked increases in the expression of all three mlp genes. These increases were reflected both at the transcript (RT-PCR) and protein (immunoblot) levels. These observations suggest that at least three mlp genes share a similar regulatory component, and thus our data support the hypothesis that the mlp genes undergo a form of global upregulation in response to elevated temperature.
In other bacterial systems, temperature-mediated regulation occurs at the levels of both transcription and translation; changes in mRNA conformation, protein conformation, and DNA supercoiling can all modulate gene expression (24). In the case of DNA supercoiling, the H-NS protein, a histone-like protein with the ability to affect DNA supercoiling (22, 23), is an important effector of transcription. Thus far, an H-NS homolog has not been identified in B. burgdorferi. Regarding thermoregulation, Schwan et al. (32) and Stevenson et al. (38) were the first to show that OspC is induced upon temperature shift from 23°C to 35 or 37°C. The decorin-binding protein A of B. burgdorferi also is upregulated by temperature (6). Of particular relevance to the present study, Stevenson et al. (36) reported that eight OspE/F-related proteins (erp genes), also encoded on the cp32 (one encoded on lp56) plasmids of B. burgdorferi B31, are expressed at various levels at 23°C but are all upregulated at 35°C. Stevenson et al. (38) and Akins et al. (1) also showed that OspE and OspF of B. burgdorferi N40 and 297 are upregulated by temperature shift from 23°C to 35 or 37°C. Although recent analyses suggest that OspE, OspF, and OspE/F-related proteins (Erps) are more distantly related than initially appreciated (2), that they are all encoded on homologs of cp32 and cp18 invites the provocative hypothesis that other genes on the cp32 or cp18 plasmids may be globally upregulated in response to temperature shift. A variation on this theme might be that selected cp32 or cp18 plasmids have overall higher basal expression levels of their genes, perhaps modulated by degrees of supercoiling, thereby giving rise to an asynchronous expression pattern of antigens encoded on certain circular plasmids. In this regard, it is noteworthy that OspF and Mlp-8, which are approximately 10 kb apart on the same cp32 plasmid (cp32-3) of B. burgdorferi 297 (2), are expressed at relatively higher levels than their homologous family members (1) (Fig. 7). p21 and Mlp-9, which are about 10 kb apart on the same cp18 plasmid (cp18-2) of B. burgdorferi 297 (2), both are expressed at very low basal levels or are undetectable (2) (Fig. 7). Additional linkage and expression data for all of the mlp and other genes encoded on the cp32 and cp18 plasmids, however, will be required before firm conclusions can be drawn regarding plasmid-based mechanisms of gene regulation.
The cultivation of B. burgdorferi in DMCs implanted intraperitoneally into rats has been an important advance toward assessing differential antigen expression by B. burgdorferi in a mammalian host-adapted state (1). As in the prior study (1), we again observed that OspA was sharply downregulated by spirochetes cultivated in DMCs. In contrast, however, both comparative RT-PCR and immunoblot procedures indicated that spirochetes cultivated in DMCs expressed even higher levels of Mlp lipoproteins (particularly Mlp-8) than those cultivated in vitro under temperature-shifted conditions. These findings underscore that temperature induction alone does not appear to account for maximal levels of mlp gene expression and that some other, as yet undefined factor(s) modulates further upregulation of these genes during mammalian infection. Although it is not yet completely clear to what extent DMC-cultivated spirochetes precisely resemble those in mammalian tissue (1), the rat peritoneal chamber model will continue to be instrumental in deciphering at least some host factors that contribute to the upregulation of mlp and other genes of B. burgdorferi replicating in a mammalian host-adapted state.
Data provided in this study prompt a working model for the temporal expression of the Mlp lipoproteins as B. burgdorferi transitions from its arthropod vector into mammalian tissues. Low levels of certain Mlp lipoproteins (e.g., Mlp-9 and Mlp-10) imply that at least some of the 2.9 lipoproteins are not essential for B. burgdorferi survival in ticks. However, during engorgement, it is likely that increasing temperature within nymph midguts induces mlp gene expression to levels that culminate in lipoprotein synthesis sufficient for a newly needed physiological function(s). The net effect of such a process(es) could be viewed as an alternative form of differential antigen expression that is not predicated on a reciprocal upregulation/downregulation process, such as that embodied in the OspC/OspA paradigm (27, 32). This hypothesis, however, remains to be tested further by examining the expression of the Mlp lipoproteins within B. burgdorferi-infected tick midguts before and after feeding. That tick-inoculated mice produced antibodies against Mlp-8 relatively early (2 weeks) postinfection, however, was consistent with the early temporal expression of at least one or more of the Mlp lipoproteins. We cannot conclude definitively which of the Mlp lipoproteins may be expressed early during mouse infection because immunoblot data for Mlp-8 were subject to possible misinterpretation; mouse antibodies elicited after tick challenge may have been cross-reactive with another Mlp, particularly one within the same antigenic class. However, spirochetes harvested from DMCs and probed with antibodies of defined specificities for Mlp-8, Mlp-9, and Mlp-10 showed conclusively that all three were expressed during the replication of B. burgdorferi within rat peritoneal cavities, with Mlp-8 being the most abundant. It is anticipated that the further use of B. burgdorferi harvested from DMCs and the development of additional antibody probes with defined specificity for each Mlp will assist in obtaining a more complete differential expression pattern for all of the Mlp lipoproteins. Last, it is noteworthy that as the infection of mice progressed to 8 and 16 weeks postinfection, antibodies to additional Mlp lipoproteins (e.g., Mlp-10 and Mlp-9) appeared, consistent with our contention that one or more of the lower-abundance Mlp lipoproteins eventually are expressed during progression of the mammalian infection.
A central question surrounding the mlp gene family is to what extent Mlp lipoproteins may be surface exposed in B. burgdorferi, thereby potentially serving as adjuncts to the current Lyme disease vaccine. The recently approved OspA human Lyme disease vaccine represents an important medical advance. However, as reviewed by Steigbigel and Benach (35), a number of unresolved issues remain concerning this first-generation monovalent vaccine. One way of potentially enhancing the efficacy of the current OspA Lyme disease vaccine would be to expand the number of vaccinogens to include one or more also expressed during the mammalian phase of infection, particularly during the early phase, thereby providing immune targets during both phases of the zoonotic life cycle of B. burgdorferi. Given its high basal expression level, upregulation by temperature shift and the mammalian environment, and evidence of an early antibody response in the murine model of Lyme borreliosis, Mlp-8 would be an excellent candidate vaccinogen. In this regard, preliminary studies recently have shown that when B. burgdorferi was cultivated in vitro at 37°C, four Mlp lipoproteins tested (Mlp-4, Mlp-5, Mlp-7A, and Mlp-8) were sensitive to proteinase K digestion under conditions which left flagellin intact, implying that at least some Mlp lipoproteins are surface exposed in B. burgdorferi. Moreover, preliminary vaccine experiments with a multivalent formulation of Mlp-2, Mlp-3, Mlp-7A, Mlp-8, and Mlp-10 (antigenic class II) gave rise to 80% protection of mice, a level comparable to what has been observed for the decorin-binding protein (20, 21). These provocative findings give impetus for assessing further the temporal expression patterns, surface exposure, and vaccinogenic potentials for all of the Mlp lipoproteins.
ACKNOWLEDGMENTS
X.Y. and T.G.P. contributed equally to this work.
We thank Deborah Bouis, Ranjit Deka, and Anette Huebner for helpful discussions and Martin Goldberg, Charis Lawrenson, and Hsiao-Ching Yen for excellent technical assistance.
We gratefully acknowledge funding for this work provided by the Centers for Disease Control and Prevention (U50/CCU614875), the Arthritis Foundation (via the Engalitcheff Research Fund), the Robert A. Welch Foundation (grant I-0940), and grants AI-45538 and AI-29735 from the Lyme disease program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Investig. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins D R, Caimano M J, Yang X, Cerna F, Norgard M V, Radolf J D. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect Immun. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins D R, Porcella S F, Popova T G, Shevchenko D, Li M, Norgard M V, Radolf J D. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol Microbiol. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- 4.Aron L, Toth C, Godfrey H P, Cabello F C. Identification and mapping of a chromosomal gene cluster of Borrelia burgdorferi containing genes expressed in vivo. FEMS Microbiol Lett. 1996;145:309–314. doi: 10.1111/j.1574-6968.1996.tb08594.x. [DOI] [PubMed] [Google Scholar]
- 4a.Borrelia burgdorferi Genome Database. 7 July 1999, revision date. [Online.] http://www.tigr.org/tdb/mdb/bbdb/bbdb.html. The Institute for Genomic Research, Rockville, Md. [24 September 1999, last date accessed].
- 5.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassatt D R, Patel N K, Ulbrandt N D, Hanson M S. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect Immun. 1998;66:5379–5387. doi: 10.1128/iai.66.11.5379-5387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Lyme disease—United States, 1996. Morbid Mortal Weekly Rep. 1997;46:531–535. [PubMed] [Google Scholar]
- 8.Champion C I, Blanco D R, Skare J T, Haake D A, Giladi M, Foley D, Miller J N, Lovett M A. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect Immun. 1994;62:2653–2661. doi: 10.1128/iai.62.7.2653-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox D L, Akins D R, Bourell K W, Lahdenne P, Norgard M V, Radolf J D. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc Natl Acad Sci USA. 1996;93:7973–7978. doi: 10.1073/pnas.93.15.7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Barthold S W, Giles S S, Montgomery R R, Telford S R, Fikrig E. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J Clin Investig. 1997;99:987–995. doi: 10.1172/JCI119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lorenzo V, Perez-Martin J. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol Microbiol. 1996;19:1177–1184. doi: 10.1111/j.1365-2958.1996.tb02463.x. [DOI] [PubMed] [Google Scholar]
- 12.de Silva A M, Zeidner N S, Zhang Y, Dolan M C, Piesman J, Fikrig E. Influence of outer surface protein A antibody on Borrelia burgdorferi within feeding ticks. Infect Immun. 1999;67:30–35. doi: 10.1128/iai.67.1.30-35.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.deSilva A M, Telford S R, Brunet L R, Barthold S W, Fikrig E. Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J Exp Med. 1996;183:271–275. doi: 10.1084/jem.183.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fikrig E, Chen M, Barthold S W, Anguita J, Feng W, Telford S R, Flavell R A. Borrelia burgdorferi erpT expression in the arthropod vector and murine host. Mol Microbiol. 1999;31:281–290. doi: 10.1046/j.1365-2958.1999.01171.x. [DOI] [PubMed] [Google Scholar]
- 16.Fikrig E, Feng W, Aversa J, Schoen R T, Flavell R A. Differential expression of Borrelia burgdorferi genes during erythema migrans and Lyme arthritis. J Infect Dis. 1998;178:1198–1201. doi: 10.1086/515684. [DOI] [PubMed] [Google Scholar]
- 17.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 18.Gilmore R D, Mbow M L. A monoclonal antibody generated by antigen inoculation via tick bite is reactive to the Borrelia burgdorferi Rev protein, a member of the 2.9 gene family locus. Infect Immun. 1998;66:980–986. doi: 10.1128/iai.66.3.980-986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guina T, Oliver D B. Cloning and analysis of a Borrelia burgdorferi membrane-interactive protein exhibiting haemolytic activity. Mol Microbiol. 1997;24:1201–1213. doi: 10.1046/j.1365-2958.1997.4291786.x. [DOI] [PubMed] [Google Scholar]
- 20.Hagman K E, Lahdenne P, Popova T G, Porcella S F, Akins D R, Radolf J D, Norgard M V. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect Immun. 1998;66:2674–2683. doi: 10.1128/iai.66.6.2674-2683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson M S, Cassatt D R, Guo B P, Patel N K, McCarthy M P, Dorward D W, Hook M. Active and passive immunity against Borrelia burgdorferi decorin binding protein A (DbpA) protects against infection. Infect Immun. 1998;66:2143–2153. doi: 10.1128/iai.66.5.2143-2153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins C F, Hinton J C D, Hulton C S J, Owen-Hughes T, Pavitt G D, Seirafi A. Protein H1: a role for chromatin structure in the regulation of bacterial gene expression and virulence? Mol Microbiol. 1990;4:2007–2012. doi: 10.1111/j.1365-2958.1990.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 23.Hulton C S J, Seirafi A, Hinton J C D, Sidebotham J M, Waddell L, Pavitt G D, Owen-Hughes T, Spassky A, Buc H, Higgins C F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 24.Hurme R, Rhen M. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol Microbiol. 1998;30:1–6. doi: 10.1046/j.1365-2958.1998.01049.x. [DOI] [PubMed] [Google Scholar]
- 25.Lahdenne P, Porcella S F, Hagman K E, Akins D R, Popova T G, Cox D L, Katona L I, Radolf J D, Norgard M V. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect Immun. 1997;65:412–421. doi: 10.1128/iai.65.2.412-421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery R R, Malawista S E, Feen K J M, Bockenstedt L K. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J Exp Med. 1996;183:261–269. doi: 10.1084/jem.183.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollack R J, Telford S R, Spielman A. Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol. 1993;31:1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Promoter Prediction by Neural Network. 22 September 1998, revision date. [Online.] http://www-hgc.lbl.gov/projects/promoter.html. [24 September 1999, last date accessed.]
- 30.Ramamoorthy R, Philipp M T. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson S M, Kettman J R, Miller J N, Norgard M V. Murine monoclonal antibodies specific for virulent Treponema pallidum (Nichols) Infect Immun. 1982;36:1076–1085. doi: 10.1128/iai.36.3.1076-1085.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinsky R J, Piesman J. Ear punch biopsy method for detection and isolation of Borrelia burgdorferi from rodents. J Clin Microbiol. 1989;27:1723–1727. doi: 10.1128/jcm.27.8.1723-1727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steere A C. Borrelia burgdorferi (Lyme disease, Lyme borreliosis) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone Inc.; 1995. pp. 2143–2155. [Google Scholar]
- 35.Steigbigel R T, Benach J L. Immunization against Lyme disease—an important first step. N Engl J Med. 1998;339:263–264. doi: 10.1056/NEJM199807233390409. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson B, Casjens S, van Vugt R, Porcella S F, Tilly K, Bono J L, Rosa P. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J Bacteriol. 1997;179:4285–4291. doi: 10.1128/jb.179.13.4285-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suk K, Das S, Sun W, Jwang B, Barthold S W, Flavell R A, Fikrig E. Borrelia burgdorferi genes selectively expressed in the infected host. Proc Natl Acad Sci USA. 1995;92:4269–4273. doi: 10.1073/pnas.92.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theisen M. Molecular cloning and characterization of nlpH, encoding a novel, surface-exposed, polymorphic, plasmid-encoded 33-kilodalton lipoprotein of Borrelia afzelii. J Bacteriol. 1996;178:6435–6442. doi: 10.1128/jb.178.22.6435-6442.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallich R, Brenner C, Kramer M D, Simon M M. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]