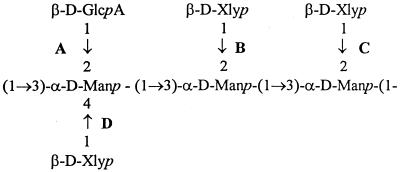Abstract
Cryptococcus neoformans strains exhibit variability in their capsular polysaccharide, cell morphology, karyotype, and virulence, but the relationship between these variables is poorly understood. A hypovirulent C. neoformans 24067A isolate, which usually produces smooth (SM) colony types, was found to undergo phenotypic switching and to produce wrinkled (WR) and pseudohyphal (PH) colony types at frequencies of approximately 10−4 to 10−5 when plated on Sabouraud agar. Cells from these colony types had large polysaccharide capsules and PH morphology, respectively. Scanning electron microscopy showed that different colony types were the result of altered cellular packing in the colony. Phenotypic switching was associated with quantitative and qualitative changes in capsular polysaccharide. Specifically, the glucuronoxylomannan (GXM) of the WR polysaccharide differed in the proportion of structural reporter groups and in increased xylose residue content linked at the 4 to 0 position. The relative virulence of the colony types was WR > PH > SM, as measured by CFU in rat lungs after intratracheal infection. Karyotype instability was observed in strain 24067A and involved primarily two chromosomes. Colonies with an alternative colony type exhibited more karyotype changes, which did not revert to the original karyotype in reverted colonies. In summary, this study revealed that phenotypic switching in C. neoformans (i) can produce WR colonies consisting of cells with either large capsule or PH morphology, (ii) is associated with production of structurally different GXM, (iii) is commonly associated with karyotype changes, (iv) can produce cells of PH morphology, and (v) can increase the virulence of a strain. Hence, phenotypic switching is an adaptive mechanism linked to virulence that can generate cell types with very different biological characteristics.
Cryptococcus neoformans is an encapsulated pathogenic fungus that is notorious for causing chronic infections, in particular chronic meningitis. Cryptococcosis is usually associated with impaired immune function but can also occur in apparently normal hosts (9, 25). In patients with AIDS, C. neoformans infections are often incurable, despite effective antifungal therapy. Studies of serial C. neoformans isolates from chronically infected patients have documented changes in virulence, capsular polysaccharide structure, and karyotype, suggesting that C. neoformans can undergo changes during chronic infection that may facilitate persistence in tissue (2, 5, 8, 13, 14). The propensity of C. neoformans to undergo phenotypic changes has also been demonstrated during in vitro passage for strain 24067, a common laboratory strain, which was noted to produce different variants ranging from avirulent to highly virulent (11). This process is referred to as microevolution.
Recently, phenotypic switching was described in three C. neoformans strains (SB4, J32, and 24067A), which resulted in various colony morphologies (17). Phenotypic switching has been described for many other pathogens and is often associated with changes in virulence (21, 22, 24, 26, 37, 39, 40). Phenotypic switching resulting in changes of the polysaccharide capsule and virulence has also been reported in the encapsulated bacterial pathogens Neisseria meningitidis and Haemophilus influenzae (22, 44). Although the molecular mechanisms mediating the switch are different among the various microorganisms, phenotypic switching is emerging as a fundamental mechanism of virulence that may allow persistence of infection in tissue by promoting the generation of new variants that successfully escape the immune response. For C. neoformans SB4, phenotypic switching is characterized by changes in colony morphology that range from smooth (SM) to wrinkled (WR). The switch between colony types is reversible and occurs at rates much higher than eukaryotic mutation rates. SB4 colony types differed in virulence for mice and rats, linking phenotypic switching and virulence in this fungus (17).
C. neoformans is unique among pathogenic fungi in having a polysaccharide capsule, which is an important virulence factor. The predominant capsular polysaccharide glucuronoxylomannan (GXM) confers the antigenic characteristics of the capsule and exhibits remarkable heterogeneity in GXM structure among serial isolates from patients and even among isolates assigned to a particular serotype (5, 6). Similarly, capsule size varies in vivo and is different in brain and lung tissue during murine infection (34). Although some of the factors involved in capsule regulation have been previously described (20, 41, 45), the relationship between capsule size, polysaccharide structure, and virulence remains a central unresolved problem in the field of C. neoformans pathogenesis. In a previous study, 24067A was reported to produce at least two colony phenotypes, SM and WR (17). In this study, we carried out a detailed analysis of strain 24067A to better understand this phenotypic switching system and its relationship to strain microevolution. Our studies indicate that strain 24067A can switch to at least two different WR colony types, composed of cells with either a large capsule or pseudohyphal (PH) morphology. Phenotypic switching was associated with a change in GXM structure and with differences in virulence and inflammatory response. Karyotype variability was observed but could not be directly linked to the colony type switch. Hence, our results suggest that phenotypic switching can provide an explanation for several unusual characteristics of C. neoformans, including its propensity toward strain microevolution, yeast-pseudohyphal transition, polysaccharide structure, and capsule size variability.
MATERIALS AND METHODS
Strains.
Strain 24067, a serotype D strain (also known as 52D), was originally isolated from a case of human cryptococcosis and can be obtained from the American Type Culture Collection (Rockville, Md.). The variant 24067A arose spontaneously during passage in vitro and was identified as different from its parent strain because it was hypovirulent for mice (11). SB4 is a serotype A strain that can switch between SM, WR, and serrated colony types (17).
Colony types, cell morphology, and capsule size measurements.
Colony type morphology was evaluated visually after growth at 30°C. The media used included Sabouraud dextrose agar (SDA) and broth (SD) (Difco Laboratories, Detroit, Mich.), chemically defined media and agar (15 mM glucose, 10 mM MgSO4, 29.4 mM KH2PO4, 3 μM thiamine), and SDA plates supplemented with 1 μM l-dopa (Sigma, Cleveland, Ohio). Cells were examined at a magnification of ×1,000 under oil in an India ink suspension. The distance from the cell wall to the outer margin capsule and the cell diameter (not including the capsule) was measured by using an eyepiece grid with a resolution of 0.5 μm.
Immunofluorescence staining.
Colonies from the various morphologies were suspended in and washed with 0.02 M phosphate-buffered saline (PBS) and air dried onto poly-l-lysine-coated slides. Cells were stained with six monoclonal antibodies (MAbs) to GXM (immunoglobulin G [IgG] 2H1, 4H3, 12A1, 13F1, 14A12, and 21D2) (3) and visualized with fluorescein isothiocyanate-labeled conjugated sheep antibody to mouse IgG or IgM. Staining patterns were categorized as annular and punctate, as previously described (7, 29). Nuclei and septa were stained with 2 μg of 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) per ml and 1 μg of calcofluor white M2R (Calcofluor; Sigma) per ml in PBS after prior fixation in 3.7% formaldehyde and washing with 0.1 M K2HPO4 (46).
SEM.
Scanning electron microscopy (SEM) of fractured colonies was performed as previously described (32). Briefly, colonies were grown on SDA plates supplemented with 1 μM l-dopa for 21 days. A scalpel was used to remove the whole yeast colony, including 5 mm of the surrounding agar from the culture plate. The sample was plunged into liquid nitrogen and then manually impact fractured with a scalpel blade in the center of the air interface surface of a colony. The colony agar pieces were lyophilized in a desiccator overnight, coated with gold palladium, and viewed at 10 kV at different magnifications with a JEOL-6400 microscope (JEOL, Peabody, Mass.).
Frequencies of alternative colony-forming phenotypes.
Frequencies of switching for the individual colony types were determined by visually scoring colonies with altered morphology after growth on SDA plates for 96 to 120 h. For frequency determination, 21 colonies of each phenotype were collected, suspended in PBS, counted with a hemacytometer, and plated at a density of 200 to 300 colonies per SDA plate.
Karyotype analysis.
Chromosomal DNA plugs were prepared from cultures derived from single colonies as previously described (14). After washing, the plugs were inserted into a 1% pulse field-certified agarose gel (Bio-Rad, Richmond, Calif.) and electrophoresis was performed in a contour-clamped homogeneous electric field DRIII variable-angle pulse field electrophoresis system (Bio-Rad) in 0.5% Tris boric acid EDTA buffer at 12°C. The electrophoresis conditions were programmed in two sequential blocks: first, a switch time of 90 s for 9 h then a switch time of 120 and 360 s for 63 h. Both blocks were run at 3.5 V/cm at an angle of 115°. Gels were stained with ethidium bromide and photographed. In addition, karyotype analysis of SM colonies was performed after prior UV and X-ray irradiation. For these experiments, cells were suspended in PBS and UV irradiated with 20,000 μJ/cm2 in a standard Stratolinker (Stratagene, La Jolla, Calif.). Cells were also irradiated with 3,000 rad in a cesium source, which delivers 1.4 rad/s. For both experiments, cell suspensions from irradiated and nonirradiated cells were plated to determine the percentage of killed cells after radiation. Southern blots of the karyotype gels were hybridized with a 1,782-bp [α-32P]dCTP-labeled probe to rRNA genes generated by PCR with the oligonucleotides MS284 and MS285 (10).
Purification and analysis of GXM.
GXM purification was done as previously described (4). Briefly, each isolate was grown at 30°C in 100 ml of SDA broth for 10 to 14 days, and the cells were removed by centrifugation. Sodium acetate per gram weight was added (10% [wt/vol]), and the pH was adjusted to 7.0 with glacial acetic acid. Polysaccharide was precipitated by adding 2.5 volumes of 95% alcohol. The precipitated polysaccharide was dissolved in 0.2 M NaCl (5 to 10 ml), and 3 mg of hexadecyltrimethyl-ammonium bromide (CTAB; Sigma) was added per milligram of polysaccharide. A 0.05% solution of CTAB was then added slowly to precipitate the GXM-CTAB complex. The precipitate was recovered by centrifugation at 10,000 × g and triturated with 10% ethanol, and the centrifugation was repeated. The pellet was dissolved in 5 ml of 1 M NaCl by stirring overnight at room temperature or until the precipitate completely dissolved. The GXM component was precipitated again by slowly adding 95% ethanol. The precipitate was dissolved in 2 M NaCl and dialyzed extensively against water before lyophilization. For nuclear magnetic resonance (NMR) spectroscopy, the GXM was exchanged in 99.66% D2O, transferred into a 5-mm NMR tube, and analyzed in a Varian Unity PLUS 500 spectroscope. 1H chemical shifts were measured relative to the methyl groups of sodium 4,4-demethyl-4-silapentane-1-sulfonate taken at 0.00 ppm. The data was processed off-line by using FELIX 2.30 software package (Biosym/Molecular Simulations, San Diego, Calif.) on a Silicon Graphics Indy workstation.
Animal studies.
Because 24067A is hypovirulent in mice, we used the rat model of intratracheal infection to study organ persistence and local inflammatory response (18). For intratracheal infection, male adult Fisher rats were anesthetized with methoxyflurane, placed on an inclined wood board, and intubated with the help of an otoscope. The vocal cords were visualized, and 0.2 ml of 107 C. neoformans cells were placed in the trachea distal to the vocal cords with a blunted 23-gauge spinal needle. Rats were killed by an overdose of pentobarbital, organs were removed, weighed, and homogenized, and dilutions were plated on SDA for CFU determination. Portions of each lung were preserved in formalin and embedded in paraffin for histological evaluation.
Histology and immunohistochemistry.
Tissue immunohistochemistry with MAb 2H1 to GXM was done to detect tissue polysaccharide as previously described (19). Hydrated tissue sections were incubated in 3% hydrogen peroxide to block exogenous peroxidase activity and then blocked with 10% goat serum. The murine antibody MAb 2H1 (10 μg/ml) was used as the primary antibody. Peroxidase-conjugated goat anti-mouse IgG1 was used as a secondary antibody and developed by incubation in diaminobenzidine. The inflammatory response was scored on the basis of the intensity and location of the cellular infiltrate as follows: minimal inflammation, consisted of occasional perivascular cuffing and minimal hyperplasia of peribronchial lymphoid tissue without granuloma formation; moderate inflammation, consisted of perivascular cuffing, moderate hyperplasia of peribronchial lymphoid tissue, and occasional small granulomas (<5% of tissue section); extensive inflammation, consisted of perivascular cuffing and hyperplasia of peribronchial lymphoid tissue with large areas of organized granuloma formation (up to 30% of tissue section). GXM deposition in tissue was graded as none, minimal, moderate, or extensive.
Statistical analysis.
The Student t test and chi-square test were used to compare cell size, capsule size, and log-transformed fungal burdens (primer; McGraw Hill, Inc., N.Y.).
RESULTS
24067A phenotypic switching system.
24067A expressed three different colony types: SM, WR, and PH (Fig. 1A). Reverted colonies from WR or PH exhibited the same smooth colony phenotype as SM and were denoted as reverted SM (RSM; RSM-WR and RSM-PH refer to RSM colonies reverted from WR or PH, respectively). A brief description of the SM and WR colony types of strain 24067A has been previously reported (17). The SM colonies were round with a smooth dome surface and smooth edges. WR colonies had a smooth dome surface and irregular edges, whereas the PH colony type exhibited wrinkled edges as well as a wrinkled dome surface. The distinction of wrinkled colonies into WR and PH was made after noting that WR colonies turned mucoid after prolonged incubation, whereas PH colonies did not. The expression of the WR colony type, not the SM or PH colony type, was dependent on nutritional factors and temperature. The WR colony type was expressed at 30°C but not at 37°C on SDA and not on minimal media. Surface properties of the WR and PH colonies were altered, and both adhered to the agar surface and exhibited flocculence (clumps) in liquid broth. Given macroscopic differences in colony types, we proceeded to study cellular and colony characteristics.
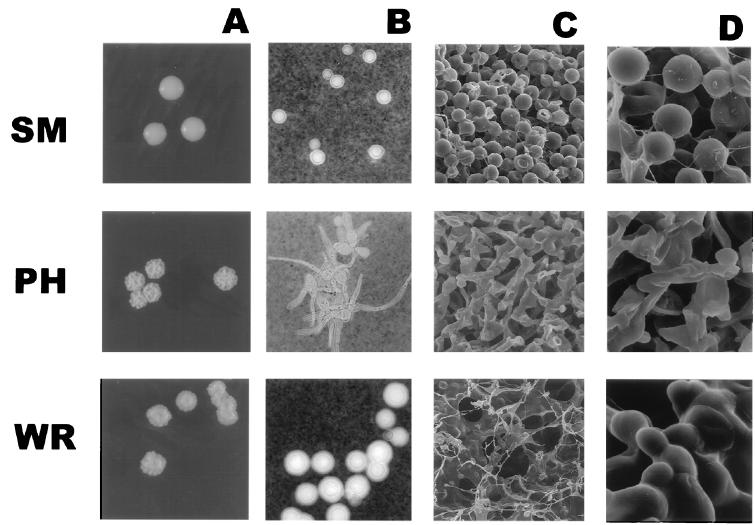
FIG. 1.
(A) SM, WR, and PH colony types. Magnification, about ×4. (B) India ink staining of SM, WR, and PH cells. Magnification, ×250. (C and D) SEM of SM, WR, and PH colonies. Magnifications, ×1,000 and ×5,000, respectively.
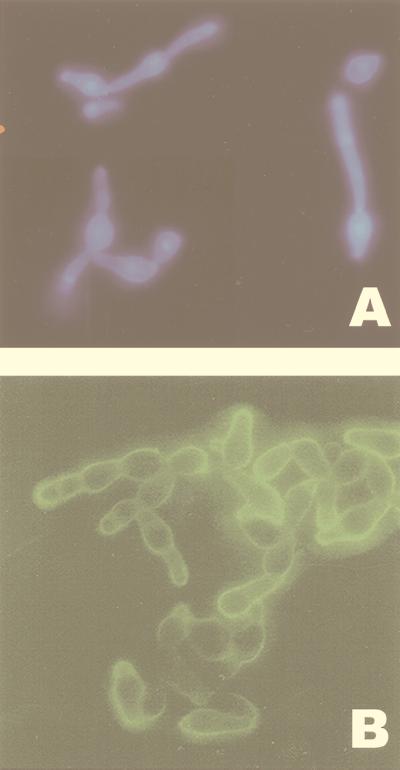
Microscopic analysis of the 24067A phenotypes revealed differences in capsule size, cell size, and morphology (Fig. 1B). At 30°C, SM and RSM cells and capsules were small compared with WR cells (P < 0.0001), whereas at 37°C, the SM, RSM, and WR cells exhibited small cells and small capsules (Table 1). In contrast, PH cells were elongated with long pseudohyphal cytoplasmic protrusions and had a thin polysaccharide capsule. PH cells, stained with DAPI, revealed one nucleus per elongated attached cell, and no clamp connections were observed (Fig. 2A). Immunofluorescence staining with GXM specific antibody demonstrated the presence of a GXM-like epitope on the surface of PH cells (Fig. 2B). Staining patterns with GXM specific antibodies between SM, PH, and WR were different, but the differences were quantitative and appeared to be related mainly to differences in capsule size (data not shown).
TABLE 1.
Summary of cellular characteristic differences of 24067A colony types
| Colony type | Cell morphology | Cell diameter | Capsule diameter | GXM SRGa |
|---|---|---|---|---|
| SM | Yeast | 4.65 ± 0.49 | 0.7 ± 0.25 | M1, M2, M5, M6 |
| WR | Yeast | 6.19 ± 1.36 | 2.0 ± 0.7 | M1, M5 |
| PH | PH | NAb | NA | M1, M5, M6 |
| RSM-WR | Yeast | 4.0 ± 1.0 | 0.7 ± 0.24 | M1, M2, M6 |
| RSM-PH | Yeast | NDc | ND | M1, M5 |
See text for the percent distribution of each SRG (M1 to M6).
NA, nonapplicable.
ND, not done.
FIG. 2.
(A) DAPI staining of cells from PH colony type. (B) Indirect immunofluorescence with MAb 2H1 of PH cells. Magnification, ×250.
SEM of fractured SM, WR, and PH colonies demonstrated differences in microarchitecture among the three colony types (Fig. 1C and D). The SM cells were round and patched in an ordered fashion and distinctly separated from each other. The PH colony was composed of elongated cells resulting in an ordered but less dense packing of the colony. In contrast, the WR colony microarchitecture was the least ordered and exhibited cells of different size matted to each other by an extracellular substance, most likely polysaccharide.
Frequencies of alternative colony-forming phenotypes.
The colony types were generally stable during in vitro and in vivo passage; however, in the first passage after freeze-thawing they were noted less stable, and occasionally additional phenotypes were observed. Frequencies of occurrence of the individual colony types on SDA plates ranged between 10−3 and 10−5 (Table 2). The SM colony type was the most stable form. Switching from the SM→PH phenotype occurred more frequently than from the SM→WR phenotype, especially in younger passage cultures. Older cultures kept at 4°C on SDA plates switched more frequently to the WR colony type (Table 2, experiments 1, 7, and 8). On one occasion, we also demonstrated a switch from the SM→WR→RSM→PH colony type. UV and X-ray radiation of SM cell suspensions resulted in 40 to 90% cell death and increased the frequency of phenotypic switching 4- to 10-fold (Table 2). Irradiation promoted switches to the PH colony type only, although several new colony morphology and sectored colonies were also observed (data not shown).
TABLE 2.
Frequencies of switching in the 24067A colony types
| Phenotypes plated or treatment | Expt no. | No. of colonies scored | No. of switches (type) | Frequency of alternative colony types |
|---|---|---|---|---|
| SM→WR or PH | ||||
| SM | 1a | 6.8 × 104 | 7 (3 WR, 4 not tested) | 1.0 × 10−4 |
| SM | 2 | 4.1 × 104 | 4 (all PH) | 9.7 × 10−5 |
| SM | 3 | 2.6 × 104 | 4 (all PH) | 2.5 × 10−4 |
| SM | 4 | 1.1 × 104 | 2 (all PH) | 1.8 × 10−4 |
| SM | 5 | 1.25 × 104 | 0 | <1.25 × 10−4 |
| SM | 6 | 2 × 104 | 4 (3 PH, 1 not tested) | 2 × 10−4 |
| SM | 7a | 5.6 × 104 | 4 (3 WR, 1 PH)b | Not doneb |
| SM | 8 | 4 × 104 | 3 (2 PH, 1 WR) | 7.5 × 10−5 |
| WR→SM | ||||
| WR1 | 1.5 × 103 | 3 (SM) | 1.9 × 10−3 | |
| WR2 | 2.5 × 103 | 7 (SM) | 2.7 × 10−3 | |
| PH→SM | ||||
| PH | 8.8 × 103 | 14 (SM) | 1.6 × 10−3 | |
| RSM→WR or PH | ||||
| RSM-WR1 | 4.1 × 103 | 1 (WR) | 2.4 × 10−4 | |
| RSM-WR2 | 1 | 1.2 × 105 | 2 (1 PH, 1 not tested) | 1.6 × 10−3 |
| RSM-WR2 | 2 | 8.8 × 103 | 4 (all WR) | 4.5 × 10−4 |
| RSM-PH | 1.6 × 104 | 4 (all PH) | 2.5 × 10−3 | |
| UV irradiatedc | ||||
| SM | 1 | 1.0 × 104 | 6 (all PH) | 6.0 × 10−4 |
| SM | 2 | 2.3 × 103 | 3 (all PH) | 1.3 × 10−3 |
| X-irradiatedd | ||||
| SM | 1 | 1.3 × 104 | 7 (all PH) | 5.4 × 10−4 |
| SM | 2 | 7.6 × 103 | 5 (all PH) | 6.6 × 10−4 |
Both experiments 1 and 7 were done with old SM colonies (>1 year) which were kept at 4°C.
Multiple sectored colonies and unstable WR phenotypes were noted; therefore, no frequency was calculated.
200 μJ of UV radiation resulted in 40 and 90% cell death and a 4- or 10-fold increase in switched colony types.
3,000-rad X-ray irradiation resulting in 50% cell death.
Karyotype changes in SM, WR, and PH colony types.
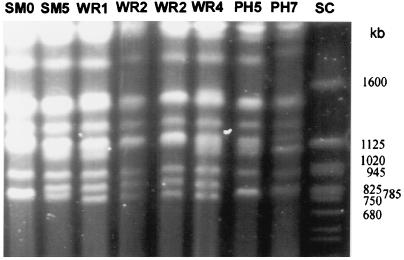
The karyotypes of 43 individual colonies (9 SM phenotypes, 12 WR phenotypes, 9 PH phenotypes, and 13 RSM phenotypes) were analyzed by pulse field electrophoresis of chromosomal DNA (Fig. 3). Colonies were selected from both independent switch events and multiple colonies of single clones. All WR, PH, and RSM colonies demonstrated changes in karyotype relative to the original karyotype (SM0 in Fig. 3). Two different karyotypes were observed among WR colonies (WR1 and WR2 in Fig. 3), and three karyotype patterns were observed among PH colonies, one of which was identical to WR2 (two shown in Fig. 3 [PH5 and PH7]). One of nine SM colonies (SM5 in Fig. 3) exhibited a change in karyotype, which was indistinguishable from WR1. This particular SM colony had been kept at 4°C for a few months but retained its SM phenotype on visual and microscopic inspection. The karyotype of most RSM-WR colonies resembled that of their original WR colony, whereas the karyotype of RSM-PH colonies was more variable. Karyotype changes in strain 24067A generally involved the same two chromosomes, which are approximately 1,100 and 780 kb. The size of the smaller chromosome was more variable. In PH7, a size change was also observed in a larger size chromosome (Fig. 3). Repeated analysis of various clones from individual colony types revealed consistent karyotypes, except for those of RSM-PH. UV or X-ray irradiation of 15 SM colonies did not result in major karyotype changes, except for minor size changes in the smallest chromosome (approximately 700 kb), which were observed in 2 SM colonies (data not shown).
FIG. 3.
Karyotypes of the SM, WR, and PH colony types. SC, Saccharomyces molecular size marker.
To determine whether rRNA genes are located on the chromosomes involved in the karyotype change, we performed Southern hybridization with a radiolabeled probe to rRNA genes and confirmed that these genes are located on a high-molecular-size chromosome (>3 Mb), as previously described (47). In summary, karyotype differences were frequent among the 24067A colony types, and reversion in colony morphology was not associated with reversion in karyotype changes.
Analysis of GXM structure.
The GXM of strain 24067A, specifically WR, PH, SM, and RSM colony types, was analyzed by NMR spectroscopy. GXM constitutes the major exopolysaccharide of the capsule and is composed of (1→3)-linked linear α-d-mannopyranan with β-d-xylopyranosyl (Xylp) and β-d-glucopyranosyluronic acid residues added to the mannose at various positions. Cherniak et al. defined six (M1 to M6) structural reporter groups (SRG) based on the amount of 2-to-0-linked and 4-to-0-linked Xylp residues and 2-to-0-linked β-d-glucopyranosyluronic acid residues by computer-assisted analysis (6) (Fig. 4). Our analysis revealed differences of GXM structure for the various colony types. Specifically, the GXM analysis of two WR colonies revealed a mixture of SRG M1 (60 and 61%) and M5 (40 and 39%), whereas the GXM structure of two RSM-WR colonies was a mixture of M1 (55 and 53%), M2 (9 and 13%), and M6 (36 and 34%).
FIG. 4.
Core repeating triad of GXM polymer. The SRG M1, M2, M6, and M5 differ in their Xlyp residue content. M1 has a Xlyp residue in the 2 to 0 position at C. M2 has a Xlyp residue in the 2 to 0 position at B and C. M3 has a Xlyp residue in the 2 to 0 position at C and B and in the 4 to 0 position at D. M5 has a Xlyp residue in the 2 to 0 position at C and also in the 4 to 0 position at D. M6 has no Xlyp residues. GlcpA, glucopyranosyluronic acid; Manp, mannopyranan.
The GXM structure of the parent 24067A strain and eight other 24067 variants was previously determined to exhibit 100% SRG M1, the predominant SRG of serotype D strains (11). In this study, the GXM structure of the 24067A SM phenotype was analyzed again and found to be a mix of M1 (48%), M2 (20%), M5 (12%), and M6 (21%). Microscopic analysis of all cultures prior to GXM purification was performed to insure that switching back to different phenotypes had not occurred during the prolonged culture in SD broth. Observing two cell populations, one with small capsule size and one with large capsules, would suggest switching in culture. The WR and the RSM-WR cultures were composed of homogenous cell populations, whereas the SM culture was composed of a heterogeneous cell population consisting of approximately 60% cells with small capsules and 40% cells with large capsules, suggesting that switching had occurred in culture during prolonged cultivation. The GXM structure of the two PH and RSM-PH colonies was more variable and did not revert to the original structure. For one PH colony, the SRG was a mix of M1 (68%), M5 (13%), and M6 (19%), and for the other PH colony, the SRG was a mix of M1 (79%) and M5 (21%) only. The GXM structure of the RSM-PH was unchanged from that of the PH colony and was found to be a mix of M1 (66%) and M5 (34%). To determine whether phenotypic switching was associated with GXM changes in other strains, we analyzed the GXM from three colony types of the serotype A strain SB4, known to switch between three phenotypes: SM, serrated, and WR (17). For SB4, the SM and WR colony had GXM with 100% SRG M2, but the serrated colony type exhibited a mix of M2 and M3 (65 and 35%, respectively). Hence, phenotypic switching was associated with GXM structure changes in two C. neoformans strains, which include serotypes A and D.
Virulence studies.
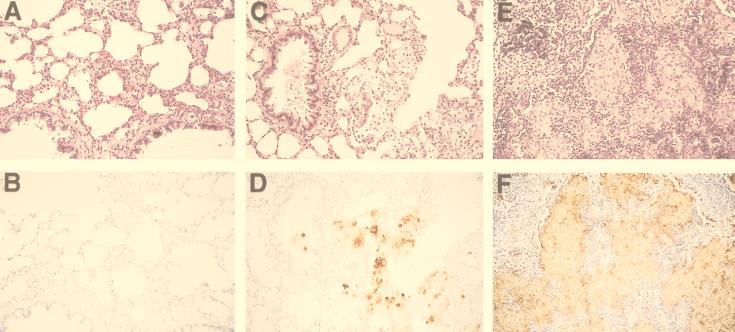
The 24067A variant exhibits attenuated virulence and does not usually cause disease in infected mice. Therefore, we relied on a pulmonary infection model in rats to evaluate the virulence of 24067A. Inoculation of C. neoformans in rats has been shown to produce chronic pneumonia. The measure of virulence was organ fungal burden and inflammatory response to infection. Rats (n = 3 or 4) were inoculated via the trachea with 107 SM, WR1, WR2, RSM-WR, WR, PH, or RSM-PH cells and killed after 2 weeks. The fungal organ burden measured in CFU differed significantly for the various colony types and was highest in rats infected with the WR phenotype and lowest in rats infected with the SM phenotype (Table 3). Rats infected with RSM-WR1 and RSM-PH cells exhibited higher CFU than rats infected with SM cells but lower CFU than rats infected with WR cells. The fungal burden in PH-infected rats was variable. The extent of the inflammatory response correlated with the immunoreactivity to GXM and fungal burden (Fig. 5). The tissue from rats infected with SM cells showed a mild inflammatory response and no granuloma formation, except for one rat, which exhibited occasional small granulomas. There was minimal to no GXM deposition in the tissue. In rats infected with RSM-WR1 and RSM-PH cells, there was minimal inflammatory response with some perivascular cuffing and hyperplasia of lymphoid tissue and rare to occasional granulomas were seen with weak GXM immunoreactivity. In rats infected with PH cells, the response was more variable. Two rats infected with PH cells (with low CFU) had minimal to no inflammatory responses, similar to rats infected with SM cells, whereas the two rats with higher CFU had minimal inflammation that was similar to that of the rats infected with RSM-WR and RSM-PH cells. The GXM deposition in rats infected with PH cells ranged from minimal to none. The most intense inflammatory response was seen in rats infected with WR cells. This included perivascular cuffing, hyperplasia of the peribronchial lymphoid tissue, and granuloma formation. GXM immunoreactivity was extensive and localized mainly to epitheloid cells within granulomas. In summary, cells from the WR colony type were more virulent than the parental SM colony type of 24067A.
TABLE 3.
Comparison of virulence of the various 24067A colony types
| Phenotype | na | Lung CFUb | Histologyc
|
Pe (phenotypes) | |
|---|---|---|---|---|---|
| Inflammation | GXM | ||||
| SM | 4 | 2.74 ± 0.86 | ± | ± | 0.035 (SM versus WR2) |
| WR2 | 3 | 4.75 ± 0.07 | +++ | +++ | |
| SM | 3 | 2.40 ± 0.53 | ± | ± | 0.06 (SM versus PH) |
| PH | 4 | 2.68 ± 0.79 | +d | + | 0.177 (PH versus RSM-PH) |
| RSM-PH | 3 | 3.49 ± 0.47 | + | + | 0.627 (RSM-PH versus SM) |
| WR1 | 3 | 4.81 ± 0.45 | +++ | +++ | 0.019 (WR1 versus RSM-WR) |
| RSM-WR1 | 4 | 3.97 ± 0.2 | + | + | |
n, number of rats in each experiment.
Log CFU per gram of lung tissue ± standard deviation.
Histology scores: ±, none or minimal; +, minimal; ++, moderate; +++, extensive.
Two rats had no evidence of infection, and two rats had minimal inflammation.
The P value was determined by the Student t test.
FIG. 5.
Inflammatory response of rat lungs infected with SM (A), PH (B), and WR (C) cells. Upper panels show HE staining of lung sections. Lower panels show GXM immunoreactivity of the corresponding sections. Brown areas contain GXM. Magnification, ×50.
DISCUSSION
We previously established that phenotypic switching occurred in C. neoformans and demonstrated this phenomenon in three genetically diverse strains, including 24067A (17). Now we extend those findings to show that strain 24067A can reversibly switch from an SM colony to two distinct wrinkled colony types composed of different cell types. The wrinkled colony types differ macroscopically and are composed of cells with either a large polysaccharide capsule (WR cells) or PH cell morphology. Since variation in capsule sizes and the sporadic description of variants with PH morphology are enigmatic aspects of C. neoformans biology, we carried out a detailed study of the 24067A strain (20, 34). The results of this study provide new insights into the relationship between phenotypic switching, capsule size, polysaccharide structure, microevolution, and virulence that help to reconcile many discrepant observations in the literature.
The observation that C. neoformans can switch to a PH cell morphology was intriguing because PH cell types have been occasionally described in clinical specimens (12). Their origin, however, is obscure. Our results suggest that these occasional reports of pseudohyphal strains in clinical specimens originate from phenotypic switching events. The biological importance of the PH morphology is not well understood, but Neilson et al. showed that pseudohyphal forms resist phagocytosis by soil amoebae which are natural predators of C. neoformans (27, 28). In Saccharomyces cerevisiae, pseudohyphal forms are induced by nutritional signals that allow the fungus to invade the agar medium and forage nutrients (16). Hence, phenotypic switching to PH cell morphology may provide C. neoformans with a survival advantage in certain environments.
One of the distinctive features of phenotypic switching in 24067A is the formation of colonies with a WR, uneven morphology. The cellular characteristics responsible for WR and SM colony types are not understood, and little is known about the microbial cellular characteristics that affect colony morphology. Investigation of colony types by SEM revealed that the altered colony morphology was the result of differences in the microarchitecture of colonies. In colonies containing PH cells, the morphology appears to be the result of disordered packing of elongated and asymmetrical cells. However, in colonies containing WR cells the morphology appears to be the result of disordered packing caused by a profuse amount of exopolysaccharide. In contrast, SM colonies consist of round symmetrical cells packed in a more orderly fashion. These findings suggest two variables that can affect colony morphology: cell shape and capsule production. SEM analysis of altered colony phenotypes of a switching Candida strain 3153A showed that differences of microarchitecture were the result of variable proportions of blastospores and hyphal cells within the colony (31, 32). In contrast, the colony types in our study are composed of homogenous cell populations, and the differences in colony types appear to be due to microscopic changes in cellular morphology and/or polysaccharide production that translate into differences in cell packing within a colony.
The observation that some WR colonies consisted of cells with large capsules packed in a mass of exopolysaccharide prompted us to examine the structure of GXM by NMR spectroscopy. This analysis showed that the GXM structure was different in colonies with alternative colony phenotypes relative to the colony they switched from. For 24067A, the GXM of WR cells was composed of the M1 and M5 SRG, whereas the GXM of RSM-WR cells was composed of the M1, M2, and M6 SRG. For SB4, the GXM of SM and WR cells was composed of M2, whereas the serrated cells were composed of M2 and M3. Most striking was the observation that the new GXM in both strains contained a mix of the M1 and M5 or M2 and M3 SRG, respectively. The synthesis of SRG M5 and M3 involves the linkage of a Xylp residue to the 4 to 0 position in the mannose backbone, which requires a different enzyme than the 2 to 0 linkage. Hence, phenotypic switching to WR and serrated colonies appears to activate a xylose transferase that adds Xylp residues to the 4 to 0 position. Furthermore, reversion of WR to RSM-WR colonies is presumably accompanied by inactivation of this enzyme, because the M5 SRG were not detected in the GXM of two RSM colonies (from WR colonies).
The GXM structure of 24067A SM, PH, and RSM-PH cells was composed of a mixture of the M1, M2, M6, and M5 SRG, and in contrast to WR cells, the reversion of PH to RSM-PH cells did not result in a reversion of the GXM structure. It is not clear whether this is the result of incomplete reversion to the original phenotype or continuous phenotypic switching in vitro. The latter interpretation is favored by our observation that in 24067A in vitro growth produced a heterogenous cell population after 2 weeks. In contrast, WR and RSM-WR colonies and the phenotypes of SB4 were homogenous cell populations after in vitro growth. Consistent with incomplete reversion of phenotypes is the observation that RSM-WR and RSM-PH cells are more virulent than SM cells (see below). The GXM structure of nine different variants of strain 24067 from different laboratories (all originally derived from the ATCC 24067 strain and all exhibiting a SM colony type) has previously been shown to be the M1 SRG. In that study, the polysaccharide was harvested from 24067 cells grown in minimal media, a condition where the phenomenon of phenotypic switching has not been observed (11). The predominance of the M1 SRG and the homogeneity of GXM in 24067 variants in an earlier study presumably reflect growth in minimal media, which does not support phenotypic switching or high polysaccharide production (17). Analysis of other nonswitching variants of strain 24067 grown in SD broth revealed a GXM composed primarily of the M1 SRG and a small percentage of M6 (data not shown).
The ratios of SRG in the GXM analysis of WR and RSM isolates were similar. We interpret this observation to indicate that the mix of SRG represents a new, unique GXM rather than distinct GXM polymers on different cell populations that are switching back and forth. A detection method to distinguish the two GXMs on a cellular level is not available at this time. GXM specific antibodies yield distinguishable staining patterns among the various cell types. However, since the capsule size is so different between RSM and WR cells, we are not convinced that these differences reflect qualitative differences in antibody binding to different SRG. GXM with various ratios of different SRG have been described in other C. neoformans strains, and the variability of SRG ratios was also noted in serial isolates from patients, including SB4 (5, 6). Hence, we propose that GXM variability in C. neoformans strains is a controlled change, such as that promoted by phenotypic switching in activation of enzymes involved in GXM synthesis.
The relationship between capsule size and virulence is not entirely understood. Granger et al. showed that the capsule size of C. neoformans is regulated in vitro by CO2 and that loss of this regulation is associated with attenuation (20). Rivera et al. demonstrated that cells in brain tissue infected with strain 24067 have small capsules, whereas cells in lung tissue exhibit large capsules (34). This work contributes additional support to the hypothesis that capsule regulation is essential for virulence. Our data demonstrates that capsule size is also regulated by phenotypic switching and involves the synthesis of a biochemically distinct GXM.
Goldman et al. previously demonstrated increased virulence in the WR colony type of the SB4 strain. The WR colony type in that switching system is more virulent presumably because it elicits a less organized inflammatory response that cannot contain the fungal infection (17). The 24067A strain is hypovirulent and does not establish an infection in murine models. In the rat model of pulmonary infection, the WR cell derived from the SM cell caused persistent infection with higher CFU and more local inflammation. Hence, phenotypic switching was associated with enhanced virulence in both the SB4 and 24067A systems. The PH colony type and both RSM colony types elicit a minimal to moderate immune response and were less virulent compared to the WR colony type. Other investigators have shown that pseudohyphal variants are less virulent in animal models and can elicit a protective immune response (15, 38). One of the limitations of defining phenotypes by colony morphology is that visual examination of colony type may not be a sensitive indication of changes in fungal cells. Hence, it is conceivable that complex phenotypes do not fully revert all the altered cellular characteristics and that our scoring system could misinterpret partially reverted colonies as completely reverted colonies, which would explain why RSM colonies did not behave exactly as SM in vivo. Nevertheless, our data supports the concept that phenotypic switching results in new variants, which are characterized by altered virulence.
Phenotypic switching in other pathogens is often mediated by a reversible rearrangement of genomic sequences (22, 39, 44), which in return could also result in karyotype changes (48). Some Candida strains that undergo phenotypic switching exhibit karyotype variability, but this could not always be correlated with morphological changes (1, 23, 33, 35, 36, 42, 43). For strain 24067A, phenotypic switching was commonly associated with karyotype changes, whereas no karyotype changes were observed in SB4 (17). We do not know the underlying molecular mechanism of the observed karyotype changes, and possibilities include rearrangement and/or breakage of chromosomes. Karyotype analysis will detect only significant chromosome size changes (>500 kb). Genomic sequences that commonly rearrange include rRNA repeats and repetitive sequences. In 24067A, the rRNA gene cluster is not located on the two chromosomes that exhibit chromosome length polymorphism (CLP). The rRNA genes were located on a large chromosome that does not appear to be involved in karyotype changes in 24067A, although small size changes of this chromosome may not be detected under these conditions. Franzot et al. demonstrated karyotype variability in variants of 24067 (11). Two of nine 24067 variants exhibited a change in one of the two chromosomes that constitute the CLP. The 24067 variant D2 exhibited CLP in the lower-weight chromosome, whereas the 24067 variant G exhibited CLP in the higher-weight chromosome. It is conceivable that these chromosomes contain a specific hot spot for rearrangement. Two observations suggest that there is no consistent cause-and-effect relationship between the observed CLP and the colony type switch. First, spontaneous CLP was observed after prolonged culture in one SM 24067A colony type and by Franzot et al. in two SM colony variants of 24067 (11). Second, the reversion of phenotypes was not accompanied by a karyotype reversion. Hence, we favor the hypothesis that the observed CLPs are a reflection of the genomic instability that mediates phenotypic switching. In that regard, the 24067A system appears to be similar to the 3153A switching system in Candida, which also generates multiple phenotypes and exhibits karyotype variability that cannot always be linked to specific colony types (33, 35). In addition, deletion of the SIR2 genes in an auxotroph derivative of a wild-type Candida strain was recently reported to result in high-frequency colony type variability and karyotype variability, suggesting that both phenotypic switching and karyotype stability are controlled by genes involved in silencing (30).
In summary, our results link phenotypic switching to several unresolved problems of cryptococcal biology and virulence, including variation in capsule size within individual strains, differences in inflammation during infection, the sporadic description of hyphal strains, heterogeneity in polysaccharide structure within a serotype, and strain microevolution. We formally propose that phenotypic switching is a mechanism responsible for the protean inflammatory responses observed during infection and organ colonization, differences in polysaccharide structure, and the observation of rapid microevolution in vivo and in vitro. Phenotypic switching for C. neoformans produces altered variants that may have a survival advantage in the environment or infection, based on altered cellular characteristics such as morphology and capsular polysaccharide expression.
ACKNOWLEDGMENTS
We thank Leslie Gunther for technical assistance in electron microscopy.
This work was supported by grants from NIH (R01-AI33774, AI3342, and HL59842 to A.C.; K08AI01300 to D.L.G.; and AI-31769 to R.C.), the Burroughs Welcome Fund (to A.C.), and the Howard Hughes Medical Institute (postdoctoral fellowship to B.C.F.).
R.J. is a senior student at Bronx Science High School.
REFERENCES
- 1.Barton R, Scherer S. Induced chromosome rearrangements and morphologic variation in Candida albicans. J Bacteriol. 1994;176:756–763. doi: 10.1128/jb.176.3.756-763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt M, Pfaller M, Hajjeh R A, Gravis E, Rees J, Spitzer E, Pinner R, Mayer L, et al. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immune deficiency virus-associated cryptococcosis. J Infect Dis. 1996;174:812–820. doi: 10.1093/infdis/174.4.812. [DOI] [PubMed] [Google Scholar]
- 3.Casadevall A, Mukherjee J, Devi S J, Schneerson R, Robbins J B, Scharff M D. Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J Infect Dis. 1992;165:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 4.Cherniak R, Morris L, Anderson B, Meyer S. Facilitated isolation purification and analysis of glucuronoxylomannan of Cryptococcus neoformans. Infect Immun. 1991;59:59–64. doi: 10.1128/iai.59.1.59-64.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherniak R, Morris L, Belay T, Spitzer E, Casadevall A. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect Immun. 1995;63:1899–1905. doi: 10.1128/iai.63.5.1899-1905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherniak R, Valafar H, Morris L C, Valafar F. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin Diagn Lab Immunol. 1998;5:146–159. doi: 10.1128/cdli.5.2.146-159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleare W, Casadevall A. The different binding patterns of two immunoglobulin M monoclonal antibodies to Cryptococcus neoformans serotype A and D strain correlate with serotype classification and differences in functional assays. Clin Diagn Lab Immunol. 1998;5:125–129. doi: 10.1128/cdli.5.2.125-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Currie B, Sanati H, Ibrahim A, Edwards J, Casadevall A, Ghannoum M. Sterol compositions and susceptibilities to amphotericin B of environmental Cryptococcus neoformans isolates are changed by murine passage. Antimicrob Agents Chemother. 1995;39:1934–1937. doi: 10.1128/aac.39.9.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Currie B P, Casadevall A. Estimation of the prevalence of cryptococcal infection among HIV infected individuals in New York City. Clin Infect Dis. 1994;19:1029–1033. doi: 10.1093/clinids/19.6.1029. [DOI] [PubMed] [Google Scholar]
- 10.Fan M, Currie B P, Gutell R R, Ragan M A, Casadevall A. The 16s-like, 5.8s and 23s-like rRNAs of the two varieties of Cryptococcus neoformans: sequence, secondary structure, phylogenetic analysis and restriction fragment polymorphisms. J Med Vet Mycol. 1994;32:163–180. doi: 10.1080/02681219480000231. [DOI] [PubMed] [Google Scholar]
- 11.Franzot S, Mukherjee J, Cherniak R, Chen L, Hamdan J, Casadevall A. Microevolution of standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E R, Duma R J, Shadomy H J, Utz J P. Meningoencephalitis due to hypha-forming Cryptococcus neoformans. Am J Clin Pathol. 1970;55:30–33. doi: 10.1093/ajcp/55.1.30. [DOI] [PubMed] [Google Scholar]
- 13.Fries B, Casadevall A. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J Infect Dis. 1998;178:1761–1766. doi: 10.1086/314521. [DOI] [PubMed] [Google Scholar]
- 14.Fries B C, Chen F, Currie B P, Casadevall A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromtling R A, Blackstock R, Hall N K, Blumer G S. Immunization of mice with an avirulent pseudohyphal form of Cryptococcus neoformans. Mycopathologia. 1979;68:179–181. doi: 10.1007/BF00578527. [DOI] [PubMed] [Google Scholar]
- 16.Gimeno J, Ljungdahl P, Styles C, Fink G. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 17.Goldman D, Fries B, Franzot S, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci USA. 1998;95:14967–14972. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman D, Lee S, Casadevall A. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect Immun. 1994;62:4755–4761. doi: 10.1128/iai.62.11.4755-4761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman D L, Lee S C, Casadevall A. Tissue localization of Cryptococcus neoformans glucuronoxylomannan in the presence and absence of specific antibody. Infect Immun. 1995;63:3448–3453. doi: 10.1128/iai.63.9.3448-3453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granger D L, Perfect J R, Durack D T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Investig. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt S, Hilse R, van Putten J, Gerady-Schahn R, Unkmair A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 22.Hood D, Deedman M, Jennings M, Biseric M, Fleischmann R, Venter J, Moxon E. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janbon G, Sherman F, Rustchenko E. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc Natl Acad Sci USA. 1998;95:5150–5155. doi: 10.1073/pnas.95.9.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lysnynansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangement. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell T, Perfect J. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myler P, Allison J, Agabian N, Stuart K. Antigenic variation in African trypanosomes by gene replacement or activation of alternate telomeres. Cell. 1984;39:203–211. doi: 10.1016/0092-8674(84)90206-x. [DOI] [PubMed] [Google Scholar]
- 27.Neilson J B, Fromtling R A, Bulmer G S. Pseudohyphal forms of Cryptococcus neoformans: decreased survival in vivo. Mycopathologia. 1981;73:57–59. doi: 10.1007/BF00443015. [DOI] [PubMed] [Google Scholar]
- 28.Neilson J B, Ivey M H, Bulmer G S. Cryptococcus neoformans pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun. 1978;20:262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nussbaum G, Cleare W, Casadevall A, Scharff M D, Valadon P. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J Exp Med. 1997;185:685–694. doi: 10.1084/jem.185.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Martin J, Uria J A, Johnson A D. Phenotypic switching in Candida albicans is controlled by SIR2 gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesti M, Sipiczki M, Pinter Y. Scanning electron microscopy characterization of colonies of Candida albicans morphological mutants. J Med Microbiol. 1999;48:167–172. doi: 10.1099/00222615-48-2-167. [DOI] [PubMed] [Google Scholar]
- 32.Radford D, Challacombe S, Walter J. A scanning electron microscopy investigation of the structure of colonies of different morphologies produced by phenotypic switching in Candida albicans. J Med Microbiol. 1994;40:416–423. doi: 10.1099/00222615-40-6-416. [DOI] [PubMed] [Google Scholar]
- 33.Ramsey H, Morrow B, Soll D R. An increase in switching frequency correlates with an increase in recombination of the ribosomal chromosomes of Candida albicans strain 3153A. Microbiology. 1994;140:1525–1531. doi: 10.1099/13500872-140-7-1525. [DOI] [PubMed] [Google Scholar]
- 34.Rivera J, Feldmesser M, Cammer M, Casadevall A. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during murine experimental infection. Infect Immun. 1998;66:5027–5030. doi: 10.1128/iai.66.10.5027-5030.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rustchenko E P, Sherman F, Hicks J B. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J Bacteriol. 1990;172:1276–1283. doi: 10.1128/jb.172.3.1276-1283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rustchenko E P, Howard D H, Sherman F. Chromosomal alterations of Candida albicans are associated with gain and loss of assimilating functions. J Bacteriol. 1994;176:3231–3241. doi: 10.1128/jb.176.11.3231-3241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan T G, Hinnebush B J. Bloodstream- versus tick-associated variants of a relapsing fever bacterium. Science. 1998;280:1938–1940. doi: 10.1126/science.280.5371.1938. [DOI] [PubMed] [Google Scholar]
- 38.Shadomy H J, Lurie H I. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraudia. 1971;9:6–9. doi: 10.1080/00362177185190031. [DOI] [PubMed] [Google Scholar]
- 39.Silverman M, Zieg J, Hilmen M, Simon M. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci USA. 1979;76:391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slutsky B, Buffo J, Soll D R. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 41.Small J, Mitchell T. Strain variation in antiphagocytic activity of capsular polysaccharide from Cryptococcus neoformans serotype A. Infect Immun. 1989;57:3751–3756. doi: 10.1128/iai.57.12.3751-3756.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soll D R. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T, Hitomi A, Magee P T, Sakaguchi S. Correlation between polyploidy and auxotrophic segregation in the imperfect yeast Candida albicans. J Bacteriol. 1994;176:3345–3353. doi: 10.1128/jb.176.11.3345-3353.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swartley L, Marfin A, Edupuganti S, Liu L, Cieslak P, Perkins B, Wenger J. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vartivarian S E, Anaissie E J, Cowart R E, Sprigg H A, Tingler M J, Jacobson E S. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- 46.Wilkes B, Mayorga M, Edman U, Edman J. Dimorphism and haploid fruiting in Cryptococcus neoformans with the a-mating type. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkes B, Moore T D, Kwong-Chung K. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology. 1994;140:543–550. doi: 10.1099/00221287-140-3-543. [DOI] [PubMed] [Google Scholar]
- 48.Zolan M. Chromosome length polymorphism in fungi. Microbiol Rev. 1995;59:686–698. doi: 10.1128/mr.59.4.686-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]