Abstract
Citrobacter rodentium is the causative agent of transmissible murine colonic hyperplasia and contains a locus of enterocyte effacement (LEE) similar to that found in enteropathogenic Escherichia coli (EPEC). EPEC espB is necessary for intimate attachment and signal transduction between EPEC and cultured cell monolayers. Mice challenged with wild-type C. rodentium develop a mucosal immunoglobulin A response to EspB. In this study, C. rodentium espB has been cloned and its nucleotide sequence has been determined. C. rodentium espB was found to have 90% identity to EPEC espB. A nonpolar insertion mutation in C. rodentium espB was constructed and used to replace the chromosomal wild-type allele. The C. rodentium espB mutant exhibited reduced cell association and had no detectable fluorescent actin staining activity on cultured cell monolayers. The C. rodentium espB mutant also failed to colonize laboratory mice following experimental inoculation. The espB mutation could be complemented with a plasmid-encoded copy of the gene, which restored both cell association and fluorescent actin staining activity, as well as the ability to colonize laboratory mice. These studies indicate that espB is necessary for signal transduction and for colonization of laboratory mice by C. rodentium.
Citrobacter rodentium (formerly C. freundii biotype 4280) and enteropathogenic Escherichia coli (EPEC) infections are characterized by epithelial cell hyperproliferation similar to that seen in human proliferative bowel disorders including Crohn's disease and ulcerative colitis (3, 35). Individuals with these diseases are known to suffer an increased risk and early onset of colorectal cancer (27). In mice experimentally infected with C. rodentium, hyperplasia is detectable as early as 4 days postinfection, with maximal hyperplasia occurring 2 to 3 weeks later. Prior to the development of maximal hyperplasia, attaching and effacing (AE) lesions are present in the descending colon (17). These lesions are characterized by dissolution of the brush border, cupping of the adherent bacteria by the epithelial cell plasma membrane, and cytoskeletal rearrangements in the underlying enterocyte cytoplasm that lead to disruption of the terminal web (17). These histologic changes are indistinguishable from AE lesions produced by EPEC (31). In adult mice, colonic hyperplasia eventually regresses, with the colon returning to normal 6 to 8 weeks postinoculation. Suckling mice experience secondary inflammation of the colon that is associated with retarded growth, soft feces, and greater than 50% mortality (4).
EPEC causes diarrhea in infants and in young children (10). Initially, adherence to epithelial cells by EPEC occurs via a plasmid-encoded bundle-forming pilus (16). This first step is followed by the triggering of epithelial signal transduction pathways that leads to the reorganization of filamentous actin (24). Signaling from EPEC to epithelial cells results in tyrosine phosphorylation of substrates that colocalize with the accumulated actin underneath adherent bacteria (33). The major phosphorylation substrate detected in EPEC-infected cells is the bacterial translocated intimin receptor (Tir), which is targeted to the host cell membrane, where it becomes the receptor for the EPEC adhesin intimin (21). EPEC also induces other signaling cascades including fluxes of inositol phosphates (15), changes in membrane potential (41), and activation of protein kinase C (8), phospholipase-Cγ (20), and NF-κB (36). The end result of these cascades is the formation of pedestal-like AE lesions composed largely of filamentous actin under the cell associated bacteria (31).
Both C. rodentium and EPEC contain a 35-kb pathogenicity island known as the locus for enterocyte effacement (LEE) (28). The nucleotide sequence of the entire LEE in EPEC has been determined and has been shown to contain 41 potential open reading frames (ORFs) (12). These include eaeA (intimin) (28); tir (21); the esc genes (12), a group of genes which encode a type III secretion system; as well as espA (22), espB (11, 14), and espD (25). Several studies have confirmed that EspB is targeted to the host cytoplasm (23, 42, 46). EspB has also been shown to be required for translocation of Tir to the host cell membrane (21). Both EspA and EspD are required for EspB translocation, and both have been implicated as components of a surface organelle involved in the delivery of EspB to the cytoplasm (23, 42, 43). The LEE has been found in enterohemorrhagic Escherichia coli (EHEC) O157:H7, rabbit EPEC strains including RDEC-1, and pathogenic strains of E. coli previously designated Hafnia alvei (13). While all of these bacterial species contain a version of the LEE, none have been shown to colonize the intestinal tract of laboratory rodents or to be associated with mucosal hyperplasia in rodents. In a previous study, a C. rodentium eaeA mutant strain was constructed. This strain does not colonize and does not cause AE lesion formation (38).
In this study we have attempted to better understand the role played by AE lesion formation in the induction of hyperplasia by C. rodentium. Whether AE activity is sufficient to induce mucosal hyperplasia or whether C. rodentium has other specialized virulence determinants required for the induction of mucosal hyperplasia is still not clear. C. rodentium espB was selected for study because EspB is required for AE lesion formation in EPEC and is the only protein secreted by EPEC that has been shown to be targeted to the host cell cytoplasm.
MATERIALS AND METHODS
Strains and plasmids.
Bacteria were stored at −80°C in Luria-Bertani (LB) broth containing 50% (vol/vol) glycerol. The bacteria were grown at 37°C in LB broth or on LB agar. For maintenance of recombinant plasmids, strains were grown in medium supplemented with appropriate antibiotics (ampicillin at a final concentration of 100 μg/ml, kanamycin at 40 μg/ml, chloramphenicol at 30 μg/ml, naladixic acid at 20 μg/ml, and tetracycline at 20 μg/ml). Colonies were assayed for β-galactosidase activity on LB agar supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) as a chromogenic substrate and isopropyl-β-d-thiogalactoside (IPTG) as an inducer, both at final concentrations of 32 μg/ml. The strains and plasmids used in this study are described in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description | Source or reference |
|---|---|---|
| Strains | ||
| DBS100 | C. rodentium ATCC 51459 | 37 |
| DBS255 | DBS100 eaeA mutant; Kanr | 38 |
| DBS506 | DBS100 espB merodiploid; Kanr | This study |
| DBS578 | DBS100 espB mutant; Kanr | This study |
| DBS586 | DBS578 espB merodiploid; Kanr | This study |
| DBS587 | DBS578 espB revertant; Kans | This study |
| JPN15 | Plasmid-cured derivative of EPEC E2348/69 | 26 |
| DH5α | F− ϕ80dlacZΔM15 recA hsdR supE thi gyrA Δ(lacZYAargF) | BRLa |
| SM10 (λpir) | thi thr leu tonA lacY supE recA::RP4-2-Tcr::Mu Kmr λpir RK6 | 30 |
| Plasmids | ||
| pDBS2 | Cosmid clone containing DBS100 DNA with espB homology; Ampr | 38 |
| pUC18K | pUC18 derivative with 850-bp nonpolar aphA-3 cassette; Ampr Kanr | 29 |
| pBAZ391 | pFSV-1 derivative with nptI-sacB-sacR cartridge; Cmr Kanr | This study |
| pBAZ463 | 1.0-kb HindIII subclone of pDBS2; Ampr | This study |
| pBAZ481 | 1.1-kb HindIII subclone of pDBS2; Ampr | This study |
| pBAZ503 | pSK(−) containing a 3-kb PvuII espB nonpolar mutant cassette; Ampr Kanr | This study |
| pBAZ504 | PvuII fragment from pBAZ503 cloned into pBAZ391; Cmr Kanr | This study |
| pJVN106 | 1,926-bp NaeI-BglII espB clone; Ampr | This study |
| pJVN109 | 1,972-bp XhoI-BglII fragment of pJVN106 cloned into pFSV-1; Cmr | This study |
| pJVN111 | 1,972-bp XhoI-BglII fragment of pJVN106 cloned into pACYC184; Cmr | This study |
| Primers | ||
| BAZ104 | 5′ GGCGCGAGTGATGTCGC 3′ | 39 |
| BAZ105 | 5′ AGCGAGCCGCTTGCCCC 3′ | 39 |
| ESPB1 | 5′ TGAGACAGTTGGCACATTGC 3′ | This study |
| ESPB2 | 5′ TGGTGGTACAACTCTTCGAGC 3′ | This study |
| aphA-3out | 5′ CGAAAACTGGGAAGAAGACAC 3′ | This study |
BRL, Bethesda Research Laboratories.
Animals.
Three-week-old outbred Swiss Webster mice (Taconic Laboratories, Germantown, N.Y.), were housed in microisolator caging within a facility approved by the Association for Assessment and Accreditation of Laboratory Animal Care. The mice were maintained on pelleted rodent chow and water ad libitum. All experiments were approved by the Massachusetts Institute of Technology Institutional Animal Care and Use Committee. Mice were orally inoculated with either 100 μl of an overnight culture of bacteria (approximately 5 × 108 CFU) or 100 μl of sterile LB broth. At predetermined time points, feces were collected from individual animals, weighed, homogenized in sterile phosphate-buffered saline, and plated on MacConkey lactose agar with or without the appropriate antibiotics. Overall colonization levels were measured by determining the number of CFU of C. rodentium per gram of feces at 3, 5, and 7 days postinoculation. At 10 days postinoculation, the animals were euthanized. The entire colon was collected aseptically and visually inspected for evidence of hyperplasia. The weight of the colon was determined, and the colon was then homogenized as described previously (37). Appropriate dilutions of the tissue homogenate were plated on differential media with and without antibiotic selection to determine the number of CFU of C. rodentium per gram of tissue. The lower limit of detection was 1 CFU/mg of tissue.
Recombinant DNA methods.
Chromosomal DNA was isolated as described previously (39). Plasmid DNA was isolated using Qiagen tips as recommended by the manufacturer (Qiagen Inc., Chatsworth, Calif.). DNA ligation, restriction endonuclease digestion, and gel electrophoresis were performed by standard methods (34). Enzymes were purchased from New England Biolabs, Inc. (Beverly, Mass.). Plasmid DNA was introduced into E. coli and C. rodentium by high-voltage electroporation with a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.).
espB probe.
PCR primers for espB were designed by using the espB sequence of EPEC E2348/69 (11, 22). The espB probe was generated with primers BAZ104 and BAZ105 and chromosomal DNA from EPEC JPN15 as previously described (39).
Cloning espB.
pDBS2 cosmid DNA (38) was isolated and digested with HindIII. Digested DNA fragments were ligated with either pBluescript SK(−) or pBluescript KS(−) (Stratagene, La Jolla, Calif.) that had been linearized with HindIII. The ligation mixture was used to transform DH5α, and bacterial clones containing recombinant plasmids were recognized as white colonies when plated on LB agar with ampicillin supplemented with X-Gal and IPTG. Plasmid DNA was isolated from a number of these clones, and Southern blot analysis was performed.
Southern blot analysis.
Digested genomic and plasmid DNA fragments were separated by electrophoresis on a 0.75% agarose gel. DNA fragments were transferred to a nylon membrane (Hybond-N+; Amersham Corp., Arlington Heights, Ill.) by the capillary method as described previously (34). Membranes were UV cross-linked (Stratalinker; Stratagene). The PCR-amplified espB fragment was directly labeled with an ECL chemiluminescence detection kit (Amersham) for use as the probe. Membranes were hybridized with probe overnight and washed at high stringency (twice for 20 min in 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at 55°C, followed by twice for 5 min in 20× SSC at room temperature). After addition of detection reagents, positive signal was detected by exposure to radiographic film (Hyperfilm-ECL; Amersham).
Nucleotide sequence determination.
The nucleotide sequence of the two HindIII fragments containing espB homology was determined. Initial DNA sequences from clones containing these inserts were determined by the dideoxy-chain termination method with Sequenase version 2.0 (U.S. Biochemicals, Naperville, Ill.). The remaining sequences were determined at the Massachusetts Institute of Technology Biopolymer Laboratory with a DyeDeoxy Terminator cycle sequencing kit and a model 373A DNA sequencer (Applied Biosystems, Foster City, Calif.).
Construction of a nonpolar insertional espB mutant.
The two HindIII fragments containing espB homology, pBAZ463 and pBAZ481, were used to construct a mutant espB allele. To avoid the polar effects associated with insertional mutations, the aphA-3 nonpolar cassette developed by Ménard et al. (29) was used to disrupt the gene. The aphA-3 cassette contains a kanamycin resistance gene preceded by translation stop codons in all three reading frames and is immediately followed by a consensus ribosomal binding site and a start codon. A 283-bp EcoRV deletion was made in the 5′ end of the espB fragment in pBAZ481 so that when the 850-bp aphA-3 cassette was cloned upstream into the SacI-PstI sites, the 3′ end of espB would be expressed by the downstream start codon of the nonpolar cassette. A 2.2-kb ScaI-SacI fragment containing the 5′ portion of espB from pBAZ463 was cloned upstream, generating the espB nonpolar insertion mutation designated pBAZ503. The construct was confirmed to contain a nonpolar insertion of aphA-3 by sequencing. A 3-kb PvuII fragment containing the espB nonpolar insertion mutation from pBAZ503 was cloned into pBAZ391, a pFSV-1 derivative containing the nptI-sacB-sacR cassette, to generate the suicide vector pBAZ504. pBAZ504 was transformed into SM10(λpir) for maintenance.
The suicide plasmid pBAZ504 was mated into C. rodentium as previously described (38). Since plasmids with the R6K replicon cannot replicate in the absence of the pir-encoded π protein (30), kanamycin-resistant exconjugates represented merodiploid strains into which the plasmid had been integrated by homologous recombination. A physical representation of the strains used in this study is shown in Fig. 1. Total bacterial DNA from an individual clone, designated DBS506, was screened by Southern blot analysis to confirm homologous recombination of the suicide plasmid into the C. rodentium chromosome (see Fig. 2). The suicide vector used, pBAZ504, contained a copy of sacB, expression of which has been shown to be lethal for some gram-negative organisms grown in the presence of 5% sucrose (29). However, no growth disadvantage of the strain was seen under these conditions. Therefore, to isolate an espB mutant, the merodiploid DBS506 was screened for homologous recombination resulting in loss of the wild-type espB allele and adjacent vector sequences. Single colonies were patched onto LB agar containing either chloramphenicol or kanamycin to identify isogenic espB mutant strains. One exconjugate, designated BAZ578, was selected for further characterization. The DNA sequence of the mutant espB allele was sequenced with primers ESPB1, ESPB2, and aphA-3out.
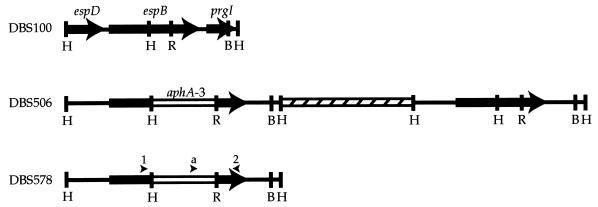
FIG. 1.
Physical maps of the espB region of the C. rodentium strains used in this study. Heavy lines represent C. rodentium DNA, the open box represents aphA-3, and the hatched box represents plasmid pBAZ391 (not to scale). The arrows represent the ORFs of espD, espB, and the prgI homolog. Arrowheads represent the binding sites of ESPB1 (arrowhead 1), ESPB2 (arrowhead 2), and aphA-3out (arrowhead a) used in sequencing. Abbreviations: B, BglII; R, EcoRV; and H, HindIII.
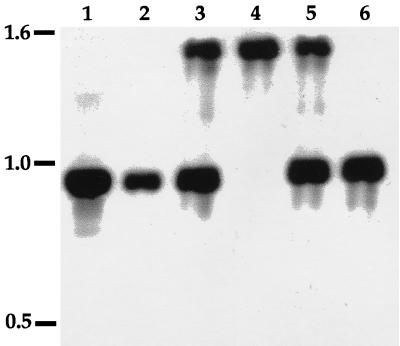
FIG. 2.
Southern analysis of C. rodentium wild-type and mutant strains, probed with the 0.9-kb BglII-HindIII fragment of pBAZ481 containing the 3′ end of the espB gene. DNA was digested with BglII and HindIII. Lanes: 1, pBAZ481; 2, wild-type DBS100; 3, espB merodiploid DBS506; 4, espB mutant DBS578; 5, espB merodiploid DBS586; 6, espB+ DBS587. Numbers on the left are sizes in kilobases.
trans complementation and replacement of the mutant allele.
The 1,020-bp NaeI-HindIII fragment of pBAZ463, containing the 5′ end and promoter sequences of espB, and the 906-bp BglII-HindIII fragment from pBAZ481, containing the 3′ end of espB, were cloned into pSP72 digested with PvuII and BglII to give pJVN106. The 1,972-bp XhoI-BglII fragment of pJVN106 was cloned into pACYC184 digested with BamHI and SalI to generate pJVN111. In addition to complementing the espB mutation in trans, the marked deletion in the mutant strain was replaced with the wild-type allele of espB. To accomplish this, the 1,972-bp XhoI-BglII fragment of pJVN106 pJVN106 was treated with the Klenow fragment of DNA polymerase I and ligated to pFSV-1 (7) which had been linearized with EcoRV, generating pJVN109. The suicide plasmid pJVN109 was transformed into espB mutant DBS578. Chloramphenicol-resistant transformants represented merodiploid strains into which the plasmid had been integrated by homologous recombination. Using the same strategy employed to generate an isogenic espB mutant, a strain carrying the espB wild-type allele was isolated and confirmed by Southern blot analysis (see Fig. 2).
SDS-PAGE analysis of secreted proteins.
A modification of the method of Kenny et al. was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (18). Briefly, C. rodentium strains were grown overnight, washed in salt-free medium, and diluted 1:100 into Dulbecco's modified Eagle's medium (DMEM) containing 0.1 M HEPES (pH 7.4). The bacteria were incubated standing at 37°C until the optical density of the culture at 600 nm reached 0.5 and were then removed by centrifugation at 16,000 × g for 5 min. Supernatant proteins were precipitated by the addition of 10% trichloroacetic acid for 60 min on ice, pelleted by centrifugation at 4°C at 16,000 × g for 30 min, and resuspended in 10 mM Tris-HCl (pH 8.0). Protein samples were diluted in Laemmli loading buffer, resolved by SDS-PAGE on a 12% polyacrylamide gel, and visualized by silver staining.
Cell association assay.
A modification of the procedure described for RDEC-1 by Abe et al. was used (1). Briefly, HEp-2 cells (104/well) were seeded in 24-well plates and grown overnight at 37°C in 5% CO2 in DMEM. Monolayers were infected with bacteria at a multiplicity of infection of 70 and incubated at 37°C in 5% CO2 for 3 h. The monolayers were washed six times with phosphate-buffered saline and stained with Diff-Quik (Dade Behring Inc. Newark, Del.). Fifty individual HEp-2 cells were scored for the number of cell-associated bacteria. Data were analyzed with Statview 5.0 and are reported as the mean ± standard deviation. Differences between groups were compared by using a two-tailed t test.
Fluorescent actin staining assay.
The fluorescent actin staining (FAS) assay was a modification of the method of Knutton et al. (24) and was performed as previously described (14). Briefly, HEp-2 cells were seeded at 5 × 104 CFU on circular coverslips in 24-well plates and grown overnight at 37°C in 5% CO2 in DMEM. Bacteria were added at a multiplicity of infection of 70 and were incubated for 6 h at 37°C in 5% CO2. Midway through the incubation period, the cell culture medium was replaced with fresh medium. Monolayers were washed six times with phosphate-buffered saline, fixed with paraformaldehyde, and extracted with ice-cold acetone. Double fluorescence labeling was performed. The presence of C. rodentium was detected by an anti-C. rodentium rabbit polyclonal antibody and Cascade blue-conjugated goat anti-rabbit immunoglobulin G (Molecular Probes, Eugene, Oreg.). Actin was labeled with Texas red-phalloidin (Molecular Probes). Coverslips were mounted and examined on a Nikon Labophot epifluorescence microscope fitted with 400- and 580-nm dichroic filters. Single labeling with the anti-C. rodentium antibody revealed no crossover when visualized with the 580-nm filter.
Nucleotide sequence accession number.
The nucleotide sequence of the C. rodentium espB gene has been submitted to GenBank and has been assigned accession no. AF177537.
RESULTS
C. rodentium espB clones and nucleotide sequence.
Seven HindIII fragments were cloned from the C. rodentium LEE region contained on the pDBS2 cosmid. Two of these inserts were found to have espB homology by Southern blot analysis. pBAZ463 contained a 1-kb insert with homology to 596 bp of the 5′ end of espB. pBAZ481 contained a 1.1-kb insert with homology to 376 bp of the 3′ end of espB. Characterizing this contig, C. rodentium espB was found to be 966 nucleotides in length and was predicted to yield a protein product with a molecular mass of 33 kDa. No hydrophobic leader sequence was apparent, indicative of a type III secretion mechanism. The gene was found to be preceded by consensus sequences for −35 and −10 promoter regions and by a strong ribosomal binding site. DNA sequence alignment of C. rodentium espB with that of EPEC E2348/69 showed 90% identity. Comparison of the C. rodentium and EPEC EspB hypothetical proteins showed them to have 85% identity and 90% similarity. An unusual sequence of 20 amino acids, 17 of which are either serine or threonine, appeared near the amino terminus of the predicted protein; this sequence was conserved between EPEC and C. rodentium (11).
The nucleotide sequence of the DNA 5′ of C. rodentium espB had homology to the EPEC espD gene (2). The DNA 3′ of C. rodentium espB contained a 222-bp ORF. The predicted protein product of this ORF was a 74-residue polypeptide with significant homology to the PrgI protein of Salmonella typhimurium. In S. typhimurium, PrgI is involved in the secretion of epithelial-cell signaling proteins (32).
Construction of a nonpolar insertional C. rodentium espB mutant.
To test the hypothesis that espB is required for cell association and signal transduction by C. rodentium, an isogenic nonpolar insertional espB mutant was constructed. The merodiploid strain DBS506 was passaged in LB broth, and clones were tested for Kanr and Cms. Of the 2,000 clones tested, 1 such clone was isolated (0.05%) and was designated DBS578. DBS578 was confirmed by Southern blot analysis to have lost vector and wild-type allele sequences (Fig. 2).
Complementation of the C. rodentium espB mutant.
DBS578 was complemented in trans with pJVN111. In addition, the marked deletion in DBS578 was replaced with the wild-type allele of espB. To accomplish this, the merodiploid strain DBS586 was passaged in LB broth and clones were screened for Kans. Of the 900 clones that were screened, 1 was found to be both Kans and Cms (0.1%). This clone was designated DBS587 and was confirmed by Southern blot analysis to have the wild-type allele of espB in place of the mutant allele (Fig. 2).
C. rodentium espB encodes a 37-kDa secreted protein.
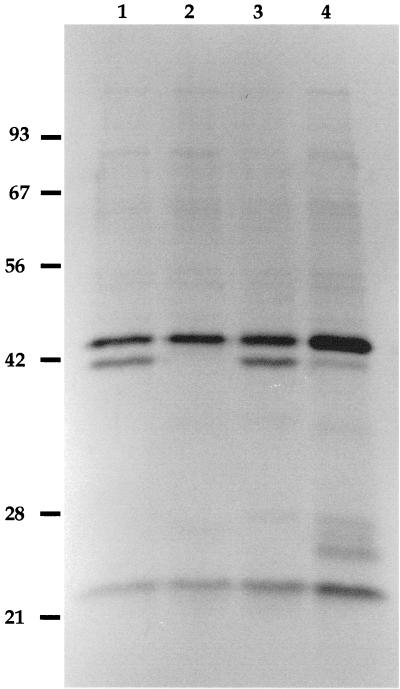
EPEC grown in cell culture medium secretes five polypeptides, including proteins of 100 kDa (EspC), 39 kDa (EspD), 37 kDa (EspB), and 24 kDa (EspA) (19). C. rodentium showed a similar secretion profile except for the absence of a protein with an apparent molecular mass similar to that of EspC (40) (Fig. 3). It has been shown that EspC is not encoded by a LEE gene and is not required for induction of host signal transduction pathways by EPEC (40). DBS100 secreted a protein similar in mobility to EPEC EspB. The mutant strain DBS578 did not secrete EspB as judged by its secretion profile (Fig. 3). The 24-kDa and 42-kDa proteins were still secreted by the espB mutant DBS578. trans complementation of the espB mutation in DBS578 with pJVN111 resulted in expression of EspB, although the amount of secreted protein appeared to be reduced. DBS587, the chromosomal revertant derived from DBS578, also expressed EspB. These findings confirm that no unlinked mutations are present in DBS578 that affect EspB secretion.
FIG. 3.
Secreted proteins of isogenic C. rodentium mutant strains. Secreted proteins were separated by SDS-PAGE on a 12% polyacrylamide gel and stained with silver. Lanes: 1, wild-type DBS100; 2, espB mutant DBS578; 3, espB+ DBS587; 4, transcomplemented DBS578/pJVN111 strain. Numbers on the left are molecular masses in kilodaltons.
espB is required for cell association of C. rodentium with cell monolayers.
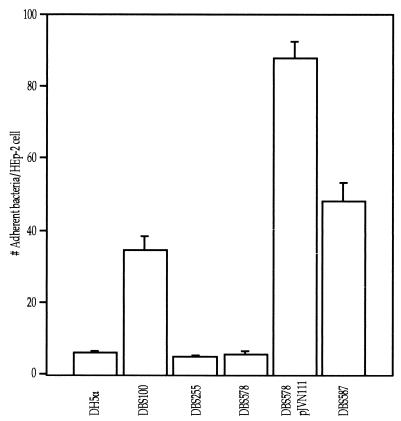
To investigate the role of C. rodentium EspB in cell association, the espB mutant strain was compared to wild-type C. rodentium in a cell association assay. The cell association of espB DBS578 was significantly reduced compared to that of wild-type DBS100 (p < 0.05) and was comparable to that seen with eaeA DBS255 (Fig. 4). The reduction in cell association seen with DBS578 could be complemented in trans or could be restored by replacing the mutant allele with a wild-type copy of the espB gene (Fig. 4). These data indicate that in C. rodentium, espB is required for cell association with cultured cell monolayers.
FIG. 4.
EspB is required for cell association by C. rodentium. The number of adherent bacteria on 50 HEp-2 cells was counted. Cell association of wild-type DBS100 was not significantly different from that of espB+ DBS587 (P = 0.05). The espB mutant DBS578, like eaeA mutant DBS255, had reduced cell adherence (P < 0.001). The trans-complemented DBS578/pJVN111 strain had increased cell association compared to wild-type DBS100 (P < 0.001).
espB is required for FAS activity by C. rodentium.
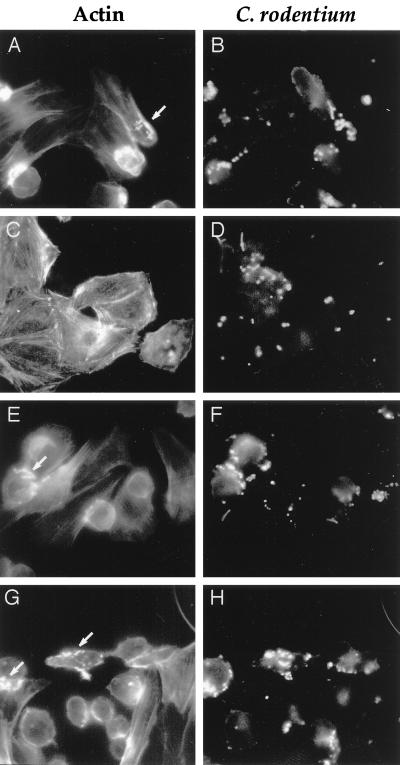
To determine whether C. rodentium espB is required for FAS activity, double fluorescence microscopy was performed. DBS100 was able to cause the accumulation of cytoskeletal actin into pedestals beneath cell-associated bacteria (Fig. 5). Both the percentage of cell-associated bacteria exhibiting actin rearrangement and the intensity of actin staining beneath cell-associated DBS100 were lower than that seen with JPN15, an EPEC strain cured of the plasmid that encodes the bfp locus (26). Cytoskeletal actin accumulation beneath cell-associated bacteria was not seen with the espB mutant DBS578. Complementation of the chromosomal espB mutation in trans or by replacement with the wild-type allele resulted in restored FAS activity (Fig. 5).
FIG. 5.
EspB is required for FAS activity. Photomicrographs of FAS assay mixtures after a 6-h incubation show filamentous actin (A, C, E, and G) and C. rodentium (B, D, F, and H). Wild-type DBS100, trans complemented DBS578/pJVN111, and espB+ DBS587 were positive for FAS activity (arrows in panels A, E, and G), while no FAS activity was seen with espB mutant DBS578 (C). Magnification, ×40.
espB is a colonization factor for C. rodentium.
When mice were challenged with the espB mutant DBS578, no kanamycin-resistant bacteria were recovered from stool after 3 days. As expected, the eaeA mutant DBS255 was also absent from the feces of mice by 3 days postinoculation. The expected levels of DBS100 CFU were recovered from mice inoculated with the wild-type strain, while no C. rodentium was recovered from any of the control mice receiving sterile broth. To confirm that it was the espB mutation that was responsible for the loss of colonization, attempts were made to complement the espB mutation in trans. While DBS578/pJVN111 CFU were recovered from infected mice at each time point, the number of CFU recovered was 2 log units lower than that of DBS100. At each time point, 100 DBS578/pJVN111 colonies from each mouse were scored for Cmr. On average, 10% of the colonies were found to be Cms, suggesting that pJVN111 was segregating in vivo. When mice were challenged with espB+ DBS587, fecal counts equivalent to DBS100 counts were seen on day 7 postinoculation.
To confirm these results, colonic colonization was also determined. No espB mutant DBS578 and eaeA mutant DBS255 CFU were isolated from total mouse colon, whereas equal numbers of wild-type DBS100 and espB+ DBS587 CFU were recovered (Table 2). DBS578/pJVN111 CFU counts were 2 log units lower than those for DBS100 and DBS587. Some mortality was seen in mice fed either wild-type DBS100 or espB+ DBS587 but not in mice infected with espB mutant DBS578, DBS578/pJVN111, or eaeA mutant DBS255. In addition, marked hyperplasia was present in the descending colons of all mice infected with DBS100 and DBS587, mild hyperplasia was present in mice infected with DBS578/pJVN111, and no hyperplasia was evident in the colons of LB-, DBS255-, or DBS578-inoculated mice (data not shown).
TABLE 2.
espB is required for colonization of mice by C. rodentiuma
| Inoculum | Log CFU/g of stool-filled colonb | nc |
|---|---|---|
| Sterile LB | 0.00 ± 0.00 | 9 |
| DBS100 | 8.95 ± 0.09 | 10d |
| DBS255 | 0.00 ± 0.00 | 6 |
| DBS578 | 0.00 ± 0.00 | 10 |
| DBS578/pJVN111 | 6.88 ± 0.14e | 6 |
| DBS587 | 9.03 ± 0.11 | 5d |
The experiment measuring the number of CFU in stool-filled colon on day 10 postinoculation was done twice, and the data presented in this table are a composite of the results of the two experiments.
Mean ± standard deviation; the lower limit of detection was 1 CFU/g.
Number of mice used.
Prior to day 10, two mice infected with DBS100 and one mouse infected with DBS587 died.
Chloramphenicol-resistant counts only.
DISCUSSION
In this study, we describe the cloning and the characterization of espB from C. rodentium. Previous characterization of the LEE in EPEC, EHEC, C. rodentium and the E. coli strains previously designated H. alvei has shown that this pathogenicity island contains homologs of eaeA, espB, and the type III secretion system encoded by the esc genes. Homologous genes are not present in nonpathogenic E. coli strains, in other species of Citrobacter (39), or in avirulent strains of H. alvei (28). Sequences homologous to espB have previously been reported to be present on pDBS2 (39), a cosmid containing both eaeA and ORFU (38). The physical organization of these genes in the chromosome of C. rodentium, human EPEC, and RDEC-1 is conserved (1, 11, 22).
The C. rodentium espB mutant strain did not secrete a 37-kDa protein. Secretion of this protein could be restored by trans complementation or by replacing the mutant allele with a wild-type copy of the espB gene. In the trans complemented strain, a number of additional protein bands appeared in the 25- to 28-kDa range. It has been previously shown that the secreted 37-kDa EspB protein in EPEC has 28-, 25-, and 16-kDa breakdown products (44). This may indicate that when espB is present in multiple copies that the EspB protein is unstable.
C. rodentium carrying an espB mutation was found to have significantly reduced cell association and no detectable FAS activity. These activities could be restored by trans complementation or by replacement of the mutant allele with a wild-type copy of the espB gene. These data demonstrate that signal transduction mediated by EspB is required for cell association and for FAS activity by C. rodentium on cultured cell monolayers. The trans complemented strain had significantly increased cell association compared to the wild type; the reasons for this remain unclear.
Our espB mutant also failed to colonize mice. An eaeA mutant has been previously reported to have the same phenotype (38), suggesting that AE lesion formation is required for colonization by C. rodentium. This distinguishes C. rodentium from the AE lesion-forming E. coli strains, which appear to utilize initial adhesins (6, 16, 26, 45). In human volunteer studies conducted with EPEC strain E2348/69 and an isogenic eaeA mutant, no overall difference was seen in the peak excretion levels of the organisms in stool, although there was significantly less severe diarrhea in subjects who had been fed the eaeA mutant (9). This demonstrates that intimate attachment is not required for colonization by EPEC. In other human volunteer studies, the initial adherence factor, bundle-forming pili, was shown to be required for full virulence but not for colonization (5, 26). We speculate that AE lesion formation may be the primary mechanism for mucosal attachment by C. rodentium. Indeed, in the absence of the initial plasmid-encoded adhesin AF/R1, AE lesion-proficient RDEC-1 colonizes only the colon and not the small bowel (44), further supporting the notion that the default site of LEE-mediated attachment is the colon. In all whole-animal studies to date, as in cultured cell systems, espB is required for AE lesion formation.
The factors responsible for inducing mucosal hyperproliferation have not been defined. It is possible that formation of AE lesions in the colon of mice is sufficient to cause transmissible colonic hyperplasia. However, a comprehensive search for virulence determinants in C. rodentium has not been undertaken. The characterization of additional genes within and outside the LEE will help define the molecular pathogenesis of transmissible colonic hyperplasia. We believe that studies with laboratory mice infected with C. rodentium will continue to provide insights into cytokinetics and cancer risk and will provide a useful model system with which to study AE activity.
ACKNOWLEDGMENTS
This work was supported by grant CA63112 and by fellowship F32 CA76716 to J.V.N. from the National Institutes of Health.
REFERENCES
- 1.Abe A, Kenny B, Stein M, Finlay B B. Characterization of two virulence proteins secreted by rabbit enteropathogenic Escherichia coli, EspA and EspB, whose maximal expression is sensitive to host body temperature. Infect Immun. 1997;65:3547–3555. doi: 10.1128/iai.65.9.3547-3555.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Barthold S W. Autoradiographic cytokinetics of colonic mucosal hyperplasia in mice. Cancer Res. 1979;39:24–29. [PubMed] [Google Scholar]
- 4.Barthold S W, Coleman G L, Jacoby R O, Livstone E M, Jonas A M. Transmissible murine colonic hyperplasia. Vet Pathol. 1978;15:223–226. doi: 10.1177/030098587801500209. [DOI] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Bliska J B, Guan K, Dixon E, Falkow S. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc Natl Acad Sci USA. 1991;88:1187–1191. doi: 10.1073/pnas.88.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantey J R, Inman L R, Blake R K. Production of diarrhea in the rabbit by a mutant of Escherichia coli (RDEC-1) that does not express adherence (AF/R1) pili. J Infect Dis. 1989;160:136–141. doi: 10.1093/infdis/160.1.136. [DOI] [PubMed] [Google Scholar]
- 8.Crane J K, Oh J S. Activation of host cell protein kinase C by enteropathogenic Escherichia coli. Infect Immun. 1997;65:3277–3285. doi: 10.1128/iai.65.8.3277-3285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Tacket C O, James S P, Losonsky G, Nataro J P, Wasermann S S, Kaper J B, Levine M M. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Investig. 1993;92:1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg M S, Tzipori S, McKee M L, O'Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliot S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y, Lai L C, MacNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic E. coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 13.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foubister V, Rosenshine I, Donnenberg M S, Finlay B B. The eaeB gene of enteropathogenic Escherichia coli is necessary for signal transduction in epithelial cells. Infect Immun. 1994;62:3038–3040. doi: 10.1128/iai.62.7.3038-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foubister V, Rosenshine I, Finlay B B. A diarrheal pathogen, enteropathogenic Escherichia coli (EPEC), triggers a flux of inositol phosphate in infected epithelial cells. J Exp Med. 1994;179:993–998. doi: 10.1084/jem.179.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giron J A, Ho A S, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 17.Johnson E, Barthold S W. The ultrastructure of transmissible murine colonic hyperplasia. Am J Pathol. 1979;97:291–301. [PMC free article] [PubMed] [Google Scholar]
- 18.Kenny B, Abe A, Stein M, Finlay B B. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny B, Finlay B B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenny B, Finlay B B. Intimin-dependent binding of enteropathogenic Escherichia coli to host cells triggers novel signaling events, including tyrosine phosphorylation of phospholipase C-gamma. Infect Immun. 1997;65:2528–2536. doi: 10.1128/iai.65.7.2528-2536.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 22.Kenny B, Lai L, Finlay B B, Donnenberg M S. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 23.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai L C, Wainwright L A, Stone K D, Donnenburg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine M M, Nataro J P, Karch H, Baldini M M, Kaper J B, Black R E, Clements M L, O'Brien A D. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 27.Lipkin M. Biology of large bowel cancer: present status and research frontiers. Cancer. 1975;36:2319–2324. doi: 10.1002/1097-0142(197512)36:6<2319::aid-cncr2820360606>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants of Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 33.Rosenshine I, Donnenberg M S, Kaper J B, Finlay B B. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 1992;11:3551–3560. doi: 10.1002/j.1460-2075.1992.tb05438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Savidge T C, Shmakov A N, Walker-Smith J A, Phillips A D. Epithelial cell proliferation in childhood enteropathies. Gut. 1996;39:185–193. doi: 10.1136/gut.39.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savkovic S D, Koutsouris A, Hecht G. Activation of NF-κB in intestinal epithelial cells by enteropathogenic Escherichia coli. Am J Physiol. 1997;273:C1160–C1167. doi: 10.1152/ajpcell.1997.273.4.C1160. [DOI] [PubMed] [Google Scholar]
- 37.Schauer D B, Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect Immun. 1993;64:2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schauer D B, Falkow S. The eaeA gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun. 1993;61:4654–4661. doi: 10.1128/iai.61.11.4654-4661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schauer D B, Zabel B A, Pedraza I F, O'Hara C M, Steigerwalt A G, Brenner D J. Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J Clin Microbiol. 1995;33:2064–2068. doi: 10.1128/jcm.33.8.2064-2068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein M, Kenny B, Stein M A, Finlay B B. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178:6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein M A, Mathers D A, Yan H, Bainbridge K G, Finlay B B. Enteropathogenic Escherichia coli markedly reduces the resting membrane potential of Caco-2 and HeLa human epithelial cells. Infect Immun. 1996;64:4820–4825. doi: 10.1128/iai.64.11.4820-4825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor K A, O'Connell C B, Luther P W, Donnenberg M S. The EspB protein of enteropathogenic Escherichia coli is targeted to the cytoplasm of infected HeLa cells. Infect Immun. 1998;66:5501–5507. doi: 10.1128/iai.66.11.5501-5507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wachter C, Beinke C, Mattes M, Schimidt M A. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1999;31:1695–1707. doi: 10.1046/j.1365-2958.1999.01303.x. [DOI] [PubMed] [Google Scholar]
- 44.Wainwright L A, Kaper J B. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol Microbiol. 1998;27:1247–1260. doi: 10.1046/j.1365-2958.1998.00771.x. [DOI] [PubMed] [Google Scholar]
- 45.Wolf M K, Andrews G P, Fritz D L, Sjogren R W, Boedeker E C. Characterization of the plasmid from Escherichia coli RDEC-1 that mediates expression of adhesin AF/R1 and evidence that AF/R1 pili promote but are not essential for enteropathogenic disease. Infect Immun. 1988;56:1846–1857. doi: 10.1128/iai.56.8.1846-1857.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolff C, Nisan I, Hanski E, Frankel G, Rosenshine I. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1998;28:143–155. doi: 10.1046/j.1365-2958.1998.00782.x. [DOI] [PubMed] [Google Scholar]