Abstract
Newcastle disease virus (NDV) causes one of the highly infectious avian diseases in poultry leading to genuine financial misfortunes around the world. Recently, there has been an increasing trend in the number of ND-associated outbreaks in commercial Jordanian poultry flocks indicating a possible complex evolutionary dynamic of NDV infections in the country. To underpin the dynamics of circulating NDV strains and to assess the vaccine-escape potential, a total of 130 samples were collected from different poultry flocks in six Jordanian Governorates during 2019–2021. Twenty positive isolates, based on real-time reverse transcriptase PCR, were used for further genetic characterization and evolutionary analysis. Our results showed that there is a high evolutionary distance between the newly identified NDV strains (genotype VII.1.1) in this study and the commercially used vaccines (genotypes I and II), suggesting that circulating NDV field strains are under constant evolutionary pressure. These mutations may significantly affect flocks that have received vaccinations as well as flocks with insufficient immunity in terms of viral immunity and disease dynamics. To assess this further, we investigated the efficacy of the heterologous inactivated LaSota or homologous genotype VII.1.1 vaccine for their protection against virulent NDV in chicken. Vaccine-induced immunity was evaluated based on the serology, and protection efficacy was assessed based on clinical signs, survival rates, histopathology, and viral shedding. Chickens vaccinated with the inactivated genotype VII.1.1 based vaccine showed 100% protection with a significant reduction in virus shedding, and ameliorated histopathology lesions compared to LaSota vaccinated chicks that showed 60% protection. These results revealed that the usage of NDV inactivated vaccine from the circulating field strains can successfully ameliorate the clinical outcome and virus pathobiology in vaccinated chicks and will serve as an effective vaccine against the threat posed by commonly circulating NDV strains in the poultry industry.
Keywords: avian orthoavulaviruses 1, evolutionary pressure, vaccine, efficacy, Jordan
1. Introduction
The Paramyxoviridae family consists of a large number of viruses which are isolated from a wide range of human and other animal species including measles, mumps, and respiratory syncytial viruses, Newcastle disease virus (NDV), canine distemper, and rinderpest viruses [1]. All paramyxoviruses are pleomorphic, enveloped, single stranded and non-segmented viruses containing a negative sense RNA genome of 10–17 Kb size. Based on structure, genomic organization, and sequence relatedness, this family is divided into two subfamilies: Paramyxovirinae and Pneumovirinae [2]. The Paramyxovirinae subfamily has five genera: Respirovirus, Rubulavirus, Morbillivirus, Henipavirus, and Avulavirus, whereas the Pneumovirinae subfamily has two genera: Pneumovirus and Metapneumovirus [3].
All paramyxoviruses isolated from avian species have been classified into two genera: Avulavirus, which represents avian paramyxoviruses (APMV), and Metapneumovirus, which represents avian pneumoviruses. Based on hemagglutination inhibition (HI) and neuraminidase inhibition (NI) assays, it has been concluded that avian avulaviruses infect a wide range of domestic and wild birds all over the world [4]. Avulaviruses’ RNA genomes encode six structural proteins (NP, P, M, F, HN, and L) as well as two non-structural proteins (V and W) via RNA editing [2,4]. The hemagglutinin-neuraminidase (HN) and fusion (F) proteins are surface glycoproteins found in avulaviruses. The new classification criterion has been proposed that involve the use of genomic sequence comparisons in the categorization of avulaviruses due to issues associated with cross-reactivity among some serotypes of avulaviruses in serologic tests. According to recent criterion, Orthoavulavirus, Paraavulavirus, and Metaavulavirus are three genera that now make up the Avulavirinae subfamily [1,5].
Avian Avulavirus serotype-1 (AAvV-1) is a member of the Avulavirus genus in the Paramyxoviridae family that causes ND in chicken. The antigenic serotypes that evolve in this group as a result of environmental or vaccination pressure elude the immune system of birds and are responsible for vaccine failure [6,7]. The fusion (F) and hemagglutinin neuraminidase (HN) genes, which encode for structural envelope proteins that play host recognition, infection, and pathogenesis roles, but also influence the antigenicity and immunogenicity of ND viruses have a high genetic and antigenic diversity within the AAvV-1 serotype [8,9,10]. Animal models (1-day-old Gallus chick) are used in infectivity assays for ND viruses, which are expensive and time consuming. Monobasic amino acid sequences at positions 112–113 and 115–116 of the C-terminus of the fusion protein cleavage site (FPCS) with leucine (L) at position 117 and/or intracerebral pathogenicity indices (ICPI) of 0.7 are used to designate low-virulent strains [11,12].
Avian Metaavulavirus has been known to cause disease, specifically mild respiratory infections in domestic poultry, including turkeys and chickens, and pose many economic effects on egg production and poultry industries [13]. The virus was first isolated from a strain in Yucaipa, California in 1956. Since then, other isolates of the virus have been isolated worldwide. Avian paramyxovirus 2 (APMV-2) has been isolated from a wide range of birds, including chickens, turkeys, racing pigeons and feral birds and appears to be circulating worldwide [1,14,15,16,17].
Furthermore, the prevalence of APMV-2 antibodies in several bird species, including commercial poultry, has been investigated [13,18,19]. Chickens, broilers, and layers from the United States, Canada, Russia, Japan, Israel, India, Saudi Arabia, the United Kingdom, and Costa Rica, as well as turkeys from the United States, Canada, Israel, France, and Italy, have all been shown to carry APMV-2 viruses. Infections with APMV-2 reduced turkey hatchability and poult output [20]. More significant illness, particularly in turkeys during subsequent infections, has been documented [21]. APMV-2 infections in turkey flocks have also been reported by virus isolation and the presence of antibodies [22]. The reason of the reduction in egg production was assumed to be APMV-2, which was identified from commercial layer farms and broiler breeder farms in Scotland [23]. APMV-2 infection in chickens via intramuscular and intratracheal routes produced no evident respiratory disease [24]. Similar findings were found in turkeys infected by intratracheal route [25].
Despite the widespread use of a LaSota and Hitchner B1-based vaccine in the poultry, ND outbreaks are still common in the Middle East, where the most commonly circulating NDV isolates are taxonomically categorized as genotype VII [26,27,28]. Because of their significant contribution to the ongoing ND pandemic, these genotype VII isolates are currently regarded as the most economically relevant NDV strains in Jordan and also frequently isolated among flocks that have been vaccinated with traditional genotype II-based ND vaccines. Because genotype mismatch between genotype II-based ND vaccines and the circulating genotype VII NDV are widely thought to be responsible for the current vaccines’ suboptimal protective efficacy. Therefore, development of new vaccines based on the currently prevalent genotype VII NDV has the potential to improve the effectiveness of ND control in the global poultry industry.
The aim of this study is to detect and characterize AAvVs using genetic and antigenic techniques to provide insights into the ecology of these viruses. We demonstrate the presence of two different avian avulavirus serotypes: one has been previously described in chicken (AOAvV-1) and one distantly related to AAvV-2/APMV-2. Further, we demonstrated the vaccine efficacy of currently deployed heterologous vaccine and a newly developed homologous vaccine in chicken. The finding warrants continued surveillance of AOAvV-1 strains in poultry and to revise vaccines and vaccination strategies trained by the ground realities.
2. Materials and Methods
2.1. Ethics Statement
Samples were collected by trained veterinarians. Samples processing and virus isolation procedures were carried out in strict accordance with the guidance and regulations of animal welfare and health that approved by the Department of Veterinary Pathology and Public Health, Faculty of Veterinary Medicine, Jordan University of Science and Technology (JUST), Jordan (JUST 387-2020).
2.2. Sampling History, Virus Isolation, and Biological Characterization
A total of 130 swab samples were collected from various Jordanian poultry flocks (Table 1). All samples were collected from vaccinated flocks except Turkey, Peacock, and Ostrich (Table 2). Individual swabs were collected in viral transport medium supplemented with antibiotics (isotonic PBS, 2000 U/mL penicillin, 2 mg/mL streptomycin, 50 μg/mL gentamycin, 50 U/mL nystatin, and 0.5% BSA). Swab samples were cleared by centrifugation for 5 min at 1700 rpm at 10 °C, and the supernatants were collected and stored at −80 °C until further use.
Table 1.
Sampling data and prevalence of Avian Avulaviruses in different geographical regions in Jordan during 2019–2021.
| Isolate | Host | Location | ICPI | Accession Number |
|---|---|---|---|---|
| NDV/chicken/Jordan/MQA-N-1/2019 | Backyard chicken | Ajloun | 1.8 | ON858785 |
| NDV/chicken/Jordan/MQA-N-2/2020 | Backyard chicken | Jarash | 1.6 | ON858786 |
| NDV/chicken/Jordan/MQA-N-3/2020 | Backyard chicken | Balqa | 1.6 | ON858787 |
| NDV/chicken/Jordan/MQA-N-4/2019 | Backyard Breeder | Amman | 1.7 | ON858788 |
| NDV/Turkey/Jordan/MQA-N-5/2020 | Turkey | Amman | 1.8 | ON858789 |
| NDV/chicken/Jordan/MQA-N-6/2021 | Layer breeder | Jarash | 1.8 | ON858790 |
| NDV/chicken/Jordan/MQA-N-7/2021 | Broiler breeder | Zaraqa | 1.7 | ON858791 |
| NDV/chicken/Jordan/MQA-N-8/2021 | Layer breeder | Zarqa | 1.8 | ON858792 |
| NDV/chicken/Jordan/MQA-N-9/2019 | Layer breeder | Zarqa | 1.7 | ON858793 |
| NDV/Peacock/Jordan/MQA-N-10/2020 | Peacock | Amman | 1.9 | ON858794 |
| NDV/chicken/Jordan/MQA-N-11/2020 | Layer breeder | Zarqa | 1.6 | ON858795 |
| NDV/chicken/Jordan/MQA-N-12/2019 | Backyard chicken | Madaba | 1.8 | ON858796 |
| NDV/chicken/Jordan/MQA-N-13/2021 | Backyard chicken | Madaba | 1.7 | ON858797 |
| NDV/chicken/Jordan/MQA-N-14/2019 | Backyard chicken | Amman | 1.7 | ON858798 |
| NDV/chicken/Jordan/MQA-N-15/2020 | Layer breeder | Amman | 1.9 | ON858799 |
| NDV/chicken/Jordan/MQA-N-16/2019 | Commercial broiler | Jarash | 1.8 | ON858800 |
| NDV/Ostrich/Jordan/MQA-N-17/2020 | Ostrich | Amman | 2.0 | ON858801 |
| NDV/chicken/Jordan/MQA-N-18/2019 | Commercial broiler | Ajloun | 1.8 | ON858802 |
| NDV/chicken/Jordan/MQA-N-19/2021 | Commercial broiler | Jarash | 1.7 | ON858803 |
| NDV/chicken/Jordan/MQA-N-20/2020 | Commercial broiler | Balqa | 1.9 | ON858804 |
| APMV2/chicken/Jordan/MQA-N-1/2020 | Commercial broiler | Jarash | 0 | ON858805 |
| APMV2/chicken/Jordan/MQA-N-2/2020 | Commercial broiler | Amman | 0 | ON858806 |
ICPI: Intracerebral Pathogenicity Index.
Table 2.
Vaccination regime used to vaccinate the Jordanian poultry flocks.
| Age | Vaccination Route | Used Vaccine |
|---|---|---|
| 0 a,b | In ovo | VAXXITEK® (HVT + IBD) |
| 1 a,b | Coarse spray | Live NDV (Avinew®) and live attenuated IBV (Poulvac IB Primer®) |
| 14 a,b | Coarse Spray | Live attenuated IBV (IBird®) + Live attenuated NDV (Clone 30) |
| 21 a,b | IM | Inactivated NDV + H9N2 + H5N1 |
| 28 a,b | Fine Spray | Live attenuated NDV LaSota |
| 49 b | Fine Spray | Live attenuated NDV LaSota |
| 65 b | Eye drop | Live attenuated ILTV |
| 77 b | IM | Inactivated NDV + H9N2 |
| 78 b | Fine Spray | Live attenuated NDV LaSota |
| 91 b | SC + IM + Fine spray | Inactivated TRT + IBV + live attenuated IBV (Poulvac IB Primer®) |
| 105 b | Fine Spray | Live attenuated ND LaSota |
| 126 b | IM | Inactivated NDV + IBV + IBDV + REO |
| 143 b | IM | Inactivated H9N2 + H5N1 |
| 175 b | Fine Spray | Live attenuated NDV (LaSota) |
a For broiler flocks; b For breeder flocks.
According to the OIE Manual of Standards for Diagnostic Tests and Vaccines, each sample was inoculated (in triplicate) into the allantoic sac of 9–10-day embryonated chicken eggs (ECEs) for viral isolation [29]. The positive HA samples were biologically characterized using intracerebral pathogenicity test in Rhode Island Red SPF chicks using the standard protocols [29]. All samples were negative for other avian respiratory viruses including influenza viruses and infectious bronchitis virus (IBV).
2.3. RNA Extraction, Genome Amplification and Sequencing
Samples were subjected to RNA extraction from the allantoic fluid using an RNA extraction kit (RNAeasy Mini Kit, Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Detection of avian avulaviruses was conducted using a real-time reverse transcriptase PCR based on the M gene of NDV and the N gene of aMPV as described previously [30]. For full length fusion (F) gene amplification, RNA was reverse transcribed into cDNA using a Superscript IV First-Strand cDNA Synthesis Kit (Invitrogen, Waltham, MA, USA) and the second strand was synthesized with Q5 DNA Polymerase (New England Biolabs, Ipswich, MA, USA) using for amplification and sequencing of the full length F gene [31,32]. Amplified PCR products were visualized by electrophoresis on a 1.2% agarose gel electrophoresis and then purified using a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The purified PCR products were sequenced bi-directionally with both sense and antisense primers that were used in the PCR amplification [31,32] using ABI PRISM BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA, USA) by Sanger sequencing method on a 3500 Applied Biosystems capillary sequencer (Source Bioscience, Cambridge, UK).
2.4. Sequence Analyses and Phylogeny
The F nucleotide and amino acid sequences were retrieved in fasta format from the NCBI GenBank and compared to those found in ViralZone UniProtKB/Swiss-Prot entries using corresponding accession numbers. These sequences were edited to the same length and aligned using ClustalW, which is included in BioEdit version 7.0. [33]. The obtained nucleotide sequences were submitted to GenBank and assigned accession numbers are outlined (Table 1). Sequence Demarcation Tool (SDT) was used to display the amino acid pairwise identity scores through a color-coded matrix [34].
Phylogenetic analyses were conducted using general time-reversible (GTR) model [35], which was selected using jModelTest [36], and maximum-likelihood trees were constructed using RaxML version 8.2.11 [37] with 1000 bootstrap replicates. The phylogenetic analysis was performed on nucleotides based on the pilot tree proposed by Dimitrov et al. [38] to maintain the tree topology and to ascertain the genotypes of avian avulaviruses isolates.
2.5. Mutations Mapping at Variable Positions and Functional Regions
The F nucleotide sequences were later translated into amino acid sequences in MEGA X program v10.1.8 to compare our AOAvV-1 isolates, and vaccine (LaSota: JF950510.1) strains at the amino acid levels. Sequence logos, graphical representations of patterns within the F protein aligned sequences, were generated using the WebLogo service (http://weblogo.threeplusone.com/create.cgi, accessed on 15 August 2022). Sequence logos give a fuller and more accurate representation of F protein sequence similarity than consensus sequences, and they can quickly expose important characteristics of the alignment that might otherwise be difficult to notice.
Sequences variations were mapped onto the protein structures and entropy calculations with the aid of Scop3D tool, which visualizes variations across multiple sequences on the protein structures [39]. The F protein numbering was based on LaSota using GenBank accession number JF950510.1. The functional regions were defined based on literature and were mapped on the structures and Jalview or Chimera-analyzed models for diversity as visualized to the predicted structure models.
2.6. DiscoTope: Structure Based Antibody Prediction
The interaction of antibodies with antigens is one of the most significant immune system strategies for removing pathogenic organisms from the host. Antibodies bind to antigens at B-cell epitopes. The precise placement of B-cell epitopes is critical in many scientific applications, including rational vaccine design, disease diagnostics, and immunotherapeutics. However, because experimental mapping of epitopes is time consuming, in silico approaches provide an interesting supplementary option [40]. Using the Discotope 2.0 online tool (https://services.healthtech.dtu.dk/service.php?DiscoTope-2.0, accessed on 11 August 2022), we tried to map the antibody binding sites within the F protein of NDV that will help to identify these residues compared to those in commercially used vaccines among Jordanian poultry sectors.
2.7. F Protein Structural Homology Analyses and Selective Pressure
To identify the conserved regions, homology models for the translated F proteins were created by matching sequences using multiple sequence alignment (MSA) with the help of ClustalW, which is included in the BioEdit software version 7.0 [33]. The consensus areas for each protein in the field isolates and vaccine were utilized in a BLAST search against the Protein Data Bank (PDB) to find known homologs or orthologs. The Synonymous-Non-Synonymous Analysis Program (SNAP) was used to predict the F gene-specific estimates of dN/dS [41]. The number of potential synonymous and non-synonymous changes as well as the number of actual synonymous and non-synonymous changes in codon between each pair were counted. Then, the dN/dS ratio was calculated by comparing the proportion of observed non-synonymous substitutions over the proportion of observed synonymous substitutions. These were then adjusted for multiple hits using the Jukes–Cantor correction [41]. A higher score than 0 indicates a more dominant diversifying positive selection while below 0 indicates negative selection.
2.8. Preparation of Inactivated Newcastle Disease Virus Vaccine
The genotype VII.1.1 seed virus (ON858797 AOAvV-1 isolate NDV/chicken/Jordan/MQA-N-13/2021) was propagated via inoculation into the allantoic sac of specific pathogen-free embryonated chicken eggs (SPF-ECE) which were incubated at 37 °C. Allantoic fluids from these inoculated eggs were harvested after overnight chilling at 4 °C and tested for hemagglutination using 1% chicken red blood cells (RBCs). The Egg Infective Dose 50 (EID50) was determined via titration in 10-day-old SPF-ECE according to the method described [42].
The titrated virus (ON858797 AOAvV-1 isolate NDV/chicken/Jordan/MQA-N-13/2021) was used as a master seed for the preparation of inactivated vaccine. Seed virus inactivation was conducted with formalin (final concentration of 0.1%) for 18 h at 37 °C. Complete inactivation of the virus was confirmed through three passages in 10-day-old embryonated SPF chicken eggs followed by HA and EID50. All SPF chicken embryos inoculated with formalin-treated virus remained alive after 120 hours, and no HA-based positivity was detected. The final dose used was 108 EID50/0.5 mL per chick. Inactivated NDV vaccine was prepared as water in oil emulsion (W/O) using Montanide ISA 70 at a ratio of 3/7 (v/v) aqueous/oil ratio. The manufacturing process was carried out according to the standard protocol of SEPPIC, France. The prepared vaccine were tested for its sterility and safety according to OIE [29]. Stability testing of emulsion involves determination of stability at long-term storage at 4 °C and 25 °C [43,44]. Velogenic Newcastle disease virus (vNDV) ON858797 AOAvV-1 isolate NDV/chicken/Jordan/MQA-N-13/2021 strain was used to challenge the vaccinated and non-vaccinated (positive control) chicks.
2.9. Vaccination and Challenge Experiments
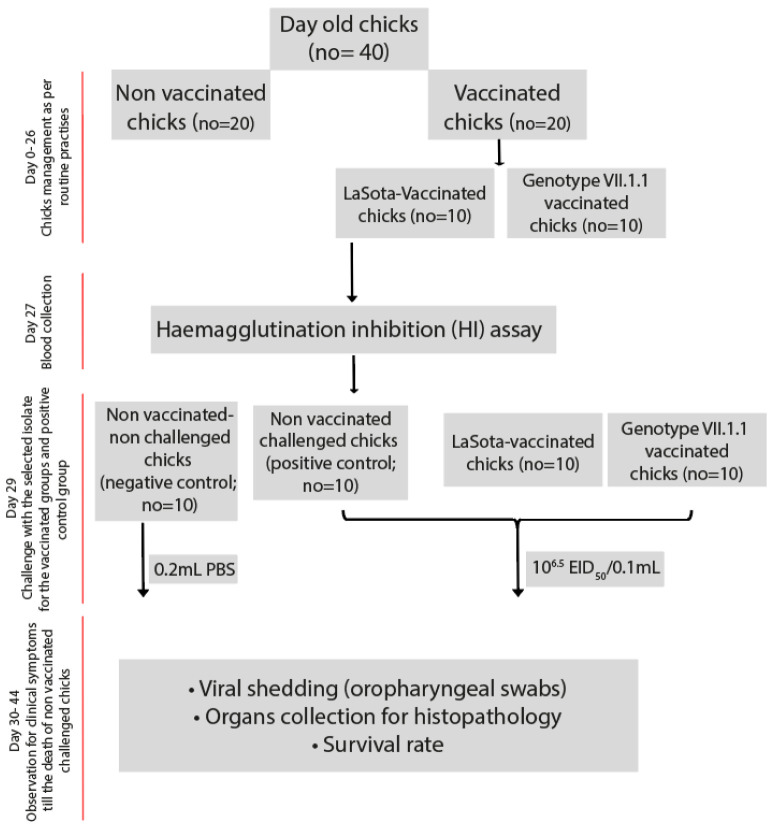
Forty SPF Rhode Island Red chicks were housed separately in two groups: vaccinated (n = 20) and unvaccinated (n = 20). The challenge experiments were conducted in accordance with all relevant guidelines and animal ethics permits issued by Department of Veterinary Pathology & Public Health, Faculty of Veterinary Medicine, Jordan University of Science and Technology (JUST), Irbid, Jordan. Chicks in the unvaccinated group were divided into three subgroups: non-vaccinated challenged (positive control, n = 10) and non-vaccinated non-challenged (negative control, n = 10). However, chicks in the vaccinated group (n = 20) were kept in two groups (10 each); LaSota vaccinated challenged group and genotype VII.1.1 vaccinated challenged group (Figure 1) and administered either the inactivated LaSota vaccine (genotype II) or inactivated genotype VII.1.1 vaccine on day 7 at a dose of 0.5 mL per chick via the subcutaneous route around the neck region (Figure 1).
Figure 1.
Experimental layout for the assessment of vaccines effectiveness in chicks.
Chicks in the vaccinated- challenged and non-vaccinated challenged groups was challenged with a dose of 100 μL of 106.5EID50 (ON858797 AOAvV-1 isolate NDV/chicken/Jordan/MQA-N-13/2021) strain through the oculonasal route on day 29. The mock-infected group served as a negative control and was inoculated with 100 μL of sterile PBS (Figure 1). For the next 10 days, chicks were monitored twice daily for any clinical signs including depression, sneezing/coughing, facial swelling, respiratory sounds, ocular/nasal discharge, ruffled feathers, dyspnea, greenish diarrhea, paralysis and tremors as well as necropsy lesions in dead chicks for pathognomic lesions of NDV including hemorrhages in the proventriculus and cecal tonsils.
2.10. Serology, Virus Shedding and Histopathology
Serum samples were obtained pre- (day 27) and post-challenge (day 38) from the vaccinated challenged groups and tested using an HI assay. The HI assay was performed using inactivated NDV antigen with 4 HAU in 0.025 mL [29]. Titers were calculated as the reciprocal of the highest serum dilution providing complete hemagglutination inhibition. Serum titers of 1:8 (23) or lower were considered negative for antibodies against NDV. Virus shedding was detected using previously described assays for identification of velogenic strains of NDV in oropharyngeal swabs [29]. Oropharyngeal swabs were collected, placed in virus transport medium, filtered through a 0.2 µm filter and then aliquoted and stored at −70 °C until all samples were collected before analysis as previously described [29].
Selected tissues including trachea and lungs were collected, fixed by immersion in 10% neutral buffered formalin at room temperature for 48 h and followed by processing and embedding in paraffin wax. Tissue sections of 5 μm were stained with Hematoxylin and Eosin and examined for microscopic lesions under a light microscope.
2.11. Statistical Analysis
Pairwise comparisons of challenged (clinical and sub-lethal doses) and control groups (positive and negative) were performed using Student’s t-test. Kaplan–Meier analysis was performed to calculate the survival rates. Two-tailed Student’s t-test and one-way analysis of variance (ANOVA) were used to determine differences between groups. Statistical significance is shown with values of p < 0.05. All data were represented as the mean ± standard deviation (SD). Statistical analyses were conducted using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. NDV Epidemiology in Jordan and Biological Characterization
We present the isolation, biological characterization and genetic analyses that map the evolution of NDV in Jordan during 2019–2021. A total of 130 samples were individually screened by RT-qPCR, followed by full length amplifications for F gene of positive samples. From this screening, 20 out of 130 samples (20/130; 15.4%) were positive among all tested swab samples for AOAvV-1 (Table 1) while only two samples were positive for avian paramyxovirus 2 (2/130; 1.5%). The ICPI was conducted for all isolated viruses individually, which calculates the mean score per bird per observation over the 8-day period. Our results revealed that ICPI values ranged between 1.6–2.0 per eight-day observation period for the 20 AOAvV-1 isolates (Table 1) indicating their velogenic nature while ICPI was zero for the two APMV-2 (Table 1) isolates indicating the lentogenic nature of these isolates.
3.2. Phylogenetic Analyses
To determine the epidemiological clustering of Jordanian NDV isolates in the current study, representative avian avulaviruses genome sequences from the National Center for Biotechnology Information (NCBI) databases were downloaded and used for phylogenetic and comparative genomic analyses. A Bayesian consensus phylogenetic analysis, which was verified using the neighbor-joining method, 20 isolates in this study clustered within avian avulavirus 1 along with previously reported isolates from Jordan, Israel, Iraq, Egypt, and China (Figure 2) while two isolates were clustered with APMV-2 along with previously reported APMV-2 isolates in Israel (Figure 2). The phylogenomic and clustering pattern of AOAvV-1 isolates revealed that 19 isolates were clustered within genotype VII.1.1 while only one isolate was allocated within genotype VII.2 (Figure 3), showed their close association within the previously reported isolates in Jordan and neighboring countries including Israel, Iraq and Egypt from both commercial and backyard flocks. Interestingly, NDV/Peacock/Jordan/MQA-N-10 isolate was isolated from wild bird indicating the close relationship between wild birds and domesticated birds in NDV epidemiology and evolution.
Figure 2.
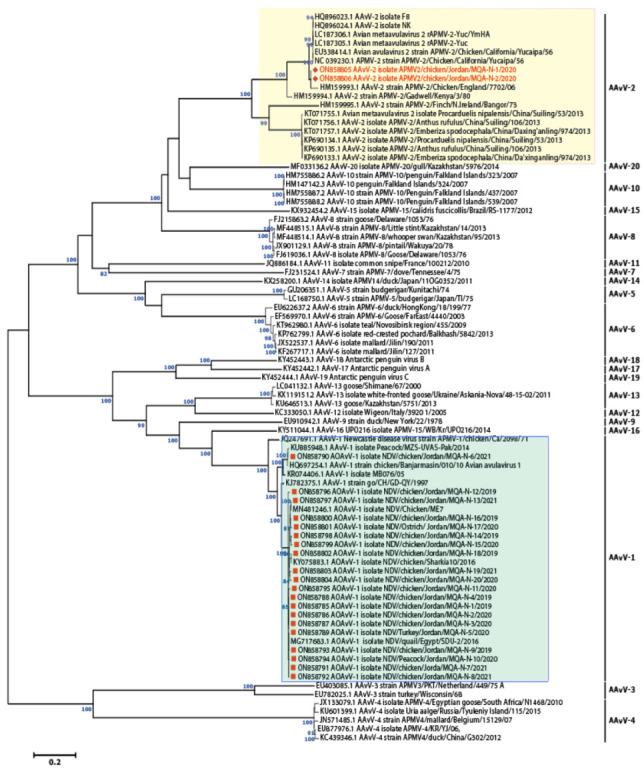
Phylogenomic revealed the clustering of 20 isolates within the AOAVv-1 while two isolates were allocated within AAvV-2/APMV-2. Unrooted phylogenetic trees were generated using the distance-based using maximum likelihood method and MEGA 6 software. Statistical support for tree branches was assessed by bootstrap analysis using 1000 replications of bootstrap re-sampling; numbers above branches indicate neighbor-joining bootstrap values that were ≥80%; the tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The reported AAvV-1 isolates in this study are marked with red square within light green box; however, AAvV-2 isolates are marked with red hexagon labelled within light yellow box.
Figure 3.
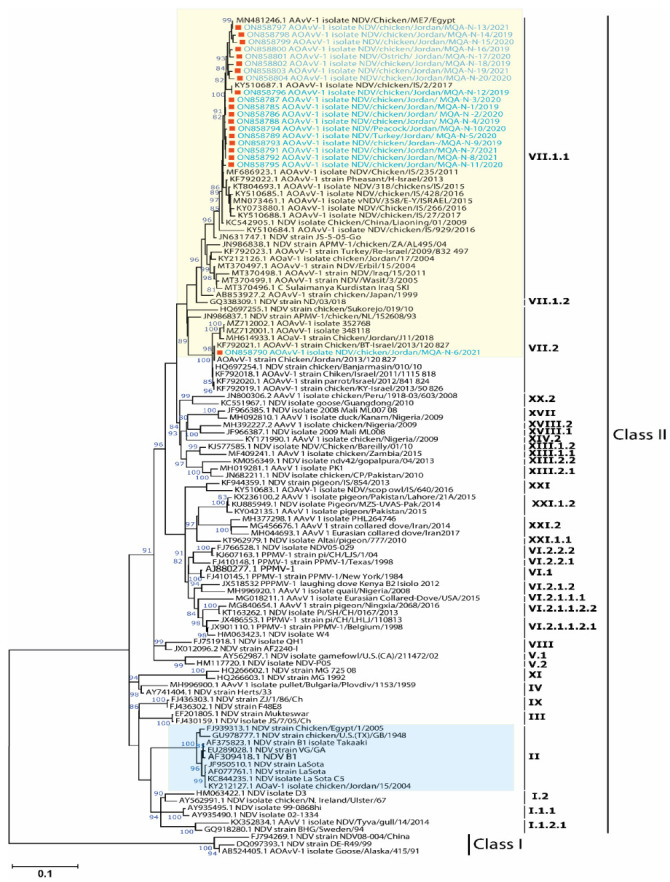
Phylogenetic analysis of the studied AOAvV-1 isolates and their clustering patterns with representative AOAvV-1 isolates. Full-length F-gene (1662 nt)-based phylogenetic analysis of our AOAvV-1 isolates with representative strains of each genotype. Reported isolates clustered in the genotype VII.1.1 of class II. Unrooted phylogenetic trees were generated using the distance-based using maximum likelihood method and MEGA 6 software. Statistical support for tree branches was assessed by bootstrap analysis using 1000 replications of bootstrap re-sampling; numbers above branches indicate neighbor-joining bootstrap values that were ≥80%; the tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The reported AOAvV-1 isolates in this study are marked with red square within yellow box, however, NDV genotype II including LaSota (commonly used vaccine) was labelled within light blue box.
3.3. Nucleotide and Amino Acid Homology
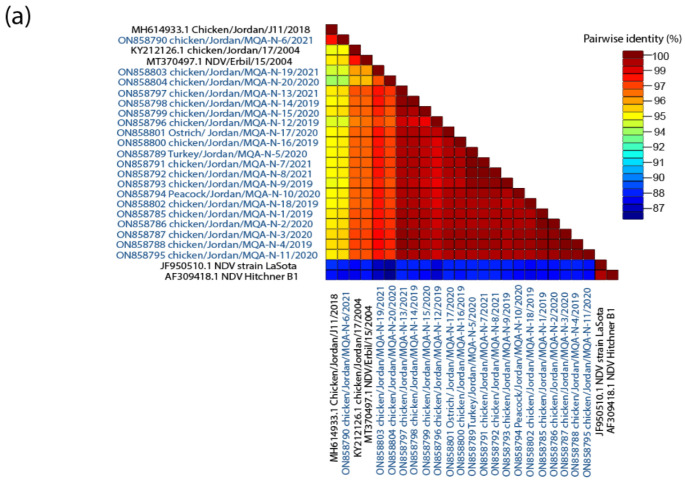
The level of nucleotide sequence identity between the AOAvV-1 studied isolates ranged between 91% and 99%, and these isolates showed varying degrees of genetic divergence from other representative genotypes of NDV (Figure 4a); however, the identity with the LaSota vaccine strain was 83% (Figure 4a). Meanwhile, all the AOAvV-1 isolates showed 11–13% amino acid difference compared to vaccines that are routinely used in the country (LaSota [genotype II]).
Figure 4.
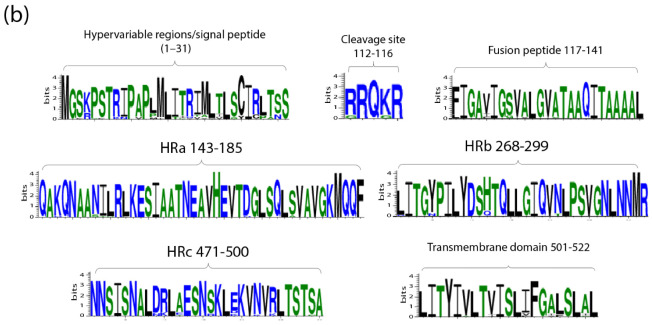
(a) The pairwise identities plot of fusion protein sequences aligned by MAFFT and displayed by Sequence Demarcation Tool (SDT) software. (b) WebLogo graphs illustrating the amino acid divergence between AOAVv-1 isolates reported in this study compared to LaSota vaccine and previously reported isolates in Jordan.
All AOAvV-1 isolates in this study exhibit multiple basic amino acid residues at the cleavage site (F0) of the F protein (Figure 4b), which is a hallmark of velogenic NDV strains [4,45]. The predicted residue analysis of F protein revealed a typical proteolytic cleavage motif 112R-R-Q-K-R116, characteristic for virulent viruses (Figure 4b). Previous studies have identified six possible glycosylation sites within the F protein that are highly conserved across the majority of AOAvV-1 genotypes. The glycosylation motif Asn-X-Ser/Thr (N-X-S/T, where X is any residue except proline [P] and aspartic acid [D]) was identified in the studied isolates. These sites were identified in the reported AOAvV-1 isolates in this study as follow; 85N-RT87, 191N-N-T193, 366N-T-S368, 471N-N-S473, and 541N-N-T543 that are key residues for receptor binding, and crucial amino acids in the hydrophobic core of the stalk [46,47]. Several substitutions were found in the transmembrane region (aa501 to aa521) of the F protein of the AOAvV-1 isolates studied in this study (Figure 4b). In addition, the AOAvV-1 isolates had different alterations in the signal and fusion peptides, and the heptad repeat (HR) regions compared to LaSota and previously reported Jordanian NDV strains (Figure 4b), which might impact on the F protein’s fusogenic activity [45,48,49,50].
3.4. Deduced Amino Acid Mutations Trend Analyses
In the pathophysiology of the ND, HN glycoprotein starts infection, whereas F glycoprotein facilitates viral attachment and penetration into host cells [2]. Both HN and F proteins stimulate the host immune response and are essential for the production of neutralizing antibodies generated by vaccinations. Antibodies against F proteins have been shown in vivo to be critical in neutralizing ND infectivity [51,52]. Previous studies showed that there are seven major F protein neutralizing epitopes involved in fusion inhibition and neutralization are shown at specific residues 72, 74, 75, 78, 79, 157–171, and 343 for epitopes A1, A2, A3, A4, and A5, respectively. Our results showed that there is an amino acid substitution (H78R) in 17 AOAvV-1 isolates reported in this study. The amino acid residues show that both F1 and F2 are involved in the formation of a single antigenic site vital in the structure and function of the active F epitopes [53].
3.5. Antibody Sites Prediction and Immune Pressure
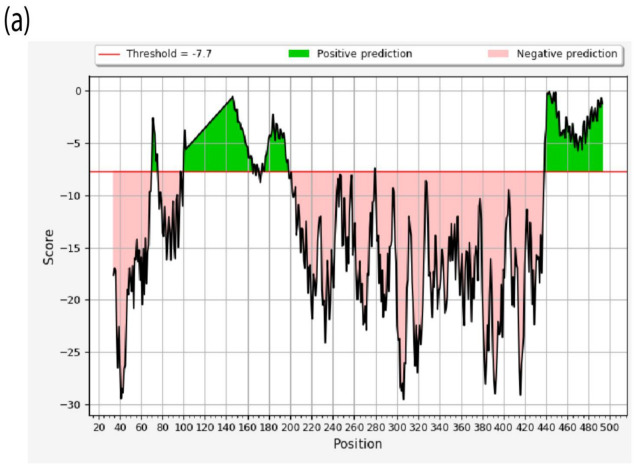
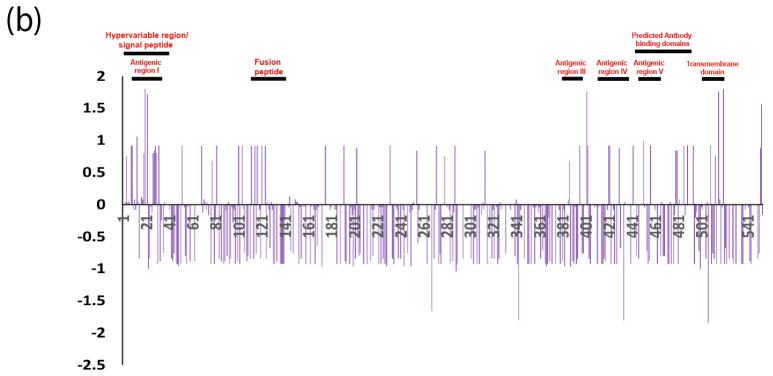
We predicted the antibody binding residues and their surface accessibility and antigenicity scores using the BepiPred linear Epitope prediction method, Emini surface accessibility tool, and Kolaskar and Tongaonkar antigenicity, which use epitope scores and immunogenicity predictions through IEDB online (www.iedb.org, accessed on 11 August 2022) facilities. Both BepiPred-2.0 prediction tool and Vaxijen 2.0 tool gave effective antigenic domains; antigenic region I, II, III, IV and V for our AOAvV-1 isolates compared to LaSota vaccine (Table 3). These domains were above antigenicity score (0.8) and surface accessibility score (0.6) suggesting that these amino acid residues could modify the effectiveness of the predicted epitopes, which are speculated for the antigenic differences between these viruses and vaccine. In addition, our analyses showed variable residues that affect the F protein hydrophobic stability (Table 4). Structure-based antibody prediction of the F protein for the AOAvV-1 isolates reported in this study showed different epitope locations (Figure 5a). Mass vaccination has a cumulative effect that plays a role in virus evolution through immune pressure. Our results demonstrated that the cumulative difference between the nonsynonymous substitution rate (dN) and the synonymous substitution rate (dS) for the Jordanian NDV strains were under positive selection at critical sites within the F protein (Figure 5b).
Table 3.
Analysis of mutations in the predicted F protein antigenic domains structure between LaSota vaccine and our AOAvV-1 isolates reported in this study.
| Domain | Antigenic Region I (7–30) |
Antigenic Region II (196–241) | Antigenic Region III (380–394) | Antigenic Region IV (413–437) | Antigenic Region V (447–460) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A.A positions | 8 | 9 | 13 | 16 | 17 | 19 | 20 | 22 | 27 | 28 | 29 | 30 | 203 | 231 | 232 | 385 | 386 | 387 | 421 | 422 | 430 | 451 | 457 |
| LaSota (JF950510.1) | K | N | M | T | I | V | A | V | C | P | A | N | A | N | K | T | I | K | K | Q | G | Q | I |
| AOAvV-1 isolates | R | I | L | I | T | I | M | I | R | L | T | S | T | T | Q | A | L | R | R | H | D | L | V |
Table 4.
Analysis of mutations in the predicted positions contribute to hydrophobic stability of the F protein in our AOAvV-1 isolates reported in this study compared to the LaSota vaccine.
| Domain | Variable Residues of Hydrophobic Stability | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A.A positions | 69 | 82 | 115 | 124 | 145 | 146 | 192 | 403 | 421 | 430 | 453 | 457 | 486 | 489 |
| LaSota (JF950510.1) | L | D | G | G | K | Q | K | N | K | G | S | I | R | D |
| AOAvV-1 isolates | M | E | K | S | K | Q | N | D | R | D | S | V | S | E/K |
Figure 5.
(a) Structure-based Antibody Prediction. X-axis contains the position of residues while the y-axis shows the propensity line indicates the threshold value. Regions above the threshold value, shown in green, are representing the residues under positive prediction. (b) The cumulative dN/dS of the average synonymous and non-synonymous substitutions moving codon by codon across F protein of AOAVv-1 isolates reported in Jordan including the reported isolates in this study with highlighting the most affected domains (high selective pressure) within the F protein.
3.6. Vaccine Sterility, Safety and Hemagglutination Inhibition Test
The prepared genotype VII.1.1 based inactivated vaccine was sterile and safe as they were free from any bacterial and fungal contaminants. No local or systemic reactions were observed. No clinical signs or mortality were recorded in vaccinated chicks and no pathological lesions were observed by postmortem examination. The post-vaccination antibody titers in the chicks’ sera were determined using HI test with homologous antigens. All chicks had no detectable NDV antibody titers just before vaccination. Similarly, the control group showed no HI antibody titers throughout the study. On the contrary, 3 weeks post vaccination; chickens vaccinated with the genotype VII.1.1 based vaccine showed increasing antibody titer log2 6.73 ± 0.50 compared to LaSota vaccinated chicks showed 4.19 ± 0.95.
3.7. Vaccines Efficacy Assessment
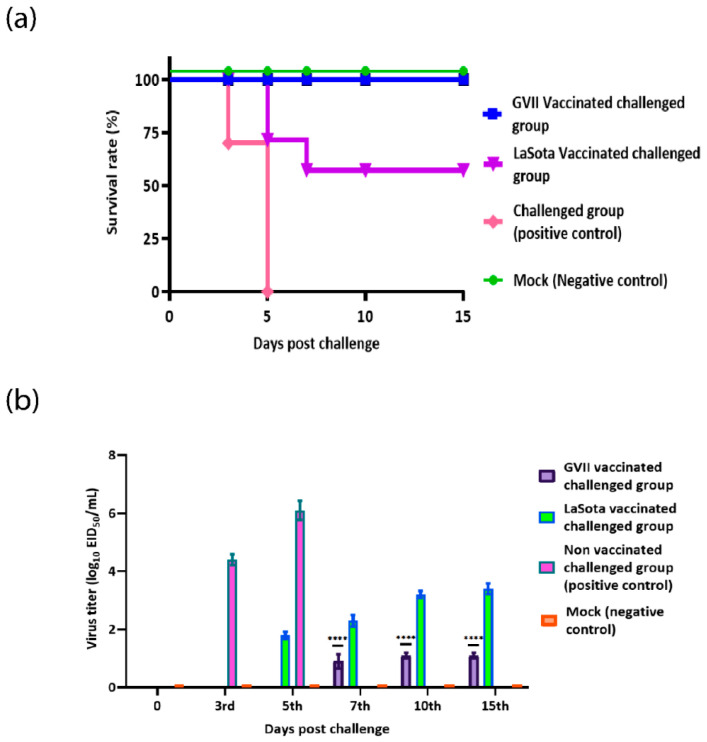
As proved to be highly immunogenic, the protective role of genotype VII.1.1 against virulent viral challenge was compared with LaSota vaccine. The inactivated genotype VII.1.1 vaccine was prepared and used to immunize SPF chicks followed by challenge with homologs virulent NDV strain. Commercial inactivated LaSota vaccine and sterile saline were used as positive and negative immunization controls. Each chick was immunized with a dose of 107EID50 via neck subcutaneous injection then challenged with 106.5EID50 dose of challenge virus. The clinical symptoms and death of the chicks were recorded every day till 15th days post-challenge. After immunization, all chicks appeared normal before challenge; chicks immunized with either inactivated genotype VII.1.1 or LaSota vaccine did not show any obvious abnormality after challenge; however, non-vaccinated challenged chicks (positive control) showed drowsiness, loss of appetite, apathetic, and row yellow-greenish dilute feces on 2nd day post challenge (dpc) and all chicks died by 5th dpc in this group. These results confirmed that vaccination with genotype VII.1.1 provide complete protection (100%) (Figure 6a) against homologous challenge while LaSota vaccination provided partial protection (60%) (Figure 6a). Clinical signs, representative of ND, started to appear in non-vaccinated challenged group on the 3rd day post-challenge including depression, anorexia, mild respiratory sounds, and oculonasal discharges. Interestingly, all chicks in the non-vaccinated challenged group were died 5 days after infection. On the other hand, clinical signs started to appear in LaSota vaccinated challenged group on the 5th day post challenge and 4 chicks (out of 10) died by the 7th day post challenge (Figure 6a).
Figure 6.
Survival rates and Evaluation of viral shedding. (a) Percentage survival rates and (b) Viral shedding from oropharyngeal swabs of genotype VII.1.1 and LaSota vaccinated challenged chicks with virulent NDV compared to negative and positive control groups. Bars represent the standard deviation means. **** indicates the level of significance at p value < 0.05.
3.8. Virus Shedding and Histopathology
The virus shedding data from oropharyngeal swabs were evaluated based on a number of shedders and amount of shedding (EID50) at 0, 3rd, 5th, 7th, 10th and 15th days post-challenge. Results of oropharyngeal viral shedding from the vaccinated chicks with genotype VII.1.1 based vaccine showed a significant reduction in the amount of virus shedding compared with LaSota vaccinated group or non-vaccinated challenged group (p ≤ 0.05) (Figure 6b), however, there was incomplete prevention for the virus shedding.
Trachea and lung organs were collected from vaccinated groups either with inactivated genotype VII.1.1 or LaSota based vaccine and non-vaccinated challenged chicks (positive control group) followed by histopathological examination compared with and non-vaccinated non-challenged chicks (negative control group) to assess the level of protection offered by vaccination in face of challenge with a virulent NDV along with the induced histopathological changes. Microscopically, trachea of control non-vaccinated non-challenged chicks and genotype VII.1.1 vaccinated chicks exhibited normal histological structure (Figure 7). On contrary, remarkable histopathological alterations were investigated in tracheal tissues of non-vaccinated challenged chicks (positive control) described by necrosis of lamina epithelialis, mucous secreting glands and mononuclear cells infiltration in lamina propria (Figure 7). Otherwise, moderate changes were noticed in tracheal tissues of LaSota vaccinated chicks; edema in the lamina propria/submucosal layer. The histopathological alterations in the trachea and lungs of different groups are summarized according to their severity in Table S1.
Figure 7.
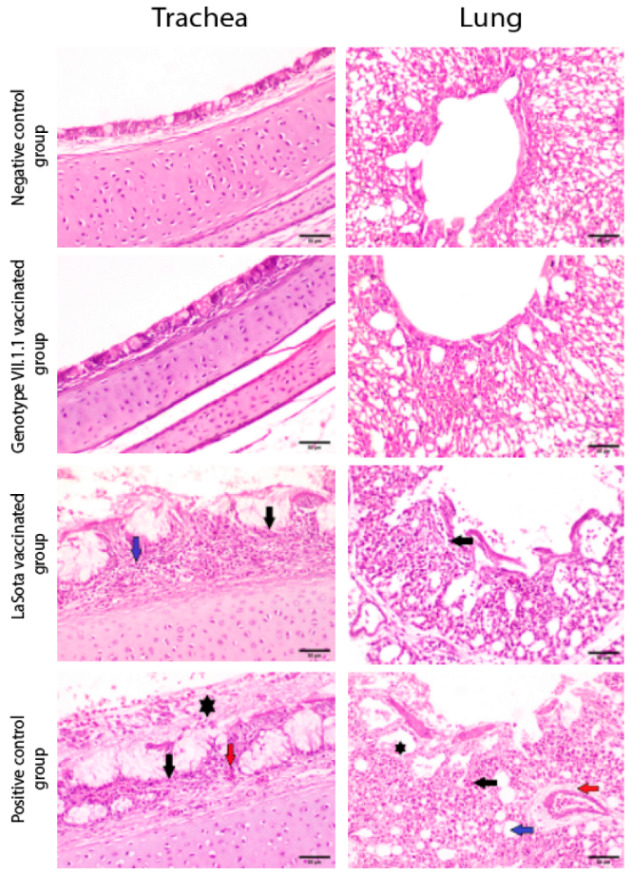
Photomicrographs representing H&E stained sections of tracheas and lungs collected from genotype VII.1.1 and LaSota vaccinated- challenged chicks with virulent NDV compared to mock chicks (negative control) and positive control groups (non-vaccinated challenged chicks). Non-vaccinated- non-challenged chicks and genotype VII.1.1 vaccinated- challenged chicks showing normal histological architecture for Tracheas and normal histological architecture of parabronchus and air capillaries in lungs. On the other hand, LaSota vaccinated challenged chicks showed necrosis of lamina epithelialis and mucosal glands (black arrow) associated with mononuclear cells infiltration in lamina propria (blue arrow) in Trachea and inflammatory cells infiltration (black arrow) in lungs. In addition, non-vaccinated-challenged chicks (positive control group) showing multifocal necrosis of lamina epithelialis (black arrow), congestion (red arrow) and accumulation of mucous exudate in the tracheal lumen (asterisk) in Trachea and showing inflammatory cells infiltration in the air capillaries (black arrow), perivascular edema (red arrow), dilatation of atria (asterisk) and dilatation of air capillaries (blue arrow) in lungs (scale bar 50 µm).
4. Discussion
In Jordan and elsewhere, the economic impact of ND on both backyard and commercial poultry is enormous. The recurrence of disease each year, vaccination failures, and potentially circulating of virulent strains in apparently healthy birds constitute a concurrent problem. Concerns about virus evolution, vaccination type utilized to protect birds, and post-vaccine assessment have been proposed several times in previous studies. The anticipated B-cell epitopes and functional domains of F protein in AOAvV-1 isolated from Jordanian birds were compared to those in vaccines to see whether there were any differences between the two groups that might explain the ND vaccination failures. Until now, the AOAvV-1 vaccine has been evaluated only on the basis of empirical cross protection in birds, whereas many of these studies show that ND vaccines with antigenically matched antigens give superior immunity [54,55]. However, such investigations are expensive and time-consuming, and they need extensive field research. Inadequate cold-chain maintenance, insufficient immunization titer, hygienic state, and other variables that might contribute to vaccination failures have all been a source of worry. In this study, we looked at the neutralizing epitopes of F-glycoproteins of enzootic wild ND viruses and vaccine (genotype II) to see if there were any virus variants or recombinants at this antigenic and surface glycoprotein in our AOAvV-1 isolates, which could lead to ineffective vaccination.
In this study, the investigated flocks had varying mortality rate (30–50%) due to putative vNDV infection symptoms such as tracheitis and proventriculus hemorrhage, which are characteristic for velogenic NDV infection [50]. Intracerebral pathogenicity index (ICPI) was used for biological characterization of our isolates reported in this study, which showed that 20 isolates (AOAvV-1) have ICPI ranged between 1.6–2.0 per eight-day observation period. However, only two isolates (APMV-2) showed zero ICPI, suggesting no morbidity or mortality. All samples were negative for other respiratory viruses including influenza viruses and infectious bronchitis virus (IBV). Molecular pathotyping was performed for the AOAvV-1 isolates using the amino acid sequences of the F0 protein cleavage site motifs (residues 112 to 117) because it is a faster and more reliable method than the mean death time (MDT), intravenous pathogenicity index (IVP), and intracerebral pathogenicity index (ICP) tests [56,57]. The majority of virulent NDV strains feature a polybasic cleavage site, which is the primary recognition site for furin (R-X-K/R-R); an intracellular protease presents in most cells that offers an efficient cleavage in a wide variety of tissues, allowing virulent strains to disseminate systemically. While avirulent NDV strains frequently have basic residues at the -1 and -4 positions relative to the cleavage site, which are cleaved by secretory protease. Because avirulent strains cannot be cleaved by furin, their replication is limited to the respiratory and intestinal routes, where secretory protease is available for cleavage.
Our findings revealed that all of the AOAvV-1 isolates had the cleavage site motif 12RRQKRF117, which is common in velogenic NDV strains. Furthermore, the presence of the phenylalanine (F) residue at position 117, which was detected in our 20 AOAvV-1 isolates, has been characterized as a probable contribution to the neurological consequences [58]. On the other hand, one or two basic residues are detected in the putative F protein cleavage site of APMV-2 isolates (DKPASR↓F), which is similar but not identical to the pattern seen in avirulent NDV strains. Previous research found that APMV-2 replicated in vitro in a wide range of cells without the addition of exogenous protease, and introducing protease did not improve the replication efficiency.
Phylogenetic study of NDV pathotypes is based on the FPCS as well as the hypervariable areas of the F protein [59]. Other ways of classifying NDV strains include genotyping and lineage analysis [60,61]. To date, two classes, I and II, have been identified and each further is classified into three sub-genotypes (1.1.1, 1.1.2, and 1.2) and 21 clades or sub-genotypes (I-XXI), respectively [38,59,60]. Notably, the class II genotype VII viruses are the most commonly reported in ND outbreaks in poultry, pet, and wild birds throughout the world, while class I, which is commonly isolated from waterfowl, shore birds, and some poultry, is less virulent and is exploited for potential vaccine candidates [60,61,62,63,64]. The two membrane-anchored glycoproteins F and HN are possible targets for the immune system response to NDV infection and are also important for cell-binding and infection [65]. ND vaccination failure has been linked to genomic and antigenic variations between field isolates and vaccine strains. These discrepancies result from many accumulated changes in the field strains’ F and HN genes as a result of vaccination pressure [6,63,66].
Phylogenetic analysis based on the F gene showed that all 20 AOAvV-1 isolates were related to velogenic strains of NDV; 19 isolates was classified as genotype VII.1.1 (class II) while only one isolate was clustered within genotype VII.2 with close relationship to previously reported isolates in Jordan, Iraq, Israel and Egypt. Moreover, the recently isolated strains are genetically distant from vaccine strains indicating the potential evolution of virulent NDV in the Jordanian poultry sector. Vaccination has been linked to viral evolution in a variety of disease affecting avian, animals and humans. Wild bird strains can spread in a new and more difficult habitat when immunization is not sterilizing. Selective pressure analysis revealed that the circulating Jordanian NDVs are under strong pressure, indicating that vaccination has a role in viral evolution as well as virus adaption in wild birds.
In the present study, the humoral immune response was assessed by HI assay for vaccinated chicks and revealed higher antibody titer; log2 6.73 ± 0.50 and 4.19 ± 0.95 for genotype VII.1.1 and LaSota vaccinated chicks, respectively 3 weeks post vaccination. In addition, our results revealed that the genotype VII.1.1 inactivated vaccine was able to protect the vaccinated chicks from the challenge virus morbidity or mortality. However, the group vaccinated with LaSota inactivated vaccine provide 60% protective efficiency (survival rate). These results were in agreement with previous studies that shown an ND inactivated vaccine must be prepared from current local circulating strains/genotypes [45]. Our results were similar to Miller et al. who observed 100% mortality for non-vaccinated chicks and 100% survival for four weeks-old SPF chicks vaccinated subcutaneously with a single dose of inactivated NDV vaccine after three weeks post-challenge with NDV [55].
While all vaccinated chicks were protected from overt clinical signs and mortality, virus shedding was noted in all the groups vaccinated with the inactivated vaccines. This indicates that these vaccines could only protect against the clinical disease but not against virulent virus infection and replication. Nevertheless, the magnitude and duration of virulent virus shedding in those groups was generally lower than those in the positive control group whose magnitude of the virus shedding was high from day 5 post-challenge. Whether genotype VII.1.1 or LaSota based inactivated ND vaccine was used, the level of virulent challenge virus shed from chicks vaccinated with a homologous vaccine (genotype VII.1.1 based) was significantly lower than that vaccinated with a heterologous vaccine (LaSota based). In consistence, our results demonstrated that the level of specific antibody response against genotype VII.1.1 is higher than that of anti-LaSota response in the vaccinated chicks, which confer a stronger protection against challenge to the immunized chicks, and lead to more efficient control of disease and reduced viral shedding.
5. Conclusions
We have identified that there are at least two types of NDV strains circulating in the country. Importantly, the F protein of the AOAvV-1 isolates were found to map numerous changes that alter the antigenic epitopes and antibody binding domains based on the deduced amino acid analyses. These mutations may significantly affect flocks that have received vaccinations as well as flocks with insufficient immunity in terms of viral immunity and disease dynamics. Therefore, it must be determined if each of these alterations, separately or together, has an impact on the virus’ antigenicity and can have major negative effects on vaccination effectiveness. Continuous genetic and phylogenetic characterization for the circulating AOAvV-1 isolates causing outbreaks are important to understand the AOAvV-1 epidemiology, evolution and to develop novel vaccines and control strategies. The results of the present study confirmed that an inactivated oil-adjuvanted vaccine from the local circulating velogenic AOAvV-1 was efficient to protect the vaccinated birds from morbidity and mortality against the challenge virus.
Acknowledgments
Mohammad Q. Al-Natour would like to thank Jordan University of Science and Technology, Irbid, Jordan for their financial support during his Sabbatical Leave at Lancaster University, UK.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/vaccines10111862/s1, Table S1: Histopathological lesion scores for tracheas and lungs of different experimental groups.
Author Contributions
Conceptualization, M.A.R. and M.M.; methodology, M.A.R., M.Q.A.-N., R.F.E.N., M.A.A., Y.M.M. and K.A.A.; software, M.A.R., M.Q.A.-N., R.F.E.N. and Y.M.M.; validation, M.A.R., M.Q.A.-N. and M.M.; formal analysis, M.A.R., M.A.A., R.F.E.N., Y.M.M. and K.A.A.; investigation, M.M.; resources, M.A.R. and M.M.; data curation, K.A.A., M.Q.A.-N. and M.M.; writing—original draft preparation, M.A.R., M.Q.A.-N., R.F.E.N., M.A.A., Y.M.M. and K.A.A.; writing—review and editing, M.A.R., M.Q.A.-N., R.F.E.N. and M.M.; visualization, M.A.R. and M.M.; supervision, M.M.; project administration, M.A.R., M.Q.A.-N. and M.M.; funding acquisition, M.A.R. and M.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
All the experimental procedures were performed in accordance with the guidance and regulations of animal welfare and health that approved by the Ethics Committee at Department of Veterinary Pathology and Public Health, Faculty of Veterinary Medicine, Jordan University of Science and Technology (JUST), Jordan.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
We would like to thank the support of the International Foundation for Science (IFS, project No. I-3-B-6270-1), The Organisation of Islamic Cooperation’s Standing Committee on Scientific and Technological Cooperation (COMSTECH). The funding sources had no role in the study design, collection or analysis of the data, writing of the manuscript or in the decision to submit the manuscript for publication.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Subbiah M., Xiao S., Khattar S.K., Dias F.M., Collins P.L., Samal S.K. Pathogenesis of two strains of avian paramyxovirus serotype 2, Yucaipa and Bangor, in chickens and turkeys. Avian Dis. 2010;54:1050–1057. doi: 10.1637/9380-041910-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb R.A., Parks G.D. Paramyxoviridae: The viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2006. pp. 1449–1496. [Google Scholar]
- 3.Mayo M.A. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 2002;147:1655–1663. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- 4.Kolakofsky D., Roux L., Garcin D., Ruigrok R.W. Paramyxovirus mRNA editing, the ‘rule of six’ and error catastrophe: A hypothesis. J. Gen. Virol. 2005;86:1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- 5.Hisanaga T., Soos C., Lewis N., Lung O., Suderman M., Berhane Y. Genetic and Antigenic Characterization of Avian Avulavirus Type 6 (AAvV-6) Circulating in Canadian Wild Birds (2005–2017) Viruses. 2021;13:543. doi: 10.3390/v13040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang R., Cao D., Li J., Chen J., Guo X., Zhuang F. Newcastle disease outbreaks in western China were caused by the genotypes VIIa and VIII. Vet. Microbiol. 2002;87:193–203. doi: 10.1016/S0378-1135(02)00050-0. [DOI] [PubMed] [Google Scholar]
- 7.Lin M., Liu H., Ke G. Genetic and antigenic analysis of Newcastle disease viruses from recent outbreaks in Taiwan. Avian Pathol. 2003;32:345–350. doi: 10.1080/0307945031000121086. [DOI] [PubMed] [Google Scholar]
- 8.Ballagi-Pordany A., Wehmann E., Herczeg J., Belak S., Lomniczi B. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F-gene. Arch. Virol. 1996;141:243–261. doi: 10.1007/BF01718397. [DOI] [PubMed] [Google Scholar]
- 9.Herczeg J., Wehmann E., Bragg R.R., Travassos Dias P.M., Hadjiev G., Werner O., Lomniczi B. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch. Virol. 1999;144:2087–2099. doi: 10.1007/s007050050624. [DOI] [PubMed] [Google Scholar]
- 10.Liu X.F., Wan H.Q., Ni X.X., Wu Y.T., Liu W.B. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. Arch. Virol. 2003;148:1387–1403. doi: 10.1007/s00705-003-0014-z. [DOI] [PubMed] [Google Scholar]
- 11.De Leeuw O.S., Koch G., Hartog L., Peeters B.P. Effect of fusion protein cleavage site mutations on virulence of Newcastle disease virus: Non-virulent cleavage site mutants revert to virulence after one passage in chicken brain. J. Gen. Virol. 2003;84:475–484. doi: 10.1099/vir.0.18714-0. [DOI] [PubMed] [Google Scholar]
- 12.Panda A., Huang Z., Elankumaran S., Rockemann D., Samal S. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2004;36:1–10. doi: 10.1016/j.micpath.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warke A., Stallknecht D., Williams S.M., Pritchard N., Mundt E. Comparative study on the pathogenicity and immunogenicity of wild bird isolates of avian paramyxovirus 2, 4, and 6 in chickens. Avian Pathol. 2008;37:429–434. doi: 10.1080/03079450802216645. [DOI] [PubMed] [Google Scholar]
- 14.Lipkind M., Weisman Y., Shihmanter E., Shoham D., Aronovici A. Isolation of Yucaipa-like avian paramyxovirus froma wild mallard duck (Anas platyrhinchos) wintering in Israel. Vet. Rec. 1982;110:15–16. doi: 10.1136/vr.110.1.15. [DOI] [PubMed] [Google Scholar]
- 15.Lipkind M., Weisman Y., Shihmanter E., Shoham D., Aronovici A. The isolation of Yucaipa-like paramyxoviruses from epizootics of a respiratory disease in turkey poultry farms in Israel. Vet. Rec. 1979;105:577–578. [PubMed] [Google Scholar]
- 16.Goodman B.B., Hanson R.P. Isolation of avian paramyxovirus-2 from domestic and wild birds in Costa Rica. Avian Dis. 1988;32:713–717. doi: 10.2307/1590989. [DOI] [PubMed] [Google Scholar]
- 17.Shihmanter E., Weisman Y., Manwell R., Alexander D.J., Lipkind M. Mixed paramyxovirus infection of wild and domestic birds in Israel. Vet. Microbiol. 1997;58:73–78. doi: 10.1016/S0378-1135(97)00147-8. [DOI] [PubMed] [Google Scholar]
- 18.Ley E.C., Morishita T.Y., Harr B.S., Mohan R., Brisker T. Serologic survey of slaughter-age ostriches (Struthio camelus) for antibodies to selected avian pathogens. Avian Dis. 2000;44:989–992. doi: 10.2307/1593077. [DOI] [PubMed] [Google Scholar]
- 19.Zhang G.Z., Zhao J., Wang M. Serological Survey on Prevalence of Antibodies to Avian Paramyxovirus Serotype 2 in China. Avian Dis. 2007;51:137–139. doi: 10.1637/0005-2086(2007)051[0137:SSOPOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.Bankowski R.A., Almquist J., Dombrucki J. Effect of paramyxovirus Yucaipa on fertility, hatchability, and poult yield of turkeys. Avian Dis. 1981;25:517–520. doi: 10.2307/1589944. [DOI] [PubMed] [Google Scholar]
- 21.Lang G., Gagnon A., Howell J. Occurrence of paramyxovirus Yucaipa in Canadian poultry. Can. Vet. J. 1975;16:233–237. [PMC free article] [PubMed] [Google Scholar]
- 22.Bradshaw G.L., Jensen M.M. The epidemiology of Yucaipa virus in relationship to the acute respiratory disease syndrome in turkeys. Avian Dis. 1979;23:539–542. doi: 10.2307/1589586. [DOI] [Google Scholar]
- 23.Weisman Y., Aronovici A., Malkinson M., Shihmanter E., Lipkind M. Isolation of paramyxoviruses from pigeons in Israel. Vet. Rec. 1984;115:605. doi: 10.1136/vr.115.23.605-a. [DOI] [PubMed] [Google Scholar]
- 24.Bankowski R.A., Corstvet R.E. Isolation of a haemagglutinating agent distinct from Newcastle disease from the respiratory tract of chickens. Avian Dis. 1961;5:253–269. doi: 10.2307/1587635. [DOI] [Google Scholar]
- 25.Bankowski R.A., Conrad R.D., Reynolds B. Avian Influenza A and paramyxoviruses complicating respiratory diagnosis in poultry. Avian Dis. 1968;12:259–278. doi: 10.2307/1588226. [DOI] [PubMed] [Google Scholar]
- 26.Roussan D.A., Haddad R., Khawaldeh G. Molecular survey of avian respiratory pathogens in commercial broiler chicken flocks with respiratory diseases in Jordan. Poult. Sci. 2008;87:444–448. doi: 10.3382/ps.2007-00415. [DOI] [PubMed] [Google Scholar]
- 27.Ababneh M., Ferreira H.L., Khalifeh M., Suarez D.L., Afonso C.L. First Genome Sequence of Newcastle Disease Virus of Genotype VIIi from Jordan. Microbiol. Resour. Announc. 2018;7:e01136-18. doi: 10.1128/MRA.01136-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ababneh M.M., Dalab A.E., Alsaad S.R., Al-Zghoul M.B., Al-Natour M.Q. Molecular characterization of a recent Newcastle disease virus outbreak in Jordan. Res. Vet. Sci. 2012;93:1512–1514. doi: 10.1016/j.rvsc.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 29.OIE, editor. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 7th ed. OIE Organisation Mondiale de la Santé Animale; Paris, France: 2012. [(accessed on 24 September 2022)]. Newcastle disease. Chapter 2.3.14. Available online: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.14_NEWCASTLE_DIS.pdf. [Google Scholar]
- 30.Mia Kim L., Suarez D.L., Afonso C.L. Detection of a broad range of class I and II Newcastle disease viruses using a multiplex real-time reverse transcription polymerase chain reaction assay. J. Vet. Diagn. Investig. 2008;20:414–425. doi: 10.1177/104063870802000402. [DOI] [PubMed] [Google Scholar]
- 31.Munir M., Linde A.M., Zohari S., Stahl K., Baule C., Engstrom B., Renström L.H., Berg M. Whole genome sequencing and characterization of a virulent Newcastle disease virus isolated from an outbreak in Sweden. Virus Genes. 2011;43:261–271. doi: 10.1007/s11262-011-0636-2. [DOI] [PubMed] [Google Scholar]
- 32.Brown P.A., Lemaitre E., Briand F.X., Courtillon C., Guionie O., Allée C., Toquin D., Bayon-Auboyer M.H., Jestin V., Eterradossi N. Molecular comparisons of full length metapneumovirus (MPV) genomes, including newly determined French AMPV-C and -D isolates, further supports possible subclassification within the MPV Genus. PLoS ONE. 2014;9:e102740. doi: 10.1371/journal.pone.0102740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall T.A. Bio-Edit: A user-friendly biological sequence alignment editor and analysis for windows 95/98/N.T. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 34.Muhire B.M., Varsani A., Martin D.P. SDT: A virus classifcation tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. In: Miura R.M., editor. Some Mathematical Questions in Biology—DNA Sequence Analysis. Amer Math Soc; Providence, RI, USA: 1986. pp. 57–86. [Google Scholar]
- 36.Guindon S., Dufayard J., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitrov K.M., Abolnik C., Afonso C.L., Albina E., Bahl J., Berg M., Briand F.X., Brown I.H., Choi K.S., Chvala I., et al. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 2019;74:103917. doi: 10.1016/j.meegid.2019.103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vermeire T., Vermaere S., Schepens B., Saelens X., Van Gucht S., Martens L., Vandermarliere E. Scop3D: Three-dimensional visualization of sequence conservation. Proteomics. 2015;15:1448–1452. doi: 10.1002/pmic.201400354. [DOI] [PubMed] [Google Scholar]
- 40.Kringelum J.V., Lundegaard C., Lund O., Nielsen M. DiscoTope 2.0: Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012;8:e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kryazhimskiy S., Plotkin J.B. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- 43.Cessi D., Nardelli L. Vaccination against Newcastle disease: Efficacy of an oil emulsion vaccine. Avian Pathol. 1974;3:247–253. doi: 10.1080/03079457409353838. [DOI] [PubMed] [Google Scholar]
- 44.El-Bagoury G.F., Nasr M.H.M., El-Habbaa A.S., Hala M.E.-M. A trial to improve stability and immunogenicity of inactivated NDV vaccine with paraffin oil adjuvant using aluminum stearate. BVMJ. 2015;28:199–209. [Google Scholar]
- 45.El Naggar R.F., Rohaim M.A., Bazid A.H., Ahmed K.A., Hussein H.A., Munir M. Biological characterization of wild-bird-origin avian avulavirus 1 and efficacy of currently applied vaccines against potential infection in commercial poultry. Arch. Virol. 2018;163:2743–2755. doi: 10.1007/s00705-018-3916-5. [DOI] [PubMed] [Google Scholar]
- 46.Hu S., Wang T., Liu Y., Meng C., Wang X., Wu Y., Liu X. Identifcation of a variable epitope on the Newcastle disease virus hemagglutinin-neuraminidase protein. Vet. Microbiol. 2010;140:92–97. doi: 10.1016/j.vetmic.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 47.Yuan P., Paterson R.G., Leser G.P., Lamb R.A., Theodore S., Jardetzky S. Structure of the ulster strain Newcastle Disease Virus hemagglutinin neuraminidase reveals auto-inhibitory interactions associated with low virulence. PLoS Pathog. 2012;8:e1002855. doi: 10.1371/journal.ppat.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Römer-Oberdörfer A., Veits J., Werner O., Mettenleiter T.C. Enhancement of pathogenicity of Newcastle disease virus by alteration of specifc amino acid residues in the surface glycoproteins F and HN. Avian Dis. 2006;50:259–263. doi: 10.1637/7471-111505R.1. [DOI] [PubMed] [Google Scholar]
- 49.Czeglèdi A., Ujvári D., Somogyi E., Wehmann E., Werner O., Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 2006;20:36–48. doi: 10.1016/j.virusres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Alexander D.J., Senne D.A. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Iowa State University Press; Ames, IA, USA: 2008. pp. 75–116. [Google Scholar]
- 51.Merz D. Importance of antibodies to the fusion glycoprotein of paramyxoviruses in the prevention of spread of infection. J. Exp. Med. 1980;151:275–288. doi: 10.1084/jem.151.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merz D.C., Scheid A., Choppin P.W. Immunological studies of the functions of paramyxovirus glycoproteins. Virology. 1981;109:94–105. doi: 10.1016/0042-6822(81)90474-8. [DOI] [PubMed] [Google Scholar]
- 53.Yusoff K., Nesbit M., McCartney H., Meulemans G., Alexander D.J., Collins M.S., Emmerson P.T., Samson A.C.R. Location of neutralizing epitopes on the fusion protein of Newcastle disease virus strains Beaudette. J. Gen. Virol. 1989;70:3105–3109. doi: 10.1099/0022-1317-70-11-3105. [DOI] [PubMed] [Google Scholar]
- 54.Kapczynski D., Afonso C.L., Miller P.J. Immune responses of poultry to Newcastle disease virus. Dev. Comp. Immunol. 2013;41:447–453. doi: 10.1016/j.dci.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 55.Miller P.J., Koch G. Newcastle disease. In: Swayne D.E., Glisson J.R., McDougald L.R., Nolan L.K., Suarez D.L., editors. Disease of Poultry. 5th ed. Wiley-Blackwell; Hoboken, NJ, USA: 2013. pp. 89–138. [Google Scholar]
- 56.De Battisti C., Salomoni A., Ormelli S., Monne I., Capua I., Cattoli G. Rapid pathotyping of Newcastle Disease Virus by pyrosequencing. J. Virol. Methods. 2013;188:13–20. doi: 10.1016/j.jviromet.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 57.Ganar K., Das M., Sinha S., Kumar S. Newcastle disease virus: Current status and our understanding. Virus Res. 2014;184:71–81. doi: 10.1016/j.virusres.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins M.S., Bashiruddin J.B., Alexander D.J. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch. Virol. 1993;128:363–370. doi: 10.1007/BF01309446. [DOI] [PubMed] [Google Scholar]
- 59.Aldous E.W., Mynn J.K., Banks J., Alexander D.J. A molecular epidemiological study of Avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 2003;32:239–256. doi: 10.1080/030794503100009783. [DOI] [PubMed] [Google Scholar]
- 60.Kim L.M., King D.J., Curry P.E., Suarez D.L., Swayne D.E., Stallknecht D.E., Slemons R.D., Pedersen J.C., Senne D.A., Winker K., et al. Phylogenetic diversity among low-virulence newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 2007;81:12641–12653. doi: 10.1128/JVI.00843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wrshana T.A., Dowidar Y.A., El-Fiky B.A., El-Rify A.M., El-Sayed W.A., Ahmed B.M. Infection of Bone Marrow-Derived Mesenchymal Stem Cells with Virulent Newcastle Disease Virus Maximizes Cytokine Production: A Step Toward vNDV Immunotherapy. Adv. Anim. Vet. Sci. 2022;10:2013–2023. doi: 10.17582/journal.aavs/2022/10.9.2013.2023. [DOI] [Google Scholar]
- 62.Naz Z., Ismat F., Saleem M., Iqbal M., Shehzad A., Rahman M. Single Step Detergent Assisted Extraction and Solubilization of the Recombinant Matrix Protein of Newcastle Disease Virus (NDV) and Development of ELISA for Detection of Anti-NDV Antibodies in Chickens. Pak. J. Zool. 2022;54:2765–2773. doi: 10.17582/journal.pjz/20210913180900. [DOI] [Google Scholar]
- 63.Fayyaz A., Zahoor M.K., Ashraf A., Zahoor M.A., Rasul A., Majeed H.N., Zulhussnain M., Ranian K., Riaz B., Khalil N. House Fly, Musca domestica (Diptera: Muscidae) Acts as a Mechanical Vector for Newcastle Disease of Poultry in Faisalabad, Pakistan. Pak. J. Zool. 2022;54:1943–1946. doi: 10.17582/journal.pjz/20190215080254. [DOI] [Google Scholar]
- 64.Snoeck C.J., Ducatez M.F., Owoade A.A., Faleke O.O., Alkali B.R., Tahita M.C., Tarnagda Z., Ouedraogo J.B., Maikano I., Mbah P.O., et al. Newcastle disease virus in West Africa: New virulent strains identified in non-commercial farms. Arch. Virol. 2009;154:47–54. doi: 10.1007/s00705-008-0269-5. [DOI] [PubMed] [Google Scholar]
- 65.Kim S.H., Chen Z., Yoshida A., Paldurai A., Xiao S., Samal S.K. Evaluation of fusion protein cleavage site sequences of Newcastle disease virus in genotype matched vaccines. PLoS ONE. 2017;12:e0173965. doi: 10.1371/journal.pone.0173965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zanetti F., Berinstein A., Carrillo E. Effect of host selective pressure on Newcastle disease virus virulence. Microb. Pathog. 2008;44:135–140. doi: 10.1016/j.micpath.2007.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.